Significance
Quiescent bacteria are intrinsically resistant to antibiotics and the host immune response. A conserved bacterial starvation survival response is the consumption of ATP to make an inorganic polymer, polyphosphate (polyP), which then forms granule superstructures. PolyP granules occur in all three domains of life, yet how and why cells form these structures is poorly understood. Through high-resolution spatiotemporal characterization of de novo granule genesis, we find that polyP granule synthesis is required for and coordinated with cell cycle exit in the opportunistic pathogen Pseudomonas aeruginosa. PolyP has also been functionally connected with the cell cycle in eukaryotes, suggesting that polyP may be a broadly conserved mediator between metabolic state and the cell cycle.
Keywords: polyphosphate, nucleoid, cell cycle, starvation, biomineralization
Abstract
Polyphosphate (polyP) granule biogenesis is an ancient and ubiquitous starvation response in bacteria. Although the ability to make polyP is important for survival during quiescence and resistance to diverse environmental stresses, granule genesis is poorly understood. Using quantitative microscopy at high spatial and temporal resolution, we show that granule genesis in Pseudomonas aeruginosa is tightly organized under nitrogen starvation. Following nucleation as many microgranules throughout the nucleoid, polyP granules consolidate and become transiently spatially organized during cell cycle exit. Between 1 and 3 h after nitrogen starvation, a minority of cells have divided, yet the total granule number per cell decreases, total granule volume per cell dramatically increases, and individual granules grow to occupy diameters as large as ∼200 nm. At their peak, mature granules constitute ∼2% of the total cell volume and are evenly spaced along the long cell axis. Following cell cycle exit, granules initially retain a tight spatial organization, yet their size distribution and spacing relax deeper into starvation. Mutant cells lacking polyP elongate during starvation and contain more than one origin. PolyP promotes cell cycle exit by functioning at a step after DNA replication initiation. Together with the universal starvation alarmone (p)ppGpp, polyP has an additive effect on nucleoid dynamics and organization during starvation. Notably, cell cycle exit is temporally coupled to a net increase in polyP granule biomass, suggesting that net synthesis, rather than consumption of the polymer, is important for the mechanism by which polyP promotes completion of cell cycle exit during starvation.
Most of our understanding of bacterial physiology comes from laboratory studies of bacteria growing under nutrient-rich conditions. However, in many environments, bacteria face dramatic fluctuations in nutrient conditions, including long periods of scarcity when they survive in a nonproliferative stationary state. Although eukaryotic cells and some bacteria have discrete cell cycle checkpoints, many fast-growing bacterial species have uncoupled DNA replication and cell division. They use multifork DNA replication to achieve a doubling time that is faster than the time required to copy the chromosome, giving them a competitive edge in nutrient-replete conditions. This strategy requires distinct regulatory mechanisms and comes at a considerable cost when there is a rapid downshift in nutrient availability: stalled open DNA replication forks are vulnerable to potentially lethal double-stranded DNA breaks (1). Therefore, the ability to reallocate resources under such conditions to prioritize completion of DNA replication is critical for survival. Prioritizing chromosome remodeling and compaction by starvation-specific nucleoid structural proteins is also important during such transitions because resources for DNA repair become limited in deep starvation (2–4). Operating an uncoupled cell cycle, where growth, DNA replication, and cell division do not function in lockstep, requires regulatory mechanisms quite different from cell cycle checkpoints in eukaryotes.
Cell cycle exit in bacteria encompass four general steps: (i) inhibition of inappropriate reinitiation of DNA replication, (ii) completion of open rounds of DNA replication, (iii) segregation and compaction of daughter chromosomes, and (iv) septation. How bacteria orchestrate efficient cell cycle exit and protect the nucleoid during starvation remains poorly understood (5, 6). To date, the starvation signaling molecule guanosine tetraphosphate [(p)ppGpp] is the only known widely conserved regulator of cell cycle exit in bacteria and functions by a variety of mechanisms in different bacterial species, ranging from inhibiting replication initiation (7, 8) to modulating replication elongation rates (9, 10) and promoting septation (11). (p)ppGpp was also recently shown to play a role in transcription-coupled DNA repair (12). A related molecule, polyphosphate (polyP), is an inorganic polymer consisting simply of phosphoryl groups and can be hundreds of protomers in length. A recent study in Caulobacter crescentus demonstrated a role for polyP in cell cycle exit by inhibiting reinitiation of DNA replication during carbon starvation (13). Studies in diverse bacteria and under a variety of starvation conditions have shown that polyP promotes fitness during starvation (14). Although some bacteria appear to make polyP granules constitutively, many bacteria make polyP granules in response to nutrient limitation. Carbon, nitrogen, phosphate, and amino acid starvation have all been shown to drive polyphosphate synthesis in bacteria (15). Escherichia coli has also been found to synthesize polyP in response to stalled DNA replication forks during exponential growth (16). Bacterial mutants that are unable to make polyP die more rapidly when starved than those that can, and are more sensitive to other stressors such as antibiotics (17). Collectively, these findings raise the question of whether polyP, like (p)ppGpp, may help protect the nucleoid under stress.
A challenge in studying polyP is that its chemical simplicity makes it difficult to verify bona fide binding interactions. However, although it lacks specificity at the primary level of organization, the polymer forms tertiary granule superstructures that can be hundreds of nanometers in diameter. Observations of tight spatial organization of polyP in diverse species has led to the hypothesis that granules may form at predetermined locations in the cell (18–20). Indeed, DNA replication and nucleoid segregation play a role in positioning polyP granules in C. crescentus (19).
However, how polyP granules form and how polyP affects the integrity of the nucleoid during starvation remain poorly understood. The study of early stages of polyP formation presents various technical obstacles. PolyP granule localization has largely relied on traditional fluorescence imaging, which precludes looking at the early stages of granule genesis due to the size of nascent granules. Although transmission electron microscopy (TEM) has also been used to image polyP granules in fixed cells, fixation has the potential to distort subcellular structures. Confounding polyP granule imaging is the need to unambiguously distinguish them from other storage granules. We reasoned that quantitative description at high spatial and temporal resolution of the dynamics underpinning the nucleation and growth of polyP could provide insight into how it promotes starvation fitness. To achieve this, we used a combination of cryo- and conventional electron microscopy without fixation and light microscopy, coupled to genetic and spectroscopic techniques.
We chose the human opportunistic pathogen Pseudomonas aeruginosa as a model organism in which to characterize granule biogenesis for two reasons. First, unlike some bacteria including C. crescentus and Campylobacter jejuni, which are thought to make polyP granules even during exponential growth, P. aeruginosa makes polyP granules conditionally, facilitating the observation of de novo granule formation (19, 20). Second, aside from E. coli, it is the organism in which the most detailed biochemical characterization of the polyphosphate biosynthetic machinery and quantification of the polyP polymer under a variety of starvation states has been done (15, 21, 22). We used nitrogen limitation as the model stress condition to study polyP granule biogenesis. Our goals in this study were (i) to characterize the de novo granule genesis process and its spatial and temporal relationship to cell cycle exit and (ii) to characterize the function of polyP and (p)ppGpp in cell cycle exit in P. aeruginosa.
Results
Characterization of polyP Granule Formation in Near-Native State in P. aeruginosa.
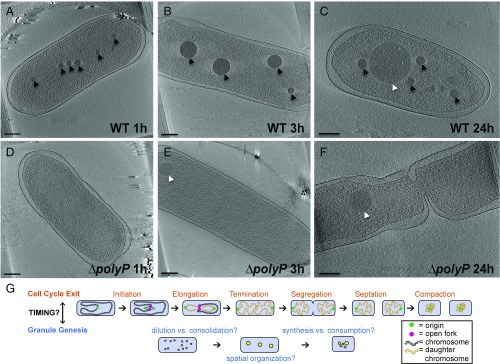
To characterize polyP granule nucleation and maturation in near native-state conditions, we imaged P. aeruginosa cells with electron cryotomography at multiple time points after shifting exponentially growing cells to nitrogen-limited media. One hour after this shift, multiple putative polyP granules form in the nucleoid region, which is defined as the area of the cell depleted of ribosomes (Fig. 1A). By 3 h, cells have fewer, larger granules (Fig. 1B). Energy dispersive X-ray spectroscopy (EDS) demonstrates that these electron-dense bodies are enriched for phosphorous and oxygen in comparison with the cytosol, consistent with these structures being polyP (SI Appendix, Fig. S1). By 24 h, a distinct class of granules with lighter intensity are visible (Fig. 1C). In addition to polyP, pseudomonads can accumulate carbon in the form of polyhydroxyalkanoate (PHA) storage granules under nitrogen starvation (23, 24). To distinguish between polyP and putative PHA granules, we imaged two strains, one each lacking the respective genes required for either polyP or PHA synthesis. The PHA mutant lacks the lighter-intensity granules, whereas the polyP mutant (∆polyP) lacks the darker granules (Fig. 1 D–F and SI Appendix, Fig. S2 A–C and E–G). We note that P. aeruginosa contains four polyphosphate kinase (ppk) genes from two protein families (one ppk1 gene, and three ppk2 genes, which we refer to as ppk2A (PA14_01730), ppk2B (PA14_33240), and ppk2C(PA14_19410), respectively), all of which are deleted in the ∆polyP mutant. Table 1 shows the phenotype of knocking out these genes in various combinations. Either Ppk1 or Ppk2A alone can drive granule genesis during nitrogen starvation (SI Appendix, Fig. S2 J–L) and expression of Ppk2A or Ppk2B is sufficient to promote granule genesis during exponential phase (Table 2). An active site mutant of Ppk2A does not drive granule synthesis in exponential phase, indicating granules form directly due to polyP synthetic activity rather than a general stress response to overexpression of the Ppk2A protein (Table 2). Together, these results allow us to confidently identify polyP granules in our experimental system.
Fig. 1.
PolyP granule formation in Pseudomonas aeruginosa under nitrogen starvation. Each panel is a 20-nm-thick tomographic slice through a 3D reconstruction of an intact cell. (Scale bar, 200 nm.) (A) Cryotomogram of a WT cell 1 h after inducing starvation. The cell shows many microgranules of polyP (black arrowheads). (B) WT cell 3 h after inducing starvation. Microgranules consolidate within the nucleoid region into regularly spaced mature granules. (C) WT cell 24 h after inducing starvation. PolyP granules lose tight spatial organization, and they are of heterogeneous size and number. PHA granules (white arrowheads) are formed late in starvation. (D) ∆polyP cell 1 h after inducing starvation. Quadruple knockout of polyphosphate kinases ∆ppk1∆ppk2a∆ppk2b∆ppk2c. (E) ∆polyP cell 3 h after inducing starvation do not form polyP granules but form PHA granules earlier than WT. (F) ∆polyP cell 24 h after inducing starvation contain PHA granules and are elongated, with some showing evidence of initiation but not completion of septation. (G) (Top) Model of steps in cell cycle exit. Reinitiation of DNA replication is inhibited. Replication elongation rates are modulated for efficiency to prevent stalled forks and fork breakage. Daughter chromosomes are segregated and compacted. Septation proceeds in the absence of stalled forks and once daughter chromosomes are segregated. (Bottom) Model for steps in polyP granule genesis. PolyP granules initiate throughout the nucleoid region. Granule number decreases, whereas size increases, by some combination of dilution of granules due to cell division and/or granule consolidation. Mature granules become transiently spatially organized. Granule number, size distribution, and positioning relax deeper into starvation.
Table 1.
Individual kinases required during nitrogen starvation
| Available kinase | Strain | PolyP granules | Cell cycle exit |
| All | WT | Yes | WT |
| None | ∆ppk1∆ppk2a∆ppk2b∆ppk2c | No | Defect |
| Ppk1 | ∆ppk2a ∆ppk2b ∆ppk2c | Yes | WT |
| Ppk2A | ∆ppk1 ∆ppk2b ∆ppk2c | Yes | Slight defect |
| Ppk2B, Ppk2C | ∆ppk1 ∆ppk2a | No | Defect |
| All | ∆relA ∆spoT | Yes | Defect |
| All | ∆(phaC1-phaC2) | Yes | Not determined |
Table 2.
Sufficiency of individual kinases to drive polyP granule formation
| Strain | PolyP granules |
| Para FLAG-Ppk2A | Yes |
| Para FLAG-Ppk2A D183A,R184A | No |
| Para 6His-TEV-Ppk2B | Yes |
Two striking observations from following polyP growth and localization as cells starve are that (i) cells become elongated in ∆polyP, raising the possibility that polyP may be required for cell cycle exit during starvation, and (ii) polyP granules decrease in number yet increase in size after nucleation. Together, these observations suggest that polyP granules may be more dynamic than previously thought and their biogenesis may be coordinated with cell cycle exit. To test this hypothesis, we used multiple approaches to track polyP localization over the course of cell cycle exit at both the population and single cell level.
Nitrogen Starvation Generates Smaller Cells with Single Chromosomes.
As described in the introduction and seen in Fig. 1G, cell cycle exit involves four steps. Despite extensive studies of the molecular machinery underpinning these processes in model organisms such as E. coli and Bacillus subtilis, very little is understood about how cells couple starvation cues to efficient cell cycle exit and nucleoid compaction. To follow cell cycle exit in our population of unsynchronized cells, we quantified total cell number, chromosomal origins per cell, open DNA replication forks per cell, and cell volume as a function of time (Fig. 2).
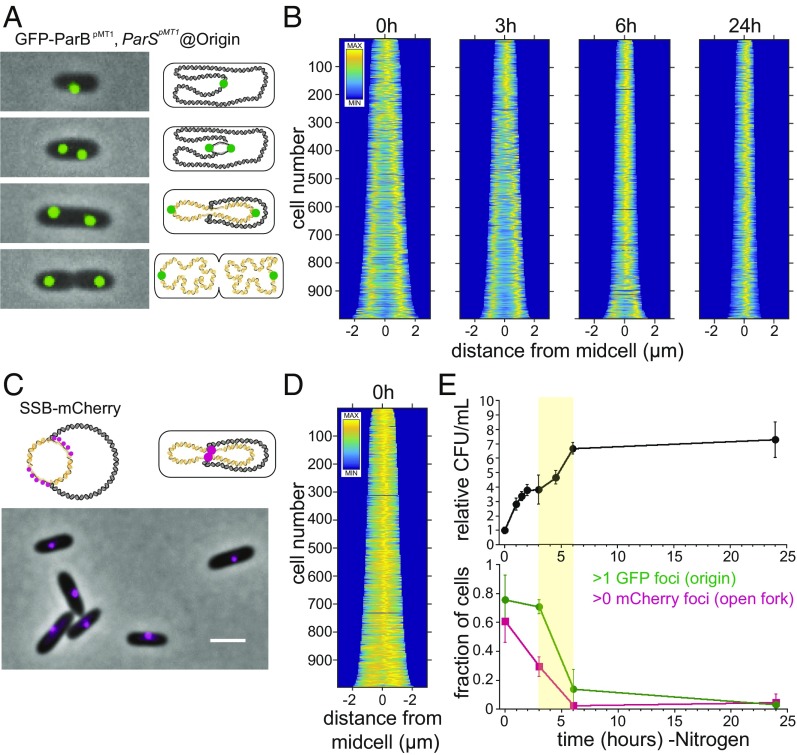
Fig. 2.
Timing of cell cycle exit during nitrogen starvation determined by counting cells, chromosomal origins of replication per cell, and open DNA replication forks per cell. (A) (Left) Examples of cells labeled using the GFP-ParBpMT1 chimeric reporter and the parSpMT1 binding site inserted at the attTn7 locus, 19.5 kb from the origin of replication. The GFP-ParBpMT1 chimera is expressed under the control of a second copy of the native P. aeruginosa AraJ-SSB promoter, also inserted at the attT7 locus. (Right) Illustration of model for organization of chromosomal origins (green), chromosome (black), and daughter chromosomes (gold). (B) Demographs of fluorescence intensity of the GFP-ParBpMT1 reporter construct in wild-type cells at 0, 3, 6, and 24 h into nitrogen starvation. (C) (Top) A translational fusion of SSB-mCherry under its native promoter was used to label open DNA replication forks, which colocalize midcell in P. aeruginosa. (Bottom) Example of exponential-phase wild-type cells with the SSB-mCherry translational fusion. (Scale bar, 2 µm.) (D) Demograph of mCherry fluorescence of exponential-phase cells with SSB-mCherry translational fusion construct. (E) (Top) Relative fold change in the number of cells (cfu/mL) after the shift to nitrogen-limited medium. (Bottom) Green circles represent the fraction of cells with >1 GFP focus, indicating the presence of more than one chromosome origin of replication after the shift to nitrogen-limited medium. Magenta squares represent the fraction of cells with >0 mCherry foci, indicating the presence of an open DNA replication fork after the shift to the nitrogen-limited medium. Highlighted yellow region between 3 and 6 h indicates the period during which the majority of cells in the population complete cell cycle exit (the fraction of cells with more than one GFP focus drops from 71 ± 17% to 14 ± 14% from 3 to 6 h, means and SDs of four independent experiments, and the fraction of cells with at least one mCherry focus drops from 29 ± 6.8% to 2.5 ± 2.1% from 3 to 6 h, means and SDs of five independent experiments).
We used the heterologous chimeric chromosome partitioning protein GFP-ParBpMT1 and its cognate DNA binding sequence parSpMT1 to label the origin of the P. aeruginosa chromosome, as has been done previously (Fig. 2A) (25, 26). We integrated one copy of the parSpMT1 DNA binding sequence at the attB locus, 19.5 kb from the origin, using a mini-Tn7 suicide vector (27). We then confirmed the specificity of the GFP-ParBpMT1 protein for the parSpMT1 binding site by the lack of foci formation in the absence of parSpMT1 (SI Appendix, Fig. S3). Over time, cells shrink in size and the number of origins converges to a single centrally localized position (Fig. 2B). Labeling of single-stranded DNA binding protein (SSB) with mCherry allows visualization of open DNA replication forks in the cell center, as previously seen in E. coli (Fig. 2 C and D) (28). Cells complete between two and three doublings between 0 and 6 h after their shift to starvation conditions (Fig. 2E, Top), during which time the number of cells with more than one origin decreases from 76 ± 17% to 3 ± 3% and the number of cells with one or more open DNA replication forks goes from 61 ± 15% to 4 ± 6% (Fig. 2E, Bottom). Collectively, these results indicate that the population completes cell cycle exit during this period.
Consolidation of Granules Without Net Consumption of polyP During Cell Cycle Exit.
Having defined the timing of cell cycle exit under nitrogen starvation conditions, we proceeded to quantify the spatiotemporal dynamics of polyP biogenesis (Fig. 3). Specifically, we sought to evaluate whether (i) net consumption or genesis of polyP occurs during cell cycle exit, (ii) the decrease in granule number results from dilution and segregation of granules during cell division or consolidation of nascent granules, and (iii) when polyP granules become spatially organized (Fig. 1G, Right). These measurements required high-resolution imaging of a large number of cells. Because commonly used aldehyde fixatives disrupt granules (SI Appendix, Fig. S2I), we used unfixed cells, rapidly dried onto TEM grids. We measured the diameter of individual granules, as well as their position along the long axis of the cell at different times after initiating starvation (Fig. 3A and SI Appendix, Fig. S2 M and N).
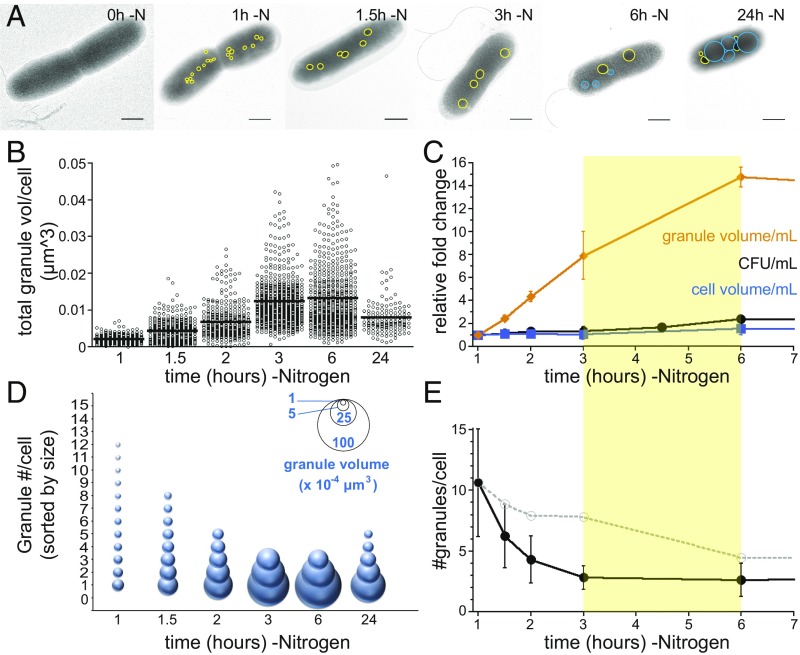
Fig. 3.
PolyP granule growth and consolidation during cell cycle exit. (A) Examples of nitrogen-starved cells as a function of time. (Scale bar, 0.5 μM.) PolyP granules are outlined in yellow, and polyhydroxyalkanoate (PHA) granules are outlined in blue. (B) The total volume of granules per cell increases throughout cell cycle exit [0.0021 ± 0.0021 µm3 at 1 h (n = 214 cells), 0.004 ± 0.003 µm3 at 1.5 h (n = 319 cells), 0.0069 ± 0.0048 µm3 at 2 h (n = 316 cells), 0.0124 ± 0.067 µm3 at 3 h (n = 542 cells), and 0.0132 ± 0.0094 µm3 at 6 h (n = 560 cells)] and then decreases to 0.008 ± 0.005 µm3 at 24 h (n = 136 cells). Global analysis of two to four independent experiments is shown. (C) The total volume of granules per mL of culture increases throughout cell cycle exit. The yellow bar between 3 and 6 h highlights the period where the population as a whole completes cell cycle exit. (D) The size distribution of granules that contribute 95% or more of total granular volume. Inserted key shows granule volume represented by bubble size. For each cell, granules were ranked by size, and their volumes were summed, starting from the largest to smallest granule, until 95% of total granule volume was reached; any additional smaller granules are excluded. At each time point, the average number of granules required to reach 95% of total granule volume is shown, with granule size depicted reflecting the average size of granules of that numerical rank. (E) The number of granules per cell that contribute to 95% of total granular volume. Black closed circles represent experimental data: 10.6 ± 4 granules per cell at 1 h, 6.3 ± 3 at 1.5 h, 4.3 ± 2 at 2 h, 2.8 ± 1 at 3 h, 2.6 ± 1 at 6 h, and 4.3 ± 2 at 24 h. Gray open circles and dotted line represent the number of granules predicted by dilution, i.e., the number of granules predicted per cell if there is no net change in the number of granules, but they are partitioned to new cells due to cell division. Cell counts were used to generate the dilution factors: 8.9 at 1.5 h, 7.9 at 2 h, 7.8 at 3 h, 4.5 at 6 h, and 4.1 at 24 h.
To determine how polyP granule biomass changes during cell cycle exit, we quantified the total volume of all granules in individual cells, as well as the volume of each cell. Volume calculations assume granules are spherical and cells cylindrical. Total granule volume per cell increases between 1 and 3 h, plateaus between 3 and 6 h, and then decreases between 6 and 24 h (Fig. 3B). Moreover, total granule biomass increases during cell cycle exit (Fig. 3C, yellow bar). We used the average cell volume, granule volume, and cfu measurements to determine the total cellular volume and polyP volume per mL of culture. Between 1 and 6 h, cell number increases 2.4-fold, but the increase in cell volume is only 1.6-fold, indicating that cell division outpaces growth during this period. In contrast, granule volume increases more than an order of magnitude during this time (Fig. 3C).
We next determined how the distribution of polyP volume in individual cells changes over time. Although total polyP volume per mL increases between 1 and 6 h, the overall number of granules per cell decreases from 13 ± 6 to 5 ± 4 (SI Appendix, Fig. S4A). We noticed at 3 and 6 h that the distribution of granule size becomes bimodal, with cells containing large granules with a diameter of 200 ± 50 nm and some cells also containing smaller satellite granules with a diameter of 59 ± 20 nm (SI Appendix, Fig. S4 E and F). To assess how the volume distribution of polyP changes over time, we ranked granules by size. Counting granules that collectively contribute 95% or more of total granular volume per cell allowed us to calculate the contribution of individual size classes as can be seen in Fig. 3D. The distribution of granule volume shifts from a large number of small volumes (10.6 ± 4 granules of diameter 67 ± 19 nm) to a small number of large volumes (2.8 ± 1 granules of diameter 190 ± 53 nm; Fig. 3D). Between 6 and 24 h, the distribution of polyP volume among granules broadens again (2.6 ± 1 granules of diameter 182 ± 77 nm to 4.3 ± 2 granules of diameter 124 ± 55 nm; Fig. 3D). The change in distribution of polyP granule biomass between 3 and 6 h cannot be explained simply by dilution due to cell division; rather, it appears to reflect granule consolidation within cells (Fig. 3E).
Cells Transiently Organize polyP Granules During Cell Cycle Exit.
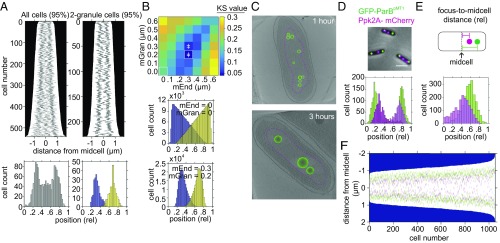
Having observed that granules consolidate by 3 h, we chose this time point to characterize the spatial organization of mature granules, using a combination of TEM and fluorescence imaging, as well as modeling. Demographs of granules from TEM imaging of 3 h-starved cells reveal a predominantly bimodal distribution (Fig. 4A, Left). Characterizing subpopulations of cells segregated by granule number reveals that two-, three-, and four-granule cells exhibit remarkably even spacing (Fig. 4A, Right, and SI Appendix, Fig. S5). To test whether this organization could simply reflect physical occlusion between granules, we simulated their localization using the granule and cell length distribution in our TEM dataset for two-granule cells (Fig. 4B and SI Appendix, Fig. S6). We generated a population of two-granule cells in silico with lengths and granule diameters reflecting the distribution of cell length and granule diameter observed in vivo and then randomly positioned both granules along the long axis of the cell. If the entire length of the cell is considered as available to granules in the simulation, their resulting distribution is different from our experimental observations (Fig. 4B, Middle). However, when we constrain the space granules can occupy by introducing a requirement of a minimum distance between granules (mGran) and between granules and the cell poles (mEnd), the simulation more closely matches our in vivo data (Fig. 4B, Bottom, and SI Appendix, Fig. S6A). We tested all combinations of mEnd and mGran constraints from 0 to 0.6 µm in 0.1-µm steps and used the nonparametric Kolmogorov–Smirnov (KS) test to compare the resulting overall distributions (both granules) to our in vivo data. We found the combination of a minimum distance of 0.3 μm from cell poles and 0.2 μm between granules to be the best fit to our experimentally observed data with this model (Fig. 4B, Top and Bottom, and SI Appendix, Fig. S6 B–G; see SI Appendix, SI Methods, for further description of modeling).
Fig. 4.
PolyP granules are spatially organized 3 h into nitrogen starvation. (A) (Top Left) Demograph of granules contributing to 95% of total granular volume per cell of all 3 h-starved cells imaged by TEM. (Bottom Left) Histogram of relative position of granules along the long axis of cells imaged by TEM as above. (Top Right) Demograph of 3 h-starved cells with two granules imaged by TEM. (Bottom Right) Histogram of relative position of granules along the long axis of cells imaged by TEM as above. First granule, 0.32 ± 0.06 (blue), and second granule, 0.68 ± 0.08 (yellow). (B) (Top) Heat map of KS test statistic from comparing the experimentally observed distribution of granules in two-granule cells to a simulation of randomly positioning granules along the long axis of the cell, with two added constraints: first, a minimum distance between granules and cell poles (mEnd) and second, a minimum distance between first and second granule (mGran). Symbol ǂ indicates parameter space where the model is statistically indistinguishable from the data. See Methods and SI Appendix, Fig. S6 and SI Methods, for more detailed description. (Middle) Histogram of relative granule position along long axis of cells from a simulation of 229,000 cells in which the only constraint is that granules cannot overlap along long axis of the cell (mEnd = 0, mGran = 0). (Bottom) As in Middle, with the added constraints that granules must be a minimum of 0.3 µm from cell poles and 0.2 µm from each other. (C) (Top) Cryotomogram of WT cell 1 h after inducing starvation. Granules are represented as green spheres, and the nucleoid region is outlined with a magenta dotted line. (Bottom) As in Top, at 3 h. (D) (Top) Example cells with nucleoid origins labeled using the GFP-ParBpMT1 chimera and the parSpMT1 DNA binding site on the chromosome at the attTn7 site (green), and polyP granules labeled with the Ppk2A-mCherry chimera (magenta), replacing the native copy of ppk2A on the chromosome. (Scale bar, 0.5 μM.) (Bottom) Histogram of relative position on long axis of the cell of GFP foci (0.25 ± 0.08 and 0.76 ± 0.09; green) and mCherry foci (0.34 ± 0.13 and 0.71 ± 0.10; magenta). (E) Histogram of relative position of GFP and mCherry foci from midcell, 0.28 ± 0.07 and 0.20 ± 0.09, respectively. P < 0.01 by Student’s t test. (F) Demograph representation of position of nucleoid origins (green) and polyP granules (magenta).
We wondered whether the colocalization of polyP granules with the nucleoid might restrict the distance of polyP granules from cell poles. The nucleoid-containing region is visible in electron cryotomography due to its lack of ribosomes (Fig. 4C). Granules appear to both nucleate and mature within this region (Fig. 4C). To test this hypothesis, we used a fluorescence technique to concurrently visualize granules and GFP-labeled chromosome origins in two-origin cells (Fig. 4 D and E). Given that the origins of daughter chromosomes are extruded toward cell poles during DNA replication, the nucleoid spans at least this region bounded by the two origins. PolyP granules have the same distribution pattern whether they are stained by DAPI or imaged by TEM (SI Appendix, Fig. S7 A–C). However, DAPI staining disrupts the fluorescence of the GFP-ParBpMT1 -parSpMT1 construct we used to label origins, precluding DAPI use. Accordingly, we used a translational fusion of one of the polyphosphate kinases (Ppk2A) with mCherry (Fig. 4D, Top). This chimera is functional in the absence of the other three kinases because cells are able to make polyP granules and mCherry foci colocalize with DAPI-stained polyP granules (SI Appendix, Fig. S7 D and E). The relative position of origins is farther from midcell than that of the outermost polyP granules (normalized position of 0.28 ± 0.07 versus 0.21 ± 0.09, respectively; P < 0.001; Fig. 4 D and E). Moreover in individual cells, 82 ± 6.8% of origins are farther from midcell than the nearest outer granule (Fig. 4F). We do not see colocalization of GFP and mCherry foci, but because the parSpMT1 DNA binding site was inserted 19.5 kb from the nucleoid origin, our data do not rule out the possibility that granules and origins colocalize. However, these data suggest that the nucleoid region demarcates a minimum distance between granules and cell poles.
PolyP and (p)ppGpp Are Required for Completion of Cell Cycle Exit After Replication Initiation.
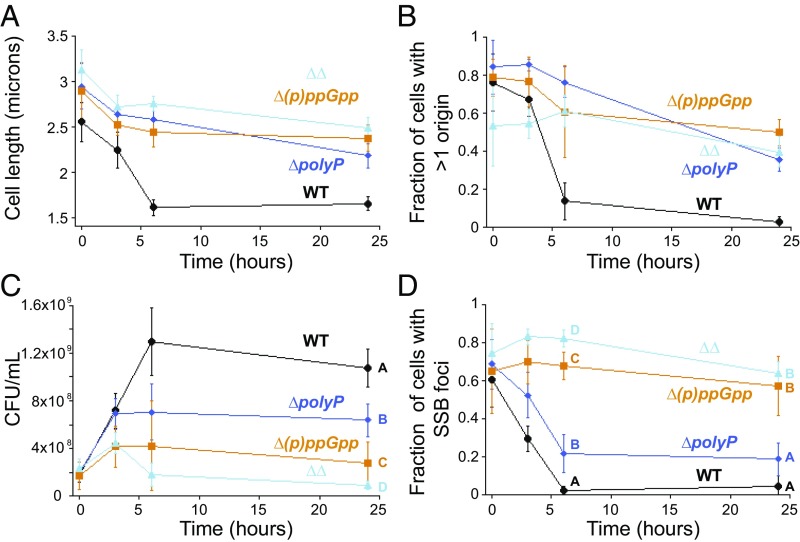
Could polyP be required for cell cycle exit during starvation? That granule consolidation is temporally coupled with cell cycle exit, together with the fact that cells lacking polyP become elongated during starvation, suggests this might be the case. To test this idea, we compared the phenotype of a polyP mutant to that of a strain lacking the conserved cell cycle regulator in bacteria, guanosine tetraphosphate [(p)ppGpp] [Δ(p)ppGpp, a ΔrelA ΔspoT strain]. Both strains become elongated during starvation (Fig. 5A and SI Appendix, Fig. S2 F and H), and their populations contain a larger fraction of cells with more than one origin compared with the WT (Fig. 5B). That either Ppk1 or Ppk2A is sufficient to promote cell cycle exit during starvation (Table 1 and SI Appendix, Fig. S8 A–C) indicates that polyP synthesis per se, rather than the action of a specific kinase, is responsible for cell cycle exit. We note that cell cycle exit is slightly slower than WT in a strain containing only Ppk2A, consistent with our observation that granules are slightly smaller in this strain at 3 h than WT (SI Appendix, Fig. S8D). To distinguish whether the persistence of cells with more than one origin is the result of inappropriate reinitiation of DNA replication versus failure to complete a later step in cell cycle exit, we measured viable cell number over time during starvation using a cfu assay. We found that both the ∆polyP and ∆(p)ppGpp mutants complete fewer doublings than WT under these conditions, consistent with a failure to finish open rounds of cell division rather than inappropriate reinitiation (Fig. 5C).
Fig. 5.
PolyP and (p)ppGpp promote cell cycle exit during starvation. WT, black circles; ∆polyP, blue diamonds; ∆(p)ppGpp, orange squares; and ∆∆, cyan triangles. (A) Average cell length as a function of time after induction of nitrogen starvation. Mean and SD from at least six independent experiments for each time point. (B) Fraction of cells with >1 GFP-ParBpMT1 focus as a function of time after induction of nitrogen starvation. Mean and SD from at least three independent experiments for each time point. (C) Cell counts (cfu) after the shift to nitrogen-limited medium. Mean and SD from at least three independent experiments, variance analyzed using a one-way ANOVA. Significant differences between strains at the same time point are marked with uppercase letters based on post hoc Tukey test. Strains at the same time point marked with different letters have significantly different means. P < 0.05. (D) Fraction of cells with at least one SSB-mCherry focus as a function of time during nitrogen starvation, mean and SD from five independent experiments, variance analyzed using a one-way ANOVA. Significant differences between strains at a given time point are marked with uppercase letters based on post hoc Tukey test. Strains at the same time point marked with different letters have significantly different means. P < 0.05.
To further delineate which step in cell cycle exit polyP affects compared with (p)ppGpp (e.g., replication elongation or postelongation effects such as chromosome segregation and septation), we looked at the number of open DNA replication forks in the mutants (Fig. 5D). Twenty-four hours into nitrogen starvation, the majority of cells in ∆(p)ppGpp still have SSB-mCherry foci, consistent with a failure to finish replication elongation. The fraction of cells with SSB-mCherry foci in the ∆polyP mutant is significantly smaller than in the ∆(p)ppGpp mutant at 3, 6, and 24 h but still larger than in WT at 6 h (22 ± 0.10 vs. 0.25 ± 0.02, respectively; P = 0.002 using one-way ANOVA analysis for multiple comparisons, as described in the legend of Fig. 5 and Methods). Together, these data indicate that polyP and (p)ppGpp both play postinitiation roles in cell cycle exit during starvation, with (p)ppGpp primarily functioning during replication elongation and polyP affecting replication elongation to a lesser extent than (p)ppGpp yet still influencing subsequent steps.
polyP and (p)ppGpp Have Additive Effects on Nucleoid Organization and Stability.
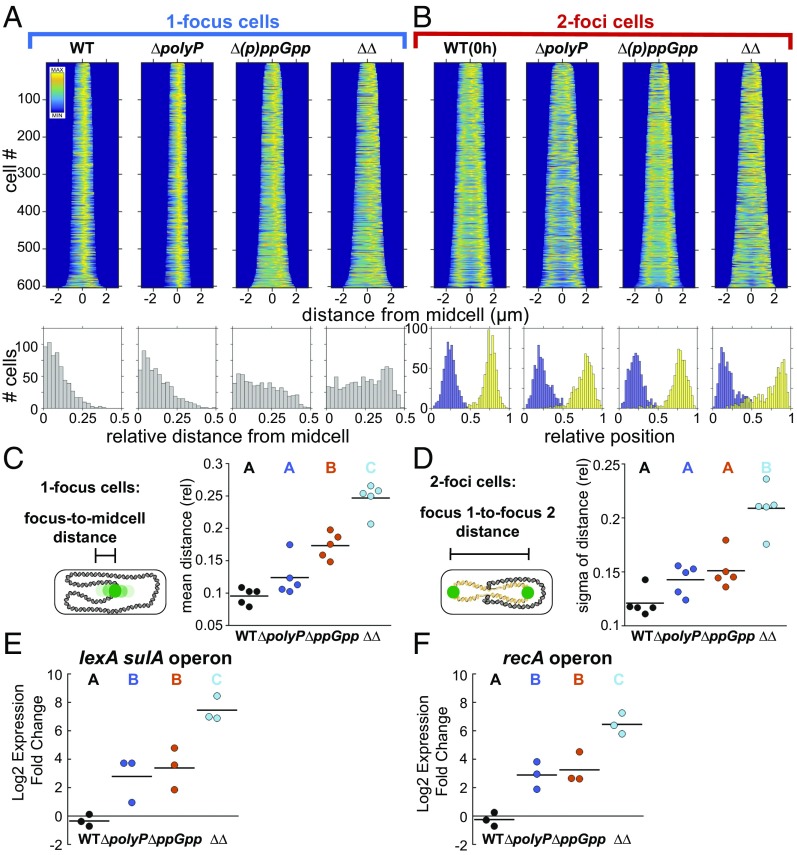
To ascertain whether (p)ppGpp and polyP function in the same pathway to regulate cell cycle exit and to further probe the differences between their effects on the nucleoid, we performed epistasis tests. A mutant lacking both (p)ppGpp and polyP (“∆∆”) is more impaired in doubling during starvation than either single mutant (Fig. 5C, 24-h time point). The ∆∆ mutant also has more persistent open DNA replication forks at 6 h than either single mutant (Fig. 5D). Together these findings suggest polyP and (p)ppGpp have at least some independent effects on cell cycle exit. Consistent with this interpretation, the (p)ppGpp mutant makes polyP, indicating that unlike in E. coli, (p)ppGpp is not required for polyP biosynthesis under these starvation conditions in P. aeruginosa (SI Appendix, Fig. S2 D and H). How then might these molecules exert independent effects on doubling? Although we do not observe additive effects on cell length or the number of cells with extra origins (Fig. 5 A and B), we noticed a dramatic change in the organization of the nucleoid in the double mutant (Fig. 6).
Fig. 6.
PolyP and (p)ppGpp have synergistic effects on nucleoid organization and the SOS DNA damage response. Variance analyzed using a one-way ANOVA. Significant differences between strains at the same time point are marked with uppercase letters based on post hoc Tukey test. Strains marked with different letters have significantly different means. P < 0.05. (A) (Top) Fluorescence demographs of cells with one GFP-ParBpMT1 focus, 24 h nitrogen starvation, using GFP-ParBpMT1 chimeric reporter under the SSB promoter and the parSpMT1 DNA binding site at the attT7 locus. (Bottom) Histograms of relative distance from midcell of GFP-ParBpMT1 foci. (B) (Top) As in A but for two-foci cells. (Bottom) Relative position of first GFP-ParBpMT1 focus (blue) and second focus (yellow) on long axis of cell, normalized to 1. (C) Relative distance from midcell of GFP-ParBpMT1 foci in one-focus cells as above. Each point represents the population mean from an independent experiment. P < 0.05 for strains marked with different letters. (D) Broadness of distribution (sigma) of relative distance of GFP-ParBpMT1 foci to each other in a population of two-foci cells. Each point represents an independent experiment. P < 0.001. (E) Relative change in expression 3 h into nitrogen starvation relative to 0 h of lexA sulA operon, mRNA quantification by qPCR. Each point represents an independent experiment. P < 0.05. (F) As in E, for recA operon. P < 0.01.
The origin in single-nucleoid cells in ∆polyP is positioned midcell, with a similar Gaussian distribution to WT (Fig. 6 A and C). Origins are further from midcell in the ∆(p)ppGpp mutant, and the positioning of origins approaches a uniform distribution in the double mutant (Figs. 6A, Bottom Right, and 6C). Origin position in cells with two foci is bimodal in both single mutants and is similar to the organization found in WT during exponential phase (Figs. 2B and 6B). The distribution of origins is significantly broader in the double mutant (Fig. 6 B and D). These data suggest a synergistic defect leading to a profound disruption in nucleoid organization. Remodeling and compacting the nucleoid during starvation is thought to be important for protecting the integrity of the chromosome. To determine whether the loss of nucleoid organization in the double mutant reflects underlying DNA damage, we looked at expression of the SOS DNA damage response genes during starvation (lexA and recA; Fig. 6 E and F). Both genes show heightened expression 3 h after starvation in the ΔpolyP and Δ(p)ppGpp single mutants relative to WT, with even greater enhancement in the double mutant. The pleiotropic nature of both molecules makes it difficult to draw simple conclusions from genetic epistasis. However, the effects we observe in the double mutant on doubling, persistent open DNA replication forks, nucleoid organization, and the SOS response collectively suggest that polyP and (p)ppGpp have at least partially distinct roles in protecting the integrity of the nucleoid during starvation.
Discussion
The mechanism(s) by which polyP help bacteria to adapt to constantly changing nutrient conditions in the environment are poorly understood. Lacking has been a high-resolution dissection of the spatiotemporal progression of polyP granule biogenesis during nutrient limitation, to help frame the problem. Our quantitative characterization of de novo polyP granule synthesis during the transition from exponential growth to stationary phase in P. aeruginosa revealed several properties of these structures: (i) granules mature and spatially organize within the nucleoid region, temporally coupled to cell cycle exit; (ii) granule number decreases due to consolidation; and (iii) there is a net increase in granule volume during cell cycle exit (Fig. 1G). These properties raise several implications for polyP function.
In P. aeruginosa, we find that polyP acts postinitiation to permit efficient completion of open rounds of DNA replication and cell division following nitrogen starvation, and polyP and (p)ppGpp have additive roles in organizing the nucleoid during starvation. The discovery in the 1950s that bacteria synthesize polyP from ATP during nutritional downshift initiated a debate that has remained unresolved for more than half a century as to whether this polymer serves as a physiologically relevant energy store (29–31). Although polyP may have functions deeper in starvation or cell cycle reentry as an energy store, our results imply that when it drives completion of open rounds of cell division during starvation, net synthesis rather than consumption of polyP is required. We propose three nonexclusive mechanisms to explain this phenomenon: (i) synthesis of polyP may be required as a phosphate sink during starvation, (ii) the polyP polymer acts as a regulatory molecule to promote cell cycle exit, and/or (iii) granule superstructures may serve as an important organizational microenvironment for specific chemical processes during starvation.
A long-standing model for polyP function has been that granule formation is a mechanism to store phosphate, rather than ATP per se. That polyP may be equally important as a phosphate sink was proposed by Harold and others in the 1960s (29) but has subsequently received less attention. Because phosphorous is limiting in many environments, cells that can hold onto it during starvation should have a growth advantage when conditions improve. Indeed, the phosphate overplus phenomenon, whereby bacteria sequester phosphate in the form of polyP when starved for other macronutrients, has long been observed in the context of wastewater treatment when carbon becomes limiting (31). However, polyP granules acting simply as a storage mechanism does not explain the defect in cell cycle exit rather than reentry. Catabolism of mRNA and rRNA and decreased demand for nucleotides for DNA and RNA synthesis might create a situation of excess free phosphate in the cell, which could act as a poison and inhibit many essential processes. PolyP as a phosphate sink may therefore be critical for cellular equilibrium. A role for polyP in nucleotide homeostasis has been recently proposed in yeast, where oscillation in the concentration of polyP measured biochemically was observed to correlate with cell cycle progression, in the absence of starvation (32). One of the functions of (p)ppGpp during starvation in B. subtilis is suppression of GTP biosynthesis, which unchecked leads to cell death (33). A role for polyP in nucleotide homeostasis could potentially explain the synergistic effects we observe on cell cycle exit and nucleoid stability between polyP and (p)ppGpp.
A second possibility is that like (p)ppGpp, polyP acts in a regulatory manner to drive cell cycle exit during starvation. PolyP, like (p)ppGpp, may directly bind to and alter the activity of cell cycle machinery. Several previous studies suggest that polyP may bind to and alter the function of macromolecular complexes such as Lon protease (34). It is also possible that although the steady state level of polyP increases during cell cycle exit, some polyP is consumed as a specialized source of phosphoryl groups for regulatory kinases, and this consumption is balanced by synthesis. A handful of kinases, including some glucokinases, have been shown to preferentially use polyP as a substrate (35). Recent findings in E. coli also suggest that polyP acts in a regulatory toxin–antitoxin mediated feedback loop that drives persistence during starvation (36).
A third explanation for the importance of polyP net synthesis during cell cycle exit is that granule superstructures could serve as microenvironments for specific enzymatic activities. Our observation that granules mature by consolidation after nucleation is reminiscent of a class of membraneless structures in cells: stress granules, P bodies, and the nucleolus are eukaryotic compartments that form phase-separated liquid droplets. At the structural level, there are some parallels between the physical properties of the proteins that drive formation of these droplets and polyP: these proteins are intrinsically disordered and have repetitive low-complexity sequences, allowing them to make many weak and transient interactions with each other and with RNA (37). Might polyP serve a similar role as an intrinsically unstructured biopolymer capable of making nonspecific interactions? In vitro studies of the polyP polymer indicate that it can demix from solution to form diverse materials, including phase-separated liquid droplets in the presence of some cations, hydrogels, amorphous glasses, and crystals (38, 39). Where on the phase spectrum polyP exits in cells is not known; the biophysical properties of polyP granules may also vary with time during starvation. At a functional level, there are parallels between the known intracellular droplets and polyP: P bodies and stress granules serve as RNA chaperone and processing hubs, and the nucleolus surrounds the rDNA loci and helps couple starvation and stress signaling to changes in rRNA synthesis, growth rates, and the cell cycle (40–42). In E. coli, the polyP polymer may serve as a protein chaperone during oxidative stress, and there is evidence that Ppk1 is a component of the RNA degradosome (43, 44). Like the low-complexity sequence-containing proteins that drive formation of phase-separated liquid droplets, polyP was recently shown in E. coli also to have a propensity to drive formation of amyloid-like protein aggregates under some conditions (45, 46). Exploring the physical properties of granules is important to understanding the possible partitioning of other cellular factors between the cytosol and granules.
In addition to the net increase in polyP granule biomass, these structures become transiently evenly spaced during cell cycle exit in P. aeruginosa. How actively cells organize granules is not clear. Passive abiotic phenomena such as diffusion-limited cluster aggregation (DLCA) can result in spacing of colloidal aggregates (47). Additionally, decreased cytoplasmic fluidity during nutritional downshift has been shown to decrease mobility of PHA carbon storage granules in C. crescentus and could similarly result in decreased polyP granule mobility (48). However, there is also evidence from work in C. crescentus that nucleoid dynamics play a role because interference with DNA replication or chromosome segregation disrupts granule positioning (19). It is also possible that other factors, such as cytoskeletal elements like the Par proteins, act on polyP granules. What benefit, if any, cells derive from evenly spaced granules is not known. In P. aeruginosa, granules are only transiently evenly spaced, suggesting that this organization matters during the transition to quiescence. Even spacing could be important to ensure equal partitioning of resources to daughter cells, as is thought to be the case for regular spacing of carboxysomes in cyanobacteria (49). Another possibility is that rather than the nucleoid driving granule positioning, polyP granules play a role in organizing the nucleoid, perhaps actively promoting global remodeling and compaction of the nucleoid. Although this process remains poorly understood, two broad types of phenomena can drive nucleoid condensation during the transition to quiescence: starvation-specific chromatin structural proteins replace those of exponential phase, and molecular crowding drives self-association of the nucleoid (50, 51). Components of polyP granules might make specific interactions that help organize the nucleoid during starvation, or formation of granule superstructures within the nucleoid region might contribute to crowding.
Although the enzymatic machinery for polyP synthesis is not conserved between bacteria and eukaryotes, the formation of granule superstructures is observed in all three domains of life, suggesting that the ability to make polyP and form these structures is ancient and important. Taken together with findings in C. crescentus that polyP inhibits DNA replication initiation (13), our observation in P. aeruginosa that polyP functions in a postreplication initiation step during starvation reveals that like (p)ppGpp, polyP uses diverse mechanisms to regulate the cell cycle in bacteria. Most eukaryotes lack (p)ppGpp, but a number of recent studies in eukaryotes suggest that polyP may be important in cell cycle regulation more broadly. In Dictyostelium discoideum, polyP is required for cytokinesis as well as spore germination; in Saccharomyces cerevisiae, polyP concentrations oscillate in a cell cycle-dependent manner and influence the rate of progression from G1 to S phase, affecting genomic stability; and in a mammalian cancer cell line, polyP was found to stimulate mTor kinase and promote cell proliferation (32, 52–54). It is thus tempting to speculate that polyP may be a universal mediator between metabolic cues and the cell cycle.
Methods
Strains, Plasmids, and Growth Conditions.
All strains, plasmids, and primers used in this study are listed in SI Appendix, Tables S1–S3, respectively. Detailed description of construction methods used to generate strains using P. aeruginosa PA14 are provided in SI Appendix.
Briefly, for nitrogen starvation experiments, strains were grown in Mops-buffered minimal media containing 22 mM ammonium chloride. From overnight cultures, subcultures were grown to log phase at 37 °C (OD500 = 0.4–0.6), then shifted into Mops-buffered media with 1 mM ammonium chloride (SI Appendix).
Growth Measurements.
The cfu measurements were done using serial dilution in Mops-buffered growth medium and plating on LB agar plates and are reported as averages of at least three independent experiments.
Transmission Electron Microscopy.
Two microliters of unfixed cells in growth media were spotted onto carbon-coated copper grids for 45 s and blotted with Whatman paper, and media salts were washed off of the grids by spotting the grids with 2 uL of water and rapidly blotting; wash step was repeated five times. Grids were imaged on a Tencai T12 electron microscope.
Electron Cryotomography.
Cells were prepared for ECT by plunge freezing in a nitrogen-cooled liquid ethane–propane mix. Images and tilt series were collected on an FEI Polara (FEI Company) 300-kV field emission gun transmission electron microscope equipped with a Gatan energy filter and a lens-coupled 4k-by-4k Ultra-Cam camera (Gatan). Samples were imaged with 200 e-/Å2, a defocus of −10 µm, 1° oscillation, and a tilt range from −60 to +60°. The 3D reconstructions and analyses were produced with the software package IMOD (55).
Energy Dispersive X-Ray Analysis.
Samples of unfixed cells were spotted onto grids using the same procedure as with conventional TEM (see above) and analyzed using a FEI Tencai F30 (TF30ST) operating in scanning transmission electron microscopy (STEM) mode. The EDS data were acquired using an Oxford Instruments INCA system. The analysis was done at 300 kV.
Fluorescence Microscopy.
Live cells were imaged on 1% agarose pads containing Mops-buffered media with 1 mM ammonium chloride on slides. Phase contrast and fluorescence images were taken on a Zeiss AxioObserver.A1 using a 100× oil immersion objective (Apo 1.4 PH3). For imaging DAPI-stained polyP granules, cells were stained for 10 min with 200 µM DAPI and imaged using a custom filter cube from Chroma Technology Corp consisting of an excitation bandpass filter (415/20 nm), long-pass dichroic (425 nm), and emission bandpass filter (555/10 nm).
Gene Expression Measurements.
RNA measurements were made by qPCR as described in SI Appendix and are reported as averages of three independent experiments.
Image Analysis and Simulations.
For fluorescent images, MicrobeTracker (56) was used both for segmentation to identify cells and for spot position analysis. Demographs are constructed by compressing each cell in a population at a given time point into a 2D object representing the position of the granules or loci along the line. These 2D objects are then sorted by cell length from shortest to longest and displayed as a stack where each row represents one cell. Demographs of fluorescence intensity were constructed using the method and script developed by Hocking et al. (57). For TEM images, cell length, width, granule diameter, and granule position were measured by hand using ImageJ (see SI Appendix, Fig. S2 M and N, for examples). See SI Appendix, SI Methods, for additional details of statistical analysis. Demographs of granule position, incorporating granule diameter, were constructed using a custom MATLAB script inspired by Hocking et al. (57). Simulated granule position distributions with different physical constraints were generated in MATLAB using the mean and SD of granule diameter and cell length from TEM experimental data at 3 h (simulation code attached, peas2pod_simulation.m; see SI Appendix for additional details).
Supplementary Material
Acknowledgments
We specially thank Dr. Alasdair McDowall, Howard Hughes Medical Institute, for electron microscopy support. We thank Carol Garland for technical assistance with Energy Dispersive X-ray Spectroscopy (EDS) data collection and analysis using California Institute of Technology (Caltech)’s Applied Physics and Materials Science Department’s Transmission Electron Microscopy Facility. We also thank Brittany J Belin and Noah Ollikainen for help with statistical analysis of the granule position modeling and gratefully acknowledge Megan Bergkessel and other D.K.N. laboratory members for constructive comments on the manuscript. The Caltech Electron Microscopy Facility is funded in part by the Gordon and Betty Moore Foundation, the Agouron Institute, and the Beckman Foundation. This work was funded in part by the Howard Hughes Medical Institute, and the National Institutes of Health (5R01HL117328-03). Lisa Racki is a Damon Runyon Fellow supported by the Damon Runyon Cancer Research Foundation (DRG-2126-12).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1615575114/-/DCSupplemental.
References
- 1.Kreuzer KN. DNA damage responses in prokaryotes: Regulating gene expression, modulating growth patterns, and manipulating replication forks. Cold Spring Harb Perspect Biol. 2013;5(11):a012674. doi: 10.1101/cshperspect.a012674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Saint-Ruf C, Pesut J, Sopta M, Matic I. Causes and consequences of DNA repair activity modulation during stationary phase in Escherichia coli. Crit Rev Biochem Mol Biol. 2007;42(4):259–270. doi: 10.1080/10409230701495599. [DOI] [PubMed] [Google Scholar]
- 3.Minsky A, Shimoni E, Frenkiel-Krispin D. Stress, order and survival. Nat Rev Mol Cell Biol. 2002;3(1):50–60. doi: 10.1038/nrm700. [DOI] [PubMed] [Google Scholar]
- 4.Frenkiel-Krispin D, Minsky A. Nucleoid organization and the maintenance of DNA integrity in E. coli, B. subtilis and D. radiodurans. J Struct Biol. 2006;156(2):311–319. doi: 10.1016/j.jsb.2006.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Wang JD, Levin PA. Metabolism, cell growth and the bacterial cell cycle. Nat Rev Microbiol. 2009;7(11):822–827. doi: 10.1038/nrmicro2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jonas K, Chen YE, Laub MT. Modularity of the bacterial cell cycle enables independent spatial and temporal control of DNA replication. Curr Biol. 2011;21(13):1092–1101. doi: 10.1016/j.cub.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schreiber G, et al. Overexpression of the relA gene in Escherichia coli. J Biol Chem. 1991;266(6):3760–3767. [PubMed] [Google Scholar]
- 8.Lesley JA, Shapiro L. SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus. J Bacteriol. 2008;190(20):6867–6880. doi: 10.1128/JB.00700-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang JD, Sanders GM, Grossman AD. Nutritional control of elongation of DNA replication by (p)ppGpp. Cell. 2007;128(5):865–875. doi: 10.1016/j.cell.2006.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denapoli J, Tehranchi AK, Wang JD. Dose-dependent reduction of replication elongation rate by (p)ppGpp in Escherichia coli and Bacillus subtilis. Mol Microbiol. 2013;88(1):93–104. doi: 10.1111/mmi.12172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nazir A, Harinarayanan R. Inactivation of cell division protein FtsZ by SulA makes Lon indispensable for the viability of a ppGpp0 strain of Escherichia coli. J Bacteriol. 2015;198(4):688–700. doi: 10.1128/JB.00693-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamarthapu V, et al. ppGpp couples transcription to DNA repair in E. coli. Science. 2016;352(6288):993–996. doi: 10.1126/science.aad6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boutte CC, Henry JT, Crosson S. ppGpp and polyphosphate modulate cell cycle progression in Caulobacter crescentus. J Bacteriol. 2012;194(1):28–35. doi: 10.1128/JB.05932-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rao NN, Kornberg A. Inorganic polyphosphate supports resistance and survival of stationary-phase Escherichia coli. J Bacteriol. 1996;178(5):1394–1400. doi: 10.1128/jb.178.5.1394-1400.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ault-Riché D, Fraley CD, Tzeng CM, Kornberg A. Novel assay reveals multiple pathways regulating stress-induced accumulations of inorganic polyphosphate in Escherichia coli. J Bacteriol. 1998;180(7):1841–1847. doi: 10.1128/jb.180.7.1841-1847.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amado L, Kuzminov A. Polyphosphate accumulation in Escherichia coli in response to defects in DNA metabolism. J Bacteriol. 2009;191(24):7410–7416. doi: 10.1128/JB.01138-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rao NN, Gómez-García MR, Kornberg A. Inorganic polyphosphate: essential for growth and survival. Annu Rev Biochem. 2009;78(1):605–647. doi: 10.1146/annurev.biochem.77.083007.093039. [DOI] [PubMed] [Google Scholar]
- 18.Tocheva EI, et al. Polyphosphate storage during sporulation in the gram-negative bacterium Acetonema longum. J Bacteriol. 2013;195(17):3940–3946. doi: 10.1128/JB.00712-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henry JT, Crosson S. Chromosome replication and segregation govern the biogenesis and inheritance of inorganic polyphosphate granules. Mol Biol Cell. 2013;24(20):3177–3186. doi: 10.1091/mbc.E13-04-0182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller A, et al. Ultrastructure and complex polar architecture of the human pathogen Campylobacter jejuni. MicrobiologyOpen. 2014;3(5):702–710. doi: 10.1002/mbo3.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Varela C, et al. New structural and functional defects in polyphosphate deficient bacteria: a cellular and proteomic study. BMC Microbiol. 2010;10(1):7–14. doi: 10.1186/1471-2180-10-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim HY, et al. Alginate, inorganic polyphosphate, GTP and ppGpp synthesis co-regulated in Pseudomonas aeruginosa: Implications for stationary phase survival and synthesis of RNA/DNA precursors. Mol Microbiol. 1998;27(4):717–725. doi: 10.1046/j.1365-2958.1998.00702.x. [DOI] [PubMed] [Google Scholar]
- 23.Anderson AJ, Dawes EA. Occurrence, metabolism, metabolic role, and industrial uses of bacterial polyhydroxyalkanoates. Microbiol Rev. 1990;54(4):450–472. doi: 10.1128/mr.54.4.450-472.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stubbe J, et al. Nontemplate-dependent polymerization processes: Polyhydroxyalkanoate synthases as a paradigm. Annu Rev Biochem. 2005;74(1):433–480. doi: 10.1146/annurev.biochem.74.082803.133013. [DOI] [PubMed] [Google Scholar]
- 25.Nielsen HJ, Youngren B, Hansen FG, Austin S. Dynamics of Escherichia coli chromosome segregation during multifork replication. J Bacteriol. 2007;189(23):8660–8666. doi: 10.1128/JB.01212-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vallet-Gely I, Boccard F. Chromosomal organization and segregation in Pseudomonas aeruginosa. PLoS Genet. 2013;9(5):e1003492. doi: 10.1371/journal.pgen.1003492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Choi K-H, Schweizer HP. mini-Tn7 insertion in bacteria with single attTn7 sites: Example Pseudomonas aeruginosa. Nat Protoc. 2006;1(1):153–161. doi: 10.1038/nprot.2006.24. [DOI] [PubMed] [Google Scholar]
- 28.Liu J, et al. Novel, fluorescent, SSB protein chimeras with broad utility. Protein Sci. 2011;20(6):1005–1020. doi: 10.1002/pro.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harold FM. Inorganic polyphosphates in biology: Structure, metabolism, and function. Bacteriol Rev. 1966;30(4):772–794. doi: 10.1128/br.30.4.772-794.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kornberg A, Rao NN, Ault-Riché D. Inorganic polyphosphate: A molecule of many functions. Annu Rev Biochem. 1999;68(1):89–125. doi: 10.1146/annurev.biochem.68.1.89. [DOI] [PubMed] [Google Scholar]
- 31.He S, McMahon KD. Microbiology of ‘Candidatus Accumulibacter’ in activated sludge. Microb Biotechnol. 2011;4(5):603–619. doi: 10.1111/j.1751-7915.2011.00248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bru S, Martínez JM, Ortega SH. Polyphosphate is involved in cell cycle progression and genomic stability in Saccharomyces cerevisiae. Mol Microbiol. 2016;101(3):367–380. doi: 10.1111/mmi.13396. [DOI] [PubMed] [Google Scholar]
- 33.Kriel A, et al. Direct regulation of GTP homeostasis by (p)ppGpp: A critical component of viability and stress resistance. Mol Cell. 2012;48(2):231–241. doi: 10.1016/j.molcel.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuroda A, et al. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science. 2001;293(5530):705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- 35.Albi T, Serrano A. Two strictly polyphosphate-dependent gluco(manno)kinases from diazotrophic Cyanobacteria with potential to phosphorylate hexoses from polyphosphates. Appl Microbiol Biotechnol. 2015;99(9):3887–3900. doi: 10.1007/s00253-014-6184-7. [DOI] [PubMed] [Google Scholar]
- 36.Maisonneuve E, Castro-Camargo M, Gerdes K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013;154(5):1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 37.Weber SC, Brangwynne CP. Getting RNA and protein in phase. Cell. 2012;149(6):1188–1191. doi: 10.1016/j.cell.2012.05.022. [DOI] [PubMed] [Google Scholar]
- 38.van Wazer JR. 1958. Chemistry, Phosphorus and Its Compounds: In Two Volumes (Interscience, New York), Vol 1.
- 39.de Azevedo MMM, Bueno M-IMS, Davanzo CU, Galembeck F. Coexistence of liquid phases in the sodium polyphosphate-chromium nitrate-water system. J Colloid Interface Sci. 2002;248(1):185–193. doi: 10.1006/jcis.2001.8201. [DOI] [PubMed] [Google Scholar]
- 40.Grummt I. The nucleolus—Guardian of cellular homeostasis and genome integrity. Chromosoma. 2013;122(6):487–497. doi: 10.1007/s00412-013-0430-0. [DOI] [PubMed] [Google Scholar]
- 41.Tsai RYL, Pederson T. Connecting the nucleolus to the cell cycle and human disease. FASEB J. 2014;28(8):3290–3296. doi: 10.1096/fj.14-254680. [DOI] [PubMed] [Google Scholar]
- 42.Hyman AA, Weber CA, Jülicher F. Liquid-liquid phase separation in biology. Annu Rev Cell Dev Biol. 2014;30(1):39–58. doi: 10.1146/annurev-cellbio-100913-013325. [DOI] [PubMed] [Google Scholar]
- 43.Gray MJ, et al. Polyphosphate is a primordial chaperone. Mol Cell. 2014;53(5):689–699. doi: 10.1016/j.molcel.2014.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blum E, Py B, Carpousis AJ, Higgins CF. Polyphosphate kinase is a component of the Escherichia coli RNA degradosome. Mol Microbiol. 1997;26(2):387–398. doi: 10.1046/j.1365-2958.1997.5901947.x. [DOI] [PubMed] [Google Scholar]
- 45.Cremers CM, et al. Polyphosphate: A conserved modifier of amyloidogenic processes. Mol Cell. 2016;63(5):768–780. doi: 10.1016/j.molcel.2016.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Courchaine EM, Lu A, Neugebauer KM. Droplet organelles? EMBO J. 2016;35(15):1603–1612. doi: 10.15252/embj.201593517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poulin P, Bibette J, Weitz DA. From colloidal aggregation to spinodal decomposition in sticky emulsions. Eur Phys J B. 1999;7(2):277–281. [Google Scholar]
- 48.Parry BR, et al. The bacterial cytoplasm has glass-like properties and is fluidized by metabolic activity. Cell. 2014;156(1-2):183–194. doi: 10.1016/j.cell.2013.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Savage DF, Afonso B, Chen AH, Silver PA. Spatially ordered dynamics of the bacterial carbon fixation machinery. Science. 2010;327(5970):1258–1261. doi: 10.1126/science.1186090. [DOI] [PubMed] [Google Scholar]
- 50.Sato YT, et al. Structural change of DNA induced by nucleoid proteins: growth phase-specific Fis and stationary phase-specific Dps. Biophys J. 2013;105(4):1037–1044. doi: 10.1016/j.bpj.2013.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frenkiel-Krispin D, et al. Nucleoid restructuring in stationary-state bacteria. Mol Microbiol. 2004;51(2):395–405. doi: 10.1046/j.1365-2958.2003.03855.x. [DOI] [PubMed] [Google Scholar]
- 52.Zhang H, Gómez-García MR, Shi X, Rao NN, Kornberg A. Polyphosphate kinase 1, a conserved bacterial enzyme, in a eukaryote, Dictyostelium discoideum, with a role in cytokinesis. Proc Natl Acad Sci USA. 2007;104(42):16486–16491. doi: 10.1073/pnas.0706847104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Livermore TM, Chubb JR, Saiardi A. Developmental accumulation of inorganic polyphosphate affects germination and energetic metabolism in Dictyostelium discoideum. Proc Natl Acad Sci USA. 2016;113(4):996–1001. doi: 10.1073/pnas.1519440113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang L, Fraley CD, Faridi J, Kornberg A, Roth RA. Inorganic polyphosphate stimulates mammalian TOR, a kinase involved in the proliferation of mammary cancer cells. Proc Natl Acad Sci USA. 2003;100(20):11249–11254. doi: 10.1073/pnas.1534805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116(1):71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 56.Sliusarenko O, Heinritz J, Emonet T, Jacobs-Wagner C. High-throughput, subpixel precision analysis of bacterial morphogenesis and intracellular spatio-temporal dynamics. Mol Microbiol. 2011;80(3):612–627. doi: 10.1111/j.1365-2958.2011.07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hocking J, et al. Osmolality-dependent relocation of penicillin-binding protein PBP2 to the division site in Caulobacter crescentus. J Bacteriol. 2012;194(12):3116–3127. doi: 10.1128/JB.00260-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.