Significance
Metastasis is the main cause of cancer death, but the underlying mechanisms are largely unknown. Here, we developed an orthotopic organoid transplantation approach and used engineered human colon tumor organoids to study the contribution of common CRC mutations to metastasis. Using this approach, we show that the combination of oncogenic mutations in Wnt, EGFR, P53, and TGF-β signaling pathways facilitates efficient tumor cell migration and metastasis. These mutations allow growth independent of stem cell niche factors, enabling cells to grow at foreign distant sites that lack these factors. Our findings suggest that metastasis is a direct consequence of acquired niche independency.
Keywords: colorectal cancer, adenoma-carcinoma sequence, organoids, metastasis, niche independence
Abstract
In the adenoma-carcinoma sequence, it is proposed that intestinal polyps evolve through a set of defined mutations toward metastatic colorectal cancer (CRC). Here, we dissect this adenoma-carcinoma sequence in vivo by using an orthotopic organoid transplantation model of human colon organoids engineered to harbor different CRC mutation combinations. We demonstrate that sequential accumulation of oncogenic mutations in Wnt, EGFR, P53, and TGF-β signaling pathways facilitates efficient tumor growth, migration, and metastatic colonization. We show that reconstitution of specific niche signals can restore metastatic growth potential of tumor cells lacking one of the oncogenic mutations. Our findings imply that the ability to metastasize—i.e., to colonize distant sites—is the direct consequence of the loss of dependency on specific niche signals.
In the early 90s, Vogelstein and coworkers proposed a model in which they defined a set of genetic alterations that facilitate colorectal cancer (CRC) progression (1, 2). In this so-called adenoma-carcinoma sequence, the evolution of a benign tumor toward a highly malignant and metastatic stage arises in a stepwise manner where each step is thought to be associated with specific genetic mutations. Constitutive activation of Wnt signaling is often found in early adenomas and, therefore, mutations in this pathway are considered to be triggering tumor initiation. Progression to the carcinoma stage is associated with activating mutations in the EGF receptor (EGFR) signaling pathway and subsequent inactivating mutations in the transforming growth factor (TGF)-β and P53 pathways (2). Although it is generally believed that the total accumulation of these mutations is more important than their order of occurrence (1, 2), this assumption could never be directly tested. Because of the genetic instability and heterogeneity of CRCs (3), it is challenging to link these specific individual genetic events to discrete steps of metastasis, including the ability of cells to migrate, disseminate, and grow at a foreign distant site (4). Moreover, all of the available genetic mouse models are not suitable to study the effect of combinations of common CRC genetic mutations in the late development of the disease, because these animals inevitably die before the tumors can progress toward the metastatic stage. Here, we describe the establishment of a CRC model based on orthotopic transplantation of human intestinal tumor organoids that leads to the development of colorectal tumors spontaneously metastasizing to distant sites. Moreover, by transplanting human colon organoids engineered to harbor different combinations of the adenoma-carcinoma sequence by using CRISPR/Cas9, we determine the contribution of defined genetic events to the different steps of CRC progression.
Results and Discussion
Generation of Human Colon Organoids with Different CRC Mutation Combinations Using CRISPR/Cas9.
Human intestinal stem cells can be grown “indefinitely” in vitro as 3D organoids in medium containing the stem cell niche factors Wnt, R-spondin, EGF, and Noggin while remaining genetically stable (5, 6). Previously, we used CRISPR/Cas9 genome editing to engineer human colon organoids harboring mutations in four of the most frequently mutated pathways in CRC (Wnt, EGFR, P53, and TGF-β), i.e., mutations in APC [APC knock out (APCKO)], KRAS (KRASG12D), P53 (P53KO), and SMAD4 (SMAD4KO) (7), from here referred to as quadruple mutant organoids. Mutants were functionally selected by changing the medium composition. Moreover, quadruple mutant organoids grow in the absence of all stem cell niche factors and in the presence of the P53-stabilizer nutlin-3 (7). To study the contribution of the different mutations to CRC progression, we additionally engineered various triple mutant human colon organoids that are wild type for P53 (TripleP53WT), APC (TripleAPCWT), KRAS (TripleKRASWT) or SMAD4 (TripleSMAD4WT), using similar selection strategies (SI Appendix, Table S1 and Fig. S1 A–C). In vitro characterization revealed that TripleP53WT, TripleAPCWT, and TripleKRASWT organoids emerged as well-organized cystic structures (SI Appendix, Fig. S2A). As described, TripleSMAD4WT organoids consisted of multilayered disorganized epithelium, whereas quadruple mutant organoids mainly appeared as disorganized solid masses (SI Appendix, Fig. S2A) (7). Cell cycle analysis did not reveal major differences in proliferation rate among TripleAPCWT, TripleSMAD4WT, TripleKRASWT, and quadruple mutant organoids in vitro (SI Appendix, Fig. S2 B and C), indicating that the niche factors present in the conditioned organoid culture medium compensate for the mutation that is lacking in these triple mutants. However, we observed a reduction in the amount of proliferating cells in TripleP53WT organoids, which is caused by an apparent increase in the percentage of cells in G1 phase of the cell cycle (SI Appendix, Fig. S2 B and C). These data, together with the elevated expression of P53 protein (SI Appendix, Fig. S1B), suggest that TripleP53WT cells induce a P53-dependent G1 arrest.
Orthotopic CRC Organoid Transplantation Leads to Formation of Metastatic Tumors.
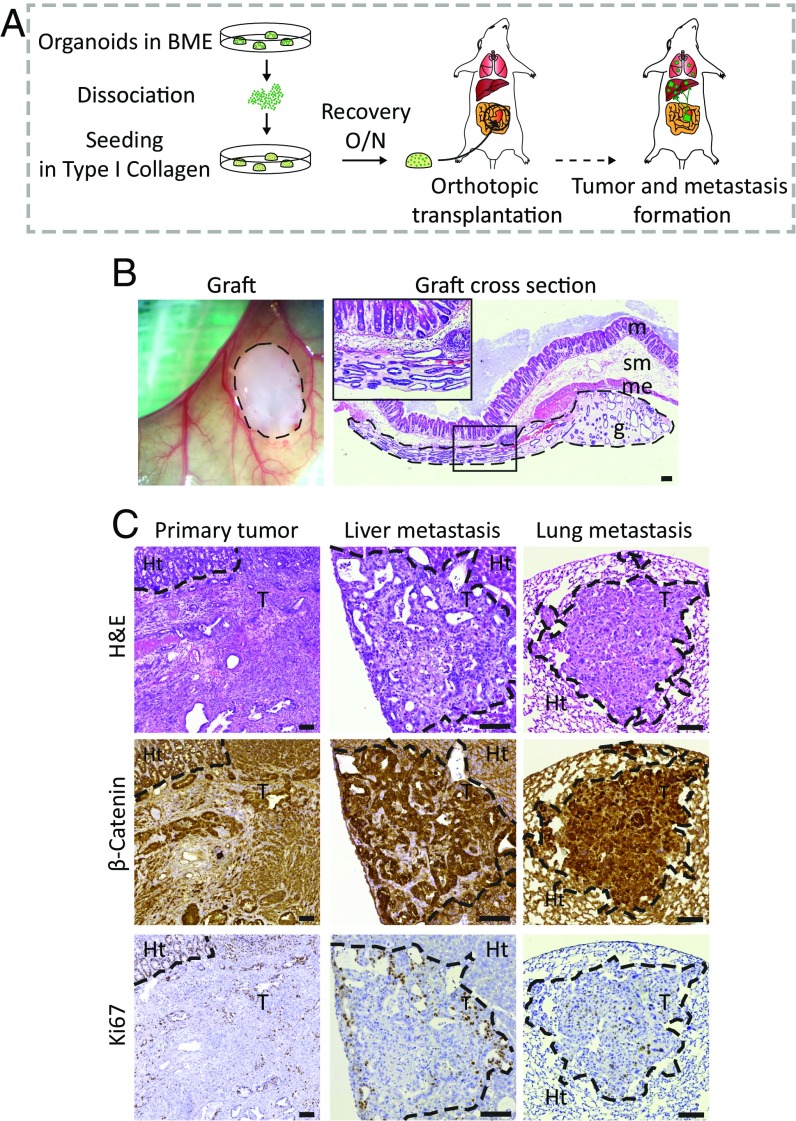
To assess which genetic alterations drive the various steps of metastasis, we developed an orthotopic organoid transplantation model, in which transplanted cells are surrounded by their natural environment. As a proof of principle, we made use of mouse small intestinal organoids (8) derived from Villin-CreERT2::APCfl/fl::KRASG12D/WT::P53fl/R172H transgenic mice. The day before transplantation, the organoids were harvested, dissociated, and plated in high concentration type-I collagen (Fig. 1A). The next day, the organoid-containing collagen drops were surgically transplanted into the submucosa of the caecal wall of immune-deficient mice (Fig. 1 A and B). Approximately 4–6 wk after transplantation, we could detect the presence of primary tumors by abdominal palpation (SI Appendix, Fig. S3A). Immunohistochemical analysis revealed highly proliferative tumors with features of poorly differentiated adenocarcinomas (Fig. 1C and SI Appendix, Fig. S3B). Strikingly, multiple macroscopic metastases were found in liver and lungs 6–8 wk after transplantation (Fig. 1C). Thus, transplantation of mouse APCfl/fl::KRASG12D/WT::P53fl/R172H organoids in their native environment gives rise to primary tumors that spontaneously metastasize to distant organs, allowing us to study the processes underlying metastasis formation.
Fig. 1.
Development of an orthotopic intestinal organoid transplantation model to study CRC progression. (A) Experimental setup of the orthotopic transplantation model. The day before transplantation, 250,000 cells were plated in type I collagen. The collagen drops with the organoids were subsequently transplanted into the caecal wall of immune-deficient mice. Approximately 6–8 wk later, mice were analyzed for tumor growth and presence of metastasis. (B) Representative merged tile scan image of a transplanted collagen drop with organoids (Left) and a cross-section of the graft 1 d after transplantation (Right). g, graft; m, mucosa; me, muscolaris externa; sm, submucosa. Dashed lines highlight the graft. (C) Representative H&E, β-catenin, and Ki-67 staining of a primary tumor, and liver and long metastases. The borders between tumors tissue (T) and healthy tissue (Ht) are indicated with a dotted line. (Scale bars: 100 μm.)
In Vivo Dissection of the Adenoma-Carcinoma Sequence Using Orthotopic Organoid Transplantations.
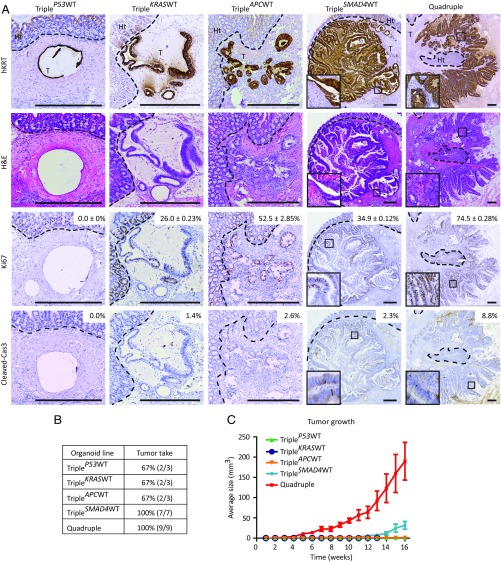
To investigate in a human system which of the adenoma-carcinoma sequence mutations are key drivers of CRC progression toward metastasis formation, we transplanted all CRISPR/Cas9-engineered mutant human colon organoids into the caecal wall of immune-deficient mice. After 16 wk, mice were analyzed for tumor development. Two-thirds (2/3) of the mice implanted with either TripleP53WT, TripleKRASWT, or TripleAPCWT organoids and all of the mice implanted with TripleSMAD4WT (7/7) and quadruple (9/9) mutant organoids developed primary tumors (Fig. 2B). Histological analysis revealed extensive proliferation in quadruple mutant-derived tumors (Fig. 2A) leading to fast growth kinetics (Fig. 2C). By contrast, moderate proliferation and slow growth kinetics were observed in the TripleAPCWT and TripleKRASWT tumors, suggesting that activating mutations in the proliferation-inducing Wnt and EGFR pathways are required for efficient in vivo growth (Fig. 2C) (9, 10). Indeed, TripleSMAD4WT tumors showed high levels of nuclear β-catenin, whereas little to none was detected in TripleAPCWT tumors (SI Appendix, Fig. S4A). Interestingly, TripleSMAD4WT tumors also displayed moderate proliferation (Fig. 2A) and growth (Fig. 2C). These tumors contained features of carcinoma in situ and locally of well-differentiated adenocarcinomas (Fig. 2A), as defined (7). By contrast, quadruple tumors displayed all features of poorly differentiated invasive adenocarcinomas, including irregular multilayered epithelium, increased nuclear-cytoplasmic ratio, pleiomorphic and hyperchromatic nuclei, and invasion of tumor aggregates into the surrounding stroma (Fig. 2A). These data, together with the observation that quadruple mutant-derived tumors showed decreased expression of CK7/8 (SI Appendix, Fig. S4C), suggest that loss of SMAD4 prevents differentiation. Combined, our results suggest that the initiating APC and KRAS mutations drive efficient proliferation and growth, whereas inactivating mutations in SMAD4 block differentiation during tumor progression.
Fig. 2.
Orthotopic transplantation of different CRC driver mutation combinations reveals contribution of the separate mutations to progression of human CRC. (A) Human-specific cytokeratin, H&E, Ki-67, and cleaved caspase-3 immunostainings on tumors isolated from mice transplanted with the indicated mutant human colon organoids. Additionally, percentages of Ki-67 positive (average and SEMs) and cleaved caspase-3-positive cells are depicted. (Scale bars: 500 µm.) (B) List of engineered mutant human colon organoid lines transplanted into the caecal wall of immune-deficient mice with relative tumor take. (C) Graph representing average tumor growth. Tumor volume was measured weekly by palpation. Error bars indicate the SEM of biological replicates, TripleP53WT, TripleKRASWT, and TripleAPCWT n = 3, TripleSMAD4WT n = 8, quadruple n = 9.
Although TripleP53WT organoids contain mutations in the Wnt, EGFR, and TGF-β pathways that promote proliferation and block differentiation, transplantation of these organoids led to tumors consisting of big cysts lined by single-layered epithelium showing no signs of proliferation (Fig. 2A) or growth (Fig. 2C). P53 controls cell cycle progression and cells, with wild-type P53 rapidly stop proliferating in vivo, most likely due to the induction of a P53-dependent cell cycle arrest, as we observed in vitro (SI Appendix, Figs. S1B and S2 B and C). Indeed, TripleP53WT tumors showed marked expression of the cell cycle inhibitor P21 (SI Appendix, Fig. S4B). Importantly, the observation that cell growth depends on the deletion of P53 has consequences for the order of occurrence of mutations during the adenoma-carcinoma sequence. We and others (7, 11) have demonstrated that loss of P53 function in intestinal stem cells induces chromosome instability (CIN). Because CIN can only lead to genetic alterations when cells divide (7, 10), our data indicates that P53 is a gatekeeper that prevents acquisition of additional mutations. Furthermore, accumulation of genetic alterations drives the adenoma-carcinoma sequence and, therefore, it is beneficial for the progression of tumors to have an early loss of P53.
Mutations in APC, KRAS, P53, and SMAD4 Are Required for Tumor Cell Migration.
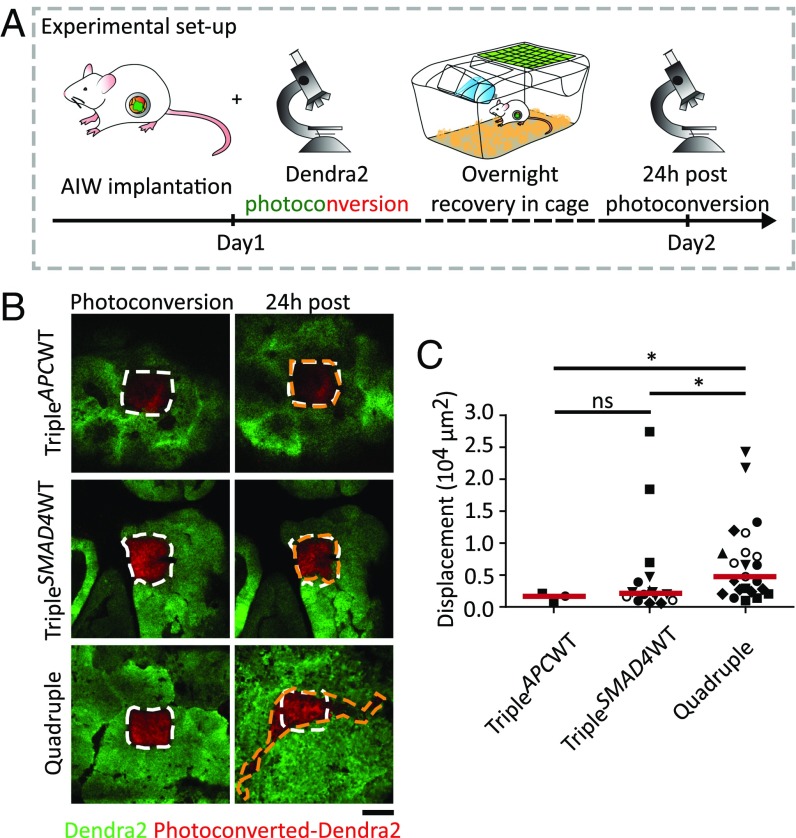
Next, we set out to study how the induced genetic alterations influence the ability of tumor cells to migrate. To fluorescently label tumor cells, we transduced all engineered mutant human colon organoids with a fluorescent photoconvertible Dendra2. By repetitive intravital imaging through an abdominal imaging window (AIW; ref. 12), we monitored tumor cell migration within the mutant organoid-derived primary tumors in living mice. First, we photoconverted from green to red approximately 100 tumor cells located in a square (Fig. 3A). Twenty-four hours after photoconversion, the imaging area was retraced and a change in the localization of the photomarked red cells was monitored as described (13) (Fig. 3A). The imaging was only performed on TripleAPCWT, TripleSMAD4WT, and quadruple mutant-derived tumors (Fig. 3B), because TripleP53WT and TripleKRASWT tumors did not develop to a sufficient size to be imaged. Tumor cells within TripleAPCWT tumors did not show any migratory behavior (Fig. 3B, Top and C and SI Appendix, Fig. S5), whereas TripleSMAD4WT tumor cells displayed a poor migratory phenotype (Fig. 3B, Middle and C and SI Appendix, Fig. S5). Strikingly, quadruple mutant-derived tumor cells showed a significant increase in migratory behavior, indicating that the combination of APC, KRAS, P53, and SMAD4 mutations is required for pronounced tumor cell migration (Fig. 3B, Bottom and C and SI Appendix, Fig. S5).
Fig. 3.
Intravital imaging shows high migratory behavior of tumor cells in quadruple organoid-derived tumors. (A) Setup of the intravital imaging experiment. After a primary tumor has developed, an abdominal imaging window (AIW) (33) was implanted onto the tumor. Photoconversion was performed in randomly picked regions within the tumor. Mice recovered overnight in cage. Twenty-four hours after photoconversion, converted regions were retraced, and the photoswitched areas were analyzed for migration. (B) Representative intravital images of Dendra2-expressing TripleAPCWT, TripleSMAD4WT, and quadruple tumors in which green represents nonswitched Dendra2 and red the photoconverted Dendra2. Images of the photoswitched areas at time point 0 h (Left) and the same imaging area 24 h after photoconversion (Right). White dashed lines highlight the photoswitched areas at beginning of the experiment. Orange dashed lines mark the edges of the red-Dendra2 areas 24 h after photoconversion. (Scale bar: 100 µm.) (C) Displacement of the red-Dendra2 areas 24 h after photoconversion per each condition. Red lines show the median; each symbol represents an imaging field and different symbol shapes represent different animals. TripleAPCWT n = 2, TripleSMAD4WT and quadruple n = 6. Ns, not significant, *P < 0.05.
Mutations in APC, KRAS, P53, and SMAD4 Are Required for Metastatic Growth.
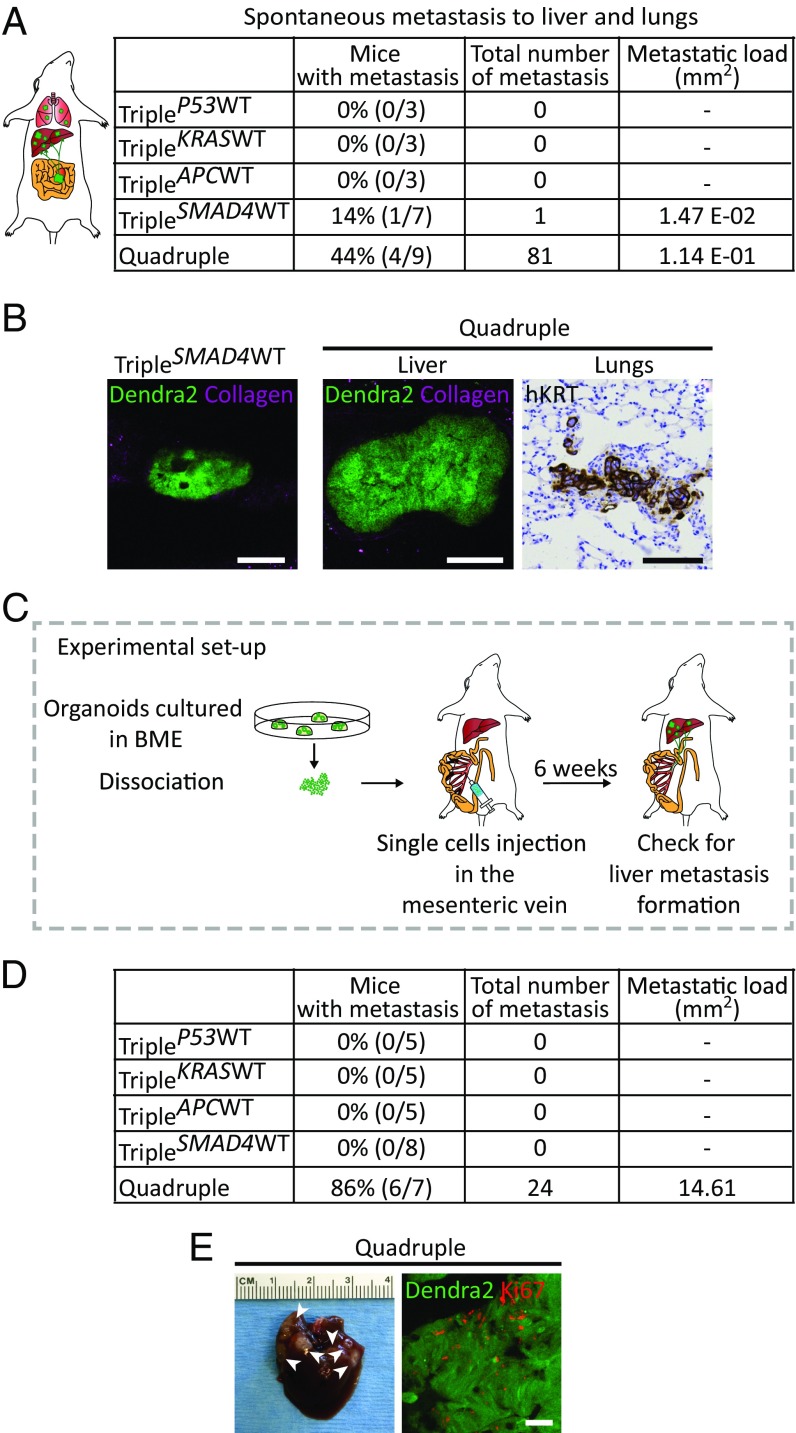
Next, we determined which genetic alterations enable cells to disseminate and grow at distant sites by analyzing the spontaneous metastatic capacity of the orthotopically transplanted organoids. None of the mice transplanted with TripleP53WT, TripleKRASWT, and TripleAPCWT organoids developed metastases. In one of seven mice transplanted with TripleSMAD4WT organoids, only one spontaneous metastatic focus was detected in the liver. Interestingly, four of nine mice transplanted with quadruple mutant organoids developed large amounts of metastases in liver and lungs with high metastatic load (Fig. 4 A and B). These data indicate that quadruple mutant organoids are most prone to metastasize to distant organs. To test the polyclonal nature of these distant metastases, we transplanted a mixture of quadruple mutant organoids expressing either low, intermediate, or high levels of Dendra2. The subsequent developed primary tumors existed of regions that express either low, intermediate, and high levels of Dendra2, which indicates polyclonal outgrowth. Importantly, if metastases are formed by the same clone, it is expected that the Dendra2 expression level is equal in all of them. However, we observed different Dendra2 expression level between the different metastatic foci. These data show that different metastases have originated from different clones present in the primary tumor, and not just from a minority of quadruple mutant organoids that acquired a particular mutation (SI Appendix, Fig. S6). To further exclude that coincident mutations introduced during culturing of the organoids are responsible for the metastatic potential of quadruple mutant organoids, we analyzed the genome-wide mutation load of the quadruple mutant culture at the time it was transplanted, after it was kept in culture for 16 wk and of a resulting metastasis, using whole-genome sequencing. Strikingly, this analysis did not reveal any additional mutations that are likely to contribute to the observed phenotype (SI Appendix, Table S2 and Fig. S7A).
Fig. 4.
Quadruple mutant human colon organoids spontaneously metastasize and efficiently colonize and grow in the liver. (A) Liver and lungs of all transplanted mice were analyzed for presence of spontaneous metastases. The second column shows percentage and number of transplanted mice that spontaneously developed metastases. The third column represents the total number of metastases found, and the last column depicts the metastatic load in squared millimeters. (B) Representative confocal images of a Dendra2-positive liver metastasis in a TripleSMAD4WT-transplanted mouse (Left) and in a quadruple-transplanted mouse (Middle). At Right, representative pictures of a quadruple spontaneous lung metastasis revealed by human-specific cytokeratin immunostaining. (C) Mutant human colon organoids were dissociated, and subsequently 250,000 cells were injected into a mesenteric vein of immune-deficient mice. After 6 wk, livers were analyzed for the presence of metastases. (D) Quantification of the experiment described in C. Second column depicts percentages and numbers of mice that developed liver metastases. Total number of metastases and metastatic load are represented in columns 3 and 4, respectively. (E, Left) Representative picture of a liver containing quadruple mutant metastasis. Arrowheads indicate the metastatic foci. (E, Right) Ki-67 staining (red) of Dendra2-positive quadruple mutant metastasis (green). (Scale bars: B, 100 µm; E, 50 µm.)
Metastatic spread is a process that can also occur upon passive migration of tumor cells into the blood circulation (14). For this reason, although triple mutant-derived tumors do not show any migratory behavior and do not efficiently disseminate, we set out to investigate whether they are still able to colonize and expand in a distant organ when infused into the blood stream. Therefore, we injected suspensions of the same number of single cells from organoid lines with the different mutation combinations in the mesenteric vein. From the mesenteric vein, cells are transported through the portal vein to the liver, which is the primary organ to which CRCs metastasize (15) (Fig. 4C). Interestingly, no metastases were detected in livers of mice injected with TripleP53WT, TripleKRASWT, TripleAPCWT, and TripleSMAD4WT cells, whereas 86% of the mice (6/7) injected with quadruple mutant organoids developed multiple metastases (Fig. 4 D and E, Left). Importantly, Ki-67 staining revealed that quadruple mutant-derived metastatic foci were highly proliferative (Fig. 4E, Right).
Reconstitution of the Stem Cell Niche Factor Noggin Enables Metastasis Formation.
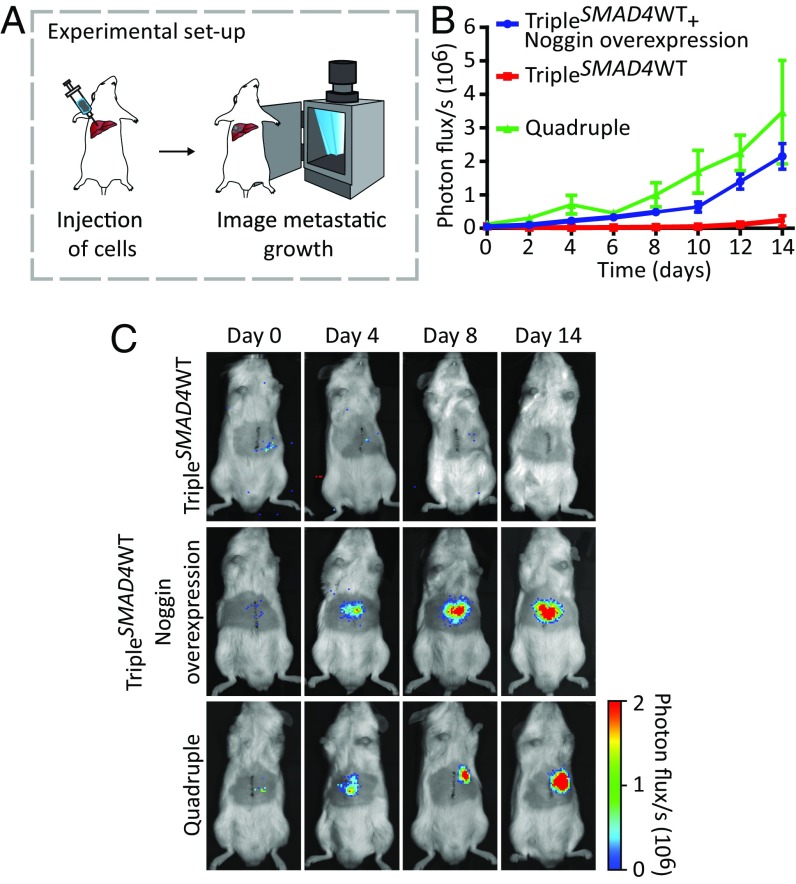
Because depletion of SMAD4 seems to drive the progression of carcinomas in situ (TripleSMAD4WT tumors in Fig. 2B) to invasive carcinomas (quadruple tumors in Fig. 2B) that can grow liver metastasis (Fig. 4), we wondered which mechanism drives this metastatic transition. In vitro, both organoids grow, but opposed to the quadruple mutant, growth of TripleSMAD4WT organoids depends on the polypeptide Noggin that binds and inactivates members of the TGF-β superfamily signaling proteins (16). To test whether niche factor independence is a key contributor to the metastatic potential, we generated TripleSMAD4WT organoids overexpressing Noggin. Importantly, these organoids were now able to grow in vitro in the absence of all of the niche factors. To test whether niche independence enables these tumor cells to grow liver metastases, we injected TripleSMAD4WT organoids overexpressing Noggin into the liver parenchyma (Fig. 5A). To be able to follow tumor growth over time, we stably expressed luciferase in the engineered mutant human colon organoids. Indeed, because of the reconstitution of the lacking niche factor, TripleSMAD4WT organoids overexpressing Noggin were able to grow liver metastasis similarly to quadruple mutant organoids, whereas the nonoverexpressing controls showed limited growth potential (Fig. 5 B and 5C and SI Appendix, Fig. S8D). To further gain direct support of our claim, we used an established approach to reconstitute the niche by intraperitoneally injection of Noggin (17) (SI Appendix, Fig. S8A). Strikingly, under these conditions, TripleSMAD4WT organoids were also able to grow as liver metastasis (SI Appendix, Fig. S8 B–D). Combined these experiments show that niche independence is a key contributor to metastatic growth, although the interplay of other affected cellular processes cannot be excluded.
Fig. 5.
Niche independence is a key contributor to metastatic growth. (A) Mutant human colon organoids were dissociated, and subsequently 50,000 cells were intrahepatically injected in mice. The niche was reconstituted by overexpression of Noggin. Tumor growth was monitored by bioluminescence. (B) The kinetics of metastatic growth. Points are averages, and error bars indicate SEM of biological replicates. Quadruple n = 3, TripleSMAD4WT and TripleSMAD4WT overexpressing Noggin n = 5. (C) Representative images at indicated timepoints.
Concluding Remarks
In this work, we presented an orthotopic approach to study the role that oncogenic CRC alterations in the Wnt, EGFR, P53, and TGF-β signaling pathways play during tumor progression. Preexisting CRC genetic and xenograft models are insufficient to uncover all of the steps of the progression of the disease. Our alternative approach offers the possibility to visualize in vivo the different steps of the metastatic cascade of CRC. Our results show that stem cell niche independent growth through the accumulation of genetic mutations in the Wnt, EGFR, P53, and TGF-β signaling pathways is required for tumor cells to efficiently migrate, disseminate, colonize, and grow at foreign, distant sites. Importantly, the differences in metastatic growth potential between the various organoids is not caused by accidental clonal variations introduced during engineering and selection processes, because similar metastatic potential differences were observed for TripleSMAD4WT and quadruple mutants engineered in an independent healthy colon organoid line (SI Appendix, Fig. S9). Importantly, our model allows studying the contribution of the single adenoma-carcinoma sequence mutations to tumor progression. In the adenoma-carcinoma sequence, it is proposed that the total accumulation of changes, rather than their order with respect to one another, is responsible for tumors to develop (1). Our data confirms and further refines this model, because it shows that P53 is a gatekeeper that prevents acquisition of additional mutations in vivo, and, therefore, early loss P53 is favorable for the progression of CRC. Loss of P53 function allows cells to keep on proliferating, while rendering them sensitive for further accumulation of genetic alternations because of the emergence of CIN (7, 11). Indeed, whole genome sequencing revealed different copy number profiles in P53-deficient organoids, derived primary tumor and corresponding spontaneous liver metastasis (SI Appendix, Fig. S7). These data suggest that additional alterations caused by CIN, such as SMAD4 loss, are most likely involved in tumor progression, which is in line with a previous study (18). Moreover, our data agrees with a recently proposed Big Bang model where only mutations providing a strong selection advantage, i.e., mutations allowing niche independent growth, can persist in a rapidly expanding population (19). Our results are further confirmed by the observation that in addition to loss of P53, SMAD4 inactivation plays a key role in late stages of CRC, such as migration and metastatic outgrowth potential. SMAD4 inactivation appeared to block differentiation, which is in line with previous findings where SMAD4 loss was associated with cell spreading, liver metastasis, and a poor disease prognosis (2, 20–23). Together, our study depicts metastasis as an extremely inefficient process where at least four genetic alterations are required for tumor cells to seed and grow out at distant sites independently of stem cell niche factors. Further research will be required to highlight other alterations specifically involved in triggering more extensive metastatic phenotypes. In this perspective, our approach of orthotopically transplanting engineered human tumor organoids represents a key tool to shed light on the molecular and cellular mechanisms underlying spontaneous progression of CRC.
Experimental Procedures
Human Material for Organoid Cultures.
Approval for this study was obtained by the ethics committees of the Diakonessen Hospital Utrecht. Written informed consent was obtained.
Constructs.
The human codon-optimized Cas9 expression plasmid and sgRNA-GFP plasmid were obtained from Addgene (41815 and 41819). The GFP targeting sequence was exchanged by inverse PCR followed by DpnI digestion and T4 ligation as described (7, 24). APC, P53, SMAD4, and KRAS sgRNA sequences were described (7).
Mouse and Human Organoid Culturing.
Small intestinal organoids were derived from Villin-CreERT2::APCfl/fl::KRASG12D/WT::P53fl/R172H transgenic mice as described (8). Derivation of human colon organoid culture was described (7). In brief, normal human colon tissue was isolated from a resected colon segment derived from a patient (female, age 60 y) diagnosed with CRC (sigmoid) and from a healthy donor (female, age 31 y) retrieved through an endoscopy. Complete human organoid culture medium contains advanced DMEM/F12 medium (Invitrogen) including B27 (Invitrogen), nicotinamide (Sigma-Aldrich), N-acetylcysteine (Sigma-Aldrich), noggin (Peprotech), R-spondin 1 (25), epidermal growth factor (Peprotech), Wnt-conditioned media [50% (vol/vol), produced using stably transfected L cells], TGF-β type I receptor inhibitor A83-01 (Tocris), and p38 inhibitor SB202190 (Sigma-Aldrich).
Transfection and Genotyping of Organoids.
The organoid lipofection and genotyping protocols were described in detail (7, 24). TripleSMAD4WT and quadruple mutant organoids were described (7). To generate TripleP53WT and TripleAPCWT organoids, single-cell suspensions of KRASG12D organoids (7) were cotransfected with Cas9-expressing plasmid and sgRNAs targeting APC and SMAD4 or P53 and SMAD4, respectively. To generate TripleKRASWT organoids, single-cell suspensions of APCKO/P53KO organoids (7) were cotransfected with Cas9-expressing plasmid and an sgRNA-targeting SMAD4. Functional selection strategies were started 2 d after transfection and are depicted for the different mutation combinations in SI Appendix, Table S1. On average, the efficiency of introducing frameshift mutations is ∼1% (7). Single organoids were picked and clonally expanded. Because of the relatively low efficiency, outgrowing organoids are in the vast majority of cases clonal. Clonality was confirmed by Sanger sequencing and for several clones by whole-genome sequencing (SI Appendix, Fig. S1; ref. 7; EGAS00001001969). TripleSMAD4WT and quadruple mutant organoids were generated in the second independent healthy human colon organoid line (female donor, age 31 y) as described (7).
RNA Isolation, cDNA Preparation, and Quantitative RT-PCR.
Organoids were cultured in the presence or absence of Wnt and R-spondin for 48 h. Organoids were harvested in RLT lysis buffer, and RNA was isolated by using the Qiagen RNeasy kit (Qiagen) according to the manufacturer’s instructions. Extracted RNA was used as a template for cDNA production by using GoScript reverse transcriptase (Promega) according to manufacturer’s instructions. Quantitative RT-PCR was performed by using IQ SYBR green mix (Bio-Rad) according to the manufacturer’s protocol. Results were calculated by using ΔΔCt method. Primer sequences: AXIN2_for 5′-AGCTTACATGAGTAATGGGG-3′, AXIN2_rev 5′-AATTCCATCTACACTGCTGTC-3′; GAPDH_for 5′-TGCACCACCAACTGCTTAGC-3′, GAPDH_rev 5′-GGCATGGACTGTGGTCATGAG-3′.
Whole-Genome Sequencing and Read Alignment.
DNA libraries for Illumina sequencing were generated by using standard protocols (Illumina) from 0.5 μg of genomic DNA isolated from organoid cultures by using genomic tips (Qiagen) or by phenol-chloroform extraction. For sequencing of primary tumor material, Dendra2-positive cells were sorted from the resected cecum tissue by FACS. Metastasized tumor cells were expanded as organoids in vitro (3–5 passages) to generate a sufficient number of cells for whole-genome sequencing. All samples were sequenced (2 × 100 bp) by using Illumina HiSeq X Ten sequencers to 30× base coverage. Sequence reads were mapped against human reference genome GRCh37 by using Burrows-Wheeler Aligner v0.5.9 mapping tool (26) with settings 'bwa mem -c 100 -M'. Sequence reads were marked for duplicates by using Sambamba v0.4.732 and realigned per donor by using Genome Analysis Toolkit (GATK) IndelRealigner v2.7.2, and sequence read quality scores were recalibrated with GATK BaseRecalibrator v2.7.2. Full pipeline description and settings also available at: https://github.com/UMCUGenetics/IAP.
In Silico Karyograms.
To detect copy number variations (CNVs), BAM files were analyzed for read-depth variations by Control-FREEC v6.7 with a bin size of 5 kb. Highly variable regions, defined as harboring germ-line CNVs in at least three control samples, were excluded from the analysis. To obtain somatic CNVs, we excluded CNVs for which there was evidence in the reference sample. Median copy number over 500-kb segments was calculated per chromosome and plotted. Regions identified as deleted or gained by FREEC are colored red and blue, respectively, according to HGVS standards (www.hgvs.org/). Odd and even chromosomes are shaded gray and black.
Point Mutation Calling and Filtering.
Raw variants were multisample-called by using the GATK HaplotypeCaller v3.4–46 (27) and GATK-Queue v3.4–46 with default settings and additional option ‘EMIT_ALL_CONFIDENT_SITES’. The quality of variant and reference positions was evaluated by using GATK VariantFiltration v3.4–46 with options ‘-snpFilterName LowQualityDepth -snpFilterExpression “QD < 2.0” -snpFilterName MappingQuality -snpFilterExpression “MQ < 40.0” -snpFilterName StrandBias -snpFilterExpression “FS > 60.0” -snpFilterName HaplotypeScoreHigh -snpFilterExpression “HaplotypeScore > 13.0” -snpFilterName MQRankSumLow -snpFilterExpression “MQRankSum < −12.5” -snpFilterName ReadPosRankSumLow -snpFilterExpression “ReadPosRankSum < −8.0” -cluster 3 -window 35’. To obtain high-quality somatic point mutation catalogs, we applied the following postprocessing filters: absence of the variant with any evidence from a paired control sample (the original bulk culture used to generate the mutant lines), a minimum depth of 10× base coverage in both test and control sample, variants passed by VariantFiltration with a GATK phred-scaled quality score ≥100, exclusion of variants that overlap a single nucleotide polymorphism (SNP) position in the Single Nucleotide Polymorphism Database v137.b37 (28) and absence of variant from panel of unmatched normal human genomes (not in study, BED-file available upon request). To obtain catalogs of mutations with potentially functional consequences, we used SnpSift -filter with options ‘ANN[*].IMPACT=’HIGH’||ANN[*].IMPACT=’MODERATE’’ (29).
Western Blotting.
Protein lysate preparation and Western blot procedure was performed as described (7). The antibodies used were as follows: P53 (DO-1; Santa Cruz Biotechnology), SMAD4 (B8; Santa Cruz Biotechnology), and GAPDH (ab-9485; Abcam).
Cell Cycle Profile Analysis.
Organoids were incubated in complete organoid culture medium containing 10 μM bromodeoxyuridine (BrdU; Sigma-Aldrich) for 6 h. Organoids were dissociated into single cells by using TrypLE (Gibco) and subsequently fixed in 70% (vol/vol) ethanol at 4 °C for 1 h. Cells were washed with PBS and treated for 30 min at 37 °C with RNase A (0.5 mg/mL). After PBS wash, cells were incubated for 20 min in 5 M HCl/0.5% Triton solution, and neutralized with 0.1 M Na2B4O7. Cells were sequentially stained with mouse anti-human BrdU (Dako) and Alexa Fluor 488 goat anti-mouse IgG (Life Technologies). Before FACS, cells were resuspended in 50 μg/mL propidium iodide in PBS.
Mice.
Eight- to 12-wk-old males NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were used as acceptors for orthotopic transplantation, mesenteric vein injection, and liver injection. All experiments were performed in accordance with the Animal Welfare Committee of the Royal Netherlands Academy of Arts and Sciences, The Netherlands. Animals were kept at the Hubrecht animal facility in Utrecht, The Netherlands.
Orthotopic Transplantation.
The day before transplantation, organoids were collected and dissociated in small clumps of approximately 3–5 cells by using TrypLE Express (Life Technologies). Approximately 250,000 cells were plated in 15-µL drops of neutralized Rat Tail High Concentrated Type I Collagen (Corning). Organoids were allowed to recover overnight at 37 °C, 5% (vol/vol) CO2. Acceptor mice were subsequently sedated by using isoflurane inhalation anesthesia [∼2% (vol/vol) isoflurane/O2 mixture]. Before surgery, the mice were treated with an s.c. dose of buprenorphine (3 mg per mouse; Temgesic, BD Pharmaceutical System). The cecum was exteriorized through a midline abdominal incision and a single collagen drop containing organoids was surgically transplanted in the caecal submucosa. The collagen keeps the organoids in place, allowing them to engraft and expand in the cecal wall. Over time, the collagen is degraded and the engrafted tissue remains. The mice were checked weekly for the presence of tumors via abdominal palpation.
Immunohistochemistry.
Tissues were fixed in 4% (wt/vol) paraformaldehyde, dehydrated, and embedded in paraffin. H&E and immunohistochemical staining were performed on 4-µm sections. The stainings were performed with the following primary antibodies: anti-β-catenin clone 14 (BD Bioscience), anti-Ki-67 clone MM1 (Monosan), anti-cleaved-Caspase3 D175 (Cell Signaling), anti-cytokeratin clone Cam5.2 (BD Bioscience), anti-P21 sc471 (Santa Cruz), anti-Dendra2 (Evrogen). Detection was realized with anti-mouse (Dako; Envision+ System- HRP labeled Polymer anti-mouse), anti-rabbit (ImmunoLogic; Poly-HRP) horseradish peroxidase labeled secondary antibodies and Alexa Fluor goat anti-rabbit 488 (Life Technologies) fluorescent secondary antibody. Membrane staining was realized with Phalloidin-Atto 647N (Sigma-Aldrich).
Intravital Imaging.
To perform intravital microscopy, human in vitro engineered organoids were infected (30) with pLKO-1.UbC.Dendra2.puro lentivirus. Ten to 12 wk after orthotopic organoid transplantation, an abdominal imaging window was surgically implanted on the primary tumor as described (12, 31). The surgical procedure was performed under isoflurane inhalation anesthesia. Before surgery, the mice were treated with an s.c. dose of buprenorphine (3 mg per mouse, Temgesic; BD Pharmaceutical System). After surgery, the mice were kept at 37 °C until fully recovered. For every imaging session, mice were sedated by using isoflurane inhalation anesthesia (∼1.5% isoflurane/O2 mixture), and placed with their head in a facemask within a custom designed imaging box. The imaging box and microscope were kept at 36.5 °C by using a climate chamber. Between the imaging sessions, mice were allowed to recover in their cage. Intravital images were acquired with an inverted Leica TCS SP5 AOBS two-photon microscope with a chameleon Ti:Sapphire pumped Optical Parametric Oscillator (Coherent). To monitor tumor cell migration, Dendra2 photoswitching was performed as described (32, 33). Briefly, the region of interest (ROI) scan option of LAS AF Lite software was used to photoconvert a chosen population of cells by using the 405-nm laser line (20% power, 40 scans). The converted-red isoform of the protein was collected to monitor cell migration. The imaging areas were retraced in subsequent imaging sessions by storing the stage coordinates of the imaging areas and tracking photomarked Dendra2 cells as described (31). Custom-designed Visual Basic and ImageJ software were used to enhance the red signal from the photoswitched cells. Noise was removed from the images and regions of interest were drawn around the Dendra2-photoconverted areas of comparable z planes of the two consecutive imaging days. The displacement of the photoconverted cells was calculated by using the XOR function. Statistical analysis was performed in GraphPad Prism by using Mann–Whitney u test.
Mesenteric Vein Injection.
Before surgery, organoids were collected and dissociated in single cells by using TrypLE Express (Life Technologies). We resuspended 250,000 cells in 100 μL of sterile PBS and injected them in the mesenteric vein of acceptor mice as described (15). The surgical procedure was performed under isoflurane inhalation anesthesia. Before surgery, the mice were treated with a s.c. dose of buprenorphine (3 mg per mouse, Temgesic; BD Pharmaceutical System). After surgery, the mice were kept at 37 °C until fully recovered. Six weeks after injection the mice were killed, and the livers were inspected for presence of metastasis under a fluorescence-stereo microscope (Leica). Metastases were stained for Ki-67 (clone SP6; Abcam) to check for cell proliferation. Detection was realized with Alexa Fluor goat anti-rabbit 594 (Life Technologies) fluorescent secondary anibody.
Intrahepatic Injection.
To perform bioluminescence imaging (BLI), human in vitro engineered organoids were infected (30) with pLKO-1.UbC.fire-fly-luciferase.blast lentivirus. For Noggin overexpression, cells were additionally infected with pLKO-1.UbC.mNog.puro lentivirus. Before surgery, organoids were collected and dissociated in single cells by using TrypLE Express (Life Technologies). We resuspended 50,000 cells in 100 μL of sterile Matrigel (Corning) and PBS mix (2.5:1) and injected into the liver parenchyma. The surgical procedure was performed under isoflurane inhalation anesthesia. Before surgery, the mice were treated with an s.c. dose of buprenorphine (3 mg per mouse, Temgesic; BD Pharmaceutical System). After surgery, the mice were kept at 37 °C until fully recovered.
Noggin Administration.
For Noggin administration experiments, mice were injected i.p. daily with 0.5 mg/kg recombinant human Noggin (Peprotech) starting from the day before intrahepatic injection until the last bioluminescence imaging session.
Bioluminescence Imaging.
Bioluminescence imaging was executed immediately after intrahepatic injection and after, every 48 h for a maximum of 8 d after injection. To perform the imaging, mice were kept under isoflurane inhalation anesthesia. Mice injected with 60 µg/kg d-Luciferin (ThermoFisher Scientific), and after a 5-min incubation, imaged in a Aequoria Dark Box (Hamamatsu) by using 4 × 4 binning, an EM gain of 1,200 and exposure times ranging from 0.5 to 300 s. The metastatic growth kinetics were measured by using HoKaWo software (Hamamatsu) by quantification of the total intensity of nonsaturated ROIs followed by a background subtraction. The photon flux/s was determined by dividing this number by the exposure time.
Supplementary Material
Acknowledgments
We thank Anko de Graaff and the Hubrecht Imaging Centre for imaging support and Stefan van der Elst for FACS support. This work was financially supported by NWO-ZonMw Veni Grant 91614138 (to J.D.), Dutch Cancer Society Fellowship BUIT-2013-5847 (to S.J.E.S.), European Research Council Grant COLONCAN and CRUK Program (to O.J.S.), European Research Council Grant CANCER-RECURRENCE 648804 (to J.v.R.), and the CancerGenomics.nl (Netherlands Organisation for Scientific Research) program (to H.C. and J.v.R.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequencing data have been deposited to the European Genome-Phenome Archive (https://www.ebi.ac.uk/ega/) under accession no. EGAS00001001969.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1701219114/-/DCSupplemental.
References
- 1.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 2.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 3.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386(6625):623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- 4.Nguyen DX, Bos PD, Massagué J. Metastasis: From dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9(4):274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 5.Sato T, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141(5):1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- 6.Jung P, et al. Isolation and in vitro expansion of human colonic stem cells. Nat Med. 2011;17(10):1225–1227. doi: 10.1038/nm.2470. [DOI] [PubMed] [Google Scholar]
- 7.Drost J, et al. Sequential cancer mutations in cultured human intestinal stem cells. Nature. 2015;521(7550):43–47. doi: 10.1038/nature14415. [DOI] [PubMed] [Google Scholar]
- 8.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 9.Korinek V, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC-/- colon carcinoma. Science. 1997;275(5307):1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 10.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 11.Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333(6051):1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- 12.Ritsma L, et al. Surgical implantation of an abdominal imaging window for intravital microscopy. Nat Protoc. 2013;8(3):583–594. doi: 10.1038/nprot.2013.026. [DOI] [PubMed] [Google Scholar]
- 13.Kedrin D, et al. Intravital imaging of metastatic behavior through a mammary imaging window. Nat Methods. 2008;5(12):1019–1021. doi: 10.1038/nmeth.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bockhorn M, Jain RK, Munn LL. Active versus passive mechanisms in metastasis: Do cancer cells crawl into vessels, or are they pushed? Lancet Oncol. 2007;8(5):444–448. doi: 10.1016/S1470-2045(07)70140-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Bij GJ, et al. Experimentally induced liver metastases from colorectal cancer can be prevented by mononuclear phagocyte-mediated monoclonal antibody therapy. J Hepatol. 2010;53(4):677–685. doi: 10.1016/j.jhep.2010.04.023. [DOI] [PubMed] [Google Scholar]
- 16.Fujii M, et al. A colorectal tumor organoid library demonstrates progressive loss of niche factor requirements during tumorigenesis. Cell Stem Cell. 2016;18(6):827–838. doi: 10.1016/j.stem.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 17.Ramasamy SK, Kusumbe AP, Wang L, Adams RH. Endothelial Notch activity promotes angiogenesis and osteogenesis in bone. Nature. 2014;507(7492):376–380. doi: 10.1038/nature13146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matano M, et al. Modeling colorectal cancer using CRISPR-Cas9-mediated engineering of human intestinal organoids. Nat Med. 2015;21(3):256–262. doi: 10.1038/nm.3802. [DOI] [PubMed] [Google Scholar]
- 19.Sottoriva A, et al. A Big Bang model of human colorectal tumor growth. Nat Genet. 2015;47(3):209–216. doi: 10.1038/ng.3214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyaki M, et al. Higher frequency of Smad4 gene mutation in human colorectal cancer with distant metastasis. Oncogene. 1999;18(20):3098–3103. doi: 10.1038/sj.onc.1202642. [DOI] [PubMed] [Google Scholar]
- 21.Papageorgis P, et al. Smad4 inactivation promotes malignancy and drug resistance of colon cancer. Cancer Res. 2011;71(3):998–1008. doi: 10.1158/0008-5472.CAN-09-3269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang B, et al. Antimetastatic role of Smad4 signaling in colorectal cancer. Gastroenterology. 2010;138(3):969–980 e961-963. doi: 10.1053/j.gastro.2009.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kodach LL, et al. The bone morphogenetic protein pathway is active in human colon adenomas and inactivated in colorectal cancer. Cancer. 2008;112(2):300–306. doi: 10.1002/cncr.23160. [DOI] [PubMed] [Google Scholar]
- 24.Schwank G, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13(6):653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 25.Kim KA, et al. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science. 2005;309(5738):1256–1259. doi: 10.1126/science.1112521. [DOI] [PubMed] [Google Scholar]
- 26.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43(5):491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherry ST, et al. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6(2):80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Koo BK, et al. Controlled gene expression in primary Lgr5 organoid cultures. Nat Methods. 2011;9(1):81–83. doi: 10.1038/nmeth.1802. [DOI] [PubMed] [Google Scholar]
- 31.Ritsma L, et al. Intestinal crypt homeostasis revealed at single-stem-cell level by in vivo live imaging. Nature. 2014;507(7492):362–365. doi: 10.1038/nature12972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gligorijevic B, Kedrin D, Segall JE, Condeelis J, van Rheenen J. Dendra2 photoswitching through the Mammary Imaging Window. J Vis Exp. 2009;28:e1278. doi: 10.3791/1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritsma L, et al. Intravital microscopy through an abdominal imaging window reveals a pre-micrometastasis stage during liver metastasis. Sci Transl Med. 2012;4(158):158ra145. doi: 10.1126/scitranslmed.3004394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.