Significance
The dormant subpopulations of Pseudomonas aeruginosa biofilms are linked to chronic infections because dormant cells tolerate antibiotic treatment and then repopulate the infections when conditions become favorable. Dormant cells must maintain cellular integrity, including preformed ribosomes, to resuscitate. The small-ribosome–binding proteins, ribosome modulation factor, and hibernation promoting factor (HPF) have evolved to maintain ribosomes in an inactive state. Using both population and single-cell–level studies, we show that HPF provides the primary mechanism used by P. aeruginosa to maintain ribosome integrity during dormancy, and that HPF is required for optimal P. aeruginosa resuscitation from dormancy. Preventing regrowth of the dormant subpopulation by targeting HPF may provide an effective means for eliminating the dormant subpopulations of P. aeruginosa infections.
Keywords: dormancy, resuscitation, hibernation promoting factor, ribosome, rRNA
Abstract
Pseudomonas aeruginosa biofilm infections are difficult to treat with antibiotic therapy in part because the biofilms contain subpopulations of dormant antibiotic-tolerant cells. The dormant cells can repopulate the biofilms following alleviation of antibiotic treatments. While dormant, the bacteria must maintain cellular integrity, including ribosome abundance, to reinitiate the de novo protein synthesis required for resuscitation. Here, we demonstrate that the P. aeruginosa gene PA4463 [hibernation promoting factor (HPF)], but not the ribosome modulation factor (PA3049), is required for ribosomal RNA preservation during prolonged nutrient starvation conditions. Single-cell–level studies using fluorescence in situ hybridization (FISH) and growth in microfluidic drops demonstrate that, in the absence of hpf, the rRNA abundances of starved cells decrease to levels that cause them to lose their ability to resuscitate from starvation, leaving intact nondividing cells. P. aeruginosa defective in the stringent response also had reduced ability to resuscitate from dormancy. However, FISH analysis of the starved stringent response mutant showed a bimodal response where the individual cells contained either abundant or low ribosome content, compared with the wild-type strain. The results indicate that ribosome maintenance is key for maintaining the ability of P. aeruginosa to resuscitate from starvation-induced dormancy and that HPF is the major factor associated with P. aeruginosa ribosome preservation.
Biofilms are communities of microorganisms that are attached to surfaces through their secreted extracellular polymeric substance material (1, 2). Biofilms are found in most aqueous environments but become problematic when associated with infectious diseases (3). In particular, bacteria growing in biofilms on host tissue or artificial implant devices are difficult to eradicate with antibiotic treatments and often result in chronic infections (4). For example, Pseudomonas aeruginosa growing in biofilms on pulmonary tissue is associated with chronic infections of cystic fibrosis (CF) patients (5). Even though the population of P. aeruginosa associated with biofilm infections can be reduced with antibiotic treatments, it is rarely eliminated. Results of longitudinal genomics studies of P. aeruginosa strains infecting CF pulmonary tissue show that strains within a patient are usually clonal over time (6, 7), suggesting that even though antibiotics reduce the bacterial loads of pulmonary biofilms, clones of the original infecting strains are able to re-emerge and establish new biofilm infections. One mechanism for enhanced tolerance of biofilm-associated bacteria to antibiotics is that biofilms contain heterogeneous populations of cells, including subpopulations of cells that are tolerant of the treatments (8, 9).
Bacterial heterogeneity in biofilms may arise by several mechanisms (10) including adaptation to local environmental conditions. Cells within regions of the biofilm with low nutrients or oxygen may enter a slow-growth or dormant state. Because antibiotics generally target active metabolic functions, dormant bacteria are tolerant of most antibiotic treatments. The dormant bacteria may then resuscitate and repopulate the biofilms following alleviation of antibiotics. Supporting this mechanism for biofilm-associated antibiotic tolerance, in prior research, we differentially labeled P. aeruginosa cells with the green fluorescent protein (GFP) and sorted them based on their metabolic activity (9). In those studies, the slow-growing P. aeruginosa cells in biofilms were tolerant to ciprofloxacin or tobramycin at concentrations 10-fold greater than their minimum inhibitory concentrations, whereas the active bacteria were killed by those antibiotics. We also used transcriptomics in combination with laser capture microdissection to identify mRNA transcripts that were abundant in the different biofilm subpopulations (9). As expected, most mRNA transcripts were in low abundance in the dormant subpopulation. However, the slow-growing antibiotic-tolerant subpopulation had a high abundance of mRNA transcripts for several genes, including PA4463 [a homolog to the Escherichia coli hibernation promoting factor (HPF)]. In E. coli, HPF along with ribosome modulation factor (RMF) inactivates ribosomes during stationary phase (11).
RMF and HPF have been well characterized in E. coli as ribosome-interacting proteins (12–15). RMF binds to the ribosome near the mRNA exit tunnel on the 30S ribosomal subunit, and HPF binds at the channel of the 30S ribosomal subunit where tRNA and mRNA bind, thereby inhibiting translation (12, 16, 17). RMF and HPF also cause conformational changes to the ribosome, which results in dimerization of two ribosomes to form an inactive 100S form (18). Ueta et al. (11) developed a model for ribosome inactivation during stationary phase of E. coli. They concluded that RMF binds the ribosome, forming an inactive 90S dimer, and that HPF stabilizes the ribosome in an inactive 100S form. E. coli also encodes an HPF paralog, YfiA, that inactivates the 70S ribosome, but inhibits the formation of the 100S dimer (11).
Homologs to RMF and HPF are found in many bacterial taxa, but vary depending on the organism. E. coli and most other gamma Proteobacteria have genes for rmf, hpf, and yfiA, whereas bacteria other than the gamma Proteobacteria lack the gene for rmf. Staphylococcus aureus does not encode rmf, but has an hpf with an extended C-terminal tail, termed long HPF (19). Long HPF results in 100S ribosome formation in stationary-phase S. aureus cells, even in the absence of an RMF homolog (19). P. aeruginosa PAO1 contains genes for rmf (PA3049) and hpf (PA4463), but does not encode the hpf paralog, yfiA (20). The involvement of HPF and RMF on maintenance of cell viability also varies among bacterial species. In E. coli, RMF, but not the HPF homologs, is required to maintain cell viability during stationary phase (11, 14). Vibrio cholerae, which has an rmf and two hpf homologs, requires at least one copy of hpf to maintain cell viability during stationary phase (21). Mycobacterium spp. and Listeria monocytogenes, which encode long HPF, require HPF for cell viability maintenance during prolonged incubation and during hypoxic conditions (22, 23).
In our prior study (9), biofilms of P. aeruginosa PAO1 with deletions of rmf or hpf had increased uptake of the membrane-impermeable stain, propidium iodide, compared with wild-type cells, suggesting that the nutrient- or oxygen-starved P. aeruginosa cells may lose viability in the absence of these ribosome-interacting factors. Here, we investigated the roles of hpf and rmf in the maintenance of viability of P. aeruginosa undergoing nutrient-deprived conditions. Under starvation conditions, wild-type P. aeruginosa PAO1 maintained the ability to resuscitate for weeks with little loss of viability. Surprisingly, the Δrmf mutant also had little loss of viability during extended starvation. However, the Δhpf mutant was impaired in its ability to resuscitate from starvation. To investigate the molecular mechanism for impaired resuscitation of the mutant strain, we analyzed the rRNAs of the starved cells both at the population level and at the single-cell level. Unlike the wild-type cells and the Δrmf mutant cells, the Δhpf mutant strain lost most of its rRNA by day 4 of starvation. In addition, by using drop-based microfluidic approach, we show that most of the Δhpf mutant cells were unable to divide and remained as single nonreplicating cells following extended starvation. Overall, the results demonstrate that, in P. aeruginosa, HPF is required to protect cells from ribosome loss during extended nutrient-deprived conditions and that ribosome protection by HPF is necessary for P. aeruginosa to resuscitate from dormancy.
Results
HPF, but Not RMF, Is Required for Maintenance of P. aeruginosa Viability Under Starvation Conditions.
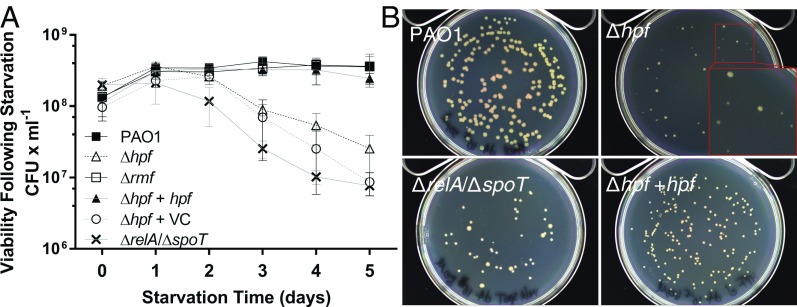
To characterize the physiological roles of HPF and RMF in P. aeruginosa, we tested the ability of P. aeruginosa PAO1 with Δhpf, Δrmf, and Δhpf/Δrmf mutations to resuscitate following prolonged nutrient deprivation. Wild-type and mutant cells were cultured to early stationary phase, washed in PBS, and then incubated in PBS with shaking at 37 °C. Aliquots of the cultures were sampled daily for their ability to form colonies on Tryptic Soy Agar (TSA) plates. The wild-type P. aeruginosa PAO1 was able to survive the starvation conditions with no apparent loss of viability (P = 0.96) (Fig. 1A). Based on results from E. coli (11, 14), RMF is predicted to play a role in ribosome inactivation. However, no observable phenotype with respect to recovery from starvation conditions (P = 0.37) was observed for P. aeruginosa Δrmf (Fig. 1A). In addition, and in contrast to published results for RMF in E. coli (24–27), the P. aeruginosa Δrmf mutant did not show an observable survival phenotype compared with the wild-type strain when exposed to osmotic shock, heat shock, acid stress, or sensitivity to gentamicin. In contrast, a deletion of PA4463 (Δhpf) resulted in a decrease in cell recovery following starvation (P < 0.0001). The Δhpf mutant strain initially produced 1.9 × 108 cfu × mL−1 after 30 min of starvation and reduced to 2.5 × 107 cfu × mL−1 by 5 d of starvation (Fig. 1A). When Δhpf was complemented in trans (Δhpf + hpf), survival under nutrient-limited conditions was restored to wild-type levels (P = 0.25), but not for the vector control strain (Δhpf + VC) (P < 0.0001). The Δhpf/Δrmf double mutant also showed loss of recovery following starvation compared with the wild-type strain (P = 0.0002) (SI Appendix, Fig. S1). The Δhpf/Δrmf double mutant could be restored to wild-type levels of resuscitation with a plasmid containing hpf alone, but not with a plasmid containing rmf alone (SI Appendix, Fig. S1).
Fig. 1.
Recovery of P. aeruginosa following extended incubation under nutrient-deprived conditions. (A) The cfu’s on TSA agar following incubation in aerated PBS. (B) Colony morphology of P. aeruginosa on TSA, following 4 d of incubation in PBS for wild-type P. aeruginosa PAO1, PAO1 Δhpf, PAO1 ΔrelA/ΔspoT, and PAO1 Δhpf + hpf. Colony morphologies of each strain and at each time point are shown in SI Appendix, Fig. S2.
The stringent response plays a role in the ability of P. aeruginiosa to survive in stationary phase (28). In the stringent response, guanidine penta- and tetra-phosphate [(p)ppGpp], produced by the activities of RelA and SpoT when RelA interacts with stalled ribosomes (29), acts as a signaling molecule that induces expression of genes required for survival during stationary phase (30). We tested a ΔrelA/ΔspoT mutant under the same starvation conditions as the Δrmf and Δhpf mutant strains. The ΔrelA/ΔspoT mutant showed a similar response to starvation as the Δhpf mutant, where cell recovery was impaired compared with the wild-type strain (P = 0.03) (Fig. 1A). The results indicate that HPF and the stringent response, but not RMF, is necessary for prolonged P. aeruginosa survival under nutrient-limited conditions.
HPF Is Required for Optimal Recovery of P. aeruginosa from Nutrient Starvation.
During the starvation experiments, we noted that, in addition to the reduced number of cfu’s for the Δhpf mutant strain, the colonies that arose were heterogeneous in morphology (Fig. 1B and SI Appendix, Fig. S2). The starved wild-type strain produced large, uniform colonies. Although some colonies of the Δhpf strain were similar to those of the wild type, many were small and required extended incubation time to become visible. The Δhpf + hpf complemented strain had smaller colonies than the wild type, but the colonies were uniform (Fig. 1B and SI Appendix, Fig. S2). The ΔrelA/ΔspoT strain had colony morphologies with heterogeneity in size, but did not require extended incubation time to become visible. When the small Δhpf colonies were restreaked onto new TSA plates, the wild-type colony morphology was restored, indicating that heterogeneity in recovered colony size was not heritable. To determine if the reduced cfu counts following starvation of the ΔrelA/ΔspoT mutant strain were due to the role of the stringent response in regulation of hpf expression, we introduced a plasmid copy of hpf under control of the isopropyl β-d-1-thiogalactopyranoside-inducible Ptrc promoter into the ΔrelA/ΔspoT strain. The ΔrelA/ΔspoT + hpf strain had a similar survival phenotype as the ΔrelA/ΔspoT strain (SI Appendix, Fig. S3), indicating that loss of cell viability in the ΔrelA/ΔspoT mutant is not associated with loss of hpf expression.
HPF Is Required for Preservation of 23S rRNA During Nutrient Starvation.
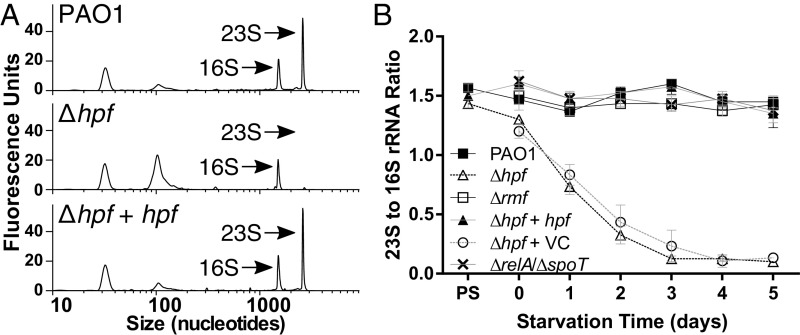
HPF and RMF are ribosome-associated proteins that in E. coli cause formation of inactive 100S ribosome dimers during stationary phase (11). To characterize the roles of RMF and HPF on ribosomes under starvation conditions of P. aeruginosa, we extracted total RNA from nutrient-deprived cultures and assayed the relative abundance of the 23S and 16S rRNAs using the Agilent Bioanalyzer. P. aeruginosa PAO1 cells cultured to stationary phase showed a 23S/16S rRNA ratio of ∼1.6 (Fig. 2 A and B). Remarkably, this ratio was maintained in P. aeruginosa PAO1 throughout the starvation period (Fig. 2B and SI Appendix, Fig. S4), indicating little loss of ribosome quality during prolonged nutrient deprivation. Similarly, the Δrmf mutant maintained a 23S/16S rRNA ratio of ∼1.5 throughout starvation (Fig. 2B and SI Appendix, Fig. S4). In contrast, the Δhpf strain had 23S/16S rRNA ratios of 1.4 before starvation, which rapidly decreased to 0.1 by day 4 of starvation (Fig. 2 A and B and SI Appendix, Fig. S4). The differences between the Δhpf mutant and the wild-type strain were significant throughout starvation, except on day 0 (P < 0.000001). When the Δhpf mutant strain was complemented with hpf, the ratio of 23S and 16S rRNA was restored to levels that were not significantly different from the wild type (P = 0.83), whereas the vector control (Δhpf + VC) did not restore the rRNA ratios (P < 0.0001) (Fig. 2 A and B and SI Appendix, Fig. S4). We also determined the 23S/16S rRNA ratios of the Δhpf/Δrmf double mutant. As with the Δhpf mutant, the Δhpf/Δrmf double mutant had loss of the 23S rRNA during nutrient starvation (SI Appendix, Fig. S5). The 23S/16S rRNA ratio for the double mutant was complemented by a plasmid copy of hpf without rmf, but not with rmf alone (SI Appendix, Fig. S5). The results indicate that HPF, but not RMF, is required for maintenance of the 23S rRNA under starvation conditions. Interestingly, although the ΔrelA/ΔspoT mutant had reduced viability during starvation compared with wild type, the strain did not show selective loss of the 23S rRNA compared with wild type (P = 0.30) (Fig. 2B and SI Appendix, Fig. S4). The results indicate that the molecular mechanism for reduced viability in the starved ΔrelA/ΔspoT mutant likely differs from that of the Δhpf mutant.
Fig. 2.
P. aeruginosa 23S-to-16S rRNA ratios following extended incubation under nutrient-deprived conditions with shaking at 37 °C. (A) Bioanalyzer traces of 23S and 16S rRNA following 4 d of starvation for wild-type strain PAO1, the Δhpf mutant, and the Δhpf mutant complemented with hpf (Δhpf + hpf). (B) P. aeruginosa 23S-to-16S rRNA ratios following extended starvation conditions. Representative bioanalyzer traces showing 23S and 16S rRNAs for all strains at each time point are shown in SI Appendix, Figs. S4 and S5.
During nutrient starvation of E. coli, ribosome degradation is initiated by site-specific endoribonuclease cleavages of the 16S and 23S rRNAs, leading to reduced 70S ribosome abundances and proteolysis of ribosomal proteins (31–33). To determine the fate of the rRNA during nutrient starvation of the P. aeruginosa Δhpf strain, we performed time-course Bioanalyzer studies over the first day of starvation. The Δhpf mutant showed reduction of the 23S peak and a concomitant increase in peak-associated small-molecular-weight RNAs (∼100 nt in length), characteristic of degraded rRNA fragments (SI Appendix, Fig. S6A). In contrast to the wild-type cells and the Δhpf + hpf cells, the small-molecular-weight RNA comprised most of the cellular RNA of the Δhpf mutant strain by day 4 of starvation (SI Appendix, Fig. S6B). The total RNA content of the Δhpf culture also decreased, compared with the wild-type control, after 4 d of starvation (P = 0.01) (SI Appendix, Fig. S6D). We next analyzed the relative amounts of 16S and 23S rRNA of the Δhpf mutant strain compared with wild-type strain following 0 d and 4 d of starvation by reverse transcription-quantitative PCR (RT-qPCR). The results showed a reduction of both rRNA subunits in the Δhpf mutant cells on day 4 (P = 0.03), but with greater reduction of the 23S rRNA subunit than the 16S subunit (SI Appendix, Fig. S6E).
Heterogeneity of HPF-Based rRNA Protection Determined at the Single-Cell Level.
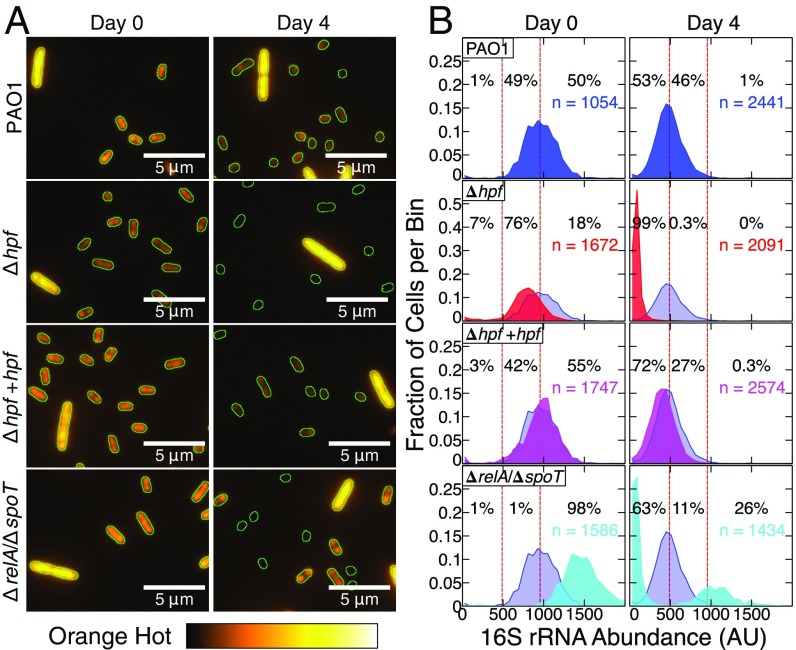
Although cell viability was reduced in the Δhpf mutant, viability was not completely eliminated during nutrient deprivation (Fig. 1). In addition, loss of the rRNA was characterized for the entire population of cells, which may have masked the amount of rRNA present in individual cells within the population. Therefore, we used FISH analysis to determine the 16S rRNA levels of individual nutrient-deprived cells. To analyze the FISH results quantitatively, we determined the average fluorescence intensity per pixel within the area of single cells using ImageJ (https://imagej.nih.gov/ij/index.html). The fluorescence intensity for each cell was background-subtracted and then normalized by the spike-in control cells, consisting of exponentially growing P. aeruginosa PAO1 (pMF230). The spike-in cells were differentiated from the starved test cells because they were larger, had higher abundances of rRNAs, and had GFP fluorescence (Fig. 3A). The results from the wild-type cells showed a gradual reduction in the average FISH fluorescence intensity per cell over time (P = 0.008) (Fig. 3 and SI Appendix, Fig. S7). However, the FISH fluorescence of the wild-type cells was maintained at high levels throughout the starvation conditions, indicating that most cells maintained abundant ribosome content during starvation (Fig. 3 and SI Appendix, Fig. S7). The FISH-fluorescence intensities for the Δhpf mutant strain were at similar levels to the wild-type cells before nutrient starvation (P = 0.17) (Fig. 3). However, the average FISH-fluorescence intensity of Δhpf decreased following 1 d of starvation (P < 0.001) and continued to decrease compared with wild-type cells with continued starvation incubation (Fig. 3B and SI Appendix, Fig. S7 and Table S1). The complemented Δhpf + hpf strain had restored FISH fluorescence, comparable to the wild type (Fig. 3 and SI Appendix, Fig. S7 and Table S2).
Fig. 3.
(A) FISH analysis of 16S rRNAs for P. aeruginsoa PAO1 and mutant derivatives, starved for 4 d in PBS at 37 °C. The16S probe labeled with Cy3 is false-colored orange. Each field contains a spike-in control cell of exponential phase PAO1 (pMF230) (the large cell with high rRNA abundance). The edges of cells were determined by using bright-field microscopy and are outlined in green. (B) The mean fluorescence intensity for 16S-Cy3 probe was quantified, normalized to the spike-in controls, and binned based on the fluorescence intensity of individual cells. The dashed vertical lines represent the average FISH fluorescence intensity for the wild-type strain before starvation and after 4 d of starvation. Data for the wild-type strain are shown on each plot in blue. Data shown are from three independent biological replicates per strain at each time point with the total number of cells quantified indicated. Histograms showing the FISH fluorescence intensities for each day of starvation are shown in SI Appendix, Fig. S7. Box-whisker plots showing the average and range of these data are shown in SI Appendix, Fig. S8.
The FISH results for the ΔrelA/ΔspoT mutant showed a strikingly different response to starvation than either the wild-type strain or the Δhpf mutant strain. During the first 2 d of starvation, the ΔrelA/ΔspoT cells had higher abundances of 16S rRNA than the wild-type cells (SI Appendix, Fig. S7). Continued incubation in nutrient-free medium resulted in ΔrelA/ΔspoT mutant cells segregating into two populations with bimodal distribution. One subpopulation of ΔrelA/ΔspoT cells continued to have higher 16S rRNA content than the wild-type cells, whereas the other subpopulation of cells had little detectable rRNA (Fig. 3 and SI Appendix, Fig. S7). By day 4 of starvation, most (∼63%) ΔrelA/ΔspoT cells had little signal for the 16S rRNA, whereas 26% had signals greater than the average of the wild-type cells.
The presence of outliers with high and low FISH fluorescence intensities were observed for all strains. The box and whisker plots (SI Appendix, Fig. S8) show a greater average decrease of 16S rRNA for the Δhpf cells than for the wild-type cells. However, all strains had outliers where either high or low abundances of rRNAs were observed, including a small fraction of Δhpf mutant cells that had fluorescence intensity as high as the average intensity of 4-d-starved wild-type cells. The observation of outliers at the single-cell level suggests that HPF may not be the sole mechanism for maintenance of ribosome integrity of P. aeruginosa.
Role of HPF in the Resuscitation of Individual Dormant Cells.
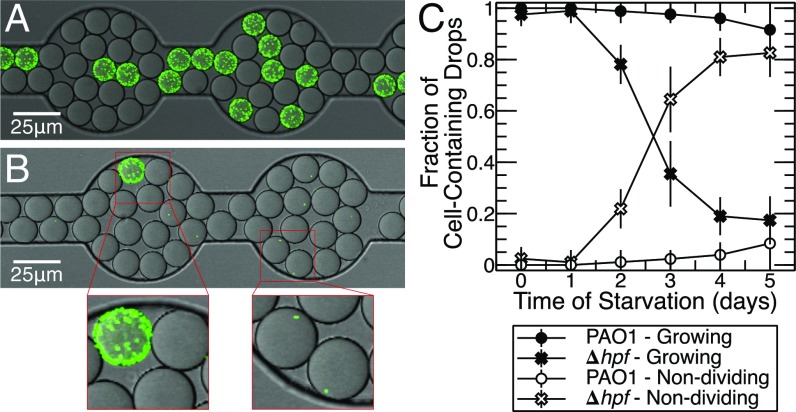
The colony size and variable rRNA levels of the starved Δhpf mutant cells suggested that there was heterogeneity in the resuscitating population. Therefore, we used a drop-based microfluidic approach (34) to quantify the heterogeneity in this population. These drop-based experiments allowed us to differentiate and quantify individual cells undergoing resuscitation from the nondividing cells within many drop-based bioreactors. We compared resuscitation of the wild-type cells and the Δhpf mutant cells, both expressing GFP, so that they could be visualized by confocal scanning laser microscopy (CSLM) (Fig. 4 A and B). Cultures were starved using the conditions described above, and then individual cells were encapsulated in 15-μm-diameter, water-in-oil drops containing TSB medium as the dispersed phase in oil. The ratio of drops containing one cell to empty drops was set at ∼1:5–1:10. The cells were then incubated in the TSB contained in each drop for 24–48 h to allow resuscitation of individual cells from the starvation-induced dormancy. Drops were then analyzed by CSLM and quantified by image analysis for cell growth (drops that were completely or partially filled with bacteria versus drops that contained only one cell) (Fig. 4). Similar to the cfu counts, there was an increase in total number of cells from day 0 to day 1 of starvation for both the wild-type (P = 0.037) and Δhpf mutant cells (P = 0.003) (Fig. 1 and SI Appendix, Fig. S9), suggesting that the cells had divided in the PBS-starvation medium. To determine if cells were capable of division during the first day of starvation, we encapsulated and incubated cells in drops containing PBS. Under these conditions, most drops contained multiple cells after 24 h (SI Appendix, Fig. S10), indicating that the cells had undergone at least one round of cell division during the first day of incubation in PBS.
Fig. 4.
Examples of cell resuscitation of individual P. aeruginosa (pMF230) cells within 15-μm-diameter oil drops with visualization by CSLM. Cells were incubated in PBS for 4 d, and encapsulated in drops containing TSB medium. (A) P. aeruginosa PAO1 following 4 d of starvation and then regrowth for 24 h in TSB. (B) P. aeruginosa PAO1 ∆hpf after 4 d of starvation and then regrowth for 24 h in TSB. (Insets) Cell regrowth and single nondividing cells in drops. (C) Percentage of cells that replicated inside of TSB-containing drops versus cells that remained as single intact nondividing cells following starvation in PBS. Cell counts of replicating and nonreplicating cells are shown in SI Appendix, Fig. S9. Regrowth of PAO1 and the ∆hpf mutant cells in drops over 24 h is shown in Movies S1 and S2.
Following the initial cell division in PBS from day 0 to day 1 of starvation, the total number of cells captured in drops remained essentially constant throughout the starvation period for both the Δhpf mutant (P = 0.41) and wild-type cells (P = 0.88) (SI Appendix, Fig. S9), indicating little cell lysis during starvation. However, the percentage of cells that remained as single nonreplicating cells versus cells that grew within the drops differed over time for PAO1 (P = 0.03) and the Δhpf mutant strains (P = 0.01) (Fig. 4C and SI Appendix, Fig. S9). Initially, ∼100% of the wild-type cells were capable of resuscitation inside of drops. The number of PAO1 cells capable of resuscitation decreased to 96% by 4 d of starvation (Fig. 4C and SI Appendix, Fig. S9). In contrast, the number of Δhpf mutant cells capable of resuscitation inside of the drops decreased over time of starvation, with most cells (81%) remaining as single nondividing cells by day 4 of starvation (Fig. 4C and SI Appendix, Fig. S9). Additional incubation to 48 h inside drops did not result in a significant increase in the number of cells that could resuscitate (P = 0.81). The results indicate that the number of nondividing P. aeruginosa cells increases during starvation in the absence of a functional HPF.
The resuscitating Δhpf mutant cells had heterogeneous colony morphology (Fig. 1), suggesting that some of the recovering Δhpf cells either had an increased lag time or a reduced growth rate. To distinguish these two possibilities, we performed time-course imaging of single cells in microfluidic drops following 4 d of starvation. Most wild-type cells (98.7%) recovered from starvation and had a relatively uniform growth rate averaging 0.56 h−1 (Movie S1 and SI Appendix, Fig. S11), whereas most of the Δhpf mutant cells (84.4%) did not recover following 4 d of starvation. The Δhpf cells that recovered had a slightly lower average growth rate (0.44 h−1) than the wild-type cells. However, the Δhpf cells had highly variable lag times (Movie S2 and SI Appendix, Fig. S11). The results indicate that the small-colony variants of the Δhpf cells as seen in Fig. 1 are likely caused by an increased lag time required for these cells to recover from starvation.
Discussion
Heterotrophic bacteria living in aqueous environments often experience conditions where nutrients are scarce, including the nutrient-depleted zones of biofilms (10). Rapidly growing bacteria contain sufficient ribosome concentrations for maximum protein biosynthesis during exponential phase (35, 36). When the cells switch to slow-growth conditions, the number of ribosomes per cell diminishes through degradation or by partitioning to daughter cells. However, slowly growing and dormant cells must have mechanisms to maintain sufficient quantities of premade macromolecules, including ribosomes, to allow de novo protein synthesis and cell regrowth when conditions become favorable (37). Mechanisms have evolved to avoid complete loss of these essential macromolecules when cells are dormant. Here, we demonstrate that the small-ribosome–interacting protein, HPF, is required for ribosome preservation during dormancy of P. aeruginosa. Surprisingly, although important for E. coli ribosome inactivation (13) and abundant in most P. aeruginosa biofilm transcriptomic experiments (38), RMF does not appear to play a significant role in ribosome preservation in P. aeruginosa under the conditions tested here. Mechanisms for starvation responses may be specific for a certain nutrient. In P. aeruginosa, hpf is located downstream of the nitrogen-stress–associated sigma factor, rpoN, and is expressed, in part, from the rpoN promoter. However, hpf also has its own promoter(s), allowing high expression under certain conditions.
In a previous study, we measured the ribosome abundance of cells at different vertical strata within P. aeruginosa biofilms by microdissecting the biofilms and then assaying 16S rRNA amounts from the different microzones using RT-qPCR (39). Because rRNA is rapidly degraded when not associated with ribosomes (40), quantification of rRNA provides an estimate of the ribosome copy number per cell. From that study, we showed that exponentially growing P. aeruginosa had ∼70,000 ribosomes per cell, whereas cells in the slowly growing subpopulation of biofilms had ∼20,000 ribosomes per cell (39). This relatively high concentration of ribosomes in the slow-growing cells was maintained even for cells with little transcriptional activity, suggesting that ribosomes are preserved in the cells in the interior of the biofilms. Identification of transcripts for the ribosomal accessory protein, hpf, as abundant in the interior of the biofilms indicated that the product of this gene may be important for ribosome preservation in the dormant subpopulation of cells (9). Here, we show that, in the absence of hpf, starved cells have reduced capacity to resuscitate following nutrient deprivation and have reduced rRNA levels. The population-level experiments of the Δhpf strain showed greater loss of the 23S rRNA than of the 16S rRNA (Fig. 2), but with a significant loss of both rRNA species (SI Appendix, Fig. S6), compared with only a modest decrease of 16S and 23S rRNA levels for starved wild-type cells. The FISH single-cell results also showed loss of the 16S rRNA in the Δhpf mutant, whereas the wild-type strain maintained relatively high levels of 16S rRNA during starvation (Fig. 3).
Ribosome degradation during nutrient starvation in E. coli is initiated by site-specific endoribonuclease cleavages of 16S and 23S rRNAs, leading to the loss of detectable free subunits as well as reduced 70S ribosome abundance (31, 33). Ribosomal proteins dissociated from rRNAs are susceptible to proteolysis and likely recycled as nutrients (32). We observed an additional peak in the Δhpf mutant in the Bioanalyzer studies, which may indicate the product of 23S rRNA cleavage (Fig. 2 and SI Appendix, Fig. S6). Cleavage of 23S rRNA may initiate rRNA degradation to small fragments as observed here in the Bioanalyzer studies. The crystal structures of ribosomes with HPF and its paralog revealed that the HPF-binding site overlaps with the binding sites of translation initiation factors on the 30S subunit (12, 41). In vitro studies also indicate that the HPF paralog YfiA competes for 30S binding with IF-3 in the presence of polyamines (41). Binding of IF-3 to the 30S subunit prevents the association of the 50S subunit until IF-2 promotes subunit joining to form the 70S initiation complex (42). Because the interaction of ribosomal subunits with HPF induces subunit joining (12, 16, 41), HPF may protect unused ribosomes from endoribonucleases through 70S-HPF complex formation, whereas IF-3 binding to the 30S subunit in the absence of HPF would leave free 50S subunits available for endonuclease cleavage.
The heterogeneity observed in microfluidic studies of resuscitating cells indicates that HPF may not provide the sole mechanism for ribosome preservation. A percentage of the Δhpf cells was capable of regrowth in microfluidic drops (Fig. 4). In addition, some Δhpf cells maintained their ribosomal content even after extended starvation (SI Appendix, Fig. S8). Therefore, an alternative strategy to ensure ribosome preservation may exist in P. aeruginosa. The microfluidic studies of wild-type cells also showed that a percentage of cells (4% by day 4 of starvation) were unable to regrow following starvation. Whether these cells still had metabolic activity and were unable to grow (e.g., viable but nonculturable cells), or whether the cells were not viable, is not known yet. In either case, the results demonstrate that, even in a clonal population of cells, heterogeneity in ribosome content and cellular capacity for regrowth exists.
The stringent response mutant had a similar loss of recovery from starvation as the Δhpf mutant. However, the mechanism for impaired recovery of the stringent response mutant differs from the Δhpf mutant strain. Our preliminary data using a transcriptional reporter of hpf indicate that the stringent response has a small modulatory effect on hpf expression, but that hpf expression is not abolished in the ΔrelA/ΔspoT mutant. A plasmid copy of hpf did not have any effect on the survival phenotype of the ΔrelA/ΔspoT strain (SI Appendix, Fig. S3). Therefore, we conclude that expression of hpf is not solely regulated through the product of the stringent response alarmone, (p)ppGpp. Overall, the results demonstrate that HPF provides the primary mechanism for ribosome preservation during nutrient starvation of P. aeruginosa and that ribosome preservation is needed for P. aeruginosa cells to resuscitate from dormancy.
Materials and Methods
Bacterial Strains and Growth Conditions.
Studies were performed on P. aeruginosa strain PAO1 and its Δrmf, Δhpf, and Δhpf/Δrmf mutant derivatives and on complemented strains as described in SI Appendix, SI Materials and Methods, and previously (9). The stringent response mutant, containing deletions of relA and spoT (28), was provided by Pradeep Singh, University of Washington, Seattle. Nutrient-starvation studies and the drop-plate method (43) used to quantify cfu’s were performed as described in SI Appendix, SI Materials and Methods.
RNA Extraction and Determination of Relative rRNA Abundances.
RNA was extracted as described previously (44) with modifications described in SI Appendix, SI Materials and Methods. RNA was analyzed using the Bioanalyzer 2100 (Agilent Technologies) as described in detail in SI Appendix, SI Materials and Methods. Total rRNA and 16S and 23S rRNAs were quantified using the NanoDrop1000 (Thermo Fisher Scientific) and by RT-qPCR as described in SI Appendix, SI Materials and Methods.
FISH for Quantification of rRNA.
FISH, as described by Brileya et al. (45), and using a Cy3-labeled 16S rRNA probe described by Hogardt et al. (46), was used to quantify the relative amounts of 16S RNA from individual cells. A detailed description of the FISH method and analysis is provided in SI Appendix, SI Materials and Methods.
Drop Encapsulation and Monitoring Growth of Single Bacterial Cells.
Single bacterial cells were encapsulated into microfluidic drops as described in detail in SI Appendix, SI Materials and Methods and in ref. 47. Drop-encapsulated cells were injected into a modified “Dropspots” immobilization device (48). Microscopic and statistical analyses of bacterial growth in drops are provided in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Phil Stewart, Joanna Borgogna, and Al Parker for their helpful discussions of this work; Geoffrey Zath for microfluidic drop growth rate analysis; Betsy Pitts, Kristen Brileya, and Amanda Richards for microscopy data acquisition; and Dr. Pradeep K. Singh for providing the ΔrelA/ΔspoT mutant. This work was funded by National Institute of Allergy and Infectious Diseases Grant AI113330 (to M.J.F.), National Institute of General Medical Sciences Grant P20GM103474 (to S.P.), and Montana State University's Graduate School (T.A.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1700695114/-/DCSupplemental.
References
- 1.Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappin-Scott HM. Microbial biofilms. Annu Rev Microbiol. 1995;49:711–745. doi: 10.1146/annurev.mi.49.100195.003431. [DOI] [PubMed] [Google Scholar]
- 2.Flemming HC, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi: 10.1038/nrmicro2415. [DOI] [PubMed] [Google Scholar]
- 3.Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: A common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 4.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet. 2001;358(9276):135–138. doi: 10.1016/s0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 5.Lyczak JB, Cannon CL, Pier GB. Lung infections associated with cystic fibrosis. Clin Microbiol Rev. 2002;15(2):194–222. doi: 10.1128/CMR.15.2.194-222.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns JL, et al. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis. 2001;183(3):444–452. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- 7.Warren AE, et al. Genotypic and phenotypic variation in Pseudomonas aeruginosa reveals signatures of secondary infection and mutator activity in certain cystic fibrosis patients with chronic lung infections. Infect Immun. 2011;79(12):4802–4818. doi: 10.1128/IAI.05282-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lewis K. Persister cells, dormancy and infectious disease. Nat Rev Microbiol. 2007;5(1):48–56. doi: 10.1038/nrmicro1557. [DOI] [PubMed] [Google Scholar]
- 9.Williamson KS, et al. Heterogeneity in Pseudomonas aeruginosa biofilms includes expression of ribosome hibernation factors in the antibiotic-tolerant subpopulation and hypoxia-induced stress response in the metabolically active population. J Bacteriol. 2012;194(8):2062–2073. doi: 10.1128/JB.00022-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stewart PS, Franklin MJ. Physiological heterogeneity in biofilms. Nat Rev Microbiol. 2008;6(3):199–210. doi: 10.1038/nrmicro1838. [DOI] [PubMed] [Google Scholar]
- 11.Ueta M, et al. Ribosome binding proteins YhbH and YfiA have opposite functions during 100S formation in the stationary phase of Escherichia coli. Genes Cells. 2005;10(12):1103–1112. doi: 10.1111/j.1365-2443.2005.00903.x. [DOI] [PubMed] [Google Scholar]
- 12.Polikanov YS, Blaha GM, Steitz TA. How hibernation factors RMF, HPF, and YfiA turn off protein synthesis. Science. 2012;336(6083):915–918. doi: 10.1126/science.1218538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wada A, Yamazaki Y, Fujita N, Ishihama A. Structure and probable genetic location of a “ribosome modulation factor” associated with 100S ribosomes in stationary-phase Escherichia coli cells. Proc Natl Acad Sci USA. 1990;87(7):2657–2661. doi: 10.1073/pnas.87.7.2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wada A, Mikkola R, Kurland CG, Ishihama A. Growth phase-coupled changes of the ribosome profile in natural isolates and laboratory strains of Escherichia coli. J Bacteriol. 2000;182(10):2893–2899. doi: 10.1128/jb.182.10.2893-2899.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yamagishi M, et al. Regulation of the Escherichia coli rmf gene encoding the ribosome modulation factor: Growth phase- and growth rate-dependent control. EMBO J. 1993;12(2):625–630. doi: 10.1002/j.1460-2075.1993.tb05695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueta M, et al. Role of HPF (hibernation promoting factor) in translational activity in Escherichia coli. J Biochem. 2008;143(3):425–433. doi: 10.1093/jb/mvm243. [DOI] [PubMed] [Google Scholar]
- 17.Wada A, Igarashi K, Yoshimura S, Aimoto S, Ishihama A. Ribosome modulation factor: Stationary growth phase-specific inhibitor of ribosome functions from Escherichia coli. Biochem Biophys Res Commun. 1995;214(2):410–417. doi: 10.1006/bbrc.1995.2302. [DOI] [PubMed] [Google Scholar]
- 18.Kato T, et al. Structure of the 100S ribosome in the hibernation stage revealed by electron cryomicroscopy. Structure. 2010;18(6):719–724. doi: 10.1016/j.str.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 19.Ueta M, Wada C, Wada A. Formation of 100S ribosomes in Staphylococcus aureus by the hibernation promoting factor homolog SaHPF. Genes Cells. 2010;15(1):43–58. doi: 10.1111/j.1365-2443.2009.01364.x. [DOI] [PubMed] [Google Scholar]
- 20.Stover CK, et al. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature. 2000;406(6799):959–964. doi: 10.1038/35023079. [DOI] [PubMed] [Google Scholar]
- 21.Sabharwal D, Song T, Papenfort K, Wai SN. The VrrA sRNA controls a stationary phase survival factor Vrp of Vibrio cholerae. RNA Biol. 2015;12(2):186–196. doi: 10.1080/15476286.2015.1017211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kline BC, McKay SL, Tang WW, Portnoy DA. The Listeria monocytogenes hibernation-promoting factor is required for the formation of 100S ribosomes, optimal fitness, and pathogenesis. J Bacteriol. 2015;197(3):581–591. doi: 10.1128/JB.02223-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trauner A, Lougheed KE, Bennett MH, Hingley-Wilson SM, Williams HD. The dormancy regulator DosR controls ribosome stability in hypoxic mycobacteria. J Biol Chem. 2012;287(28):24053–24063. doi: 10.1074/jbc.M112.364851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El-Sharoud WM, Niven GW. The influence of ribosome modulation factor on the survival of stationary-phase Escherichia coli during acid stress. Microbiology. 2007;153(Pt 1):247–253. doi: 10.1099/mic.0.2006/001552-0. [DOI] [PubMed] [Google Scholar]
- 25.Garay-Arroyo A, Colmenero-Flores JM, Garciarrubio A, Covarrubias AA. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem. 2000;275(8):5668–5674. doi: 10.1074/jbc.275.8.5668. [DOI] [PubMed] [Google Scholar]
- 26.McKay SL, Portnoy DA. Ribosome hibernation facilitates tolerance of stationary-phase bacteria to aminoglycosides. Antimicrob Agents Chemother. 2015;59(11):6992–6999. doi: 10.1128/AAC.01532-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niven GW. Ribosome modulation factor protects Escherichia coli during heat stress, but this may not be dependent on ribosome dimerisation. Arch Microbiol. 2004;182(1):60–66. doi: 10.1007/s00203-004-0698-9. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen D, et al. Active starvation responses mediate antibiotic tolerance in biofilms and nutrient-limited bacteria. Science. 2011;334(6058):982–986. doi: 10.1126/science.1211037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol. 2015;13(5):298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Durfee T, Hansen AM, Zhi H, Blattner FR, Jin DJ. Transcription profiling of the stringent response in Escherichia coli. J Bacteriol. 2008;190(3):1084–1096. doi: 10.1128/JB.01092-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basturea GN, Zundel MA, Deutscher MP. Degradation of ribosomal RNA during starvation: Comparison to quality control during steady-state growth and a role for RNase PH. RNA. 2011;17(2):338–345. doi: 10.1261/rna.2448911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Petersen C. Escherichia coli ribosomal protein L10 is rapidly degraded when synthesized in excess of ribosomal protein L7/L12. J Bacteriol. 1990;172(1):431–436. doi: 10.1128/jb.172.1.431-436.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zundel MA, Basturea GN, Deutscher MP. Initiation of ribosome degradation during starvation in Escherichia coli. RNA. 2009;15(5):977–983. doi: 10.1261/rna.1381309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo MT, Rotem A, Heyman JA, Weitz DA. Droplet microfluidics for high-throughput biological assays. Lab Chip. 2012;12(12):2146–2155. doi: 10.1039/c2lc21147e. [DOI] [PubMed] [Google Scholar]
- 35.Nomura M, Gourse R, Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- 36.Sarmientos P, Cashel M. Carbon starvation and growth rate-dependent regulation of the Escherichia coli ribosomal RNA promoters: Differential control of dual promoters. Proc Natl Acad Sci USA. 1983;80(22):7010–7013. doi: 10.1073/pnas.80.22.7010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Madar D, et al. Promoter activity dynamics in the lag phase of Escherichia coli. BMC Syst Biol. 2013;7:136. doi: 10.1186/1752-0509-7-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Folsom JP, et al. Physiology of Pseudomonas aeruginosa in biofilms as revealed by transcriptome analysis. BMC Microbiol. 2010;10:294. doi: 10.1186/1471-2180-10-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pérez-Osorio AC, Williamson KS, Franklin MJ. Heterogeneous rpoS and rhlR mRNA levels and 16S rRNA/rDNA (rRNA gene) ratios within Pseudomonas aeruginosa biofilms, sampled by laser capture microdissection. J Bacteriol. 2010;192(12):2991–3000. doi: 10.1128/JB.01598-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Deutscher MP. Degradation of stable RNA in bacteria. J Biol Chem. 2003;278(46):45041–45044. doi: 10.1074/jbc.R300031200. [DOI] [PubMed] [Google Scholar]
- 41.Vila-Sanjurjo A, Schuwirth BS, Hau CW, Cate JH. Structural basis for the control of translation initiation during stress. Nat Struct Mol Biol. 2004;11(11):1054–1059. doi: 10.1038/nsmb850. [DOI] [PubMed] [Google Scholar]
- 42.Schmeing TM, Ramakrishnan V. What recent ribosome structures have revealed about the mechanism of translation. Nature. 2009;461(7268):1234–1242. doi: 10.1038/nature08403. [DOI] [PubMed] [Google Scholar]
- 43.Herigstad B, Hamilton M, Heersink J. How to optimize the drop plate method for enumerating bacteria. J Microbiol Methods. 2001;44(2):121–129. doi: 10.1016/s0167-7012(00)00241-4. [DOI] [PubMed] [Google Scholar]
- 44.Guragain M, et al. The Pseudomonas aeruginosa PAO1 two-component regulator carsr regulates calcium homeostasis and calcium-induced virulence factor production through its regulatory targets CarO and CarP. J Bacteriol. 2016;198(6):951–963. doi: 10.1128/JB.00963-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brileya KA, Camilleri LB, Fields MW. 3D-fluorescence in situ hybridization of intact, anaerobic biofilm. Methods Mol Biol. 2014;1151:189–197. doi: 10.1007/978-1-4939-0554-6_13. [DOI] [PubMed] [Google Scholar]
- 46.Hogardt M, et al. Specific and rapid detection by fluorescent in situ hybridization of bacteria in clinical samples obtained from cystic fibrosis patients. J Clin Microbiol. 2000;38(2):818–825. doi: 10.1128/jcm.38.2.818-825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holtze C, et al. Biocompatible surfactants for water-in-fluorocarbon emulsions. Lab Chip. 2008;8(10):1632–1639. doi: 10.1039/b806706f. [DOI] [PubMed] [Google Scholar]
- 48.Schmitz CH, Rowat AC, Köster S, Weitz DA. Dropspots: A picoliter array in a microfluidic device. Lab Chip. 2009;9(1):44–49. doi: 10.1039/b809670h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.