Significance
Retroviruses copy their RNA genome into complementary DNA, which is then inserted into the host chromosomal DNA as an obligatory part of their life cycle. Such integrated viral sequences, called proviruses, are passed to the infected cell progeny on cellular division. If germline cells are targeted, the proviruses become vertically inherited as other host genes and are called endogenous retroviruses. Deltaretroviruses, which include important human and veterinary pathogens (HTLV-1 and BLV), are the last retroviral genus for which endogenous forms were not known. We have identified a case of endogenous Deltaretrovirus, which entered the genome of long-fingered bat ancestors more than 20 million years ago. This finding opens the way for elucidating the deep evolutionary history of deltaretroviruses.
Keywords: Deltaretroviruses, endogenous retroviruses, Chiroptera
Abstract
Retroviruses can create endogenous forms on infiltration into the germline cells of their hosts. These forms are then vertically transmitted and can be considered as genetic fossils of ancient viruses. All retrovirus genera, with the exception of deltaretroviruses, have had their representation identified in the host genome as a virus fossil record. Here we describe an endogenous Deltaretrovirus, identified in the germline of long-fingered bats (Miniopteridae). A single, heavily deleted copy of this retrovirus has been found in the genome of miniopterid species, but not in the genomes of the phylogenetically closest bat families, Vespertilionidae and Cistugonidae. Therefore, the endogenization occurred in a time interval between 20 and 45 million years ago. This discovery closes the last major gap in the retroviral fossil record and provides important insights into the history of deltaretroviruses in mammals.
Deltaretroviruses are a highly unusual genus of retroviruses (family Retroviridae) that have only been identified in a restricted subset of mammalian species. They include the primate T-cell lymphotropic viruses that infect apes (including humans) and Old World monkeys, as well as the bovine leukemia virus (BLV) that infects cattle. Deltaretrovirus infections are usually asymptomatic, but can cause inflammatory and malignant disease over the longer term. For example, in humans, infection with human T-lymphotropic virus type 1 (HTLV-1) can cause adult T-cell lymphoma/leukemia or HTLV-1-associated myelopathy/tropical spastic paraparesis (1, 2). In cattle, BLV can cause persistent lymphocytosis or leukemia/lymphoma (3, 4).
Understanding of deltaretroviruses is limited by the lack of an endogenous fossil record for this genus (5, 6). Retroviruses are distinguished by a replication strategy in which a DNA copy of the viral genome (a form called a provirus) is integrated into the nuclear genome of the host cell. As a consequence, retroviral infection of germline cells can lead to retroviral proviruses being vertically inherited as host alleles, called endogenous retroviruses (ERVs). Vertebrate genomes typically contain thousands of ERVs, many of which are derived from retroviruses that circulated millions of years ago. These sequences constitute a partial historical record of the retroviruses that have been encountered by vertebrate species during their evolution (7). However, despite many vertebrate genomes having been sequenced, ERVs derived from deltaretroviruses have yet to be identified. Here we describe an endogenous Deltaretrovirus, identified in the genome of long-fingered bats (Miniopteridae).
Results
While systematically screening mammalian genomes for ERVs (8), we detected a sequence in the genome of the Natal long-fingered bat (Miniopterus natalensis) (9) that disclosed highly significant similarity to Deltaretrovirus group-specific antigen (Gag) proteins. This sequence was identified in a single large contig (GenBank accession no. LDJU01000221, 2.6 megabase long) and was flanked by the paired long terminal repeat (LTR) sequences characteristic of retroviral proviruses. A 6-bp target site duplication sequence (GCCCCC) was identified immediately upstream and downstream of the proviral insertion. We performed manual analysis of raw reads from published Miniopterus natalensis sequencing projects to accurately recover this proviral locus (Methods). In addition, we used PCR to confirm the presence of the provirus in the M. natalensis genome, as well as in four other Miniopterus species. Complete proviruses from all five Miniopterus species were sequenced and submitted to GenBank (accession numbers KY250075–KY250079). We named the provirus Miniopterus endogenous retrovirus (MINERVa).
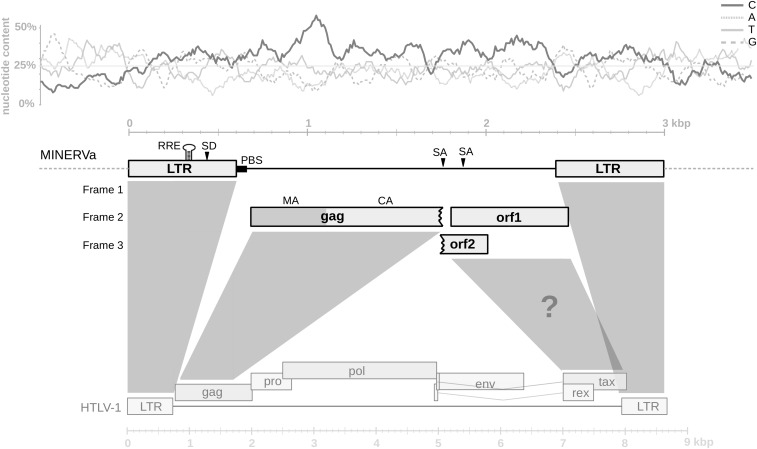
The orthologous proviruses obtained from the five miniopterid species were almost identical, differing only by several substitutions and indels. For description of the provirus, we generated a majority rule consensus sequence. The consensus MINERVa genome comprises a 1,789-bp internal region flanked by 604-bp LTRs (Fig. 1). Notably, the entire MINERVa sequence exhibits the characteristic nucleotide composition bias of deltaretroviruses (10), with cytosine (C) strongly overrepresented (Fig. 1). A primer binding site specific for proline tRNA is present in the internal region, immediately downstream of the 5′ LTR. The internal region encodes three relatively long ORFs, the first of which is 376 amino acids (aa) in length and encodes a truncated gag gene containing putative matrix (MA) and capsid (CA) proteins (Fig. 1 and Fig. S1). The predicted MA and CA amino acid sequences showed 30.8% and 45.4% identity (44.5% and 60.2% similarity), respectively, to those of HTLV-1. The entire putative gag ORF was translated and the resulting sequence aligned with Gag proteins of representative retroviruses. In phylogeny constructed using this alignment, the MINERVa Gag grouped robustly within the Deltaretrovirus clade, forming a basal branch separate from both the BLV and the primate T-cell lymphotropic virus groups (Fig. 2).
Fig. 1.
Genome organization of MINERVa. The consensus sequence of MINERVa is shown schematically in scale. The position of ORFs and other genomic features are indicated. The comparison with HTLV-1 genome structure is shown (for clarity, some HTLV-1 genes were omitted). The question mark indicates the putative accessory gene region. Nucleotide composition of MINERVa sequence and of 500-bp flanking regions is plotted in scale above the proviral scheme (calculated in 100-bp-long windows with 10% overlaps). RRE, rex response element; PBS, primer binding site; SD, splice donor site; SA, splice acceptor site.
Fig. S1.
MINERVa consensus sequence annotation. The annotation is based on sequence similarities to other deltaretroviruses; the splice sites were predicted using the NNSPLICE 0.9 algorithm (www.fruitfly.org/seq_tools/splice.html); results with score >0.80 are shown. The 3′ part of putative gag ORF with undetectable homology was not annotated. ORFs 1/2 do not begin with start codons because 5′ ends of deltaretroviral accessory genes are known to be located in a distant region of the proviral genome and intact ORFs are formed by splicing.
Fig. 2.
Phylogenetic relationship of MINERVa to other retroviruses. The ML phylogeny of partial Gag amino acid sequences is shown. Bootstrap supports for each branch are depicted. An arrow highlights the position of MINERVa consensus sequence. Branches corresponding to the viruses of particular retroviral genera are collapsed, with the exception of deltaretroviruses. Scale bar indicates number of amino acid substitutions per site.
The second and third long ORFs (ORF1 and ORF2 in Fig. 1) are 220 and 88 aa long, respectively, both beginning immediately downstream of the truncated gag ORF. They overlap each other, and ORF1 is extending a short distance (76 bp) into the 3′ LTR. We could not detect homology to any of the genetic elements that would be expected to occur immediately downstream of capsid [nucleocapsid (NC), protease (PR), polymerase (pol) and envelope (env)], or indeed to any known proteins. One way this result might be expected would be if a region spanning from the end of gag, through PR and pol and up to the end of env, had been deleted (Fig. 1). A deletion event of this nature would leave behind a truncated gag gene plus the 3′ region of the provirus genome, which is situated downstream of env and contains multiple overlapping accessory genes. The absence in MINERVa of any homology to the relatively conserved transmembrane domain, which occurs toward the C terminus of the Env protein, is consistent with this having occurred. In all previously characterized deltaretroviruses, the 3′ region of the genome encodes accessory genes, including the regulators of gene expression tax and rex, in addition to other, less well characterized genes (Fig. 1) (11).
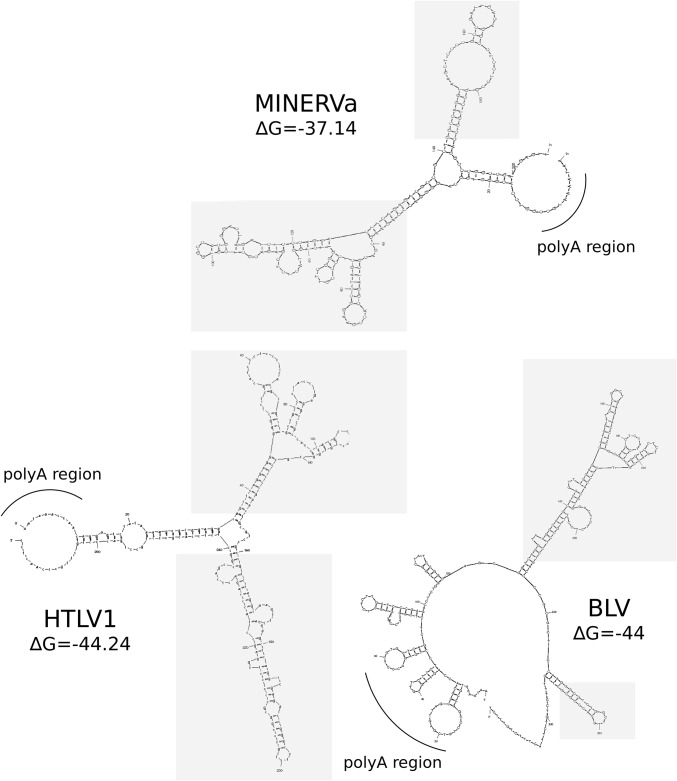
Additional, independent lines of evidence support the inference that ORF1/ORF2 are accessory genes derived from the 3′ region of the ancestral MINERVa genome. First, we detected the characteristic nucleotide bias of deltaretroviruses across the entire MINERVa genome, including the region that encodes ORF1/ORF2, consistent with this region being derived from an ancestral Deltaretrovirus (as opposed to insertion of genome-derived sequence into the MINERVa provirus after its integration, for example). The MINERVa genome also exhibited features that were consistent with ORF1/ORF2 being bona fide retroviral genes with a role in regulation of gene expression. These included the presence of a leucine-rich region toward the C terminus of the predicted orf1 gene product, as is found in the tax gene of primate and bovine deltaretroviruses (12, 13); the presence of a predicted stem-loop (Fig. S2) in the region of the LTR that would be expected to encode a Rex-responsive element (presuming that MINERVa replication was regulated in a similar posttranscriptional manner to other deltaretroviruses); and the fact that ORF1/ORF2 overlap, resembling the tax/rex overlap in other deltaretroviruses (12, 14).
Fig. S2.
Secondary structure prediction of MINERVa putative RRE. Stem loop prediction in the 5′ LTR in MINERVa is compared with other deltaretroviruses. The sequences used for secondary structure prediction in mfold (unafold.rna.albany.edu/) were MINERVa consensus sequence, HTLV-1 (GenBank accession no. M37299), and BLV (K02120). The predicted RRE (RexRE) stem-loop structures are highlighted in gray.
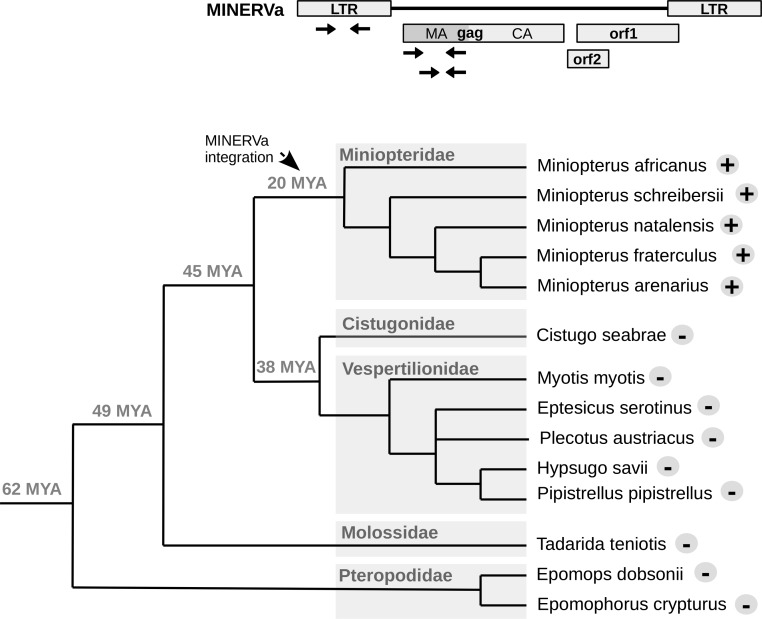
We performed a series of analyses to determine the distribution of the MINERVa sequences in bats. From our in silico genome screening, it was already clear that the sequence was absent in more distantly related bat groups, such as Old World fruit bats (family Pteropodidae), as well in some more closely related lineages (i.e., Myotis spp.). To investigate the presence of MINERVa more thoroughly, we performed a PCR-based screen of the phylogenetically closest bat species. In addition to the Miniopterus specimens already mentioned, samples were obtained from nine bat species from other families (Vespertilionidae, Cistugonidae, Molossidae and Pteropodidae; Table S1). The species designation of these samples was confirmed by amplification and phylogenetic analysis of cytB or rag2 genes (Methods). PCR primers designed to target three different amplicons in provirus (Table S2) confirmed that the MINERVa insertion is present in all miniopterid bat species examined (Fig. 3). MINERVa could be detected neither in species belonging to the most closely related bat families (Vespertilionidae and Cistugonidae) nor in more distant species.
Table S1.
List of the bat species analyzed
| Scientific name | Common name | Family | Origin |
| Miniopterus schreibersii | Schreibers’ long-fingered bat | Miniopteridae | Greece |
| Miniopterus natalensis | Natal long-fingered bat | Miniopteridae | Namibia |
| Miniopterus fraterculus | Lesser long-fingered bat | Miniopteridae | South Africa |
| Miniopterus arenarius | Sandy long-fingered bat | Miniopteridae | Ethiopia |
| Miniopterus africanus | African long-fingered bat | Miniopteridae | Ethiopia |
| Cistugo seabrae | Angolan wing-gland bat | Cistugonidae | Namibia |
| Myotis myotis | Greater mouse-eared myotis | Vespertilionidae | Spain |
| Eptesicus serotinus | Common serotine | Vespertilionidae | Spain |
| Hypsugo savii | Savi’s pipistrelle | Vespertilionidae | Spain |
| Pipistrellus pipistrellus | Common pipistrelle | Vespertilionidae | Italy |
| Plecotus austriacus | Gray long-eared bat | Vespertilionidae | Spain |
| Tadarida teniotis | European free-tailed bat | Molossidae | Cyprus |
| Epomops dobsonii | Dobson’s fruit bat | Pteropodidae | Namibia |
| Epomophorus crypturus | Peters’ epauletted fruit bat | Pteropodidae | Namibia |
Table S2.
Primer sequences used in this study
| Primer designation | Sequence (5′→3′) | Primer localization |
| F1 | GACAAGGGTCGAGTCACCTCCTAA | MINERVa gag |
| F2 | AATCTCTCCTTCTGGCCTCTCACA | MINERVa gag |
| F6 | ATTCATGAGGTGCACGTTTAAGCA | 5′flanking region of MINERVa provirus |
| F8 | TATGTTTCCCCATACCTTGCCATCA | MINERVa LTR |
| R1 | GAGGTCGCAGGGTTATATGGAGGT | MINERVa gag |
| R4 | GGCATCAAAAGGTAAACAGAAGCA | 3′flanking region of MINERVa provirus |
| R5 | CATGGTTCCACTGGTTATCATTTACA | 3′flanking region of MINERVa provirus |
| R6 | CAATCGGCGGGGAGCTTAC | MINERVa LTR |
| F5 | GGTGCACGTTTAAGCACATACTCG | 5′flanking region of MINERVa provirus |
| CYTB1 | GTTGCTCCTCAGAAAGATATTTGTCCTC | Miniopterus cytochrome B locus |
| CYTB2 | ATGACCTGTGATATGAAAAACCACTGTTG | Miniopterus cytochrome B locus |
| F4 | GTTGGTTGCTCTCTTGCC TAGTCG | MINERVa LTR |
| F10 | GGAATACCCGTTTCAGAGAGCAGA | Miniopterus genomic locus 1 |
| R9 | TGATCCCTGAGATGACAGAAGTCG | Miniopterus genomic locus 1 |
| F9 | TTCAGTATTGTGAAAGGGCTCTGC | Miniopterus genomic locus 2 |
| R8 | TCACTCTCTGGCTTTAGAGTCCTTCA | Miniopterus genomic locus 2 |
| F7 | TCATGTAAATGATAACCAGTGGAACC | Miniopterus genomic locus 3 |
| R7 | TGCAATGTGAGTTGTTGAAAGTGAAA | Miniopterus genomic locus 3 |
Fig. 3.
The presence of MINERVa in various species of bats. Three primer pairs used for PCR screening of selected bat DNA samples are depicted in MINERVa schematics. Results of the screen are shown in a phylogenetic tree of bat species. Plus sign next to the species name indicates positivity for all three MINERVa amplicons; minus sign indicates negativity for all three amplicons. Dating of split of selected species are shown next to the branch nodes [timetree.org estimates (15)].
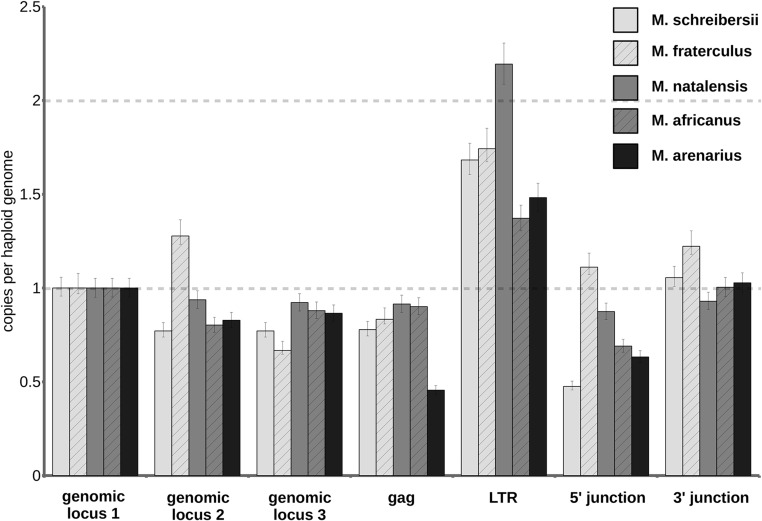
To confirm that only a single MINERVa integration is present in all five of the miniopterid species examined (i.e., two alleles per diploid genome), we used digital droplet PCR (ddPCR; Fig. S3). All these single-copy MINERVa insertions in distinct Miniopterus species are clearly orthologous, sharing >124 bp of homologous flanking sequence on either site of the proviral integration site. The five Miniopterus bats included in our study are estimated to have diverged ∼20 million years ago (MYA) (15, 16), establishing that genomic infiltration occurred before this date. The absence of related virus sequences in all members of the sister families Vespertilionidae and Cistugonidae examined indicates that invasion is unlikely to have occurred more than 45 MYA (17). Thus, we estimate that MINERVa entered the germline of miniopterid bat ancestors at some point in the period spanning 45–20 MYA (Fig. 3).
Fig. S3.
MINERVa copy number determination in miniopterid species. The chart shows copy numbers of four loci in MINERVa provirus (gag, LTR, virus–host junctions), and three control Miniopterus genomic regions. Copy numbers were determined by ddPCR absolute quantification, and all values were normalized to genomic locus 1. The error bars represent Poisson 95% confidence intervals of ddPCR analysis. Approximately one copy of gag and two copies of LTR amplicon per haploid genome equivalent indicate that a single copy of MINERVa sequence is present in all miniopterid species. Genomic locus 1, distant region in the MINERVa-containing contig (LDJU01000221); genomic locus 2, region close to the MINERVa 5′ end; genomic locus 3, region close to the MINERVa 3′ end.
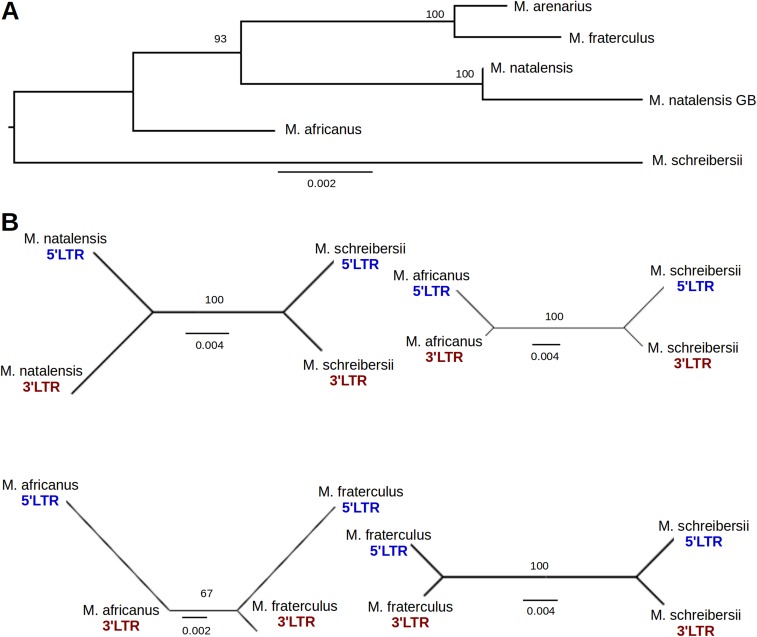
An alternative approach for estimating the age of proviral insertions is to determine the divergence between paired LTRs (which are identical at the time of integration) and apply a molecular clock (18). Taking in account the fact that ERV integration precedes the split of the host species, each proviral LTR pair should contain more changes (which accumulate from the time of integration) than orthologous LTR sequences from different species (which accumulate mainly after the species divergence). Analysis of MINERVa sequence from individual miniopterid species, however, showed that the divergence of LTR pairs in each proviral sequence is much lower than divergence between orthologous LTR sequences from some of the species analyzed (e.g., 5′ LTR form Miniopterus schreibersii is more similar to its 3′ LTR counterpart than to 5′ LTR of Miniopterus fraterculus) (Fig. S4). This observation provides compelling evidence that multiple gene conversion events, a phenomenon that has previously been described in ERVs (19, 20), have occurred between the 5′ and 3′ LTRs of individual proviruses. This fact is precluding the LTR-based approach of age estimation. In addition to phylogenetic evidence for gene conversion, we identified a 5-bp insertion that was unique to M. schreibersii MINERVa provirus, but present in both LTRs. This pattern of variation is extremely unlikely to be accounted for by any process other than gene conversion between the 5′ and 3′ LTRs.
Fig. S4.
Gene conversion between MINERVa 5′ and 3′ LTR sequences. (A) ML phylogeny of MINERVa internal sequence (LTRs were excluded). The phylogeny contains six MINERVa orthologous nucleotide sequences. M. natalensis GB represents sequence obtained from M. natalensis genome assembly. (B) Examples of MINERVa orthologs with evidence of gene conversion between 5′ and 3′ LTR sequences. ML trees of MINERVa LTRs are shown for four orthologous provirus pairs. The tree topology where 5′ and 3′ LTRs of the same provirus group together is indicative of gene conversion. Bootstrap supports are shown for each tree; scale bar indicates number of nucleotide substitutions per site.
The divergence of internal proviral sequences (excluding both LTRs) cannot yield a time estimate for virus integration. However, similar to other genomic loci, it should reflect the changes accumulated since the split of the miniopterid species analyzed. Miniopterus africanus split from the other miniopterid species around 20 MYA (Fig. 3). The average sequence divergence of M. africanus MINERVa provirus to its orthologs in other miniopterid species was found to be 1.31 ± 0.35% (mean ± SD), which corresponds to a substitution rate of 0.66 ± 0.18 substitutions/nucleotide/year. This falls within the range of mammalian neutral substitution rate estimates (21, 22).
Given the predicted age of the insertion, it was intriguing that the gag ORF was intact in five of the six MINERVa alleles. However, multiple simulations of MINERVa gag neutral evolution recapitulated this situation in 20% of cases (1,000,000 replicates; average number of sequences with at least one stop-codon = 2.52/6). Thus, there is no strong evidence of selection for gag coding sequence conservation, although this approach only considers nucleotide substitutions and not indels (23).
The genomic locus in M. natalensis in which MINERVa integrated does not contain any predicted genes. Orthologous loci, without the provirus, could be detected in several of the published bat genomes. The chromosomal location of the MINERVa locus could not be determined because none of the bat genomes is yet mapped to chromosomes (24).
Discussion
Exogenous retroviruses have been grouped into seven genera, only five of which are known to infect mammals. The discovery of MINERVa means that endogenous fossils have now been identified in mammalian genomes for all five of these retroviral genera. However, the representation of these five genera in the retroviral fossil record is very uneven. ERVs derived from retroviruses with simpler genome structures (Gammaretrovirus, Betaretrovirus) are relatively common, whereas only a handful have been identified for the Lentivirus, Spumavirus, and (now) Deltaretrovirus genera (5, 6). This could reflect inefficient entry of these viruses into germline cells, or inherent barriers to their germline replication (e.g., toxicity of gene products) (7). Notably, only a single MINERVa copy was identified. One possibility is that MINERVa was generated with the same, highly deleted genome structure that we see in all present-day copies and, being effectively “dead on arrival,” was fixed without any virus-driven increase in germline copy number (as is presumed to occur for endogenous sequences derived from nonretroviral viruses).
Deltaretrovirus is perhaps the most enigmatic of the five retroviral genera that infect mammals, and the discovery of MINERVa is therefore particularly illuminating. First, it provides unequivocal evidence that this genus has a truly ancient origin in mammals. We identified orthologous copies in miniopterid species that are estimated to have diverged ∼20 MYA, establishing that deltaretroviruses have been infecting mammals for at least this long. Previous studies have demonstrated that Deltaretrovirus infection in humans likely predates the last Ice Age (25), but MINERVa provides unequivocal evidence that Deltaretrovirus infection has affected mammals during a substantial part of their evolution.
The calibration of Deltaretrovirus evolution through the identification of a fossil sequence reveals that the characteristic features of this genus had already evolved by the early Miocene (∼23–16 MYA). These include the marked nucleotide-bias that is a hallmark of Deltaretrovirus genomes (10). Nucleotide biases are a feature of many retroviral genomes, but deltaretroviruses stand apart from all other retroviral genera in having C-rich genomes. The biological significance of these biases is uncertain, but the stability of this feature across Deltaretrovirus evolution suggests it represents an adaptation of some kind.
Complex regulation of genome expression is another characteristic feature of deltaretroviruses. Analysis of the MINERVa genome indicated that the ancestral progenitor likely encoded a region with accessory genes. The putative ORFs we identified in this region did not disclose homology to the Tax and Rex proteins of exogenous deltaretroviruses, but as these genes are relatively poorly conserved, this might be expected.
The discovery of MINERVa extends the known host range of the Deltaretrovirus genus to a new mammalian order (Chiroptera). It also raises the questions about the role of bats in Deltaretrovirus evolution. Traits associated with movement capacity are especially pronounced in miniopterids. Their most apparent apomorphy, elongated wings, presumably enabled them to colonize almost all tropic and subtropic regions of the Old World and become one of the most widespread mammalian genera (26, 27). They also concentrate in mass roosts of thousands of individuals in caves with high humidity, which could facilitate virus transmission. Conceivably, deltaretroviruses may infect bats in the present day. Searches of available metagenome and transcriptome datasets did not reveal any matches to MINERVa or other deltaretroviruses, but these data represent a relatively limited sample.
In conclusion, the identification MINERVa provides important insights into Deltaretrovirus evolution. It also fills a major gap in ERV record by eliminating the last retrovirus genus for which endogenous forms were not known.
Methods
Next-Generation Sequence Data Analysis.
Sequence datasets available from the Sequence Read Archive at the National Center for Biotechnology Information from miniopterid species genome or transcriptome (PRJNA270665, PRJNA270639, and PRJNA218524) were queried by BLAST (28) or downloaded and analyzed using CLC genomics workbench 9.5 (www.clcbio.com) or DNASTAR Lasergene 10.0.0 (www.dnastar.com). This initial analysis was used to correct errors in the original MINERVa-containing contig from the M. natalensis genome assembly.
Samples from Bats.
The bat tissue samples were obtained from museum specimens (National Museum Prague) as parts of the pectoral muscles and from released bats caught during various molecular ecology studies as wing punch biopsies stored in genetic bank (Charles University, Prague). The bat species were identified with respect to their external morphological traits and confirmed by amplification and sequencing of cytochrome b (cytb) or recombination activating gene (rag2) loci. Total DNA from the ethanol-preserved specimens was isolated using phenol-chloroform extraction method.
PCR and Sequencing.
The complete MINERVa provirus sequence was PCR-amplified using two strategies (primers listed in Table S2): a nested PCR approach with primers anchored in genomic flanking regions (primers F6 and R4 in first round, primers F5 and R5 second round), or in two overlapping parts using one primer anchored in the genomic flanking region and the second primer in the provirus sequence (5′ provirus part amplified using primers F6 and R1, 3′ provirus part using seminested PCR with primers F6 and R4 in first round and F1 and R4 in second round). PCR products were isolated from agarose gels and directly sequenced. The cytb locus was amplified using primers cytBMVZ04 and cytBMVZ05 (29), the rag2 locus using primers 968R and 428F (30). In some Miniopterus specimens, the cytb locus was amplified using primers CYTB1 and CYTB2 (Table S2). To assess the presence of MINERVa sequence in various bat species, two amplicons in the gag gene (primers F1 and R1, or F2 and R1) and one amplicon in LTR (F8 and R6) were used.
Provirus Copy-Number Determination.
ddPCR system QX200 (Bio-Rad) was used to accurately quantify the MINERVa proviral copies in miniopterid samples. Template genomic DNAs were first digested with SacI restriction endonuclease to prevent the occurrence of two LTR sequences in one molecule. The reactions containing 10 ng DNA were then treated for droplet generation according to the manufacturer's manual and PCR-amplified. The amplified samples were analyzed by droplet reader and QuantaSoft program (Bio-Rad) with thresholds set manually. Primers used for ddPCR (Table S2) were F2 and R1 (MINERVa gag region), F8 and R6 (MINERVa LTR), F5 and R6 (5′ provirus-genome junction), F4 and R5 (3′ provirus-genome junction), F10 and R9 (genomic locus 1), F9 and R8 (genomic locus 2), and F7 and R7 (genomic locus 3).
Phylogenetic Analysis.
Translated nucleotide sequences of the MINERVa gag consensus and other retroviral gag sequences were aligned using the MAFFT v7.271 with l-INS-i algorithm (31). Columns containing more than 80% of gaps were discarded, resulting in an alignment with a total of 644 positions. Maximum likelihood (ML) phylogeny was generated using PhyML v3.0 (32). LG model with gamma distribution (four categories) of rates among sites was used as a best-fitting substitution model (according to the Akaike Information Criterion calculated in Smart Model Selection module of PhyML). The SPR operations in an optimized BioNJ starting tree were used for searching of the final tree. Bootstrap support for each node was evaluated with 1,000 replicates. The accession numbers of gag sequences used are: RSV (NP_056886), ALV (BAK64245), MPMV (NP_056893), JSRV (AAD45224), SIV2 (AAA47561), FLV (NP_955576), MLV (NP_057933), BLV (NP_056897), HTLV-1 (BAA02929), HTLV-2 (AAB59884), HTLV-3 (ACF40912), HTLV-4 (YP_002455784), STLV-1 (AAU34008), STLV-2 (YP_567048), STLV-3 (CAA68892), STLV-4 (AHH34968), VISNA (NP_040839), FIV (NP_040972), HIV (AAB50258), WDSV (AAC82607), WEHV-1 (AAD30047), SFV (NP_056802), and BFV (AFR79238). The same software was used for phylogenetic inference of MINERVa LTRs and internal nucleotide sequences. Kimura 2-parameter (K80) model with gamma distribution (four categories) of rates among sites was used as a substitution model. The transition/transversion ratio was assumed to be 4. SPR operations in optimized BioNJ starting tree were used for searching of the final tree. Bootstrap support for each node was evaluated with 1,000 replicates.
Simulation of Neutral Evolution.
The probability of MINERVa gag ORF disruption was evaluated by simulating the gag sequence evolution, using Seq-Gen v1.3.3 (33). The gag ancestral sequence and ML phylogeny was inferred from six MINERVa orthologous copies in Miniopteridae. The transition/ transversion ratio was assumed to be 4. The presence of premature stop-codons in simulated gag orthologs was counted for 1,000,000 iterations.
Acknowledgments
We are grateful to Vladimír Pečenka for advice on PCR amplifications. We thank Eastern Cape Parks and Tourism Agency (ECPTA) for assistance in the field. This work was supported by the Czech Ministry of Education, Youth, and Sports under the program NÁVRAT (LK11215) and by Medical Research Council (MRC) Grant MC_UU_12014/12. The work was also institutionally supported by RVO:68378050. Access to computing and storage facilities was provided by ELIXIR CZ research infrastructure project (MEYS Grant LM2015047).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession nos. KY250075–KY250079).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1621224114/-/DCSupplemental.
References
- 1.Poiesz BJ, et al. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77(12):7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coffin JM. The discovery of HTLV-1, the first pathogenic human retrovirus. Proc Natl Acad Sci USA. 2015;112(51):15525–15529. doi: 10.1073/pnas.1521629112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barez PY, et al. Recent advances in BLV research. Viruses. 2015;7(11):6080–6088. doi: 10.3390/v7112929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Miller JM, Miller LD, Olson C, Gillette KG. Virus-like particles in phytohemagglutinin-stimulated lymphocyte cultures with reference to bovine lymphosarcoma. J Natl Cancer Inst. 1969;43(6):1297–1305. [PubMed] [Google Scholar]
- 5.Hayward A, Cornwallis CK, Jern P. Pan-vertebrate comparative genomics unmasks retrovirus macroevolution. Proc Natl Acad Sci USA. 2015;112(2):464–469. doi: 10.1073/pnas.1414980112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayward A, Grabherr M, Jern P. Broad-scale phylogenomics provides insights into retrovirus-host evolution. Proc Natl Acad Sci USA. 2013;110(50):20146–20151. doi: 10.1073/pnas.1315419110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson WE. Endogenous retroviruses in the genomics era. Annu Rev Virol. 2015;2(1):135–159. doi: 10.1146/annurev-virology-100114-054945. [DOI] [PubMed] [Google Scholar]
- 8.Hron T, Fábryová H, Pačes J, Elleder D. Endogenous lentivirus in Malayan colugo (Galeopterus variegatus), a close relative of primates. Retrovirology. 2014;11:84. doi: 10.1186/s12977-014-0084-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eckalbar WL, et al. Transcriptomic and epigenomic characterization of the developing bat wing. Nat Genet. 2016;48(5):528–536. doi: 10.1038/ng.3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kypr J, Mrázek J, Reich J. Nucleotide composition bias and CpG dinucleotide content in the genomes of HIV and HTLV 1/2. Biochim Biophys Acta. 1989;1009(3):280–282. doi: 10.1016/0167-4781(89)90114-0. [DOI] [PubMed] [Google Scholar]
- 11.Yoshida M. Discovery of HTLV-1, the first human retrovirus, its unique regulatory mechanisms, and insights into pathogenesis. Oncogene. 2005;24(39):5931–5937. doi: 10.1038/sj.onc.1208981. [DOI] [PubMed] [Google Scholar]
- 12.Currer R, et al. HTLV tax: A fascinating multifunctional co-regulator of viral and cellular pathways. Front Microbiol. 2012;3:406. doi: 10.3389/fmicb.2012.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aida Y, Murakami H, Takahashi M, Takeshima SN. Mechanisms of pathogenesis induced by bovine leukemia virus as a model for human T-cell leukemia virus. Front Microbiol. 2013;4:328. doi: 10.3389/fmicb.2013.00328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pavesi A, Magiorkinis G, Karlin DG. Viral proteins originated de novo by overprinting can be identified by codon usage: Application to the “gene nursery” of Deltaretroviruses. PLOS Comput Biol. 2013;9(8):e1003162. doi: 10.1371/journal.pcbi.1003162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hedges SB, Dudley J, Kumar S. TimeTree: A public knowledge-base of divergence times among organisms. Bioinformatics. 2006;22(23):2971–2972. doi: 10.1093/bioinformatics/btl505. [DOI] [PubMed] [Google Scholar]
- 16.Lack JB, Roehrs ZP, Stanley CE, Ruedi M, Van Den Bussche RA. Molecular phylogenetics of Myotis indicate familial-level divergence for the genus Cistugo (Chiroptera) J Mammal. 2010;91:976–992. [Google Scholar]
- 17.Miller-Butterworth CM, et al. A family matter: Conclusive resolution of the taxonomic position of the long-fingered bats, miniopterus. Mol Biol Evol. 2007;24(7):1553–1561. doi: 10.1093/molbev/msm076. [DOI] [PubMed] [Google Scholar]
- 18.Johnson WE, Coffin JM. Constructing primate phylogenies from ancient retrovirus sequences. Proc Natl Acad Sci USA. 1999;96(18):10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kijima TE, Innan H. On the estimation of the insertion time of LTR retrotransposable elements. Mol Biol Evol. 2010;27(4):896–904. doi: 10.1093/molbev/msp295. [DOI] [PubMed] [Google Scholar]
- 20.Zhuo X, Feschotte C. Cross-species transmission and differential fate of an endogenous retrovirus in three mammal lineages. PLoS Pathog. 2015;11(11):e1005279. doi: 10.1371/journal.ppat.1005279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc Natl Acad Sci USA. 2002;99(2):803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perelman P, et al. A molecular phylogeny of living primates. PLoS Genet. 2011;7(3):e1001342. doi: 10.1371/journal.pgen.1001342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katzourakis A, Gifford RJ. Endogenous viral elements in animal genomes. PLoS Genet. 2010;6(11):e1001191. doi: 10.1371/journal.pgen.1001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang J, Wang X, Mu S, Zhang S, Dong D. BGD: A database of bat genomes. PLoS One. 2015;10(6):e0131296. doi: 10.1371/journal.pone.0131296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Switzer WM, et al. Ancient, independent evolution and distinct molecular features of the novel human T-lymphotropic virus type 4. Retrovirology. 2009;6:9. doi: 10.1186/1742-4690-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miller-Butterworth CM, Jacobs DS, Harley EH. Strong population substructure is correlated with morphology and ecology in a migratory bat. Nature. 2003;424(6945):187–191. doi: 10.1038/nature01742. [DOI] [PubMed] [Google Scholar]
- 27.Miller-Butterworth CM, Eick G, Jacobs DS, Schoeman MC, Harley EH. Genetic and phenotypic differences between south African long-fingered bats, with a global miniopterine phylogeny. J Mammal. 2005;86:1121–1135. [Google Scholar]
- 28.Johnson M, et al. NCBI BLAST: A better web interface. Nucleic Acids Res. 2008;36(Web Server issue):W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Smith MF, Patton JL. Variation in mitochondrial cytochrome b sequence in natural populations of South American akodontine rodents (Muridae: Sigmodontinae) Mol Biol Evol. 1991;8(1):85–103. doi: 10.1093/oxfordjournals.molbev.a040638. [DOI] [PubMed] [Google Scholar]
- 30.Stadelmann B, Lin LK, Kunz TH, Ruedi M. Molecular phylogeny of New World Myotis (Chiroptera, Vespertilionidae) inferred from mitochondrial and nuclear DNA genes. Mol Phylogenet Evol. 2007;43(1):32–48. doi: 10.1016/j.ympev.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 31.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30(4):772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59(3):307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 33.Rambaut A, Grassly NC. Seq-Gen: An application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput Appl Biosci. 1997;13(3):235–238. doi: 10.1093/bioinformatics/13.3.235. [DOI] [PubMed] [Google Scholar]