Abstract
Amyotrophic lateral sclerosis is a neurodegenerative disease predominantly affecting upper and lower motor neurons, resulting in progressive paralysis and death from respiratory failure within 2 to 3 years. The peak age of onset is 55 to 70 years, with a male predominance. The causes of amyotrophic lateral sclerosis are only partly known, but they include some environmental risk factors as well as several genes that have been identified as harbouring disease-associated variation. Here we review the nature, epidemiology, genetic associations, and environmental exposures associated with amyotrophic lateral sclerosis.
Keywords: amyotrophic lateral sclerosis, neurodegenerative disease, motor neuron disease
Introduction
Amyotrophic lateral sclerosis (ALS) is an incurable condition, characterised by progressive degeneration of upper and lower motor neurons, resulting in paralysis and death from respiratory failure in a median of 2–3 years 1. Despite the poor prognosis, there is considerable variation in the survival rate, and up to 10% of people with ALS live for more than 8 years from first symptoms 2. Understanding what causes ALS or influences survival is crucial for the development of effective treatments.
The causes of ALS are largely unknown. Significant advances have been made in understanding the genetic and environmental components of the disease. In this report, we will explore what is known about why some people develop ALS and how the risk factors work together to cause the disease.
Epidemiological studies of ALS
The incidence of ALS is about 2 per 100,000 person-years, and the prevalence is about 5 per 100,000 persons 3. Because of the low prevalence, the average primary care physician will see 1 person in their lifetime, a typical UK neurologist will diagnose about 2 people a year, while a tertiary referral centre will see more than 100 people. Despite the low incidence, however, ALS is not particularly infrequent. The lifetime risk is about 1 in 300 by the age of 85, with the risk increasing steadily, at least until about the eighth decade of life 4, 5. This is very similar to the risk for multiple sclerosis in the UK 6.
Tertiary referral centres see sufficient numbers of people that research studies will have adequate power for statistical analysis. However, there is a significant diagnostic delay in ALS, typically about a year, which seems to be independent of healthcare system and is probably related to low recognition by primary care physicians 7. As a result, those attending specialist centres tend to be those with a better prognosis, who are younger, and who are more motivated 8, 9. In contrast, population-based registers capture all cases in a defined catchment population, regardless of attendance at a specialist clinic. Such registers have provided valuable insights into the epidemiology of ALS and offer an unbiased view of the condition 10.
ALS can affect people at any age. The mean age of onset is 56 in clinic registers but 70 in population-based registers. In clinic registers, ALS is more frequent in men, with a male:female ratio of about 3:2, and the ratio becomes more equal with increasing age. In population registers, although the male preponderance is still seen, the ratio may be closer to 1:1, an effect that can be attributed to the greater capture of older people with ALS 3.
What is ALS?
ALS presents in many different ways ( Table 1), and it has been recognised for many years that the different clinical presentations correspond with differences in survival 11, 12. Bulbar palsy, in which dysarthria followed by swallowing difficulty is the main presentation, is associated with the worst prognosis, and flail arm or flail leg syndrome, in which there is symmetrical, predominantly flaccid weakness of the limbs, is associated with the best prognosis 13. Perhaps surprisingly, statistical methods such as latent class cluster analysis can analyse the same data and identify different clinical subtypes that predict prognosis with far more discrimination than can neurologist classifications 13. Most cases of ALS are focal in onset and relentlessly progressive, often to contiguous regions, although there are some exceptions 14. The spread could be the result of a “prion-like” spread of toxic proteins through phagocytosis (consumption of cells by other cells) or possibly through a time-to-failure model 15– 17. Lower motor neuron failure is the main cause of weakness in ALS and can be measured non-invasively to provide data to assess cellular patterns of spread 18. Understanding the mechanisms of spread will aid the development of novel therapeutics and may aid models of prognosis.
Table 1. Clinical presentations of amyotrophic lateral sclerosis.
| Classifying
feature |
Name of phenotype | Description |
|---|---|---|
| Motor neuron
involvement |
Amyotrophic lateral
sclerosis (ALS) |
Mixture of upper and lower motor neuron signs on clinical examination.
Degree of certainty of diagnosis based on El Escorial criteria. May involve up to all regions. |
| Primary lateral sclerosis
or upper motor neuron predominant ALS |
Clinical signs limited to upper motor neuron features. Generally slowly
progressive but involving up to all regions. This phenotype is usually confirmed if there have been no lower motor neuron signs after 4 years. |
|
| Progressive muscular
atrophy or lower motor neuron predominant ALS |
Clinical signs limited to lower motor neuron features. Slightly slower
progression but can involve all regions. This phenotype is usually confirmed if there have been no upper motor neuron signs after 4 years. |
|
| Site of onset | Bulbar onset | Site of onset may be included in the description of ALS, as different
disease onset patterns have different rates of progression. The two categories are bulbar and spinal. |
| Spinal onset | ||
| Disease
focality |
Progressive bulbar palsy | Condition involving the bulbar region and predominantly lower motor
neurons. May progress to other regions. |
| Pseudobulbar palsy | Condition involving the bulbar region and predominantly upper motor
neurons. May progress to other regions. |
|
| Flail arm | Predominantly lower motor neuron proximal symmetrical involvement in
the upper limbs. Some upper motor neuron signs may be seen in the lower limbs. |
|
| Flail leg | Lower motor neuron distal symmetrical involvement restricted to the lower
limbs. May affect one side only. |
|
| Cognitive
involvement |
ALS with cognitive
impairment |
ALS with some cognitive involvement below the threshold criteria for
frontotemporal dementia. |
| ALS with frontotemporal
dementia (ALS-FTD) |
ALS with frank frontotemporal dementia. |
The diagnosis of ALS is clinical, with the support of electrophysiological studies and the exclusion of mimics. In some cases, early diagnosis can be challenging, particularly if weakness is confined to one region for some time or is confined to a subset of motor neurons (upper only or lower only). A sensitive set of electrodiagnostic criteria, the Awaji criteria, can be particularly useful in the early diagnosis of people with bulbar onset disease, which is important because of the need for sooner gastrostomy when swallowing is affected early 19– 22.
ALS is classified for research purposes by the El Escorial criteria and their revisions, which improve homogeneity in recruitment for clinical trials and other clinical studies 23– 26. ALS progression is measured functionally using the ALS Functional Rating Scale – Revised, which uses 12 questions scored between 0 (no function) and 4 (full function) to generate a summary score 27. The scale is widely used but has some limitations, since the subscores correlate more accurately with progression in different clinical subtypes 28.
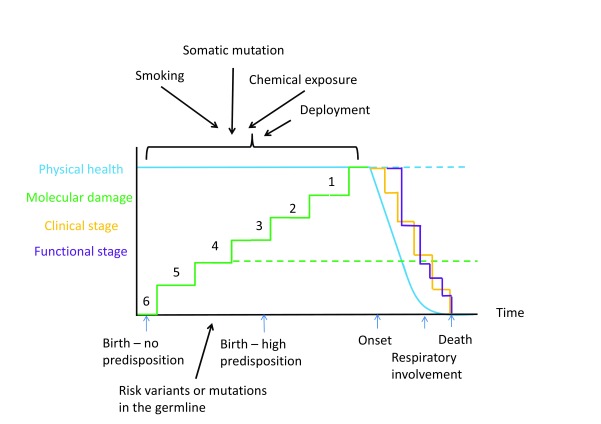
Disease staging allows a simple description of the extent of physical or functional involvement in an affected person and guides management. Such systems have been in widespread use in cancer for years. In ALS, two recent staging systems have been proposed: King’s clinical staging and Milano-Torino staging (MiToS) 29, 30. The King’s system is similar to cancer staging in that the clinical spread of disease is used to infer the extent of disease progression. Spread is defined as involvement producing signs or symptoms in the El Escorial domains (1 domain is stage 1, 2 domains is stage 2, and 3 domains is stage 3), with respiratory or nutritional failure characterising stage 4. The ALS functional rating scale can be used to estimate the King’s stage with 92% correlation 31. MiToS uses the ALS functional rating scale subscores to define functional stage 29. Each system has benefits in describing ALS stage succinctly. The two disease staging systems are complementary 32. King’s staging summarises the clinical or anatomical spread of disease. Mapping disease progression to clinical stage rather than survival could be used as a secondary endpoint in clinical trials, which would shorten trial durations and provide meaningful information on which stage of the disease is prolonged by an effective therapy 33. MiToS summarises the functional burden of disease. It would therefore be useful in showing a functional benefit in clinical trials. Comparison of the systems shows that functional stage lags behind clinical stage, reflecting the functional reserve available in an affected limb, and it has been proposed that a combined stage is used, as is standard in cancer, along the lines of K3M2, which would mean King’s stage 3, MiToS stage 2 32, 34 ( Figure 1).
Figure 1. The time course of amyotrophic lateral sclerosis (ALS).
Time is represented along the x-axis; physical health and molecular damage are represented along the y-axis. With time, molecular damage increases in a step-wise way until it reaches a threshold, at which point physical health declines, representing disease onset. People with a family history of ALS may have a large genetic predisposition to ALS and so need fewer steps to reach the level of molecular damage that causes disease, corresponding to a younger age of onset. Lack of exposure to sufficient risk factors means that the disease does not manifest, even if a genetic cause is present, explaining reduced penetrance. There is not a 1:1 mapping of risk factors and steps, as the steps represent molecular hits that lead to cellular damage rather than actual exposures. Once physical symptoms have started, progression shows a log-linear decline until the onset of respiratory symptoms, where decline is exponential. Clinical and functional involvement can be measured by the King’s clinical staging and Milano-Torino staging (MiToS) systems. A dotted line represents the hypothetical trajectory in an unaffected individual. Black arrows represent genetic and environmental risk factors. Numbers indicate remaining molecular hits until disease onset.
It is now recognised that ALS involves non-motor systems 35. Between 30 and 50% of people have cognitive impairment detectable on formal testing, resulting from involvement of the frontotemporal circuits 36, 37. Frank frontotemporal dementia occurs in about 5%, and in some families, people may have ALS, frontotemporal dementia, or both 36, 38, 39. The clinical impact of frontotemporal impairment in ALS is now more easily recognised because of recent advances in the tools available to detect it, such as the Edinburgh cognitive assessment score (ECAS) 40, 41. Other neurodegenerative diseases have also been linked to ALS, including spinocerebellar ataxia, in which case studies have reported the co-occurrence of ALS and cerebellar degeneration 42. Schizophrenia may be more frequent in families with ALS, and there may also be an increased frequency of multiple sclerosis 43, 44. In many of these cases, genetic factors are responsible for some of the risk. For example, pathological expansion of a repeat sequence in the C9orf72 gene is associated with ALS, frontotemporal dementia, or both, and the same mutation may increase the risk of schizophrenia, Parkinson’s disease, and multiple sclerosis 45. Expansion of a repeat sequence in the ATXN2 gene is associated with ALS or, if more than 30 repeats are involved, with spinocerebellar ataxia 46. Autonomic, skin, and eye movement changes are also seen. Thus, ALS is a neurodegenerative disease in which the brunt falls on the motor system, but, as for many other neurodegenerations, the clinical syndrome is also dispersed through other anatomical and physiological systems.
Understanding prognostic factors in ALS
Respiratory impairment is usually an end-stage event in ALS. Despite this, because respiratory function is difficult to measure reliably with non-invasive methods, measurement of respiratory function is generally used as a guide to the use of respiratory support rather than prognostication 47. There have been many attempts at prognostic modelling, using either clinical features alone or biological markers such as albumin, creatinine, or neurofilament levels 48, 49. Most studies find that longer survival is associated with younger age at symptom onset, presentation with limb dysfunction rather than swallowing or speech disturbance, and specific forms of ALS such as symmetrical patterns (e.g. flail arm syndrome) or upper motor neuron predominant forms 50. Conversely, cognitive impairment comprising executive dysfunction, rapid weight loss, and respiratory involvement at first examination, although not necessarily respiratory onset, predict a poor prognosis 51– 58. The best predictor of slow progression, however, appears to be a long interval between symptom onset and diagnosis, probably because this reflects the rate of disease progression overall 59. Genetic variations have been associated with survival duration, with the best studied being variation in the UNC13A gene 60, 61. Variation in the CAMTA1 gene has also been associated with survival 62. Furthermore, some risk genes harbour variants that are themselves predictors of prognosis. For example, the p.Asp91Ala variation of the SOD1 gene is associated with very slow progression 63, 64, while the p.Ala5Val variant is associated with aggressive disease 65. Statistical models can be used to provide clinically useful information for patients, the strongest message being that survival is extremely unreliably predicted in individuals, even though patterns can be seen in the data 54, 57, 66– 68.
Genetics and ALS
There are now more than 25 genes in which an association with ALS has been replicated, with the rate of gene discovery doubling every 4 years ( http://alsod.iop.kcl.ac.uk) 69. In up to 10% of people, there is a family history of ALS in a first-degree relative, but detailed genealogical studies extending to more distant relatives and including related diagnoses suggest that more than 20% have a relevant family history. The genes responsible for familial ALS have now been identified for about 70% of all cases, but there is a significant genetic component, even in those without a family history. Twin studies suggest the heritability is about 60%, and nearly every familial ALS gene has also been implicated in apparently sporadic ALS 70, 71. Furthermore, statistical analysis shows that the distinction between familial and sporadic ALS is not clear-cut, and large-scale genome-wide association studies (GWAS) show that the genetic architecture of sporadic ALS is one in which rare variation, more usually associated with familial disease, is disproportionately important 72, 73.
The most recent GWAS of ALS identified four new associations, three of which were successfully replicated 73. An interesting feature of the study was that even though this was a study of people with apparently sporadic ALS, there were associations in genes previously identified from family-based studies – C9orf72, TBK1, and NEK1 – further supporting the notion that familial and sporadic ALS are not mutually exclusive categories but rather a spectrum 74– 76. These three genes all harbour variants that are moderately penetrant. In other words, carrying a disease-associated variant does not mean ALS will inevitably follow. Current thinking is that common diseases are the consequence of the additive effects of small increases in risk from multiple common variations (polygenic), and rare diseases are the consequence of single gene variants that are themselves rare but have a large effect on the probability of disease (monogenic). For example, height and schizophrenia are polygenic traits, while Huntington’s disease and Kennedy’s disease are monogenic diseases. ALS sits somewhere between these two extremes, with a lifetime prevalence that is far greater than is typical for a monogenic disease but far less than a common disease, and it is perhaps, therefore, to be expected that its genetic architecture also seems to sit somewhere between polygenic effects and monogenic high-penetrance disease.
There are three genes that have had a major impact on our understanding of ALS. ALS-linked dominant mutations in the superoxide dismutase gene SOD1 were first identified in 1993, and since then mutations have been found in every exon of the gene 77. The SOD1 protein is a free radical scavenger, and loss of function, increasing free radical damage in cells, is a logical hypothesis to consider. However, several well-characterised SOD1 variants do not lead to a reduction in dismutase activity, and the evidence instead supports a toxic gain of function 78. Transgenic SOD1 mice develop a motor neuron degeneration and have been used to model the disease for treatment development 79. The second important ALS gene is TARDBP, which codes for TDP-43, a protein regulating RNA expression and the major component of intracellular inclusions in ALS. The discovery of ALS-linked mutations in this gene was the first of many showing RNA processing defects to be important in ALS pathogenesis and, importantly, showed that the TDP-43 inclusions were not simply a passive marker of neuronal death but a crucial part of the disease pathway 80– 82. The third important genetic finding in ALS was of linkage 83, 84 and then association 85– 87 of a locus on chromosome 9, which led researchers to the identification of a massive expansion of a hexanucleotide repeat in intron 1 of the C9orf72 gene 88, 89. This is the most frequent cause of ALS, being responsible for about 30% of familial and up to 10% of sporadic cases.
The focus of genetic research in ALS in the immediate future is therefore on rare variation. This is best discovered through high-throughput sequencing, and this technique has already identified several familial ALS genes. The major challenge facing researchers is how to interpret the findings, since the identification of a rare variant in an ALS gene is not in itself strong evidence of relevance in that individual, and over-representation of rare variation in cases over controls in a particular gene does not provide sufficient information for genetic counselling on a specific variant 90. The amount of heritability explained by genetic information captured on genome-wide microarrays is about 12%, implying that the remainder is in rare variants and other types of genetic variation such as copy number variation, microsatellite repeats, post-transcriptional RNA editing, and epigenetic changes 91. These are likely to be the next targets of ALS genetics research and are reliant on international research consortia. Project MinE is one such global collaboration that aims to analyse DNA from at least 15,000 people with ALS and 7,500 controls ( https://www.projectmine.com/).
Environmental risk factors
In contrast to genetics, environmental risk factors for ALS have been more difficult to identify. Such studies are expensive to perform but difficult to fund and are heavily reliant on recall 92. As a result, they are susceptible to bias. Furthermore, unlike genome-wide analyses, in which a hypothesis-generating approach can be taken, it is not straightforward to assay all possible environmental factors, and so a selected subset of assumed risk factors is tested. Smoking has been associated with increased risk of ALS in some studies and may hold a higher risk in some subgroups 93. Occupation, particularly military service with deployment, has been associated with risk of ALS, but the evidence mainly comes from the US, where there are large military datasets 94. Physical activity is another widely studied risk factor, partly because of a number of high-profile sports players who have had ALS and because of people with ALS having a low BMI on presentation and higher levels of leisure sports participation 95. It is not clear whether having higher levels of physical activity raises the risk of ALS and, if it does, whether it is the activity itself or being genetically predisposed to high sporting prowess that is the mechanism 96. Similarly, electric shock is not a risk factor in some analyses but is in others 97, 98. There is mixed evidence for the involvement of chemicals, such as heavy metals, ambient aromatic hydrocarbons, pesticides, and cyanotoxins 99– 103. Trauma, including head injury, also appears to be a risk factor in meta-analysis 104.
Inflammation and ALS
Evidence of an immuno-inflammatory component in ALS pathogenesis is compelling 105, 106. A pathological hallmark of the neuroinflammation is prominent microglia activation at involved sites. T-regulatory lymphocytes (Tregs) are important immunomodulatory cells that regulate the balance between activation and suppression of the immune response and control the microglia in the central nervous system: specifically, pushing them towards a state in which remodeling and repair activities are activated. Defects in Treg levels or function have been found in ALS patients, becoming more frequent as the disease progresses. Treg levels are inversely correlated with disease severity, so that lower levels are seen in more severe disease, and survival is worse in those with Treg defects 105– 109. Studies are now underway to explore immune therapies that might improve Treg function and therefore improve survival.
Retroviruses and ALS
Poliovirus and other enteroviruses can cause a post-infectious myelitis with subsequent paralysis, and HIV infection can result in an ALS-like syndrome. Studies of serum and cerebrospinal fluid from ALS patients suggested that an activated endogenous retrovirus was associated with ALS 110. Recently, the sequence has been identified as possibly HERV-K, an endogenous retrovirus that exists as an open reading frame in the human genome 111. In mice, the env protein component of HERV-K is toxic to motor neurons. There is no evidence that HERV-K is causative of the disease in humans, but studies are now underway to explore if antiretrovirals might slow progression and improve survival in ALS.
Conclusion
The apparently homogeneous phenotype of predominantly motor degeneration that is ALS can result from many different causes: genetic, epigenetic, environmental, and internal. Thus, many different pathways converge on the final outcome of upper and lower motor neuron death. Careful analysis of incidence data in European population registers shows that, on average, each pathway comprises six molecular steps 112 (see Figure 1). The model explains many otherwise enigmatic features of ALS, such as the increasing risk with age, genetic pleiotropy (the same gene variation can result in different diseases), age-dependent penetrance of disease genes, the difficulty in identifying a single environmental cause, the observation that ALS appears to start in one region and spread, and that it is specific to motor neurons but can affect other cell types. The next challenge is to understand the extent to which the pathways overlap and therefore might be amenable to a common treatment strategy. Although ALS remains a uniformly fatal diagnosis, accelerating advances in our understanding bring the hope that an effective treatment can be found for this devastating disease.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Mamede de Carvalho, Institute of Physiology, Faculty of Medicine, University of Lisbon, Lisbon, Portugal; Department of Neurosciences, Hospital de Santa Maria, Lisbon, Portugal
Richard W Orrell, University College London Institute of Neurology, London, UK
Robert P. Bowser, Barrow Neurological Institute, St. Joseph's Hospital and Medical Center, Phoenix, Pheonix, USA
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; referees: 3 approved]
References
- 1. Talbot K: Motor neuron disease: the bare essentials. Pract Neurol. 2009;9(5):303–09. 10.1136/jnnp.2009.188151 [DOI] [PubMed] [Google Scholar]
- 2. Pupillo E, Messina P, Logroscino G, et al. : Long-term survival in amyotrophic lateral sclerosis: a population-based study. Ann Neurol. 2014;75(2):287–97. 10.1002/ana.24096 [DOI] [PubMed] [Google Scholar]
- 3. Chiò A, Logroscino G, Traynor BJ, et al. : Global epidemiology of amyotrophic lateral sclerosis: a systematic review of the published literature. Neuroepidemiology. 2013;41(2):118–30. 10.1159/000351153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alonso A, Logroscino G, Jick SS, et al. : Incidence and lifetime risk of motor neuron disease in the United Kingdom: a population-based study. Eur J Neurol. 2009;16(6):745–51. 10.1111/j.1468-1331.2009.02586.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Johnston CA, Stanton BR, Turner MR, et al. : Amyotrophic lateral sclerosis in an urban setting: a population based study of inner city London. J Neurol. 2006;253(12):1642–43. 10.1007/s00415-006-0195-y [DOI] [PubMed] [Google Scholar]
- 6. Alonso A, Hernán MA: Temporal trends in the incidence of multiple sclerosis: a systematic review. Neurology. 2008;71(2):129–35. 10.1212/01.wnl.0000316802.35974.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Mitchell JD, Callagher P, Gardham J, et al. : Timelines in the diagnostic evaluation of people with suspected amyotrophic lateral sclerosis (ALS)/motor neuron disease (MND)--a 20-year review: can we do better? Amyotroph Lateral Scler. 2010;11(6):537–41. 10.3109/17482968.2010.495158 [DOI] [PubMed] [Google Scholar]
- 8. Lee JR, Annegers JF, Appel SH: Prognosis of amyotrophic lateral sclerosis and the effect of referral selection. J Neurol Sci. 1995;132(2):207–15. 10.1016/0022-510X(95)00154-T [DOI] [PubMed] [Google Scholar]
- 9. Sorenson EJ, Mandrekar J, Crum B, et al. : Effect of referral bias on assessing survival in ALS. Neurology. 2007;68(8):600–2. 10.1212/01.wnl.0000254501.58158.e7 [DOI] [PubMed] [Google Scholar]
- 10. Logroscino G, Traynor BJ, Hardiman O, et al. : Descriptive epidemiology of amyotrophic lateral sclerosis: new evidence and unsolved issues. J Neurol Neurosurg Psychiatry. 2008;79(1):6–11. 10.1136/jnnp.2006.104828 [DOI] [PubMed] [Google Scholar]
- 11. Al-Chalabi A, Hardiman O, Kiernan MC, et al. : Amyotrophic lateral sclerosis: moving towards a new classification system. Lancet Neurol. 2016;15(11):1182–94. 10.1016/S1474-4422(16)30199-5 [DOI] [PubMed] [Google Scholar]
- 12. Wolf J, Safer A, Wöhrle JC, et al. : Variability and prognostic relevance of different phenotypes in amyotrophic lateral sclerosis - data from a population-based registry. J Neurol Sci. 2014;345(1–2):164–67. 10.1016/j.jns.2014.07.033 [DOI] [PubMed] [Google Scholar]
- 13. Wijesekera LC, Mathers S, Talman P, et al. : Natural history and clinical features of the flail arm and flail leg ALS variants. Neurology. 2009;72(12):1087–94. 10.1212/01.wnl.0000345041.83406.a2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ravits JM, La Spada AR: ALS motor phenotype heterogeneity, focality, and spread: deconstructing motor neuron degeneration. Neurology. 2009;73(10):805–11. 10.1212/WNL.0b013e3181b6bbbd [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bidhendi EE, Bergh J, Zetterström P, et al. : Two superoxide dismutase prion strains transmit amyotrophic lateral sclerosis-like disease. J Clin Invest. 2016;126(6):2249–53. 10.1172/JCI84360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Clarke G, Lumsden CJ, McInnes RR: Inherited neurodegenerative diseases: the one-hit model of neurodegeneration. Hum Mol Genet. 2001;10(20):2269–75. [DOI] [PubMed] [Google Scholar]
- 17. Liu G, Fiala M, Mizwicki MT, et al. : Neuronal phagocytosis by inflammatory macrophages in ALS spinal cord: inhibition of inflammation by resolvin D1. Am J Neurodegener Dis. 2012;1(1):60–74. [PMC free article] [PubMed] [Google Scholar]
- 18. Nandedkar SD, Barkhaus PE, Stalberg EV: Motor unit number index (MUNIX): principle, method, and findings in healthy subjects and in patients with motor neuron disease. Muscle Nerve. 2010;42(5):798–807. 10.1002/mus.21824 [DOI] [PubMed] [Google Scholar]
- 19. Atassi N, Cudkowicz ME, Schoenfeld DA: Advanced statistical methods to study the effects of gastric tube and non-invasive ventilation on functional decline and survival in amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2011;12(4):272–77. 10.3109/17482968.2011.577786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Costa J, Swash M, de Carvalho M: Awaji criteria for the diagnosis of amyotrophic lateral sclerosis:a systematic review. Arch Neurol. 2012;69(11):1410–16. 10.1001/archneurol.2012.254 [DOI] [PubMed] [Google Scholar]
- 21. Geevasinga N, Menon P, Loy C, et al. : Revisiting early diagnosis in ALS. Clin Neurophysiol. 2016;127(3):e13–e14. 10.1016/j.clinph.2015.11.031 [DOI] [Google Scholar]
- 22. Geevasinga N, Menon P, Scherman DB, et al. : Diagnostic criteria in amyotrophic lateral sclerosis: A multicenter prospective study. Neurology. 2016;87(7):684–90. 10.1212/WNL.0000000000002988 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 23. Brooks BR: El Escorial World Federation of Neurology criteria for the diagnosis of amyotrophic lateral sclerosis. Subcommittee on Motor Neuron Diseases/Amyotrophic Lateral Sclerosis of the World Federation of Neurology Research Group on Neuromuscular Diseases and the El Escorial "Clinical limits of amyotrophic lateral sclerosis" workshop contributors. J Neurol Sci. 1994;124(Suppl):96–107. 10.1016/0022-510X(94)90191-0 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 24. de Carvalho M, Dengler R, Eisen A, et al. : Electrodiagnostic criteria for diagnosis of ALS. Clin Neurophysiol. 2008;119(3):497–503. 10.1016/j.clinph.2007.09.143 [DOI] [PubMed] [Google Scholar]
- 25. Miller RG, Munsat TL, Swash M, et al. : Consensus guidelines for the design and implementation of clinical trials in ALS. World Federation of Neurology committee on Research. J Neurol Sci. 1999;169(1–2):2–12. 10.1016/S0022-510X(99)00209-9 [DOI] [PubMed] [Google Scholar]
- 26. Schrooten M, Smetcoren C, Robberecht W, et al. : Benefit of the Awaji diagnostic algorithm for amyotrophic lateral sclerosis: a prospective study. Ann Neurol. 2011;70(1):79–83. 10.1002/ana.22380 [DOI] [PubMed] [Google Scholar]
- 27. Cedarbaum JM, Stambler N, Malta E, et al. : The ALSFRS-R: a revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase III). J Neurol Sci. 1999;169(1–2):13–21. 10.1016/S0022-510X(99)00210-5 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Rooney J, Burke T, Vajda A, et al. : What does the ALSFRS-R really measure? A longitudinal and survival analysis of functional dimension subscores in amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2016. pii: jnnp-2016-314661. 10.1136/jnnp-2016-314661 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 29. Chiò A, Hammond ER, Mora G, et al. : Development and evaluation of a clinical staging system for amyotrophic lateral sclerosis. J Neurol Neurosurg Psychiatry. 2015;86(1):38–44. 10.1136/jnnp-2013-306589 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Roche JC, Rojas-Garcia R, Scott KM, et al. : A proposed staging system for amyotrophic lateral sclerosis. Brain. 2012;135(Pt 3):847–52. 10.1093/brain/awr351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Balendra R, Jones A, Jivraj N, et al. : Estimating clinical stage of amyotrophic lateral sclerosis from the ALS Functional Rating Scale. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(3–4):279–84. 10.3109/21678421.2014.897357 [DOI] [PubMed] [Google Scholar]
- 32. Fang T, Al Khleifat A, Stahl DR, et al. : Comparison of the King’s and MiToS staging systems for ALS. Amyotroph Lateral Scler Frontotemporal Degener. 2017.1–6. 10.1080/21678421.2016.1265565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Balendra R, Jones A, Jivraj N, et al. : Use of clinical staging in amyotrophic lateral sclerosis for phase 3 clinical trials. J Neurol Neurosurg Psychiatry. 2015;86(1):45–9. 10.1136/jnnp-2013-306865 [DOI] [PubMed] [Google Scholar]
- 34. Ferraro D, Consonni D, Fini N, et al. : Amyotrophic lateral sclerosis: a comparison of two staging systems in a population-based study. Eur J Neurol. 2016;23(9):1426–32. 10.1111/ene.13053 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 35. Swinnen B, Robberecht W: The phenotypic variability of amyotrophic lateral sclerosis. Nat Rev Neurol. 2014;10(11):661–70. 10.1038/nrneurol.2014.184 [DOI] [PubMed] [Google Scholar]
- 36. Montuschi A, Iazzolino B, Calvo A, et al. : Cognitive correlates in amyotrophic lateral sclerosis: a population-based study in Italy. J Neurol Neurosurg Psychiatry. 2015;86(2):168–73. 10.1136/jnnp-2013-307223 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Rippon GA, Scarmeas N, Gordon PH, et al. : An observational study of cognitive impairment in amyotrophic lateral sclerosis. Arch Neurol. 2006;63(3):345–52. 10.1001/archneur.63.3.345 [DOI] [PubMed] [Google Scholar]
- 38. Lomen-Hoerth C, Anderson T, Miller B: The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59(7):1077–9. 10.1212/WNL.59.7.1077 [DOI] [PubMed] [Google Scholar]
- 39. Strong MJ: The syndromes of frontotemporal dysfunction in amyotrophic lateral sclerosis. Amyotroph Lateral Scler. 2008;9(6):323–38. 10.1080/17482960802372371 [DOI] [PubMed] [Google Scholar]
- 40. Abrahams S, Newton J, Niven E, et al. : Screening for cognition and behaviour changes in ALS. Amyotroph Lateral Scler Frontotemporal Degener. 2014;15(1–2):9–14. 10.3109/21678421.2013.805784 [DOI] [PubMed] [Google Scholar]
- 41. Niven E, Newton J, Foley J, et al. : Validation of the Edinburgh Cognitive and Behavioural Amyotrophic Lateral Sclerosis Screen (ECAS): A cognitive tool for motor disorders. Amyotroph Lateral Scler Frontotemporal Degener. 2015;16(3–4):172–79. 10.3109/21678421.2015.1030430 [DOI] [PubMed] [Google Scholar]
- 42. Tazen S, Figueroa K, Kwan JY, et al. : Amyotrophic lateral sclerosis and spinocerebellar ataxia type 2 in a family with full CAG repeat expansions of ATXN2. JAMA Neurol. 2013;70(10):1302–4. 10.1001/jamaneurol.2013.443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Etemadifar M, Abtahi SH, Akbari M, et al. : Multiple sclerosis and amyotrophic lateral sclerosis: is there a link? Mult Scler. 2012;18(6):902–4. 10.1177/1352458511427719 [DOI] [PubMed] [Google Scholar]
- 44. Howland RH: Schizophrenia and amyotrophic lateral sclerosis. Compr Psychiatry. 1990;31(4):327–36. 10.1016/0010-440X(90)90039-U [DOI] [PubMed] [Google Scholar]
- 45. Cooper-Knock J, Shaw PJ, Kirby J: The widening spectrum of C9ORF72-related disease; Genotype/phenotype correlations and potential modifiers of clinical phenotype. Acta Neuropathol. 2014;127(3):333–45. 10.1007/s00401-014-1251-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Neuenschwander AG, Thai KK, Figueroa KP, et al. : Amyotrophic lateral sclerosis risk for spinocerebellar ataxia type 2 ATXN2 CAG repeat alleles: a meta-analysis. JAMA Neurol. 2014;71(12):1529–34. 10.1001/jamaneurol.2014.2082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Polkey MI, Lyall RA, Yang K, et al. : Respiratory Muscle Strength as a Predictive Biomarker for Survival in Amyotrophic Lateral Sclerosis. Am J Respir Crit Care Med. 2017;195(1):86–95. 10.1164/rccm.201604-0848OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Chiò A, Calvo A, Bovio G, et al. : Amyotrophic lateral sclerosis outcome measures and the role of albumin and creatinine: a population-based study. JAMA Neurol. 2014;71(9):1134–42. 10.1001/jamaneurol.2014.1129 [DOI] [PubMed] [Google Scholar]
- 49. Lu CH, Macdonald-Wallis C, Gray E, et al. : Neurofilament light chain: A prognostic biomarker in amyotrophic lateral sclerosis. Neurology. 2015;84(22):2247–57. 10.1212/WNL.0000000000001642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Calvo A, Moglia C, Lunetta C, et al. : Factors predicting survival in ALS: a multicenter Italian study. J Neurol. 2016;264(1):54–63. 10.1007/s00415-016-8313-y [DOI] [PubMed] [Google Scholar]
- 51. Elamin M, Phukan J, Bede P, et al. : Executive dysfunction is a negative prognostic indicator in patients with ALS without dementia. Neurology. 2011;76(14):1263–9. 10.1212/WNL.0b013e318214359f [DOI] [PubMed] [Google Scholar]
- 52. Marin B, Arcuti S, Jesus P, et al. : Population-Based Evidence that Survival in Amyotrophic Lateral Sclerosis is Related to Weight Loss at Diagnosis. Neurodegener Dis. 2016;16(3–4):225–34. 10.1159/000442444 [DOI] [PubMed] [Google Scholar]
- 53. Moura MC, Novaes MR, Eduardo EJ, et al. : Prognostic Factors in Amyotrophic Lateral Sclerosis: A Population-Based Study. PLoS One. 2015;10(10):e0141500. 10.1371/journal.pone.0141500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wolf J, Safer A, Wöhrle JC, et al. : Factors Predicting Survival in ALS Patients--Data from a Population-Based Registry in Rhineland-Palatinate, Germany. Neuroepidemiology. 2015;44(3):149–55. 10.1159/000381625 [DOI] [PubMed] [Google Scholar]
- 55. Wolf J, Safer A, Wöhrle JC, et al. : Factors predicting one-year mortality in amyotrophic lateral sclerosis patients--data from a population-based registry. BMC Neurol. 2014;14(1):197. 10.1186/s12883-014-0197-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Czaplinski A, Yen AA, Appel SH: Forced vital capacity (FVC) as an indicator of survival and disease progression in an ALS clinic population. J Neurol Neurosurg Psychiatry. 2006;77(3):390–2. 10.1136/jnnp.2005.072660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Knibb JA, Keren N, Kulka A, et al. : A clinical tool for predicting survival in ALS. J Neurol Neurosurg Psychiatry. 2016;87(12):1361–1367. 10.1136/jnnp-2015-312908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Shoesmith CL, Findlater K, Rowe A, et al. : Prognosis of amyotrophic lateral sclerosis with respiratory onset. J Neurol Neurosurg Psychiatry. 2007;78(6):629–31. 10.1136/jnnp.2006.103564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Czaplinski A, Yen AA, Appel SH, et al. : Amyotrophic lateral sclerosis: early predictors of prolonged survival. J Neurol. 2006;253(11):1428–36. 10.1007/s00415-006-0226-8 [DOI] [PubMed] [Google Scholar]
- 60. Diekstra FP, van Vught PW, van Rheenen W, et al. : UNC13A is a modifier of survival in amyotrophic lateral sclerosis. Neurobiol Aging. 2012;33(3):630.e3–8. 10.1016/j.neurobiolaging.2011.10.029 [DOI] [PubMed] [Google Scholar]
- 61. Gaastra B, Shatunov A, Pulit S, et al. : Rare genetic variation in UNC13A may modify survival in amyotrophic lateral sclerosis. Amyotroph Lateral Scler Frontotemporal Degener. 2016;17(7–8):593–99. 10.1080/21678421.2016.1213852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fogh I, Lin K, Tiloca C, et al. : Association of a Locus in the CAMTA1 Gene With Survival in Patients With Sporadic Amyotrophic Lateral Sclerosis. JAMA Neurol. 2016;73(7):812–20. 10.1001/jamaneurol.2016.1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Andersen PM, Al-Chalabi A: Clinical genetics of amyotrophic lateral sclerosis: what do we really know? Nat Rev Neurol. 2011;7(11):603–15. 10.1038/nrneurol.2011.150 [DOI] [PubMed] [Google Scholar]
- 64. Andersen PM, Nilsson P, Ala-Hurula V, et al. : Amyotrophic lateral sclerosis associated with homozygosity for an Asp90Ala mutation in CuZn-superoxide dismutase. Nat Genet. 1995;10(1):61–6. 10.1038/ng0595-61 [DOI] [PubMed] [Google Scholar]
- 65. Cudkowicz ME, McKenna-Yasek D, Sapp PE, et al. : Epidemiology of mutations in superoxide dismutase in amyotrophic lateral sclerosis. Ann Neurol. 1997;41(2):210–21. 10.1002/ana.410410212 [DOI] [PubMed] [Google Scholar]
- 66. Taylor AA, Fournier C, Polak M, et al. : Predicting disease progression in amyotrophic lateral sclerosis. Ann Clin Transl Neurol. 2016;3(11):866–75. 10.1002/acn3.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Sato Y, Nakatani E, Watanabe Y, et al. : Prediction of prognosis of ALS: Importance of active denervation findings of the cervical-upper limb area and trunk area. Intractable Rare Dis Res. 2015;4(4):181–9. 10.5582/irdr.2015.01043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Carreiro AV, Amaral PM, Pinto S, et al. : Prognostic models based on patient snapshots and time windows: Predicting disease progression to assisted ventilation in Amyotrophic Lateral Sclerosis. J Biomed Inform. 2015;58:133–44. 10.1016/j.jbi.2015.09.021 [DOI] [PubMed] [Google Scholar]
- 69. Olubunmi A, Al-Chalabi A: ALSoD: Amyotrophic Lateral Sclerosis Online Genetics Database. [Google Scholar]
- 70. Al-Chalabi A, Fang F, Hanby MF, et al. : An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry. 2010;81(12):1324–26. 10.1136/jnnp.2010.207464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Al-Chalabi A, Lewis CM: Modelling the effects of penetrance and family size on rates of sporadic and familial disease. Hum Hered. 2011;71(4):281–88. 10.1159/000330167 [DOI] [PubMed] [Google Scholar]
- 72. Majounie E, Renton AE, Mok K, et al. : Frequency of the C9orf72 hexanucleotide repeat expansion in patients with amyotrophic lateral sclerosis and frontotemporal dementia: a cross-sectional study. Lancet Neurol. 2012;11(4):323–30. 10.1016/S1474-4422(12)70043-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. van Rheenen W, Shatunov A, Dekker AM, et al. : Genome-wide association analyses identify new risk variants and the genetic architecture of amyotrophic lateral sclerosis. Nat Genet. 2016;48(9):1043–48. 10.1038/ng.3622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Chiò A, Borghero G, Restagno G, et al. : Clinical characteristics of patients with familial amyotrophic lateral sclerosis carrying the pathogenic GGGGCC hexanucleotide repeat expansion of C9orf72. Brain. 2012;135(Pt 3):784–93. 10.1093/brain/awr366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Kenna KP, van Doormaal PT, Dekker AM, et al. : NEK1 variants confer susceptibility to amyotrophic lateral sclerosis. Nat Genet. 2016;48(9):1037–42. 10.1038/ng.3626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Williams KL, McCann EP, Fifita JA, et al. : Novel TBK1 truncating mutation in a familial amyotrophic lateral sclerosis patient of Chinese origin. Neurobiol Aging. 2015;36(12):3334.e1–34.e5. 10.1016/j.neurobiolaging.2015.08.013 [DOI] [PubMed] [Google Scholar]
- 77. Rosen DR, Siddique T, Patterson D, et al. : Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362(6415):59–62. 10.1038/362059a0 [DOI] [PubMed] [Google Scholar]
- 78. Kaur SJ, McKeown SR, Rashid S: Mutant SOD1 mediated pathogenesis of Amyotrophic Lateral Sclerosis. Gene. 2016;577(2):109–18. 10.1016/j.gene.2015.11.049 [DOI] [PubMed] [Google Scholar]
- 79. Cleveland DW, Bruijn LI, Wong PC, et al. : Mechanisms of selective motor neuron death in transgenic mouse models of motor neuron disease. Neurology. 1996;47(4 Suppl 2):S54–61; discussion S61–2. 10.1212/WNL.47.4_Suppl_2.54S [DOI] [PubMed] [Google Scholar]
- 80. Lagier-Tourenne C, Polymenidou M, Hutt KR, et al. : Divergent roles of ALS-linked proteins FUS/TLS and TDP-43 intersect in processing long pre-mRNAs. Nat Neurosci. 2012;15(11):1488–97. 10.1038/nn.3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Van Deerlin VM, Leverenz JB, Bekris LM, et al. : TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7(5):409–16. 10.1016/S1474-4422(08)70071-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Sreedharan J, Blair IP, Tripathi VB, et al. : TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319(5870):1668–72. 10.1126/science.1154584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Vance C, Al-Chalabi A, Ruddy D, et al. : Familial amyotrophic lateral sclerosis with frontotemporal dementia is linked to a locus on chromosome 9p13.2–21.3. Brain. 2006;129(Pt 4):868–76. 10.1093/brain/awl030 [DOI] [PubMed] [Google Scholar]
- 84. Morita M, Al-Chalabi A, Andersen PM, et al. : A locus on chromosome 9p confers susceptibility to ALS and frontotemporal dementia. Neurology. 2006;66(6):839–44. 10.1212/01.wnl.0000200048.53766.b4 [DOI] [PubMed] [Google Scholar]
- 85. Laaksovirta H, Peuralinna T, Schymick JC, et al. : Chromosome 9p21 in amyotrophic lateral sclerosis in Finland: a genome-wide association study. Lancet Neurol. 2010;9(10):978–85. 10.1016/S1474-4422(10)70184-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Shatunov A, Mok K, Newhouse S, et al. : Chromosome 9p21 in sporadic amyotrophic lateral sclerosis in the UK and seven other countries: a genome-wide association study. Lancet Neurol. 2010;9(10):986–94. 10.1016/S1474-4422(10)70197-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. van Es MA, Veldink JH, Saris CG, et al. : Genome-wide association study identifies 19p13.3 ( UNC13A) and 9p21.2 as susceptibility loci for sporadic amyotrophic lateral sclerosis. Nat Genet. 2009;41(10):1083–7. 10.1038/ng.442 [DOI] [PubMed] [Google Scholar]
- 88. Renton AE, Majounie E, Waite A, et al. : A hexanucleotide repeat expansion in C9orf72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–68. 10.1016/j.neuron.2011.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. : Expanded GGGGCC hexanucleotide repeat in noncoding region of C9orf72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–56. 10.1016/j.neuron.2011.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kosmicki JA, Churchhouse CL, Rivas MA, et al. : Discovery of rare variants for complex phenotypes. Hum Genet. 2016;135(6):625–34. 10.1007/s00439-016-1679-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. McLaughlin RL, Vajda A, Hardiman O, et al. : Heritability of Amyotrophic Lateral Sclerosis: Insights From Disparate Numbers. JAMA Neurol. 2015;72(8):857–57. 10.1001/jamaneurol.2014.4049 [DOI] [PubMed] [Google Scholar]
- 92. Al-Chalabi A, Hardiman O: The epidemiology of ALS: a conspiracy of genes, environment and time. Nat Rev Neurol. 2013;9(11):617–28. 10.1038/nrneurol.2013.203 [DOI] [PubMed] [Google Scholar]
- 93. Alonso A, Logroscino G, Hernán MA: Smoking and the risk of amyotrophic lateral sclerosis: a systematic review and meta-analysis. J Neurol Neurosurg Psychiatry. 2010;81(11):1249–52. 10.1136/jnnp.2009.180232 [DOI] [PubMed] [Google Scholar]
- 94. Beard JD, Kamel F, et al. : Military Service, Deployments, and Exposures in Relation to Amyotrophic Lateral Sclerosis Etiology and Survival. Epidemiol Rev. 2015;37(1):55–70. 10.1093/epirev/mxu001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Scarmeas N, Shih T, Stern Y, et al. : Premorbid weight, body mass, and varsity athletics in ALS. Neurology. 2002;59(5):773–5. 10.1212/WNL.59.5.773 [DOI] [PubMed] [Google Scholar]
- 96. Lacorte E, Ferrigno L, Leoncini E, et al. : Physical activity, and physical activity related to sports, leisure and occupational activity as risk factors for ALS: A systematic review. Neurosci Biobehav Rev. 2016;66:61–79. 10.1016/j.neubiorev.2016.04.007 [DOI] [PubMed] [Google Scholar]
- 97. Abhinav K, Al-Chalabi A, Hortobagyi T, et al. : Electrical injury and amyotrophic lateral sclerosis: a systematic review of the literature. J Neurol Neurosurg Psychiatry. 2007;78(5):450–53. 10.1136/jnnp.2006.104414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Fischer H, Kheifets L, Huss A, et al. : Occupational Exposure to Electric Shocks and Magnetic Fields and Amyotrophic Lateral Sclerosis in Sweden. Epidemiology. 2015;26(6):824–30. 10.1097/EDE.0000000000000365 [DOI] [PubMed] [Google Scholar]
- 99. Bozzoni V, Pansarasa O, Diamanti L, et al. : Amyotrophic lateral sclerosis and environmental factors. Funct Neurol. 2016;31(1):7–19. 10.11138/FNeur/2016.31.1.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Delzor A, Couratier P, Boumédiène F, et al. : Searching for a link between the L-BMAA neurotoxin and amyotrophic lateral sclerosis: a study protocol of the French BMAALS programme. BMJ Open. 2014;4(8):e005528. 10.1136/bmjopen-2014-005528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Rooney J, Vajda A, Heverin M, et al. : No association between soil constituents and amyotrophic lateral sclerosis relative risk in Ireland. Environ Res. 2016;147:102–7. 10.1016/j.envres.2016.01.038 [DOI] [PubMed] [Google Scholar]
- 102. Sutedja NA, Veldink JH, Fischer K, et al. : Exposure to chemicals and metals and risk of amyotrophic lateral sclerosis: a systematic review. Amyotroph Lateral Scler. 2009;10(5–6):302–9. 10.3109/17482960802455416 [DOI] [PubMed] [Google Scholar]
- 103. Malek AM, Barchowsky A, Bowser R, et al. : Exposure to hazardous air pollutants and the risk of amyotrophic lateral sclerosis. Environ Pollut. 2015;197:181–6. 10.1016/j.envpol.2014.12.010 [DOI] [PubMed] [Google Scholar]
- 104. Wang MD, Little J, Gomes J, et al. : Identification of risk factors associated with onset and progression of amyotrophic lateral sclerosis using systematic review and meta-analysis. Neurotoxicology. 2016; pii: S0161-813X(16)30116-4. 10.1016/j.neuro.2016.06.015 [DOI] [PubMed] [Google Scholar]
- 105. Evans MC, Couch Y, Sibson N, et al. : Inflammation and neurovascular changes in amyotrophic lateral sclerosis. Mol Cell Neurosci. 2013;53:34–41. 10.1016/j.mcn.2012.10.008 [DOI] [PubMed] [Google Scholar]
- 106. Zhao W, Beers DR, Appel SH: Immune-mediated mechanisms in the pathoprogression of amyotrophic lateral sclerosis. J Neuroimmune Pharmacol. 2013;8(4):888–99. 10.1007/s11481-013-9489-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Henkel JS, Beers DR, Wen S, et al. : Regulatory T-lymphocytes mediate amyotrophic lateral sclerosis progression and survival. EMBO Mol Med. 2013;5(1):64–79. 10.1002/emmm.201201544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Mantovani S, Garbelli S, Pasini A, et al. : Immune system alterations in sporadic amyotrophic lateral sclerosis patients suggest an ongoing neuroinflammatory process. J Neuroimmunol. 2009;210(1–2):73–79. 10.1016/j.jneuroim.2009.02.012 [DOI] [PubMed] [Google Scholar]
- 109. Rentzos M, Evangelopoulos E, Sereti E, et al. : Alterations of T cell subsets in ALS: A systemic immune activation? Acta Neurol Scand. 2012;125(4):260–64. 10.1111/j.1600-0404.2011.01528.x [DOI] [PubMed] [Google Scholar]
- 110. McCormick AL, Brown RH, Jr, Cudkowicz ME, et al. : Quantification of reverse transcriptase in ALS and elimination of a novel retroviral candidate. Neurology. 2008;70(4):278–83. 10.1212/01.wnl.0000297552.13219.b4 [DOI] [PubMed] [Google Scholar]
- 111. Li W, Lee MH, Henderson L, et al. : Human endogenous retrovirus-K contributes to motor neuron disease. Sci Transl Med. 2015;7(307):307ra153. 10.1126/scitranslmed.aac8201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Al-Chalabi A, Calvo A, Chio A, et al. : Analysis of amyotrophic lateral sclerosis as a multistep process: a population-based modelling study. Lancet Neurol. 2014;13(11):1108–13. 10.1016/S1474-4422(14)70219-4 [DOI] [PMC free article] [PubMed] [Google Scholar]