Significance
It is widely appreciated that carcinoma cells exhibiting certain mesenchymal traits are enriched for cancer stem cells (CSCs) and can give rise to tumors with aggressive features. Whereas it has been proposed that mesenchymal carcinoma cell populations are internally heterogeneous, the field has made little progress in resolving the specific subtypes of mesenchymal carcinoma cells that pose the greatest risk for patients. We demonstrate the utility of integrin-β4 (ITGB4) in segregating these cells into distinct subpopulations with differing tumor-initiating abilities and pathological behaviors. In addition, we identified mechanistic links between ZEB1 (zinc finger E-box binding homeobox 1) and TAp63α (tumor protein 63 isoform 1) as regulators of ITGB4 expression and demonstrate that ITGB4 can be used as a marker to determine which patients are more likely to relapse after treatment.
Keywords: cancer, heterogeneity, EMT, mesenchymal, ITGB4
Abstract
Neoplastic cells within individual carcinomas often exhibit considerable phenotypic heterogeneity in their epithelial versus mesenchymal-like cell states. Because carcinoma cells with mesenchymal features are often more resistant to therapy and may serve as a source of relapse, we sought to determine whether such cells could be further stratified into functionally distinct subtypes. Indeed, we find that a basal epithelial marker, integrin-β4 (ITGB4), can be used to enable stratification of mesenchymal-like triple-negative breast cancer (TNBC) cells that differ from one another in their relative tumorigenic abilities. Notably, we demonstrate that ITGB4+ cancer stem cell (CSC)-enriched mesenchymal cells reside in an intermediate epithelial/mesenchymal phenotypic state. Among patients with TNBC who received chemotherapy, elevated ITGB4 expression was associated with a worse 5-year probability of relapse-free survival. Mechanistically, we find that the ZEB1 (zinc finger E-box binding homeobox 1) transcription factor activity in highly mesenchymal SUM159 TNBC cells can repress expression of the epithelial transcription factor TAp63α (tumor protein 63 isoform 1), a protein that promotes ITGB4 expression. In addition, we demonstrate that ZEB1 and ITGB4 are important in modulating the histopathological phenotypes of tumors derived from mesenchymal TNBC cells. Hence, mesenchymal carcinoma cell populations are internally heterogeneous, and ITGB4 is a mechanistically driven prognostic biomarker that can be used to identify the more aggressive subtypes of mesenchymal carcinoma cells in TNBC. The ability to rapidly isolate and mechanistically interrogate the CSC-enriched, partially mesenchymal carcinoma cells should further enable identification of novel therapeutic opportunities to improve the prognosis for high-risk patients with TNBC.
During the multistep formation of carcinomas, the epithelial cells from which carcinomas arise are known to acquire multiple changes in cell phenotype that provide various types of selective advantage during tumor progression (1). Whereas many of these changes derive from alterations of their genomes, others are acquired through the expression of previously latent cell-biological programs; such programs may be activated in response to signals that carcinoma cells receive from their microenvironment (1–3). In particular, as occurs in a number of tissues, epithelial carcinoma cells are able to acquire mesenchymal-like traits through activation of the cell-biological program termed the epithelial-to-mesenchymal transition (EMT) (1). Carcinoma cells that have activated an EMT program often exhibit increased tumor-initiating capacity as well as enhanced migratory and invasive abilities (1, 4, 5). Furthermore, these more mesenchymal carcinoma cells often exhibit increased metastatic abilities and elevated resistance to radiation and chemotherapy (1, 5).
A complex signaling network regulates the induction and subsequent maintenance of EMT programs (1–3, 6). These signals ultimately converge on a relatively small number of genes encoding transcription factors (EMT-TFs) that operate as master regulators of the EMT program, ushering cells from epithelial into more mesenchymal states (1–3, 6). When expressed experimentally, the EMT-TFs, such as TWIST (TWIST1; twist family bHLH transcription factor 1), SNAIL (SNAI1; snail family transcriptional repressor 1), SLUG (SNAI2; snail family transcriptional repressor 2), and ZEB1 (zinc finger E-box binding homeobox 1), have been shown to activate EMT programs in a variety of normal and neoplastic epithelial cell types (1–3, 6).
Increasing evidence makes it apparent that the EMT program does not operate as a binary switch in which individual cells—both normal and neoplastic—are caused to reside in either an epithelial or a mesenchymal state (1). Instead, ongoing research from a number of groups indicates that individual carcinoma cells can dwell in intermediate states along the epithelial–mesenchymal spectrum and thus can coexpress epithelial and mesenchymal characteristics in various proportions (1, 7, 8). Although carcinoma cells with mesenchymal traits are often more invasive, resistant to chemotherapy, and more likely to be sources of clinical relapse, we currently lack the ability to clearly identify the more mesenchymal carcinoma cells that pose the greatest risk to cancer patients, notably those endowed with increased tumor-initiating potential.
The CD44 and CD24 cell-surface markers have been widely used to resolve epithelial and mesenchymal carcinoma cells (1, 4, 9). Among other uses, these markers have proven useful in identifying more mesenchymal carcinoma cell populations that are enriched in the representation of tumor-initiating cells (TICs), often termed cancer stem cells (CSCs) (1, 4, 9). However, these markers have not proven useful in further resolving the more mesenchymal cells into distinct subpopulations, leaving unanswered the precise nature of the mammary CSCs, which usually reside as minority cell populations within larger populations of the more mesenchymal cells. For this reason, additional markers have been sought to complement CD44 and CD24 in the fractionation of carcinoma cell subpopulations (10, 11).
Triple-negative breast cancer (TNBC) represents a subset of tumors that constitute only 10–15% of all breast cancer, but accounts disproportionally for greater than one-third of breast-cancer–related deaths (12). A critical feature of TNBC is abundant inter- and intratumoral heterogeneity, including mesenchymal phenotypes revealed by routine histopathological analyses. Unfortunately, our ability to resolve these heterogeneous, more mesenchymal carcinoma cells into distinct subpopulations has been limited. In light of these challenges, we undertook to identify additional cell-surface markers that would make it possible to efficiently resolve functionally important subpopulations from heterogeneous populations of the more mesenchymal TNBC cells.
In an effort to better resolve the carcinoma cell heterogeneity often observed in TNBC, we determined that integrin-β4 (ITGB4) can be used as a marker to identify CSC-enriched populations of partially mesenchymal carcinoma cells. This result was consistent with previous mechanistic work, which demonstrated that ITGB4 can promote activation of the MET protooncogene receptor tyrosine kinase, epidermal growth factor receptor, focal adhesion kinase, SRC protooncogene nonreceptor tyrosine kinase, phosphoinositide 3-kinase, AKT serine/threonine kinase (AKT), mitogen-activated protein kinase kinase 1/2, mitogen-activated protein kinase (ERK) 1/2, and ras homolog family member A signaling pathways that are known to enhance tumor progression (13–15). Importantly, ITGB4 has been previously shown to be enriched in TNBC breast cancer patient tissues (16). Moreover, an ITGB4-related 65-gene classifier has been shown to have prognostic value when used to generally analyze survival of breast cancer patients (16). We now demonstrate that ITGB4 mRNA alone can be used to stratify survival for TNBC patients that require chemotherapy; patients with tumors exhibiting high levels of ITGB4 mRNA had a significantly worse prognosis. These results were similar to those that have been reported in pancreatic ductal adenocarcinoma (17) and non-small cell lung cancer (18). Together, our results demonstrate that ITGB4 can be used to identify CSC-enriched TNBC cells and that high levels of ITGB4 mRNA can be used clinically to identify patients that may benefit from more aggressive therapeutic strategies.
Results
Expression of ITGB4 on the Surface of Epithelial and Mesenchymal Subtype Triple-Negative Breast Cancer Cells.
Multiple distinct subtypes of TNBC cells have been described based on global gene expression analyses (19, 20); however, little information existed in the literature regarding how such subtypes of TNBC cells could be grouped or physically segregated based on their cell-surface marker profiles. To address this issue, we performed a series of analyses using highly epithelial immortalized mammary epithelial cells (HMLEs) (21), and their derived naturally arising mesenchymal epithelial cell (NAMEC) population, NAMEC8 (22), which led to the identification of ITGB4 as a cell surface protein that could be used for fluorescence-activated cell sorting (FACS) to segregate highly epithelial and mesenchymal-like mammary epithelial cells according to their relative epithelial versus mesenchymal cell states (SI Appendix, Fig. S1, and Datasets S1–S4). Moreover, segregating mesenchymal-like mammary epithelial cells using ITGB4 via FACS enabled resolution of two distinct classes of mesenchymal subtype mammary epithelial cells: an ITGB4hi fraction that was determined to be more epithelial and an ITGB4lo fraction that was more highly mesenchymal, despite the fact that both populations displayed similar overt morphological and canonical molecular mesenchymal-like phenotypes (SI Appendix, Fig. S2, and Dataset S4). Importantly, using unsupervised hierarchical clustering of their gene expression profiles, the HMLE and NAMEC8 cells used for these analyses were determined to be more closely related to triple-negative breast cancer cell lines than other common breast cancer subtypes (SI Appendix, Fig. S3).
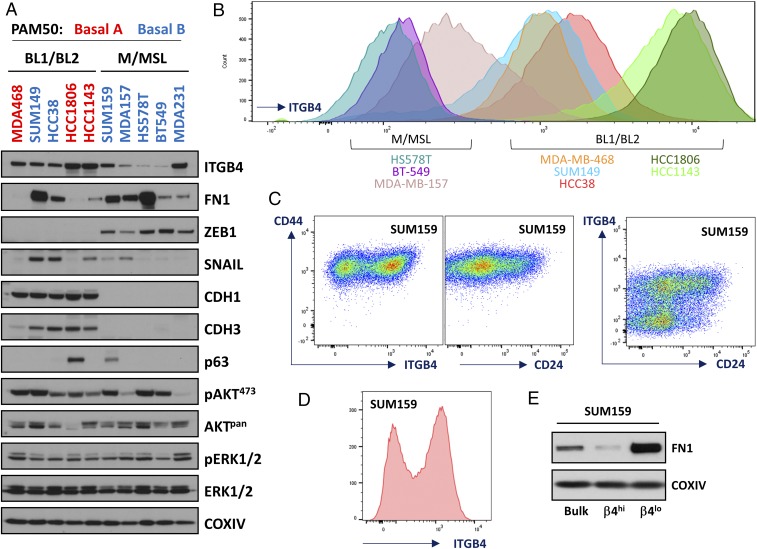
To determine whether ITGB4 would serve as a useful cell-surface marker for resolving distinct subtypes of TNBC cells, we performed Western blot and FACS analyses of 10 human TNBC cell lines (Fig. 1 A–C and SI Appendix, Fig. S4B). Our results indicated that, similar to results obtained using the highly epithelial HMLE cells (SI Appendix, Fig. S1), ITGB4 was abundantly expressed by the more epithelial TNBC cell lines (MDA-MB-468, HCC1143, HCC1806, HCC38, and SUM149), despite their lack of canonical epithelial morphological characteristics (SI Appendix, Figs. S4C and S8B). Conversely, ITGB4 was expressed at relatively low levels on the surface of cells from three of the more mesenchymal TNBC cell lines (MDA-MB-157, HS578T, and BT549) (Fig. 1 and SI Appendix, Fig. S4).
Fig. 1.
Characterization of ITGB4 expression in a panel of TNBC cell lines and isolation of SUM159 ITGB4hi and ITGB4lo cell populations. (A) Western blot analyses for 10 triple-negative breast cancer (TNBC) cell lines that have been previously characterized using the PAM50 strategy into basal A and basal B subtypes or according to the strategy outlined by Lehmann et al. (19, 20) into basal-like (BL1/BL2) and mesenchymal/mesenchymal stem-like (M/MSL) subtypes. (B) FACS histograms of ITGB4 in eight TNBC cell lines. (C) CD44, CD24, and ITGB4 FACS profiles of the SUM159 cell line. (D) Typical histogram of the SUM159 cells used to isolate ITGB4hi and ITGB4lo subpopulations. (E) Western blots of fibronectin (FN1) and cytochrome c oxidase subunit 4I1 (COXIV) for SUM159 cells and the derived ITGB4hi and ITGB4lo subpopulations.
Importantly, similar to the mesenchymal NAMEC8 cells (SI Appendix, Figs. S1 and S2), we readily detected ITGB4 expression on the surface of cells present in two mesenchymal-subtype TNBC lines, SUM159 and MDA-MB-231 (Fig. 1 C and D and SI Appendix, Fig. S4B and see Fig. S8A). As revealed by FACS analyses, the difference of ITGB4 cell-surface abundance between ITGB4hi and ITGB4lo cells within each of these two TNBC populations ranged between 10- and 100-fold (Fig. 1 C and D and SI Appendix, Fig. S4B and see Fig. S8A). This broad spectrum of expression by different cells within the SUM159 and MDA-MB-231 mesenchymal-subtype TNBC populations was sufficient in each case for isolation of distinct ITGB4hi and ITGB4lo subpopulations by FACS (Figs. 1E, 2, and 4).
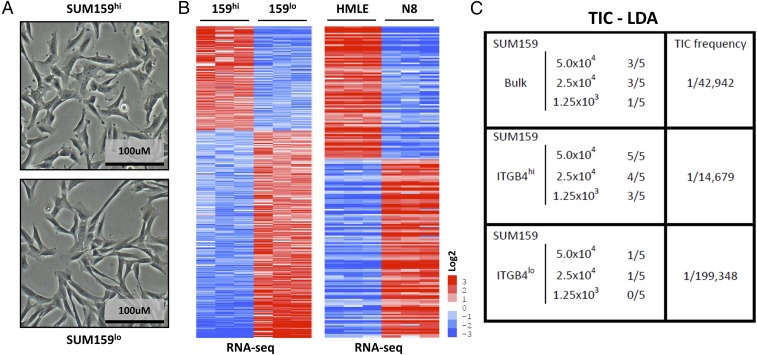
Fig. 2.
Molecular and functional characterization of isolated ITGB4hi and ITGB4lo subpopulations of SUM159 TNBC cells. (A) Morphological appearance of SUM159 ITGB4hi and ITGB4lo cells. (B) Heat maps of genes differentially expressed in the SUM159 ITGB4hi and ITGB4lo cells and the genes that were coordinately differentially expressed in the HMLE vs. NAMEC8 comparisons used to classify the relative epithelial vs. mesenchymal status of the SUM159 ITGB4hi and ITGB4lo cells. (C) Tumor-initiating cell (TIC) limiting-dilution assays (LDAs) using the SUM159 cell line and derived SUM159 ITGB4hi or ITGB4lo subpopulations.
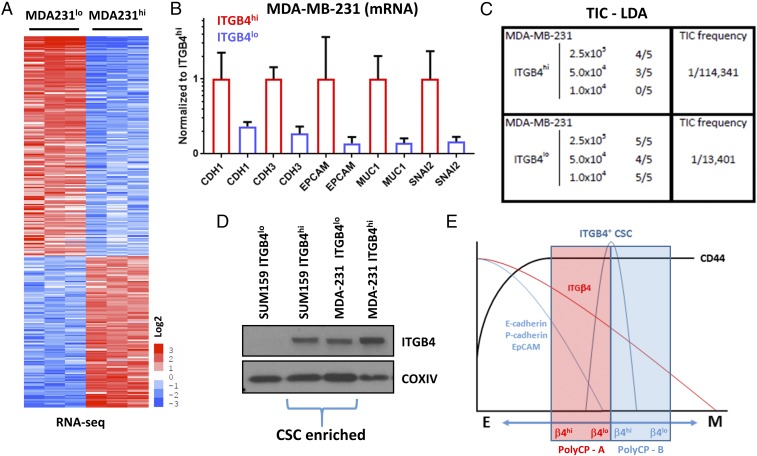
Fig. 4.
Molecular and functional characterization of ITGB4hi and ITGB4lo MDA-MB-231 cells and comparison with ITGB4hi and ITGB4lo SUM159 cell populations. (A) Heat map of genes differentially expressed in MDA-MB-231 ITGB4hi and ITGB4lo cell populations. (B) Fold change of RNA-seq values comparing MDA-MB-231 ITGB4hi and ITGB4lo cell populations. (C) Tumor-initiating cell (TIC) limiting-dilution assays (LDAs) using the MDA-MB-231 ITGB4hi and ITGB4lo cell populations. (D) Western blot analyses comparing SUM159 ITGB4hi and ITGB4lo cell populations to MDA-MB-231 ITGB4hi and ITGB4lo cell populations. (E) Working model representing TNBC partially mesenchymal carcinoma cell enrichment of cancer stem cell (CSC) populations relative to polyclonal bulk cell populations (PolyCP) that span different regions along the epithelial–mesenchymal cell state spectrum.
ITGB4 Identifies CSC-Enriched Mesenchymal TNBC Cells.
Of the mesenchymal-subtype TNBC cell lines that were analyzed by FACS, cells of the SUM159 line exhibited the greatest degree of segregation between ITGB4hi and ITGB4lo expression levels, with two distinct ITGB4hi and ITGB4lo peaks (Fig. 1 C and D). Nonetheless, all of the cells comprising this cell line shared a common CD44hi phenotype (Fig. 1C). This finding caused us to focus in more detail on the SUM159 cells and the possibility that their outwardly mesenchymal morphology concealed an underlying variability in the expression of epithelial and mesenchymal genes and corresponding protein markers—similar to the nonneoplastic polyclonal NAMEC8 mammary epithelial cells (MECs) we had used in our previous analyses.
We used FACS to fractionate the heterogeneous SUM159 cells into ITGB4hi and ITGB4lo subpopulations (Figs. 1E and 2A). These two populations were cultured independently and did not spontaneously interconvert over a period of 2 mo in continuous culture. Canonical markers of the epithelial and mesenchymal state revealed minor differences between the cells from the perspective of mRNA expression [less than a fourfold difference for SNAIL, SLUG, TWIST1, TWIST2, ZEB1, ZEB2, CDH2 (cadherin 2; also known as N-cadherin), VIM (vimentin), and FN1 (fibronectin)] (SI Appendix, Fig. S5 A and B). However, whereas FN1 mRNA differed only by twofold when comparing the two populations, FN1 protein expression was significantly elevated in the ITGB4lo population (Fig. 1E). During the first 48 h after each in vitro passage, both populations exhibited a lack of cell–cell junctions and clustering, the presence of a front–back polarity, and displayed an elongated rather than cobblestone morphology (Fig. 2A). However, the ITGB4lo cells adopted a more spindle-like morphological phenotype compared with the ITGB4hi cells (Fig. 2A).
As judged by FACS analyses, the SUM159 ITGB4hi and ITGB4lo cells differed in their respective CD24 profiles (Fig. 1C). The ITGB4hi population was composed of both CD24hi and CD24lo cells, whereas the ITGB4lo population was primarily CD24lo. These results indicated that the CD24 and CD44 markers alone could not be used to isolate the same SUM159 subpopulations that had been identified using ITGB4. To determine how the SUM159 ITGB4hi and ITGB4lo populations of mesenchymal carcinoma cells related to one another using an unbiased approach, we performed RNA-sequencing (RNA-seq) analyses (Fig. 2B and SI Appendix, Figs. S5 A and D and S6A and Dataset S5), and compared the differentially expressed genes with an EMT-associated gene expression profile identified using the highly epithelial HMLE and more mesenchymal NAMEC8 cells (Fig. 2B and SI Appendix, Fig. S5D and Datasets S1, S5, and S6).
The SUM159 ITGB4lo mesenchymal carcinoma cells exhibited levels of EMT-associated gene expression that were higher (ranging from 3- to >500-fold) than the ITGB4hi mesenchymal carcinoma cells (SI Appendix, Figs. S5 C and D and S6A and Datasets S5 and S6). These results confirmed the utility of ITGB4 as a marker to separate more epithelial from more mesenchymal subpopulations of CD44hi mesenchymal-like human mammary carcinoma cells and suggested that it could be used to determine whether the distinct subpopulations differ from one another in their relative tumor-initiating abilities.
As a prelude to tumor initiation studies, we determined that the ITGB4hi and ITGB4lo cells had equivalent proliferation rates and tumorsphere-forming abilities in culture (SI Appendix, Fig. S6E). We proceeded to determine whether either of the two SUM159 subpopulations was enriched in traits associated with CSCs by implanting these cells at limiting dilutions in nonobese diabetic (NOD)/severe combined immunodeficient (SCID) host mice to gauge their relative tumor-initiating abilities (Fig. 2C). The parental, unfractionated SUM159 cells exhibited a calculated TIC frequency of 1/42,942 cells. Cells of the ITGB4hi subpopulation were more efficient in tumor initiation, with a TIC frequency of 1/14,679 in contrast to the TIC frequency of 1/199,348 for the ITGB4lo cell population, i.e., essentially a >10-fold difference in the representation of TICs. This finding indicated that, in addition to its display of multiple mesenchymal traits, the TIC-enriched fraction expressed a significant level of an epithelial marker—ITGB4—and accordingly, resided in an intermediate state between fully epithelial and fully mesenchymal.
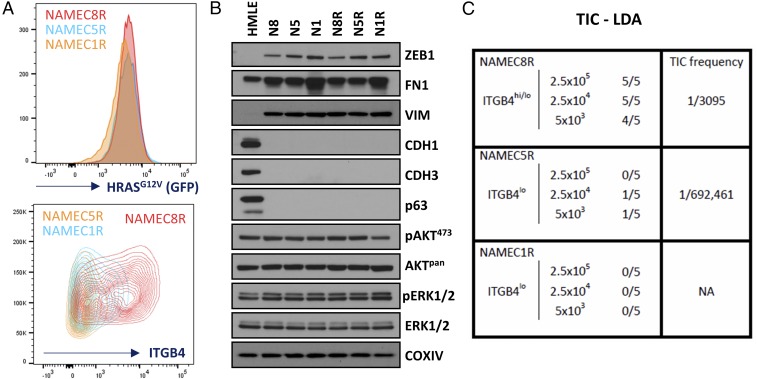
We next determined whether the behavior of the SUM159 cells, as described above, was echoed by that of other neoplastically transformed cell lines. Thus, we performed similar experiments using variants of mesenchymal-like epithelial cells that had been derived from transformation of three different NAMEC lines through introduction of an HRAS oncogene (NAMECR; Fig. 3 and SI Appendix, Fig. S7). Because it was known that expression of this oncogene can itself alter the epithelial versus mesenchymal morphological and molecular phenotype of transformed cells (SI Appendix, Fig. S7) (23), we normalized the cells for expression of the HRAS oncogene-expressing retroviral construct, which also included an internal ribosome entry site (IRES) to drive green fluorescent protein (GFP) expression (IRES-GFP), doing so via FACS for GFP and monitoring the levels of ITGB4 expression following transformation (Fig. 3A). We also monitored the expression of several EMT markers [ZEB1, FN1, VIM, CDH1 (cadherin 1; also known as E-cadherin), CDH3 (cadherin 3; also known as P-cadherin)], and TP63 (tumor protein 63) and RAS-activated proteins (AKT and ERK1/2) by Western blot analyses (Fig. 3B), which demonstrated negligible differences between the NAMEC1R, NAMEC5R, and NAMEC8R cell lines in vitro. Using these NAMECR cell lines, we determined that the NAMEC8R line, which contained a large proportion of cells with a CD44hiCD24loITGB4hi cell-surface profile, was significantly better at initiating tumors than its more highly mesenchymal CD44hiCD24loITGB4lo counterparts (Fig. 3C). Specifically, the highly mesenchymal CD44hiCD24loITGB4lo NAMEC1 and NAMEC5 cells (Fig. 3B and SI Appendix, Fig. S7 F–H and Dataset S7), when transformed by RAS (NAMEC1R and NAMEC5R; SI Appendix, Fig. S7I), were poorly tumorigenic (TIC frequency of 1/692,461 cells) and nontumorigenic, respectively, whereas the heterogeneous mesenchymal NAMEC8R population exhibited a TIC frequency of 1/3,095 cells (Fig. 3C). Once again, cells coexpressing both epithelial and mesenchymal markers exhibited an elevated tumor-initiating ability, in this instance via a factor of >200 relative to the more mesenchymal ITGB4lo cells.
Fig. 3.
ITGB4 identifies cancer stem cell-enriched populations of engineered mesenchymal mammary carcinoma cells. (A) FACS profiles of HRASG12V and ITGB4 expression for the NAMEC1R, NAMEC5R, and NAMEC8R cell lines. (B) Western blots of EMT- and RAS-signaling–associated markers. (C) Tumor-initiating cell (TIC) limiting-dilution assays (LDAs) using the NAEMC1R, NAMEC5R, and NAMEC8R cell lines.
Taken together, these results indicated that the more epithelial ITGB4hi subpopulation of outwardly mesenchymal epithelial cells exhibited a far higher tumor-initiating ability than the more highly mesenchymal ITGB4lo cells. Hence, our data demonstrated that use of ITGB4 as a marker, on its own, allowed stratification of distinct subpopulations of mesenchymal-subtype TNBC cells and experimentally transformed populations of naturally arising mesenchymal epithelial cells. Importantly, these analyses identified TIC-enriched subpopulations that could not be clearly identified by use of the CD44 and CD24 markers alone. Moreover, these results further support and extend less direct findings of others, indicating that carcinoma cells that reside in an excessively mesenchymal state are actually less efficient in tumor initiation than their more epithelial counterparts (1, 24).
Intermediate Levels of ITGB4 Identify CSC-Enriched TNBC Cells When Comparing Mesenchymal MDA-MB-231 and SUM159 Carcinoma Cell Lines.
To begin, we subjected ITGB4hi and ITGB4lo MDA-MB-231 subpopulations to RNA-seq analyses and limiting-dilution tumor initiation assays (Fig. 4 A–C and SI Appendix, Fig. S8 and Dataset S8). Similar to the observations described above, the mesenchymal MDA-MB-231 CD44+CD24loITGB4hi cells were more epithelial than their CD44+CD24loITGB4lo counterparts (Fig. 4B and SI Appendix, Fig. S8D and Datasets S8 and S9). Of note, the ITGB4hi MDA-MB-231 cells expressed higher levels of SNAI2 (SLUG) mRNA, reminiscent of normal differentiated human and mouse basal mammary epithelial cells (25, 26), whereas the more mesenchymal ITGB4lo cells expressed nearly undetectable levels of SLUG (Fig. 4B). This finding supported the notion that endogenous SLUG expression was associated with the more differentiated basal epithelial rather than more mesenchymal cell state (26). The ITGB4hi MDA-MB-231 cells also expressed higher mRNA levels of other more epithelial genes, including CDH1, CDH3, EpCAM (epithelial cell adhesion molecule), and MUC1 (mucin 1, cell surface associated) (Fig. 4B), further indicating that they exhibited a more epithelial molecular phenotype than their more mesenchymal ITGB4lo MDA-MB-231 counterparts.
We proceeded to perform limiting-dilution tumor initiation assays to determine the TIC frequencies of the ITGB4hi and ITGB4lo MDA-MB-231 cell populations (Fig. 4C). Here we found that the ITGB4lo MDA-MB-231 cells were more tumorigenic than the more epithelial ITGB4hi mesenchymal MDA-MB-231 cells (TIC frequencies of 1 in 13,401 and 1 in 114,341 cells, respectively). Hence, in this particular case, and in contrast with our previous experience with SUM159 cells described above, the more mesenchymal cells exhibited a significantly higher degree of tumorigenicity.
To determine how the absolute abundance of ITGB4 from isolated SUM159 and MDA-MB-231 populations compared with one another, we performed Western blot analyses (Fig. 4D). When comparing these four subpopulations (i.e., the two SUM159 and the two MDA-MB-231 subpopulations), the SUM159 ITGB4lo cells had the least ITGB4 protein, whereas the MDA-MB231 ITGB4hi cells had the most (Fig. 4D). The remaining two populations, SUM159 ITGB4hi and MDA-MB-231 ITGB4lo, which carried in each case the great bulk of the CSCs, expressed intermediate levels of ITGB4 (Fig. 4D). Taken together, the SUM159 and MDA-MB-231 data indicated that, in the case of these two polyclonal mesenchymal populations, cells with an intermediate level of ITGB4 expression exhibited a hybrid epithelial/mesenchymal phenotype and were more tumorigenic than their isogenic counterparts that resided in either a more complete epithelial or mesenchymal cell state (illustrated in Fig. 4E).
Correlation of ITGB4 mRNA Expression with Decreased Relapse-Free Survival in Patients with TNBC.
TNBC tumors are defined clinically by the absence of immunohistochemical staining for the estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER2). TNBCs are highly overlapping with the molecular basal-like subtype, defined through mRNA expression profiling and use of the PAM50 classification strategy (27, 28). To assess the potential clinical impact of ITGB4 marker expression in patients with TNBC, we evaluated ITGB4 mRNA expression in tumor biopsies obtained at the time of diagnosis and determined the association of this expression with relapse-free survival (RFS) in clinically defined TNBC and molecular basal-subtype patient cohorts, doing so using previously reported global gene expression patterns determined by mRNA microarray analyses (29, 30).
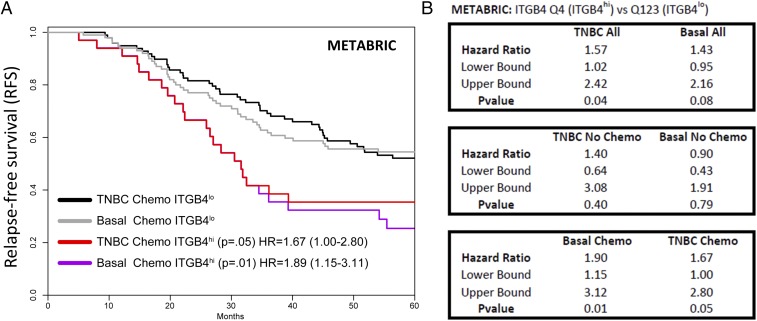
Among patients with TNBC who received chemotherapy, those whose tumors exhibited high levels of ITGB4 RNA (biopsied at the time of diagnosis) experienced a significantly lower probability of 5-year relapse-free survival relative to those whose tumors displayed low levels of ITGB4 RNA (Fig. 5 and SI Appendix, Fig. S9). This finding was true in both the clinically defined TNBC and molecular basal-subtype cohorts. These correlations were based on analyses conducted using two independent datasets (29, 30). In the Molecular Taxonomy of Breast Cancer International Consortium (METABRIC) dataset (Fig. 5 A and B and SI Appendix, Dataset S10) (29), the hazard ratio (HR)—a metric that describes the fold increase in risk for relapse over time—between the two groups was 1.67 [95% confidence intervals (CI) 1.0–2.8] with a log rank P value of <0.05, and a HR 1.89 (95% CI 1.15–3.11) with a log rank P value of <0.01, for the clinically defined and molecular basal-subtype cohorts, respectively. As a sensitivity analysis, we performed a multivariate analysis of ITGB4 expression and RFS, including primary tumor stage, histologic grade, and patient age, which resulted in nearly identical hazard ratios in clinically defined TNBC (HR 1.70; 95% CI 0.93–3.10, P = 0.086) and basal-subtype (HR 1.97; 95% CI 1.11–3.52, P = 0.01) cohorts, despite lower statistical power. In the dataset generated by Györffy, et al. (30), the hazard ratio was 3.3 (95% CI 1.16–9.4), with a log rank P value of <0.02, for the clinically defined chemotherapy-treated TNBC cohort (SI Appendix, Fig. S9A). Hence, in two independent datasets, high levels of ITGB4 correlated with a poor outcome in patients whose tumors required chemotherapy. In contrast, within cohorts of TNBC and molecular basal-subtype patients whose tumors did not receive or require chemotherapy, there was no significant difference in RFS between those that exhibited high or low levels of ITGB4 mRNA in tumor biopsies prepared at the time of diagnosis (Fig. 5B).
Fig. 5.
Clinical correlations between ITGB4 mRNA expression and patient relapse-free survival in triple-negative and molecular basal subtype breast cancer. (A) Kaplan–Meier survival curves for patients in the upper quartile of ITGB4 expression compared with those in the lower three quartiles. (B) Statistical details related to survival correlations conducted for ITGB4 expression in the METABRIC analyses. HR, hazard ratio; p, log rank P value.
The results demonstrating stratification of patients with TNBC based on ITGB4 mRNA into groups that differed in their probability of relapse-free survival led us to test whether this correlation was observed in other solid tumor types. Specifically, we found that higher levels of ITGB4 mRNA also correlated with a decreased probability of progression-free survival in patients with lung adenocarcinoma, stage 4 serous ovarian cancer, and gastric cancer (SI Appendix, Fig. S9 B–D). These results provide initial evidence suggesting that the results of our TNBC analyses may prove to be applicable to carcinomas arising in other tissues.
The observed difference in clinical outcome, for ITGB4hi and ITGB4lo chemotherapy-treated breast cancer patients, prompted us to determine whether the two classes of CD44hi mesenchymal carcinoma cells, ITGB4hi and ITGB4lo, exhibited a differential cell-intrinsic response in culture to a broad range of common chemotherapeutic and antineoplastic small molecule compounds. To do so, we performed a five-point dose–response screen of 392 compounds, including many common chemotherapeutics as well as other small molecule inhibitors of cancer-associated pathways to compare the viability of SUM159 ITGB4hi and ITGB4lo mesenchymal carcinoma cells after a 72-h period of treatment (SI Appendix, Dataset S11). The results indicated no clinically significant difference in sensitivity to the tested compounds by the SUM159 ITGB4hi or ITGB4lo populations. When taken together with the survival outcome analyses, these data suggested that one possible explanation of the poor prognosis for patients with ITGB4hi tumors, supported by our data in TNBC, is that among cells that escape therapy, the increased frequency of CSCs present in mesenchymal ITGB4hi TNBC populations can contribute to the higher frequency of relapse observed in the clinic.
ITGB4hi and ITGB4lo Mesenchymal Carcinoma Cells Harbor CSCs That Give Rise to Distinct Histopathological Phenotypes During Tumor Progression.
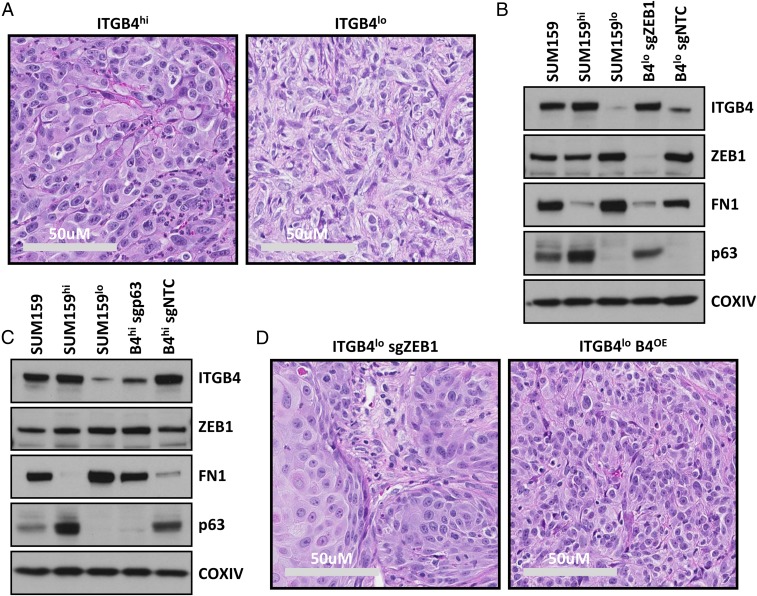
The above observations indicated that residence of carcinoma cells in a highly mesenchymal ITGB4lo cell state was negatively correlated with tumor-initiating ability. However, clinically, the histopathology of a tumor, rather than an estimation of CSC frequency, is used as a parameter to inform decisions related to therapeutic strategies aimed at managing disease progression. Hence, we examined histological sections of the tumors generated above and found that the SUM159 cell line demonstrated the most robust difference in features that could distinguish the ITGB4hi and ITGB4lo subtypes. Specifically, tumors derived from the ITGB4hi SUM159 cells were composed predominantly of carcinoma cells with large hyperchromatic nuclei (Fig. 6A), whereas the rarely observed tumors derived from ITGB4lo cells had relatively small pleomorphic nuclei (Fig. 6A). The ITGB4hi carcinoma cells primarily existed in clusters with sparse matrix deposition, whereas the ITGB4lo tumors exhibited interwoven fascicles of cells with a more mesenchymal, spindle-like morphology and abundant matrix deposition (Fig. 6A). The tumors derived from ITGB4hi cells also exhibited an increase in the abundance of polymorphonuclear immune cell infiltration compared with those derived from ITGB4lo cells (Fig. 6A).
Fig. 6.
Histopathological analyses and identification of a ZEB1–TAp63α–ITGB4 axis in SUM159 ITGB4hi and ITGB4lo cell populations. (A) H&E-stained histological sections of tumors derived from SUM159 ITGB4hi and ITGB4lo cells. (B and C) Western blots for ITGB4, ZEB1, FN1, TAp63α, and COXIV in the sorted and CRISPR-Cas9-sg engineered cell lines. (D) H&E-stained histological sections of tumors derived from SUM159 ITGB4lo cells that either lacked ZEB1 or ectopically expressed ITGB4.
The observed differences in matrix deposition and immune infiltration correlated well with RNA-seq data generated from analyses of these two populations growing in monolayer culture (SI Appendix, Dataset S5). Thus, the more mesenchymal ITGB4lo cells expressed higher levels of mesenchymal-like extracellular-matrix–associated genes, whereas the ITGB4hi cells were enriched for expression of immune-associated cytokine genes. Accordingly, the distinct histopathology of these two classes of tumors reflected a profound difference in underlying gene expression.
Mechanistic Analyses of a ZEB1–TAp63α–ITGB4 Axis Reveal an Essential Role for ZEB1 and ITGB4 in Dictating the Pathological Behavior of Highly Mesenchymal CSCs.
Based on our results demonstrating unique pathological behaviors of CSCs present in the SUM159 ITGB4hi and ITGB4lo mesenchymal carcinoma cell populations, we examined whether certain molecular mechanisms could be identified that underlay the highly mesenchymal histopathological phenotype of tumors derived from the more mesenchymal carcinoma cells compared with those derived from the more epithelial subtype of mesenchymal carcinoma cells. Thus, we noted that ZEB1 was the only EMT-TF commonly expressed in all five of the mesenchymal subtype TNBC cell lines that we had analyzed (Fig. 1A). This finding suggested its essentiality in maintaining the residence of these cells in the highly mesenchymal ITGB4lo cell state and the possibility that it might contribute to the histopathological phenotype of tumors derived from these cells in vivo.
To test this notion, we performed tumor induction assays using SUM159 ITGB4lo cells that had undergone CRISPR-mediated deletion of ZEB1 (Fig. 6 B and D and SI Appendix, Fig. S10A). Indeed, loss of ZEB1 in these ITGB4lo TNBC cells, propagated in vitro, resulted in their conversion to a more epithelial ITGB4hi FN1lo cell state (Fig. 6B and SI Appendix, Fig. S10A). The shift from a more mesenchymal to a more epithelial state was accompanied by induction of the epithelial marker TAp63α (tumor protein 63 isoform 1) (Fig. 6B), a transcription factor that has previously been characterized as a positive regulator of ITGB4 expression (31). This finding was consistent with our observations that, prior to ablation of ZEB1, the expression of TAp63α was robustly expressed by the ITGB4hi cells, whereas the ITGB4lo population had almost no detectable expression (Fig. 6 B and C). Thus, we tested the relevance of TAp63α expression on the regulation of ITGB4 and FN1 expression in the SUM159 ITGB4hi cells (Fig. 6C and SI Appendix, Fig. S10B). Indeed, ablation of TAp63α in the SUM159 ITGB4hi cells had no impact on ZEB1 expression, but significantly decreased ITGB4 expression (>10-fold) and increased the expression of the mesenchymal marker, FN1 (Fig. 6C). Together, these results indicated that loss of ZEB1 shifted the highly mesenchymal ITGB4lo cells toward a more epithelial cell state and that this was in part due to the loss of ZEB1-dependent TAp63α repression.
When injected into mice, the ZEB1 knockout ITGB4lo cells initiated tumors that were compared with those derived from the ITGB4hi, ITGB4lo, and nontargeting CRISPR control ITGB4lo cells to determine their relative histopathological phenotypes. In tumors derived from ITGB4lo cells that lacked ZEB1, the carcinoma cells exhibited a broad range of more epithelial histomorphological phenotypes (Fig. 6D). These results indicated that endogenous ZEB1 was required to maintain ITGB4lo TNBCs in a highly mesenchymal cell state, and indeed, was a critical regulator of the highly mesenchymal histopathological phenotype of the resulting tumors.
Based on the molecular and phenotypic alterations that we had observed and the known role of ZEB1 as a centrally acting EMT-TF, we presumed that the loss of ZEB1 would lead to a broad spectrum of alterations within these cells. Indeed, ITGB4 itself has been shown to be transcriptionally repressed by ZEB1 (32), and ITGB4 has been shown to promote tumor progression in a number of previously published studies (13). Accordingly, we speculated that ectopic expression of ITGB4 in the SUM159 ITGB4lo cells might, on its own, alter the histopathological phenotype of tumors arising from these carcinoma cells. To test this notion, we constitutively expressed ITGB4 in the SUM159 ITGB4lo cells (SUM159 ITGB4OE) at a similar level to that observed in the SUM159 ITGB4hi cells and used these SUM159 ITGB4OE cells to generate tumors (Fig. 6D and SI Appendix, Fig. S10 C and D). Notably, forced expression of ITGB4 in the SUM159 ITGB4lo cells on its own caused them to form tumors displaying a histopathological phenotype that closely resembled the more epithelial phenotype observed in the naturally arising SUM159 ITGB4hi tumors (Fig. 6D).
Taken together, these observations indicated that levels of ITGB4 expression can serve as a useful marker in revealing residence of cells in more epithelial or mesenchymal states, and that its expression could contribute to the histopathological phenotype observed during tumor progression. Moreover, because ITGB4 mRNA could stratify TNBC patient cohorts into groups that differed in their probability of relapse, ITGB4 could be considered a mechanistically driven prognostic biomarker, which is by definition, a prognostic biomarker with the ability to regulate the pathogenesis of disease progression.
Discussion
Intratumoral carcinoma cell heterogeneity has been widely observed and studied in various ways to identify CSC-enriched subpopulations and determine their differential susceptibilities to various treatment modalities (5). In the case of carcinomas, one major source of heterogeneity, which has been the focus of a large body of research over the past decade, derives from differences between carcinoma cells that reside in epithelial versus mesenchymal cell states (1, 11, 24, 33, 34). Indeed, conversions between these states have been shown to occur as a result of activating cell-biological programs that result in epithelial-to-mesenchymal and mesenchymal-to-epithelial transitions. As we and others have shown, activation of an EMT program fosters entrance of both normal and neoplastic mammary epithelial cells into stem-cell states (1, 25, 26, 35). Importantly, carcinoma cells within the mesenchymal-like CD44hiCD24lo state are generally more tumorigenic and more resistant to radiation and chemotherapy than those in a more epithelial CD44loCD24hi cell state (4, 5, 9, 11, 22, 36). However, very little has been reported about the tumorigenicity or therapeutic susceptibility of different subtypes of mesenchymal-like mammary carcinoma cells that reside within the larger population of these more mesenchymal carcinoma cells.
In the case of breast cancer, a growing number of cell-surface and intracellular markers have been used for the identification of different types of CSCs (1, 5, 11). Whereas many of the identified markers can enrich for stem-like carcinoma cell populations, additional markers are needed to enable more precise, focused analyses of CSCs. Here we identify a marker, ITGB4, otherwise known as CD104, that serves both in immortalized basal-like human MECs and in TNBC cells as a readout, indicating the extent to which the mesenchymal-like cells have proceeded through EMT programs. This utility may be due, in part, to the fact that the ITGB4 gene promoter is directly targeted and repressed by several of the EMT-TFs, which are known to be responsible for orchestrating EMT programs (32, 37, 38). Using this indicator function of ITGB4 has allowed us to resolve distinct subtypes of mesenchymal-like cell subpopulations, which share in common a CD44hi marker state and exhibit certain mesenchymal characteristics. As our evidence shows, this outward similarity in phenotype obscures an underlying heterogeneity. Thus, we were able to resolve in one case (MDA-MB-231) cells with an intermediate level of ITGB4 from their more epithelial, less tumorigenic ITGB4hi counterparts and, in other cases, SUM159 and NAMECR, cells with an intermediate level of ITGB4 from their more mesenchymal, less tumorigenic ITGB4lo counterparts.
These findings provide a direct demonstration that an important class of CSCs present in heterogeneous basal mammary carcinoma populations resides in a partially epithelial/partially mesenchymal state rather than the fully mesenchymal state that might be inferred simply from their CD44hiCD24lo cell-surface profile. Nonetheless, use of this ITGB4/CD104 marker only allowed us to enrich for the presence of tumor-initiating cells, i.e., CSCs, and further stratification of CD44hiCD24lo mesenchymal carcinoma cells exhibiting an intermediate level of ITGB4 expression will be required to isolate pure populations of mammary CSCs.
These observations hold implications for the nature of CSCs. Thus, activation of an EMT program appears to be productive for generating CSCs if it permits carcinoma cells to acquire certain mesenchymal traits, while retaining certain epithelial characteristics (1, 11, 24, 34, 39). This finding would explain why forced entrance of epithelial cells into a fully mesenchymal state is actually counterproductive for the formation of CSCs; we suspect that similar rules apply to normal nonneoplastic epithelial cells and their corresponding subpopulations of SCs. Together these observations, as summarized in Fig. 4E, indicate that an important class of TNBC CSCs resides in an intermediate state between highly epithelial and highly mesenchymal and that residence in either a highly epithelial or a highly mesenchymal state is less compatible with tumor-initiating capability.
Despite remarkable progress over the past decade in defining inter- and intratumoral heterogeneity in TNBC, few prognostic biomarkers are routinely used in clinical practice. Clinicopathologic characteristics of TNBC (stage, grade, and patient age) and response to neoadjuvant chemotherapy are routinely used without additional immunohistochemical, genomic, or expression-based biomarkers. Importantly, high levels of ITGB4 mRNA expression in biopsy samples at the time of diagnosis significantly correlated with reduced relapse-free survival in patients with TNBC who received chemotherapy. These findings are consistent with our observations that ITGB4 is expressed by mesenchymal TNBC cell populations containing high numbers of CSCs and suggest that ITGB4 should be investigated as a mechanistically driven prognostic biomarker in patients with high-risk TNBC.
To determine whether these results could be applicable to other types of cancer, we performed analyses using ITGB4 mRNA expression levels and progression-free survival in patients with lung adenocarcinoma, stage 4 serous ovarian cancer, and gastric cancer. These results, similar to those in TNBC, indicated that in all three types of cancer, higher levels of ITGB4 mRNA correlated with decreased progression-free survival for patients. Thus, the results reported in TNBC might be conceptually and fundamentally important for identifying and interrogating CSC-enriched populations of carcinoma cells that arise in other tissues and organs. With the identification of proteins and pathways that critically regulate the functional abilities of CSC-enriched populations, including and beyond those related to the ZEB1, p63, and ITGB4 proteins that we have initially assessed, it may be possible to design more effective strategies for therapeutically targeting these cells in patients.
Materials and Methods
Cell Lines and Culture Conditions.
HMLE and NAMEC based cell lines were cultured essentially as previously described using MEGM medium (21). MDA-MB-157, MDA-MB-231, HCC38, HCC1806, HCC1143, HS578T, BT549, MDA-MB-468, SUM159, and SUM149 cell lines were cultured using media outlined in SI Appendix. All cell lines were maintained in subconfluent conditions and media were replenished every 48 h.
Plasmid Constructs and Virus Production.
pLenti-CRISPR-Cas9 V2 (Addgene 52961) constructs were produced as previously described (40) using sequences outlined in SI Appendix. Due to the altered expression of ITGB4 resulting from use of the sgZEB1 and sgTP63 constructs, cells were FACS sorted on ITGB4 to obtain the respective knockout populations used for subsequent analyses. The pLXSN-ITGB4 construct used to create the SUM159lo ITGB4OE cell line was generously provided by the laboratory of Arthur M. Mercurio (University of Massachusetts, Worcester, MA). pLenti-based constructs were packaged with the pMD2.G (VSVG) and psPAX2 plasmids (Addgene 12259 and 12260, respectively). pLXSN-ITGB4 was packaged with pUMVC (Addgene 8449) and pMD2.G. Viral infections were performed using 6 μg/mL protamine sulfate for 8 h.
FACS Analyses and Sorting.
Cells were prepared for sorting following trypsinization and quenching in Dulbecco's modified Eagle's medium (DME) supplemented with 10% (vol/vol) inactivated fetal calf serum (IFS). For FACS analyses and sorting, cells were resuspended in ice-cold PBS− + 2% IFS at 1 × 106 cells per 100 μL. FACS antibodies were added with a 1:100 dilution. All FACS-sorted cell populations were obtained by starting with the top and bottom 2.5% of the ITGB4 FACS histograms for the first round, followed by 5 and 25% cutoffs for the second and third rounds of sorting. Cell lines were allowed a period of at least two passages after the final sort to minimize nonspecific differences between the populations attributed to the process of sorting. Antibodies used for FACS sorting and analyses are outlined in SI Appendix. Aldefluor assays were conducted essentially as described by the manufacturer (Stemcell Technologies; 01700).
Proliferation and Tumorsphere Assays.
Proliferation assays were conducted in 96-well plates using CyQuant (Thermo Fisher Scientific, C7026), according to the manufacturer’s recommendations, to measure DNA content in each well during a 4-d time course. The first day after seeding was counted as T = 0 and used for normalization of values obtained from plates collected at subsequent time points. Tumorsphere assays were conducted using the MammoCult Medium Kit (Stemcell Technologies, 05620) supplemented with 4 μg/mL heparin, 0.48 μg/mL hydrocortisone, penicillin/streptomycin, and 1% methylcellulose.
Tumor Induction and Limiting Dilution Analyses.
Orthotopic xenografts were performed via injection into the fourth inguinal mammary fat pad of NOD/SCID mice using cells resuspended in a 50% growth factor reduced Matrigel and PBS solution. Mice were monitored twice a week for a total of 8 wk following the procedure. Tumor initiating cell frequencies were calculated using the extreme limiting dilution analysis (ELDA) algorithm as previously described (41). Tumors were collected and sliced into ∼2.5-mm sections and fixed in 10% neutral buffered formalin. H&E-stained 5-μM sections of fixed tumor tissues were imaged using a Leica Biosystems Aperio digital slide scanner.
Western Blot Analyses.
To prepare protein, cells were washed twice with ice-cold PBS− and placed on ice. Ice-cold lysis buffer [50 mM Tris pH 7.5, 150 mM NaCl, 10 mM EDTA pH 8.0, 0.2% sodium azide, 50 mM NaF, 0.5% Nonidet P-40, proteinase inhibitor mixture (1:100; Sigma P8340), phosphatase mixture 2 (1:100; Sigma P5726), and phosphatase inhibitor mixture 3 (1:100; Sigma P0044)] was used to prepare protein lysates. Western blots were run using 1× MOPS buffer and NuPAGE Novex 4–12% Bis-Tris gels as described by the manufacturer (Thermo Fisher Scientific) and transferred to PVDF membranes. Antibodies and conditions are outlined in SI Appendix.
RNA Preparation, Quantitative Real-Time PCR, and RNA-Seq.
Total RNA was prepared using a modified TRIzol (Thermo Fisher Scientific) protocol. To prepare total RNA, 1 mL of TRIzol reagent was added to a subconfluent 10-cm dish of cells, scraped, then placed in a microfuge tube. Microfuge tubes were flash frozen on dry ice. Samples were then thawed and 200 μL of chloroform was added to each tube, shaken vigorously by hand for 15 s, and incubated at room temperature for 3 min, then centrifuged at 13,000 × g for 15 min. The aqueous phase was removed and placed into a gDNA elimination column from the RNeasy Plus kit (Qiagen). The eluate was mixed at a 1:1 ratio with 70% RNase-free EtOH and purified following the remaining steps outlined in the RNeasy Plus kit. cDNA was prepared using the Applied Biosystems kit (4368814) as described by the manufacturer with an RNase inhibitor (New England Biolabs, M0297L), and using OligodT primers (Thermo Fisher Scientific, NC9564171). Quantitative real-time PCR was performed using a Roche Diagnostics LightCycler 480 II and SYBR Green Mastermix (Roche Diagnostics, 04887352001). Primers used for analysis are outlined in SI Appendix. RNA-seq libraries were prepared using the TruSeq-stranded polyA mRNA kits as described by the manufacturer (Illumina, RS-122-2101). Libraries were pooled and sequenced using a HiSEq 2500. RNA-seq reads from Illumina 1.5 encoding were aligned using TopHat (v 2.0.13) (42) to the human genome (GRCh37) with Ensembl annotation (GRCh37.75) in gtf format. Differential expression was assayed using HTSeq count (43) with parameters: -m intersection-strict, - -strand=reverse, and DESeq2 (44). Cluster3 was used to perform unsupervised hierarchical clustering and the results were visualized using Java Treeview (45, 46).
Bioinformatic Analyses.
Primary patient survival correlations for TNBC and molecular basal subtype breast cancer were performed using normalized gene expression data from METABRIC (29) and obtained from the publicly available European Genome-Phenome Archive (IDs EGAD00010000210 and EGAD0001000021) (47). Detailed analytical methods for the METABRIC comparisons are outlined in SI Appendix. A secondary validation of METABRIC data and analyses of survival in lung adenocarcinoma, stage 4 ovarian cancer, and gastric cancer were performed using the kmplot tool (30). Detailed analytical methods for the conditions used for these comparisons are outlined in SI Appendix.
Small Molecule Screening.
Small molecule screening was conducted essentially as previously described (48). Briefly, cells were plated at a density of 1 × 103 cells per well in 384-well opaque, white assay plates (Corning) with a volume of 50 μL per well. Cells were incubated overnight and compounds were added the next day. Compound stocks from the Cambridge Cancer Collection (Selleck Chemicals) were plated in the 384-well format using five-point, 10-fold concentration ranges, starting at 10 μM. A total of 50 nL of compounds were pin transferred (V&P Scientific, pin tool mounted on a Tecan Freedom Evo 150 MCA96 head, Tecan) into duplicate assay plates and incubated for 72 h. DMSO content was 0.1% within each well. On each plate, 16 wells of DMSO vehicle control were used for normalization. After 3 d of incubation, 10 μL of CellTiter-Glo (Promega) was added to each well, incubated for 10 min and the luminescence output was read on an M1000 Infinite Pro plate reader (Tecan). The entire experiment was independently repeated twice (total of four compound plate replicates for each cell line) to compile the results as reported.
Supplementary Material
Acknowledgments
We thank Richard Goldsby, Arthur Lambert, Jordan Krall, Anushka Dongre, Asaf Spiegel, Sonia Iyer, Tsukasa Shibue, Xin Ye, Julia Frose, Jasmine DeCock, and Yun Zhang for essential comments and suggestions during the course of this work, as well as the Flow Cytometry Core Facility at the Whitehead Institute for Biomedical Research. This work was funded by grants from the US Department of Defense Breast Cancer Research Program (W81XWH-10-1-0647 to B.B.), C. J. Martin Overseas Biomedical Fellowship from the National Health and Medical Research Council of Australia (NHMRC APP1071853 to D.R.P.), K99/R00 Pathway to Independence Award [NIH/National Cancer Institute (NCI) 1K99CA201574-01A1 to D.R.P.], and NCI (5R01CA078461-17 and 5P01CA080111-18 to R.A.W.). R.A.W. is an American Cancer Society Research Professor and a Daniel K. Ludwig Foundation Cancer Research Professor.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE95540).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1618298114/-/DCSupplemental.
References
- 1.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166(1):21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 2.Peinado H, Olmeda D, Cano A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat Rev Cancer. 2007;7(6):415–428. doi: 10.1038/nrc2131. [DOI] [PubMed] [Google Scholar]
- 3.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6(5):392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 4.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100(7):3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pattabiraman DR, Weinberg RA. Tackling the cancer stem cells: What challenges do they pose? Nat Rev Drug Discov. 2014;13(7):497–512. doi: 10.1038/nrd4253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zadran S, Arumugam R, Herschman H, Phelps ME, Levine RD. Surprisal analysis characterizes the free energy time course of cancer cells undergoing epithelial-to-mesenchymal transition. Proc Natl Acad Sci USA. 2014;111(36):13235–13240. doi: 10.1073/pnas.1414714111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JM, Dedhar S, Kalluri R, Thompson EW. The epithelial-mesenchymal transition: New insights in signaling, development, and disease. J Cell Biol. 2006;172(7):973–981. doi: 10.1083/jcb.200601018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mani SA, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medema JP. Cancer stem cells: The challenges ahead. Nat Cell Biol. 2013;15(4):338–344. doi: 10.1038/ncb2717. [DOI] [PubMed] [Google Scholar]
- 11.Vivanco MdM . Mammary Stem Cells: Methods and Protocols. Humana Press; New York: 2015. pp x, 275 pp. [Google Scholar]
- 12.Bauer KR, Brown M, Cress RD, Parise CA, Caggiano V. Descriptive analysis of estrogen receptor (ER)-negative, progesterone receptor (PR)-negative, and HER2-negative invasive breast cancer, the so-called triple-negative phenotype: A population-based study from the California cancer Registry. Cancer. 2007;109(9):1721–1728. doi: 10.1002/cncr.22618. [DOI] [PubMed] [Google Scholar]
- 13.Stewart RL, O’Connor KL. Clinical significance of the integrin α6β4 in human malignancies. Lab Invest. 2015;95(9):976–986. doi: 10.1038/labinvest.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leng C, et al. An integrin beta4-EGFR unit promotes hepatocellular carcinoma lung metastases by enhancing anchorage independence through activation of FAK-AKT pathway. Cancer Lett. 2016;376(1):188–196. doi: 10.1016/j.canlet.2016.03.023. [DOI] [PubMed] [Google Scholar]
- 15.Bertotti A, Comoglio PM, Trusolino L. Beta4 integrin activates a Shp2-Src signaling pathway that sustains HGF-induced anchorage-independent growth. J Cell Biol. 2006;175(6):993–1003. doi: 10.1083/jcb.200605114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu S, Simin K, Khan A, Mercurio AM. Analysis of integrin beta4 expression in human breast cancer: Association with basal-like tumors and prognostic significance. Clin Cancer Res. 2008;14(4):1050–1058. doi: 10.1158/1078-0432.CCR-07-4116. [DOI] [PubMed] [Google Scholar]
- 17.Masugi Y, et al. Upregulation of integrin β4 promotes epithelial-mesenchymal transition and is a novel prognostic marker in pancreatic ductal adenocarcinoma. Lab Invest. 2015;95(3):308–319. doi: 10.1038/labinvest.2014.166. [DOI] [PubMed] [Google Scholar]
- 18.Stewart RL, et al. Elevated integrin α6β4 expression is associated with venous invasion and decreased overall survival in non-small cell lung cancer. Hum Pathol. 2016;54:174–183. doi: 10.1016/j.humpath.2016.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehmann BD, et al. Refinement of triple-negative breast cancer molecular subtypes: Implications for neoadjuvant chemotherapy selection. PLoS One. 2016;11(6):e0157368. doi: 10.1371/journal.pone.0157368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lehmann BD, et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest. 2011;121(7):2750–2767. doi: 10.1172/JCI45014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elenbaas B, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15(1):50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tam WL, et al. Protein kinase C α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell. 2013;24(3):347–364. doi: 10.1016/j.ccr.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Morel AP, et al. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One. 2008;3(8):e2888. doi: 10.1371/journal.pone.0002888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jolly MK, et al. Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front Oncol. 2015;5:155. doi: 10.3389/fonc.2015.00155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo W, et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148(5):1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ye X, et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature. 2015;525(7568):256–260. doi: 10.1038/nature14897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen TO, et al. A comparison of PAM50 intrinsic subtyping with immunohistochemistry and clinical prognostic factors in tamoxifen-treated estrogen receptor-positive breast cancer. Clin Cancer Res. 2010;16(21):5222–5232. doi: 10.1158/1078-0432.CCR-10-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parker JS, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Curtis C, et al. METABRIC Group The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486(7403):346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Györffy B, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 31.Carroll DK, et al. p63 regulates an adhesion programme and cell survival in epithelial cells. Nat Cell Biol. 2006;8(6):551–561. doi: 10.1038/ncb1420. [DOI] [PubMed] [Google Scholar]
- 32.Drake JM, et al. ZEB1 coordinately regulates laminin-332 and beta4 integrin expression altering the invasive phenotype of prostate cancer cells. J Biol Chem. 2010;285(44):33940–33948. doi: 10.1074/jbc.M110.136044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grigore AD, Jolly MK, Jia D, Farach-Carson MC, Levine H. Tumor budding: The name is EMT. Partial EMT. J Clin Med. 2016;5(5):E51. doi: 10.3390/jcm5050051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jolly MK, et al. Stability of the hybrid epithelial/mesenchymal phenotype. Oncotarget. 2016;7(19):27067–27084. doi: 10.18632/oncotarget.8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Taube JH, et al. Core epithelial-to-mesenchymal transition interactome gene-expression signature is associated with claudin-low and metaplastic breast cancer subtypes. Proc Natl Acad Sci USA. 2010;107(35):15449–15454. doi: 10.1073/pnas.1004900107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pattabiraman DR, et al. Activation of PKA leads to mesenchymal-to-epithelial transition and loss of tumor-initiating ability. Science. 2016;351(6277):aad3680. doi: 10.1126/science.aad3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chang C, Yang X, Pursell B, Mercurio AM. Id2 complexes with the SNAG domain of Snai1 inhibiting Snai1-mediated repression of integrin β4. Mol Cell Biol. 2013;33(19):3795–3804. doi: 10.1128/MCB.00434-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Turner FE, et al. Slug regulates integrin expression and cell proliferation in human epidermal keratinocytes. J Biol Chem. 2006;281(30):21321–21331. doi: 10.1074/jbc.M509731200. [DOI] [PubMed] [Google Scholar]
- 39.Ginestier C, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1(5):555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods. 2014;11(8):783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hu Y, Smyth GK. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J Immunol Methods. 2009;347(1-2):70–78. doi: 10.1016/j.jim.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 42.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Anders S, Pyl PT, Huber W. HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31(2):166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20(9):1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 46.Saldanha AJ. Java Treeview: Extensible visualization of microarray data. Bioinformatics. 2004;20(17):3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- 47.Lappalainen I, et al. The European Genome-Phenome Archive of human data consented for biomedical research. Nat Genet. 2015;47(7):692–695. doi: 10.1038/ng.3312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Basu A, et al. An interactive resource to identify cancer genetic and lineage dependencies targeted by small molecules. Cell. 2013;154(5):1151–1161. doi: 10.1016/j.cell.2013.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.