Significance
Plant leaves form at the flanks of the shoot apical meristem in response to cues provided by the phytohormone auxin. Auxin signals determine the sites of leaf initiation and bulging, a process followed by gradual and ongoing differentiation of leaf tissues. We show here that the tomato ethylene response factor-type transcription factor LEAFLESS is induced by, and necessary for, auxin-triggered leaf initiation. These mechanisms provide a localized and transient developmental context for a specific morphogenetic output generated by general regulators such as auxin.
Keywords: DRN/DRNL, extended GCC-box, lateral organ formation, SAM, TIBA pins
Abstract
Lateral plant organs, particularly leaves, initiate at the flanks of the shoot apical meristem (SAM) following auxin maxima signals; however, little is known about the underlying mechanisms. Here, we show that tomato leafless (lfs) mutants fail to produce cotyledons and leaves and grow a naked pin while maintaining an active SAM. A similar phenotype was observed among pin-like shoots induced by polar auxin transport inhibitors such as 2,3,5-triiodobenzoic acid (TIBA). Both types of pin-like shoots showed reduced expression of primordia markers as well as abnormal auxin distribution, as evidenced by expression of the auxin reporters pPIN1:PIN1:GFP and DR5:YFP. Upon auxin microapplication, both lfs meristems and TIBA-pin apices activated DR5:YFP expression with similar kinetics; however, only lfs plants failed to concurrently initiate leaf primordia. We found that LFS encodes the single tomato ortholog of Arabidopsis DORNRONSCHEN (DRN) and DRN-like (DRNL) genes and is transiently expressed at incipient and young primordia, overlapping with auxin response maxima. LFS is rapidly induced by auxin application, implying feed-forward activity between LFS and auxin signals. However, driving LFS at auxin response maxima sites using the DR5 promoter fails to fully rescue lfs plants, suggesting that additional, auxin-independent regulation is needed. Indeed, extended GCC-box elements upstream of LFS drove primordia-specific expression in a LFS-dependent but auxin-independent manner. We thus suggest that LFS transiently acts at the site of primordia initiation, where it provides a specific context to auxin response maxima culminating in leaf primordia initiation.
Lateral aerial plant organs form at the flanks of the shoot apical meristem (SAM) in a species-specific order termed phyllotaxis. Phyllotactic patterns tightly correlate with local maxima of the phytohormone auxin at the SAM periphery (1–3). These maxima are achieved by active transport (4). At these sites, auxin is required for leaf initiation, and when polar auxin transport is chemically impaired by polar auxin transport inhibitors (PATI), leaves fail to form (5–7), resulting in a pin-like shoot. When auxin is externally applied onto the meristem of these pins or to the meristem in a wild-type (WT) background, leaf primordia are initiated (5). Genetic analyses have also demonstrated that auxin response maxima, dictated by the activity of the auxin efflux carrier PINFORMED1 (PIN1), can be detected before, and are required for, leaflet initiation in compound leaves (6). However, similar evidence for leaf initiation is lacking (7).
Like other classical hormones, such as cytokinin (CK), florigen, and gibberellin, auxin has many functions, which are largely dictated by its levels and which vary within and among plants. In young leaf primordia, auxin triggers primordia bulging at P0 by stimulating local cell divisions. At P1 and P2, auxin elicits provasculature development (8). Thus, in the same group of cells, auxin cues are differentially translated to cell division (P0) or to differentiation (P1) within a 1- to 2-d gap. The mechanisms directing such context-specific responses are poorly understood.
In this work, we investigate leaf initiation at the SAM periphery and show that the tomato leafless (lfs) mutant, which is disrupted in a single DRN/DRNL homolog, fails to initiate cotyledons, leaves, and leaflets. LFS expression largely overlapped with auxin response maxima during primordia initiation, yet exogenous auxin application did not stimulate leaf initiation when applied to bare lfs shoots. We suggest that LFS responds to auxin response maxima and sets the stage for the context-specific response that culminates in leaf initiation. The transient LFS expression, which is terminated shortly after leaf initiation, may prevent rapid differentiation by promoting, among others, CK signals.
Results
The leafless Mutants Produce Pin-Like Shoots with Active SAM.
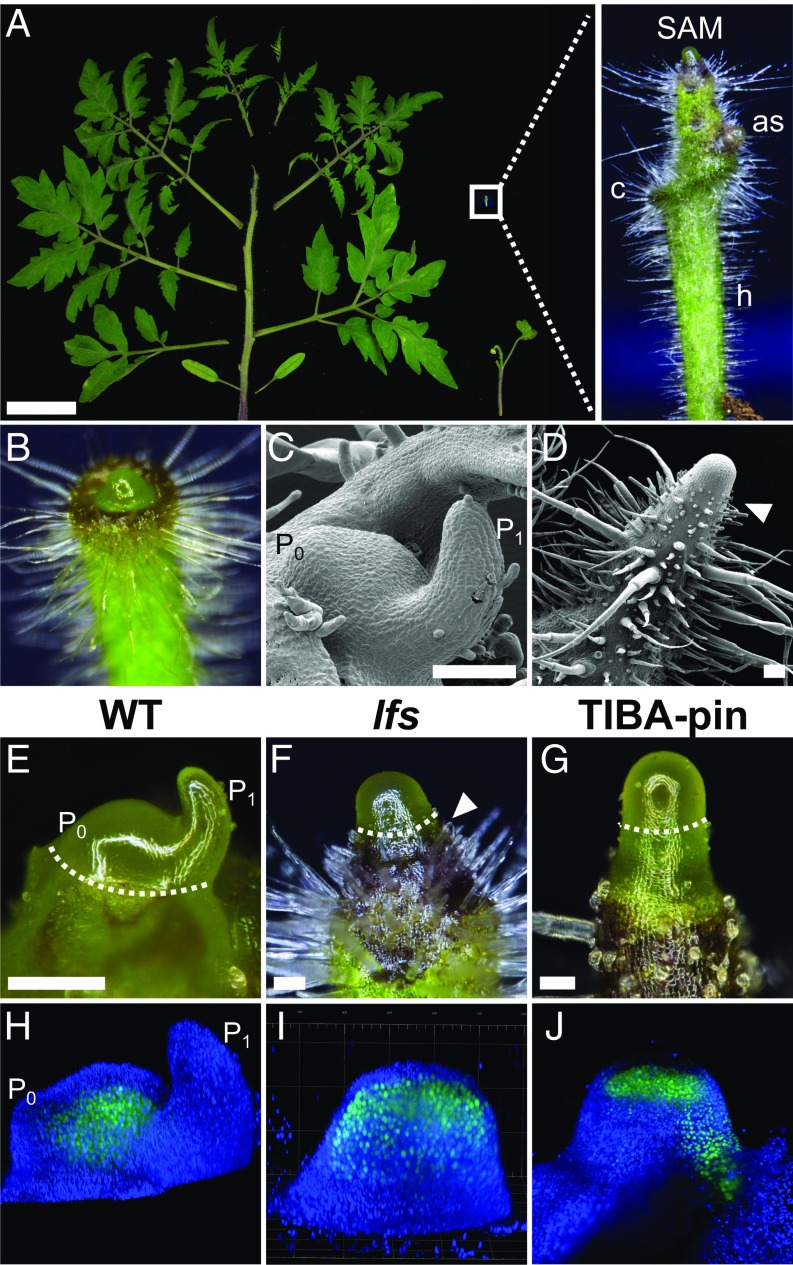
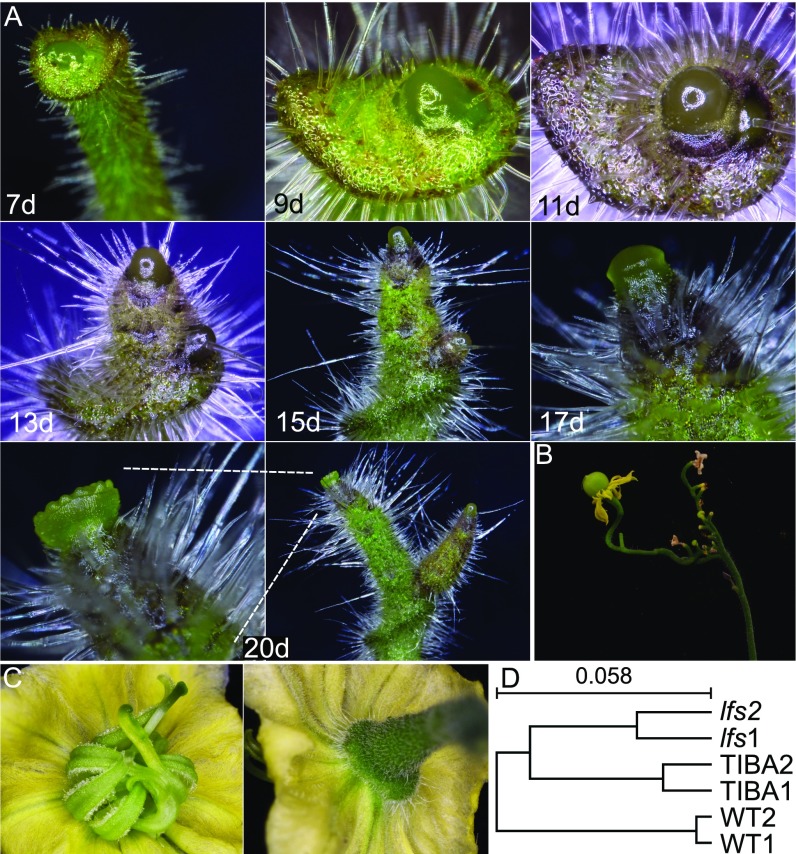
Five recessive lfs mutants (Fig. 1A) were identified in a screen for tomato plants that fail to properly produce leaves; complementation tests demonstrated that they were all allelic. The lfs seedlings lacked cotyledons, but after germination, a broad collar-like tissue surrounded the SAM, likely representing the vestigial cotyledons (Fig. 1A and Fig. S1A). As the seedlings matured, the miniature plant shoot appeared as an elongated pin. Although the meristems at the top of these pins featured a typical smooth and glossy appearance, the immediately adjacent stem tissue underwent rapid differentiation, as was inferred by the presence of large and dense trichomes just below the SAM dome (Fig. 1 A–F). The bare lfs plant SAMs occasionally formed one to three simple leaves, with zero to three leaflets and entire margins with abnormal phyllotaxis (Fig. 1A). These leaves hosted axillary buds that carried pin-like shoots in their axils; however, additional pin-like shoots also initiated from areas that corresponded to cryptic axillary buds (Fig. 1A and Fig. S1A). The naked meristems topping the elongated pin mutant shoots turned into a flower at about the same time as their WT siblings (Fig. S1A). The flowers were deformed, with sepals missing or small, petals nearly normal, stamens malformed and sterile and with two or more carpels, which rarely gave rise to parthenocarpic fruit (Fig. S1 B and C). Floral organs were often fused. All of these defects were similar in the five tested alleles. In the WT background, we could not differentiate lfs/+ plants from +/+ sibs.
Fig. 1.
LFS and auxin response maxima direct tomato leaf initiation. (A) A 5.5-wk-old WT (Left) and lfs (Bottom Right) tomato. (Scale bar: 5 cm.) (Inset) A close-up of the apex of a 15-d-old lfs seedling. as, axillary shoot; c, collar-like structure; h, hypocotyl. (B) A 9-d-old lfs seedling in which the SAM starts to protrude from the collar-like structure surrounding it. (C and D) SEM images of vegetative WT (C) and lfs (D) apices. (E) WT apical meristem bearing one incipient and one visible primordia (P0 and P1, respectively). (F) lfs meristem. (G) TIBA-induced pin. Dashed lines in E–G mark the apex tissue, sampled for gene expression analyses. Note the proximity of trichomes to the SAM in D and F, marked by the arrowhead. (H–J) Three-dimensional reconstructions of live TCS:YFP apices captured by a light sheet microscope. (H) WT. (I) lfs. (J) TIBA pin. (Scale bars in C–G: 100 µm.)
Fig. S1.
lfs developmental stages. (A) Time series of an lfs apex of the same plant at increasing time [days (d)] from seeding. (B) A mature lfs plant bearing a parthenocarpic fruit. (C) An lfs flower, viewed from the top (Left) and bottom (Right). (D) Hierarchal clustering of RNAseq samples using the Expander software. Analysis was done with log2 values of genes for which value was >100 counts under at least one condition, using average linkage and the Pearson correlation as a similarity measurement.
Apices of TIBA-Induced and lfs Pins Do Not Express Primordia Markers.
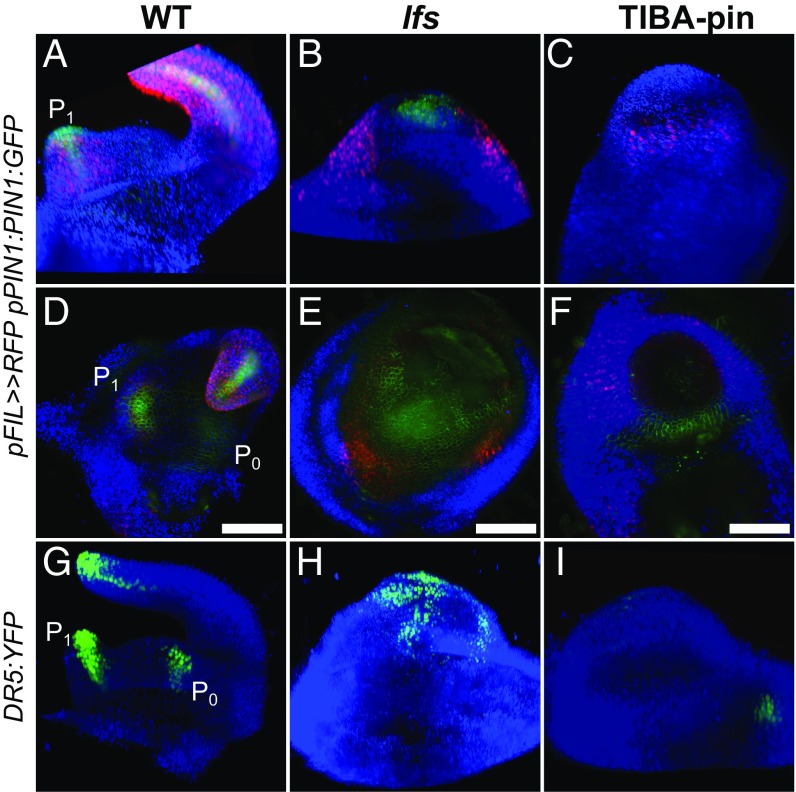
Naked pin-like apices can be induced to initiate leaves in tomato by foliar application of 2,3,5-triiodobenzoic acid (TIBA) (9). Such “TIBA-pins” can be obtained by different application regimes (0.1–1 mM, applied two to five times) and are used here as a reference system to assess and compare leaf initiation processes in lfs and in WT apices. Following repeated external application of TIBA on young tomato seedlings, plants underwent growth arrest. Soon after the last application, the apical meristem ceased to produce leaves, elongated, and produced a smooth, leaf-less naked pin, somewhat similar to the lfs pin (Fig. 1G). To determine whether the SAMs of the pin-like shoots retained their function, we monitored the expression of the two-component sensor (TCS) that marks cytokinin signaling (10). In WT, this reporter is expressed in the SAM proper, is excluded from the outermost two cell layers, and is weakly expressed in young leaf primordia (Fig. 1H). The same apical expression was maintained in lfs but was excluded from L1 in mutant plants (Fig. 1I). In TIBA pins, cytokinin expression was weaker and appeared deeper in the meristem, distributed in a lens-shaped pattern (Fig. 1J). To determine whether TIBA-treated and lfs tomato pins express young organ primordia genes, we examined the expression pattern of the primordia marker FILAMENTOUS FLOWER (FIL) (11). In WT, pFIL>>NLS:RFP signals were regularly detected at the meristem periphery, as leaf primordia initiated and progressed into later stages of leaf development (Fig. 2A). In lfs plants, we detected, albeit randomly, pFIL>>RFP expression at the meristem periphery. This expression was found at abnormal phyllotaxis and was associated with the rare primordia formation (Fig. 2B), whereas in TIBA pins, weak, uniform expression was observed around the meristem (Fig. 2C). To obtain a better perspective of primordia marker expression in naked pins, we compared the transcriptomes of apical lfs and TIBA-pin meristems to that of the WT plant (Fig. 1 E–G), using cDNA next-generation sequencing (NGS). The expression profiles of the two pins showed significant clustering and were substantially distinct from the WT profile (Fig. S1D). Of the ∼35,000 tomato genes, 256 showed fivefold higher expression levels in WT relative to lfs apices (Dataset S1, Table S1). These genes included several known primordia initiation and patterning markers (Table 1), such as six of the nine tomato YABBY genes (12), ASYMMETRIC LEAVES2 (AS2) (13), WOX1 (14), and PETROSELINUM (PTS) (15). The levels of these markers were similarly low in TIBA pins, suggesting that, much like auxin, LFS acts at a very early stage of primordia initiation.
Fig. 2.
Auxin distribution is impaired in lfs and in TIBA pins. (A–F) Expression profiles in pFIL>>NLS:RFP pPIN1:PIN1:GFP apices, as captured by a light sheet microscope. Similar analyses by confocal imaging are presented in Fig. S2. (A–C) Three-dimensional reconstructed projections. (D–F) Top view, obtained using maximum-intensity projection. (G–I) Three-dimensional reconstruction of DR5:YFP apices. Note that the patchy pattern in lfs (H) does not correspond with the discrete primordia. (Scale bars in D–F: 100 µm.)
Table 1.
Leaf primordia marker expression in WT, lfs, and TIBA-pin apices
| Gene | Annotation | WT* | lfs* | TIBA pins* |
| Solyc03g044300 | AP2 | 168 | 15 | 44 |
| Solyc11g008830 | AS2 | 175 | 0 | 0 |
| Solyc03g006900 | Crinckler | 1,550 | 62 | 580 |
| Solyc06g072480 | PTS | 3,516 | 76 | 243 |
| Solyc06g069240 | TCP18 | 140 | 1 | 16 |
| Solyc03g118770 | WOX1 | 130 | 4 | 4 |
| Solyc01g091010 | YABBY | 7,743 | 39 | 174 |
| Solyc06g073920 | YABBY | 413 | 8 | 17 |
| Solyc07g008180 | YABBY | 1,708 | 53 | 68 |
| Solyc08g079100 | YABBY | 3,674 | 6 | 12 |
| Solyc11g071810 | YABBY | 217 | 9 | 22 |
| Solyc12g009580 | YABBY | 189 | 20 | 254 |
Expression after dseq normalization (n = 2).
Auxin Signaling Is Impaired in lfs Apices.
A hallmark of primordia initiation is the preceding formation of auxin response maxima (3). Therefore, expression distribution of the auxin markers DR5:YFP and pPIN1:PIN1:GFP in WT, lfs, and TIBA pins was examined (Fig. 2 and Fig. S2). In WT, expression of the DR5:YFP marker was clearly observed at the incipient primordia (P0), as well as in distinct foci at the tips of the young leaf and leaflet primordia. In each primordium, the expression domain looked like an inverted cone, with its wide end at the tip of the primordia and the narrow region close to its base (Fig. 2G and Movie S1). In both lfs and TIBA pins, expression was abnormal; in lfs apices, expression was recorded in a large, dispersed, nonuniform patch over the meristem (Fig. 2H). Marker expression size, shape, and location varied between individuals and sampling times and did not seem to correspond with the rarely formed primordia. In contrast, DR5:YFP expression in the TIBA pins was very weak (Fig. 2I) and, when detected, seemed to uniformly cover the meristem (Fig. S2F), indicating weak residual auxin activity. WT pPIN1:PIN1:GFP expression was seen in the membranes of all of the cells of the meristem epidermis, but was enriched at the sites of the incipient and young primordia (Fig. 2 A and D). In contrast, in both lfs and TIBA pins, expression was overall weaker and lacked a discrete maxima (Fig. 2 B, C, E, and F). Taken together, the two sources of pin-like apices showed a similar absence of primordia markers, including auxin response maxima.
Fig. S2.
Expression of auxin markers, as imaged by confocal microscope. (A–C) pPIN1:PIN1:GFP apices. (D–F) DR5:YFP apices. (A and D) WT. (B and E) lfs. (C and F) TIBA pins.
LFS, but Not Auxin, Inhibits Differentiation at the Apex Periphery.
The two types of pin-like shoots failed to initiate leaf primordia. However, their stems, just below the SAM dome, were markedly different; the stem of the naked TIBA pin was glossy and lacked trichomes, whereas many trichomes covered the basal domain of the lfs apex (Fig. 1 D and F, arrowheads). In WT, all stem parts just below the apex lacked trichomes (Fig. 1C), suggesting that the lfs apex periphery undergoes rapid differentiation. To identify processes associated with this precocious differentiation, we searched for genes that were modified (more than twofold) in lfs apices but were not or only mildly modified in TIBA pins (Dataset S1, Table S2). Of 909 genes matching this criterion, 13 are involved in different aspects of CK signaling (Dataset S1, Table S2, highlighted in yellow), including three CYTOKININ OXIDASEs (CKXs) and five type A RESPONSE REGULATOR (RR). Other members of these gene groups showed similar trends (Dataset S1, Table S3), indicating reduced CK levels and responses in lfs apices. Thus, rapid differentiation of lfs compared with TIBA pins can be attributed, at least partly, to altered CK signaling.
lfs Meristems Fail to Initiate Leaf Primordia Following Auxin Microapplication.
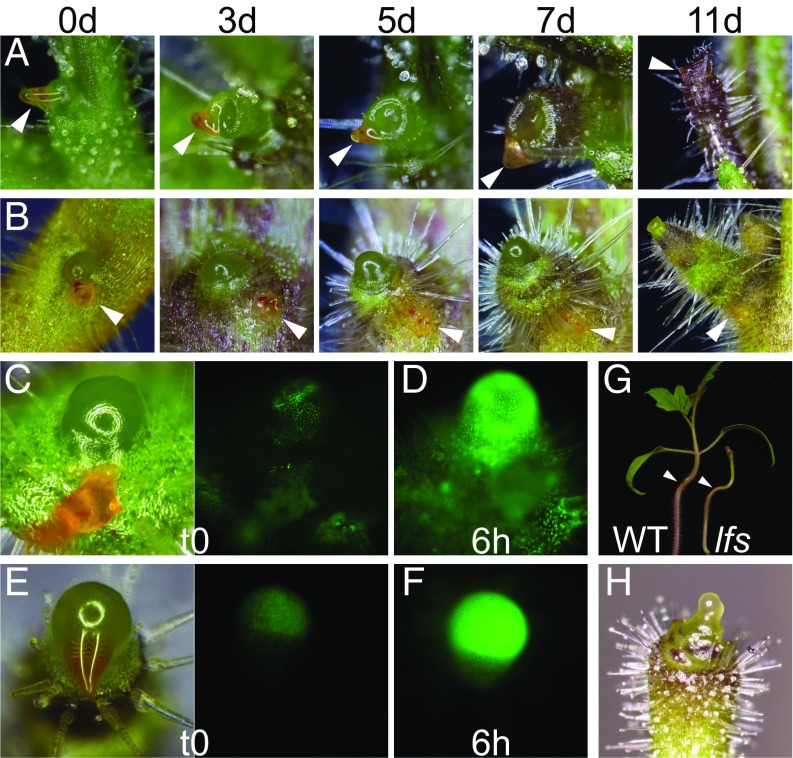
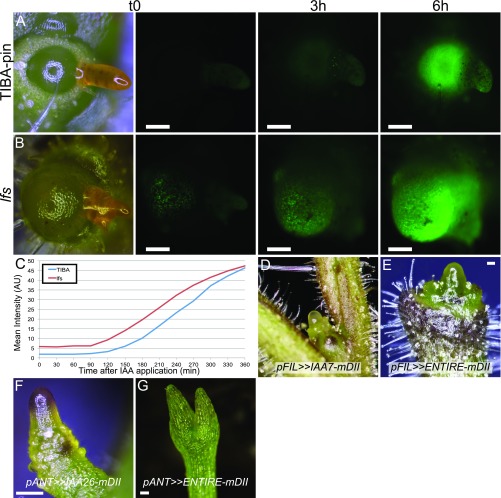
Previous studies have demonstrated that microapplication of auxin is sufficient to initiate leaf primordia outgrowths in PATI-induced pins (5, 16). The loss of normal expression of auxin signaling and transport markers may indicate loss of auxin sensitivity in lfs or failure to respond to auxin. To test whether auxin could still initiate leaf primordia in lfs apices, a lanolin paste with 10–100 mM indoleacetic acid (IAA) was applied to its apices. IAA-treated lfs pins did not generate any leaf primordia even after 2 wk and seemed morphologically unaffected by the treatment. In contrast, in TIBA pins, leaf primordia initiated at and beyond the application site, typically surrounding the meristem like a sleeve (Fig. 3 A and B). Despite the morphological differences, DR5 expression markedly increased and displayed similar dynamics (Fig. S3 A–C) in both lfs DR5:YFP plants and TIBA-pin controls following auxin treatment (Fig. 3 C–F). Furthermore, application of 20 mM IAA to the hypocotyl of 10-d-old plants induced bending in lfs seedlings, as it did in WT plants (14/14 and 16/16 plants, respectively; Fig. 3G), demonstrating that the lfs mutant is still auxin-sensitive. To test if inhibited auxin signaling at the organ primordia can indeed inhibit leaf initiation, proteolysis-resistant AUX/IAA genes were expressed at the primordia domain under the FIL promoter. To this end, IAA7 and IAA26, two genes up-regulated in lfs, and ENTIRE/IAA9, a gene that is not up-regulated (Dataset S1, Table S4, highlighted), were mutated at their proteolysis-defining domain, DII (17, 18). Transactivation of the three genes with the pFIL driver resulted in various degrees of primordial arrest, peaking at formation of pin-like shoots (Fig. 3H and Fig. S3 D and E). Primordial expression of the same mutated genes in Arabidopsis under the pANT promoter also inhibited leaf formation, resulting, in extreme cases, in pin-like shoots (Fig. S3 F and G).
Fig. 3.
Auxin-induced leaf formation is LFS-dependent. (A) TIBA pin. (B) lfs at the indicated time points after microapplication of 20 mM IAA. Arrowheads show the site of application (orange paste). (C–F) Response of DR5:YFP plants to 50 mM IAA, 6 h after application. (C and D) TIBA pins, imaged at 300 ms exposure. (E and F) lfs meristems imaged at 1,500 ms exposure. (G) WT and lfs plant bending in response to application of 20 mM IAA on the hypocotyl (marked by arrowheads); images were taken 2 d after application. (H) A pFIL>>IAA26-mDII seedling with no cotyledons and rudimentary leaves.
Fig. S3.
Attributes of different pin-like shoots. (A–C) Kinetics of response to auxin (20 mM IAA) microapplied to TIBA pin (A) and lfs (B), imaged at the same settings every 30 min. The t0, 3 h, and 6 h images are presented, as well as quantification of the meristem fluorescence (C), measured with ImageJ. AU: arbitrary units. (D) Apex of tomato pFIL>>IAA7-mDII. (E) Tomato pFIL>>ENTIRE-mDII. Note that, like TIBA pins, naked pins lack trichomes at their basal domain. (F) Arabidopsis pANT>>IAA26-mDII. (G) Arabidopsis pANT>>ENTIRE-mDII comprising short radial cotyledons and an arrested meristem. (Scale bars: 100 µm.)
LFS Is the Single Tomato Ortholog of the Arabidopsis DRN and DRNL Genes.
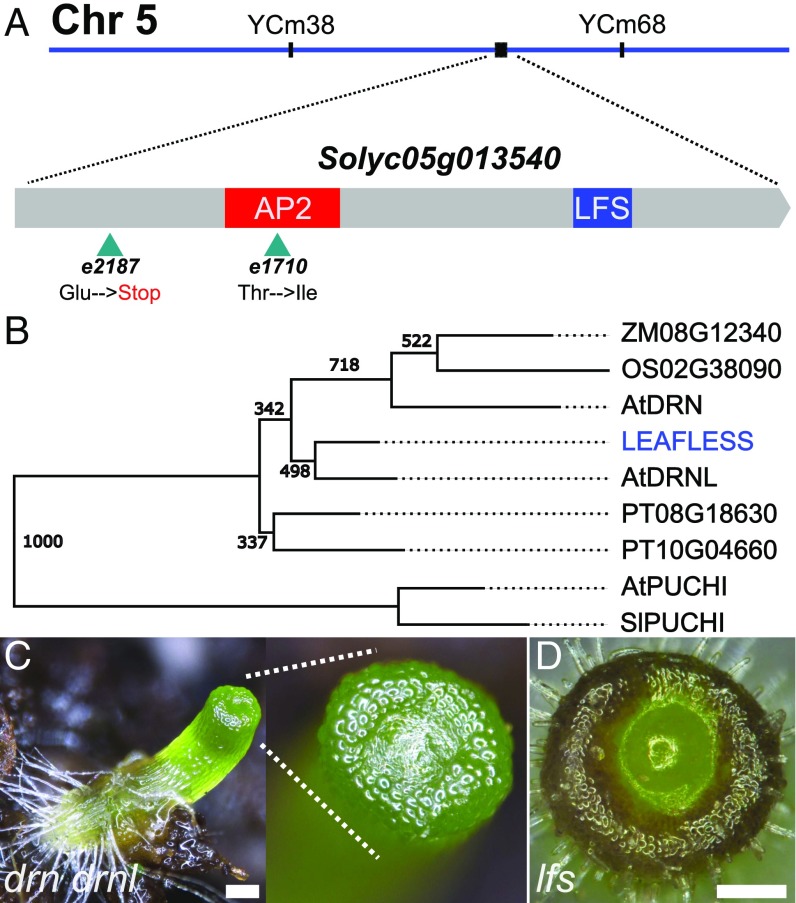
To determine which gene encodes LFS, a mapping population of 214 F2 plants from a cross between Solanum pimpinellifolium and an M82 line heterozygous for lfs was used. Informative recombinants mapped the gene to a 440-kbp region on chromosome 5. The region included a candidate gene, Solyc05g013540, which was sequenced from available mutant alleles. Two lfs alleles, e2187 and e1710, had single-base-pair changes that caused an early nonsense mutation and a missense mutation within the conserved AP2 domain of the protein, respectively (Fig. 4A). In three other alleles, amplification of the genomic region of Solyc05g013540 failed, most likely due to genomic rearrangements resulting from their generation using fast neutrons. These data, combined with the partial complementation of lfs obtained by expressing its WT cDNA under its endogenous promoter (Fig. S4), indicate that Solyc05g013540 encodes LFS.
Fig. 4.
LFS is the single tomato ortholog of DRN and DRNL. (A) Recombination-based mapping placed LFS between the PCR markers YCm34 and YCm68 (Dataset S1, Table S5). Two identified lesions in Solyc05g013540 are indicated by arrowheads. The AP2 box and a LFS-specific box are color-coded (SI Appendix). (B) Phylogenetic relationship of closest LFS homologs in select mono- and dicotyledonous plants. Nodes show bootstrap values. (C) Arabidopsis drn-1 drnl-2 whole seedling. (Inset) The SAM. (D) Top view of a tomato lfs plant. (Scale bars in C and D: 250 µM.)
Fig. S4.
Rescue of lfs by pLFS>>LFS expression. (A) A mature, fertile lfs pLFS>>LFS plant. (B) Pin-like (triangle) and regular (spearheads) flowers exist on the same lfs pLFS>>LFS branch. (C) Ectopic structures are formed beneath the sepals in lfs pLFS>>LFS plants. Sepals of mature green fruit are shown after removal of the fruit. pLFS was constructed based on the homology of upstream and downstream regions to potato and comprised 5 kb of the upstream and 1 kb of the downstream regions of LFS. (D) WT (Left) and pFIL>>LFS (Right) plants. Note epinasty in cotyledons and leaves in pFIL>>LFS.
LFS encodes for a single AP2-domain transcription factor, a member of the large ethylene response factor (ERF) family. Cladistic analysis (Fig. 4B and SI Appendix) showed that LFS is the single tomato ortholog of the Arabidopsis DORNRONSCHEN (DRN) and DRN-like (DRNL), showing a closer relation to DRNL than to DRN. PUCHI, the closest family member among all ERFs, clustered as an outgroup. In Arabidopsis, drn-1 mutants develop normally, but exhibit a low incidence of defects in cotyledon development (19). drnl-2 plants exhibit a similarly low frequency of cotyledon defects, as well as impaired anther development (20). Progenies of drn-1 drnl-2/+ parents featured a short hypocotyl, topped by a bare tip, surrounded by what may be vestigial cotyledons (Fig. 4C). However, unlike in tomato (Fig. 4D), these plants did not support a viable SAM throughout their lives, and thus organ initiation could not be assessed. Notably, when the weaker allele drnl-1 is used to generate double-mutant drn drnl plants, their phenotype is much weaker and most plants have rather normal shoots (19, 21). Both tomato lfs and Arabidopsis drn drnl seeds had a short shelf life, and their germination rate decreased with time, leading to non-Mendelian segregation ratios. Ectopic expression of either LFS or DRNL under the FIL promoter in tomato caused epinasty in cotyledons and leaves (Fig. S4D), similar to the effects of DRNL overexpression in tobacco (22). These results suggest that at the protein level the tomato and Arabidopsis gene products have similar potentials. Like LFS, DRN and DRNL have also been implicated in cotyledon initiation (23, 24) and in auxin signaling (25).
LFS Expression Is Transient in the Incipient Leaf Primordia and Overlaps with DR5 Expression.
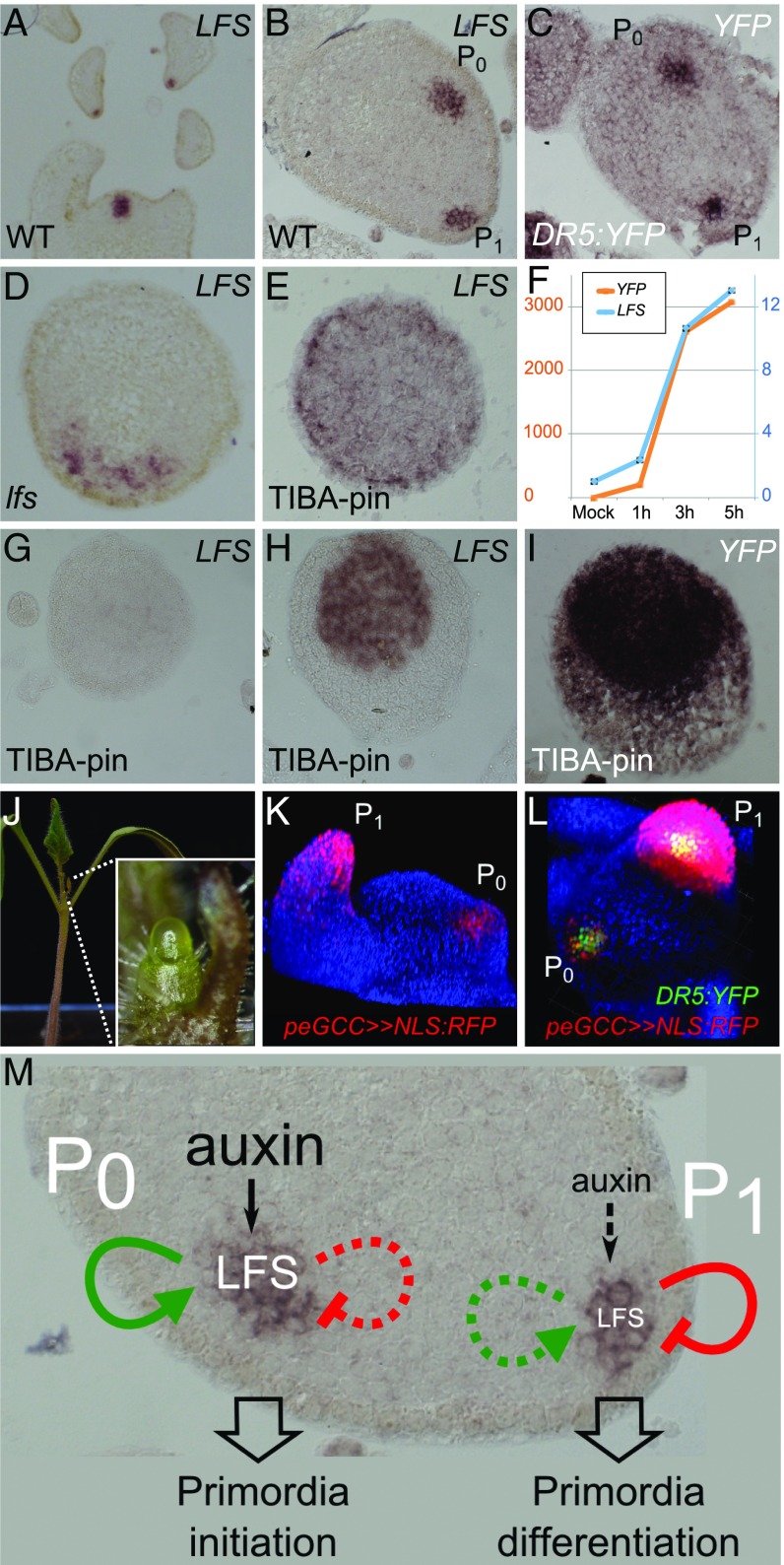
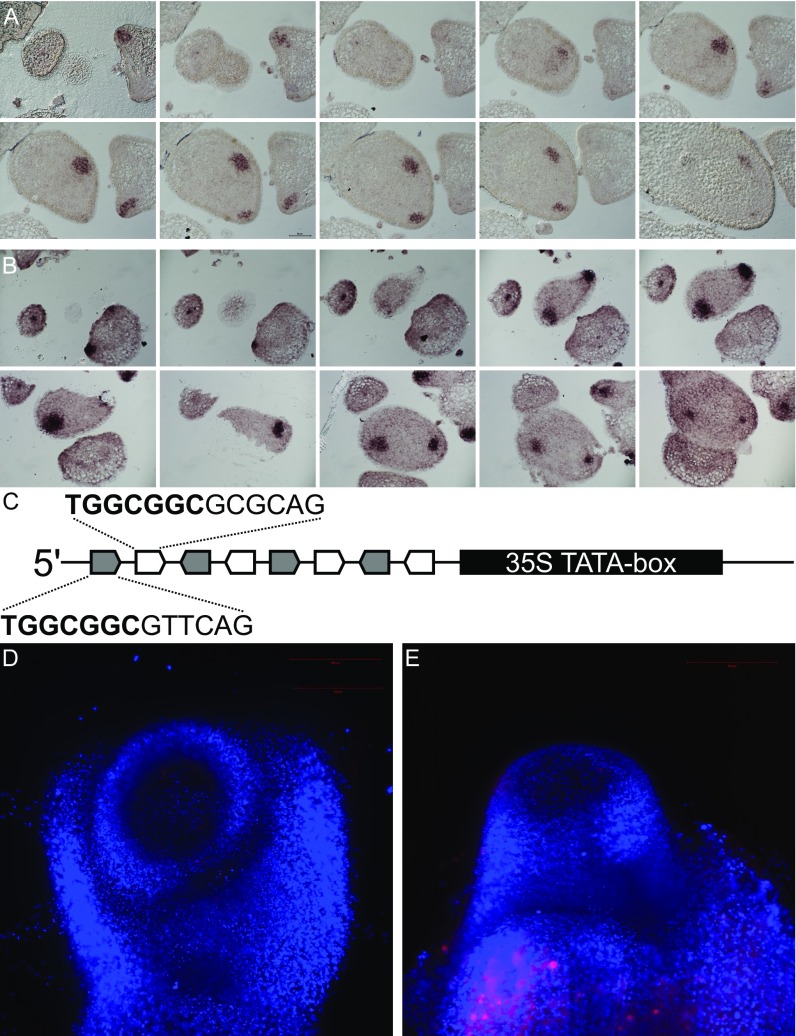
To assess the role of LFS in leaf initiation, its expression pattern in the shoot apex was characterized by RNA in situ hybridization (Fig. 5 A and B). LFS mRNA was first detected at primordia anlagen (P0), several cell layers below the epidermis. In P1 primordia, it was expressed only in the cell layer beneath the epidermis. Later, its expression was seen at the tips of P1–2 leaf primordia, at sites of initiating leaflet primordia, as well as in axillary buds. At all stages, the expression domain had an inverted cone shape, which was similar to that of the DR5 expression domain (Figs. 2G and 5C and Movie S1). Detailed comparison of the two expression domains, performed using serial sections (Fig. S5 A and B), demonstrated a significant overlap between the LFS and DR5 expression domains, with both showing early, subepidermal expression at the P0 primordia, although the DR5 expression domain was slightly broader and included one to two outer cell layers. The spatial and temporal LFS expression pattern was congruent with reports in Arabidopsis (22) and maize (26), where expression of LFS orthologs precedes the formation of newly formed leaves (P0) and is transiently maintained until primordia initiation (P1–2).
Fig. 5.
Complex regulation of LFS expression in leaf primordia. (A–E) In situ hybridization of LFS or YFP probes on WT, DR5:YFP, and lfs apices (genotypes are indicated at the Bottom Left of each panel; probes are at the Top Right). All sections were stained overnight, except E, which was stained for 4 d. (F) Relative quantification of LFS and YFP expression in response to auxin microapplication to TIBA pins. (G–I) In situ hybridization of LFS or YFP probes on DR5:YFP TIBA pins in response to microapplication of 20 mM IAA at t0 (G) and after 6 h (H and I). Probes in G–I were stained for 16 h. (J) A lfs pDR5>>LFS plant. (Inset) The pin-like apex. (K) Tomato apex with peGCC>>NLS:RFP marking young leaf primordia. (L) peGCC>>NLS:RFP DR5:YFP, viewed from the top. (M) A model for transient LFS action. Green arrows indicate GCC-dependent activation, and red arrows indicate GCC-independent inhibition.
Fig. S5.
Expression of LFS coincides with the DR5 auxin expression maxima. (A and B) In situ hybridization of an LFS probe in 8-µm serial sections of a WT apex (A) and of a YFP probe in DR5:YFP apex (B). (C) Schematic presentation of the synthetic peGCC promoter composed of four repeats of the LFS (gray arrows) and the DRNL (white arrows) eGCC-box elements, followed by a 35S TATA-box (black rectangle). See Dataset S1, Table S5, for the full sequence. (D) lfs peGCC>>NLS:RFP. The presence of the transgenes was confirmed by PCR. (E) peGCC>>NLS:RFP TIBA pin. Red fluorescence was confirmed before TIBA application.
LFS Expression Is Induced by Auxin.
To understand the relations between primordia formation, LFS expression, and auxin signaling, we examined LFS RNA distribution in the absence of primordia. In lfs apices, a large patch of LFS expression was evident (Fig. 5D), similar to the expression of DR5-YFP in lfs apices (Fig. 2G), and differing from the defined domains observed in a typical phyllotaxis. In TIBA pins, LFS was expressed in a weak ring-like pattern, primarily at the L2 layer of the two to three most distal apex cells (Fig. 5E). In agreement, LFS expression levels, as determined by NGS, were lower in TIBA pins compared with lfs apices (TIBA pins: 460; WT: 709; lfs: 3,459) (Dataset S1, Table S2), further demonstrating the differences between the two kinds of pins and suggesting complex control on LFS expression by polar auxin transport and by itself.
To characterize the temporal hierarchy between LFS and auxin signaling, we compared the dynamics of LFS and DR5:YFP activation following microapplication of IAA onto TIBA pins (Fig. 5 F–I), as measured by quantitative RT-PCR (qRT-PCR). Up-regulation of both genes began after 1 h and continued to increase at a similar rate, albeit at a two orders of magnitude difference, for at least 6 h (Fig. 5F). Indeed, 6 h after IAA application, strong expression of LFS RNA was detected at the central region of the pin, adjacent to one of the sides, likely where IAA was applied (Fig. 5H). Similarly, DR5:YFP TIBA pins showed strong YFP RNA expression in the entire meristem in response to IAA (Fig. 5I), consistent with the fluorescent signals (Fig. 3 E and F and Fig. S3 A–C). These results suggest that auxin response maxima at incipient leaf primordia promote LFS expression, which then subsequently facilitates auxin-induced primordia initiation.
Extended GCC-Box Elements Are Sufficient for Primordia Expression.
If auxin is the primary driver of LFS expression, we set out to assess whether lfs can be rescued by LFS expression driven by a DR5 promoter. pDR5>>LFS lfs plants showed only partial rescue of the lfs defects, as manifested by normal cotyledons and simpler first two leaves than usual, alongside a meristem that ceased to generate leaves and eventually flowered on top of an elongated pin (Fig. 5J). We thus searched for additional elements that could regulate LFS expression.
DRN can positively regulate a reporter comprising GCC-box elements (27), and DRNL positively regulates STY gene expression through GCC-box elements in their promoters (28). The upstream regions of both LFS and DRNL include conserved and extended GCC boxes (eGCC; 25 and 3.1 kbp upstream of LFS and 845 bp upstream of DRNL), suggesting that both genes may control their own expression. A synthetic peGCC promoter, comprising six repeats of the extended GCC boxes of LFS and DRNL promoter regions (Fig. S5C and Dataset S1, Table S5), effectively drove NLS:RFP expression in incipient and young leaf primordia (Fig. 5 K and L). Unlike DR5:YFP, the peGCC reporter did not show expression levels above those measured in the background in lfs apices (Fig. S5D), suggesting that it is positively regulated by LFS. In TIBA pins, too, the reporter was not detected (Fig. S5E), suggesting that low LFS levels in these pins (Fig. 5 E and G) were insufficient to trigger its expression. The high LFS expression levels in lfs compared with WT (3,459 and 709 normalized counts, respectively; Dataset S1, Table S2) suggest that other mechanisms contribute to LFS down-regulation after the initial activation, and these are likely to be eGCC-independent (Fig. 5M).
Discussion
The tight association between auxin expression maxima and initiating primordia and restoration of primordia formation in both PATI-treated shoots and mutants lacking auxin transport components, such as pin and pid mutants (2), by external application of auxin, implicates the role of auxin in primordia initiation. Mathematical models have further supported the suggested role of auxin maxima, dictated by auxin transporter activity, as a mechanism for achieving ordered primordia initiation patterns (3). But how auxin, a broad-acting hormone, specifies primordia cells, has remained an enigma. The results of this and previous studies (3, 5, 21) provide a framework (Fig. 5M) for understanding the specification process. Briefly, we suggest that auxin response maxima define the primordia initiation site and activate LFS expression. LFS then licenses incipient primordia cells to divide and bulge in response to auxin signals, while inhibiting differentiation of the bulging primordia and the underneath stem tissue. Later down-regulation of LFS in the expanding primordia allows for spatially defined differentiation of the various leaf domains, for example, auxin-induced provasculature formation.
Auxin acts at the SAM periphery by removing AUX/IAA suppressors that inhibit factors such as MONOPTEROS (29). In PATI-induced pins, auxin-induced primordia initiation is not restricted to the radial dimension of the meristem but rather only to a certain distance from the SAM summit (5). Thus, a ring domain surrounding the meristem periphery is adequate for auxin-induced primordia initiation and coincides with the LFS-expressing domain in TIBA-induced tomato pins (Fig. 5E). Of note, although auxin failed to stimulate primordia initiation in lfs (Fig. 3B) and auxin transport and distribution were abnormal (Fig. 2), the hormone still successfully induced molecular responses at the shoot apex as well as shoot bending at the hypocotyl (Fig. 3 C–G).
Sassi et al. (30) showed that chemical or genetic perturbation of cortical microtubules restores some primordia initiation in pin1 mutants as well as in naphtylphtalamic acid (NPA)-treated Arabidopsis apices, suggesting that auxin is not the only factor directing organ formation and that, although it is sufficient for initiation, it might not be necessary. Indeed, in Arabidopsis pin1 and pid mutants, abolishment of lateral organ initiation is restricted to the bolting stem, whereas rosette leaves develop quite normally (4, 7, 31), questioning the unique contribution of auxin regulatory mechanisms to primordia initiation. Similarly, when flowers are formed on lfs shoots, they produce nearly all organs. Thus, additional mechanisms may contribute to primordia initiation, even if these are not required by theoretical models.
We suggest that primordia initiation is induced by auxin maxima and requires LFS activity, which both enhances and later moderates auxin responses. LFS expression peaks at incipient primordia, where it allows auxin to specify leaf initiation while transiently preventing differentiation. LFS positively regulates itself, presumably through an eGCC-box element that can largely mimic the primordia-specific LFS expression. However, LFS also negatively regulates itself through a yet-unknown mechanism, as evident by the high LFS transcript levels in lfs meristems. Such transient expression is common in plants with diverse leaves, such as Arabidopsis, tomato, and maize (20, 26), suggesting that LFS defines a unique step in the conversion of SAM cells into an organ. Genetically, LFS promotes CK biosynthesis/signaling (Dataset S1, Table S2), thereby maintaining a hormonal balance at the meristem periphery that delays differentiation. This is consistent with activities of DRN and DRNL in tissue culture assays, the overexpression of which can substitute for cytokinin supplementation in shoot regeneration assay (23). As leaves are believed to evolve from branched meristem systems (reviewed in ref. 32), LFS expression may define a SAM program change and not necessarily activation of a leaf program.
Materials and Methods
Light-sheet imaging was carried out using a Zeiss Z1 microscope and processed by ZEN and ARIVIS. Apices were dissected under water and embedded in 1.5% low-melting-point agarose column before imaging. Detailed information on plant materials, imaging methods, RNA expression analyses, and cladistics is described in SI Materials and Methods.
SI Materials and Methods
Plant Lines and DNA.
All tomato lines were prepared in the CV. M82 (SP) background. Mutants [generated by either ethyl methanesulfonate (EMS) or fast neutron] were obtained from Menda et al. (33) mutagenesis screen. All lfs analyses were performed using the e2187 allele. Plants were grown in a greenhouse or growth room.
Primers for cloning and genotyping are listed in Dataset S1, Table S5. All constructs were subcloned into pART27 and introduced into the Agrobacterium tumefaciens strain GV3101 by electroporation. Transgenic lines were generated by cotyledon transformation. For most transgenes, the transactivation system was used, generating either constructs with a promoter of interest driving LhG4 or constructs that express a cDNA of interest under the control of 10 operator sequences (11). The DR5:YFP construct and the pPIN1:PIN1:GFP line were kind gifts from Marcus Heisler, European Molecular Biology Laboratory (EMBL), Heidelberg, Germany, and Cris Kuhlemeier, University of Bern, Bern, Switzerland, respectively. Arabidopsis double mutants were obtained by crossing drn-1 and drnl-2 plants, both a kind gift from John Chandler, University of Cologne, Cologne, Germany, that were backcrossed to Landsberg (ER).
Microscopy and Imaging.
Live-plant imaging was performed using a Nikon D3200 SLR camera, Nikon SMZ18 fluorescent binocular, Nikon Eclipse Ti microscope with an A1 confocal unit, or a Zeiss Z1 light-sheet microscope. For confocal imaging, meristems were dissected and placed on agar pads [a drop of melted 1% agarose was placed on a concave slide (Bar-Naor, catalog #BN2411KH) and allowed to solidify] with a drop of water and then covered with a coverslip. Scanning electron microscopy was used as described (12) after fixing samples in 70% ethanol. Images were analyzed using ImageJ, Arivis, and NIS-elements softwares. For some images, the NIS-elements Extended Depth of Focus module was used.
High-Throughput RNA Sequencing and Data Analysis.
Shoot apices from 20 to 50 plants were collected in acetone, in two biological repeats, and dissected as previously described (34). Total RNA was extracted using the RNeasy kit (QIAGEN, catalog #74004), according to the manufacturer’s instructions, and processed by the Weizmann Institute Israel National Center for Personalized Medicine (INCPM) unit. At the INCPM, libraries were prepared using the Truseq RNA Sa prep-Illumina V2 Guide, Part #15026495, and 21- to 34-million single-end 50-bp reads were sequenced using an Illumina Hi-sEq. 2500 v3 instrument. TopHat (v2.0.10) was used to align the reads to the tomato genome sequence SL2.50. On average, 92% of the reads were uniquely mapped. Reads on ITAG2.4 genes were counted using HTSeq-count (version 0.6.1p1). Differential analysis was performed using DESEq. (1.6.3). Other analyses and data filtering were conducted using Excel. The RNAseq data were deposited in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) (35) and are accessible through the GEO accession no. GSE93540.
Hormonal Manipulations.
To generate TIBA pins, germinated seedlings with spread cotyledons (usually 1 wk from seeding) were sprayed twice with 1 mM TIBA, with a 1-d interval between applications. TIBA pins formed after several days. Microapplication experiments were performed using a lanolin paste containing 10–100 mM IAA and colored with Orange G (Sigma, catalog #O3756-25G).
Cladistic Analysis.
The LFS protein was first used to identify close homologs among the mono- and dicotyledonous species defining the DRN/DRNL clade described (26) (SI Appendix). The AP2 domains of these proteins were then used to generate a phylogenetic tree, in which the LFS clade was depicted, using PUCHI, the closest DRN/DRNL homolog, as an outgroup. Phylogenetic trees were constructed using ClustalX (36) and NJplot. Domain analysis of all LFS clade members was performed using the MeMe suite (37).
In Situ Hybridization.
In situ hybridization RNA probes were synthesized from a PCR product (Dataset S1, Table S5), with addition of the T7 promoter on the forward primer, and were labeled with digoxigenin (Roche, catalog #11–209-256-910) using an RNA in vitro reverse transcription kit (CellScript, catalog #C-AS2607). Experiments were carried out according to a standard protocol (20). Selected images are representatives of at least five repeats.
qRT-PCR.
A lanolin paste containing 20 mM IAA was microapplied on TIBA pins of DR5:YFP plants. Approximately 10 apices were collected at each time point in two biological replicates. IAA-treated samples were collected after 1, 3, or 5 h, and samples without IAA (mock) were collected at 5 h and fixed in acetone, as described above. Meristems were then dissected, and total RNA was extracted using a Quick-RNA MicroPrep kit (Zymo Research, catalog #R1050). cDNA was synthesized using SuperScript II reverse transcriptase (Invitrogen, catalog #18064022). qRT-PCR was carried out in technical triplicates, using the primers listed in Dataset S1, Table S5 and a Platinum SYBR-green mix (Invitrogen, catalog #11744500) in an Applied Biosystems StepOnePlus device according to a standard protocol. The relative quantification was normalized to actin and calculated according to the comparative ΔΔCt method. Results were analyzed using StepOne v2.3 and DataAssist softwares.
Supplementary Material
Acknowledgments
We thank Naomi Ori, John Alvarez, Idan Efroni, and Chihiro Furumizu for their helpful discussions; members of the Y.E. laboratory for their valuable input; and John Chandler, Hadas Ben-Gera, Chris Kuhlemeier, and Marcus Heisler for materials. We also acknowledge the technical assistance of the following Weizmann Institute units and staff: The Electron Microscopy Unit, Yoseph Addadi from the de Picciotto-Lesser Cell Observatory of the Moross Integrated Cancer Center, Gilgi Friedlander from the Israel National Center for Personalized Medicine, and Ziva Amsellem and Shilo Rosenwasser. This work was supported by Israel Science Foundation Research Grants 1552/13 and 1788/14.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The expression dataset reported in this paper has been deposited in the Gene Expression Omnibus database (accession no. GSE93540).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1617146114/-/DCSupplemental.
References
- 1.Heisler MG, et al. Patterns of auxin transport and gene expression during primordium development revealed by live imaging of the Arabidopsis inflorescence meristem. Curr Biol. 2005;15(21):1899–1911. doi: 10.1016/j.cub.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 2.Reinhardt D, et al. Regulation of phyllotaxis by polar auxin transport. Nature. 2003;426(6964):255–260. doi: 10.1038/nature02081. [DOI] [PubMed] [Google Scholar]
- 3.Kuhlemeier C. Phyllotaxis. Trends Plant Sci. 2007;12(4):143–150. doi: 10.1016/j.tplants.2007.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the auxin polar transport system in early stages of Arabidopsis floral bud formation. Plant Cell. 1991;3(7):677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reinhardt D, Mandel T, Kuhlemeier C. Auxin regulates the initiation and radial position of plant lateral organs. Plant Cell. 2000;12(4):507–518. doi: 10.1105/tpc.12.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barkoulas M, Hay A, Kougioumoutzi E, Tsiantis M. A developmental framework for dissected leaf formation in the Arabidopsis relative Cardamine hirsuta. Nat Genet. 2008;40(9):1136–1141. doi: 10.1038/ng.189. [DOI] [PubMed] [Google Scholar]
- 7.Guenot B, et al. Pin1-independent leaf initiation in Arabidopsis. Plant Physiol. 2012;159(4):1501–1510. doi: 10.1104/pp.112.200402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scarpella E, Barkoulas M, Tsiantis M. Control of leaf and vein development by auxin. Cold Spring Harb Perspect Biol. 2010;2(1):a001511. doi: 10.1101/cshperspect.a001511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gorter C. 1949. The influence of 2, 3, 5-triiodobenzoic acid on the growing points of tomatoes. Proc Kon Ned Akad Wet:1185–1193.
- 10.Steiner E, et al. The putative O-linked N-acetylglucosamine transferase SPINDLY inhibits class I TCP proteolysis to promote sensitivity to cytokinin. Plant Physiol. 2016;171(2):1485–1494. doi: 10.1104/pp.16.00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lifschitz E, et al. The tomato FT ortholog triggers systemic signals that regulate growth and flowering and substitute for diverse environmental stimuli. Proc Natl Acad Sci USA. 2006;103(16):6398–6403. doi: 10.1073/pnas.0601620103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sarojam R, et al. Differentiating Arabidopsis shoots from leaves by combined YABBY activities. Plant Cell. 2010;22(7):2113–2130. doi: 10.1105/tpc.110.075853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Iwakawa H, et al. The ASYMMETRIC LEAVES2 gene of Arabidopsis thaliana, required for formation of a symmetric flat leaf lamina, encodes a member of a novel family of proteins characterized by cysteine repeats and a leucine zipper. Plant Cell Physiol. 2002;43(5):467–478. doi: 10.1093/pcp/pcf077. [DOI] [PubMed] [Google Scholar]
- 14.Nakata M, et al. Roles of the middle domain-specific WUSCHEL-RELATED HOMEOBOX genes in early development of leaves in Arabidopsis. Plant Cell. 2012;24(2):519–535. doi: 10.1105/tpc.111.092858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kimura S, Koenig D, Kang J, Yoong FY, Sinha N. Natural variation in leaf morphology results from mutation of a novel KNOX gene. Curr Biol. 2008;18(9):672–677. doi: 10.1016/j.cub.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 16.Stieger PA, Reinhardt D, Kuhlemeier C. The auxin influx carrier is essential for correct leaf positioning. Plant J. 2002;32(4):509–517. doi: 10.1046/j.1365-313x.2002.01448.x. [DOI] [PubMed] [Google Scholar]
- 17.Ramos JA, Zenser N, Leyser O, Callis J. Rapid degradation of auxin/indoleacetic acid proteins requires conserved amino acids of domain II and is proteasome dependent. Plant Cell. 2001;13(10):2349–2360. doi: 10.1105/tpc.010244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ben-Gera H, et al. ENTIRE and GOBLET promote leaflet development in tomato by modulating auxin response. Plant J. 2012;70(6):903–915. doi: 10.1111/j.1365-313X.2012.04939.x. [DOI] [PubMed] [Google Scholar]
- 19.Chandler JW, Cole M, Flier A, Grewe B, Werr W. The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development. 2007;134(9):1653–1662. doi: 10.1242/dev.001016. [DOI] [PubMed] [Google Scholar]
- 20.Nag A, Yang Y, Jack T. DORNROSCHEN-LIKE, an AP2 gene, is necessary for stamen emergence in Arabidopsis. Plant Mol Biol. 2007;65(3):219–232. doi: 10.1007/s11103-007-9210-7. [DOI] [PubMed] [Google Scholar]
- 21.Chandler JW, Jacobs B, Cole M, Comelli P, Werr W. DORNRÖSCHEN-LIKE expression marks Arabidopsis floral organ founder cells and precedes auxin response maxima. Plant Mol Biol. 2011;76(1-2):171–185. doi: 10.1007/s11103-011-9779-8. [DOI] [PubMed] [Google Scholar]
- 22.Marsch-Martinez N, et al. BOLITA, an Arabidopsis AP2/ERF-like transcription factor that affects cell expansion and proliferation/differentiation pathways. Plant Mol Biol. 2006;62(6):825–843. doi: 10.1007/s11103-006-9059-1. [DOI] [PubMed] [Google Scholar]
- 23.Ikeda Y, Banno H, Niu Q-W, Howell SH, Chua N-H. The ENHANCER OF SHOOT REGENERATION 2 gene in Arabidopsis regulates CUP-SHAPED COTYLEDON 1 at the transcriptional level and controls cotyledon development. Plant Cell Physiol. 2006;47(11):1443–1456. doi: 10.1093/pcp/pcl023. [DOI] [PubMed] [Google Scholar]
- 24.Chandler JW, Cole M, Jacobs B, Comelli P, Werr W. Genetic integration of DORNRÖSCHEN and DORNRÖSCHEN-LIKE reveals hierarchical interactions in auxin signalling and patterning of the Arabidopsis apical embryo. Plant Mol Biol. 2011;75(3):223–236. doi: 10.1007/s11103-010-9721-5. [DOI] [PubMed] [Google Scholar]
- 25.Cole M, et al. DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development. 2009;136(10):1643–1651. doi: 10.1242/dev.032177. [DOI] [PubMed] [Google Scholar]
- 26.Zimmermann R, Werr W. Transcription of the putative maize orthologue of the Arabidopsis DORNROSCHEN gene marks early asymmetry in the proembryo and during leaf initiation in the shoot apical meristem. Gene Expr Patterns. 2007;7(1-2):158–164. doi: 10.1016/j.modgep.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 27.Matsuo N, Banno H. The Arabidopsis transcription factor ESR1 induces in vitro shoot regeneration through transcriptional activation. Plant Physiol Biochem. 2008;46(12):1045–1050. doi: 10.1016/j.plaphy.2008.07.007. [DOI] [PubMed] [Google Scholar]
- 28.Eklund DM, et al. Expression of Arabidopsis SHORT INTERNODES/STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box-like regulatory element. Plant Physiol. 2011;157(4):2069–2080. doi: 10.1104/pp.111.182253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandler JW, Werr W. Cytokinin-auxin crosstalk in cell type specification. Trends Plant Sci. 2015;20(5):291–300. doi: 10.1016/j.tplants.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 30.Sassi M, et al. An auxin-mediated shift toward growth isotropy promotes organ formation at the shoot meristem in Arabidopsis. Curr Biol. 2014;24(19):2335–2342. doi: 10.1016/j.cub.2014.08.036. [DOI] [PubMed] [Google Scholar]
- 31.Christensen SK, Dagenais N, Chory J, Weigel D. Regulation of auxin response by the protein kinase PINOID. Cell. 2000;100(4):469–478. doi: 10.1016/s0092-8674(00)80682-0. [DOI] [PubMed] [Google Scholar]
- 32.Kenrick P, Crane PR. The Origin and Early Diversification of Land Plants: A Cladistic Study. Smithsonian Institution Press; Washington, DC: 1997. [Google Scholar]
- 33.Menda N, Semel Y, Peled D, Eshed Y, Zamir D. In silico screening of a saturated mutation library of tomato. Plant J. 2004;38(5):861–872. doi: 10.1111/j.1365-313X.2004.02088.x. [DOI] [PubMed] [Google Scholar]
- 34.Park SJ, Jiang K, Schatz MC, Lippman ZB. Rate of meristem maturation determines inflorescence architecture in tomato. Proc Natl Acad Sci USA. 2012;109(2):639–644. doi: 10.1073/pnas.1114963109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25(24):4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bailey TL, et al. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009;37(Web Server issue):W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.