Significance
Most DNA polymerases initiate DNA synthesis by extending a preexisting primer. Exceptions to this dogma are recently characterized bifunctional primase–polymerases (prim–pols) that resemble archaeal primases in their structure and initiate DNA synthesis de novo using only NTPs or dNTPs. We report here a DNA polymerase encoded by a phage NrS-1 from deep-sea vents. NrS-1 has a genome organization unlike any other known phage. Although this polymerase does not contain a zinc-binding motif typical for primases, it is nonetheless able to initiate DNA synthesis from a specific DNA sequence exclusively using dNTPs. Thus, it represents a unique de novo replicative DNA polymerase that possesses features found in DNA polymerases, primases, and RNA polymerases.
Keywords: NrS-1, primase, prim–pol, helicase, ssDNA-binding protein
Abstract
A DNA polymerase is encoded by the deep-sea vent phage NrS-1. NrS-1 has a unique genome organization containing genes that are predicted to encode a helicase and a single-stranded DNA (ssDNA)-binding protein. The gene for an unknown protein shares weak homology with the bifunctional primase–polymerases (prim–pols) from archaeal plasmids but is missing the zinc-binding domain typically found in primases. We show that this gene product has efficient DNA polymerase activity and is processive in DNA synthesis in the presence of the NrS-1 helicase and ssDNA-binding protein. Remarkably, this NrS-1 DNA polymerase initiates DNA synthesis from a specific template DNA sequence in the absence of any primer. The de novo DNA polymerase activity resides in the N-terminal domain of the protein, whereas the C-terminal domain enhances DNA binding.
DNA polymerases play a pivotal role in maintaining genetic information in living organisms by catalyzing the synthesis of cDNA strands on existing DNA templates (1). A long-held dogma was that DNA polymerases are unable to synthesize DNA de novo; rather, a preexisting primer bound to the template strand is required to extend the existing DNA strand. In nature, such primers are usually provided by DNA primases (2), a special group of polymerases that assemble short oligonucleotides, usually RNA, at certain sequences on the template DNA. The rationale to require two kinds of polymerases to fulfill de novo DNA synthesis is not clear (3). One possibility is that in contrast to the fast extension of an existing primer, the condensation step of the initial nucleotides and the maintenance of the unstable short oligonucleotide on the template pose severe challenges when using just a single active site. Although RNA polymerases synthesize RNA de novo from NTPs during transcription, drastic conformational changes occur that often lead to abortion of synthesis during the transition between initiation and elongation (4), reflecting the challenge of using a single polypeptide for both initiation and elongation of polynucleotide synthesis.
A group of enzymes have been characterized that are capable of polymerizing long DNA directly from dNTPs and thus are technically de novo DNA polymerases. These are archaeal primases, which can use dNTPs as well as NTPs for primer synthesis and, remarkably, have extraordinary processivity compared with primases from other organisms, synthesizing several thousand nucleotides without dissociating (5–8). However, the function of these primases is thought to be limited to initiation of DNA synthesis and repair. Recently, more specialized polymerases called primase–polymerase (prim–pol), encoded by extrachromosomal plasmids, have been discovered in archaea (9–14). These polymerases also catalyze de novo synthesis of long DNA and, unlike the archaeal primases, are thought to use their high processivity to carry out the replication of the entire plasmid DNA (9, 10). The structure of the active domain of pRN1 prim–pol, the best characterized member of this family, shows that its overall structure resembles that of an archaeal primase (10). Thus, these prim–pols should also be grouped into the archaeo-eukaryotic primase (AEP) superfamily (14, 15). More recently, prim–pols have been found in other organisms including bacteriophage and human (16–18), indicating the importance of such de novo DNA synthesis across bacterial, archaeal, and eukaryotic kingdoms.
The DNA polymerases from bacteriophage have provided systems for basic studies of the mechanism of DNA replication, in large part because of their simplicity. The explosion in the pace of genome sequencing has revealed a vast phage world, harboring the largest genetic diversity on earth (19). A major portion of those unknown genes are located in the region of the phage genomes responsible for nucleic acid metabolism, likely encoding numerous novel enzymes responsible for the replication of DNA, including DNA polymerases. Because enzymes from phage systems tend to be much simpler and have higher efficiency than those of host systems, they have played very important roles as tools for molecular biology. We are particularly interested in characterizing novel phage enzymes involved in nucleic acid metabolism, especially those from special environments such as the ocean (20). In the present study, we have characterized a DNA polymerase from the newly discovered deep-sea vent phage NrS-1 (21). We report that it is a self-priming DNA polymerase that synthesizes long DNA strands de novo exclusively with dNTPs; NTPs can be incorporated to a limited extent. The NrS-1 polymerase active site shares weak homology to those of prim–pols from archaeal plasmids. However, in contrast to those enzymes, the NrS-1 polymerase does not have a zinc-binding motif that is typical for primases, and it recognizes a relatively long DNA template sequence to initiate polymerization. Interestingly, during the de novo DNA synthesis, NrS-1 polymerase produces short abortive oligonucleotides, a feature that mimics that of RNA polymerases during their transition from transcription initiation to elongation.
Results
A DNA Polymerase Identified from the Deep-Sea Hydrothermal Vent Phage NrS-1.
NrS-1 is the first phage to be isolated and characterized that infects deep-sea vent Epsilonproteobacteria (21). Epsilonproteobacteria are among the predominant primary producers in the deep-sea hydrothermal vent ecosystems. The temperate phage has been assigned to the Siphoviridae family based on morphology, although its DNA sequence and genomic organization are distinct from those of any other known members of that family (21). Among its approximately 50 genes, DNA sequence homology predicts that one encodes a helicase and another single-stranded DNA (ssDNA)-binding protein, suggesting that the phage uses its own enzymes for the replication of its genome. However, bioinformatic analysis failed to predict any replicative polymerase, based on the lack of homology to any known DNA polymerase. We noticed that one gene, referred to as gene 28, encoded for a putative protein that was designated as a primase based on its weak homology to the active sites of the prim–pols found in archaeal plasmids (Fig. 1A). However, the other structural features found in these prim–pols were missing (21). Based on this limited homology, we suspected that this protein might be a unique replicative DNA polymerase that is part of the phage replisome. We cloned, overexpressed, and purified the predicted NrS-1 primase and various truncated and mutant forms expressing the potential N-terminal catalytic domain as well as the gene products for the putative NrS-1 helicase and ssDNA-binding protein (Fig. 1B).
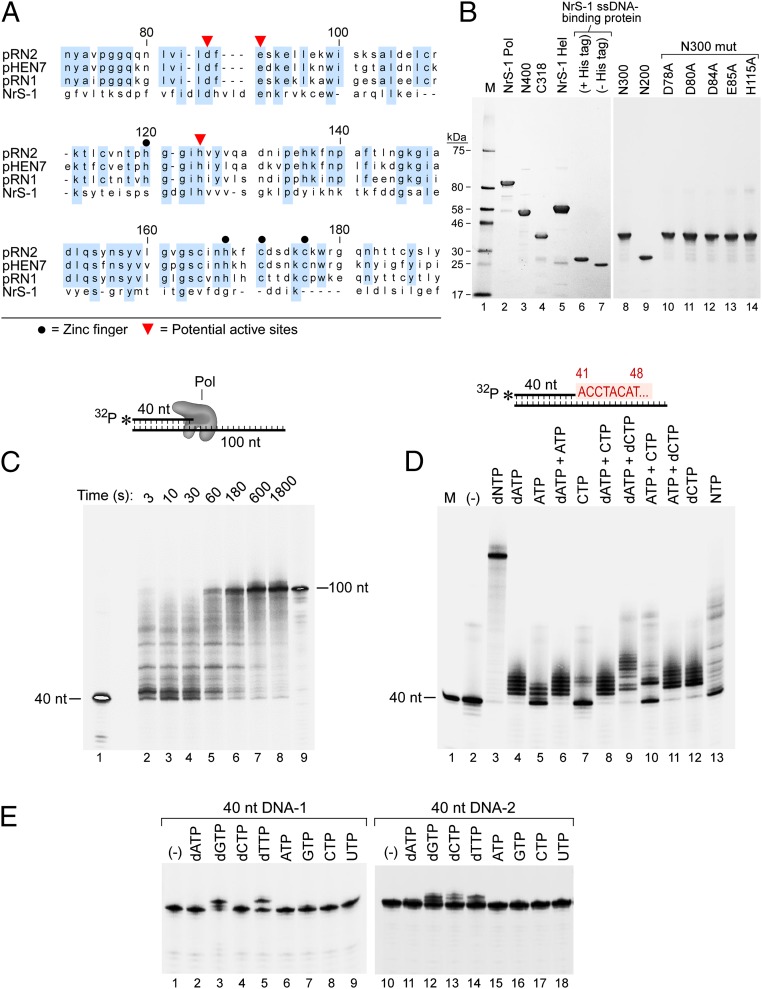
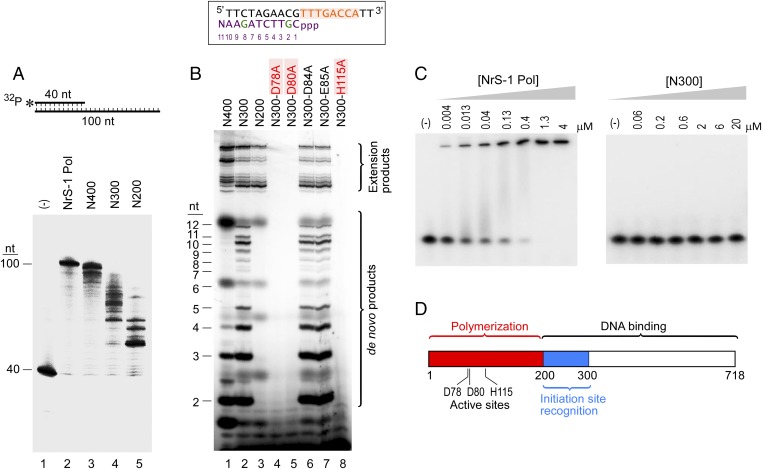
Fig. 1.
An unusual DNA polymerase from deep-sea vent phage NrS-1. (A) NrS-1 gene protein 28 shares weak homology to archaeal prim–pols. Alignment between NrS-1 gene protein 28 and three prim–pols encoded by archaeal plasmids (10) was made using CLC Sequence Viewer 6, and the regions containing homology are shown. Identical residues among archaeal prim–pols and those among prim–pols and NrS-1 gene protein 28 are highlighted by blue background. (B) SDS/PAGE gels showing purified proteins analyzed in this work. Lane 1, protein size marker; lane 2, NrS-1 polymerase; lane 3, N-terminal 400 residues of NrS-1 polymerase (N400); lane 4, C-terminal 318 residues of NrS-1 polymerase; lane 5, NrS-1 helicase; lane 6, NrS-1 ssDNA-binding protein; lane 7, NrS-1 ssDNA-binding protein (His-tag removed); lane 8, N-terminal 300 residues of NrS-1 polymerase (N300); lane 9, N-terminal 200 residues of NrS-1 polymerase (N200); lane 10, N300-D78A; lane 11, N300-D80A; lane 12, N300-D84A; lane 13, N300-E85A; lane 14, N300-H115A. All proteins carry an N-terminal His-tag except that in lane 7. (C) Extension of a primer by NrS-1 polymerase. A primed-template (40/100 nt) substrate in which the primer was 5′-32P–labeled and was incubated at 100 nM with 200 nM NrS-1 polymerase and 0.5 mM four dNTPs at 50 °C. Aliquots were removed at increasing time and analyzed on a 10% (wt/vol) TBE-urea gel (lanes 2–8). Lanes 1 and 9 contain 5′-32P–labeled 40 nt (primer) and the 100 nt complete complement to the template strand, respectively. (D) Nonspecific incorporation by NrS-1 polymerase. A primed-template (40/100 nt) substrate in which the primer was 5′-32P–labeled was incubated at 100 nM with 200 nM NrS-1 polymerase and 0.5 mM of various (d)NTPs at 50 °C for 30 min and then analyzed on a 10% (wt/vol) TBE-urea gel. Lane 1 is the 5′-32P–labeled 40 nt marker. Shown in red is the 5′ part of the DNA sequence to be synthesized downstream of the primer. (E) 100 nM each of the two 5′-32P–labeled 40-nt DNA strand (40 nt DNA-1 sequence 5′-TTTAGGTACCGGTGCCTAGCAGAAGGCCTAATTCTGCAAA-3′; 40 nt DNA-2 sequence 5′-TTTGCAGAATTAGGCCTTCTGCTAGGCACCGGTACCTAAA-3′) was incubated with 200 nM NrS-1 polymerase and 0.5 mM various (d)NTPs at 50 °C for 30 min and reaction products analyzed on a 10% (wt/vol) TBE-urea gel.
To determine whether the purified gene 28 “primase” could catalyze the polymerization of nucleotides, we incubated it with a 40-nt DNA primer labeled at its 5′ end that was annealed to a 100-nt DNA template in the presence of the four dNTPs. The primer sequence is 5′-TTTAGGTACCGGTGCCTAGCAGAAGGCCTAATTCTGCAAA-3′ and the template sequence is 5′-TAGACTGAATAGTTAAATAGGCAGATATAAAATGGTCAAACGTTCTAGAACTATGTAGGTTTTGCAGAATTAGGCCTTCTGCTAGGCACCGGTACCTAAA-3′. Under these conditions, the primer is extended from 40 nt to 100 nt, the length of the template, showing that indeed this protein has DNA polymerase activity (Fig. 1C). Thus, in the remainder of this study, we will refer to this protein as NrS-1 DNA polymerase. The apparent processivity of NrS-1 DNA polymerase is low, as abortive products can be observed at short times (Fig. 1C), despite the fact that there is a twofold excess of protein molecules over primer-template molecules in this experiment.
NrS-1 DNA polymerase is able to incorporate several mismatching deoxyribonucleotides in the absence of the correct ones (Fig. 1D, lanes 4, 9, and 12). Interestingly, even ribonucleotides can be incorporated in the absence of deoxyribonucleotides (Fig. 1D, lanes 5, 7, 10, and 13), albeit with lower efficiency. When both dNTPs and NTPs are present, the enzyme shows a preference for the dNTPs, as the products are similar compared with those with only dNTPs (Fig. 1D, lane 4 vs. 6, and lane 11 vs. 12). To test whether the nonspecific incorporation observed was the result of template-dependent misincorporation or rather due to a template-independent terminal transferase activity of the enzyme, we incubated the labeled 40 nt primer strand in the absence of template with the enzyme in the presence of various nucleoside triphosphates (Fig. 1E, Left). In contrast to the results shown in Fig. 1D in the presence of template, when the primer alone is incubated with enzyme, no dAMP or dCMP incorporation could be observed, confirming that it is a template-dependent event. The incorporation of dGMP and dTMP shown in Fig. 1E is likely caused by the formation of a primed template-like secondary structure of the ssDNA, which was confirmed by testing ssDNA with various sequences (Fig. 1E, Right and Fig. S1). As shown in Fig. S1A, the 37 nt ssDNA substrate tends to form a primed-template structure that favors the incorporation of dTMP, and consistently only dTMP could be efficiently incorporated among four dNMPs. The efficiency of dTMP incorporation was not improved by increasing dTTP (Fig. S1B) or NrS-1 DNA polymerase concentration (Fig. S1C), which further excluded the possibility that such incorporation resulted from terminal transferase activity. In the absence of any dNTP or NTP, the enzyme does not degrade the ssDNA (Fig. 1E and Fig. S1). Thus, NrS-1 polymerase is a DNA polymerase with low fidelity that lacks exonuclease or terminal transferase activity.
Fig. S1.
DNA terminal extension by NrS-1 DNA polymerase. (A) Each of the two DNA strands [5′-ACTGGGATCCTTTGCACACTCAAGTATCCAACTTTCAGAACC-3′ (lanes 2–6) and 5′-ACTGGGATCCTTTAGAAAGTAAAGCTTGGATGAAAGC-3′ (lanes 7–11)] was labeled at its 5′ end with 32P. We mixed 100 nM of each template with 200 nM NrS-1 polymerase in the presence of 0.5 mM of the following dNTPs: none (lanes 2 and 7), dATP (lanes 3 and 8), dGTP (lanes 4 and 9), dCTP (lanes 5 and 10), and dTTP (lanes 6 and 11). Reaction mixtures were incubated at 50 °C for 30 min, and then the products were analyzed on a 10% (wt/vol) polyacrylamide TBE-urea gel. Lane 1 contains a 5′-32P–labeled 40-nt DNA, same as that used in Fig. 1E, Right. A potential intermolecular secondary structure formed by the 37-nt DNA (template for lanes 7–11) was shown, in which the complementary sequences are indicated in red and the templates for dTMP incorporation are indicated in blue. (B) Terminal extension of two DNA strands. Each of two DNA strands [5′-TTTGCAGAATTAGGCCTTCTGCTAGGCACCGGTACCTAAA-3′ (lanes 2–7) and 5′-ACTGGGATCCTTTAGAAAGTAAAGCTTGGATGAAAGC-3′ (lanes 8–13)] was labeled at its 5′ end with 32P. We mixed 100 nM of each template with 200 nM of NrS-1 DNA polymerase in the presence of the following amounts of dTTP: 0 mM (lanes 2 and 8), 0.1 mM (lanes 3 and 9), 0.4 mM (lanes 4 and 10), 1.1 mM (lanes 5 and 11), 3.3 mM (lanes 6 and 12), and 10 mM (lanes 7 and 13). Lane 1 is the same as lane 1 in A. Reaction mixtures were incubated at 50 °C for 30 min, and then the products were analyzed on a 10% (wt/vol) polyacrylamide TBE-urea gel. (C) Terminal extension of two DNA strands. Each of two DNA strands [5′-TTTGCAGAATTAGGCCTTCTGCTAGGCACCGGTACCTAAA-3′ (lanes 2–7) and 5′-ACTGGGATCCTTTAGAAAGTAAAGCTTGGATGAAAGC-3′ (lanes 8–13)] was labeled at its 5′ end with 32P. We mixed 100 nM of each template with 1 mM dTTP and the following amounts of NrS-1 DNA polymerase: 0 nM (lanes 2 and 8), 12 nM (lanes 3 and 9), 37 nM (lanes 4 and 10), 111 nM (lanes 5 and 11), 333 nM (lanes 6 and 12), and 1,000 nM (lanes 7 and 13). Reaction mixtures were incubated at 50 °C for 30 min, and then the products were analyzed on a 10% (wt/vol) polyacrylamide TBE-urea gel. Lane 1 is the same as lane 1 in A.
NrS-1 DNA Polymerase Initiates DNA Synthesis with a dNTP.
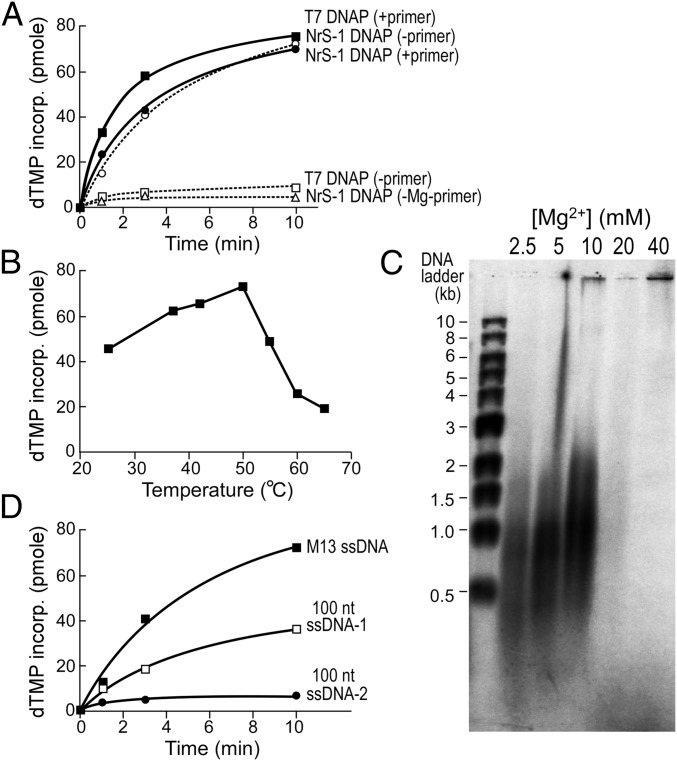
Because the predicted NrS-1 primase is in fact a DNA polymerase, an interesting question is what enzyme provides for the primase activity for this phage. In light of the weak homology between a small region in the NrS-1 DNA polymerase and the archaeal prim–pols (Fig. 1A), which can synthesize long DNA de novo, we examined if the NrS-1 DNA polymerase is able to prime DNA synthesis in the absence of a primer. When a typical DNA polymerase such as T7 DNA polymerase is incubated with an M13mp18 ssDNA template and only dATP, dGTP, dCTP, and dTTP, there is essentially no DNA synthesis in the absence of a preexisting primer annealed to the DNA template (Fig. 2A). Under the same conditions, in the absence of a primer, NrS-1 DNA polymerase catalyzes robust DNA synthesis, as reflected by the incorporation of radioactively labeled dNMPs. If a primer was preannealed onto the M13 ssDNA, the level of DNA synthesis by T7 DNA polymerase was boosted whereas that by NrS-1 DNA polymerase was not affected significantly (Fig. 2A). The optimal reaction temperature for de novo DNA synthesis by NrS-1 DNA polymerase is 50 °C (Fig. 2B). We analyzed the lengths of the products of de novo synthesis on a denaturing alkaline agarose gel (Fig. 2C). The products range from a few hundred nucleotides to about 2,000 nucleotides in length. Like all known DNA polymerases, DNA synthesis by NrS-1 DNA polymerase requires Mg2+ (Fig. 2A), with an optimized concentration between 5 and 10 mM. Higher concentrations of Mg2+ inhibit the reaction (Fig. 2C). The efficiency of de novo DNA synthesis is highly variable depending upon the sequence of the template. For example, in Fig. 2D, we compare de novo DNA synthesis on the 100-mer template used in the above studies (ssDNA-1) with a template in which the sequence is antiparallel to it (ssDNA-2; 5′-TTTAGGTACCGGTGCCTAGCAGAAGGCCTAATTCTGCAAAACCTACATAGTTCTAGAACGTTTGACCATTTTATATCTGCCTATTTAACTATTCAGTCTA-3′). Synthesis on ssDNA-1 is about eightfold higher than that observed on ssDNA-2. For comparison, de novo DNA synthesis on the much longer and circular M13 ssDNA template is about twice that observed on ssDNA-1. These results suggest that some sequences are used preferentially for initiation of de novo DNA synthesis.
Fig. 2.
NrS-1 DNA polymerase synthesizes DNA de novo. (A) 10 nM M13 ssDNA or primed-M13 ssDNA (annealed with M13 primer M2, 5′-CCCAG TCACG ACGTT-3′) was incubated with 100 nM T7 DNA polymerase (solid square, with primer; open square, without primer) or NrS-1 DNA polymerase (solid circle, with primer; open circle, without primer) in the presence of 250 μM each of dATP, dGTP, and dCTP and 10 μM 3H-dTTP; [Mg2+] was 5 mM for all assays except that no Mg2+ was added for the assay shown by an open triangle, and the incorporation of dTMP into DNA at various time points at 37 °C (for T7 DNA polymerase) or 50 °C (for NrS-1 DNA polymerase) was measured by DE81 (Whatman) filter-binding assay. (B) Effect of temperature on de novo DNA synthesis by NrS-1 DNA polymerase. Reaction conditions were as described for A using 10 nM M13 ssDNA template and 100 nM NrS-1 DNA polymerase. Reactions were carried out for 10 min at the indicated temperatures, and then the incorporation of dTMP into DNA at various time points was measured by DE81 (Whatman) filter-binding assay. (C) De novo DNA synthesis products catalyzed by NrS-1 DNA polymerase on M13 ssDNA were analyzed on 0.8% alkaline agarose gel. Reaction conditions were as described for A, except that nucleotide mixture was 250 μM each of dATP, dCTP, and dTTP and 25 μM α-32P-dGTP, and the indicated amounts of MgCl2 were used. Reactions were carried out for 10 min at 50 °C. (D) De novo DNA synthesis by NrS-1 DNA polymerase on the templates M13 ssDNA (solid square), 100 nt template-1 (open square), and -2 (solid circle) (ssDNA-1 sequence 5′-TAGACTGAATAGTTAAATAGGCAGATATAAAATGGTCAAACGTTCTAGAACTATGTAGGTTTTGCAGAATTAGGCCTTCTGCTAGGCACCGGTACCTAAA-3′; ssDNA-2 sequence 5′-TTTAGGTACCGGTGCCTAGCAGAAGGCCTAATTCTGCAAAACCTACATAGTTCTAGAACGTTTGACCATTTTATATCTGCCTATTTAACTATTCAGTCTA-3′). Reaction conditions were as described for A, except for the DNA templates used.
The Template Recognition Sequence for Initiation of de Novo DNA Synthesis by NrS-1 DNA Polymerase.
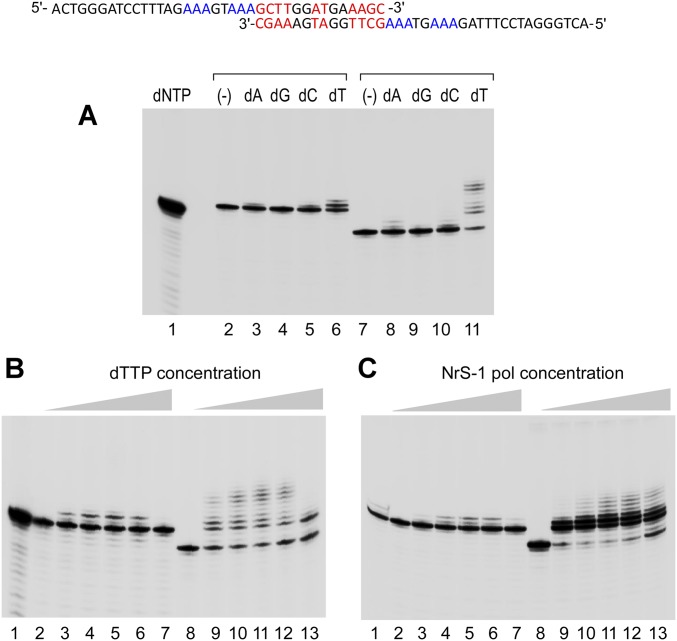
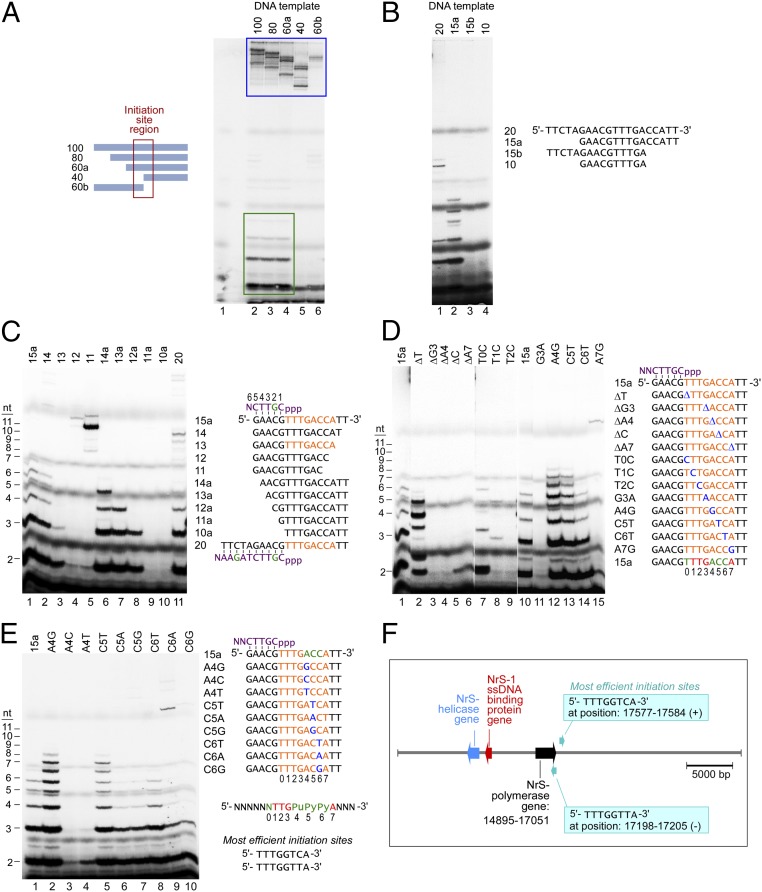
The wide variation in the de novo synthesis efficiency on different DNA templates suggests that there must be preferential template sequences recognized by the NrS-1 DNA polymerase to initiate de novo DNA synthesis. To determine the sequences responsible, we analyzed de novo DNA synthesis products by NrS-1 polymerase on various truncated versions of the 100-nt DNA template shown in Fig. 2D that support extensive de novo DNA synthesis (ssDNA-1). For all of the DNA templates analyzed, we observed the incorporation of radioactively labeled dNMPs into some long DNA product beyond the resolution of the gel (Fig. 3A, outlined by blue box). These large products are likely formed by extension of the 3′ end of the template similar to that shown in Fig. 1E. However, on three of the truncated ssDNA templates, all of which contain the 3′ 60-nt region of the full-length template, some short products of two to several nucleotides were observed (Fig. 3A, outlined by green box). Because all known de novo RNA syntheses by RNA polymerases and primases all produce abortive short oligonucleotides during the initiation of synthesis (2–4), these short DNA oligonucleotides are suggestive of similar abortive products from sequence-dependent initiation of de novo DNA synthesis.
Fig. 3.
NrS-1 DNA polymerase initiates DNA synthesis at specific template sequences. (A) Gel analysis of products synthesized by NrS-1 DNA polymerase on ssDNA-1 (described in Fig. 2D) and templates that are truncated forms of ssDNA-1. We incubated 10 μM 100 nt ssDNA-1 (or its 80, 60, or 40 nt 3′ fragments or its 60 nt 5′ fragment) with 200 nM NrS-1 polymerase in the presence of 250 μM each of dATP, dCTP, and dTTP and 25 μM α-32P-dGTP at 50 °C for 30 min. The products were separated on a 25% (wt/vol) TBE-urea gel and analyzed by phosphoimager. Extended DNA templates are indicated by the blue box, and the abortive 2–5-nt products are indicated by the green box. The region that contains the potential initiation site is indicated by the red box. (B) We incubated 10 μM of either 20-nt, 15-nt, or 10-nt DNA fragments (derived from the sequence within the red box of A) with 200 nM NrS-1 polymerase in the presence of 250 μM each dATP dCTP, and dTTP and 25 μM α-32P-dGTP at 50 °C for 30 min. And the products were separated on a 25% (wt/vol) TBE-urea gel and analyzed by phosphoimager. (C) DNA templates derived from template 15a in B were tested for whether they could support de novo synthesis by NrS-1 polymerase. Assay conditions were the same as that described for B. The sequence in orange is the deduced recognition sequence for initiation of de novo synthesis by NrS-1 polymerase. The sequence of the synthesized product is shown in purple (with the radioactively labeled nucleotide shown in green). Nucleotides in the products are numbered from 5′ to 3′. (D) Same assay as that described in C, except that various DNA templates were used that are derived from template 15a, with one nucleotide deletion or replacement (shown in blue) within the recognition region (shown in orange), were tested for their efficiency to support de novo synthesis. Based on these results, the strictly required sequence for template recognition for de novo DNA synthesis is shown at the bottom in red, and those sequences that do not show strict specificity are shown in green. The 8 nt of the recognition sequence are numbered from 5′ to 3′ as 0–7. (E) Same assay as that described in C, except that various DNA templates were used that are derived from template 15a, with one nucleotide replacement (shown in blue) within the recognition region (shown in orange), were tested for their efficiency to support de novo synthesis. Based on these results, the strictly required sequence for template recognition site for de novo DNA synthesis is shown at the bottom in red, whereas those sequences that are important but do not show strict specificity are shown in green. (F) Position of the NrS-1 DNA polymerase gene and the two most efficient initiation sites in NrS-1 genome.
Neither the truncated DNA templates containing the 5′ 60 nt or 3′ 40 nt of the original template supported the synthesis of these short fragments (Fig. 3A, lanes 5 and 6), indicating that the recognition sequence must be near the interface between these two regions. We further truncated the DNA templates to narrow down the location of recognition sequence to a 15-nt DNA template (template 15a; Fig. 3B, lane 2), on which short products of 2–6 nt were produced by NrS-1 polymerase. When we further shortened this 15-nt DNA from the 5′ or 3′ ends, we found that the minimum template sequence to support the de novo DNA synthesis is an 8-nt sequence, 5′-TTTGACCA-3′ (indicated in orange in Fig. 3C). Templates missing any nucleotides from either end of this region do not support the synthesis of the short fragments (Fig. 3C, lanes 4, 5, 9, and 10). Removal of 5′ nucleotides flanking the template recognition sequence results in shortening of the products (Fig. 3C, lanes 6–8), indicating that the synthesis is initiated immediately downstream of the 8-nt recognition sequence in a template-dependent manner, like that observed for primases and RNA polymerases. In addition to abortive products ranging from 2 to 5 nt and also run-off products (Fig. 3C, lane 1, in which there is a 5-nt runoff product from the 15-nt template, and Fig. 3C, lane 11, in which there is a 10-nt runoff product from the 20-nt template), the NrS-1 DNA polymerase also produces products with a single overextended nucleotide (Fig. 3C, lanes 1 and 11, 6 nt and 11 nt “N+1” products, respectively).
To further investigate the sequence specificity of de novo initiation of DNA synthesis by NrS-1 DNA polymerase, we compared 15-nt DNA templates carrying variations in the 8-nt recognition region for their efficiency to support de novo DNA synthesis (Fig. 3D). For the template sequence 5′-T0T1T2G3A4C5C6A7-3′, deletion of any one nucleotide of G3A4C5C6A7 eliminated de novo DNA synthesis (Fig. 3D, lanes 3–6). C or G at position T0 supports synthesis, although the patterns of fragments produced are different depending upon which nucleotide is at the T0 position (Fig. 3D, lanes 2 and 7). Positions T1, T2, G3, and A7 are stringent, as even pyrimidine-to-pyrimidine or purine-to-purine transversions at these positions prevent initiation of DNA synthesis (Fig. 3D, lanes 8, 9, 11, and 15). Positions A4, C5, and C6 can tolerate purine-to-purine or pyrimidine-to-pyrimidine transversions (Fig. 3D, lanes 12–14) but not purine-to-pyrimidine or pyrimidine-to-purine transversions (Fig. 3E) to remain efficient templates for de novo DNA synthesis.
In summary, the recognition sequence for NrS-1 DNA polymerase to initiate de novo DNA synthesis at this site is 5′-NTTGPuPyPyA-3′. Interestingly, a G at position 4 or a T at position 5 results in a more efficient template than the original one (Fig. 3E, compare lanes 2 and 5 to lane 1), whereas at position 6 either a C or T provides templates equally efficient in promoting de novo initiation (Fig. 3E, comparing lane 8 to lane 1). Based on these results, we conclude that the most efficient sites for de novo initiation of DNA by NrS-1 DNA polymerase are 5′-TTTGGTCA-3′ and 5′-TTTGGTTA-3′. A search for these sequences in the NrS-1 genome reveals two sites, each present once in the genome, one in the plus strand and the other in the minus strand. Intriguingly, both sites are just downstream of the NrS-1 DNA polymerase gene (Fig. 3F), suggesting that perhaps they may have coevolved with the polymerase gene and serve as the origin for NrS-1 genome replication.
NrS-1 DNA Polymerase Function Domains.
Because bioinformatics suggested that both the N- and C-terminal regions of NrS-1 DNA polymerase share weak homology to different types of primases (21), it was of interest whether this enzyme uses just a single active site or two active sites to catalyze de novo DNA synthesis. We analyzed the activities of truncated versions of NrS-1 DNA polymerase, specifically polypeptides consisting of the N-terminal 400, 300, and 200 amino acid residues (N400, N300, and N200, respectively) and the C-terminal 318 amino acid residues (C318). All of the truncated N-terminal peptides retain primer extension activity, even the 200 amino acid residue peptide, although there is a gradual decrease in the apparent processivity of DNA synthesis (Fig. 4A). The N300 fragment has similar de novo synthesis activity to that of the full-length enzyme, as shown by the abortive and run-off products produced on the 15 nt template containing the initiation site (Fig. 4B, lane 2). In contrast, no de novo synthesis could be detected using the N200 fragment (Fig. 4B, lane 3), even though it retains its ability to extend the terminus of the template, presumably due to secondary structures (Fig. 4B, lane 3, upper region of the gel labeled “Extension Products”). This observation suggests that the C-terminal 100 amino acids of N300 are involved in the recognition of the initiation site. The N400 fragment, although still able to catalyze de novo synthesis, has reduced ability to support DNA synthesis compared with that of N300 (Fig. 4B, lane 1), perhaps due to the interference by its improperly structured C-terminal region. In the region of limited homology between NrS-1 DNA polymerase and the well-studied archaeal plasmid pRN1 prim–pol, the residues Asp111, Glu113, and His145 (indicated by red arrows in Fig. 1A) are crucial for the activity of the pRN1 prim–pol; substitution of any of these residues by alanine abolishes polymerase activity (10). Consequently, we changed each of the four acidic residues in this region, as well as His115, to alanine, in the gene encoding the truncated N300 fragment of NrS-1 DNA polymerase. Mutations in two of the acidic residues, Asp78 and Asp80, and in His115 completely abolished any detectable DNA synthesis by the enzyme (Fig. 4B, lanes 4, 5, and 8), indicating the similarity in active site architecture between NrS-1 DNA polymerase and pRN1 prim–pol. This result is surprising, as there is a zinc stem close to the pRN1 prim–pol active site (indicated by the black arrows in Fig. 1A) that is not found in the NrS-1 DNA polymerase. A single alanine mutation of any of the three crucial residues abolishes both de novo DNA synthesis and primer extension (Fig. 4B, lanes 4, 5, and 8), suggesting that a single active site is used for polymerization during both initiation and elongation. Although the N300 fragment retains the ability to support extensive de novo DNA synthesis activity, its DNA binding activity is decreased dramatically compared with the full-length enzyme (Fig. 4C). In a gel mobility-shift assay, the N300 fragment does not show any stable binding to the 15-nt DNA template containing the initiation site (Fig. 4C), in contrast to the full-length enzyme that shows tight binding to DNA with a Kd of ∼20 nM (Fig. 4C). Based on these results, we propose that the N-terminal active site is responsible for polymerization, whereas the C-terminal domain enhances the binding of the enzyme to the DNA template to increase its processivity (Fig. 4D).
Fig. 4.
Active site and functional subdomains of NrS-1 DNA polymerase. (A) Conditions were as in Fig. 1C, except that each reaction was carried out using either 200 nM full-length NrS-1 DNA polymerase (lane 2) or 500 nM of either the N-terminal fragment, N400 (lane 3), N300 (lane 4), or N200 (lane 5). The ability of each of these enzymes to extend 100 nM of a 40/100 nt primed-template was determined. The 40-mer primer is labeled at its 5′ end with 32P. Lane 1 shows the reaction mixture in the absence of enzyme. (B) 10 μM of the 20-nt DNA template containing the NrS-1 polymerase recognition sequence (shown in orange) was incubated with 500 nM of either NrS-1 polymerase fragment N400 (lane 1), N300 (lane 2), N200 (lane 3), or N300 mutants (D78A, lane 4; D80A, lane 5; D84A, lane 6; E85A, lane 7; H115A, lane 8) in the presence of 250 μM each of dATP, dCTP, and dTTP and 25 μM α-32P-dGTP at 50 °C for 30 min. The products were separated on a 25% (wt/vol) TBE-urea gel and analyzed by phosphoimager. Products larger than 20 nt are the result of extension of the templates (Extension Products), whereas those smaller than 13 nt represent abortive and runoff products synthesized de novo (“de novo products”). The runoff product sequence is shown in purple (with the two radioactively labeled G’s indicated in green). Numbering of the nucleotides in the product is 5′ to 3′. Mutations that abolish NrS-1 polymerase activities are shown in red. (C) In a DNA mobility shift assay, 5 nM of 5′-32P–labeled 15 nt DNA (15a) was incubated with the indicated amounts of either full-length NrS-1 polymerase or N300 fragment in the absence of dNTPs for 20 min, followed by loading on a 10% (wt/vol) TBE native acrylamide gel to detect the mobility shift. After electrophoresis, the gel was analyzed using a phosphoimager. (D) A schematic showing the active site and proposed functional subdomains of NrS-1 DNA polymerase.
Initiation of DNA Synthesis by NrS-1 DNA Polymerase.
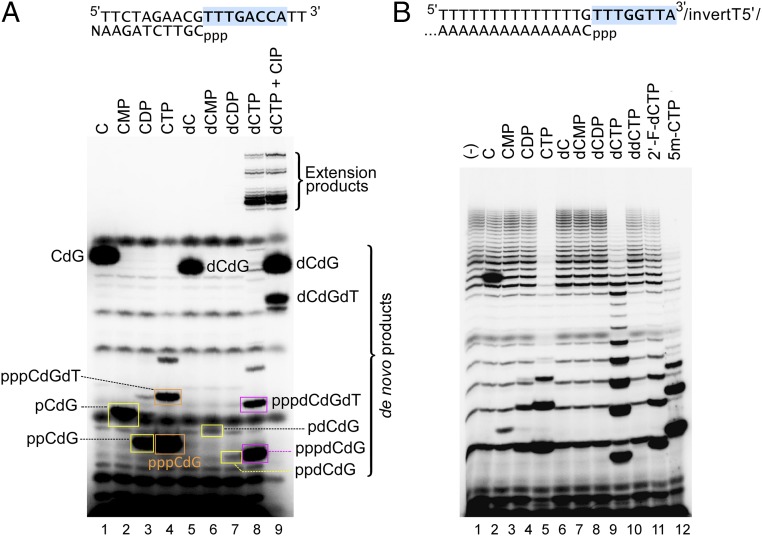
To gain more insight into the de novo DNA synthesis by NrS-1 DNA polymerase, we investigated the specificity of the first nucleotide to be incorporated. On the 20-nt template containing the initiation site used in Fig. 3, the first nucleotide to be incorporated into the de novo product by the NrS-1 DNA polymerase N300 fragment is predicted to be a cytidine (Fig. 5A). We replaced the dCTP in the four dNTP mixture (dATP, dGTP, dCTP, and dTTP) with either C, CMP, CDP, CTP, dC, dCMP, or dCDP and then tested the ability of the mixture to promote de novo synthesis. If the substituted nucleotide efficiently replaced dCTP for initiation, the product should be at least 5 nt, before the template contains a second G (Fig. 5A). Most of the analogs tested (C, CMP, dC, dCMP, and dCDP) only support the synthesis of a dinucleotide (Fig. 5A, lanes 1, 2, and 5–7). CDP and CTP can each be extended into a trimer and tetramer, respectively (Fig. 5A, lanes 3 and 4). In contrast, in the presence of dCTP, NrS-1 DNA polymerase synthesizes both abortive products and full-length run-off products (10 and 11 nt, Fig. 5A, lane 8). When treated with calf-intestinal alkaline phosphatase, the dephosphorylated dimer product initiated with dCTP migrated the same as the dimer product initiated with dC (Fig. 5A, compare lanes 8, 9, and 5), confirming that in the presence of dCTP DNA synthesis was initiated with a nucleoside triphosphate.
Fig. 5.
NrS-1 DNA polymerase initiates DNA synthesis exclusively with dNTP. (A) The sequence of the 20-nt DNA template used in this experiment is shown at the top, with the NrS-1 polymerase recognition sequence in blue background and the product sequence shown below the template. We mixed 10 μM of this template with 500 nM NrS-1 DNA polymerase N300 fragment, 250 μM dATP, 250 μM dTTP, and 50 μM α-32P-dGTP. The reaction was initiated by the addition of 250 μM of either C (lane 1), CMP (lane 2), CDP (lane 3), CTP (lane 4), dC (lane 5), dCMP (lane 6), dCDP (lane 7), or dCTP (lane 8). Reaction mixtures were incubated at 50 °C for 30 min. The products then were separated on a 25% (wt/vol) TBE-urea polyacrylamide gel. The gel was then analyzed using a phosphoimager. Lane 9 contains the same reaction mixture as that in lane 8, except that it was treated with 1 U/μL calf-intestinal alkaline phosphatase at 37 °C for 15 min before loading on the gel. The identities of the major product bands on the gel are annotated. (B) The sequence of the 23-nt DNA template used in this experiment is shown at the top, with the NrS-1 polymerase recognition sequence in blue background and the product sequence shown below the template. The template was designed for the specificity of NrS-1 polymerase to produce a product initiated with dCTP and followed by a run of dAMPs (5′-CAAAAAAAAAAAAAA-3′). Reactions were prepared and analyzed as in A above, with the mixtures containing 50 μM α-32P-dATP and 50 μM of either C (lane 2), CMP (lane 3), CDP (lane 4), CTP (lane 5), dC (lane 6), dCMP (lane 7), dCDP (lane 8), dCTP (lane 9), ddCTP (lane 10), 2’-F-dCTP (lane 11), or 5m-CTP (lane 12). Lane 1 contains the control reaction mixture carried out in the absence of any dCTP analogs.
We carried out a similar assay using a different DNA template, consisting of the most efficient initiation site 5′-TTTGGTTA-3′ and a template sequence 5′-TTTTTTTTTTTTTTG-3′ encoding a 15-nt runoff product containing only one dCTP (as the first nucleotide) and a string of dAMPs (Fig. 5B). Using this template, we tested the various cytidine analogs at 50 μM, five times lower than that used in the experiment shown in Fig. 5A. At this concentration, the cytidine derivatives C, CMP, CDP, CTP, 2’-F-dCTP, and 5m-CTP can all support the synthesis of abortive products 2–5 nt in length (Fig. 5B, lanes 2–5, 11, and 12), but none of them lead to synthesis of runoff products. The deoxycytidine derivatives dC, dCMP, and dCDP all fail to initiate DNA synthesis by NrS-1 DNA polymerase N300 at this concentration (Fig. 5B, lane 6–8); however, only dCTP initiated the de novo synthesis of both abortive and elongated products (Fig. 5B, lane 9). In many of the reactions, a strong background synthesis of poly-dA was observed, especially in the absence of an efficient initiator like CTP and dCTP, suggesting that the enzyme was able to skip the first position downstream of the initiation site during synthesis. Highly heterogeneous termini of products were observed in these cases, likely due to template slippage during poly-dA synthesis.
Coordination Between NrS-1 DNA Polymerase and Other Phage Replication Proteins.
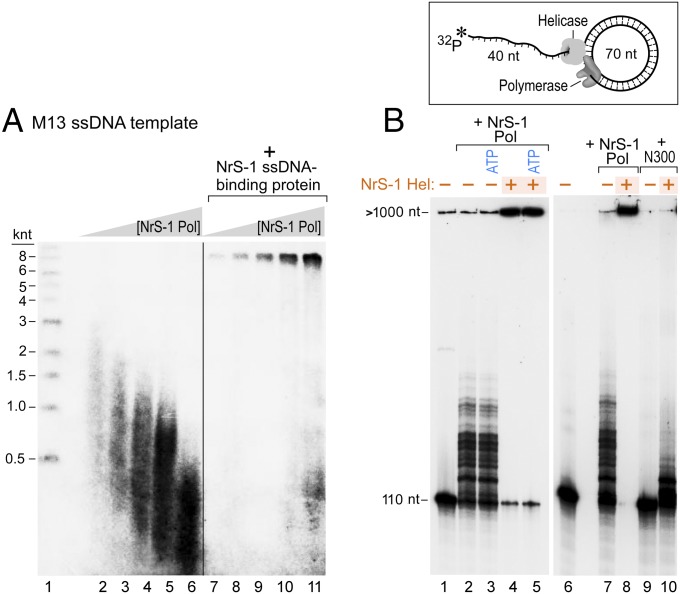
The interaction between NrS-1 DNA polymerase and its recognition sequences on the DNA template may interfere with its processivity and the replication of the whole phage genome. It is likely that NrS-1 DNA polymerase interacts with other NrS-1 proteins to form a replisome (22). The DNA sequence of the NrS-1 genome predicts that two of the genes encode a helicase and a ssDNA-binding protein (21). We overproduced these two gene products that we have designated helicase and ssDNA-binding protein and tested whether they improved the ability of NrS-1 DNA polymerase to replicate long single- and double-stranded DNA templates. When incubated with M13mp18 ssDNA and the four dNTPs, NrS-1 DNA polymerase was able to synthesize DNA up to about 3,000 nt in length (Fig. 2C and Fig. 6A, lane 2). Interestingly, the length of the products synthesized decreases with increasing polymerase concentration (Fig. 6A, lanes 2–6). This result is consistent with the strong affinity of the enzyme for DNA; it is likely that the excess DNA polymerase molecules bind to the ssDNA template and impede the movement of the polymerase extending the primer. At high polymerase concentration, molecular collision may also occur between elongating polymerases and initiating polymerases at sequences mimicking the recognition sites. In M13mp18 ssDNA, there are at least five initiation sites for NrS-1 DNA polymerase including 5′-TTTGATTA-3′, 5′-ATTGACCA-3′, 5′-ATTGGTTA-3′, 5′-GTTGGTCA-3′, and 5′-GTTGGCCA-3′.
Fig. 6.
Coordination between NrS-1 replication proteins. (A) We incubated 10 nM of M13 ssDNA with 250 µM of dATP, dCTP, and dTTP and 25 μM α-32P-dGTP in the absence (lanes 2–6) or presence (lanes 7–11) of 30 μM NrS-1 ssDNA-binding protein (His-tag removed, as shown in Fig. 1B, lane 7). Reactions were carried out with increasing concentrations of NrS-1 DNA polymerase as follows: 12 nM (lanes 2 and 7), 37 nM (lanes 3 and 8), 111 nM (lanes 4 and 9), 333 nM (lanes 5 and 10), and 1,000 nM (lanes 6 and 11). Reactions were carried out at 50 °C for 30 min. The products were separated on a 0.8% alkaline agarose gel, and then the gel was analyzed by a phosphoimager. Lane 1 contains the 5′-32P–labeled DNA size marker. (B) DNA synthesis on a minicircle template to measure leading strand DNA synthesis. The primer template consists of a 110 nt 5′-32P–labeled DNA template annealed to a 70-nt circular template, shown schematically on top of the gel (23). We incubated 50 nM of this template with 200 nM NrS-1 DNA polymerase (lanes 2–5, 7, and 8) and 250 μM four dNTPs in the absence (lanes 2, 3, and 7) or presence (lanes 4, 5, and 8) of 1 μM NrS-1 helicase (shown in Fig. 1B, lane 5). Reaction mixtures were incubated at 50 °C for 30 min. The products were analyzed on a 10% (wt/vol) polyacrylamide TBE-urea gel, and then the gel was analyzed by a phosphoimager. The reaction mixtures in lanes 3 and 5 also contain 1 mM ATP. The reaction mixtures in lanes 9 and 10 contained 500 nM NrS-1 polymerase N300 fragment in place of the full-length NrS-1 polymerase. Lanes 1 and 6 contain the 5′-32P–labeled 110 nt DNA as a marker for no extension.
If the putative NrS-1 ssDNA-binding protein is present at a concentration sufficient to cover the entire ssDNA template, most of the products are extended to the full length of the template (Fig. 6A, lanes 7–11). This result suggests that coordination between NrS-1 DNA polymerase and NrS-1 ssDNA-binding protein improves the processivity of DNA elongation by removing secondary structures in the ssDNA and the polymerase molecules bound to the template ahead of the primer being synthesized. Such coordination enables the NrS-1 DNA polymerase to perform lagging strand synthesis (22).
Efficient leading strand synthesis requires the coordination between a DNA polymerase and a helicase to unwind the duplex DNA ahead of the replication fork (22). We mimicked conditions for leading strand DNA synthesis using a minicircle template described previously (23) (Fig. 6B). On such a template, NrS-1 DNA polymerase demonstrated limited strand-displacement DNA synthesis (Fig. 6B, lanes 2, 3, and 7). However, when the putative NrS-1 DNA helicase is present, the labeled primer can be extended rapidly and extensively by strand-displacement DNA synthesis (Fig. 6B, lanes 4, 5, and 8). Interestingly, the helicase is active in the absence of ATP; presumably the energy for translocation and unwinding of the DNA is provided by one of the four deoxynucleoside triphosphates as in the case of the T7 helicase where the hydrolysis of dTTP provides the energy (Fig. 6B, lane 4). The NrS-1 DNA helicase failed to coordinate with the N300 fragment of NrS-1 DNA polymerase to carry out efficient strand-displacement DNA synthesis, suggesting that the C-terminal subdomain of NrS-1 DNA polymerase is involved in the interaction between the two proteins (Fig. 6B, lane 10).
Discussion
The replicative DNA polymerase from the deep-sea vent phage NrS-1 does not share sequence homology with any of the known replicative DNA polymerase families. Lacking the characteristic finger and thumb subdomains, its N-terminal 200 amino acid subdomain is capable of polymerizing DNA (Fig. 4A, lane 5). The N-terminal 300 amino acid subdomain (Fig. 4B, lane 2) catalyzes de novo DNA synthesis from a specific sequence in the DNA template without exogenous primers. This activity has not been observed previously in DNA polymerases and is similar to the activity observed in primases and RNA polymerases. Polymerization of nucleotides by the enzyme is likely catalyzed by the two-metal ion mechanism typical for all polymerases; we have identified by mutagenesis the two crucial and conserved aspartic acids (24) located in its active site (Fig. 4B, lanes 4 and 5). The accuracy of incorporation by NrS-1 DNA polymerase is relatively low, as both NMPs and mismatching dNMPs can be incorporated in the absence of the correct dNTP (Fig. 1D). However, such misincorporation cannot exceed several nucleotides, suggesting that an imperfect base pair can destabilize the primed template in the enzyme active site. The abortion of polymerization after misincorporation would provide the opportunity for an enzyme, such as an exonuclease, to repair the 3′ terminus of the elongating DNA.
The NrS-1 DNA polymerase is likely to form a replisome with the phage-encoded helicase and ssDNA-binding protein to enhance processivity and coordinate synthesis of the leading and lagging strands at the replication fork. Such a complex would be similar to the replisome described for bacteriophage T7. The T7 replisome consists of T7 DNA polymerase, Escherichia coli thioredoxin as processivity factor, a bifunctional phage-encoded primase–helicase, and a ssDNA-binding protein (22). However, in the NrS-1 replisome, the primase and DNA polymerase are harbored in a single polypeptide.
Sequence homology of the active site residues of NrS-1 polymerase to other proteins suggests that the only known protein group to which it is related is the prim–pols from archaeal plasmids. Such a relationship is intriguing, as phage replication systems are usually closely related to prokaryotic systems. Indeed, these archaeal prim–pols, as well as some archaeal primases and eukaryotic prim–pols, have also been shown to be de novo polymerases, as they can synthesize long polynucleotides using dNTP or NTP or both in the absence of any primer (5–18). The well-studied prim–pol ORF904 from archaeal plasmid pRN1 uses ATP to initiate DNA synthesis (11). In contrast, NrS-1 polymerase catalyzes de novo DNA synthesis in the presence of only dNTPs. One characteristic common to primases is a zinc-binding motif that is involved in template binding and the interaction of the enzyme with its recognition site (2). Prim–pols also possess such a zinc-stem structure (10). Like with primases, the recognition sites on which prim–pols initiate polymerization are short sequences 3–4 nt in length, suggesting that the zinc-binding domains in the two protein families may play a similar role (11, 18). The absence of any zinc-binding motif in NrS-1 polymerase indicates that this enzyme must use a different mechanism to recognize its initiation site on a DNA template. The recognition site for initiation by the NrS-1 polymerase is 8 nt, significantly longer than that found in primase recognition sites (2). With such a long recognition sequence, the specificity of NrS-1 polymerase to initiate polymerization is much higher than that observed with known primases.
Another group of polymerases that initiate de novo synthesis at highly specific template sequences, designated promoters, are DNA-dependent RNA polymerases. The strong binding by RNA polymerases to promoters benefits the assembly of the initial ribonucleotides; however, such binding does not support processive RNA elongation. A drastic conformational change must occur for an RNA polymerase to release from its promoter and transition from initiation mode to elongation mode. As a consequence, a large amount of abortive products ranging from 2 to 12 nt are generated (4). We also observe this phenomenon for NrS-1 polymerase, with abortive products 2–5 nt in length consistently produced during de novo synthesis. In addition, considering the long template recognition sequence observed for NrS-1 polymerase, it is reasonable to propose that a similar conformation change occurs for this enzyme between initiation and elongation stages as that observed for RNA polymerases.
In summary, NrS-1 polymerase shares features of the three classic polymerase families: DNA polymerases, primases, and RNA polymerases. Considering its deep-sea origin, these shared features could reflect an ancient status in polymerase evolution, possibly being a common ancestor for all present polymerases. This suggestion is consistent with the theory that replication enzymes evolved from phages or viruses (25) and that life originated in the ocean (26). Recently, the vast amount of genomic analysis has led to the proposal that marine phage harbor the greatest gene and protein diversity on earth (19). There likely are a great number of novel polymerases in this reservoir that will have evolved novel mechanisms and provide clues for the evolution of genome replication.
Materials and Methods
DNA encoding the wild-type NrS-1 polymerase and its truncated or mutated versions were amplified by PCR from NrS-1 genomic DNA and cloned into plasmid pET28b between the NheI and NotI sites. DNA encoding the NrS-1 helicase and ssDNA-binding protein were also cloned between the NdeI and NotI sites of pET28b. The resulting proteins overproduced from these constructs have a His-tag moiety at their N terminus. Enzymes used for cloning were from New England Biolabs. E. coli BL21(DE3) cells harboring each of the plasmids were grown in 2 L of LB medium containing 50 μg/mL kanamycin at 37 °C until they reached an OD600 of 1.2. Protein expression was induced by the addition of 0.5 mM IPTG at 25 °C, and incubation continued for 5 h. The cells were harvested; resuspended in 50 mM sodium phosphate, pH 8.0, and 100 mM NaCl; and then lysed by three cycles of freeze-thaw in the presence of 0.5 mg/mL lysozyme. NaCl was added to the lysed cells to a final concentration of 1 M, and then the cleared lysate was collected after centrifugation. We added 2 mL Ni-NTA agarose to the clear lysate and gently mixed it at 4 °C overnight. The resin was loaded and collected in a column and washed with 60 mL of 50 mM sodium phosphate, pH 8.0, 1 M NaCl, and 10 mM imidazole. Proteins were eluted from the column using 15 mL of 50 mM sodium phosphate, pH 8.0, 1 M NaCl, and 100 mM imidazole. Eluted fractions were concentrated to 1 mL using an Amicon Ultra-15 centrifugal filter unit (Millipore), and the concentrated sample was loaded directly onto a 200 mL preparative Superdex 200 column. The gel filtration buffer contained 20 mM Tris-HCl, pH 7.5, 1 M NaCl, 0.5 mM DTT, and 0.5 mM EDTA. Fractions were analyzed on SDS/PAGE gels, and those fractions that contained homogenous target proteins were pooled. The pooled fractions were concentrated using an Amicon Ultra-15 centrifugal filter unit followed by dialysis at 4 °C against 50 mM potassium phosphate, pH 7.5, 0.1 mM DTT, 0.1 mM EDTA, and 50% (vol/vol) glycerol and then stored at –20 °C. We have removed the His-tag from NrS-1 polymerase N300 using thrombin cleavage and checked the effect of His-tag on the de novo DNA synthesis activity and found that the N300 with or without His-tag showed similar activity. Thus, we used N-terminal His-tagged version for all enzymes in this work except for the NrS-1 ssDNA-binding protein. ssDNA-binding protein is relatively small in size, and thus, a His-tag may have a larger effect on its activity, so we used thrombin cleavage to remove the His-tag, and the NrS-1 ssDNA-binding protein without His-tag was used in this work. M13mp18 ssDNA was from New England Biolabs. DNA oligonucleotides used as primers and templates were synthesized by Integrated DNA Technologies. The sequences of DNA substrates are shown in the relevant figures and figure legends. Minicircle DNA was prepared as previously described (23). DNA with 5′-32P label was prepared using γ-[32P]ATP (PerkinElmer) and T4 polynucleotide kinase (New England Biolabs). Nucleotides and nucleosides all with a purity >99% were from Sigma-Aldrich, except that 2’-F-dCTP and 5m-CTP had a purity >95% from TriLink. NrS-1 polymerase reactions all contained 20 mM Tris-HCl, pH 8.8, 10 mM (NH4)2SO4, 10 mM KCl, 0.1% Triton X-100, and 5 mM MgSO4 unless stated otherwise. We incubated 10 μL reaction mixtures at 50 °C for 30 min unless stated otherwise. Nucleotides, DNA, and enzymes used in each reaction, as well as the methods used for analysis of the results, are described in the relevant figures and figure legends.
Acknowledgments
We thank Steven Moskowitz (Advanced Medical Graphics) for illustrations. This work was supported by Harvard University, Natural Science Foundation of China Grant 31670175, and the 1000 Young Talent Program of China.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700280114/-/DCSupplemental.
References
- 1.Johansson E, Dixon N. Replicative DNA polymerases. Cold Spring Harb Perspect Biol. 2013;5(6):a012799. doi: 10.1101/cshperspect.a012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frick DN, Richardson CC. DNA primases. Annu Rev Biochem. 2001;70:39–80. doi: 10.1146/annurev.biochem.70.1.39. [DOI] [PubMed] [Google Scholar]
- 3.Kuchta RD, Stengel G. Mechanism and evolution of DNA primases. Biochim Biophys Acta. 2010;1804(5):1180–1189. doi: 10.1016/j.bbapap.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cheetham GM, Steitz TA. Insights into transcription: Structure and function of single-subunit DNA-dependent RNA polymerases. Curr Opin Struct Biol. 2000;10(1):117–123. doi: 10.1016/s0959-440x(99)00058-5. [DOI] [PubMed] [Google Scholar]
- 5.Liu L, et al. The archaeal DNA primase: Biochemical characterization of the p41-p46 complex from Pyrococcus furiosus. J Biol Chem. 2001;276(48):45484–45490. doi: 10.1074/jbc.M106391200. [DOI] [PubMed] [Google Scholar]
- 6.Bocquier AA, et al. Archaeal primase: Bridging the gap between RNA and DNA polymerases. Curr Biol. 2001;11(6):452–456. doi: 10.1016/s0960-9822(01)00119-1. [DOI] [PubMed] [Google Scholar]
- 7.Matsui E, et al. Distinct domain functions regulating de novo DNA synthesis of thermostable DNA primase from hyperthermophile Pyrococcus horikoshii. Biochemistry. 2003;42(50):14968–14976. doi: 10.1021/bi035556o. [DOI] [PubMed] [Google Scholar]
- 8.Lao-Sirieix SH, Bell SD. The heterodimeric primase of the hyperthermophilic archaeon Sulfolobus solfataricus possesses DNA and RNA primase, polymerase and 3′-terminal nucleotidyl transferase activities. J Mol Biol. 2004;344(5):1251–1263. doi: 10.1016/j.jmb.2004.10.018. [DOI] [PubMed] [Google Scholar]
- 9.Lipps G, Röther S, Hart C, Krauss G. A novel type of replicative enzyme harbouring ATPase, primase and DNA polymerase activity. EMBO J. 2003;22(10):2516–2525. doi: 10.1093/emboj/cdg246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lipps G, Weinzierl AO, von Scheven G, Buchen C, Cramer P. Structure of a bifunctional DNA primase-polymerase. Nat Struct Mol Biol. 2004;11(2):157–162. doi: 10.1038/nsmb723. [DOI] [PubMed] [Google Scholar]
- 11.Beck K, Lipps G. Properties of an unusual DNA primase from an archaeal plasmid. Nucleic Acids Res. 2007;35(17):5635–5645. doi: 10.1093/nar/gkm625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Prato S, et al. Molecular modeling and functional characterization of the monomeric primase-polymerase domain from the Sulfolobus solfataricus plasmid pIT3. FEBS J. 2008;275(17):4389–4402. doi: 10.1111/j.1742-4658.2008.06585.x. [DOI] [PubMed] [Google Scholar]
- 13.Soler N, et al. Two novel families of plasmids from hyperthermophilic archaea encoding new families of replication proteins. Nucleic Acids Res. 2010;38(15):5088–5104. doi: 10.1093/nar/gkq236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gill S, et al. A highly divergent archaeo-eukaryotic primase from the Thermococcus nautilus plasmid, pTN2. Nucleic Acids Res. 2014;42(6):3707–3719. doi: 10.1093/nar/gkt1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iyer LM, Koonin EV, Leipe DD, Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: Structural insights and new members. Nucleic Acids Res. 2005;33(12):3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Halgasova N, Mesarosova I, Bukovska G. Identification of a bifunctional primase-polymerase domain of corynephage BFK20 replication protein gp43. Virus Res. 2012;163(2):454–460. doi: 10.1016/j.virusres.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 17.Wan L, et al. hPrimpol1/CCDC111 is a human DNA primase-polymerase required for the maintenance of genome integrity. EMBO Rep. 2013;14:1104–1112. doi: 10.1038/embor.2013.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.García-Gómez S, et al. PrimPol, an archaic primase/polymerase operating in human cells. Mol Cell. 2013;52(4):541–553. doi: 10.1016/j.molcel.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suttle CA. Viruses in the sea. Nature. 2005;437(7057):356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 20.Suttle CA. Marine viruses--Major players in the global ecosystem. Nat Rev Microbiol. 2007;5(10):801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida-Takashima Y, Takaki Y, Shimamura S, Nunoura T, Takai K. Genome sequence of a novel deep-sea vent epsilonproteobacterial phage provides new insight into the co-evolution of Epsilonproteobacteria and their phages. Extremophiles. 2013;17(3):405–419. doi: 10.1007/s00792-013-0529-5. [DOI] [PubMed] [Google Scholar]
- 22.Hamdan SM, Richardson CC. Motors, switches, and contacts in the replisome. Annu Rev Biochem. 2009;78:205–243. doi: 10.1146/annurev.biochem.78.072407.103248. [DOI] [PubMed] [Google Scholar]
- 23.Lee J, Chastain PD, 2nd, Kusakabe T, Griffith JD, Richardson CC. Coordinated leading and lagging strand DNA synthesis on a minicircular template. Mol Cell. 1998;1(7):1001–1010. doi: 10.1016/s1097-2765(00)80100-8. [DOI] [PubMed] [Google Scholar]
- 24.Steitz TA. DNA polymerases: Structural diversity and common mechanisms. J Biol Chem. 1999;274(25):17395–17398. doi: 10.1074/jbc.274.25.17395. [DOI] [PubMed] [Google Scholar]
- 25.Forterre P. The origin of DNA genomes and DNA replication proteins. Curr Opin Microbiol. 2002;5(5):525–532. doi: 10.1016/s1369-5274(02)00360-0. [DOI] [PubMed] [Google Scholar]
- 26.Nisbet EG, Sleep NH. The habitat and nature of early life. Nature. 2001;409(6823):1083–1091. doi: 10.1038/35059210. [DOI] [PubMed] [Google Scholar]