Abstract
Epstein-Barr virus (EBV) is a common human herpes virus known to infect the majority of the world population. Infection with EBV is often asymptomatic but can manifest in a range of pathologies from infectious mononucleosis to severe cancers of epithelial and lymphocytic origin. Indeed, in the past decade, EBV has been linked to nearly 10% of all gastric cancers. Furthermore, recent advances in high-throughput next-generation sequencing and the development of humanized mice, which effectively model EBV pathogenesis, have led to a wealth of knowledge pertaining to strain variation and host-pathogen interaction. This review highlights some recent advances in our understanding of EBV biology, focusing on new findings on the early events of infection, the role EBV plays in gastric cancer, new strain variation, and humanized mouse models of EBV infection.
Keywords: EBV, Herpesvirus, Herpes simplex virus, gastric cancer
Introduction
Epstein-Barr virus (EBV), also known as human herpes virus 4, is a gamma-herpes virus that infects the majority of the world’s population. Initial infection with EBV is often asymptomatic but can also manifest as infectious mononucleosis. Following acute lytic replication in epithelial cells, EBV infects B cells where a distinct set of latency-associated genes and transcripts are expressed 1. EBV was first identified in 1964 from cultured tumor cells derived from a patient with Burkitt’s lymphoma (BL) 2. Early studies have demonstrated EBV’s ability to transform resting human B cells into lymphoblastoid cell lines (LCLs), further supporting the oncogenic potential of this virus 3, 4. Since then, EBV infection has been associated with a number of different malignancies of both lymphoid and epithelial origin and accounts for 1.8% of all cancer-related deaths worldwide 5.
In vivo, EBV infection begins in the oral mucosa. Replication in epithelial cells is typically lytic; however, latent infection of epithelial cells can result in nasopharyngeal carcinoma or gastric cancer (as discussed in more detail later). After replication in the epithelia, virus is primed for entry into B cells, where a transient growth program is thought to mimic a germinal center reaction, ultimately promoting maturation of the infected cell into the peripheral memory B-cell compartment. Advances in next-generation sequencing and the development of humanized mice have led to better ways to identify and understand the natural strain variation that occurs with EBV. New strain variations, particularly with mutations in latency-associated genes, have been identified in various malignancies. Harnessing these new humanized mice enables studies modeling latent infection and pathogenesis of host-restricted pathogens like HIV and EBV. This review will focus on the recent advances in EBV biology and primarily on understanding events in early B-cell infection, the role of EBV in gastric cancer, the breadth of EBV strain variation revealed by next-generation sequencing, and recent discoveries made using humanized mice.
Early events
Initial events of infection
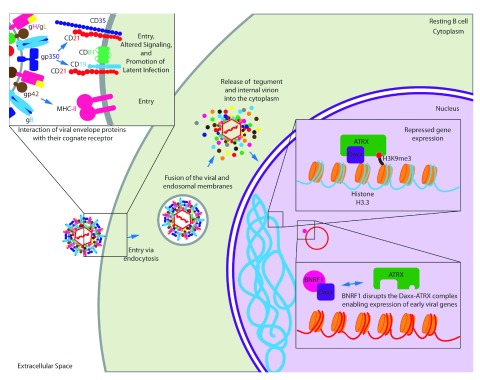
EBV entry into epithelial cells occurs by direct fusion of the viral envelope with the cell plasma membrane; however, entry into B cells requires the virus to be endocytosed before membrane fusion to escape the endosome 6, 7. B-cell entry requires five viral glycoproteins: gp350/220 allows for attachment by binding to CD21, gp42 binds to major histocompatibility complex (MHC) class II to initiate entry 8, and the core herpes-virus fusion machinery consisting of gB and the heterodimer gH/gL ( Figure 1) 1. Interaction of gp350/220 with the cell surface molecule CD21 results in the alteration of major signaling pathways believed to prime the cell to stable latent EBV infection. In particular, specific transcriptional profiles are involved in the evasion of apoptosis and there is evidence that EBV/CD21 binding alters the expression of specific histone transcripts from clusters 1 and 2 (H2AFC, H2AFM, H2BF, H2BFG, H2BI, H3FA, H3FB, H3FL, H4FL, H4FK, H4FI, H4FK, H4F2, H1F3, and H1F4) ( Figure 1) 9.
Figure 1. Initial events of Epstein-Barr virus (EBV) infection.
The EBV membrane glycoprotein gp42 binds to its cell surface receptor major histocompatibility complex class II (MHC-II) to initiate entry into the cell. Also, gp350/220 binds to its cell surface receptor CD21 for entry. Interaction with CD21 initiates signaling cascades that prime resting B cells for persistent latent infection. Following endocytosis, the virion and packaged tegument proteins are released into the cytoplasm following fusion of the virion membrane with endosomal membrane. In particular, BNRF1 disrupts the Daxx/ATRX repressor complex to facilitate viral gene expression.
Upon entry into B cells, the virion is endocytosed and is released into the cytoplasm following fusion of the virion membrane with endosomal membrane. This process releases the viral tegument proteins into the host cell. One such tegument protein, BNRF1, binds the cellular protein Daxx and disrupts the Daxx-ATRX complex 10. This complex is known to suppress transcription through histone methylation 11. Upon deposition into the nucleus, the viral DNA is associated with cellular histones 12. Daxx-ATRX might normally support methylation of this new EBV chromatin to suppress transcription of viral genes. However, BNRF1 disruption of the Daxx-ATRX complex allows early viral latent gene expression ( Figure 1) 10.
Packaged, virally encoded RNA is also released upon fusion of the virion membrane with endosomal membrane. In particular, BZLF1 transcripts have been shown to be packaged into viral particles and are translated immediately upon release into the infected cell. These immediately translated proteins then function to transactivate viral promoters initiating the pre-latent phase of EBV infection. EBV also counters T-cell responses through the delivery of BNLF2a mRNA and non-coding EBV-encoded RNA transcripts that induce cellular cytokine synthesis 13. EBV is known to encode at least 44 microRNAs (miRNAs). Though many of the miRNAs have no known function, it has recently been shown that these virally encoded miRNAs function in immune evasion by specifically suppressing the release of interleukin-12 (IL-12), disrupting CD4 + T-cell differentiation into type 1 T helper (Th1) cells, and reducing antigen presentation to CD4 + and CD8 + T cells. These miRNAs function by interfering with peptide processing, by directly targeting the TAP2 subunit, and by disrupting antigen presentation on MHC-II and MHC-I molecules 14, 15.
Pre-latent gene expression
Pre-latent gene expression occurs immediately upon deposition of the viral genome into the nucleus of newly infected B cells. Promiscuous expression of both lytic and latent genes occurs at this time with the majority of infected B cells initially expressing EBV immediate-early genes 16. Others have shown that BZLF1, the major transcriptional activator of lytic gene expression, is expressed as early as 1.5 hours post infection in the absence of protein expression, implicating BZLF1 as an immediate-early gene being expressed immediately following B-cell infection 17. This initial burst of lytic gene expression could be essential to the production of progeny virus competent for infecting new B cells 18, or immediate expression of lytic genes could be essential for the survival of latently infected B cells through inactivation of p53 19. However, it is important to note that during this pre-latent phase, genes essential for DNA replication and structural proteins of the virion are not readily detectable 16.
EBV encodes two bcl-2 proteins: BHRF1 and BALF1. Viral mutants lacking both bcl-2 proteins are unable to initiate proliferation and die from immediate apoptosis. Peak expression of these transcripts is detected at 24 hours post infection, implicating BHRF1 and BALF1 in the initial events prior to cell proliferation 20. Also, it has recently been shown that BHRF1 is constitutively expressed as a latent protein in BamHI W promoter (Wp)-restricted BL cell lines and LCLs 21. These findings implicate BHRF1 and BALF1 proteins as playing an important role in the evasion of apoptosis during latency; however, virally encoded miRNAs also cluster at the BHRF1 locus. Following induction of viral replication in latency I restricted Akata cells, these miRNAs are detectable at 24 hours post stimulation and have been shown to drive proliferation and aid in the evasion of apoptosis 22– 24.
Hyper-proliferation
Following the pre-latent phase, the initial Epstein-Barr nuclear antigen (EBNA) latency promoter, Wp, is active promoting expression of EBNA-LP and EBNA2. Subsequently, these proteins transactivate the viral C promoter, Cp, to initiate expression of the EBNA3s and EBNA1 along with their own transcripts to high levels. This EBNA-only gene expression state is associated with a period of rapid proliferation with the first three or four divisions occurring once every 8 to 12 hours 25. This period lasting approximately the first 2 weeks following resting B-cell infection is termed latency IIb 26, 27. At this time, the virus expresses all of the EBNA proteins and minimally expresses latent membrane proteins (LMPs) 1, 2A, and 2B 26. LMP1 is expressed as early as 2 days post infection; however, during this period, inhibition of early nuclear factor-kappa B (NFκB) activation does not affect transformation, supporting the distinction of this latency phase from the LCL state, which requires LMP1-mediated NFκB activity for survival 26.
As a consequence of this rapid proliferation, EBV-infected B cells are susceptible to growth arrest induced by hyper-proliferation-associated DNA damage response 26, 28. Cells then transition from a period of rapid proliferation and high Myc activity to the steady-state proliferation (about 24 hours per cycle) observed in LCLs with lower Myc and high NFκB activity 26, 29, 30. The high Myc/low NFκB state that occurs during latency IIb might play a role in immune evasion as elevated Myc and low NFκB as observed in BL have been implicated in downregulation of MHC class I and II (MHC-I and MHC-II) and avoidance of T-cell recognition and killing 29, 30.
Epstein-Barr virus infection in gastric cancer
Viral entry into epithelial cells is primarily mediated by three CD21-independent mechanisms. First, EBV can enter into epithelial cells by close membrane-to-membrane contact of EBV-infected lymphocytes to uninfected epithelial cells. Second, cell-free virus can enter polarized epithelial cells through their basolateral membranes which is mediated in part by interaction between BMRF2 and beta1 and alpha5beta1 integrins. The third mechanism is by lateral spread through the epithelium from infected to uninfected epithelial cells 31. EBV-associated gastric carcinomas (EBVaGCs) are epithelial in origin and make up approximately 9% of all gastric carcinomas worldwide 32. EBVaGCs characteristically acquire mutations within the cellular PIK3CA gene and display extreme cellular DNA hyper-methylation. Specifically, mutations in PIK3CA identified in intestinal-type gastric cancers were associated with an increased tumor incidence in the lower third of the stomach compared with those without 33. Also, PIK3CA mutations in diffuse-type gastric cancer were associated with an increased tumor incidence in the upper third of the stomach and an increased association with hematogenous metastasis. Tumors identified with PIK3CA mutation in the middle third of the stomach had an increased association with EBV infection and increased peritoneal recurrence; however, PIK3CA mutations did not demonstrate a significant effect on patient outcomes.
EBVaGCs are also known to have increased expression of JAK2, programmed death ligand 1 (PD-L1), and PD-L2 34. PD-L1 is known to interact with programmed death receptor 1 found on the surface of T cells. This interaction causes the inhibition of T-cell proliferation, cytokine secretion, and cytotoxic activity (reviewed in 35). Also, EBVaGCs have been shown to express BNLF2a, which functions in immune evasion by inhibiting the transporter associated with antigen-processing transport of antigenic peptides. Though this transcript is typically associated with lytic replication, in gastric cancers it is expressed latently and has the potential to protect the infected cell from immunosurveillance 36. Despite the immunologically evasive nature of EBVaGC, patients with diagnosed EBVaGC had longer survival post diagnosis as opposed to EBV-negative gastric carcinoma 37.
Infection of an EBV-negative GC cell line (AGS) with Akata EBV results in robust expression of virally encoded BART miRNAs with minimal protein expression 38. Importantly, these infected AGS cells displayed a more transformed phenotype than their uninfected counterparts. The prototypical transforming EBV strain, B95-8, readily infects and immortalizes human B cells. However, this virus is deleted for most of the BART miRNAs, and infection of B cells with viral variants encoding these miRNAs results in minimal BART expression 39, 40. This tissue-specific BART expression suggests that these miRNAs are likely to play a significant role in the transformed growth properties of EBVaGC.
Recently, it has been shown that CRISPR/Cas9-mediated cleavage for bacterial artificial chromosome (BAC) insertion into EBV episomal DNA in gastric carcinoma (GC) cell lines has facilitated the cloning of these viral genomes with unprecedented efficiency 41. Subsequent infection of epithelial cells with the BAC clone reconstituted viruses induced resistance to oncogene-induced cell death, providing important clues concerning EBV-mediated epithelial carcinogenesis. Establishing this new state-of-the-art technique will enable future investigation into new strain variations and their relationship with EBV-associated disease.
Epstein-Barr virus strain variation
Recent advances in next-generation whole genome sequencing (NGS) have changed the landscape surrounding the analysis of EBV-type differences. Historically, the major distinction in EBV strains has been the delineation of type 1/type 2. Currently, the largest distinguishing factors between EBV type 1 and type 2 rely on differences observed in the EBNA2 and EBNA3A, EBNA3B, and EBNA3C genes. Indeed, it has been shown that a single amino acid change in the transactivation domain of EBV-2 EBNA2 (S442D) can drastically alter EBV-2 B-cell transformation efficacy similar to that observed with EBV-1 and increase induction of LMP1 expression with a higher affinity for the LMP1 promoter 42. However, a number of other factors may contribute to the underlying strain variation, including immunological pressures, skewed cell tropism, and geographic isolation 43. Indeed, a recently described strain of EBV derived from a nasopharyngeal carcinoma case, M81, displays high epithelial tropism and also contains a polymorphism in the promoter of the lytic transactivator BZLF1 leading to elevated lytic replication 44.
It has been proposed that the prevalence of MHC haplotypes within specific geographic regions induces immunological pressures that can contribute to strain variation within immunologically dominant epitopes of particularly immunogenic proteins 45. However, recent sequence analyses demonstrate that the large numbers of non-synonymous mutations observed in the EBNA3 proteins are outside of known cytotoxic T-cell epitopes. More work is needed to identify alternative cytotoxic T lymphocyte (CTL) epitopes within the EBNA3s to explain this variation, or alternatively another selective pressure could be driving this variation perhaps regarding EBNA3 function 43. For example, a recent study found that EBNA3B, an immunodominant latency protein, actually serves as a tumor suppressor and can be found deleted in EBV strains associated with diffuse large B-cell lymphomas (DLBCLs) 46.
Recently, a provocative study implicated EBV-2 as having unique cell tropism skewing toward CD8 + T cells 47. EBV has also been commonly detected in non-B cells in the blood of patients with EBV-positive lymphoproliferative disorder (LPD), including patients with HIV, post-transplant, anaplastic anemia, chronic active EBV (CA-EBV), and others 48, 49. While CA-EBV patients often had EBV+ T cells in the blood, other EBV+ LPD patients contained EBV in monocytes as well as non-B, non-T, non-monocyte cell types based on surface staining 49. Although this population is certainly skewed from the norm with elevated viral loads and altered EBV immune responses, these findings suggest that EBV infection of T cells may be clinically relevant in some instances. Indeed, the detection of EBV in natural killer (NK)/T lymphomas 50 and a high percentage of T cells in EBV-associated hemophagocytic lymphohistiocytosis (HLH) 51 suggest that lack of control of EBV infection might be associated with a broadening of cellular tropism. Interestingly, cases of CA-EBV are most commonly reported as being of T and NK cell origin in Asia 52 and almost entirely B-cell origin in the United States 53. Information gained through NGS studies coupled with further virus-host interaction work in vitro and clinical observation will lead to a greater understanding of how different EBV strains might achieve these drastic differences in cellular tropism and maintenance of latency in various cell types.
Humanized mouse models of Epstein-Barr virus infection
EBV infection had been restricted to in vitro systems until the breakthrough of the scid-hu PBL mouse. Scid-hu PBL mice are based on the C.B-17 severe combined immunodeficient (SCID) mouse, which lack both B and T cells 54. These mice are injected with human peripheral blood mononuclear cells and, after infection with EBV, effectively model the LPD observed in immunocompromised humans (reviewed in 55). However, these mice have several drawbacks, including frequently observed graft-versus-host disease caused by the human T cells attacking mouse tissue, the transient nature of the engrafted human immune system, and a relatively low level of engraftment. Most importantly, these mice are unable to mount adaptive immune responses with their engrafted immune systems.
In order to overcome the obstacles of the scid-hu PBL mouse model, a new suite of humanized mice was generated by transplantation of non-obese diabetic/SCID (NOD/SCID) animals with hematopoietic stem cells. These NOD/SCID mice have a complete null mutation of the common IL-2 cytokine receptor gamma chain—NOD/LtSz-scid/ IL-2 receptor gamma null (NSG), NOD/Shi-scid/ IL-2 receptor gamma null (NOG)—and, once transplanted, display a humanized immune system that persists for more than 24 weeks post transplant (reviewed in 56). In this model, the CD34 + hematopoietic stem cells are able to differentiate into various mature blood cells, including myelomonocytes, dendritic cells, erythrocytes, platelets, and lymphocytes. B cells undergo normal class switching, produce normal immunoglobulins, and even infiltrate into mucosal tissues in these mice. However, it is important to note that circulating IgG is approximately 1,000 times lower than that observed in immunocompetent humans and that infiltration into mucosal tissues has been demonstrated to be severely attenuated. Differentiated T cells display human MHC-I/HLA-restricted cytotoxic functions: a vast improvement over scid-hu PBL mice 57. The introduction of the human HLA A2 allele into NSG mice transplanted with CD34 + hematopoietic stem cells (NSG- HLA-A2) resulted in mice capable of reproducing adaptive immune responses known to occur after EBV infection of HLA A2-expressing individuals 58. These NSG- HLA-A2 mice have been used to demonstrate the essential contribution of NK cells in controlling EBV infection with NK depletion resulting in the development of disseminated EBV + lymphomas 59. Further still, the BLT-NOD mice were developed by transplantation of autologous human hematopoietic fetal liver CD34 + cells into NOD/SCID mice previously implanted with human fetal thymic and liver tissues. This resulted in long-term, systemic human T-cell homeostasis capable of mounting anti-EBV MHC-I and MHC-II restricted adaptive immune responses 60. Given the vast improvements in small animal models of EBV infection, we now have the tools to study post-transplant LPD in the context of a human immune system, adaptive immune responses to EBV infection, and an experimental model to understand the in vivo effects of strain variation and other important biological questions.
Humanized mice have been shown to demonstrate the cardinal features of EBV-associated diseases developing B-cell LPD, EBV-associated HLH, and erosive arthritis resembling rheumatoid arthritis (RA). NOG humanized mice injected with 10 3 50% transforming dose (TD50) of EBV develop B-cell LPD. This LPD models the histological and viral gene expression signature observed in immunocompromised patients. Lower dose infection of less than or equal to 10 TD50 in NOG humanized mice resulted in a persistent asymptomatic infection with adaptive CD8 + T-cell responses and virus-specific IgM detectable in the serum of infected animals 61. Infection of NOG humanized mice has also been shown to result in the cardinal features of HLH with infected animals developing hemophagocytosis, erythrocytopenia, thrombocytopenia, hypercytokinemia, histiocyte proliferation and infiltration of activated CD8 + T cells into the spleen 62. EBV has been implicated in the pathogenic manifestation of RA. Patients with this disorder demonstrate elevated EBV reactive antibody titers and impaired lymphocyte responses to EBV, and EBV has been identified in the synovial fluid of patients with RA, indirectly implicating EBV in RA pathogenesis 63– 65. Modeling this pathological phenotype, humanized NOG mice infected with EBV develop an erosive arthritis. However, these findings are purely morphological and require in-depth molecular characterization to further validate this model 66. A detailed description of recent publications involving the use of humanized mice in EBV research can be found in Table 1.
Table 1. Epstein-Barr virus humanized mouse studies.
| Mouse | Epstein-Barr virus
strain |
Year | Findings | References |
|---|---|---|---|---|
| NSG+CD34-depleted human
cord blood mononuclear cells |
M81 BAC and p2089
B95-8 LMP1-KO |
2016 | Blocking PD-1/CTLA-4 inhibits Epstein-Barr virus
(EBV)-induced lymphoma growth. |
67 |
| NSG+purified CD34-positive
cells from individual fetal liver samples |
GFP-EBV B95-8 WT | 2016 | Leukocytes lacking cognate HLA ligands interfere with
KIR + natural killer (NK) recognition of HLA- tumors but improve NK-mediated control of EBV infection. |
68 |
| NSG-A2tg (expressing
HLA -A2)+purified CD34-positive cells from two fetal liver samples |
M81BAC,
M81BACΔC1, M81BACΔC2, M81BACΔC1C2, M81BACΔb2, and M81BACΔAll |
2015 | BART microRNAs repress tumorigenesis
in vivo and
likely facilitate long-term persistence in the infected host. |
69 |
| Rag2
−/− γC
−/− double
knockout+human hematopoietic stem cells injected into the liver |
293EBV
+ and
293EBVdelta (BPLF1-KO) |
2015 | BPLF1 contributes to EBV oncogenicity. | 70 |
| NSG+purified human
cord blood CD34-positive hematopoietic stem cells injected into the liver |
B95-8 | 2015 | EBV-associated Hodgkin’s lymphoma develops
exclusively in mice with activated T-cell conditions and EBV-associated non-Hodgkin’s lymphoma develops in mice with a largely suppressed T-cell condition predominantly characterized with an abundance of immature B cells. |
71 |
| NSG-A2tg +purified human
cord blood CD34-positive hematopoietic stem cells injected into the liver |
B95-8 BAC, EBER1
or EBER2 deletion mutants, and revertant viruses |
2015 | Wild-type and EBER-deleted mutant viruses
demonstrate equal ability to persist in vivo. |
72 |
| NSG+purified human fetal liver
CD34-positive hematopoietic stem cells injected into the liver |
B95-8 GFP + | 2015 | The human SAP-dependent 2B4 receptor is required for
CD8 + T cell-mediated control of EBV infection. |
73 |
| NSG+purified CD34-positive
cells from individual fetal liver samples and fetal thymus from the same donor |
p2089 B95-8 BAC
and p2089 B95-8 BAC LMP1-KO |
2015 | LMP1 is not essential for EBV-induced lymphomas
in vivo, and T cells supply signals that substitute for LMP1 in EBV-positive B-cell lymphomagenesis. |
74 |
| NSG-A2tg +purified human
cord blood CD34-positive hematopoietic stem cells injected into the liver |
Wild-type B95-8 and
BZLF1 knockout |
2014 | T cells specific for the lytic EBV antigen BMLF1 can
effectively control lytically replicating EBV + B cells in vivo. |
75 |
| Rag2
−/− γC
−/− double
knockout+human peripheral blood mononuclear cells (PBMCs) or Vγ9Vδ2-T cell- depleted PBMCs |
B95-8 and
B95.8EBfaV- GFP |
2014 | Vγ9Vδ2-T cells contribute to EBV immunity. | 76 |
| NSG+purified CD34-positive
human cord blood mononuclear cells |
B95-8 | 2014 | CD4
+ T cells are necessary for the generation/
maintenance of cells with latency I/IIa phenotype in humanized mice and contribute to this process through expression of CD40L. |
77 |
Concluding remarks
The recent advances described in this review address many of the key questions facing the EBV field today. With the advent of NGS and the development of humanized mice to better model EBV disease in vivo, we now have the tools to better understand the effects of strain variation on the development of EBV-associated diseases. Future research will benefit from further refinement of the humanized mouse models to better model the full spectrum of the human immune response to EBV infection with the aim of developing effective EBV-specific prophylactics and therapeutics. Further studies of the early period after B-cell infection and its contribution to tumorigenesis and immune evasion will be important to study in the humanized mouse. Finally, the role of EBV in epithelial malignancies and other diseases outside of the B-cell compartment is ripe for study in this post-genomic era of EBV biology.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Bo Zhao, Brigham and Women’s Hospital, Harvard Medical School, Boston, USA
Richard M Longnecker, Northwestern University, Chicago, Chicago, IL, USA
Alan K Chiang, Department of Paediatrics and Adolescent Medicine, The University of Hong Kong, Hong Kong, Hong Kong
Christian Münz, Department of Viral Immunology, Institute of Experimental Immunology, University of Zürich, Zurich, Switzerland
Funding Statement
This work received funding support from the National Cancer Institute (grant R01CA140337) and the National Institute of Dental and Craniofacial Research (grant R01DE025994).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; referees: 4 approved]
References
- 1. Knipe DM, Howley PM: Fields virology.6th ed. Philadelphia, PA: Wolters Kluwer/Lippincott Williams & Wilkins Health.2013; 2 volumes. Reference Source [Google Scholar]
- 2. Epstein MA, Achong BG, Barr YM: Virus Particles In Cultured Lymphoblasts From Burkitt's Lymphoma. Lancet. 1964;1(7335):702–3. 10.1016/S0140-6736(64)91524-7 [DOI] [PubMed] [Google Scholar]
- 3. Henle W, Diehl V, Kohn G, et al. : Herpes-type virus and chromosome marker in normal leukocytes after growth with irradiated Burkitt cells. Science. 1967;157(3792):1064–5. 10.1126/science.157.3792.1064 [DOI] [PubMed] [Google Scholar]
- 4. Pope JH, Horne MK, Scott W: Transformation of foetal human keukocytes in vitro by filtrates of a human leukaemic cell line containing herpes-like virus. Int J Cancer. 1968;3(6):857–66. 10.1002/ijc.2910030619 [DOI] [PubMed] [Google Scholar]
- 5. Khan G, Hashim MJ: Global burden of deaths from Epstein-Barr virus attributable malignancies 1990–2010. Infect Agent Cancer. 2014;9(1):38. 10.1186/1750-9378-9-38 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 6. Miller N, Hutt-Fletcher LM: Epstein-Barr virus enters B cells and epithelial cells by different routes. J Virol. 1992;66(6):3409–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nemerow GR, Cooper NR: Early events in the infection of human B lymphocytes by Epstein-Barr virus: the internalization process. Virology. 1984;132(1):186–98. [DOI] [PubMed] [Google Scholar]
- 8. Mullen MM, Haan KM, Longnecker R, et al. : Structure of the Epstein-Barr virus gp42 protein bound to the MHC class II receptor HLA-DR1. Mol Cell. 2002;9(2):375–85. 10.1016/S1097-2765(02)00465-3 [DOI] [PubMed] [Google Scholar]
- 9. Arredouani MS, Bhasin MK, Sage DR, et al. : Analysis of host gene expression changes reveals distinct roles for the cytoplasmic domain of the Epstein-Barr virus receptor/CD21 in B-cell maturation, activation, and initiation of virus infection. J Virol. 2014;88(10):5559–77. 10.1128/JVI.03099-13 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 10. Tsai K, Thikmyanova N, Wojcechowskyj JA, et al. : EBV tegument protein BNRF1 disrupts DAXX-ATRX to activate viral early gene transcription. PLoS Pathog. 2011;7(11):e1002376. 10.1371/journal.ppat.1002376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Xue Y, Gibbons R, Yan Z, et al. : The ATRX syndrome protein forms a chromatin-remodeling complex with Daxx and localizes in promyelocytic leukemia nuclear bodies. Proc Natl Acad Sci U S A. 2003;100(19):10635–40. 10.1073/pnas.1937626100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shaw JE, Levinger LF, Carter CW, Jr: Nucleosomal structure of Epstein-Barr virus DNA in transformed cell lines. J Virol. 1979;29(2):657–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jochum S, Ruiss R, Moosmann A, et al. : RNAs in Epstein-Barr virions control early steps of infection. Proc Natl Acad Sci U S A. 2012;109(21):E1396–404. 10.1073/pnas.1115906109 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Albanese M, Tagawa T, Bouvet M, et al. : Epstein-Barr virus microRNAs reduce immune surveillance by virus-specific CD8 + T cells. Proc Natl Acad Sci U S A. 2016;113(42):E6467–E6475. 10.1073/pnas.1605884113 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 15. Tagawa T, Albanese M, Bouvet M, et al. : Epstein-Barr viral miRNAs inhibit antiviral CD4 + T cell responses targeting IL-12 and peptide processing. J Exp Med. 2016;213(10):2065–80. 10.1084/jem.20160248 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 16. Kalla M, Schmeinck A, Bergbauer M, et al. : AP-1 homolog BZLF1 of Epstein-Barr virus has two essential functions dependent on the epigenetic state of the viral genome. Proc Natl Acad Sci U S A. 2010;107(2):850–5. 10.1073/pnas.0911948107 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Wen W, Iwakiri D, Yamamoto K, et al. : Epstein-Barr virus BZLF1 gene, a switch from latency to lytic infection, is expressed as an immediate-early gene after primary infection of B lymphocytes. J Virol. 2007;81(2):1037–42. 10.1128/JVI.01416-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Halder S, Murakami M, Verma SC, et al. : Early events associated with infection of Epstein-Barr virus infection of primary B-cells. PLoS One. 2009;4(9):e7214. 10.1371/journal.pone.0007214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang Q, Gutsch D, Kenney S: Functional and physical interaction between p53 and BZLF1: implications for Epstein-Barr virus latency. Mol Cell Biol. 1994;14(3):1929–38. 10.1128/MCB.14.3.1929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Altmann M, Hammerschmidt W: Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol. 2005;3(12):e404. 10.1371/journal.pbio.0030404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kelly GL, Long HM, Stylianou J, et al. : An Epstein-Barr virus anti-apoptotic protein constitutively expressed in transformed cells and implicated in burkitt lymphomagenesis: the Wp/BHRF1 link. PLoS Pathog. 2009;5(3):e1000341. 10.1371/journal.ppat.1000341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feederle R, Linnstaedt SD, Bannert H, et al. : A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 2011;7(2):e1001294. 10.1371/journal.ppat.1001294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seto E, Moosmann A, Grömminge S, et al. : Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6(8):e1001063. 10.1371/journal.ppat.1001063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xing L, Kieff E: Epstein-Barr virus BHRF1 micro- and stable RNAs during latency III and after induction of replication. J Virol. 2007;81(18):9967–75. 10.1128/JVI.02244-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nikitin PA, Yan CM, Forte E, et al. : An ATM/Chk2-mediated DNA damage-responsive signaling pathway suppresses Epstein-Barr virus transformation of primary human B cells. Cell Host Microbe. 2010;8(6):510–22. 10.1016/j.chom.2010.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Price AM, Tourigny JP, Forte E, et al. : Analysis of Epstein-Barr virus-regulated host gene expression changes through primary B-cell outgrowth reveals delayed kinetics of latent membrane protein 1-mediated NF-κB activation. J Virol. 2012;86(20):11096–106. 10.1128/JVI.01069-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Klein E, Nagy N, Rasul AE: EBV genome carrying B lymphocytes that express the nuclear protein EBNA-2 but not LMP-1: Type IIb latency. Oncoimmunology. 2013;2(2):e23035. 10.4161/onci.23035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nikitin PA, Price AM, McFadden K, et al. : Mitogen-induced B-cell proliferation activates Chk2-dependent G1/S cell cycle arrest. PLoS One. 2014;9(1):e87299. 10.1371/journal.pone.0087299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. God JM, Cameron C, Figueroa J, et al. : Elevation of c-MYC disrupts HLA class II-mediated immune recognition of human B cell tumors. J Immunol. 2015;194(4):1434–45. 10.4049/jimmunol.1402382 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Staege MS, Lee SP, Frisan T, et al. : MYC overexpression imposes a nonimmunogenic phenotype on Epstein-Barr virus-infected B cells. Proc Natl Acad Sci U S A. 2002;99(7):4550–5. 10.1073/pnas.072495599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tugizov SM, Berline JW, Palefsky JM: Epstein-Barr virus infection of polarized tongue and nasopharyngeal epithelial cells. Nat Med. 2003;9(3):307–14. 10.1038/nm830 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. Shibata D, Weiss LM: Epstein-Barr virus-associated gastric adenocarcinoma. Am J Pathol. 1992;140(4):769–74. [PMC free article] [PubMed] [Google Scholar]
- 33. Fang WT, Huang KH, Lan YT, et al. : Mutations in PI3K/AKT pathway genes and amplifications of PIK3CA are associated with patterns of recurrence in gastric cancers. Oncotarget. 2016;7(5):6201–20. 10.18632/oncotarget.6641 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 34. Cancer Genome Atlas Research Network: Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513(7517):202–9. 10.1038/nature13480 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 35. Blank C, Mackensen A: Contribution of the PD-L1/PD-1 pathway to T-cell exhaustion: an update on implications for chronic infections and tumor evasion. Cancer Immunol Immunother. 2007;56(5):739–45. 10.1007/s00262-006-0272-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Strong MJ, Laskow T, Nakhoul H, et al. : Latent Expression of the Epstein-Barr Virus (EBV)-Encoded Major Histocompatibility Complex Class I TAP Inhibitor, BNLF2a, in EBV-Positive Gastric Carcinomas. J Virol. 2015;89(19):10110–4. 10.1128/JVI.01110-15 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Camargo MC, Kim W, Chiaravalli AM, et al. : Improved survival of gastric cancer with tumour Epstein-Barr virus positivity: an international pooled analysis. Gut. 2014;63(2):236–43. 10.1136/gutjnl-2013-304531 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 38. Marquitz AR, Mathur A, Shair KH, et al. : Infection of Epstein-Barr virus in a gastric carcinoma cell line induces anchorage independence and global changes in gene expression. Proc Natl Acad Sci U S A. 2012;109(24):9593–8. 10.1073/pnas.1202910109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Raab-Traub N, Dambaugh T, Kieff E: DNA of Epstein-Barr virus VIII: B95-8, the previous prototype, is an unusual deletion derivative. Cell. 1980;22(1 Pt 1):257–67. 10.1016/0092-8674(80)90173-7 [DOI] [PubMed] [Google Scholar]
- 40. Edwards RH, Marquitz AR, Raab-Traub N: Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing. J Virol. 2008;82(8):9094–106. 10.1128/JVI.00785-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kanda T, Furuse Y, Oshitani H, et al. : Highly Efficient CRISPR/Cas9-Mediated Cloning and Functional Characterization of Gastric Cancer-Derived Epstein-Barr Virus Strains. J Virol. 2016;90(9):4383–93. 10.1128/JVI.00060-16 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Tzellos S, Correia PB, Karstegl CE, et al. : A single amino acid in EBNA-2 determines superior B lymphoblastoid cell line growth maintenance by Epstein-Barr virus type 1 EBNA-2. J Virol. 2014;88(16):8743–53. 10.1128/JVI.01000-14 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 43. Palser AL, Grayson NE, White RE, et al. : Genome diversity of Epstein-Barr virus from multiple tumor types and normal infection. J Virol. 2015;89(10):5222–37. 10.1128/JVI.03614-14 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Tsai MH, Raykova A, Klinke O, et al. : Spontaneous lytic replication and epitheliotropism define an Epstein-Barr virus strain found in carcinomas. Cell Rep. 2013;5(2):458–70. 10.1016/j.celrep.2013.09.012 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 45. Midgley RS, Bell AI, Yao QY, et al. : HLA-A11-restricted epitope polymorphism among Epstein-Barr virus strains in the highly HLA-A11-positive Chinese population: incidence and immunogenicity of variant epitope sequences. J Virol. 2003;77(21):11507–16. 10.1128/JVI.77.21.11507-11516.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. White RE, Ramer PC, Naresh KN, et al. : EBNA3B-deficient EBV promotes B cell lymphomagenesis in humanized mice and is found in human tumors. J Clin Invest. 2012;122(4):1487–502. 10.1172/JCI58092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Coleman CB, Wohlford EM, Smith NA, et al. : Epstein-Barr virus type 2 latently infects T cells, inducing an atypical activation characterized by expression of lymphotactic cytokines. J Virol. 2015;89(4):2301–12. 10.1128/JVI.03001-14 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Kimura H, Hoshino Y, Kanegane H, et al. : Clinical and virologic characteristics of chronic active Epstein-Barr virus infection. Blood. 2001;98(2):280–6. 10.1182/blood.V98.2.280 [DOI] [PubMed] [Google Scholar]
- 49. Calattini S, Sereti I, Scheinberg P, et al. : Detection of EBV genomes in plasmablasts/plasma cells and non-B cells in the blood of most patients with EBV lymphoproliferative disorders by using Immuno-FISH. Blood. 2010;116(22):4546–59. 10.1182/blood-2010-05-285452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kimura H, Ito Y, Kawabe S, et al. : EBV-associated T/NK-cell lymphoproliferative diseases in nonimmunocompromised hosts: prospective analysis of 108 cases. Blood. 2012;119(3):673–86. 10.1182/blood-2011-10-381921 [DOI] [PubMed] [Google Scholar]
- 51. Fox CP, Shannon-Lowe C, Gothard P, et al. : Epstein-Barr virus-associated hemophagocytic lymphohistiocytosis in adults characterized by high viral genome load within circulating natural killer cells. Clin Infect Dis. 2010;51(1):66–9. 10.1086/653424 [DOI] [PubMed] [Google Scholar]
- 52. Kimura H, Morishima T, Kanegane H, et al. : Prognostic factors for chronic active Epstein-Barr virus infection. J Infect Dis. 2003;187(4):527–33. 10.1086/367988 [DOI] [PubMed] [Google Scholar]
- 53. Cohen JI, Jaffe ES, Dale JK, et al. : Characterization and treatment of chronic active Epstein-Barr virus disease: a 28-year experience in the United States. Blood. 2011;117(22):5835–49. 10.1182/blood-2010-11-316745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Mosier DE, Gulizia RJ, Baird SM, et al. : Transfer of a functional human immune system to mice with severe combined immunodeficiency. Nature. 1988;335(6187):256–9. 10.1038/335256a0 [DOI] [PubMed] [Google Scholar]
- 55. Fujiwara S, Matsuda G, Imadome K: Humanized mouse models of epstein-barr virus infection and associated diseases. Pathogens. 2013;2(1):153–76. 10.3390/pathogens2010153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Fujiwara S, Imadome K, Takei M: Modeling EBV infection and pathogenesis in new-generation humanized mice. Exp Mol Med. 2015;47(1):e135. 10.1038/emm.2014.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ishikawa F, Yasukawa M, Lyons B, et al. : Development of functional human blood and immune systems in NOD/SCID/IL2 receptor {gamma} chain null mice. Blood. 2005;106(5):1565–73. 10.1182/blood-2005-02-0516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shultz LD, Saito Y, Najima Y, et al. : Generation of functional human T-cell subsets with HLA-restricted immune responses in HLA class I expressing NOD/SCID/IL2r gamma null humanized mice. Proc Natl Acad Sci U S A. 2010;107(29):13022–7. 10.1073/pnas.1000475107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chijioke O, Müller A, Feederle R, et al. : Human natural killer cells prevent infectious mononucleosis features by targeting lytic Epstein-Barr virus infection. Cell Rep. 2013;5(6):1489–98. 10.1016/j.celrep.2013.11.041 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 60. Melkus MW, Estes JD, Padgett-Thomas A, et al. : Humanized mice mount specific adaptive and innate immune responses to EBV and TSST-1. Nat Med. 2006;12(11):1316–22. 10.1038/nm1431 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 61. Yajima M, Imadome K, Nakagawa A, et al. : A new humanized mouse model of Epstein-Barr virus infection that reproduces persistent infection, lymphoproliferative disorder, and cell-mediated and humoral immune responses. J Infect Dis. 2008;198(5):673–82. 10.1086/590502 [DOI] [PubMed] [Google Scholar]
- 62. Sato K, Misawa N, Nie C, et al. : A novel animal model of Epstein-Barr virus-associated hemophagocytic lymphohistiocytosis in humanized mice. Blood. 2011;117(21):5663–73. 10.1182/blood-2010-09-305979 [DOI] [PubMed] [Google Scholar]
- 63. Toussirot E, Roudier J: Pathophysiological links between rheumatoid arthritis and the Epstein-Barr virus: an update. Joint Bone Spine. 2007;74(5):418–26. 10.1016/j.jbspin.2007.05.001 [DOI] [PubMed] [Google Scholar]
- 64. Erre GL, Mameli G, Cossu D, et al. : Increased Epstein-Barr Virus DNA Load and Antibodies Against EBNA1 and EA in Sardinian Patients with Rheumatoid Arthritis. Viral Immunol. 2015;28(7):385–90. 10.1089/vim.2015.0035 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 65. Mahabadi M, Faghihiloo E, Alishiri GH, et al. : Detection of Epstein-Barr virus in synovial fluid of rheumatoid arthritis patients. Electron Physician. 2016;8(3):2181–6. 10.19082/2181 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 66. Kuwana Y, Takei M, Yajima M, et al. : Epstein-Barr virus induces erosive arthritis in humanized mice. PLoS One. 2011;6(10):e26630. 10.1371/journal.pone.0026630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ma SD, Xu X, Jones R, et al. : PD-1/CTLA-4 Blockade Inhibits Epstein-Barr Virus-Induced Lymphoma Growth in a Cord Blood Humanized-Mouse Model. PLoS Pathog. 2016;12(5):e1005642. 10.1371/journal.ppat.1005642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Landtwing V, Raykova A, Pezzino G, et al. : Cognate HLA absence in trans diminishes human NK cell education. J Clin Invest. 2016;126(10):3772–82. 10.1172/JCI86923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lin X, Tsai MH, Shumilov A, et al. : The Epstein-Barr Virus BART miRNA Cluster of the M81 Strain Modulates Multiple Functions in Primary B Cells. PLoS Pathog. 2015;11(12):e1005344. 10.1371/journal.ppat.1005344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Whitehurst CB, Li G, Montgomery SA, et al. : Knockout of Epstein-Barr virus BPLF1 retards B-cell transformation and lymphoma formation in humanized mice. mBio. 2015;6(5):e01574–15. 10.1128/mBio.01574-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Lee EK, Joo EH, Song K, et al. : Effects of lymphocyte profile on development of EBV-induced lymphoma subtypes in humanized mice. Proc Natl Acad Sci U S A. 2015;112(42):13081–6. 10.1073/pnas.1407075112 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 72. Gregorovic G, Boulden EA, Bosshard R, et al. : Epstein-Barr Viruses (EBVs) Deficient in EBV-Encoded RNAs Have Higher Levels of Latent Membrane Protein 2 RNA Expression in Lymphoblastoid Cell Lines and Efficiently Establish Persistent Infections in Humanized Mice. J Virol. 2015;89(22):11711–4. 10.1128/JVI.01873-15 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 73. Chijioke O, Marcenaro E, Moretta A, et al. : Role of the 2B4 Receptor in CD8 + T-Cell-Dependent Immune Control of Epstein-Barr Virus Infection in Mice With Reconstituted Human Immune System Components. J Infect Dis. 2015;212(5):803–7. 10.1093/infdis/jiv114 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Ma SD, Xu X, Plowshay J, et al. : LMP1-deficient Epstein-Barr virus mutant requires T cells for lymphomagenesis. J Clin Invest. 2015;125(1):304–15. 10.1172/JCI76357 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 75. Antsiferova O, Muller A, Ramer PC, et al. : Adoptive transfer of EBV specific CD8 + T cell clones can transiently control EBV infection in humanized mice. PLoS Pathog. 2014;10(8):e1004333. 10.1371/journal.ppat.1004333 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 76. Xiang Z, Liu Y, Zheng J, et al. : Targeted activation of human Vγ9Vδ2-T cells controls epstein-barr virus-induced B cell lymphoproliferative disease. Cancer Cell. 2014;26(4):565–76. 10.1016/j.ccr.2014.07.026 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 77. Heuts F, Rottenberg ME, Salamon D, et al. : T cells modulate Epstein-Barr virus latency phenotypes during infection of humanized mice. J Virol. 2014;88(6):3235–45. 10.1128/JVI.02885-13 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation