Significance
Monitoring human organ-on-a-chip systems presents a significant challenge, where the capability of in situ continual monitoring of organ behaviors and their responses to pharmaceutical compounds over extended periods of time is critical in understanding the dynamics of drug effects and therefore accurate prediction of human organ reactions. In this work, we report a fully integrated modular physical, biochemical, and optical sensing platform, interfaced through a fluidics-routing breadboard with a multi–organ-on-a-chip system to achieve in situ, continual, and automated sensing of microenvironment biophysical and biochemical parameters. It is anticipated that our platform technology that is conveniently compatible with existing organ-on-a-chip models will potentially enhance their performance in drug screening by providing a multitude of sensing data not previously available.
Keywords: organ-on-a-chip, microbioreactor, electrochemical biosensor, physical sensor, drug screening
Abstract
Organ-on-a-chip systems are miniaturized microfluidic 3D human tissue and organ models designed to recapitulate the important biological and physiological parameters of their in vivo counterparts. They have recently emerged as a viable platform for personalized medicine and drug screening. These in vitro models, featuring biomimetic compositions, architectures, and functions, are expected to replace the conventional planar, static cell cultures and bridge the gap between the currently used preclinical animal models and the human body. Multiple organoid models may be further connected together through the microfluidics in a similar manner in which they are arranged in vivo, providing the capability to analyze multiorgan interactions. Although a wide variety of human organ-on-a-chip models have been created, there are limited efforts on the integration of multisensor systems. However, in situ continual measuring is critical in precise assessment of the microenvironment parameters and the dynamic responses of the organs to pharmaceutical compounds over extended periods of time. In addition, automated and noninvasive capability is strongly desired for long-term monitoring. Here, we report a fully integrated modular physical, biochemical, and optical sensing platform through a fluidics-routing breadboard, which operates organ-on-a-chip units in a continual, dynamic, and automated manner. We believe that this platform technology has paved a potential avenue to promote the performance of current organ-on-a-chip models in drug screening by integrating a multitude of real-time sensors to achieve automated in situ monitoring of biophysical and biochemical parameters.
Drug discovery is a lengthy process associated with tremendous cost and high failure rate (1). On average, less than 1 in 10 drug candidates are eventually approved by the Food and Drug Administration (2), during which successful transition ratios to next phases are roughly 65, 32, and 60% for phase I, phase II, and phase III, respectively (3). Among the primary causes of failure, nonclinical/clinical safety (>50%) and efficacy (>10%) stand out in the front, more than all other factors (e.g., strategic, commercial, operational) combined (3, 4). These two major causes not only contribute to the low success rates during the drug development but also lead to the withdrawal of approved drugs from the market. Among all of the drug attritions, it is estimated that safety liabilities related to the cardiovascular system account for 45%, whereas 32% are due to hepatotoxicity (5, 6). These high failure/attrition ratios of drugs in human patients are mainly attributed to the inefficient validation based on planar static cell cultures and preclinical animal models for establishing drug efficacy and safety (7). Therefore, there is an urgent need for the development of human-based 3D organ models that precisely recapitulate the important physiology and function of their counterparts in humans as viable platforms for drug screening (8–10).
To this end, the recently emerged organ-on-a-chip systems that combine advanced microfluidic technologies and tissue engineering approaches to simulate both the biology and physiology of human organs have been developed (8, 11–30). These miniaturized human organ models have several advantages over conventional models, such as more accurate prediction of human responses and, in particular, multiorgan interactions when different organ modules are assembled in a single fluid circuit (7, 31). Although a majority of focus in the field has been placed on the construction of biomimetic organ models (7, 15, 31–33), it is increasingly recognized that incorporating biosensing would allow for in situ monitoring of the status of these miniaturized organs (9, 34). Such a need originates from the fact that many drugs can trigger chronic cellular reactions, whereas others may induce delayed cell responses. Conventional analytical methods require manual sample collection from the microfluidic system, large working volumes, and frequent system disturbance, and thus are not suitable for miniaturized organ-on-a-chip platforms (9). Rather, an integrated system that seamlessly combines not only organ models and microfluidic units but also biosensors, which ideally runs in a continuous and fully automated manner over extended periods of time, would be very much desired. Several efforts have been initiated toward the accomplishment of this ambitious goal. For example, prototype microphysiometers have been developed for on-line measurement of glucose and lactate (19, 35), oxygen (35), transepithelial electrical resistance (24, 36, 37), ions (38), extracellular acidification rate (pH), and protein biomarkers (39) in organ-on-a-chip systems (40–46). However, these sensing units were either built upon a single chip and limited in system-level integration and full automation (19), or were designed for measurements over relatively short periods of time in the range of minutes to hours. In addition, despite that the available commercial sensors are able to monitor a wide range of cellular parameters, such as the Multiparametric BioChip-H (for measuring pH, O2, temperature, and electrical signals) by Cellasys, GLU.K.OMETERPRO (for glucose) and LACPRO (for lactate) by BST, and VisiSens (for 2D visualization of pH, O2, or CO2 distributions) by PreSens, the integration of these sensors with organ-on-a-chip platforms cannot be easily achieved due to their incompatibility with microfluidic systems.
Here, we report the development of a fully integrated modular platform that includes a microfluidic controlling breadboard for timed routing of fluids, physical sensors for monitoring extracellular microenvironment parameters (e.g., pH, O2, temperature), electrochemical sensors for measuring soluble protein biomarkers, and miniature microscopes for observation of organoid morphologies. All of the sensing could be performed through uninterrupted in situ monitoring. The integrated system was controlled by a computer and operated in a completely automated manner for at least 5 d. We analyzed the performance of the automated microfluidic control as well as characterized the capacity of the integrated physical and biochemical sensors. We further investigated the possibility to adapt such an integrated platform for monitoring drug-induced organ toxicity using dual-organ human liver-and-heart-on-a-chip and liver-cancer-and-heart-on-a-chip model systems, to demonstrate long-term assessment of chronic drug responses and short-term evaluation of acute toxicity, respectively. We believe that the platform technology described in this paper will pave an avenue for interfacing existing biomimetic organ-on-a-chip models to achieve automated in situ monitoring of biophysical and biochemical parameters.
Results and Discussions
Modular Design of the Integrated Organ-on-a-Chip Platform.
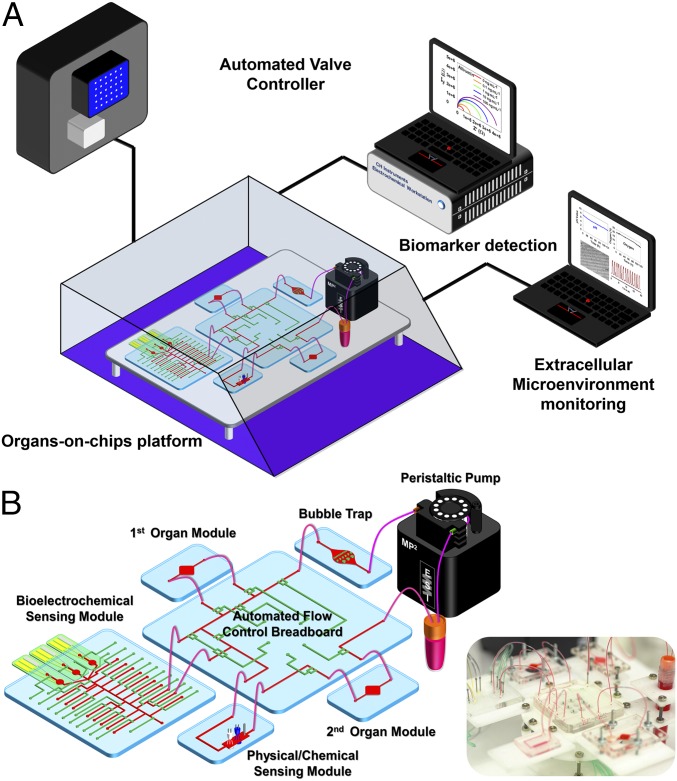
This organ-on-a-chip platform was contained within a custom-designed benchtop incubator capable of maintaining appropriate temperature and CO2 level (Fig. 1A). The on-chip valving was powered by nitrogen pressure applied through a programmable Wago controller and Festo valves (SI Appendix, Fig. S1), using a set of MATLAB codes. These codes were written to also drive the electrochemical station, which was annexed to a multiplexed detector, for automated electrochemical measurements at predetermined time points. Physical sensing was achieved using a data acquisition (DAQ) card connected to a LabVIEW program, and was continuous during the entire course of experiments at a desired data-sampling rate. A flow sensor was also incorporated into the system to monitor the flow rate and potential channel blockage and leakage.
Fig. 1.
Integrated automated multiorgan-on-a-chip and sensing platform. (A) Schematic of a full system where the multiorgan-on-a-chip platform was encased in an in-house designed benchtop incubator, and of automated pneumatic valve controller, electronics for operating physical sensors, potentiostat for measuring electrochemical signals, and computer for central programmed integration of all of the commands. (B) Schematic of the integrated microfluidic device consisting of modular components including microbioreactors, breadboard, reservoir, bubble trap, physical sensors, and electrochemical biosensors. Inset shows the photograph of an integrated platform.
Our integrated organ-on-a-chip platform was designed to be modular, including a breadboard for microfluidic routing via built-in pneumatic valves, microbioreactors for housing organoids, a physical sensing suite for measurement of microenvironment parameters, one or multiple electrochemical sensing units for detection of soluble biomarkers secreted by the organoids, a medium reservoir, and bubble traps (Fig. 1B and SI Appendix, Fig. S2). Individual modules were interconnected with Teflon tubes to allow for fluid flow. Each module in the circuit may also be individually replaced on demand, such as the medium reservoir, the bubble traps, and the electrochemical sensing chips.
Breadboard for Fluid Routing and Bubble Traps.
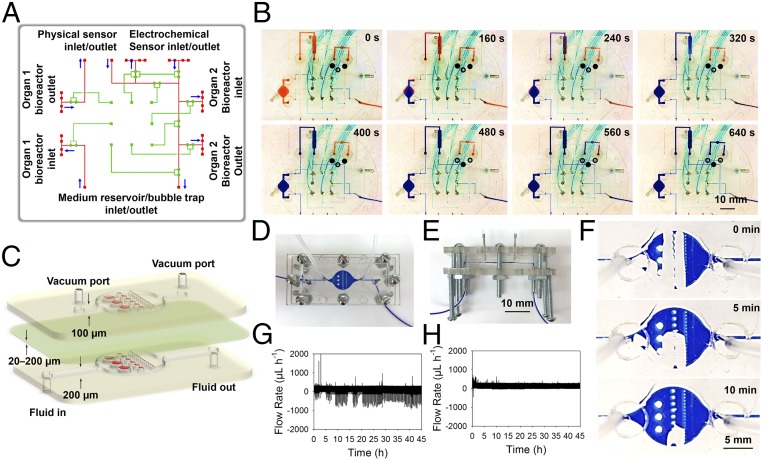
To control the routing of the medium to different modules at predetermined time points, we designed a unique breadboard system with built-in microchannels and pneumatic valves that allowed programmable fluid manipulation (Fig. 2A). The fluid microchannels were processed into hemicylindrical shapes to allow for complete sealing when the thin-membrane valves were actuated (SI Appendix, Fig. S3). A double-valve mechanism was also adopted to secure the fluid sealing and prevent leakage due to the potentially high pressures at the transition points between the breadboard and modules. In a typical routing example, the microbioreactor(s), the physical sensing unit, and the medium reservoir/bubble trap were constantly perfused through the breadboard connections to achieve organoid culture and continuous monitoring of the microenvironment, whereas the electrochemical sensing unit was only accessed when necessary for biomarker measurements. Such a process could be precisely automated by programmed actuation of the valves on the breadboard (Fig. 2B), where the fluidic flow to the electrochemical sensing module was suspended during regular perfusion through the microbioreactors and the physical sensing unit, but restored by activating the corresponding valve set. Additionally, the fluidic channels of the breadboard could be coated with a layer of endothelial cells (SI Appendix, Fig. S4). The endothelial cells were able to proliferate to become confluent within the channels in 14 d, where the perfusion seemed to have aligned the cells better along the direction of flow (SI Appendix, Fig. S4 E and F). These endothelialized breadboards were not integrated for further experiments in the current work, but rather indicated potential future function as its own vascular module for interconnecting multiple organoids without the need for additional vascular microbioreactors (47).
Fig. 2.
Microfluidic-based fluid routing and operations. (A) Schematic diagram showing design of the breadboard module consisting of a microfluidic layer (red) and a pneumatic valve controlling layer (green) for microfluidic routing of different modules. (B) Photographs showing the constant routing (e.g., the microbioreactor and mimic physical sensing unit) and timed routing (e.g., the mimic electrochemical sensing unit) via controlled valve operations. For the constant routing, the two valves of the channels connecting to the mimic electrochemical sensing unit were closed (see signs in the panels), whereas that for the constant routing was open, resulting in the red dye gradually being washed out from the microbioreactor, the mimic physical sensing unit, and the breadboard by the infused blue dye (0–400 s); for the timed routing, the two valves of the mimic electrochemical sensing unit were released, whereas that for the constant routing was closed, and therefore the blue dye was forced to flow into this unit to exchange the red dye (400–640 s). (C) Schematic diagrams showing design of the bubble trap, which is composed of three layers: (i) a fluidic layer at the bottom with a thickness of 200 µm composed of arrays of micropillars with different sizes (1,000, 500, and 250 µm); (ii) a thin PDMS membrane in the middle with a thickness of ∼20–200 µm; and (iii) a vacuum layer at the top with a thickness of 100 µm composed of the same arrangement of micropillars. (D and E) Photographs showing the assembly of the bubble trap. (F) Time-lapse images showing trapping and removal of a large air bubble with a volume of around 10 µL. (G and H) Plots showing flow rate data obtained from a flow rate sensor in the (G) absence and (H) presence of an upstream bubble trap.
Due to the gas permeability of the polydimethylsiloxane (PDMS) and the presence of junction points, air bubbles of different sizes could be generated inside the microfluidics. Therefore, a miniaturized trapping device to capture and remove these air bubbles was designed to prevent potential interference with sensors and optics, which is critical for proper functioning of the integrated platform reported in this work. The bubble trap featured a 10-mm single chamber containing micropillars with a series of uniform diameters of 1,000, 500, and 250 µm (with spacing identical to micropillar diameter), and a height of 200 µm, resulting in a total chamber volume of ∼20 µL (SI Appendix, Fig. S5A). The aligned micropillars in the fluidic layer were expected to prevent the bubbles from passing downstream, acting both as physical and hydrophobic barriers. We further added a vacuum chamber on top of the fluidic chamber featuring the same arrangement of microposts but a height of 100 µm and reduced side-channel length (SI Appendix, Fig. S5B). Fig. 2C shows a schematic of the device, where each vacuum chamber and the fluidic chamber were bonded via a thin PDMS membrane (20-µm thick). The micropillars in the vacuum chamber were expected to prevent the collapse of the membrane under negative pressure. The PDMS-based devices were sandwiched between a pair of poly(methyl methacrylate) (PMMA) clamps tightened by a set of screws/bolts to completely seal the device (Fig. 2 D and E). The negative pressure was realized by utilization of vacuum lines. The operational principle of the bubble trap is illustrated in SI Appendix, Fig. S6. The bubble trapping and removal in the device were then visually validated using a dye (Fig. 2F, SI Appendix, Fig. S7, and Movie S1). Flow sensor data further demonstrated successful bubble removal from the microfluidic circuitry when the bubble trap was integrated, as indicated by significantly reduced amounts (>99%) of spikes caused by bubbles of different sizes traveling through the flow sensor (Fig. 2 G and H, and SI Appendix, Table S1 and Fig. S8). In addition to improving sensor performance, the utilization of the bubble traps also contributed to enhanced morphological observation of cells in the microbioreactor by preventing bubble accumulation (SI Appendix, Fig. S9). The inclusion of bubble traps into the platform was also shown not to induce measurable evaporation and thus loss of water for up to 7 d.
Integration of Noninvasive Biochemical, Physical, and Optical Sensors for On-Line Organoid Monitoring.
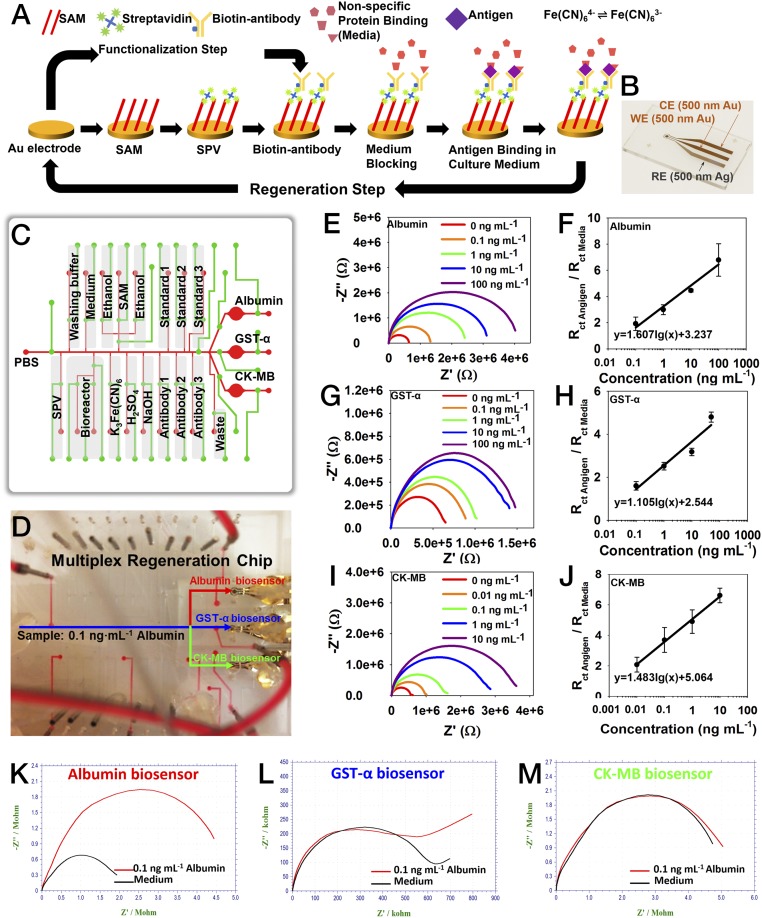
We developed label-free electrochemical immunobiosensors for continual in situ monitoring of soluble biomarkers secreted by the organoids (48, 49). Here, we have further integrated these immunobiosensors with a set of specifically designed microfluidic chips to achieve fully automated operations of the sensors for high-sensitivity and long-term organoid monitoring in our platform. One critical feature of our approach lies in the fact that the electrode surface could not only be functionalized with specific antibodies for biomarker binding and detection but also be regenerated upon saturation by captured antigens (Fig. 3A). In a typical procedure, the surface of the gold (Au) microelectrode is first coated with a self-assembled monolayer (SAM) using 11-mercaptoundecanoic acid (11-MUA), followed by immobilization of streptavidin (SPV) by covalent bonding with 11-MUA through the carbodiimide reaction (50). Afterward, desired biotinylated antibodies are attached to SPV via strong interaction between SPV and biotin. This strong interaction was able to maintain the stability of the functionalized antibodies under flow conditions during the subsequent detection process. The multistep functionalization process consumes ∼174 min.
Fig. 3.
Integration of microfluidic electrochemical and physical sensors. (A) Schematic diagram showing the functionalization and regeneration process of the electrode for measurement of soluble antigens. (B) Photograph showing a fabricated microelectrode set containing an Au WE, an Au CE, and an Ag RE. (C) Schematic showing the design of the multiplexed microfluidic chip for precisely timed injections of the chemicals for electrochemical detection. (D) Photograph of automated multiplexed regeneration microfluidic chip. (E and F) EIS plot of the calibration impedance measurements obtained from 0 to 100 ng⋅mL−1 albumin and the corresponding calibration curve for albumin. (G and H) EIS plot of the calibration impedance measurements obtained from 0 to 100 ng⋅mL−1 GST-α and the corresponding calibration curve for GST-α. (I and J) EIS plot of the calibration impedance measurements obtained from 0 to 100 ng⋅mL−1 CK-MB and the corresponding calibration curve for CK-MB. Three measurements on three individual electrodes were used to plot each data point on each calibration curve. (K) The response of albumin biosensor to albumin (0.1 ng⋅mL−1). (L) The response of albumin biosensor to GST-α (0.1 ng⋅mL−1). (M) The response of albumin biosensor to CK-MB (0.1 ng⋅mL−1).
The label-free detection mechanism of this electrochemical immunobiosensor is based on the change of interfacial electron-transfer kinetics of redox probe [Fe(CN)6]4−/3− upon antibody–antigen binding events (51). The antigen binding and measurement processes take ∼58 min. The amount of biomarkers captured on the functionalized electrode surface is proportional to the concentration in the solution, which would saturate after a few binding events (SI Appendix, Fig. S10). To address this issue, we further adapted a mechanism based on a dual-step cleaning process to regenerate the electrode surface upon saturation with captured antigens (32) (Fig. 3A and SI Appendix, Fig. S11). The first cleaning step (10 mM H2SO4, potential, 0–1.8 V; scan rate, 100 mV⋅s−1) could efficiently remove the entire layer of antibody/antigen complexes by breaking the thiol-Au bonds and peeling off Au atoms from the surface of the electrode, whereas the second cleaning step [50 mM K3Fe(CN)6, potential, −1.2 to 1.2 V; scan rate, 200 mV⋅s−1] would restore the electrical current to the level of pristine electrode surfaces. Regeneration of the electrode surface requires 18 min.
To realize these procedures, we further fabricated an in-house microelectrode system for easy integration with microfluidic chips (48, 49). The system consisted of three microelectrodes, a reference electrode (RE), a working electrode (WE), and a counter electrode (CE). The CE and WE were made of Au, whereas the RE was made of silver (Ag) (Fig. 3B and SI Appendix, Fig. S12 A and B). Au was selected as the material for WE due to its relatively good stability, favorable electron transfer kinetics with high in-plane conductivity, biocompatibility, and its ability to readily create covalent bonding for generating stable immobilization of receptors onto its surface (48, 49). The annealing process allowed us to create robust and smooth microelectrodes (SI Appendix, Fig. S12C), enabling minimal damage to the electrodes under harsh environments such as low and high pH solutions, corrosive solvents, and application of high electrical potentials. Representative cyclic voltammetry (CV) and electrochemical impedance spectroscopy (EIS) Nyquist plots are shown in SI Appendix, Fig. S13. Both Au and Ag layers were fabricated in 500-nm thickness to allow for repeated regeneration of the electrode surface for up to four cycles (thus enabling five total measurements) without significant changes in the electrode performance, including CV current peaks and charge transfer resistance (Rct) ratios (SI Appendix, Fig. S14 A–C). Satisfactory regeneration of the electrode surface should show peak current (IpA) at 0.14 V within the range of 1.4 × 10−4 ± 2.2 × 10−6 during CV measurement (SI Appendix, Fig. S14B), which typically leads to consistent electrochemical signals across multiple regeneration cycles (<3% of variance; SI Appendix, Fig. S14C). Only electrodes that met this standard were used for electrochemical sensing. Atomic force microscopy images further confirmed that the functionalized surface of the electrode could be reversed back to its original state following the regeneration process (SI Appendix, Fig. S14 D–F).
According to the required reagents and procedure for the functionalization and regeneration of the microelectrode, we further designed a microfluidic chip containing a series of inlet channels for long-term monitoring of organoid-secreted biomarkers, where the electrode was irreversibly bound to the bottom of the detection chamber with a sampling volume of 7 µL (SI Appendix, Fig. S15A). This microfluidic chip allowed for automated switches of desired channels through programmed operation of built-in pneumatic valves for accessing a series of reservoirs (Movie S2), achieving sequential injection of different agents for automated functionalization of the electrode surface (SI Appendix, Fig. S16) and subsequent regeneration. A miniature bubble trap was built directly on top of the microelectrode chamber to efficiently eliminate any bubbles that could potentially enter the chamber and interfere with the electrochemical measurements (SI Appendix, Fig. S17).
The microfluidic immunobiosensor chip could be further multiplexed to measure several biomarkers (Movie S3). For example, we designed a chip simultaneously hosting three microelectrode sets and associated microfluidic channels/valves to achieve automated functionalization and regeneration for detection of liver biomarkers albumin and glutathione S-transferase α (GST-α) as well as cardiac biomarker creatine kinase MB (CK-MB) (Fig. 3 C and D). The calibration curves of the three biomarkers, albumin (Fig. 3 E and F), GST-α (Fig. 3 G and H), and CK-MB (Fig. 3 I and J), were obtained using the microfluidic electrochemical immunobiosensors to determine their performances. The three biosensors achieved high sensitivities of 1.607, 1.105, and 1.483 log(ng⋅mL−1)−1, respectively, leading to ultralow limit of detections for these biosensors with wide dynamic detection ranges (SI Appendix, Table S2). Moreover, the selectivity of the electrochemical immunobiosensors was evaluated. A multiplexed chip was functionalized with albumin, GST-α, and CK-MB antibodies on the three microelectrode sets, respectively, after which albumin (0.1 ng⋅mL−1) was introduced to the detection chambers. Albumin incubation induced the signal response of only the albumin antibody-functionalized electrode set (Fig. 3K), whereas both GST-α and CK-MB biosensors did not generate any significant signals in response to albumin (Fig. 3 L and M). Similarly, GST-α and CK-MB biosensors only responded to GST-α and CK-MB, respectively (SI Appendix, Fig. S18).
One additional type of microfluidic chip was designed for the integrated platform, where the single detection chamber was developed for single biomarker detection. This simple microfluidic chip contained only the number of channels that were sufficient for sampling, electrochemical detection [K3Fe(CN)6], and washing [phosphate-buffered saline (PBS)] (SI Appendix, Fig. S15B and Movie S4), which was suitable for off-line functionalization, single-shot in-line detection, and disposability postmeasurement. It should be emphasized that, during the antigen binding and electrochemical signal measurement, the valves on any of these three different types of sensing chips are closed to ensure no flow interference on the electrode surface. In addition, all of the solutions involved in functionalization, detection, and regeneration processes including K3Fe(CN)6 are isolated from the main circulation and are confined only within the sensing chip with the only connection being a port sampling the medium from the breadboard (Fig. 3C and SI Appendix, Fig. S15); there is no direct interaction between these solutions and the organoids, eliminating any potential toxic effects.
Significantly, the immunobiosensing mechanism developed here is universal. By using corresponding antibodies, these functionalized electrochemical biosensors were capable of measuring a wide range of soluble biomarkers secreted by the liver organoid (e.g., α-1 antitrypsin, transferrin, and ceruloplasmin), cardiac organoid (e.g., troponin I), and vascular organoid (e.g., von Willebrand factor) (SI Appendix, Fig. S19). The stability and sensitivity of the sensors could be further tuned by switching the antibodies to desired aptamers (49), which are short-chain nucleic acids that generally known to possess higher specificity toward antigens and stability under a wide range of conditions than protein-based antibodies. In addition, our impedance-based immunobiosensors measure the charge transfer from the electrode and antigen-captured bioreceptors in the presence of K3Fe(CN)6 solution, which are potentially less affected by the interferrants present in the medium.
A physical sensing unit integrated with optical pH and oxygen sensors and a temperature probe was used to monitor the microenvironment of the organoid platform (SI Appendix, Fig. S20A). The optical pH sensor was designed to detect the absorption (>515 nm) of the culture medium supplemented with phenol red that indicated the pH value (SI Appendix, Fig. S20B). The optical pH sensor module was calibrated with media at different pH values (6.5–8.0), which generated a linear response with a sensitivity of 0.159 V⋅pH−1 (SI Appendix, Fig. S20 C and D). An optical oxygen sensor was developed based on the oxygen-sensitive fluorescence of a ruthenium dye (SI Appendix, Fig. S20E) (52). Characterization of the optical oxygen sensor was performed in media containing a series of nitrogen and air concentrations. The oxygen sensor presented a relatively rapid response to oxygen level changes, and a linear regression with a sensitivity of 7 mV⋅O2%−1 (SI Appendix, Fig. S20 F and G). Optical methods were chosen due to their suitability for on-line monitoring of the microenvironment over extended periods of time. The temperature sensor also showed a stable measurement of the temperature in the benchtop incubator over a period of 7 d (SI Appendix, Fig. S20H). Minimicroscopes positioned at the bottom of the microbioreactors fabricated by our established method (53) were used to monitor the status of hepatic and cardiac organoids in situ by recording their morphologies in real time (SI Appendix, Fig. S20I). A HepG2 liver microbioreactor showed progressive proliferation of the hepatocytes (SI Appendix, Fig. S20J), whereas the contraction of the cardiac organoids in the microbioreactor could also be monitored (SI Appendix, Fig. S20 K and L, and Movie S5). The beating signals were subsequently used in analyzing drug-induced cardiotoxicity in our organ-on-a-chip platforms.
Microbioreactors for Organoid Construction.
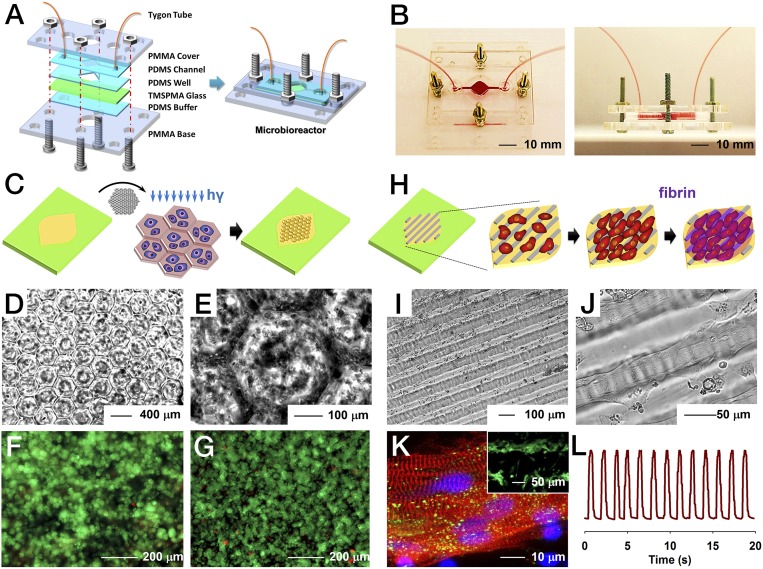
A resealable microbioreactor for perfusion culture of organoids was developed by modifying our recently published protocols (48, 54, 55). The microbioreactor was designed to possess two hemichambers embedded within a pair of PDMS (top) and PDMS/glass (bottom) pieces, which were then sandwiched between two rigid supports made of PMMA and a thin PDMS cushion layer to ensure hydraulic tightness (Fig. 4A and SI Appendix, Fig. S21). The pair of PDMS layers together formed the microbioreactor chamber at a thickness of ∼1.5 mm when secured together under slight compression (Fig. 4 A and B). The thickness of the bottom chamber was ∼250 µm, whereas that of the top chamber was 1.5 mm. The main chamber was square (7 × 7 mm2) with tapered edges, and connected with the inlet and outlet channels on two sides, resulting in a total working volume of ∼120 µL. Turbulent or stagnation zones and bubble formation were minimized by such a design due to the progressive increasing and decreasing width of the chamber, as well as the presence of rounded corners. In addition, the microbioreactor featured a circular opening in the center of the PMMA support at the bottom to enable direct microscopic monitoring of the behavior and morphology of the organoids without the need of disassembling the microbioreactor (Fig. 4A).
Fig. 4.
Construction of the microbioreactor and hepatic/cardiac organoids. (A) Schematic diagrams showing the fabrication of resealable microfluidic microbioreactor. (B) Photographs showing top view and side view of the microbioreactor. (C) Schematics indicating the construction of the hepatic organoid via direct photopatterning of hepatocytes encapsulated inside GelMA. (D and E) Optical micrographs showing the lobule-like patterns generated for the hepatic organoid in the microbioreactor. (F and G) Live/dead analysis showing the viability of the patterned human primary hepatocytes at day 1 and day 5 of culture, respectively. (H) Schematics indicating the construction of the cardiac organoid, where iPSC-CMs were first seeded on top of aligned GelMA patterns followed by coverage by a layer of fibrin to protect the cells from shear stress. (I and J) Optical micrographs showing the generated cardiac organoid featuring highly aligned human iPSC-CMs. (K) Sarcomeric α-actinin (red), connexin-43 (green), and nuclei (blue) staining of the iPSC-CMs at day 3 of culture indicating the alignment of the cells and well-developed contractile phenotype and intercellular junctions. The Inset is live/dead analysis showing high viability of the iPSC-CMs. (L) Beating analysis of the cardiac organoid inside the microbioreactor.
Specifically, we demonstrated the construction of two prototypes of human organoids in these bioreactors, liver and heart. Although these organoid models were simplified and did not recapitulate the full biological functions, they, however, were sufficient to validate the capability of our in situ biosensors. The liver organoid was constructed using a micropatterning technique through a photomask containing arrays of lobule-like structures with a diameter of 400 µm (Fig. 4C and SI Appendix, Fig. S22A). Human primary hepatocytes were encapsulated in 5 wt% gelatin methacryloyl (GelMA) and loaded into the bottom chamber of the microbioreactor, which was subsequently photo–cross-linked against the mask to generate a 3D human primary liver organoid. The patterning was clearly transferred from the photomask (Fig. 4 D and E), leading to the formation of hepatic lobules resembling those in vivo. Live/dead staining performed immediately after the photopatterning showed a high viability (>90%) of the primary hepatocytes (Fig. 4F), indicating that the photo–cross-linking procedure was not harmful to the cells. The hepatocytes could maintain a viability of >85% over a culture period of up to 5 d (Fig. 4G). This culture time induced an aggregation of the primary hepatocytes without an increase in cell number. The liver organoids remained functional, indicating sustained secretion of albumin measured by electrochemical biosensors (SI Appendix, Fig. S23).
The cardiac organoid was also fabricated using the micropatterning technique where a photomask possessing parallel lines of 50 µm in width with 50-µm spacing was used to pattern the 5 wt% GelMA at the bottom of the microbioreactor chamber (56) (Fig. 4H and SI Appendix, Fig. S22B). To enhance the adhesion of the cells, the patterned GelMA surface was further coated with a layer of fibronectin. Then human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) were seeded onto the patterns. Another layer of fibrin gel with a thickness of ∼200 µm was gently placed on top of the seeded cardiomyocytes to enhance the spreading of iPSC-CMs and prevent direct exposure to fluid flow. The iPSC-CMs started to align themselves with the GelMA pattern as early as day 1 postseeding; alignment and stretching of the cells were observed at days 3–4 (Fig. 4 I and J). The iPSC-CMs maintained high viability (Fig. 4K, Inset), which also showed strong expression of the contractile marker sarcomeric α-actinin and junction protein connexin-43 (Fig. 4K), indicating maturation of the cardiomyocytes on the patterns. The chosen feature size of the patterns allowed the aligned cardiomyocytes to form cellular bridges between the ridges (56), leading to strong and synchronized beating of the cardiac organoids at a rate of ∼60 beats⋅min−1 (Fig. 4L and Movie S6), where the beating rates within a single batch of cells were uniform. The cardiac organoids remained functional showing consistent beating for up to 5 d in culture in the bioreactors (SI Appendix, Fig. S24).
Fully Integrated Human Liver (Cancer)- and Heart-on-Chips for Automated Drug Screening.
Automation is strongly preferred to minimize human labor especially when samples require high-throughput or long-term analysis. After validating the functions of individual components including the breadboard, bubble trap, electrochemical immunobiosensor, physical sensors, minimicroscope, and microbioreactors/organoids, we subsequently assembled these modular units into a fully integrated system that supported complete automation (Fig. 1). Specifically, two dual-organ platforms were assembled, human heart-and-liver-on-chips, and human heart-and-liver-cancer-on-chips, to demonstrate long-term monitoring of chronic drug responses and short-term evaluation of acute toxicity, respectively. The functionality of the dual-organ platform was first validated using capecitabine, a prodrug that can undergo enzymatic activation by hepatocytes to the active form 5-fluorouracil (5-FU), which is a thymidylate synthase inhibitor thus inhibiting DNA synthesis (57). Once converted to 5-FU, it would exert increased cardiotoxicity compared to capecitabine (58). Indeed, we were able to reproduce such a process using our human heart-and-liver-on-chips platform (SI Appendix, Fig. S25A) and human heart-on-chip platform (SI Appendix, Fig. S25B), where pronounced enhancement of cardiotoxicity could only be observed when the liver organoid was present in the dual-organ system (SI Appendix, Fig. S25C).
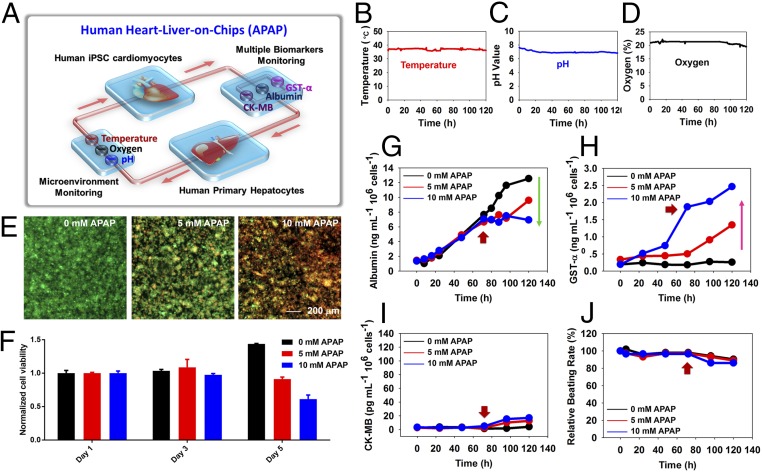
In the healthy group (Fig. 5A), human cardiac organoids derived from iPSC-CMs and human liver organoids derived from primary hepatocytes were linked together for up to 120 h, where acetaminophen (APAP) was used as a model pharmaceutical compound. APAP is a nonsteroidal antiinflammatory drug and the primary cause of induced acute liver failure in the United States (2), where it causes hepatotoxicity via necroapoptotic mechanisms (59). Doses of 0, 5, and 10 mM, as determined in our previous work (54), were introduced into the dual-organoid system at 72 h, to demonstrate the capability of our integrated sensors for detecting dose-dependent APAP toxicity toward the organoids. All three physical parameters (pH, O2, and temperature) were constantly monitored, which showed stable values over the entire period without much disturbance from drug administrated (Fig. 5 B–D). The relatively stable O2 levels observed could be attributed to the gas permeability of the PDMS devices; because the flow rate used for perfusion was relatively low (200 µL⋅h−1), by the time that the medium passing by the liver organoid reached the oxygen sensor it would have been recalibrated to the ambient oxygen level. In addition, a conventional end-point cell viability assessment at 120 h presented dose-dependent toxic effect of APAP on the survival of the hepatocytes (Fig. 5 E and F, and SI Appendix, Fig. S26). More interestingly, the noninvasive biomarker analysis achieved by electrochemical immunobiosensors revealed that the secreted levels of albumin and GST-α decreased and increased, respectively, in a dose-dependent manner (Fig. 5 G and H). This is consistent with the hepatotoxicity induced by APAP measured by the viability assay. CK-MB secretion by the cardiac organoid, however, showed only minor elevation (Fig. 5I), corresponding to their slightly reduced beating rates upon APAP treatment (Fig. 5J), as reported previously (60).
Fig. 5.
Automated in-line sensing of APAP-induced organoid toxicity from normal human heart-liver-on-chips. (A) Schematic diagram of biomimetic human heart-liver-on-chips. (B–D) Continual measurements of temperature, pH, and O2 concentration within the integrated heart-liver-on-chips. (E–I) Integrated primary hepatic and iPSC-cardiac dual-organoid platform: (E) live/dead staining of the hepatic organoids post treatment with 0, 5, and 10 mM APAP, at the end of 120 h. The drugs were introduced into the system at 72 h. (F) Normalized cell viability in the presence of APAP from day 1 to day 5. (G–I) In-line automated electrochemical measurements of albumin and GST-α secreted from the hepatic organoids as well as CK-MB from the cardiac organoids. (J) Beating analysis of the cardiac organoids. Red arrows indicate the time of drug addition (72 h).
In the liver cancer group (Fig. 6A), human cardiac organoids derived from iPSC-CMs and human hepatic organoids derived from HepG2/C3A hepatocellular carcinoma cells were linked together for up to 24 h to investigate the acute toxicity upon drug treatment. Doxorubicin (DOX), a well-known chemotherapeutic drug, was used to ablate the liver cancer organoid (61). Using toxic doses of 5 and 10 µM, we expected a detrimental effect on HepG2 cells while also inducing severe cardiotoxic side effects (62). Indeed, the liver cancer organoid underwent significant death upon drug treatment (Fig. 6 B and C, and SI Appendix, Fig. S27), accompanied by diminished secretion of albumin with a significant release of cytoplasmic GST-α (Fig. 6 D and E). Contrary to APAP treatment, administration of DOX induced pronounced cardiotoxicity, as demonstrated by rounding, detachment, and death of the iPSC-CMs (Fig. 6F). Caspase-3 staining further indicated apoptosis of the iPSC-CMs with elevated dose of DOX (SI Appendix, Fig. S28). The level of CK-MB, as measured by the electrochemical biosensor, surged to extremely high values (Fig. 6G), which was associated with arrhythmic beating of the cardiac organoid recorded by the minimicroscope (Fig. 6H).
Fig. 6.
Automated in-line sensing of DOX-induced organoid toxicity from heart-liver-cancer-on-chips. (A) Schematic diagram of biomimetic human heart-liver-cancer-on-chips. (B) Live/dead staining of the liver cancer organoids post treatment with 0, 5, and 25 µM DOX, at the end of 24 h. The drugs were introduced into the system from time 0. (C) Normalized cell viability in the presence of DOX from 6 to 24 h. (D and E) In-line automated electrochemical measurements of albumin and GST-α secreted from the liver cancer organoids. (F) Live/dead staining of the cardiac organoids at the end of 24 h. (G) In-line automated electrochemical measurements of CK-MB from the cardiac organoids. (H) Beating analysis of the cardiac organoids.
It should be noted that, due to the noninvasiveness of our electrochemical biomarker analyses, we were able to continually monitor individual dual-organ platforms in an automated mode by dynamically sampling the media at predefined time points (Figs. 5 G–J and 6 D, E, G, and H). As such, the measurements would be continuous and accurate for an individual organoid pair as opposed to the conventional end-point assays where average values from multiple organoids need to be obtained. However, results derived from parallel systems might still be averaged to understand the collective statistical effects of the pharmaceutical compounds applied to the organoids (SI Appendix, Figs. S29 and S30).
To further prove the ability of physical sensors to monitor cellular responses upon microenvironmental stimulation, they were integrated with an additional platform containing a liver cancer bioreactor (SI Appendix, Fig. S31A). Hyperthermia treatments of the liver cancer organoids were subsequently conducted, where the temperatures in parallel groups were rapidly increased to 41, 43, and 45 °C, respectively, held for 30 min, and allowed to reverse to the baseline at 37 °C (SI Appendix, Fig. S31B) (63). The changes in the pH and oxygen levels in the medium due to the metabolism of the hepatocellular carcinoma cells were continuously measured (SI Appendix, Fig. S31 C and D). We observed reduced metabolic rate as the hyperthermic temperature was elevated to 45 °C possibly due to the initiation of apoptosis/necrosis of the cells (64), where temperature increases of up to 43 °C did not seem to have significant effect on the metabolism of the cells. The bioreactors were completely sealed to prevent gas exchange through the PDMS.
Conclusions and Perspectives
We have developed an automated modular design platform that used a microfluidics-controlling breadboard for timed routing of fluids to interface physical sensors for monitoring microenvironmental parameters (e.g., pH, O2, temperature), electrochemical immunobiosensors for measuring soluble biomarkers, and miniature microscopes for observation of the organoid morphologies. All of the sensing was performed in situ in an uninterrupted and automated manner, allowing for long-term monitoring of drug-induced organ toxicity in a dual-organ human liver-and-heart-on-a-chip platform insulted with APAP for up to 5 d and a dual-organ human liver-cancer-and-heart-on-a-chip platform challenged with DOX for up to 24 h. We believe that our integrated modular in-line fluid routing and sensing platform will be compatible with existing organ-on-a-chip models and will promote their performances in drug screening by providing the capability of real-time, in situ monitoring of their microenvironment biophysical and biochemical parameters.
Meanwhile, we acknowledge several limitations of our current integrated multisensor-and-organ-on-chips platform. First, the material for making the microfluidic chips was PDMS at this prototyping stage, which is not optimal for organs-on-chip applications due to their potential adsorption and absorption of hydrophobic small molecules and drugs (65, 66). For example, it was found that DOX at higher concentrations could be partially adsorbed/absorbed by PDMS during the 24-h period of drug testing on the human heart-and-liver-cancer-on-chips platform (SI Appendix, Fig. S32). To solve this issue, we have recently developed a method to fabricate thermoplastic microfluidic chips with built-in channels as well as pneumatic valves for large-scale fluid manipulation, which will be reported separately. Additionally, it is realized that the current platform features relatively large sizes requiring nonstandard manual assemblies as well as overly high complexity. We believe, however, at the initial prototyping stage such a platform has allowed us to validate our approach of multisensor integration. Efforts have been initiated toward further compacting and simplifying the platform by adopting LEGO and/or cartridge assemblies as recently reported for the fabrication of organs-on-a-chip models (67–71). Further efforts on improved scaling of our platform should, in principle, allow for more accurate modeling of the human system and therefore achieve better prediction of drug efficacy/toxicity (33).
Methods
Fabrication of Breadboard.
The breadboard was built using PDMS using a standard photolithography technique. Subsequently, the microfluidic layer master mold was converted to a convex hemicylindrical secondary mold for generating the microchannels (SI Appendix, Fig. S3) (72). The detailed procedures are described in SI Appendix.
Fabrication of Microbioreactor.
The microbioreactor was fabricated with PDMS (SI Appendix, Fig. S21), consisting of a microfluidic perfusion layer at the top, and a chamber layer at the bottom bonded to a piece of glass. The detailed procedures are described in SI Appendix.
Fabrication of Bubble Trap.
The bubble trap featured a 10-mm single chamber containing micropillars with a series of uniform diameters of 1,000, 500, and 250 µm (with spacing identical to micropillar diameter), and a height of 200 µm, leading to a total chamber volume of ∼20 µL (SI Appendix, Fig. S5A). A vacuum chamber was further added on top of the fluidic chamber featuring the same array of micropillars but a height of 100 µm and reduced length of side channels (SI Appendix, Fig. S5B). The detailed procedures are described in SI Appendix.
Fabrication of Microelectrode Set.
The microelectrode set was composed of three electrodes: Au WE, Au CE, and Ag RE. The diameter of the WE was 800 μm, the width of CE and RE were 150 μm, and gap between WE and other two electrodes was 200 μm, and the diameter of detection area was about 1,500 μm (SI Appendix, Fig. S12). The detailed procedures are described in SI Appendix.
Integration of the Microelectrode Set with Microfluidic Electrochemical Sensing Chip.
The microelectrode set was bonded to the chip using air plasma with the detection area aligning with the chamber. The three leads of the microelectrode set were coated with silver epoxy (MG Chemicals) to make electrical contact with copper wires for measurement. The detailed procedures are described in SI Appendix.
Regeneration of Microelectrode Set-Integrated PDMS Microfluidic Chip.
To achieve long-term monitoring, the saturated microelectrode surface was regenerated via a two-step electrochemical cleaning process: 10 mM H2SO4 followed by 50 mM K3Fe(CN)6. The H2SO4 cleaning was performed between the electrical sweep potentials of 0.0 and 1.8 V, where the scan rate, sensitivity, and sampling interval were set to 100 mV⋅s−1, 10−2 A/V, and 0.001 V, respectively. The K3Fe(CN)6 cleaning was carried out between potentials of 1.0 and 0.0 V at scan rate of 200 mV⋅s−1, sensitivity of 10−4 A/V, and sampling interval of 0.001 V. During the cleaning procedure, the solutions were kept flowing at a rate of 1,000 µL⋅h−1. The oxidation peak current of K3Fe(CN)6 as well as Rct of the bare microelectrode were compared before and after regeneration to assess the stability and sensitivity of the regenerated microelectrode.
Physical Sensors.
The physical module housed the integrated pH and oxygen sensors (SI Appendix, Fig. S20A). The pH sensor detected changes in the light absorption of phenol red (Sigma-Aldrich) in the medium to translate into a voltage change (SI Appendix, Fig. S20B). The oxygen-quenchable luminescent dye [Ru(dpp)3]2+Cl2–tris(4,7-diphenyl-1,10-phenanthroline)ruthenium(II) chloride (Sigma-Aldrich), with an excitation wavelength at 463 nm and a maximum emission at 618 nm, was used for the oxygen sensor (53). The detailed procedures are described in SI Appendix.
Construction of the Automation Controller.
The automation controller was constructed using a set of WAGO controllers and FESTO solenoid valves connected to a nitrogen gas source, where each of the valves could be individually actuated using custom-coded MATLAB (Mathworks) programs to control injection or microvalve function on the microfluidic chips (SI Appendix, Fig. S1) (73). The detailed procedures are described in SI Appendix.
Cell Culture and Patterning.
Human primary hepatocytes (Triangle Research Labs) were used to fabricate the liver organoids, and human iPSC-CMs (Stem Cell Theranostics) were used to generate the cardiac organoids. The detailed procedures are described in SI Appendix.
Minimicroscope.
To monitor the behaviors of the organoids inside the microbioreactors in situ, a webcam (Logitech C160)-based minimicroscope was fabricated and fixed at the bottom of the microbioreactors (SI Appendix, Fig. S20I). The detailed procedures are described in SI Appendix.
Drug Treatment.
Two drugs were selected to test the biomimetic human dual-organ-on-chip systems according to their target toxicity and differences among their adverse effects in the human body, which are detailed in the main text (Figs. 5 and 6). APAP (Sigma-Aldrich), a liver toxic drug, was administrated to the heart-and-liver-on-chip system at dosages of 0, 5, and 10 mM based on results obtained from our previous work (54). DOX (Sigma-Aldrich), a common chemotherapeutic drug, was used to evaluate toxicity in the heart-and-liver-cancer-on-chip system (62, 74). In addition, capecitabine (Sigma-Aldrich) at 80 µM was used to demonstrate the functionality of the human liver-and-heart-on-chips platform, whereas hyperthermia treatment via elevation of temperature of the incubation system to 41, 43, or 45 °C allowed for validation of the utility of the physical sensors to monitor environmental stimuli-induced changes in cell behaviors and consequent alteration in physical parameters. The detailed procedures are described in SI Appendix.
Statistical Analysis.
Sigmaplot 12.0 and Graphpad 6.0 were used to perform statistical analysis of the experimental results. At least three samples were analyzed per group, and a power analysis was used to determine the sample size before each experiment.
Code Availability.
MATLAB codes for measuring cardiac beating are included in SI Appendix and Datasets S1 and S2.
Supplementary Material
Acknowledgments
We thank Dr. Rafael Gómez-Sjöberg for providing advice on the construction of the pneumatic controller system, and Drs. Šeila Selimović and Ljupcho Prodanov for their assistance in designing the breadboard. H.A. sincerely thanks Eskisehir Osmangazi University and Dr. M. Bahaddin Acat. We gratefully acknowledge funding by the Defense Threat Reduction Agency under Space and Naval Warfare Systems Center Pacific Contract N66001-13-C-2027. We also acknowledge funding from the Office of Naval Research Young National Investigator Award, the National Institutes of Health (Grants EB012597, AR057837, DE021468, HL099073, R56AI105024, AR068258, AR066193, EB022403, and EB021148), and the Presidential Early Career Award for Scientists and Engineers. Y.S.Z. acknowledges the National Cancer Institute of the National Institutes of Health K99/R00 Pathway to Independence Award (K99CA201603). T.K. and H.A. acknowledge Scientific and Technological Research Council of Turkey. The publication of this material does not constitute approval by the government of the findings or conclusions herein.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.B. is a Guest Editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612906114/-/DCSupplemental.
References
- 1.Paul SM, et al. How to improve R&D productivity: The pharmaceutical industry’s grand challenge. Nat Rev Drug Discov. 2010;9(3):203–214. doi: 10.1038/nrd3078. [DOI] [PubMed] [Google Scholar]
- 2.US Food and Drug Administration 2015 Drug-Induced Liver Toxicity. Available at www.fda.gov/Drugs/ScienceResearch/ResearchAreas/ucm071471.htm. Accessed March 30, 2016.
- 3.Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. 2014;32(1):40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 4.Arrowsmith J, Miller P. Trial watch: Phase II and phase III attrition rates 2011–2012. Nat Rev Drug Discov. 2013;12(8):569. doi: 10.1038/nrd4090. [DOI] [PubMed] [Google Scholar]
- 5.Ferri N, et al. Drug attrition during pre-clinical and clinical development: Understanding and managing drug-induced cardiotoxicity. Pharmacol Ther. 2013;138(3):470–484. doi: 10.1016/j.pharmthera.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 6.Stevens JL, Baker TK. The future of drug safety testing: Expanding the view and narrowing the focus. Drug Discov Today. 2009;14(3-4):162–167. doi: 10.1016/j.drudis.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Esch EW, Bahinski A, Huh D. Organs-on-chips at the frontiers of drug discovery. Nat Rev Drug Discov. 2015;14(4):248–260. doi: 10.1038/nrd4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bhise NS, et al. Organ-on-a-chip platforms for studying drug delivery systems. J Control Release. 2014;190:82–93. doi: 10.1016/j.jconrel.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang YS, Khademhosseini A. Seeking the right context for evaluating nanomedicine: From tissue models in petri dishes to microfluidic organs-on-a-chip. Nanomedicine (Lond) 2015;10(5):685–688. doi: 10.2217/nnm.15.18. [DOI] [PubMed] [Google Scholar]
- 10.Jang HL, Zhang YS, Khademhosseini A. Boosting clinical translation of nanomedicine. Nanomedicine (Lond) 2016;11(12):1495–1497. doi: 10.2217/nnm-2016-0133. [DOI] [PubMed] [Google Scholar]
- 11.Ebrahimkhani MR, Neiman JAS, Raredon MSB, Hughes DJ, Griffith LG. Bioreactor technologies to support liver function in vitro. Adv Drug Deliv Rev. 2014;69-70:132–157. doi: 10.1016/j.addr.2014.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wikswo JP. The relevance and potential roles of microphysiological systems in biology and medicine. Exp Biol Med (Maywood) 2014;239(9):1061–1072. doi: 10.1177/1535370214542068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esch MB, King TL, Shuler ML. The role of body-on-a-chip devices in drug and toxicity studies. Annu Rev Biomed Eng. 2011;13:55–72. doi: 10.1146/annurev-bioeng-071910-124629. [DOI] [PubMed] [Google Scholar]
- 14.Ebrahimkhani MR, Young CL, Lauffenburger DA, Griffith LG, Borenstein JT. Approaches to in vitro tissue regeneration with application for human disease modeling and drug development. Drug Discov Today. 2014;19(6):754–762. doi: 10.1016/j.drudis.2014.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moraes C, Mehta G, Lesher-Perez SC, Takayama S. Organs-on-a-chip: A focus on compartmentalized microdevices. Ann Biomed Eng. 2012;40(6):1211–1227. doi: 10.1007/s10439-011-0455-6. [DOI] [PubMed] [Google Scholar]
- 16.Knowlton S, Yenilmez B, Tasoglu S. Towards single-step biofabrication of organs on a chip via 3D printing. Trends Biotechnol. 2016;34(9):685–688. doi: 10.1016/j.tibtech.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 17.Lee H, Cho D-W. One-step fabrication of an organ-on-a-chip with spatial heterogeneity using a 3D bioprinting technology. Lab Chip. 2016;16(14):2618–2625. doi: 10.1039/c6lc00450d. [DOI] [PubMed] [Google Scholar]
- 18.Maschmeyer I, et al. Chip-based human liver–intestine and liver–skin co-cultures—a first step toward systemic repeated dose substance testing in vitro. Eur J Pharm Biopharm. 2015;95(Part A):77–87. doi: 10.1016/j.ejpb.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Misun PM, Rothe J, Schmid YRF, Hierlemann A, Frey O. Multi-analyte biosensor interface for real-time monitoring of 3D microtissue spheroids in hanging-drop networks. Microsyst Nanoeng. 2016;2:16022. doi: 10.1038/micronano.2016.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khetani SR, Bhatia SN. Microscale culture of human liver cells for drug development. Nat Biotechnol. 2008;26(1):120–126. doi: 10.1038/nbt1361. [DOI] [PubMed] [Google Scholar]
- 21.Bale SS, et al. A novel low-volume two-chamber microfabricated platform for evaluating drug metabolism and toxicity. Technology (Singap World Sci) 2015;3(4):155–162. doi: 10.1142/S2339547815200034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Benam KH, et al. Small airway-on-a-chip enables analysis of human lung inflammation and drug responses in vitro. Nat Methods. 2016;13(2):151–157. doi: 10.1038/nmeth.3697. [DOI] [PubMed] [Google Scholar]
- 23.Kim HJ, Li H, Collins JJ, Ingber DE. Contributions of microbiome and mechanical deformation to intestinal bacterial overgrowth and inflammation in a human gut-on-a-chip. Proc Natl Acad Sci USA. 2016;113(1):E7–E15. doi: 10.1073/pnas.1522193112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huh D, et al. Reconstituting organ-level lung functions on a chip. Science. 2010;328(5986):1662–1668. doi: 10.1126/science.1188302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bettinger CJ, Langer R, Borenstein JT. Engineering substrate topography at the micro- and nanoscale to control cell function. Angew Chem Int Ed Engl. 2009;48(30):5406–5415. doi: 10.1002/anie.200805179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mathur A, et al. Human iPSC-based cardiac microphysiological system for drug screening applications. Sci Rep. 2015;5:8883. doi: 10.1038/srep08883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huh D, et al. Acoustically detectable cellular-level lung injury induced by fluid mechanical stresses in microfluidic airway systems. Proc Natl Acad Sci USA. 2007;104(48):18886–18891. doi: 10.1073/pnas.0610868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee JS, et al. Placenta-on-a-chip: A novel platform to study the biology of the human placenta. J Matern Fetal Neonatal Med. 2016;29(7):1046–1054. doi: 10.3109/14767058.2015.1038518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oleaga C, et al. Multi-organ toxicity demonstration in a functional human in vitro system composed of four organs. Sci Rep. 2016;6:20030. doi: 10.1038/srep20030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esch MB, Ueno H, Applegate DR, Shuler ML. Modular, pumpless body-on-a-chip platform for the co-culture of GI tract epithelium and 3D primary liver tissue. Lab Chip. 2016;16(14):2719–2729. doi: 10.1039/c6lc00461j. [DOI] [PubMed] [Google Scholar]
- 31.Bhatia SN, Ingber DE. Microfluidic organs-on-chips. Nat Biotechnol. 2014;32(8):760–772. doi: 10.1038/nbt.2989. [DOI] [PubMed] [Google Scholar]
- 32.Huh D, Hamilton GA, Ingber DE. From 3D cell culture to organs-on-chips. Trends Cell Biol. 2011;21(12):745–754. doi: 10.1016/j.tcb.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wikswo JP, et al. Scaling and systems biology for integrating multiple organs-on-a-chip. Lab Chip. 2013;13(18):3496–3511. doi: 10.1039/c3lc50243k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wikswo JP, et al. Engineering challenges for instrumenting and controlling integrated organ-on-chip systems. IEEE Trans Biomed Eng. 2013;60(3):682–690. doi: 10.1109/TBME.2013.2244891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bavli D, et al. Real-time monitoring of metabolic function in liver-on-chip microdevices tracks the dynamics of mitochondrial dysfunction. Proc Natl Acad Sci USA. 2016;113(16):E2231–E2240. doi: 10.1073/pnas.1522556113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Griep LM, et al. BBB on chip: Microfluidic platform to mechanically and biochemically modulate blood-brain barrier function. Biomed Microdevices. 2013;15(1):145–150. doi: 10.1007/s10544-012-9699-7. [DOI] [PubMed] [Google Scholar]
- 37.Maschmeyer I, et al. Chip-based human liver-intestine and liver-skin co-cultures—a first step toward systemic repeated dose substance testing in vitro. Eur J Pharm Biopharm. 2015;95(Pt A):77–87. doi: 10.1016/j.ejpb.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taurino I, et al. Platinum nanopetal-based potassium sensors for acute cell death monitoring. RSC Advances. 2016;6(46):40517–40526. [Google Scholar]
- 39.Cho S, Islas-Robles A, Nicolini AM, Monks TJ, Yoon J-Y. In situ, dual-mode monitoring of organ-on-a-chip with smartphone-based fluorescence microscope. Biosens Bioelectron. 2016;86(15):697–705. doi: 10.1016/j.bios.2016.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eklund SE, et al. Modification of the Cytosensor microphysiometer to simultaneously measure extracellular acidification and oxygen consumption rates. Anal Chim Acta. 2003;496(1-2):93–101. [Google Scholar]
- 41.Eklund SE, et al. Multianalyte microphysiometry as a tool in metabolomics and systems biology. J Electroanal Chem. 2006;587(2):333–339. [Google Scholar]
- 42.Lima EA, et al. Multichamber multipotentiostat system for cellular microphysiometry. Sens Actuators B Chem. 2014;204:536–543. doi: 10.1016/j.snb.2014.07.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eklund SE, Taylor D, Kozlov E, Prokop A, Cliffel DE. A microphysiometer for simultaneous measurement of changes in extracellular glucose, lactate, oxygen, and acidification rate. Anal Chem. 2004;76(3):519–527. doi: 10.1021/ac034641z. [DOI] [PubMed] [Google Scholar]
- 44.Weltin A, et al. Cell culture monitoring for drug screening and cancer research: A transparent, microfluidic, multi-sensor microsystem. Lab Chip. 2014;14(1):138–146. doi: 10.1039/c3lc50759a. [DOI] [PubMed] [Google Scholar]
- 45.Eklund SE, et al. Metabolic discrimination of select list agents by monitoring cellular responses in a multianalyte microphysiometer. Sensors (Basel) 2009;9(3):2117–2133. doi: 10.3390/s90302117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKenzie JR, Cognata AC, Davis AN, Wikswo JP, Cliffel DE. Real-time monitoring of cellular bioenergetics with a multianalyte screen-printed electrode. Anal Chem. 2015;87(15):7857–7864. doi: 10.1021/acs.analchem.5b01533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang W, et al. Elastomeric free-form blood vessels for interconnecting organs on chip systems. Lab Chip. 2016;16(9):1579–1586. doi: 10.1039/c6lc00001k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Riahi R, et al. Automated microfluidic platform of bead-based electrochemical immunosensor integrated with bioreactor for continual monitoring of cell secreted biomarkers. Sci Rep. 2016;6:24598. doi: 10.1038/srep24598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shin SR, et al. Aptamer-based microfluidic electrochemical biosensor for monitoring cell secreted cardiac biomarkers. Anal Chem. 2016;88(20):10019–10027. doi: 10.1021/acs.analchem.6b02028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chivers CE, et al. A streptavidin variant with slower biotin dissociation and increased mechanostability. Nat Methods. 2010;7(5):391–393. doi: 10.1038/nmeth.1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bertassoni LE, et al. Direct-write bioprinting of cell-laden methacrylated gelatin hydrogels. Biofabrication. 2014;6(2):024105. doi: 10.1088/1758-5082/6/2/024105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sud D, et al. Optical imaging in microfluidic bioreactors enables oxygen monitoring for continuous cell culture. J Biomed Opt. 2006;11(5):050504. doi: 10.1117/1.2355665. [DOI] [PubMed] [Google Scholar]
- 53.Zhang YS, et al. A cost-effective fluorescence mini-microscope for biomedical applications. Lab Chip. 2015;15(18):3661–3669. doi: 10.1039/c5lc00666j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bhise NS, et al. A liver-on-a-chip platform with bioprinted hepatic spheroids. Biofabrication. 2016;8(1):014101. doi: 10.1088/1758-5090/8/1/014101. [DOI] [PubMed] [Google Scholar]
- 55.Zhang YS, et al. Google Glass-directed monitoring and control of microfluidic biosensors and actuators. Sci Rep. 2016;6:22237. doi: 10.1038/srep22237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Annabi N, et al. Highly elastic micropatterned hydrogel for engineering functional cardiac tissue. Adv Funct Mater. 2013;23(39):4950–4959. doi: 10.1002/adfm.201300570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Twelves C. Vision of the future: Capecitabine. Oncologist. 2001;6(Suppl 4):35–39. doi: 10.1634/theoncologist.6-suppl_4-35. [DOI] [PubMed] [Google Scholar]
- 58.Sorrentino MF, Kim J, Foderaro AE, Truesdell AG. 5-Fluorouracil induced cardiotoxicity: Review of the literature. Cardiol J. 2012;19(5):453–458. doi: 10.5603/cj.2012.0084. [DOI] [PubMed] [Google Scholar]
- 59.Zhang YF, et al. Role of receptor interacting protein (RIP)1 on apoptosis-inducing factor-mediated necroptosis during acetaminophen-evoked acute liver failure in mice. Toxicol Lett. 2014;225(3):445–453. doi: 10.1016/j.toxlet.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 60.Abassi YA, et al. Dynamic monitoring of beating periodicity of stem cell-derived cardiomyocytes as a predictive tool for preclinical safety assessment. Br J Pharmacol. 2012;165(5):1424–1441. doi: 10.1111/j.1476-5381.2011.01623.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Menna P, Salvatorelli E, Minotti G. Cardiotoxicity of antitumor drugs. Chem Res Toxicol. 2008;21(5):978–989. doi: 10.1021/tx800002r. [DOI] [PubMed] [Google Scholar]
- 62.Sadeghi-Aliabadi H, Minaiyan M, Dabestan A. Cytotoxic evaluation of doxorubicin in combination with simvastatin against human cancer cells. Res Pharm Sci. 2010;5(2):127–133. [PMC free article] [PubMed] [Google Scholar]
- 63.Xie X, et al. In vitro hyperthermia studied in a continuous manner using electric impedance sensing. RSC Advances. 2015;5(76):62007–62016. [Google Scholar]
- 64.Basile A, Biziato D, Sherbet GV, Comi P, Cajone F. Hyperthermia inhibits cell proliferation and induces apoptosis: Relative signaling status of P53, S100A4, and Notch in heat sensitive and resistant cell lines. J Cell Biochem. 2008;103(1):212–220. doi: 10.1002/jcb.21401. [DOI] [PubMed] [Google Scholar]
- 65.Toepke MW, Beebe DJ. PDMS absorption of small molecules and consequences in microfluidic applications. Lab Chip. 2006;6(12):1484–1486. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- 66.Wang JD, Douville NJ, Takayama S, ElSayed M. Quantitative analysis of molecular absorption into PDMS microfluidic channels. Ann Biomed Eng. 2012;40(9):1862–1873. doi: 10.1007/s10439-012-0562-z. [DOI] [PubMed] [Google Scholar]
- 67.Fujii T, et al. A plug and play microfluidic device. Lab Chip. 2003;3(3):193–197. doi: 10.1039/b301410j. [DOI] [PubMed] [Google Scholar]
- 68.Yuen PK. SmartBuild–A truly plug-n-play modular microfluidic system. Lab Chip. 2008;8(8):1374–1378. doi: 10.1039/b805086d. [DOI] [PubMed] [Google Scholar]
- 69.Bhargava KC, Thompson B, Malmstadt N. Discrete elements for 3D microfluidics. Proc Natl Acad Sci USA. 2014;111(42):15013–15018. doi: 10.1073/pnas.1414764111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Loskill P, et al. μOrgano: A Lego®-like plug & play system for modular multi-organ-chips. PloS One. 2015;10(10):e0139587. doi: 10.1371/journal.pone.0139587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuen PK. A reconfigurable stick-n-play modular microfluidic system using magnetic interconnects. Lab Chip. 2016;16(19):3700–3707. doi: 10.1039/c6lc00741d. [DOI] [PubMed] [Google Scholar]
- 72.Kang E, Shin SJ, Lee KH, Lee SH. Novel PDMS cylindrical channels that generate coaxial flow, and application to fabrication of microfibers and particles. Lab Chip. 2010;10(14):1856–1861. doi: 10.1039/c002695f. [DOI] [PubMed] [Google Scholar]
- 73.Gómez-Sjöberg R. 2012 Rafael’s Microfluidics Site. Available at https://sites.google.com/site/rafaelsmicrofluidicspage/. Accessed November 11, 2013.
- 74.Alami N, et al. Comparative analysis of xanafide cytotoxicity in breast cancer cell lines. Br J Cancer. 2007;97(1):58–64. doi: 10.1038/sj.bjc.6603829. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.