Significance
Because structures in the developing embryo are organized by secreted signals, embryonic cells must integrate multiple inputs to turn on the target genes necessary for proper development. Little is known about how multiple signals can work together to regulate such target genes in an embryological context. In this work, we use cultured limb bud mesenchymal cells to investigate how two such signals, Sonic hedgehog (Shh) and fibroblast growth factor 8 (Fgf8), work together to control the activity of Hoxd genes, a set of transcription factors necessary for the patterning of developing tetrapod limbs.
Keywords: Sonic hedgehog, fibroblast growth factor, Hox genes, limb development
Abstract
During embryonic development, fields of progenitor cells form complex structures through dynamic interactions with external signaling molecules. How complex signaling inputs are integrated to yield appropriate gene expression responses is poorly understood. In the early limb bud, for instance, Sonic hedgehog (Shh) is expressed in the distal posterior mesenchyme, where it acts as a mediator of anterior to posterior (AP) patterning, whereas fibroblast growth factor 8 (Fgf8) is produced by the apical ectodermal ridge (AER) at the distal tip of the limb bud to direct outgrowth along the proximal to distal (PD) axis. Here we use cultured limb mesenchyme cells to assess the response of the target Hoxd genes to these two factors. We find that they act synergistically and that both factors are required to activate Hoxd13 in limb mesenchymal cells. However, the analysis of the enhancer landscapes flanking the HoxD cluster reveals that the bimodal regulatory switch observed in vivo is only partially achieved under these in vitro conditions, suggesting an additional requirement for other factors.
The developing vertebrate limb bud has long been a model system for the study of the emergence of pattern in developing tissues. Beginning as a hemisphere of undifferentiated mesenchymal progenitor cells surrounded by an epithelial encasement (ectoderm), the limb bud develops and grows from the tip to establish the familiar limb skeletal pattern. The anterior–posterior (AP) axis demarcates the line between the little finger and the thumb of the hand (or autopod), whereas the proximo–distal (PD) axis demarcates the line between the shoulder and the digits. Classic experiments established that the posterior portion of the limb bud mesenchyme, the zone of polarizing activity (ZPA), is essential for developing the asymmetry in digits observed across the AP axis (1). They also established that the distal lip of the epithelium, the apical ectodermal ridge (AER), is critical for limb bud outgrowth and proper establishment of the PD axis (2). Sonic hedgehog (Shh), the ligand secreted by ZPA cells, serves as a molecular activator of posterior limb pattern (3, 4). On the other hand, fibroblast growth factor (Fgf) ligands secreted by the AER are required for the outgrowth and patterning of the PD axis (5–9).
The ZPA and AER signaling centers are linked to each other through mutual cross-regulation; Fgf signaling in the AER is maintained through the positive feedback of Shh signaling from the ZPA and vice versa (10–13). In addition, these signaling centers need to be integrated at the level of their target genes. Although direct targets of Shh such at Ptch1 and Gli1 require only Shh (14, 15) and, likewise, direct targets of Fgf such as Sprouty require only Fgf activity (16), other targets including Bmp2 and Hoxd genes require simultaneous activation of both pathways (8, 11). The dual regulation of these genes by Shh and Fgf is independent of the endogenous feedback loops within the limb bud, as seen following ectopic application of these ligands in limb buds where endogenous signaling centers had been removed.
Hox gene members of both HoxA and HoxD clusters are necessary to properly pattern the limbs (17). Hoxd genes are expressed in two distinct phases in limb buds, an early proximal phase during the patterning of the zeugopod with a strong transcription of Hoxd9, Hoxd10, and Hoxd11 (phase I) and, subsequently, a second transcription wave during the establishment of the autopod with Hoxd13 as the most strongly expressed gene (phase II) (18–20). Genetic studies have shown that during phase I, Hox genes are required for the transcription of Shh (21), whereas Shh is necessary for their expression in phase II, highlighting the importance of this signaling pathway for Hox gene regulation in the distal limb bud (22). The switch in Hoxd gene regulation between phases I and II involves a change in utilization of the enhancer landscapes flanking the gene cluster. Phase I expression depends on contacts with the telomeric regulatory landscape, whereas phase II transcription depends upon enhancers located in the centromeric landscape (23, 24). The switch between these distinct regulations is at least partially dependent upon the activity of the Hoxd13 and Hoxa13 genes (25).
Previous experiments showing that Hoxd13 requires both Shh and Fgf for its expression were carried out in vivo, relying on the delivery of signals via protein-loaded beads or retroviral vectors. More recently, however, a culture system was developed allowing in vitro culture of limb bud mesenchyme while maintaining, at least for several days, correct patterning information and capacity for morphogenesis (26). This provided the opportunity to revisit the question of how Fgf and Shh signaling are integrated to direct proper Hox gene expression, using a more quantitative approach.
Results
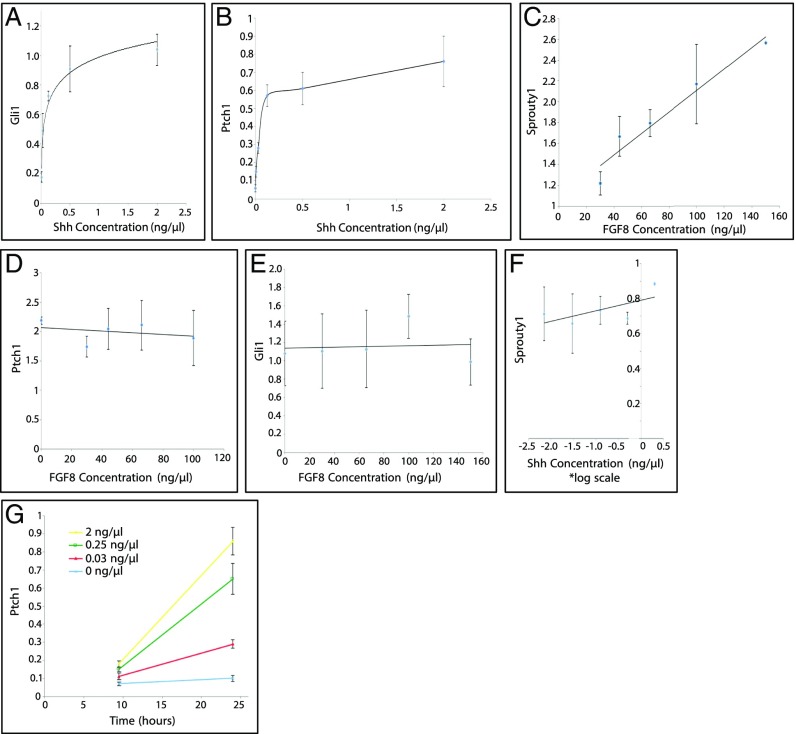
We first verified the responses of the known direct targets of Shh and Fgf signaling to single ligands in vitro. Limb mesenchymal progenitor cells were incubated in the presence of Wnt3a, which is normally secreted from the ectoderm, enabling cells to maintain proliferative and undifferentiated status (e.g., ref. 27). Limb progenitors were then treated with increasing doses of Shh (active N-terminal fragment) and assayed for their Gli1 or Ptch1 transcript levels by quantitative PCR (qPCR). In response to increasing Shh dose, both Ptch1 and Gli1 displayed increasing steady-state levels of mRNAs (Fig. 1 A and B). Similarly, when limb progenitors were treated with increasing doses of Fgf8, Sprouty1 gene expression levels increased with respect to dose (Fig. 1C). Therefore, the direct targets of both signals can be activated by their ligands in vitro. Moreover, the level of activation of Shh targets was not changed with addition of Fgf8 (Fig. 1 D and E), and Fgf8 target response did not change with the addition of Shh (Fig. 1F).
Fig. 1.
Responses of target genes to either Shh or Fgf ligands. (A and B) Shh ligand dose and Gli1/Ptch1 expression titration curve. Naïve limb progenitors were dosed with varying concentrations of Shh for 24 h. Fgf was present at a fixed concentration of 120 ng/µL. (C) Fgf ligand dose and Sprouty1 expression titration curve. Limb progenitors were treated with increasing doses of Fgf8 for 12 h. (D and E) Fgf ligand dose and Ptch1/Gli1 expression titration curve. Limb progenitors were treated with increasing doses of Fgf8 for 12 h. (F) Shh ligand dose and Sprouty1 expression titration curve. Limb progenitors were treated with increasing doses of Shh for 24 h. (G) Ptch1 temporal titration curves with varying levels of Shh ligand. Limb progenitors were treated for either 10 or 24 h at one of four doses of Shh ligand (0, 0.03, 0.25, and 2 ng/µL). The expression level for each gene was normalized to Actb and depicted as fold change relative to the baseline (no ligand) in A, B, and G. Error bars indicate the SD of three biological replicates of each concentration dose or temporal course.
Noteworthy, differences were observed in the shape of response curves to Shh and Fgf8. Fgf exhibited a linear dose–response, consistent with it being an activator of gene transcription. The more activator present, the higher the level of transcription within the range of conditions examined. In contrast, the Shh dose–response increased over a range of 0–0.25 or 0–0.5 ng/mL and then reached a plateau, consistent with the fact that the primary transcriptional mediator of Shh signal transduction in the limb is Gli3—that is, a transcriptional repressor (28). Acting via derepression may result in a plateau, as additional Shh signaling would not have any further effect whenever all Gli3R is inactivated.
Although the dose–response of Ptch1 activation to Shh reached a plateau at high concentrations, conceivably reflecting a full derepression, treated cells required 24–40 h of exposure for a maximal level of Ptch1 expression. This increase in Ptch1 expression over the first 24 h of Shh exposure was scored with all Shh concentrations examined (Fig. 1G).
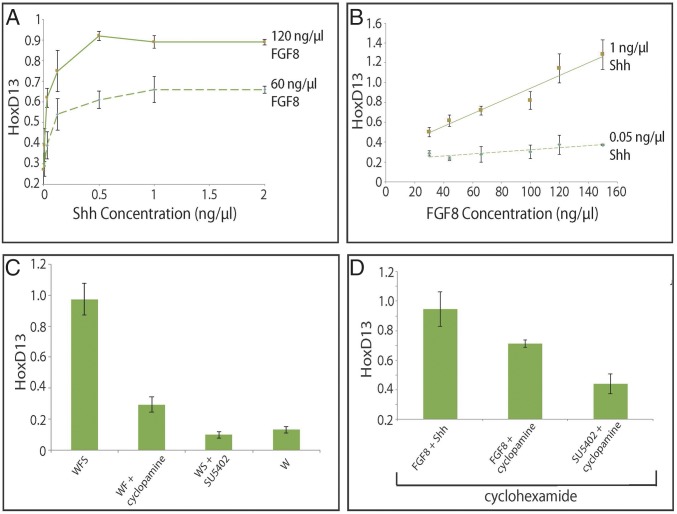
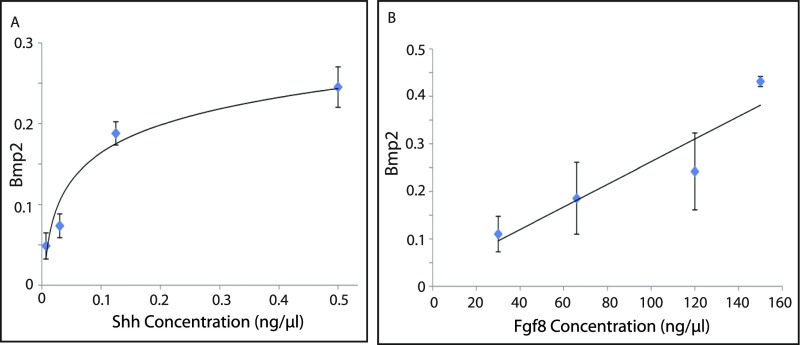
We next looked at Hoxd13, a target of both Shh and Fgf8 in the developing distal limb bud. As both inputs are required for induction of Hox13 genes in vivo, in vitro cultured limb progenitor cells were assessed for their dose–response to Shh in the context of a fixed, relatively high concentration of Fgf8. Conversely, the response to Fgf8 was assayed in the context of a fixed, relatively high level of Shh. Both signals could activate Hoxd13 in a manner similar to their direct targets: Shh induced Hoxd13 over similar concentration ranges as Ptch1 and Gli1, followed by plateau, whereas Hoxd13 had a linear dose–response to Fgf8 (Fig. 2 A and B). Bmp2, another target coregulated by Shh and Fgf in the limb bud, displayed a similar response profile when limb progenitors were treated with either Shh or Fgf ligand (Fig. S1 A and B). When the Shh response was analyzed with a lower level of Fgf8, an increase in Hoxd13 expression occurred over the same concentration range but plateauing at a lower level (Fig. 2A). Likewise, reducing the concentration of Shh decreased both the level and the slope of the Fgf response curve for Hoxd13 (Fig. 2B).
Fig. 2.
Expression of Hoxd13 in response to Shh/Fgf. (A) Shh ligand and Hoxd13 titration curve at two different doses of Fgf8. Limb progenitors were treated with increasing amounts of Shh at one of two concentrations of Fgf8. (B) Fgf8 ligand and Hoxd13 titration curve at two different doses of Shh. Limb progenitors were treated with increasing amounts of Fgf8 at one of two concentrations of Shh. (C) Hoxd13 gene expression in limb progenitors in response to Shh and/or Fgf ligand. Limb progenitors were treated with (i) Fgf8 and Shh, (ii) Fgf8 and Shh antagonist cyclopamine, (iii) Shh and Fgf antagonist SU5402, or (iv) no signal for 12 h. (D) Response of Hoxd13 gene expression in limb progenitors to Shh and/or Fgf ligand in the presence of cyclohexamide. Limb progenitors were treated with cyclohexamide in conjunction with one of the following three conditions for 12 h: (i) Fgf8 and Shh, (ii) Fgf8 and cyclopamine, or (iii) SU5402 and cyclopamine. The expression level of each gene was normalized to Actb. Error bars are as for Fig. 1.
Fig. S1.
Bmp2 response to Shh and Fgf ligand. (A) Shh ligand dose and Bmp2 gene expression titration curves. Limb progenitors were treated for 24 h with varying levels of Shh ligand. Fgf8 ligand was supplied at a fixed dose of 120 ng/µL. (B) Fgf ligand dose and Bmp2 expression titration curve. Limb progenitors were treated with increasing doses of Fgf8 for 12 h. Shh was supplied at a fixed dose of 1 ng/µL. The expression level of the Bmp2 gene was normalized to Actb and was depicted as fold change relative to the baseline (lowest) level/condition in A. Error bars indicate the SD of three biological replicates of each dose.
Next, we examined the requirement of both Fgf8 and Shh in concert to activate Hoxd13 expression. When limb progenitors were exposed to Shh in the absence of Fgf8, no activation of Hoxd13 was seen relative to control cultures. However, Fgf8 was able to activate Hoxd13 expression, although to a slight level, in the absence of Shh. When both signals were supplied, a synergistic effect was scored with levels of Hoxd13 far above the sum of the levels produced by Fgf8 (Fig. 2C). In the presence of cycloheximide, a pharmacological inhibitor of translation, Fgf8 was still able to activate slight levels of Hoxd13. The combination of Shh and Fgf8 produced a higher level of Hoxd13 than Fgf8 alone, but the synergistic increase was heavily dampened in the absence of protein synthesis (Fig. 2D). Therefore, although Hoxd13 expression can be induced in the absence of translation, a full Hoxd13 response likely requires a protein-dependent transcriptional feedback from this initial stimulus.
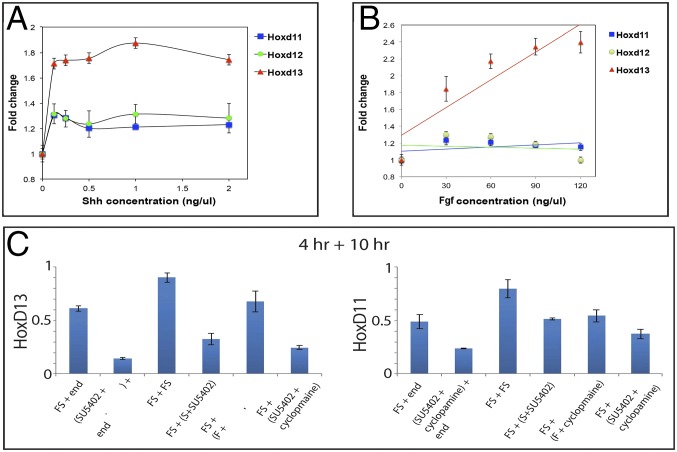
During the start of phase II, the coordinated expression of Hoxd11, Hoxd12, and Hoxd13 is scored in a posterior–distal cellular population, including Shh-positive cells and more distal cells located underneath the AER (21, 29). Also, grafts of Shh-secreting cells induce ectopic expression of all these genes in the developing limb (30), suggesting a common effect of both Fgf and Shh upon the transcription of these target genes. We therefore examined the response of Hoxd11 and Hoxd12 to Shh, to compare with the observed activation of Hoxd13. Both Hoxd11 and Hoxd12 were induced by Shh, however with a dose–response significantly below that of Hoxd13 (Fig. 3A). Moreover, variations in the dose of Fgf had only a minimal (if any) effect on the expression of Hoxd11 and Hoxd12 (Fig. 2B). These data supported Hoxd13 as the major Hoxd gene involved during phase II (31) and suggest that the quantitative differences observed among these three genes in distal limb bud cells may be in part determined by differences in their response to the Shh and Fgf signals, in addition to regulatory constraints due to gene topology (24, 32).
Fig. 3.
Differences in Hoxd13, Hoxd12, and Hoxd11 responses. (A) Shh ligand dose and Hoxd gene expression titration curves. Limb progenitors were treated for 24 h with varying levels of Shh ligand. Fgf8 ligand was supplied at a fixed dose of 120 ng/µL. (B) Fgf8 ligand dose and Hoxd gene expression titration curves. Limb progenitors were treated for 24 h with varying levels of Fgf8 ligand. Shh ligand was supplied at a fixed dose of 1 ng/μL. (C) Limb progenitors were treated in a two-step protocol. In the first step, progenitors were treated with Fgf8 and Shh for 4 h. In the second step, progenitors were treated for 10 h under one of four possible conditions: (i) Fgf8 and Shh, (ii) Fgf antagonist SU5402 and Shh, (iii) Fgf8 and Shh antagonist cyclopamine, or (iv) Fgf antagonist SU5402 and Shh antagonist cyclopamine. The expression levels of each gene were normalized to Actb and depicted as fold change relative to the baseline (no ligand) in A and B. Error bars are as for Fig. 1.
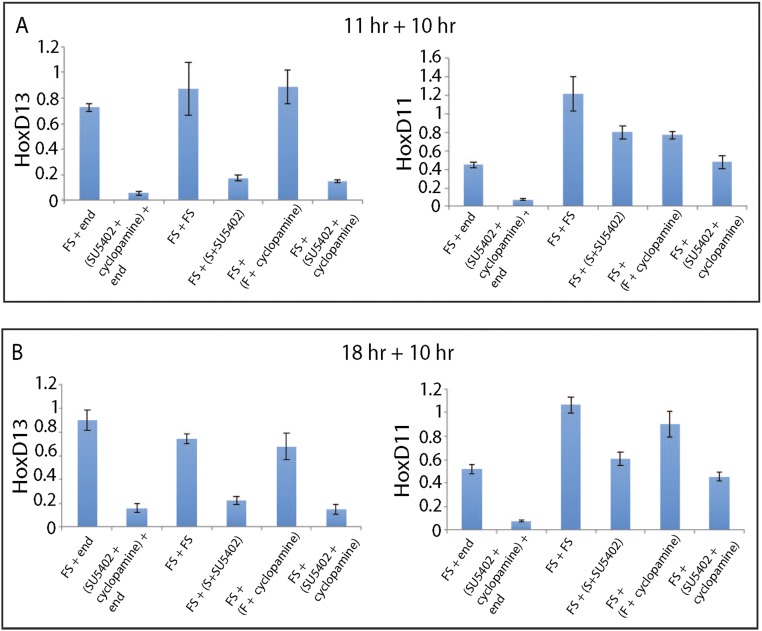
We also examined the maintenance of Hoxd13 expression, following its initial activation by Shh and Fgf signaling. Limb progenitors were treated with both Shh and Fgf for 11 h to allow Hoxd13 activation. After this treatment period, cells were exposed to Shh or to Fgf8 independently or to both signals for 10 h (Fig. 3C). Strikingly, Hoxd13 expression was lost after pretreatment in cells cultured either with no signal or with Shh alone. In contrast, Hoxd13 expression was maintained at similar levels with Fgf alone or when both Fgf and Shh were added, indicating that Fgf is sufficient to maintain Hoxd13 levels by itself, once the gene is activated by the combination of Fgf and Shh. A similar set of responses was seen when cells were pretreated for either 4 or 18 h (Fig. S2 A and B).
Fig. S2.
Temporal dependency of Hoxd13 and Hoxd11 expression on Fgf and Shh ligands. (A and B) Limb progenitors were treated in two distinct steps. In the first step, progenitors were treated with Fgf8 and Shh for either 11 or 18 h. In the second step, progenitors were treated for 10 h under one of four possible conditions: (i) Fgf8 and Shh, (ii) Fgf antagonist SU5402 and Shh, (iii) Fgf8 and Shh antagonist cyclopamine, and (iv) Fgf antagonist SU5402 and Shh antagonist cyclopamine. Each distinct combination was carried out in biological triplicate. The expression level of the Hoxd13 and Hoxd11 gene was normalized to Actb.
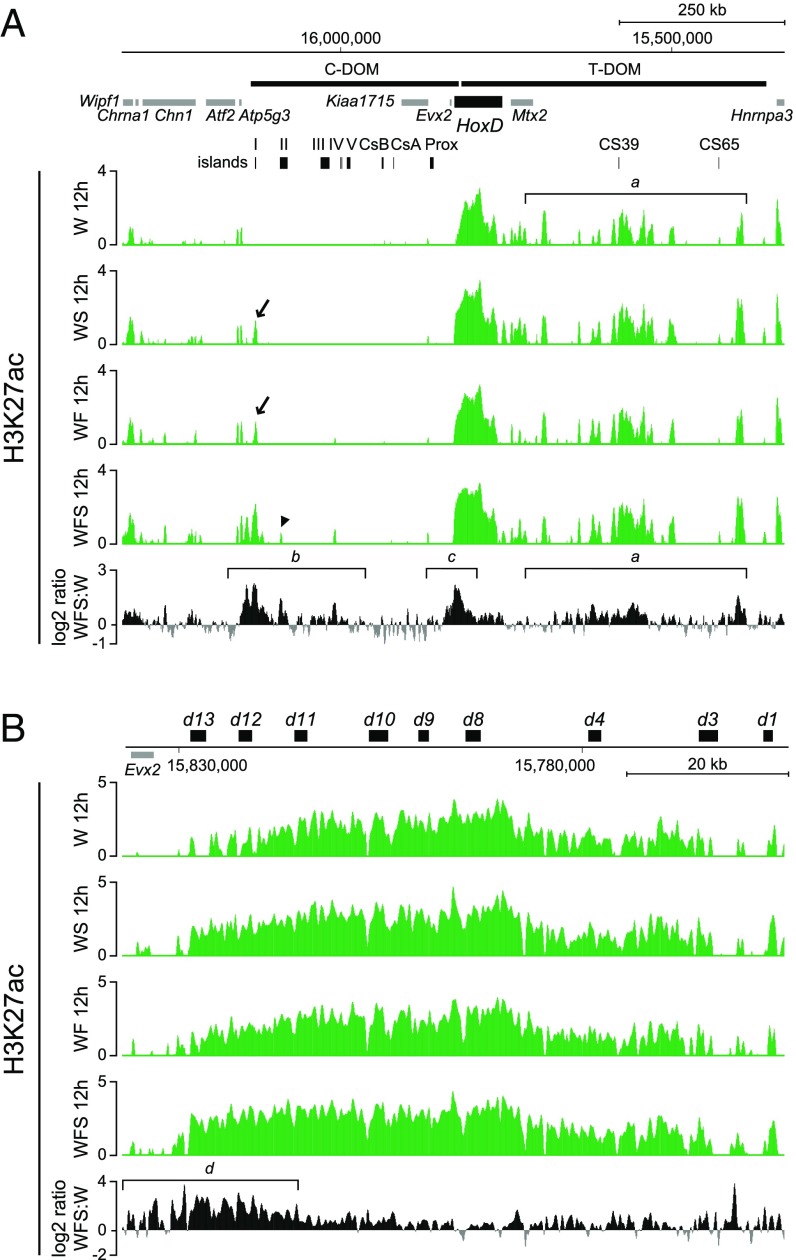
The successive implementation of phase I and phase II Hoxd gene regulations in the limb bud depends upon two distinct and opposite regulatory landscapes, matching topologically associating domains (TADs) (24, 25). In the incipient limb bud, Hoxd9, Hoxd10, and Hoxd11 transcription is controlled by T-DOM, a regulatory landscape containing several enhancers with a proximal specificity. Subsequently, Hoxd13, Hoxd12, Hoxd11, and Hoxd10 are activated by distal enhancers located within the opposite C-DOM regulatory landscape (Fig. 4). We asked whether treatment of cultured progenitors with Shh and Fgf could recapitulate this characteristic switch in regulations observed in vivo. To this aim, we monitored the activities of both regulatory landscapes by mapping the acetylation of lysine 27 on histone H3 tail (H3K27ac), a chromatin mark associated with active transcription and enhancer sequences.
Fig. 4.
Fgf8 and Shh act together to initiate C-DOM regulation in distal–anterior limb mesenchymal cells in culture. (A) Comparison of H3K27ac profiles in limb mesenchymal cells cultured with basic condition (Wnt3a; W), treated with either Shh (WS) or Fgf8 (WF), or both Fgf8 and Shh (WFS). All treatments increase the enrichment of H3K27ac not only within the T-DOM but also in and around the C-DOM–located island I. (B) Comparison of H3K27ac at the HoxD cluster. All treatments induced the strong activation of Hoxd11 to Hoxd13. Enrichment (y axis) is shown as the log2 ratio of the normalized number of reads between ChIP and input samples, except for each fifth track. The log2 ratio of the normalized number of reads between H3K27ac of Fgf8 and Shh-treated cells and the cells under basic condition (only Wnt3a) is shown by positive (black) and negative (gray) values, respectively.
We used ChIP-seq (chromatin immunoprecipitation combined with high-throughput sequencing) analysis of cultured progenitor cells after 12 h of treatment with either Shh alone, Fgf alone, or both molecules together. The control profile revealed a strong acetylation over the T-DOM landscape, in agreement with an “early” and “proximal” regulation of Hoxd genes in these cells in the absence of any treatment (Fig. 4A, track 1, bracket a). Treatment involving either Shh or Fgf did not show any major difference, except for the appearance of a significant peak within the C-DOM landscape, exactly matching island I—that is, one of the sequences previously defined in mice in vivo as a digit enhancer regulating Hoxd13 (23) (Fig. 4A, arrow in tracks 2 and 3). When both Shh and Fgf were used concomitantly, the amount of H3K27ac at this sequence and immediately around substantially increased, whereas at the same time another peak appeared at the position of island II, another “digit-specific” region mapping within C-DOM (Fig. 4A, arrowhead in track 4) (24).
The log2/ratio (33, 34) between this latter profile and that of untreated cells (Fig. 4A, Bottom) confirmed that a gain of acetylation occurred in C-DOM in cells treated with both Shh and Fgf (Fig. 4A, bracket b). This gain, however, was relatively minor compared with the switch normally observed in distal limb cells under in vivo conditions (e.g., ref. 24). This absence of clear regulatory switch was confirmed by the increase in acetylation observed over T-DOM, as if the proximal enhancers were also reinforced by these treatments (Fig. 4A, bracket a). Consistent with both increases in acetylation in T-DOM and C-DOM, acetylation was also found increased over the “posterior” part of the HoxD cluster, supporting the increase in mRNA shown previously (Fig. 4A, bracket c). This increase in acetylation mostly concerned Hoxd13 and the surrounding sequences (Fig. 4B, bracket d), again matching the preferential activation of this gene under these in vitro conditions as reported above.
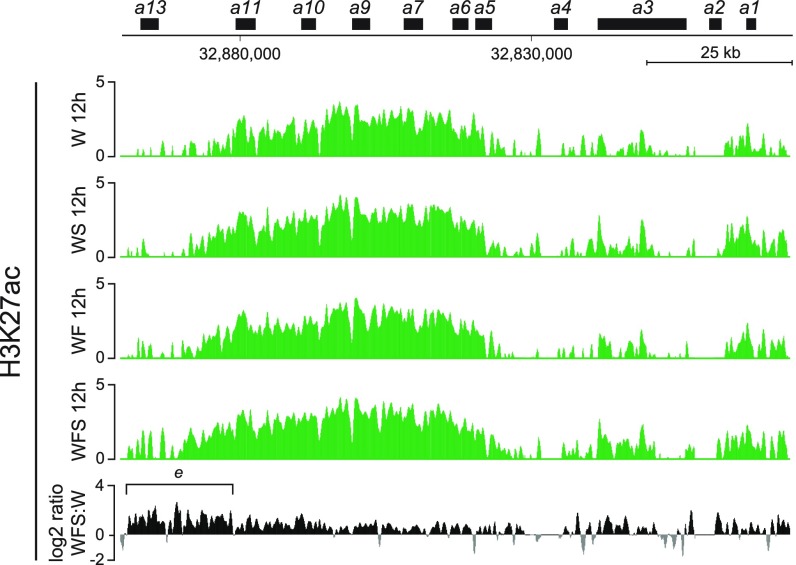
We concluded that treatment of limb progenitor cells with Shh and Fgf in vitro both reinforced the proximal regulation (the early phase I) and started to elicit the regulatory switch by showing signs of activation of the distal (phase II) landscape. The progressive and global distalization of these cells upon treatment was further assayed by looking at the H3K27ac distribution over the HoxA cluster, which revealed a robust increase over the Hoxa13 gene (Fig. S3, bracket e), another Hox gene often used as a marker of distal limb bud cells (e.g., refs. 35, 36).
Fig. S3.
H3K27ac ChIP-seq on the HoxA cluster. Shown are the H3K27ac ChIP-seq profiles covering the HoxA gene cluster in cultured limb progenitor cells treated either with Shh (WS), Fgf8 (WF), or Fgf8 and Shh (WFS). The latter condition induced a robust activation from the Hoxa11 to the Hoxa13 genes. Enrichment (y axis) is shown as the log2 ratio of the normalized number of reads between ChIP and input samples, except for each fifth track. The log2 ratio of the normalized number of reads between H3K27ac of Fgf8 and Shh-treated cells and control cells (only Wnt3a) is shown by positive (black) and negative (gray) values, respectively.
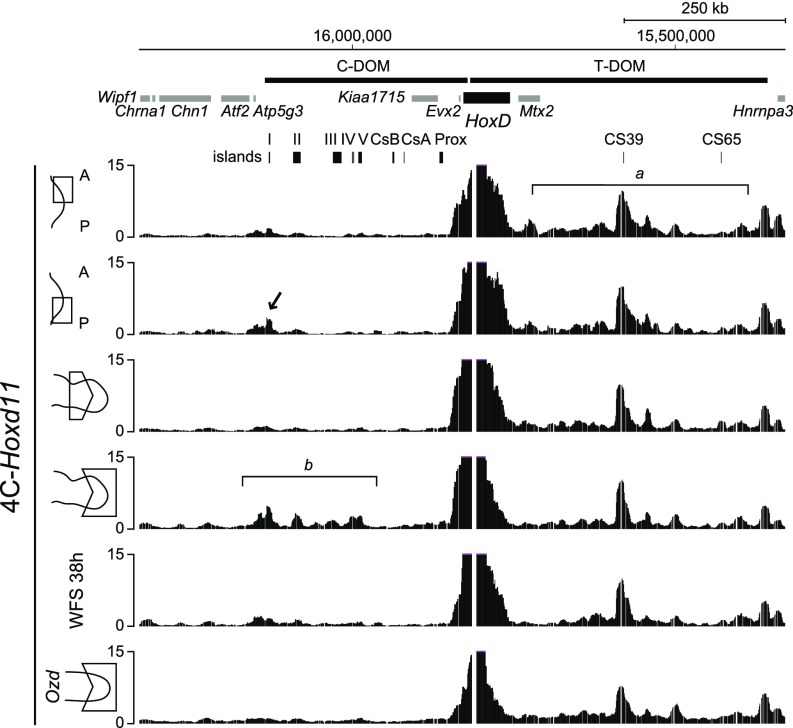
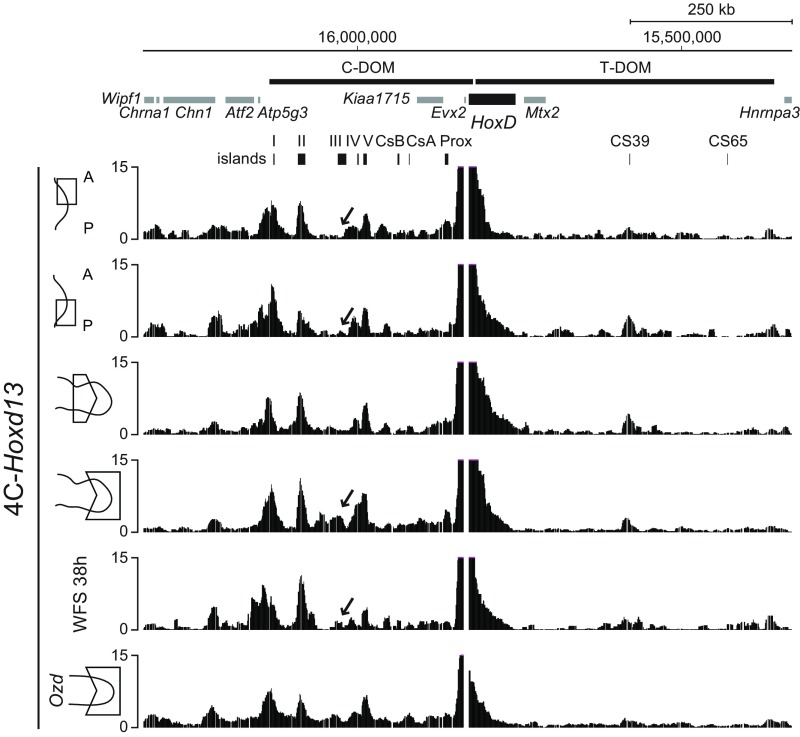
We verified this conclusion by looking at the physical interactions established either by Hoxd11 or by Hoxd13 before and after treatment with Shh and Fgf. We used 4C-seq, a derivative of chromosome conformation capture (e.g., refs. 37, 38), to determine which of the two regulatory landscapes were contacted by these two genes under various conditions. In both anterior and posterior early limb bud cells, Hoxd11 contacted strongly the T-DOM, as previously shown in mice, due to a mix of both productive and constitutive contacts (Fig. 5, bracket a) (24). In posterior limb bud cells, however, slightly increased contacts were observed with island I of the C-DOM (Fig. 5, arrow), suggesting the onset of the switch in regulations in these cells, which start to be exposed to both Shh and Fgf signaling. Signs of the switch were best seen in distal cells of older limb buds at stage 27, where Hoxd11 started to interact with most of the regulatory islands located within C-DOM (Fig. 5, bracket b). The interaction profile obtained with cells after 38 h of treatment resembled that of early limb bud cells, lacking most contacts with C-DOM and thus confirming that the switch had not fully occurred in these cells.
Fig. 5.
4C-seq interaction profiles using Hoxd11 as a viewpoint. Interaction profiles between Hoxd11 and the C-DOM regions using either the distal–anterior or distal–posterior part of forelimb bud, the proximal or distal part of forelimb bud or progenitor limb cells cultured with Fgf8 and Shh, or the distal part of forelimb bud from Ozd mutant at HH stages 25–26 (51). The interaction profile between Hoxd11 and C-DOM regions using cells treated with Shh and Fgf8 (WFS; track 5) is similar to that of the limb anterior–distal part (track 1). In distal cells from mutant Ozd chick limb bud lacking Shh signaling, the interaction profile between Hoxd11 and C-DOM lacks all specific interaction as seen in the wild-type control (track 6, compared with track 4, bracket b), indicating that the regulatory switch had not occurred.
This conclusion was supported by the interaction profile established by Hoxd13 under the same conditions. Unlike Hoxd11, Hoxd13 always displays strong constitutive contacts with C-DOM, due to its position at the extremity of the gene cluster and its belonging to another TAD (24). Consequently, the switch in regulation cannot be observed in 4C profiles based on this viewpoint. However, the transcriptional activation of Hoxd13 is systematically associated with the appearance of specific contacts within C-DOM, which can thus be used as hallmarks for C-DOM being transcriptionally active, such as island III (24). Accordingly, contacts with island III could not be detected in early limb bud cells whenever Hoxd13 was used as a viewpoint (Fig. S4, tracks 1 and 2, arrow), whereas it peaked along with the surrounding region in older distal cells (Fig. S4, track 4, arrow). There again, contacts with island III were very weak (if any) in limb progenitor cells cultured for 38 h with both compounds, indicating that C-DOM was not fully functional in these cells, despite this treatment (Fig. S4, track 5, arrow).
Fig. S4.
4C-seq interaction profiles using Hoxd13 as a viewpoint. Interaction profiles between Hoxd13 and the C-DOM regions, using either the distal–anterior or distal–posterior part of forelimb bud, the proximal or distal part of forelimb bud or progenitor limb cells cultured with Fgf8 and Shh, or the distal part of forelimb bud from Ozd mutant. Treatment by the two factors increased the contacts between Hoxd13 and island II, similar to what is seen in distal limb bud cells. However, some hallmarks of C-DOM activity such as island III (track 5, arrow) do not appear in the in vitro system, indicating that the regulatory switch had not occurred.
The Hoxd11 interaction profile observed in progenitor cells after 38 h of treatment resembled very much the pattern obtained when processing distal limb bud cells dissected out from chicken carrying the olygozeugodactyly (Ozd) mutation (Fig. 5, compare tracks 5 and 6) (22). Ozd mutant chickens lack several postaxial bones and do not implement the late phase of Hoxd gene transcription due to the lack of Shh signaling (22) as a result of a deletion within an essential enhancer sequence (39). The similarity between these two interaction profiles further indicated that Shh-dependent activation of the Hoxd13 gene in treated progenitor cells was weak at best.
Discussion
Hox genes are important regulators of embryonic patterning. Although the complex and dynamic patterns of collinear Hoxd gene expression during limb development have recently found a general explanatory framework (40), the relationships between Hox gene activation and the two ZPA and AER signaling centers remain unclear. These signaling centers organize the early limb bud by releasing specific factors (e.g., ref. 41), and the link between such factors and the control of Hox gene transcription has not yet been clearly established. Here we interrogate the role of early limb signaling pathways in initiating and maintaining Hox gene expression.
Signaling and Localization of Hox Expression in the Limb.
Our results confirm and expand the observation that Shh and Fgf signaling in the limb is integrated at the level of coregulation of key target genes. In addition to the well-established positive feedback loop wherein Shh and Fgf activities are each required for maintaining expression of the other, the production of Shh and Fgf8 by the ZPA and AER, leading to complex quantitative distributions of ligands, may initiate and secure the expression of Hoxd genes during phase II within the correct posterior–distal domain. It has also been shown that the expression of posterior Hoxd genes during phase I is required for the initiation of Shh (21) and Fgf8 (42), representing additional positive feedback loops operating within the early limb bud to control the transition from a proximal to a distal context.
We find that Hoxd11, Hoxd12, and Hoxd13 are induced synergistically by Shh and Fgf. Hoxd13, however, is activated to a significantly greater extent, supporting Hoxd13 as the major target of these signaling pathways (31) and suggesting that a differential response to Shh and Fgf could both contribute to the distinct strengths in expression of the various Hoxd genes in distal limb cells during phase II (19) and to the maintenance of the nested Hoxd expression domains after their initial organization during phase I (18, 20).
Hoxd11 and Hoxd13 seem to differ in their requirement for Shh and Fgf to maintain their expression. Once activated, Hoxd11 does not require either ligand for its active transcription, consistent with the necessity to maintain Hoxd11 expression in zeugopod cells despite their progressively increased distance to the AER by continued growth at the distal tip. Conversely, Hoxd13 needs continued exposure to Fgf8, thus securing its expression in the most distal autopod cells. These observations contrast with those reported in ref. 8, where Hoxd11, Hoxd12, and Hoxd13 all require sustained Shh for maintaining their expression. As both sets of experiments were carried out at similar stages of limb bud development, the reason for this difference is unclear.
Shh as a Morphogen in Regulating Hox Gene Expression?
The dose–response to Shh is graded over a range from 0 to 0.25 ng/mL (Ptch1) or 0–0.5 ng/mL (Gli1) but remains flat at a maximal level from 0.25 to 2.5 ng/mL. From a biological perspective, this could either be viewed as a sharp switch, where any amount of Shh above 0.25 ng/mL triggers a full response, or alternatively as a graded response that could yield a series of threshold-based changes in gene regulation between 0 and 0.25 ng/mL. Which is the case in the limb bud depends entirely on the effective concentration of Shh in vivo, which is difficult to ascertain. Also, this concentration cannot be directly related to the in vitro concentration due to differences in specific activity of the recombinant protein. Nonetheless, it is worth noting that the concentration range of Shh inducing the morphogen-like response in neural tube explants (43, 44) is similar to the lower range giving a concentration-dependent Hox gene response in our experiments.
The dose–response curves suggest that Shh may act as both a morphogen and a switch during limb development, at two distinct stages. Although a brief expression of Shh may specify digit patterns as a potential morphogen, it is also required at a sustained level for growth and expansion through most of limb bud development (45, 46). Initially, the lower levels of Shh activity might be in the linear part of the dose–response curve, thus allowing it to act as a morphogen. Subsequently, its sustained expression might expose cells to a level high enough to act as a switch, allowing full Hox expression in conjunction with Fgf activity.
Lack of Temporal Adaptation to Shh and Fgf Signaling in the Limb.
Ptch1 expression was increased in response to Shh over the first 24 h in a way proportional to the concentration of ligand at both early and late time points. In contrast, in the neural tube, there is a high initial pathway response that is concentration independent, followed by a loss of pathway response over time such that cells exposed to a lower dose of Shh lose responsiveness faster than those exposed to higher doses (44). This phenomenon, dubbed temporal adaptation, may reflect an accumulation of Ptch1 protein over time in the responding cells. In the limb, the predominant mode of gene regulation by Shh is derepression through inactivation of Gli3, whereas in the neural tube multiple Gli proteins are present and may regulate Ptch1 gene activity through activation. At low dose, by 18 h of exposure, the neural tube cells are no longer responsive to Shh (44). In contrast, limb buds exposed in vivo to low levels of Shh continue to respond for up to 36 h (47). Also, in the chick limb but not in neural tube, 24-h exposure to a low level of Shh gives an equivalent phenotypic change as a shorter exposure (16 h) at a higher concentration of Shh (48), similar to what we see in vitro where 15 h of exposure to 0.25 ng/mL gives a similar level of response of Ptch1 as 24 h of exposure to 0.03 ng/mL. The difference in temporal response to Shh in the limb and neural tube is surprising and may reflect the versatility of the system, displaying fundamentally different properties depending on which signal transduction machinery is present.
Within the concentration range tested, Fgf signaling does not plateau with increasing amounts of Fgf protein. This may be seen as surprising, as the Fgf target genes of the Sprouty (Spry) family are themselves Fgf signaling antagonists. Similarly, in vivo, up-regulation of Spry genes in the distal mesenchyme must have the effect of dampening the response to Fgf signaling in this domain. Nonetheless, it is also clear that Fgf signaling remains active in the distal limb mesenchyme throughout limb development (AER removal or genetic ablation of Fgf activity within the AER has dramatic effects well after Spry genes are induced); thus, the level of Fgf signaling must exceed the inhibitory effect of the Spry proteins. Based on our dose–response curves, we would conclude that additional Fgf signaling is able to induce still strong target gene responses at higher concentrations, even in the context of the inhibitory activity of the Spry genes.
Target Gene Activation in the Limb.
The primary transcription factor downstream of Shh in the limb is the Gli3 repressor. A number of potential target genes in the limb have putative Gli3 binding sites (49), many of which were verified as genuine Shh targets (8), including Hoxd11, Hoxd12, and Hoxd13, suggesting that Shh directly regulates Hox genes. In contrast, transcription factors downstream of Fgf-regulating Hox genes are unknown. However, we do see synergistic effects with Shh and Fgf in the presence of cyclohexamide, suggesting an at least partial direct effect of these two factors. On the other hand, the synergistic effect was greatly reduced in the presence of cyclohexamide levels, suggesting that one or both of these factors also feeds into Hox regulation through a second, indirect pathway.
Effects of Shh and Fgf on Hox Gene Transcriptional Activation.
The distribution of H3K27ac marks in cells treated with both Shh and Fgf suggests that it reinforced phase I transcription, with a strengthening of T-DOM enhancer activity. In the meantime, however, signs of enhancer activity within C-DOM were clearly detected, indicating the start of the regulatory switch toward C-DOM and thus the beginning of phase II. Most of the previously described regulatory islands (23) nevertheless remained silent, indicating that under these in vitro conditions, the switch does not fully occur. This was verified by using chromosome conformation capture, which revealed mostly constitutive contacts between either Hoxd11 or Hoxd13 with C-DOM, rather than the expected contacts normally labeling C-DOM regulatory activity. This interaction profile resembled that of Ozd mutant distal limb cells, where Shh is inactivated, suggesting again that, as in such mutant limb buds, cultured progenitor cells did not respond to the Shh/Fgf treatment by a full switch in their regulation.
It remains, however, that Hoxd13 was activated by Shh and Fgf treatment, a property normally restricted to the C-DOM enhancers (24), thus supporting a weak but functionally significant switch in regulations. The occurrence of the switch is supported by the increase in H3K27ac over Hoxa13, a marker of distal limb cells, and could remain undetected by 4C analysis, should it happen in a relatively low number of cells. Alternatively, it is possible that, under these conditions, Hoxd13 expression was controlled by enhancers localized within T-DOM. Although this does not seem to normally occur in vivo, at least with a high efficiency (24), T-DOM enhancers were shown recently to have this capacity in the absence of both Hoxd13 and Hoxa13 products (25). Therefore, in our in vitro system, we cannot rule out the possibility that the amounts of HOX13 products may not be high enough to implement the regulatory switch. Consequently, the increase in Hoxd13 mRNAs may reflect the reinforcement of H3K27 acetylation over the T-DOM landscape rather than the appearance of such chromatin marks over C-DOM. However, because acetylation and 4C interactions over C-DOM appear exactly at the positions of previously defined distal enhancers, we do not favor the latter possibility.
Materials and Methods
The ChIP-seq and 4C-seq experiments were performed as described in ref. 25. De-multiplexing and mapping were carried out using the HTSstation (htsstation.epfl.ch) (50). Datasets are available from the NCBI Gene Expression Omnibus repository under accession no. GSE92557. An extended description of the materials and methods is provided in SI Materials and Methods.
SI Materials and Methods
Chicken Embryos.
Fertilized white leghorn chicken eggs (Charles River Laboratories) and Ozd mutant eggs, kindly provided by John F. Fallon, University of Wisconsin, Madison WI, were incubated at 38 °C. Chicken embryos were staged according to Hamburger and Hamilton (50).
Culture of Limb Progenitor Cells.
Tissues from the distal anterior portion of forelimb buds were dissected from stage 20–21 embryos (4 d postfertilization). In the current context, this tissue is referred to as naïve limb bud mesenchyme, to reflect the fact that it has not been exposed to Shh signaling. After dissection, forelimb buds were incubated in 0.25% trypsin on ice for 5–10 min to loosen attached ectodermal tissue. After trypsin incubation, tissue was transferred to 10% (vol/vol) chick serum (Life Technologies) in PBS. Loosened ectodermal tissues were removed, and the remaining mesenchyme was mechanically dissociated with manual pipetting. Cells were plated in 96-well plates at high density (1 × 107 cells per mL). Baseline media in culture contained 250 ng/mL Wnt3a protein (R&D Systems) in DMEM/F12 media (Life Technologies). Additional ligands added to media include Shh (1 ng/µL unless otherwise noted, N-terminal fragment, R&D Systems), Fgf8 (120 ng/µL unless otherwise noted, Human Fgf8 isoform B, R&D Systems), SU5402 (Tocris), cyclopamine (Tocris), and cycloheximide solution (R&D Systems).
Quantitative Real-Time PCR.
RNA was extracted using the RNeasy Mini Kit (Qiagen) and then reverse-transcribed with Oligo(dT) primers (Invitrogen) using SuperScript III (Invitrogen). cDNA was analyzed by qPCR on the Stratagene MX3000P cycler. Each sample was run in triplicate and quantified by comparison with a standard curve (10-fold serial dilutions of a sample or synthesized gene fragment). Primers for Hoxd11, Hoxd12, Hoxd13, Ptch1, Gli1, and Sprouty1 (below) were designed to span intronic regions and also tested for amplification efficiency as well as the capacity to produce a single amplification product.
Primers for RT-qPCR.
Hoxd11
Forward: CAGCAAGGCACCTTCCGGCC
Reverse: CCTCGATCGCTGAGGAACTGCTG
Hoxd12
Forward: AGTTCAGCCCGCGGCAGTCCA
Reverse: CGATCTCCCCTGGGTGGGG
Hoxd13
Forward: AGACATGGTCTCAACGTTTGGC
Reverse: GCACATCTCGGGCTGGTTTAG
Ptch1
Forward: TTTTTCTTTTCCTGGGCTTACTT
Reverse: CATCTCTCACCCGGGTAGTTC
Gli1
Forward: ACTGAGCACCCGGGTGGTGC
Reverse: GGAGTTGGGCGAGGTGCGGA
Sprouty1
Forward: AGCCAGCACGGCAGTGGTGG
Reverse: GCTCCGCGGATCGCCTTGA
Bmp2
Forward: GCCTCATGCCGGAGGTGGGA
Reverse: TGTTCGGGCTGAAGCGGCGG
Actb
Forward: TCTGACTGACCGCGTTACTC
Reverse: CCATCACACCCTGATGTCTG
Chromosome Conformation Capture.
Chromosome conformation capture (4C-seq) was performed as previously described (25). Tissues from the distal–anterior portion of 144 pairs of forelimb buds were dissected from stage 20–21 embryos. After dissection, limb buds were incubated in 0.25% trypsin on ice for 5–10 min to loosen attached ectodermal tissue. After trypsin incubation, tissue was transferred to 10% (vol/vol) chick serum (Life Technologies) in PBS. Loosened ectodermal tissues were removed, and the remaining mesenchyme was mechanically dissociated with manual pipetting. The cell suspension was evenly divided into four tubes and centrifuged at 2,000 rpm in a C5 swinging bucket centrafuge (Carolina Biological Supply Co.) for 5 min. After removal of supernatants, the cells were resuspended with 2 mL of DMEM/F12 containing WFS (250 ng/mL of Wnt3a, R&D Systems; 120 ng/mL of Fgf8b, R&D Systems; and 1 ng/µL of N-terminal fragment of Shh, R&D Systems). We plated 400 μL of cell suspension in each well of 24-well plates (5 × 106 cells per mL) and cultured them for 38 h at 37 °C. Dissected 144 pairs of distal–anterior or distal–posterior forelimb buds at stage 20, 16 pairs of distal or 11 pairs of proximal forelimb buds at stage 27, 48 pairs of distal forelimb at stage 25–26 from Ozd mutant, and the cultured distal–anterior limb mesenchymal cells for 38 h were individually fixed with 2% formaldehyde, lysed, and stored at –80 °C.
Primers Used for Amplification of the Hoxd11 and Hoxd13 Regions.
Hoxd11_inverse_forward: 5′-AATGATACGGCGACCACCGAACACTCTTTCCCTACACGACGCTCTTCCGATCTGAGTCGCCGGCTCTTTCTATG-3′
Hoxd11_inverse_reverse:
5′-CAAGCAGAAGACGGCATACGAAGGACTTTGGCGTGCAGAC-3′
Hoxd13_inverse_forward:
5′-AATGATACGGCGACCACCGAACACTCTTTCCCTACACGACGCTCTTCCGATCTAGAAATTCACATTTCAAAGGGTTGAC-3′
Hoxd13_inverse_reverse:
5′-CAAGCAGAAGACGGCATACGAGACGAAGTTTACTCAGTTGCTCCT-3′.
PCR products were multiplexed and sequenced, followed by postprocessing (demultiplexing, mapping, and 4C analysis) using the HTS station (htsstation.epfl.ch) (49). Fragment scores were normalized to the mean score of fragments falling into a region defined as the bait coordinated ± 1 Mb, and the data were smoothed by using a running mean with a window size of 11 fragments. During the procedure, fragments at chr7, 15,807,716–15,815,518 (galGal4), at Hoxd11 viewpoint and those at chr7, 15,828,450–15,833,591 (galGal4), at Hoxd13 viewpoint were excluded from the analysis.
ChIP-Seq.
ChIP experiments were performed as described in ref. 25. Tissues from the distal–anterior portion of 170 forelimb buds were dissected from stage 21. After dissection, limb buds were incubated in 0.25% trypsin/PBS for 10 min at room temperature to loosen attached ectodermal tissue. Then, tissues were transferred to 10% FBS/DMEM to quench trypsin reaction. Loosened ectodermal tissues were removed, and the remaining mesenchyme was mechanically dissociated with pipetting. The cell suspension was evenly divided into four tubes and centrifuged at 2,000 rpm for 5 min. After removal of supernatants, the cells were resuspended with 500 μL of DMEM/F12 containing W (250 ng/mL of Wnt3a, R&D Systems), WF (Wnt3a and 120 ng/mL Fgf8b, R&D Systems), WS (Wnt3a and 1 ng/µL of N-terminal fragment of Shh, R&D Systems), or WFS (Wnt3a, Fgf8b, and Shh). We plated 100 μL of cell suspension in each well of 96-well plates (5 × 106 cells per mL) and cultured it for 12 h at 37 °C. After rinsing with PBS, the cultured cells were trypsinized for 5 min at 37 °C and were harvested. Dissociated cells were cross-linked with 1% formaldehyde/PBS for 15 min at room temperature, and the cross-link reaction was quenched by adding Glycine, followed by washing with cold PBS containing proteinase inhibitor. Chromatin was sheared and used for each immunoprecipitation with antibody against H3K27ac (ab4729, Abcam). For sequencing, at least 5 ng of purified DNA was used to make libraries according to Illumina’s protocol. The material was sequenced with 100 bp single-end reads on Illumina HiSeq. Mapping of demultiplexed ChIP-seq reads onto the International Chicken Genome Reference Consortium Gallus_gallus-4.0 (galGal4) and removal of unmapped regions and PCR duplicates were done with Galaxy webserver (https://usegalaxy.org/), as previously described (25). Input data from W, WF, and WFS were concatenated and used for further analysis in this experiment. The merged input data and all ChIP data from W, WF, WS, and WFS were normalized and compared with compute the log2 of the number of reads ratio (33, 34). The ChIP data from W and WFS were directly compared with the log2 of the number of reads.
Acknowledgments
We thank Drs. John F. Fallon for providing the chicken Ozd mutant embryos and Jean-Marc Matter for sharing materials. This work was supported by NIH Grant HD032443 (to C.J.T.) and Swiss National Science Foundation Grant 310030B_138662 (to D.D.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE92557).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1620767114/-/DCSupplemental.
References
- 1.Saunders JW, Gasseling MT. Ectodermal-Mesodermal Interactions in the Origin on Limb Asymmetry. Williams & Wilking; Baltimore: 1968. pp. 78–97. [Google Scholar]
- 2.Saunders JW., Jr The proximo-distal sequence of origin of the parts of the chick wing and the role of the ectoderm. J Exp Zool. 1948;108(3):363–403. doi: 10.1002/jez.1401080304. [DOI] [PubMed] [Google Scholar]
- 3.Riddle RD, Johnson RL, Laufer E, Tabin C. Sonic hedgehog mediates the polarizing activity of the ZPA. Cell. 1993;75(7):1401–1416. doi: 10.1016/0092-8674(93)90626-2. [DOI] [PubMed] [Google Scholar]
- 4.Litingtung Y, Dahn RD, Li Y, Fallon JF, Chiang C. Shh and Gli3 are dispensable for limb skeleton formation but regulate digit number and identity. Nature. 2002;418(6901):979–983. doi: 10.1038/nature01033. [DOI] [PubMed] [Google Scholar]
- 5.Niswander L, Martin GR. Fgf-4 expression during gastrulation, myogenesis, limb and tooth development in the mouse. Development. 1992;114(3):755–768. doi: 10.1242/dev.114.3.755. [DOI] [PubMed] [Google Scholar]
- 6.Niswander L, Tickle C, Vogel A, Booth I, Martin GR. FGF-4 replaces the apical ectodermal ridge and directs outgrowth and patterning of the limb. Cell. 1993;75(3):579–587. doi: 10.1016/0092-8674(93)90391-3. [DOI] [PubMed] [Google Scholar]
- 7.Sun X, Mariani FV, Martin GR. Functions of FGF signalling from the apical ectodermal ridge in limb development. Nature. 2002;418(6897):501–508. doi: 10.1038/nature00902. [DOI] [PubMed] [Google Scholar]
- 8.Lewandowski JP, et al. Spatiotemporal regulation of GLI target genes in the mammalian limb bud. Dev Biol. 2015;406(1):92–103. doi: 10.1016/j.ydbio.2015.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moon AM, Capecchi MR. Fgf8 is required for outgrowth and patterning of the limbs. Nat Genet. 2000;26(4):455–459. doi: 10.1038/82601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Niswander L, Jeffrey S, Martin GR, Tickle C. A positive feedback loop coordinates growth and patterning in the vertebrate limb. Nature. 1994;371(6498):609–612. doi: 10.1038/371609a0. [DOI] [PubMed] [Google Scholar]
- 11.Laufer E, Nelson CE, Johnson RL, Morgan BA, Tabin C. Sonic hedgehog and Fgf-4 act through a signaling cascade and feedback loop to integrate growth and patterning of the developing limb bud. Cell. 1994;79(6):993–1003. doi: 10.1016/0092-8674(94)90030-2. [DOI] [PubMed] [Google Scholar]
- 12.Zúñiga A, Haramis AP, McMahon AP, Zeller R. Signal relay by BMP antagonism controls the SHH/FGF4 feedback loop in vertebrate limb buds. Nature. 1999;401(6753):598–602. doi: 10.1038/44157. [DOI] [PubMed] [Google Scholar]
- 13.Capdevila J, Tsukui T, Rodríquez Esteban C, Zappavigna V, Izpisúa Belmonte JC. Control of vertebrate limb outgrowth by the proximal factor Meis2 and distal antagonism of BMPs by Gremlin. Mol Cell. 1999;4(5):839–849. doi: 10.1016/s1097-2765(00)80393-7. [DOI] [PubMed] [Google Scholar]
- 14.Marigo V, Johnson RL, Vortkamp A, Tabin CJ. Sonic hedgehog differentially regulates expression of GLI and GLI3 during limb development. Dev Biol. 1996;180(1):273–283. doi: 10.1006/dbio.1996.0300. [DOI] [PubMed] [Google Scholar]
- 15.Marigo V, Scott MP, Johnson RL, Goodrich LV, Tabin CJ. Conservation in hedgehog signaling: Induction of a chicken patched homolog by Sonic hedgehog in the developing limb. Development. 1996;122(4):1225–1233. doi: 10.1242/dev.122.4.1225. [DOI] [PubMed] [Google Scholar]
- 16.Minowada G, et al. Vertebrate Sprouty genes are induced by FGF signaling and can cause chondrodysplasia when overexpressed. Development. 1999;126(20):4465–4475. doi: 10.1242/dev.126.20.4465. [DOI] [PubMed] [Google Scholar]
- 17.Zakany J, Duboule D. The role of Hox genes during vertebrate limb development. Curr Opin Genet Dev. 2007;17(4):359–366. doi: 10.1016/j.gde.2007.05.011. [DOI] [PubMed] [Google Scholar]
- 18.Nelson CE, et al. Analysis of Hox gene expression in the chick limb bud. Development. 1996;122(5):1449–1466. doi: 10.1242/dev.122.5.1449. [DOI] [PubMed] [Google Scholar]
- 19.Dollé P, Izpisúa-Belmonte JC, Falkenstein H, Renucci A, Duboule D. Coordinate expression of the murine Hox-5 complex homoeobox-containing genes during limb pattern formation. Nature. 1989;342(6251):767–772. doi: 10.1038/342767a0. [DOI] [PubMed] [Google Scholar]
- 20.Tarchini B, Duboule D. Control of Hoxd genes’ collinearity during early limb development. Dev Cell. 2006;10(1):93–103. doi: 10.1016/j.devcel.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 21.Tarchini B, Duboule D, Kmita M. Regulatory constraints in the evolution of the tetrapod limb anterior-posterior polarity. Nature. 2006;443(7114):985–988. doi: 10.1038/nature05247. [DOI] [PubMed] [Google Scholar]
- 22.Ros MA, et al. The chick oligozeugodactyly (ozd) mutant lacks sonic hedgehog function in the limb. Development. 2003;130(3):527–537. doi: 10.1242/dev.00245. [DOI] [PubMed] [Google Scholar]
- 23.Montavon T, et al. A regulatory archipelago controls Hox genes transcription in digits. Cell. 2011;147(5):1132–1145. doi: 10.1016/j.cell.2011.10.023. [DOI] [PubMed] [Google Scholar]
- 24.Andrey G, et al. A switch between topological domains underlies HoxD genes collinearity in mouse limbs. Science. 2013;340(6137):1234167. doi: 10.1126/science.1234167. [DOI] [PubMed] [Google Scholar]
- 25.Beccari L, et al. A role for HOX13 proteins in the regulatory switch between TADs at the HoxD locus. Genes Dev. 2016;30(10):1172–1186. doi: 10.1101/gad.281055.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cooper KL, et al. Initiation of proximal-distal patterning in the vertebrate limb by signals and growth. Science. 2011;332(6033):1083–1086. doi: 10.1126/science.1199499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.ten Berge D, Brugmann SA, Helms JA, Nusse R. Wnt and FGF signals interact to coordinate growth with cell fate specification during limb development. Development. 2008;135(19):3247–3257. doi: 10.1242/dev.023176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100(4):423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 29.Zákány J, Kmita M, Duboule D. A dual role for Hox genes in limb anterior-posterior asymmetry. Science. 2004;304(5677):1669–1672. doi: 10.1126/science.1096049. [DOI] [PubMed] [Google Scholar]
- 30.Izpisúa-Belmonte JC, Tickle C, Dollé P, Wolpert L, Duboule D. Expression of the homeobox Hox-4 genes and the specification of position in chick wing development. Nature. 1991;350(6319):585–589. doi: 10.1038/350585a0. [DOI] [PubMed] [Google Scholar]
- 31.Montavon T, Le Garrec J-F, Kerszberg M, Duboule D. Modeling Hox gene regulation in digits: Reverse collinearity and the molecular origin of thumbness. Genes Dev. 2008;22(3):346–359. doi: 10.1101/gad.1631708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kmita M, Fraudeau N, Hérault Y, Duboule D. Serial deletions and duplications suggest a mechanism for the collinearity of Hoxd genes in limbs. Nature. 2002;420(6912):145–150. doi: 10.1038/nature01189. [DOI] [PubMed] [Google Scholar]
- 33.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: A flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42(Web Server issue):W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramírez F, et al. deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44(W1):W160–W165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mercader N, et al. Conserved regulation of proximodistal limb axis development by Meis1/Hth. Nature. 1999;402(6760):425–429. doi: 10.1038/46580. [DOI] [PubMed] [Google Scholar]
- 36.Scotti M, Kherdjemil Y, Roux M, Kmita M. A Hoxa13:Cre mouse strain for conditional gene manipulation in developing limb, hindgut, and urogenital system. Genesis. 2015;53(6):366–376. doi: 10.1002/dvg.22859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Wit E, de Laat W. A decade of 3C technologies: Insights into nuclear organization. Genes Dev. 2012;26(1):11–24. doi: 10.1101/gad.179804.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49(5):773–782. doi: 10.1016/j.molcel.2013.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maas SA, Suzuki T, Fallon JF. Identification of spontaneous mutations within the long-range limb-specific Sonic hedgehog enhancer (ZRS) that alter Sonic hedgehog expression in the chicken limb mutants oligozeugodactyly and silkie breed. Dev Dyn. 2011;240(5):1212–1222. doi: 10.1002/dvdy.22634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Andrey G, Duboule D. SnapShot: Hox gene regulation. Cell. 2014;156(4):856.e1. doi: 10.1016/j.cell.2014.01.060. [DOI] [PubMed] [Google Scholar]
- 41.Tabin C, Wolpert L. Rethinking the proximodistal axis of the vertebrate limb in the molecular era. Genes Dev. 2007;21(12):1433–1442. doi: 10.1101/gad.1547407. [DOI] [PubMed] [Google Scholar]
- 42.Raines AM, Magella B, Adam M, Potter SS. Key pathways regulated by HoxA9,10,11/HoxD9,10,11 during limb development. BMC Dev Biol. 2015;15:28. doi: 10.1186/s12861-015-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dessaud E, et al. Dynamic assignment and maintenance of positional identity in the ventral neural tube by the morphogen sonic hedgehog. PLoS Biol. 2010;8(6):e1000382. doi: 10.1371/journal.pbio.1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dessaud E, et al. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature. 2007;450(7170):717–720. doi: 10.1038/nature06347. [DOI] [PubMed] [Google Scholar]
- 45.Towers M, Mahood R, Yin Y, Tickle C. Integration of growth and specification in chick wing digit-patterning. Nature. 2008;452(7189):882–886. doi: 10.1038/nature06718. [DOI] [PubMed] [Google Scholar]
- 46.Zhu J, et al. Uncoupling Sonic hedgehog control of pattern and expansion of the developing limb bud. Dev Cell. 2008;14(4):624–632. doi: 10.1016/j.devcel.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harfe BD, et al. Evidence for an expansion-based temporal Shh gradient in specifying vertebrate digit identities. Cell. 2004;118(4):517–528. doi: 10.1016/j.cell.2004.07.024. [DOI] [PubMed] [Google Scholar]
- 48.Yang Y, et al. Relationship between dose, distance and time in Sonic Hedgehog-mediated regulation of anteroposterior polarity in the chick limb. Development. 1997;124(21):4393–4404. doi: 10.1242/dev.124.21.4393. [DOI] [PubMed] [Google Scholar]
- 49.Vokes SA, Ji H, Wong WH, McMahon AP. A genome-scale analysis of the cis-regulatory circuitry underlying sonic hedgehog-mediated patterning of the mammalian limb. Genes Dev. 2008;22(19):2651–2663. doi: 10.1101/gad.1693008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.David FP, et al. HTSstation: A web application and open-access libraries for high-throughput sequencing data analysis. PLoS One. 2014;9(1):e85879. doi: 10.1371/journal.pone.0085879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hamburger V, Hamilton HL. A series of normal stages in the development of the chick embryo. 1951. Dev Dyn. 1992;195(4):231–272. doi: 10.1002/aja.1001950404. [DOI] [PubMed] [Google Scholar]