Significance
The spatial organization of chromatin within the nucleus regulates genomic functions including DNA repair, transcription, and replication. For example, replication origins cluster prior to initiating replication, likely to aggregate the many necessary factors, but the mechanism is poorly understood. We recently discovered yeast “Forkhead Box” (Fox) DNA binding proteins, Forkhead 1 (Fkh1) and Forkhead 2 (Fkh2), as required for this origin clustering and regulation of initiation timing. This study reveals that Fkh1 and Fkh2 share a structural motif that allows dimerization to bring distal DNA binding sites into close proximity. Mutation that disrupts dimerization ablates origin clustering and deregulates origin activation, suggesting causality between origin clustering and initiation control. We propose that Fkh1 and Fkh2 and related Fox proteins in metazoans establish chromatin architecture.
Keywords: DNA replication timing, chromatin, nuclear organization, Fox proteins, DNA binding protein
Abstract
Forkhead Box (Fox) proteins share the Forkhead domain, a winged-helix DNA binding module, which is conserved among eukaryotes from yeast to humans. These sequence-specific DNA binding proteins have been primarily characterized as transcription factors regulating diverse cellular processes from cell cycle control to developmental fate, deregulation of which contributes to developmental defects, cancer, and aging. We recently identified Saccharomyces cerevisiae Forkhead 1 (Fkh1) and Forkhead 2 (Fkh2) as required for the clustering of a subset of replication origins in G1 phase and for the early initiation of these origins in the ensuing S phase, suggesting a mechanistic role linking the spatial organization of the origins and their activity. Here, we show that Fkh1 and Fkh2 share a unique structural feature of human FoxP proteins that enables FoxP2 and FoxP3 to form domain-swapped dimers capable of bridging two DNA molecules in vitro. Accordingly, Fkh1 self-associates in vitro and in vivo in a manner dependent on the conserved domain-swapping region, strongly suggestive of homodimer formation. Fkh1- and Fkh2-domain-swap-minus (dsm) mutations are functional as transcription factors yet are defective in replication origin timing control. Fkh1-dsm binds replication origins in vivo but fails to cluster them, supporting the conclusion that Fkh1 and Fkh2 dimers perform a structural role in the spatial organization of chromosomal elements with functional importance.
Fundamental processes of DNA repair, recombination, transcription, and replication often occur in specific subnuclear domains or in localized foci (reviewed in refs. 1–4). In yeast for example, hundreds of highly expressed tRNA genes coalesce into multiple foci, each containing several active tRNA genes (reviewed in ref. 5). Similarly, hundreds of replication origins coalesce into foci containing several origins each, which become bidirectional replisomes that remain colocalized as DNA is spooled through during replication (reviewed in ref. 6). Such spatial organization is thought to contribute to the efficiency of these processes by increasing the local concentration of the involved factors, which may consequently also exclude competing or interfering factors or processes. How distal DNA sequences are assembled into these structures is poorly understood.
We recently identified the Saccharomyces cerevisiae transcription factors Forkhead 1 (Fkh1) and Forkhead 2 (Fkh2) as being required for the clustering of a subset of replication origins in G1 phase and for the early initiation of these origins in the ensuing S phase (7). How Fkh1 and Fkh2 promote clustering is unclear; however, their binding near origins might promote origin–origin interactions through binding to other proteins at origins, such as the origin recognition complex (ORC) (7). Fkh1 has also been implicated as a regulator of mating-type switching, which involves homologous recombination between distal chromosomal loci (reviewed in ref. 8). Fkh1 and Fkh2 have been most extensively characterized as transcription factors that control groups of cell cycle control genes, particularly the “cyclin B 2 (CLB2) cluster” genes (reviewed in ref. 9). Similarly, related Forkhead Box (Fox) proteins in other organisms, including humans, have been primarily characterized as functioning in transcriptional control (reviewed in ref. 10). Whereas most forkhead domains bind DNA as monomers, crystallographic and biochemical studies of members of the human FoxP family have revealed that the forkhead domains of FoxP2 and FoxP3 form homodimers, in vitro and likely in vivo, through an exchange of corresponding domains (or subdomains) between monomers, referred to as domain-swapped dimers (11–13). Because Fkh1 and/or Fkh2 (Fkh1/2) have been shown to regulate origin firing through binding in cis to origins, Fkh1/2 dimerization would potentially provide a mechanism for clustering two or more Fkh1/2-bound origins in trans (14).
Results
Fkh1 and Fkh2 Share Amino Acids Specifically Required for FoxP2 and FoxP3 Dimerization.
To address the possibility that the forkhead domains of Fkh1/2 dimerize, we began by comparing their amino acid sequences to those of the other S. cerevisiae forkhead domains from the high-copy suppressor of calmodulin 1 (Hcm1) and Forkhead-like 1 (Fhl1) proteins and those of representative mammalian Fox proteins including the dimerizing FoxP subfamily (Fig. 1A and Fig. S1A). We observed that yeast Fkh1/2 share substantial amino acid identity and similarity with human forkhead domains (Fig. S1B). In fact, these domains are about as similar to the human domains as they are to their yeast counterparts in Hcm1 and Fhl1 (Fig. S1B). Most notably we found that in the positions that have been identified as essential for domain-swap dimerization (11, 12), the Fkh1/2 residues are similar or identical to those of the FoxP subfamily and not to those of the classical monomeric Fox proteins from mammals or yeast (Fig. 1A and Fig. S1A).
Fig. 1.
Fkh1 and Fkh2 share key residues and predicted structure with human FoxP family proteins. (A) Multiple sequence alignment of several human (hs) and budding yeast (sc) forkhead domain regions. Amino acid numbers are based on Fkh1, and specific residues discussed in the text are highlighted. (B) The predicted structure of Fkh1 forkhead domain configured as domain-swapped dimer bound to DNA based on modeling and optimization using the solved crystal structure of the corresponding region of human FoxP3. Individual monomers are colored differently, and the DNA is gray. The arrow indicates the position of A338. (C) Zoomed-in view of domain-swap stabilizing residues Q314 and N326 with salt-bridge interactions depicted as dashed lines.
Fig. S1.
Fkh1 and Fkh2 share key residues and predicted structure with human FoxP family proteins. (A) Multiple sequence alignment of additional human and fission yeast (sp) forkhead domains. Amino acid numbers are based on Fkh1, and specific residues discussed in the text are highlighted. (B) Amino acid identities and similarities in Forkhead proteins across species. (C) Modeled forkhead domain structures of Fkh1 (orange) and Fkh2 (pink) overlaid on the solved structure of FoxP3 domain-swapped dimer (red).
The most important of these domain-swapping related positions is 338 in Fkh1 (375 in Fkh2, 539 in FoxP2, and 372 in FoxP3) (11, 12) (Fig. 1A). The yeast Fkh1/2 and FoxP proteins have an alanine residue at this position while almost all other yeast and human forkhead domains have a proline (Fig. 1A and Fig. S1A). Unlike proline, alanine at this position enables the formation of the extended α-helix that is required for domain swapping (Fig. 1B) (12). Mutation of this alanine to proline ablates in vitro dimerization of FoxP2 and FoxP3 forkhead domains (Fig. 1B) (11, 12).
To further examine the possibility that the Fkh1/2 forkhead domains can dimerize by domain swapping as in the FoxP subfamily, we modeled the structures of Fhk1/2 forkhead domains based on the crystallographic structure of domain-swapped FoxP3 forkhead domain (Fig. 1B and Fig. S1C) (11). Using these models, we evaluated the potential for the formation of a stable domain-swapped interface that would be required for dimerization. At positions 314 and 336 in Fkh1 (351 and 373 in Fkh2, 515 and 537 in FoxP2, 348 and 370 in FoxP3), the yeast Fkh1/2 proteins have glutamine and asparagine residues, respectively (Fig. 1A). These residues can stabilize the dimer interface at the core of the domain-swapped protein by forming salt bridges with their counterparts from the other monomer (Fig. 1C). Such dimer-stabilizing residues are absent from other yeast and human forkhead domains (Fig. 1A and Fig. S1A). These findings imply that S. cerevisiae Fkh1 and Fkh2 have the potential to form domain-swapped dimers like FoxP2 and FoxP3, whereas Hcm1 and Fhl1 do not.
Fkh1 Self-Interaction in Vitro and in Vivo Depends on Conserved Domain-Swapping Residues.
To address whether Fkh1 dimerizes in vivo, we used a coimmunoprecipitation (co-IP) strategy to test for physical interaction between distinctly epitope-tagged, endogenously expressed Fkh1. Immunoprecipitation of Myc-tagged Fkh1 (Fkh1-Myc) resulted in co-IP of Flag-tagged Fkh1 (Fkh1-Flag), consistent with dimer formation (Fig. 2A). Similar pull-down strategies using Escherichia coli-expressed and purified His-tagged or GST-tagged Fkh1 also resulted in coprecipitation of Fkh1-Myc from yeast extracts, again consistent with dimer formation (Fig. S2). Similar strategies to examine Fkh2 were unsuccessful because Fkh2 was unstable in the yeast extract.
Fig. 2.
Fkh1 self-interacts in vivo. (A) Strains OAy1100 (FKH1-3FLAG) harboring pGAL-FKH1-MYC9 and OAy1101 (fkh1-dsm-3FLAG) harboring pGAL-fkh1-dsm-MYC9 were grown to induce expression of Fkh1-Myc or Fkh1-dsm-Myc, and harvested for co-IP analysis. -, control strain lacking epitope tag. (B) Quantification of results from two experiments as in A showing mean (±SD) quantity of Fkh1-Flag pulled down by Fkh1-dsm-Myc versus Fkh1-Myc (the amount of each protein was first normalized relative to its input amount). (C) Cells expressing Fkh1-VC plus either VN, VN-Fkh1, or VN-Fkh1-dsm were analyzed by fluorescence microscopy. Total fluorescence equal to 21, 157, and 21 (arbitrary units) was detected for VN, VN-Fkh1, and VN-Fkh1-dsm, respectively. (D) Zoomed-in and weaker exposure of cells in white box in C coexpressing Fkh1-VC plus VN-Fkh1.
Fig. S2.
Fkh1 dimerizes in vitro. (A) Coomassie-stained SDS/PAGE of E. coli-expressed and purified His-Fkh1-FD (lane 1). (B) Pull-down assay with Fkh1-Myc or Fkh1-dsm-Myc isolated from yeast whole-cell extracts (strains ZOy46 and ZOy50, respectively) and immobilized on beads, incubated with E. coli-expressed and purified His-Fkh1-FD. Bound material was analyzed by immunoblotting with a polyclonal anti-Fkh1 antibody. (C) Quantification of results of three experiments as in B showing the average (±SD) ratio of His-Fkh1-FD pulled down by Fkh1-dsm-Myc versus Fkh1-Myc. (D) Bacterially produced GST, GST-Fkh1, and GST-Fkh1-dsm were immobilized on Glutathione Sepharose beads and incubated with yeast lysate containing Fkh1-Myc (lanes 1–4) or Fkh1-dsm-Myc (lanes 5–8). Pull-downs (Upper) and Input (Lower) were analyzed by immunoblot analysis using anti-Myc antibody. (E) Quantification of data in D showing the ratio of Fkh1-dsm-Myc pulled down by GST-Fkh1-dsm to Fkh1-Myc pulled-down by GST-Fkh1.
Because mutation of the aforementioned amino acids in FoxP2 and FoxP3 ablates dimerization, we tested whether the corresponding mutations in Fkh1 would similarly disrupt its self-interaction, suggestive of dimerization. We constructed a strain in which we replaced the epitope-tagged FKH1 genes with a “domain-swap minus” (dsm) allele, in which we mutated A338 to P to destabilize the essential α-helix (Fig. 1B) and Q314 to E and N336 to D to create electrostatic repulsion rather than attraction at the modeled domain-swap interface (Fig. 1C). As predicted, co-IP of Fkh1-dsm-Flag with Fkh1-dsm-Myc was significantly reduced compared with the wild-type interaction; Fkh1-Flag and Fkh1-dsm-Flag were present at similar levels in the extracts (Fig. 2 A and B). Similar diminution in the in vitro interaction between E. coli-expressed and purified His-tagged or GST-tagged Fkh1-dsm and Fkh1-dsm-Myc from yeast extracts was observed (Fig. S2). These results support the conclusion that Fkh1 dimerizes by domain-swapping through a mechanism analogous to FoxP2/3. The high sequence similarity of the Fkh1 and Fkh2 forkhead domains suggests that Fkh2 is also likely to dimerize (Fig. 1A and Fig. S1B).
To further test for dimerization of Fkh1, we used bimolecular fluorescence complementation (BiFC). The C-terminal half of Venus was fused to the C terminus of Fkh1 (Fkh1-VC, expressed from the endogenous locus), and the N-terminal half of Venus was fused to the N terminus of either Fkh1 or Fkh1-dsm (VN-Fkh1 or VN-Fkh1-dsm, expressed from the GPD promoter on a plasmid). Coexpression of Fkh1-VC and VN-Fkh1 gave rise to yellow fluorescence in the nucleus, indicative of physical proximity of the proteins, and consistent with dimerization (Fig. 2C). In contrast, little or no such interaction was detected in cells expressing Fkh1-VC and VN alone, indicating that Fkh1 is required for the interaction between VC and VN (Fig. 2C). Furthermore, little or no interaction was detected in cells expressing Fkh1-VC and VN-Fkh1-dsm, strongly suggesting that domain-swap dimerization is the mechanism of Fkh1 self-interaction (Fig. 2C). Closer examination of the cells coexpressing Fkh1-VC and VN-Fkh1 revealed an uneven distribution of the BiFC signal (Fig. 2D), which has also been observed with analysis of Fkh1-GFP (15, 16). This uneven distribution of Fkh1 signal is consistent with the idea that Fkh1 molecules cluster together, with dimerization providing the mechanism. Altogether, our data strongly support the conclusion that Fkh1 dimerizes through domain swapping.
fkh1-dsm and fkh2-dsm Are Separation-of-Function Alleles Defective in Origin Regulation.
To examine the functional consequence of the dsm mutations in Fkh1 and Fkh2, we tested the ability of the dsm alleles to complement as effectively as the corresponding wild-type (WT) alleles. We introduced the WT or dsm allele of FKH1 or FKH2 (under native regulation) into fkh1∆ fkh2∆ cells, which display phenotypes not observed in the single mutant cells. These phenotypes include elongated cell morphology, pseudohyphal growth, which is characterized by chains of cells due to delayed cytokinesis, and agar scarring due to invasive growth (17–20); all of these phenotypes are associated with misregulation of Fkh1/2 target genes, particularly cyclin CLB2 (17). The presence of WT FKH1 and FKH2 suppressed all three phenotypes as expected (Fig. 3 A and B). The individual fkh1-dsm and fkh2-dsm alleles also suppressed these phenotypes, indicating that these alleles remain functional in regulation of CLB2.
Fig. 3.
Fkh1-dsm and Fkh2-dsm complement transcriptional defects caused by deletion of FKH1 and FKH2. (A) Phase-contrast images of strains CVy43 (WT), CVy139 (fkh1∆ fkh2∆), ZOy44 (fkh1-dsm fkh2∆), and ZOy45 (fkh1∆ fkh2-dsm); the chains of elongated cells are indicative of pseudohyphal growth. (B) Agar-scarring assay with strains in A showing growth on solid agar prewashing and postwashing; invasive growth results in the remnants in the agar after washing. (C) RNA-seq analysis of 23 CLB2 cluster gene expression levels in logarithmically growing strains ZOy20 (fkh2∆), ZOy21 (fkh1∆), ZOy10 (fkh1-dsm fkh2∆), ZOy12 (fkh1∆ fkh2-dsm), and SKy1 (fkh1∆ fkh2∆). Values were calculated as the ratio between the indicated strains: fkh1∆ and fkh1-dsm alleles were compared with FKH1 in fkh2∆ background, and fkh2∆ and fkh2-dsm alleles were compared with FKH2 in fkh1∆ background. Results of paired t tests are indicated. The 23 CLB2 cluster genes analyzed here are ACE2, AIM20, ALK1, APC1, BUD3, BUD4, BUD8, CDC5, CDC20, CHS2, CLB1, CLB2, HST3, IQG1, IRC8, KIP2, MOB1, MYO1, NUM1, SHE2, SWI5, TEM1, and VAC17.
To examine the function of the fkh1-dsm and fkh2-dsm alleles in greater detail, we performed RNA-sequencing (RNA-seq) analysis of strains bearing WT, dsm, or deletion alleles of FKH1 and FKH2. Analyses of these FKH1 and FKH2 alleles were performed in fkh2∆ and fkh1∆ strains, respectively, to eliminate cross-complementation between FKH1 and FKH2 and establish a robust phenotype with complete genetic dependence on the allele under examination. We focused analysis of the RNA-seq data on the FKH1/2-regulated CLB2 cluster genes. Changes in expression of CLB2 cluster genes were determined by calculating the ratio of RNA levels in the dsm and null stains to WT. For most genes, the ratio of fkh1-dsm/FKH1 and of fkh2-dsm/FKH2 is close to 1, indicating relatively minor changes in expression of CLB2 cluster genes in cells bearing either dsm allele versus its WT equivalent (Fig. 3C). In contrast, more substantial changes in CLB2 cluster gene expression were observed in cells bearing a deletion allele of either FKH1 or FKH2 relative to its WT equivalent (Fig. 3C). Together with the phenotypic analyses above, these results show that Fkh1-dsm and Fkh2-dsm are essentially functional as transcription factors and similar to WT in regulation of CLB2-target genes.
To determine the effect of the dsm mutations on origin regulation by Fkh1 and Fkh2, we generated early S phase replication profiles by using BrdU immunoprecipitation analyzed by DNA sequencing (BrdU-IP-seq) of G1-synchronized cells released into S phase in the presence of hydroxyurea. Plots of replication profiles on representative chromosomes III and VI show BrdU incorporation patterns in WT and fkh1∆ fkh2∆ cells using published data (7) (Fig. 4A). Origins are indicated below the plots and defined as Fkh-activated or Fkh-repressed, if activity in the double mutant was decreased or increased, respectively, as previously determined (7). Consistent with previous results, fkh2∆ cells produced a replication profile nearly identical to WT cells, showing that Fkh1 alone fully maintains normal origin regulation (Fig. 4A). In contrast, fkh1-dsm fkh2∆ cells exhibited a replication profile similar to fkh1∆ fkh2∆ cells, indicating that fkh1-dsm is defective in origin regulation to a degree approaching that of a null mutation. Two-way correlation analyses to quantitatively compare similarity between the genome-wide replication profiles objectively support this conclusion (Fig. 4B).
Fig. 4.
Early replication profiles show that fkh1-dsm and fkh2-dsm are defective in replication timing control. Strains CVy43 (WT), ZOy20 (fkh2∆), ZOy21 (fkh1∆), ZOy10 (fkh1-dsm fkh2∆), ZOy12 (fkh1∆ fkh2-dsm), and SKy1 (fkh1∆ fkh2∆) were analyzed by BrdU-IP-seq after G1 block-and-release into hydroxyurea. (A) Replication profiles are shown for representative chromosomes III and VI; the plots for WT and fkh1∆fkh2∆ were generated from published data. Spheres below the chromosomal plots denote replication origins, with red indicating Fkh-activated and green indicating Fkh-repressed. (B) Heatmap of correlation coefficients of BrdU-IP-seq counts for 5-kb regions surrounding origins. (C) Heatmaps of BrdU-IP-seq counts averaged for 5-kb regions aligned on the indicated origin classes.
The fkh2-dsm allele also exhibited a null-like phenotype with respect to origin regulation as fkh1∆ fkh2-dsm cells yielded replication profiles more like fkh1∆ fkh2∆ than fkh1∆ cells (Fig. 4 A and B). Note that fkh1∆ cells exhibit a phenotype intermediate to WT and fkh1∆ fkh2∆ cells, consistent with Fkh2 only partially compensating for absence of Fkh1, as shown (7). Closer examination shows that the dsm mutations affect the same sets of origins as the null mutations, with decreased activity of Fkh-activated origins, and slightly increased activity of Fkh-repressed origins (Fig. 4C). Together, these results demonstrate that Fkh1-dsm and Fkh2-dsm are defective in origin regulation while functioning to complement the transcription-related growth defects of fkh1∆ fkh2∆ cells.
Fkh1-dsm Binds Origin DNA in Vivo.
The finding that the dsm mutations specifically deregulate origin firing suggests that some aspect of dimerization is important for origin regulation by Fkh1/2. The ability of Fkh1-dsm and Fkh2-dsm to suppress defects associated with CLB2 transcriptional deregulation in fkh1∆ fkh2∆ cells indicates that both proteins retain DNA binding as transcription factors. Indeed, the dsm residues are present on the forkhead domain surface distal to the DNA binding surface (Fig. 1B) (12). Furthermore, previous analysis of the corresponding dsm mutations in FoxP2 and FoxP3 showed no defect in DNA binding in vitro (11, 12). Nevertheless, Fkh1/2 show cell cycle-specific binding to replication origins, which might involve dimerization or some other function dependent on the domain-swapping residues (21). Any such influences on DNA binding would have important consequences for the mechanism of action, so we analyzed chromatin binding of Fkh1-dsm by chromatin immunoprecipitation (ChIP).
We began by analyzing Fkh1-Myc and Fkh1-dsm-Myc binding at Fkh-activated origins ARS305, ARS607, and ARS714, and at CLB2 cluster gene BUD4, using quantitative PCR (qPCR) (Fig. 5A). The results show similar levels of Fkh1 and Fkh1-dsm binding at all of these loci, despite the significantly delayed activation of these origins in S phase. Thus, deregulation of origin timing in fkh1-dsm cells does not appear to reflect defective DNA binding by Fkh1-dsm.
Fig. 5.
Fkh1-dsm binds Fkh-activated origins yet fails to activate them early. Strains ZOy48 (FKH1-MYC9 fkh2∆) and ZOy50 (fkh1-dsm-MYC9 fkh2∆) were analyzed by ChIP-qPCR of unsynchronized cells (A) and by ChIP-chip of G1-arrested cells (B and C). (A) ChIP-qPCR of the indicated loci. Fold enrichment was calculated relative to a low occupancy locus; the error bars represent SD for three experimental samples. (B) Heatmaps of average ChIP-chip signal for Fkh1-Myc and Fkh1-dsm-Myc is shown for 5-kb regions centered on Fkh-activated origins (n = 94), Fkh-repressed origin (n = 80), and ORFs (n = 6607). (C) Box-and-whisker plots of ChIP-chip and BrdU-IP-seq signals are shown for Fkh-activated origins in the top quartiles of both Fkh1-Myc and Fkh1-dsm-Myc binding (ARS216, ARS305, ARS523, ARS729, ARS914, ARS1019, ARS1103, ARS1213, ARS1415, XV-398, and ARS1623). The red “+” indicates an outlier in the distributions; the outliers on the ChIP-chip plots off the scale (M values of 1.8 and 3.1, for Fkh1-Myc and Fkh1-dsm-Myc, respectively) represent the same origin.
To examine Fkh1 and Fkh1-dsm binding genome-wide, we analyzed ChIP with tiling DNA microarrays (ChIP-chip). We analyzed the ChIP signal at Fkh-activated origins by generating heatmaps showing the averaged ChIP signal across 5-kb regions, centered on these origins as well as open reading frames (ORFs) for comparison (Fig. 5B). The analysis shows binding of Fkh1-dsm to Fkh-activated origins with similar average signal as Fkh1. As expected, little enrichment of Fkh1 and Fkh1-dsm was observed at Fkh-repressed origins and at total ORFs. These results counter the idea that defective DNA binding by Fkh1-dsm is responsible for the loss of replication origin regulation. To further support the conclusion that Fkh1-dsm disrupts origin regulation independently of a defect in origin DNA binding, we examined replication origin function at those origins that exhibited the most robust Fkh1 and Fkh1-dsm binding. This analysis shows a strong reduction of early origin activation despite Fkh1-dsm occupancy levels equivalent to WT at this subset of origins (Fig. 5C). Together, these results indicate that Fkh1-dsm deregulates origin function without general loss of DNA binding.
Fkh1-dsm Disrupts 3D Origin Clustering.
The subnuclear localization of replication origins has been correlated with their initiation timing (reviewed in ref. 22), and early-firing origins in yeast have been shown to cluster in the nuclear space during G1 phase using high-throughput chromosome conformation capture analysis (23). We showed that Fkh1/2 are required for this clustering of early replication origins, suggesting that the localization of origins by Fkh1/2 is part of the mechanism of origin timing specification (7). Because we had also demonstrated physical association between Fkh1/2 and ORC, we previously hypothesized that this interaction might mediate the observed interorigin clustering (7). Thus, we considered the possibility that the dsm mutations altered origin timing by altering this physical interaction with ORC. Immunoprecipitation analysis confirms the previously reported interaction between ORC and Fkh1 and further demonstrates that Fkh1-dsm interacts with ORC with similar avidity, indicating that the dsm mutation does not affect interaction with ORC and that interaction between ORC and Fkh1/2 is insufficient for normal origin timing control (Fig. S3).
Fig. S3.
Fkh1 interaction with ORC is unaffected by dsm mutation. Immunoprecipitation of Fkh1-Myc and Fkh-dsm-Myc from cell extracts of strains ZOy46 and ZOy50, respectively, was analyzed by immunoblotting with anti-ORC polyclonal antibody.
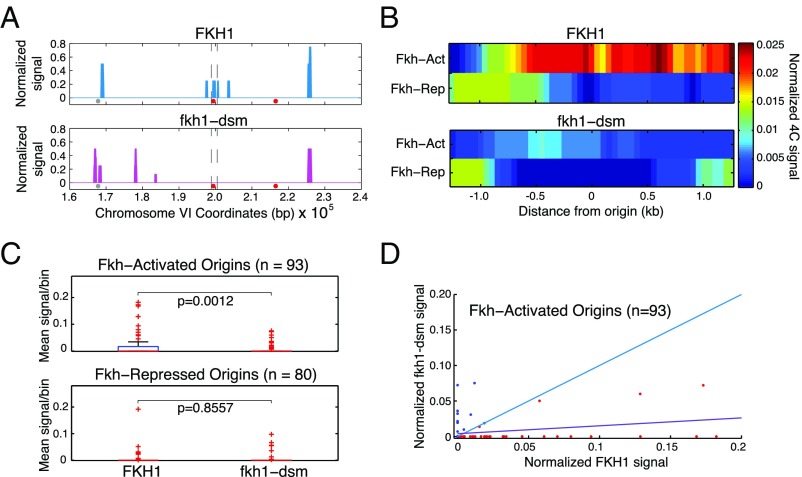
The results presented above indicating potential Fkh1/2 dimerization led us to revise our previous model and to propose that dimerization between origin-bound Fkh1/2 mediates origin-origin clustering, thereby stimulating origin activation. To test this proposition, we performed circular chromosome conformation capture (4C) analyzed by sequencing of fkh2∆ and fkh1-dsm fkh2∆ strains using Fkh-activated origin ARS305 as the bait. These 4C datasets were validated by analysis demonstrating expected features of the data including discrete enrichment of specific XbaI restriction fragments and similarity between experimental replicates (Fig. 6A and Fig. S4). Furthermore, a plot of a region of chromosome VI harboring Fkh-activated origin ARS607, which has been previously captured by ARS305 using 4C analysis, shows interaction with the ARS607-containing XbaI fragment in fkh2∆ cells as expected, due to the presence of FKH1 (Fig. 6A). This ARS305–ARS607 interaction was lost in fkh1-dsm fkh2∆ cells (Fig. 6A), suggesting that the ability of Fkh1 to dimerize is required for this origin–origin interaction. We further analyzed ARS305 interactions by generating heatmaps of the interaction signals captured at other replication origins. ARS305 captured enriched interactions specifically with other Fkh-activated origins but not with Fkh-repressed origins in fkh2∆ cells (Wilcoxon test: P = 2.2886e−5) (Fig. 6B). However, the majority of ARS305–origin interactions were not captured in fkh1-dsm fkh2∆ cells (Fig. 6B). We also examined the distributions of signals at individual origins by generating boxplots and scatter plots comparing the results between fkh2∆ and fkh1-dsm fkh2∆ cells (Fig. 6 C and D). The results show statistically distinct distributions between the Fkh-activated origins interacting with ARS305 in fkh2∆ versus fkh1-dsm fkh2∆ cells (Fig. 6 C and D). Thus, mutations that specifically disrupt the ability of Fkh1 to self-interact also disrupt interchromosomal interactions of Fkh-activated origins, supporting the conclusion that the mechanism by which Fkh1 regulates origins involves dimerization-mediated clustering of replication origins.
Fig. 6.
Origin–origin interactions are lost in fkh1-dsm cells. Strains ZOy20 (fkh2∆) and ZOy10 (fkh1-dsm fkh2∆) were synchronized in G1 phase and subjected to 4C analysis with ARS305 as bait; only the FKH1 genotype is indicated on the images. (A) Data for a region of chromosome VI is plotted, with vertical dashed lines indicating the position of the XbaI fragment containing ARS607. Spheres below the chromosomal plots denote replication origins, with red indicating Fkh-activated origins. (B) Heatmaps show mean, normalized 4C signal of a 2.5-kb region centered on replication origins of the indicated classes. (C) The 4C signals for origins in the indicated classes are plotted as quartile boxplot distributions; red “+” indicate outliers in the distributions; P values were calculated by using a Wilcoxon test. (D) Two-dimensional scatter plot shows normalized 4C signals for each Fkh-activated origin in fkh2∆ and fkh2∆ fkh1-dsm cells. The blue diagonal line indicates equal signals in fkh2∆ versus fkh2∆ fkh1-dsm; the purple line indicates the least-squares regression fit to the data.
Fig. S4.
ARS305 4C interactions along chromosome XII. Strains ZOy20 (fkh2∆) and ZOy10 (fkh1-dsm fkh2∆) were synchronized in G1 phase and subjected to 4C analysis with ARS305 as bait; only the FKH1 genotype is indicated on the images. Spheres below the chromosomal plots denote replication origins, with red indicating Fkh-activated and green indicating Fkh-repressed.
Discussion
Fkh1 and Fkh2 as Genome Architecture Proteins.
The spatial organization of chromosomal elements and related nuclear processes is important for proper chromosomal maintenance and function, however, relatively little is understood about how genome architecture is determined. We recently identified Fkh1 and Fkh2 as novel mediators of early replication origin clustering, required also for their proper initiation timing (7). In this study, we provide further evidence for this role in chromosomal architecture and reveal a potential mechanism to cluster Fkh1/2-bound DNA loci, such as replication origins, involving dimerization through domain-swapping of Fkh1/2. Because FKH1 is sufficient (in fkh2∆ cells) and FKH2 is partly sufficient (in fkh1∆ cells) for normal origin regulation in the absence of the other gene, we can infer that homodimers are involved, especially of Fkh1 which is most enriched at replication origins (21).
This investigation was motivated by the convergent findings that Fkh1/2 regulate replication origin clustering and initiation timing (7), and that the related human FoxP2 and FoxP3 proteins dimerize in vitro (11, 12). Further analysis now shows a remarkable conservation of the residues involved in domain-swapping between Fkh1/2 and FoxP2/3, and an equally remarkable conservation of the helix-breaking proline residue in the other yeast Forkhead proteins Hcm1 and Fhl1 as found in virtually all other human Fox proteins. These findings suggest that diversification of Forkhead proteins into members that can dimerize and others that cannot has long been extant. This diversification of a fundamental, structural capability clearly expands the regulatory possibilities of Fox proteins across species. Intriguingly, Fkh2 in Schizosaccharomyces pombe carries the proline residue (Fig. S1A), suggesting a lack of dimerization potential in this organism, which lacks the highly defined replication timing observed in S. cerevisiae (24).
The dsm mutations that eliminate the potential for dimerization, but not DNA binding, consequently disrupt origin clustering and early initiation timing of bound origins. In contrast, the dsm alleles retain function as transcriptional regulators. This finding might imply that the structural role of Fkh1/2 in genome architecture is more critical for replication timing control than for control of gene expression. However, we note that dsm mutant cells complete genome replication with similar kinetics as WT cells despite the deregulation of individual origin timing, and although gross defects associated with CLB2 cluster gene deregulation are suppressed, transcriptional regulation may not be completely normal (Fig. 3C). Indeed, it seems likely that a role of Fkh1/2 in genome architecture would impact transcriptional regulation at some level for at least a subset of genes.
We have presented in silico, in vivo, and in vitro evidence supporting the conclusion that Fkh1 (and likely Fkh2) homodimerizes through domain swapping, including structural modeling, biochemical pull-down, co-IP, and BIFC. Interestingly, the BIFC analysis exhibited an uneven distribution of the fluorescent signal, consistent with the idea that Fkh1 forms clusters of bound loci. At this point, we cannot rule out that origin clustering and regulation of initiation timing are independent of each other; however, given the specific nature of the dsm mutations, a mechanistic link between the two phenotypes seems likely. Such a link is also consistent with reported correlations between subnuclear origin localization and subsequent replication timing (3). Because Fkh1/2-dependent origin clustering occurs before initiation, in G1 phase when origin timing is established, the results are consistent with a causal relationship. In the case of FoxP3, Chen et al. showed using 4C that expression of FoxP3 in FoxP3-negative T cells induces changes in the 3D interactome of a FoxP3 target gene (13). These results imply that FoxP3 dimers between target genes play a role in coordinating their expression. Together, these results support the conclusion that budding yeast Fkh1 and Fkh2 and human FoxP2 and FoxP3 mediate the 3D organization of chromosomal elements to regulate genome function.
Regulation of Fkh1/2 Dimerization and Control of Genome Structure.
It will be of great interest to determine whether and how Fkh1/2 dimerization is regulated. Fkh1/2 binding at replication origins is distinctly cell cycle regulated (21). Given the specificity of cell cycle regulation for certain loci, we imagine that dimerization might regulate binding at certain loci and/or that certain binding sites, perhaps due to their associated factors such as replication proteins, might preferentially form dimers between DNA-bound loci and, thereby, form intrachromosomal loops or interchromosomal contacts. Dimerization could act as a switch between transcription and replication modes or between activation and repression of such activities. Also, in this regard, dimerization might change the affinity of Fkh1/2 for certain sites, thereby acting selectively to organize certain chromosomal elements in time and space. Phosphorylation or other modifications might regulate dimerization as well. Dissolution of dimers may also be an important point of regulation. For example, clusters of replication origins and associated replicons may require disassembly to permit replication progression or termination, resolution of topological constraints, or chromosome segregation. Finally, alterations in chromosome structure incurred by regulation of Fkh1/2 binding or dimerization are likely to induce secondary effects on the localization and, hence, function of other DNA loci and elements.
Experimental Procedures
Homology Modeling of the Yeast Forkhead Proteins.
Multiple sequence alignments were carried out by using Clustal Omega (25). The structures of yeast Fkh1 and Fkh2 forkhead domains were modeled based on the published structure of FoxP3 domain-swapped dimer (PDB ID code 3QRF) and optimized by using the SWISS-MODEL workspace (26, 27). Molecular graphics and analyses were performed with the University of California, San Francisco Chimera package (28).
Strain and Plasmid Constructions and Other Methods.
Most yeast strains are congenic with W303 and most are derived from BrdU-incorporating strain CVy43 (29); BY4741 was the parent for the strains used in the BiFC analysis. Complete strains genotypes are given in Table 1. Construction of pGAL-FKH1-MYC9 has been described (30). FKH1 and FKH2 from pCF480 and pCF403 (from C. Fox, University of Wisconsin, Madison, WI), respectively, were subcloned by using XhoI and NotI into integrating vector pRS405, yielding p405-FKH1 and p405-FKH2, respectively. The dsm mutations were introduced into the above plasmids by using QuikChange Lightning Mutagenesis Kit (Agilent Technologies), yielding pGAL-fkh1-dsm-MYC9, p405-fkh1-dsm, and p405-fkh2-dsm. Fkh1-dsm was subcloned into pRS306 to yield p306-fkh1-dsm. The pRS405-based FKH1- and FKH2-bearing plasmids were integrated into the genome, after digestion by BsaBI and HpaI, respectively, by lithium acetate transformation (31). Genetic modifications to the genome were confirmed by PCR analysis and/or sequencing as appropriate. Flag-tagging of FKH1 was performed by PCR amplification using plasmid p2L-3Flag-TRP1 (K.l.) (from T. Tsukiyama, Fred Hutchinson Cancer Research Center, Seattle) for transformation into yeast. Plasmid pFA6a-Venus-C (accession no. EF210810; ref. 32) was used to PCR amplify the Fkh1-Venus-C fusion cassette for transformation into yeast. Construction of plasmids p426GPD-VN and p426GPD-VN-Fkh1 has been described (33); p426GPD-VN-Fkh1-dsm was similarly constructed by using p306-fkh1-dsm as a template. Table 2 lists primer sequences used for mutagenesis, plasmid, and strain constructions. For the GST pull-down experiments, the plasmids pGEX6p2-Fkh1 and pGEX6p2-Fkh1-dsm encoding the ORF of the respective genes fused to GST were generated by subcloning the respective DNA fragments treated with the restriction endonucleases SalI and NotI into the SalI/NotI sites of the plasmid pGEX6p2 (Phamarcia Biotech).
Table 1.
Yeast strains used in this study
| Strain | Genotype | Source |
| Strains share the SSy161 genotype except as noted | ||
| SSy161 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 bar1Δ::hisG | 29 |
| CVy43 | ura3::BrdU-Inc (URA3) | 29 |
| CVy138 | fkh2Δ::HIS3MX ura3::BrdU-Inc (URA3) | 7 |
| CVy139 | fkh1Δ::KanMX fkh2Δ::HIS3MX ura3::BrdU-Inc (URA3) | 7 |
| SKy1 | fkh1Δ::KanMX fkh2Δ::HIS3MX ura3::BrdU-Inc (URA3) pfkh2∆C | 7 |
| ZOy10 | fkh1Δ::KanMX::fkh1-dsm (LEU2) fkh2Δ::HIS3MX ura3::BrdU-Inc (URA3) pfkh2∆C | Present study |
| ZOy12 | fkh1Δ::KanMX fkh2Δ::HIS3MX leu2::fkh2-dsm (LEU2) ura3::BrdU-Inc (URA3) pfkh2∆C | Present study |
| ZOy20 | fkh2Δ::HIS3MX ura3::BrdU-Inc (URA3) pfkh2∆C | Present study |
| ZOy21 | fkh1Δ::KanMX fkh2Δ::HIS3MX leu2::FKH2 (LEU2) ura3::BrdU-Inc (URA3) pfkh2∆C | Present study |
| ZOy22 | FKH1-MYC9 (TRP1) fkh2Δ::HIS3MX ura3::BrdU-Inc (URA3) | 7 |
| ZOy44 | fkh1Δ::KanMX::fkh1-dsm (LEU2) fkh2Δ::HIS3MX ura3::BrdU-Inc (URA3) | Present study |
| ZOy45 | fkh1Δ::KanMX fkh2Δ::HIS3MX leu2::fkh2-dsm (LEU2) | Present study |
| ZOy48 | FKH1-MYC9 (TRP1) fkh2Δ::HIS3MX | Present study |
| ZOy50 | fkh1Δ::KanMX::fkh1-dsm (LEU2)::MYC9 (TRP1) fkh2Δ::HIS3MX | Present study |
| Strains below BY4741 share its genotype except as noted | ||
| BY4741 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | Euroscarf |
| Fkh1-VC/VN | MATa FKH1-VC::kanMX6 p426GPDpr-VN | Present study |
| Fkh1-VC/VN-Fkh1 | MATa FKH1-VC::kanMX6 p426GPDpr-VN-FKH1 | Present study |
| Fkh1-VC/VN-Fkh1dsm | MATa FKH1-VC::kanMX6 p426GPDpr-VN-fkh1-dsm | Present study |
Table 2.
Oligonucleotide DNA sequences used in this study
| Primer name | Sequence |
| Fkh1 dsm Q->E | tatgcatcgatgatcaccgaagccattctttcaacac |
| Fkh1 dsm A,N->P,D | gatatttacaaatttatctctgacgactacccattttacaggttttctcaaatgg |
| Fkh2 dsm Q->E | tcattcatacgcaactatgataacagaggccatactgtc |
| Fkh2 dsm A,N->P,D | gatatctacaagtatatttcttccgattatccatactacaggtttgctaaatccg |
| ARS305-4C-us-XbaI-ap | ctaagtgtcctgtttcggaac |
| ARS305-4C-us-MseI | caggccgctcttataaaatga |
| ARS714-for | atacttccccgctatttg |
| ARS714-rev | actatagcagaacgaggta |
| ARS305-for | ttactttgtagttcttaaagc |
| ARS305-rev | cgtgtaagttactttaatgag |
| ARS607-for | ccattagagacagagaaactattcattg |
| ARS607-rev | ctactgtgccgaataatgtgtaag |
| BUD4 promoter-for | cacacatacacacaaatctc |
| BUD4 promoter-rev | ggtcatctattcgttcctc |
| BUD3 upstream-for | tcatctttgctacgtgag |
| BUD3 upstream-rev | cgtcccatatatcttgacat |
Cells were grown in YEPD unless otherwise noted; synchronization methods have been described (34). Myc-tagging, ChIP, ChIP-chip, and BrdU-IP-seq were performed as described (7), except that sonication was performed with a Covaris S2 Instrument. For the agar-scarring assay, cells were patched onto YEPD and grown ∼3 d at 30 °C. Plates were washed with a gentle stream of water to remove cells on the agar surface. For co-IP, log cells were grown in YEP-2% (wt/vol) raffinose were resuspended in YEP-2% (wt/vol) galactose for 3 h at 25 °C and harvested for co-IP as described (7), with anti-Myc 9E10 (Covance MMS150P) or 9E11 (Biolegend 904401), and anti-Flag M2 (Sigma F1805). For RNA-seq analysis, total RNA was isolated from 10-mL cultures grown in YEPD to OD600 ∼ 1 by using Ribopure Yeast Kit (ThermoFisher AM1926). cDNA libraries were created by using SuperScript III First-Strand Synthesis (ThermoFisher 18080051) with oligo dT primers followed by NEBNext Second Strand Synthesis (NEB E6111S). cDNA libraries were prepared for sequencing and analyzed as described for BrdU-IP-Seq. The 4C analysis was performed as described (7), with the following modifications: 50-mL cultures were used, and the protocol was scaled accordingly. Primary digestion was incubated without shaking. ARS305 PCR material was processed for sequencing as described for BrdU-IP-Seq. Sequence data preprocessing was performed as described for BrdU-IP-Seq with the following differences: reads were 5′ trimmed to remove primer sequence up to MseI cut site before alignment, aligned reads were binned into genomic XbaI fragments, and binned data were normalized against total read counts for each experiment. Primers for 4C are listed in Table 2.
ChIP-Quantitative PCR.
The experiment was performed in triplicate for each strain; a technical replicate of each qPCR reaction was performed. Primers were designed around the loci of interest to amplify ∼170-bp fragments. IP DNA was diluted 1:5, Total DNA was diluted 1:500; 1 μL of diluted DNA was used for each qPCR reaction. Fifty-microliter qPCR reactions were performed by using Phusion High-Fidelity DNA Polymerase (NEB), following manufacturer’s instructions, plus 26 μM SYBR green (Thermo Fisher) and 2.6 μM ROX solution (Thermo Fisher), using an MJ Research Opticon 2 QPCR instrument. qPCR results were analyzed by percent of input (35), then normalized against an internal control with low occupancy signal (upstream BUD3). Table 2 gives the sequences of primers used.
Expression and Purification of Fkh1-FD from E. coli.
Fkh1-FD (amino acids 243–484) cloned into pET28 (generously provided by C. Fox) was transformed into Rosetta 2 DE3 cells. IPTG (0.2 mM) was added to induce expression; after overnight growth at 16 °C, cells were harvested by centrifugation at 4 °C. The following steps on ice or at 4 °C: Cells were suspended in MCAC-0 buffer [20 mM Tris-Cl pH 8.0, 0.5 M NaCl, 10% (vol/vol) glycerol, 1× Protease Inhibitor Mixture (Roche), 1 mM PMSF] and broken by sonication; the lysate was clarified by centrifugation and incubated with Ni-NTA resin (Qiagen) preequilibrated in MCAC-0. After binding, resin was washed sequentially with MCAC-10, -20, -30, and -40 mM imidazole, followed by elution with MCAC-250 mM imidazole. Buffer change was performed with Amicon Ultra Centrifugal Filters (EMD Millipore) for subsequent analysis.
Pull-Down Assays.
Cells from 50-mL culture (OD600 ∼ 1) were harvested by centrifugation (∼1,500 × g, 3 min). Hereon, all steps were performed at 4 °C or on ice by using prechilled reagents, materials, and instruments unless noted otherwise. Bead incubations and washings were performed with gentle rotation. Cells were resuspended in 20 mL of 50 mM Hepes-KOH (pH 7.6), pelleted again, and resuspended in 1 mL of 50 mM Hepes-KOH (pH 7.6). Cells were transferred to a 2-mL Fastprep tube, pelleted, supernatant removed, frozen in dry ice/ethanol bath, and stored at −80 °C. Pellets were thawed on ice, 300-μL glass beads (∼600 μm), and 300 μL of HMGI-0 buffer [50 mM Hepes-KOH (pH 7.6), 1 mM MgCl2, 5% (vol/vol) glycerol, 0.15% Tween-20, 1× protease inhibitor mixture (Roche), 50 mM NaF, 25 mM β-glycero-phosphate] was added. Cells were broken by bead-beating (Speed: 5.5, 45 s) in the Fastprep instrument (MP Biomedicals) two times, each followed by 1 min on ice. Two hundred microliters of HMGI-0 buffer was added and bead-beating was performed once more. Glass beads and cell debris were pelleted in a microcentrifuge (∼1,000 × g, 5 min) and the supernatant decanted to a new microcentrifuge tube. Extract was clarified by centrifugation (∼15,000 × g, 5 min), and soluble extract was decanted to a new tube. Protein concentration was determined by Bradford assay. Protein G Dynabeads (Invitrogen) were prepared by washing twice with 1 mL of HMGI-250 buffer (HMGI-0 plus 250 mM imidazole), incubating in HMGI-250 for 1 h, followed by washing three times with HMGI-0 buffer. Four miligrams of protein was diluted to 400 μL with HMGI-0 buffer and 20 μL of prepared Protein G Dynabeads was added and incubated for 15 min. Supernatant was transferred to a new tube and 4 μL of 9E10 monoclonal antibody (Covance) was added and incubated 30 min. Beads were washed three times, 3 min each, with 1 mL of HMGI-0. Beads were resuspended in 150 µL of HMGI-0 buffer and 500 ng E. coli-purified Fkh1-FD was added and incubated 15 min. Beads were washed three times, 3 min each, with HMGI-0 buffer. After removal of final wash buffer, beads were resuspended in 36 μL of denaturing sample buffer, boiled 3 min, and 14 μL was loaded onto 10% (wt/vol) SDS/PAGE gel. Immunoblotting was performed with semidry transfer and detection with polyclonal anti-Fkh1 (1:500), a gift from C. Fox. GST pull-down assays were performed as described (36).
BiFC Assay.
Haploid yeast cells expressing the C-terminal region of the Venus protein fused to the C-terminal region of Fkh1 (Fkh1-VC) were transformed either with plasmid p426GPD-VN, p426GPD-VN-Fkh1, or p426GPD-VN-Fkh1-dsm encoding fusion proteins between the N-terminal region of Venus and the N-terminal region of the selected proteins. Transformed clones were isolated and cultured in liquid SC-URA media (OD600 ∼ 0.6). Cells were washed once with 1× PBS and monitored for a Venus-dependent fluorescent (BiFC) signal by using a Zeiss Axio Scope A1 microscope (Carl Zeiss AG) with a Plan-Apochromat 100×/1.4 N.A. Oil Ph3 immersion objective. Fluorescence images were taken by using a standard fluorescein isothiocyanate filter set (scar range 450–650 nm; 25 mm excitation and emission filters; 25.5 × 36 × 1 mm beamsplitter), and recorded on a Zeiss AxioCam ICm1 (Carl Zeiss AG).
Array and DNA Sequence Data Processing.
Sequence data were preprocessed and microarray data scanned as described (21, 37). Processing and normalization of slide data were performed with MA2C (38), and resulting .wig files were converted to .bed format. Data were then converted into 50-bp binned .bed files by using BEDtools (39). To construct a replication profile correlation matrix, 5-kb windows around origins identified in ref. 7 were analyzed to produce correlation coefficients specific to replication profiles in each strain. Heat maps of signal around origins and CLB2 cluster gene promoters were created as described (21). Selection of top quartiles of ChIP-chip binding signal was done by ranking Fkh-activated origins (7) from highest to lowest based on ChIP-chip M values for Fkh1-Myc and Fkh1-dsm-Myc. The top quartile of each respective list was chosen, and the overlap between the two selections was used to represent the top-bound origins in both Fkh1-Myc and Fkh1-dsm-Myc strains. The same list of origins was then analyzed for signal in BrdU-IP-seq experiments.
Acknowledgments
We thank Drs. Irene Chiolo, Simon Knott, and Jared Peace for helpful discussions and critical reading of the manuscript; Drs. Stephen Bell (Massachusetts Institute of Technology), Toshio Tsukiyama (Fred Hutchinson Cancer Research Center), and Catherine Fox (University of Wisconsin) for plasmids and/or antibodies; Dr. Jordan Eboreime for assistance with qPCR; Dr. Justin Dalton for assistance with microscopy; and Selene Tyndale and Dr. Charles Nicolet of the NextGen Sequencing Core of the Norris Comprehensive Cancer Center at USC Keck School of Medicine, Dr. Roger Mercer of the Translational Science Laboratory at Florida State University College of Medicine, and Dr. Michelle Arbeitman at Florida State University College of Medicine for assistance with high-throughput DNA sequencing. We also thank Max Planck Institute for Molecular Genetics, Berlin, Germany, for hosting C.L. and M.B. during the generation of the plasmids pGEX6p2-Fkh1, p426GPDp2-VN, and p426GPDp2-VN-Fkh1 and Markus Ralser (University of Cambridge) for the plasmid pGEX6p2. This study was supported by NIH Grants 2R01-GM065494 (to O.M.A.) and Cancer Center Support Grant P30CA014089 from the National Cancer Institute, and by the SILS Starting Grant of the University of Amsterdam (to M.B.). A.Z.O. also received support from NIH Grant P50-HG002790.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE94796).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1612422114/-/DCSupplemental.
References
- 1.Misteli T. Beyond the sequence: Cellular organization of genome function. Cell. 2007;128(4):787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- 2.Cavalli G, Misteli T. Functional implications of genome topology. Nat Struct Mol Biol. 2013;20(3):290–299. doi: 10.1038/nsmb.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Taddei A, Gasser SM. Structure and function in the budding yeast nucleus. Genetics. 2012;192(1):107–129. doi: 10.1534/genetics.112.140608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bickmore WA, van Steensel B. Genome architecture: Domain organization of interphase chromosomes. Cell. 2013;152(6):1270–1284. doi: 10.1016/j.cell.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Haeusler RA, Engelke DR. Genome organization in three dimensions: Thinking outside the line. Cell Cycle. 2004;3(3):273–275. [PMC free article] [PubMed] [Google Scholar]
- 6.Natsume T, Tanaka TU. Spatial regulation and organization of DNA replication within the nucleus. Chromosome Res. 2010;18(1):7–17. doi: 10.1007/s10577-009-9088-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knott SR, et al. Forkhead transcription factors establish origin timing and long-range clustering in S. cerevisiae. Cell. 2012;148(1-2):99–111. doi: 10.1016/j.cell.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haber JE. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191(1):33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murakami H, Aiba H, Nakanishi M, Murakami-Tonami Y. Regulation of yeast forkhead transcription factors and FoxM1 by cyclin-dependent and polo-like kinases. Cell Cycle. 2010;9(16):3233–3242. doi: 10.4161/cc.9.16.12599. [DOI] [PubMed] [Google Scholar]
- 10.Lalmansingh AS, Karmakar S, Jin Y, Nagaich AK. Multiple modes of chromatin remodeling by Forkhead box proteins. Biochim Biophys Acta. 2012;1819(7):707–715. doi: 10.1016/j.bbagrm.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 11.Bandukwala HS, et al. Structure of a domain-swapped FOXP3 dimer on DNA and its function in regulatory T cells. Immunity. 2011;34(4):479–491. doi: 10.1016/j.immuni.2011.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stroud JC, et al. Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 2006;14(1):159–166. doi: 10.1016/j.str.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, et al. DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions. Nucleic Acids Res. 2015;43(2):1268–1282. doi: 10.1093/nar/gku1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aparicio OM. Location, location, location: It’s all in the timing for replication origins. Genes Dev. 2013;27(2):117–128. doi: 10.1101/gad.209999.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425(6959):686–691. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 16.Tkach JM, et al. Dissecting DNA damage response pathways by analysing protein localization and abundance changes during DNA replication stress. Nat Cell Biol. 2012;14(9):966–976. doi: 10.1038/ncb2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hollenhorst PC, Bose ME, Mielke MR, Müller U, Fox CA. Forkhead genes in transcriptional silencing, cell morphology and the cell cycle. Overlapping and distinct functions for FKH1 and FKH2 in Saccharomyces cerevisiae. Genetics. 2000;154(4):1533–1548. doi: 10.1093/genetics/154.4.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kumar R, et al. Forkhead transcription factors, Fkh1p and Fkh2p, collaborate with Mcm1p to control transcription required for M-phase. Curr Biol. 2000;10(15):896–906. doi: 10.1016/s0960-9822(00)00618-7. [DOI] [PubMed] [Google Scholar]
- 19.Pic A, et al. The forkhead protein Fkh2 is a component of the yeast cell cycle transcription factor SFF. EMBO J. 2000;19(14):3750–3761. doi: 10.1093/emboj/19.14.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu G, et al. Two yeast forkhead genes regulate the cell cycle and pseudohyphal growth. Nature. 2000;406(6791):90–94. doi: 10.1038/35017581. [DOI] [PubMed] [Google Scholar]
- 21.Ostrow AZ, et al. Fkh1 and Fkh2 bind multiple chromosomal elements in the S. cerevisiae genome with distinct specificities and cell cycle dynamics. PLoS One. 2014;9(2):e87647. doi: 10.1371/journal.pone.0087647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rhind N, Gilbert DM. DNA replication timing. Cold Spring Harb Perspect Biol. 2013;5(8):a010132. doi: 10.1101/cshperspect.a010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan Z, et al. A three-dimensional model of the yeast genome. Nature. 2010;465(7296):363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patel PK, Arcangioli B, Baker SP, Bensimon A, Rhind N. DNA replication origins fire stochastically in fission yeast. Mol Biol Cell. 2006;17(1):308–316. doi: 10.1091/mbc.E05-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sievers F, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: A web-based environment for protein structure homology modelling. Bioinformatics. 2006;22(2):195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 27.Bordoli L, et al. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4(1):1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 28.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 29.Viggiani CJ, Aparicio OM. New vectors for simplified construction of BrdU-Incorporating strains of Saccharomyces cerevisiae. Yeast. 2006;23(14-15):1045–1051. doi: 10.1002/yea.1406. [DOI] [PubMed] [Google Scholar]
- 30.Peace JM, Villwock SK, Zeytounian JL, Gan Y, Aparicio OM. Quantitative BrdU immunoprecipitation method demonstrates that Fkh1 and Fkh2 are rate-limiting activators of replication origins that reprogram replication timing in G1 phase. Genome Res. 2016;26(3):365–375. doi: 10.1101/gr.196857.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2(1):31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 32.Sung MK, Huh WK. Bimolecular fluorescence complementation analysis system for in vivo detection of protein-protein interaction in Saccharomyces cerevisiae. Yeast. 2007;24(9):767–775. doi: 10.1002/yea.1504. [DOI] [PubMed] [Google Scholar]
- 33.Linke C, Klipp E, Lehrach H, Barberis M, Krobitsch S. Fkh1 and Fkh2 associate with Sir2 to control CLB2 transcription under normal and oxidative stress conditions. Front Physiol. 2013;4:173. doi: 10.3389/fphys.2013.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aparicio JG, Viggiani CJ, Gibson DG, Aparicio OM. The Rpd3-Sin3 histone deacetylase regulates replication timing and enables intra-S origin control in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24(11):4769–4780. doi: 10.1128/MCB.24.11.4769-4780.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nagaki K, et al. Chromatin immunoprecipitation reveals that the 180-bp satellite repeat is the key functional DNA element of Arabidopsis thaliana centromeres. Genetics. 2003;163(3):1221–1225. doi: 10.1093/genetics/163.3.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barberis M, et al. Sic1 plays a role in timing and oscillatory behaviour of B-type cyclins. Biotechnol Adv. 2012;30(1):108–130. doi: 10.1016/j.biotechadv.2011.09.004. [DOI] [PubMed] [Google Scholar]
- 37.Ostrow AZ, Viggiani CJ, Aparicio JG, Aparicio OM. ChIP-Seq to analyze the binding of replication proteins to chromatin. Methods Mol Biol. 2015;1300:155–168. doi: 10.1007/978-1-4939-2596-4_11. [DOI] [PubMed] [Google Scholar]
- 38.Song JS, et al. Model-based analysis of two-color arrays (MA2C) Genome Biol. 2007;8(8):R178. doi: 10.1186/gb-2007-8-8-r178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Quinlan AR, Hall IM. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26(6):841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]