Abstract
Malaria is caused by parasites of the genus Plasmodium. All human-infecting Plasmodium species can establish long-lasting chronic infections1–5, creating an infectious reservoir to sustain transmission1,6. It is widely accepted that maintenance of chronic infection involves evasion of adaptive immunity by antigenic variation7. However, genes involved in this process have been identified in only two of five human-infecting species: P. falciparum and P. knowlesi. Furthermore, little is understood about the early events in establishment of chronic infection in these species. Using a rodent model we demonstrate that only a minority of parasites from among the infecting population, expressing one of several clusters of virulence-associated pir genes, establishes a chronic infection. This process occurs in different species of parasite and in different hosts. Establishment of chronicity is independent of adaptive immunity and therefore different from the mechanism proposed for maintainance of chronic P. falciparum infections7–9. Furthermore, we show that the proportions of parasites expressing different types of pir genes regulate the time taken to establish a chronic infection. Since pir genes are common to most, if not all, species of Plasmodium10, this process may be a common way of regulating the establishment of chronic infections.
Long-lasting Plasmodium falciparum infections are thought to be maintained by cytoadherence to avoid splenic clearance and antigenic variation of the adherent proteins to avoid clearance by antibodies. We wanted to know how chronic infections are established by malaria parasites lacking genes known to be involved in these processes (var and sicavar). We used, as an example, the rodent malaria parasite Plasmodium chabaudi chabaudi AS that gives rise to chronic infections in laboratory mice (Figure 1A). The acute infection is defined by a peak of parasitaemia (5-10% iRBC) approximately ten days after mosquito bite, and clearance of the majority of parasites by day fifteen. A chronic infection is then established, giving rise to several episodes of patent parasitemia (>0.01%) for up to eighty days. Comparison of the transcriptomes of parasite populations from the acute and chronic phases of infection showed that among the multigene families identified in the P. chabaudi AS genome, the pir family is the most differentially expressed (Figure 1A, Supplementary Tables 1 and 2). This multigene family is present in most Plasmodium genomes11 and has been associated with virulence12. More than half of the pir genes were differentially expressed between acute and chronic phases. The majority of these genes had significantly higher expression in the acute phase of infection, whereas only a few had higher expression in the chronic phase.
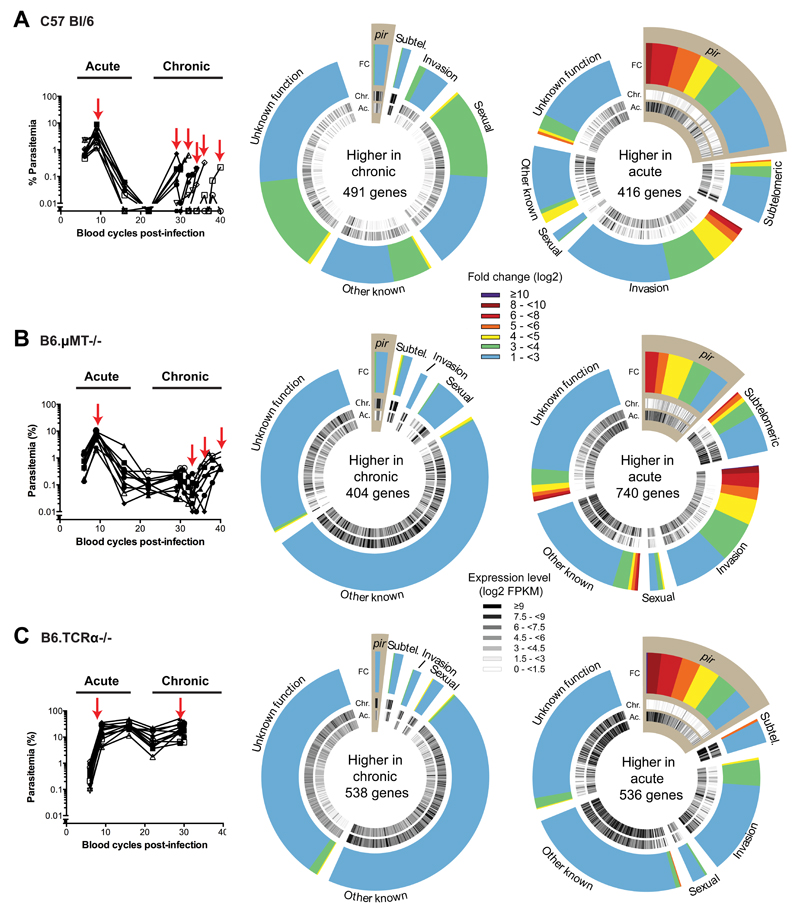
Figure 1. Chronic infections modify the transcriptome of P. chabaudi independently of the adaptive immune response.
Parasitaemias over the course of infection in (A) 10 wild-type C57Bl/6 mice, (B) 10 B6.μMT-/- mice and (C) 10 TCRα-/- mice are shown on the left panels. Parasite mRNAs were collected from 9 wild-type C57Bl/6 mice, 5 B6.μMT-/- mice and 3 TCRα-/- mice selected randomly at the time points indicated by the red arrows. On the right panels, hot pie diagrams show expression levels (black and white inner circles) and fold changes (coloured outer circles) for genes expressed more highly (FDR <= 0.01, fold change >=2) in the chronic and acute phases. Genes are classified into groups according to several categories including: subtelomeric genes (subtel.); genes associated with red blood cell invasion (invasion); genes associated with gametocytogenesis (sexual) and pir genes (highlighted in brown).
A Plasmodium-specific antibody response is detectable from day seven of infection13 and thus the change in pir expression might be the result of adaptive immunity-dependent selection of parasites expressing certain pir genes. However, we observed the same change in mice lacking B cells and antibodies (μMT; Figure 1B) or lacking T cells able to promote an antibody response (TCRα-/-; Figure 1C; Supplementary Figure 1; Supplementary Table 1). Therefore the transcriptomic changes observed between acute and chronic phases are independent of adaptive immunity and distinct from the mechanism suggested to be involved in maintenance of chronic infection in P. falciparum9.
In the genomes of most malaria parasites, pir genes are located in subtelomeric regions that are difficult to resolve based on the de novo assembly of short sequencing reads. Using Single Molecule Real-Time (SMRT) sequencing, we generated a new genome assembly for P. chabaudi AS comprising a complete set of fourteen chromosomes with no gaps (Supplementary Table 3). We observed that some subtelomeres had similar sets of pir genes arranged in clusters (Supplementary Figures 2 and 3). In recent work, pir genes were classified into several short and long forms (S1-7 and L1-4, respectively)14. We found eight clusters with common structures: three were enriched for the L1 subfamily and five were enriched for the S7 subfamily (Figure 2; Supplementary Table 4). Interestingly, we found that the majority of pir expression in our samples came from this small number of clusters, and that genes within clusters were co-expressed during infection (Supplementary Figure 4). While the S7-rich clusters were highly expressed during the acute phase, the chronic phase was dominated by expression of L1-rich loci (Figure 2C; Supplementary Table 5). This pattern was replicated in the αβT-cell receptor- and B-cell knockout mice (Figure 2C) reinforcing our earlier conclusion that changes in pir gene expression are independent of selection by the adaptive immune response. Individual L1- or S7-rich loci are hereafter termed Chronicity-Associated Pir Locus (ChAPL), and Acute-Associated Pir Locus (AAPL), respectively. We identified an intergenic motif, commonly occurring upstream of most ChAPL genes, that might serve as a promoter sequence (Supplementary Figure 5). However, as only a subgroup of ChAPL genes are expressed in each mouse (Figure 2C), it is likely that epigenetic mechanisms regulate access of transcription factors to this motif, as shown for subtelomeric gene families in P. falciparum15. Of the three ChAPLs, we observed a preference for clusters on chromosome 6.
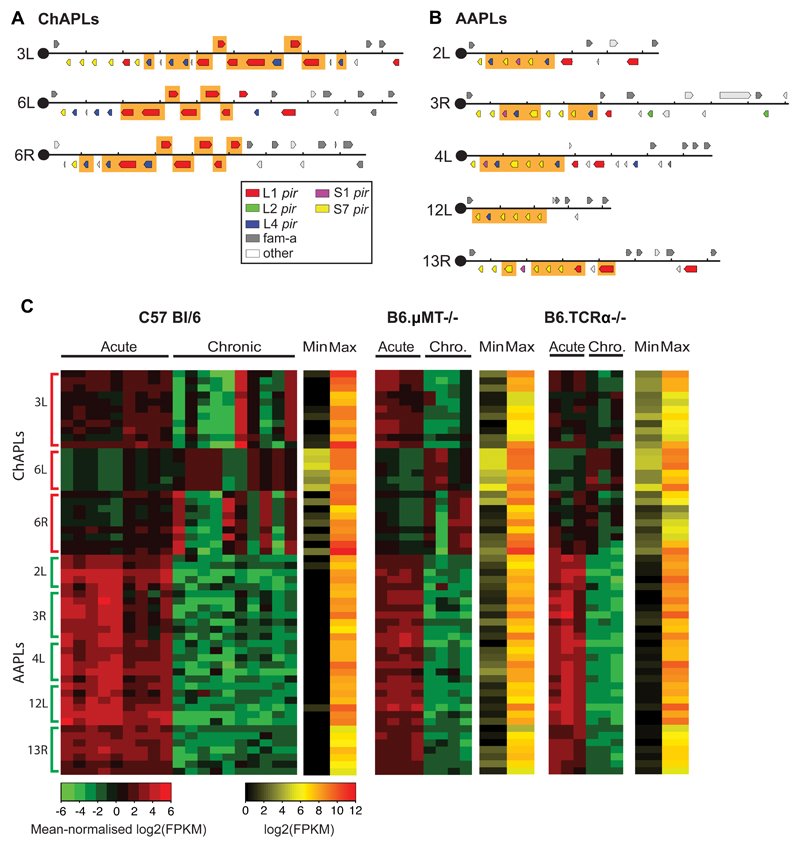
Figure 2. Chronic infections are characterized by the expression of distinctive clusters of pir genes in P. chabaudi.
(A) Chronic Associated Pir Loci (ChAPL), and (B) Acute Associated Pir Loci (AAPL) are shown in the context of the subtelomeric region from the telomeric repeats to the last pir or fam-a gene. Those genes co-expressed across mice over the course of infection are highlighted in orange. (C) Heatmap of pir gene expression at ChAPL and AAPL loci, showing mean-normalised RPKM values for 10 wild type C57Bl/6 mice, four B6.μMT-/- mice and three TCRα-/- mice infected by mosquito bites. Each column represents values obtained from an individual mouse. Samples were collected from the same mice during the acute and chronic (chro.) phases of infection. On the side, numbers indicate chromosomes and letters indicate the location on that chromosome: L for left hand end, R for right hand end. ChAPLs and AAPLs are highlighted. For each gene, red represents its maximal expression, green its minimal expression. The absolute maximum and minimum values for each gene are shown in a separate heatmap.
We recently showed that increased P. chabaudi virulence during serial blood passage (SBP) is associated with a reduced repertoire of pir gene transcripts compared with parasites transmitted by mosquito (MT)12. The gene most highly upregulated in SBP parasites was a pir from the L1 subfamily (PCHAS_1100300). Although not found in a ChAPL, this gene is from the same subfamily that is most common in these loci. We therefore hypothesized that chronic phase parasites expressing ChAPLs would have a similar phenotype. To test this, we injected acute and chronic phase parasites into naïve mice. Chronic parasites gave rise to significantly greater parasitaemias and pathology than parasites derived from the acute phase of infection (Figure 3A; Supplementary Figure 6A). This suggests that parasite populations derived from the chronic phase expand better in the face of the response of a naive mouse than parasites derived from the acute phase. However, as chronic parasites only reached high parasitaemias when injected into naïve mice, our data also indicate that they are better controlled when the host has previously experienced an acute infection. This pattern was also observed when chronic parasites were collected from B-cell knockout mice (Supplementary Figure 6B). Chronic infections therefore involve the emergence of phenotypically distinct parasites expressing ChAPLs, independently of adaptive immunity.
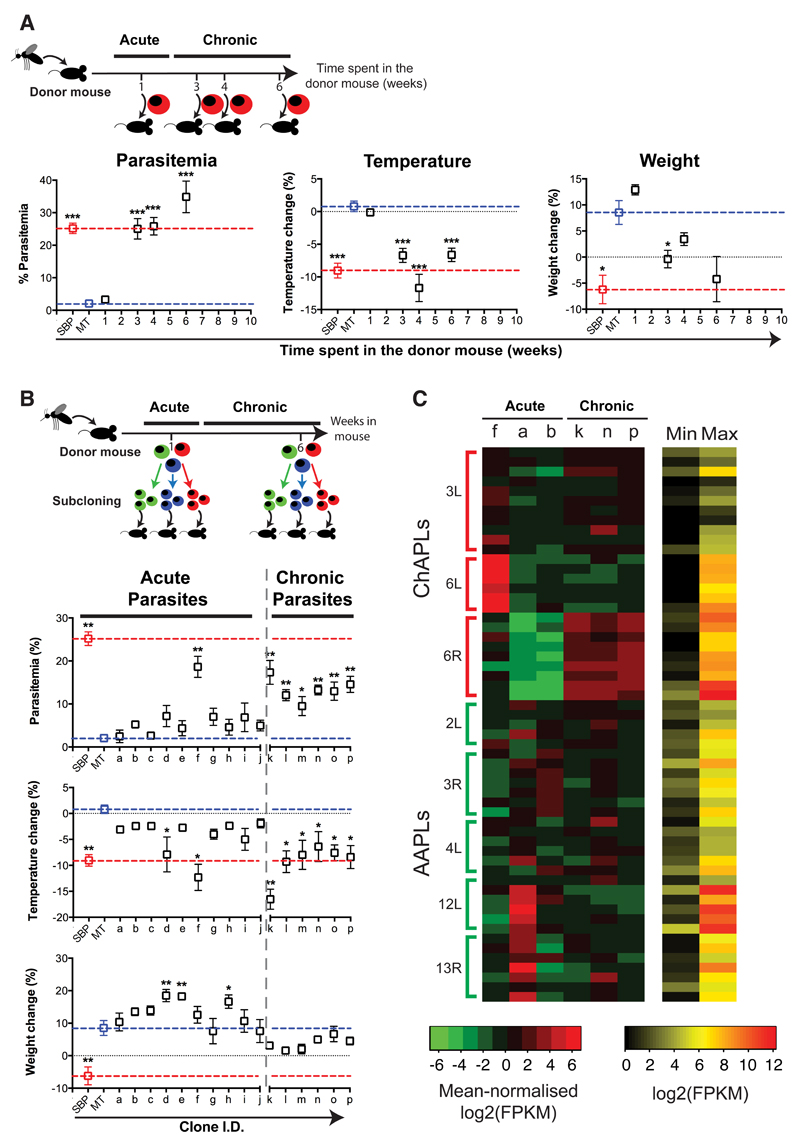
Figure 3. Chronic infections select for virulent P. chabaudi parasites expressing distinctive clusters of pir genes.
Donor mice were infected by mosquito bites. (A) Parasites were subsequently passaged into naïve recipient mice after one, three, four or six weeks (n=15 mice per group). (B) Individual parasites were cloned during the acute and chronic phases of infection. After their expansion, clonal populations of parasites were used to infect recipient mice (n= 5 mice per clone). In (A) and (B), we show maximum parasitaemia, temperature change and weight change for recipient mice compared to mice directly infected by mosquito bites (MT; blue dotted line) or with serially blood passaged parasites (SBP; red dotted line). Each dot represents the mean ± standard error of the mean (SEM) of 5 individual mice. Each group has been compared to mice infected with 105 MT parasites (*P<0,05; **P<0,01; ***P<0,001, two-sided Mann Whitney Test). (C) Heatmap of pir gene expression at ChAPLs and AAPLs in three acute and three chronic clones. The first three columns represent cloned parasite lines derived from from the acute phase (f, a, b), the next three columns represent cloned lines from the chronic phase (k, n, p) from 3 individual mice. On the side, chromosomes numbers are indicated as well as the location on that chromosome: L for left hand, R for right hand. ChAPLs and AAPLs are highlighted. For each gene, red represents its maximal expression, green its minimal expression. The absolute maximum and minimum values for each gene are shown in a separate heatmap.
Thus we have established that there are transcriptional and phenotypic changes observed during chronic Plasmodium infections and that they do not stem from classical antigenic variation. We then sought to determine whether these were due to a transcriptional switch at the population level, or to a selection of pre-existing parasites from the acute phase. We isolated single parasites from the acute and chronic phases of infection, initiated clonal infections in C57Bl/6 mice and performed RNA sequencing analyses of these cloned populations of parasites (Figure 3B). We found that one out of ten acute-phase clones was more virulent than the population as a whole (Figure 3B; Supplementary Figure 7). While avirulent clones expressed AAPLs, the virulent clone expressed a ChAPL, as predicted by our finding that ChAPL-expressing parasites are more virulent in naïve mice (Figure 3C; Supplementary Figure 8; Supplementary Table 6). As expected, the clones from the chronic phase were all virulent and expressed ChAPLs (Figures 3B & 3C, Supplementary Figures 8 & 9). This suggests that the establishment of chronic infection involves selection of a small proportion of parasites expressing ChAPLs from a clonally variant population largely expressing AAPLs.
On the basis of these observations, we hypothesized that the proportion of virulent parasites already in the acute phase might affect the success of establishing a chronic infection. In our initial transcriptome experiment, we found that high expression of ChAPLs during the acute phase was associated with a more rapid recrudescence (Supplementary Table 7). We then tested whether chronic-like parasites, expressing L1 pir genes, outgrew acute parasites largely expressing AAPLs. We used SBP parasites as a proxy for chronic parasites as they express the L1 pir PCHAS_1100300, and have a similar phenotype to ChAPL-expressing chronic parasites. We performed mixed infections in C57BL/6 mice with different ratios of SBP and acute MT (expressing AAPLs) parasites tagged with different fluorescent markers (Figure 4, Supplementary Figure 10 A-C; Supplementary Table 8). SBP parasites rapidly outcompeted MT parasites (Figure 4; Supplementary Figure 10D), confirming that the more virulent, chronic-like parasites had a selective advantage, as has been shown previously for different parasite isolates16–20. The selection of a minority of iRBCs, expressing virulence-associated pir genes (Figures 3B & 3C), can therefore explain the change in expression from S- to L-pir genes observed in the parasite population between the acute and chronic phases of infection (Figures 1 & 2). As we had hypothesized, higher doses of L1-expressing parasites resulted in an earlier recrudescence confirming that these parasites have a better capacity to establish a chronic infection (Figure 4, Supplementary Figure 11).
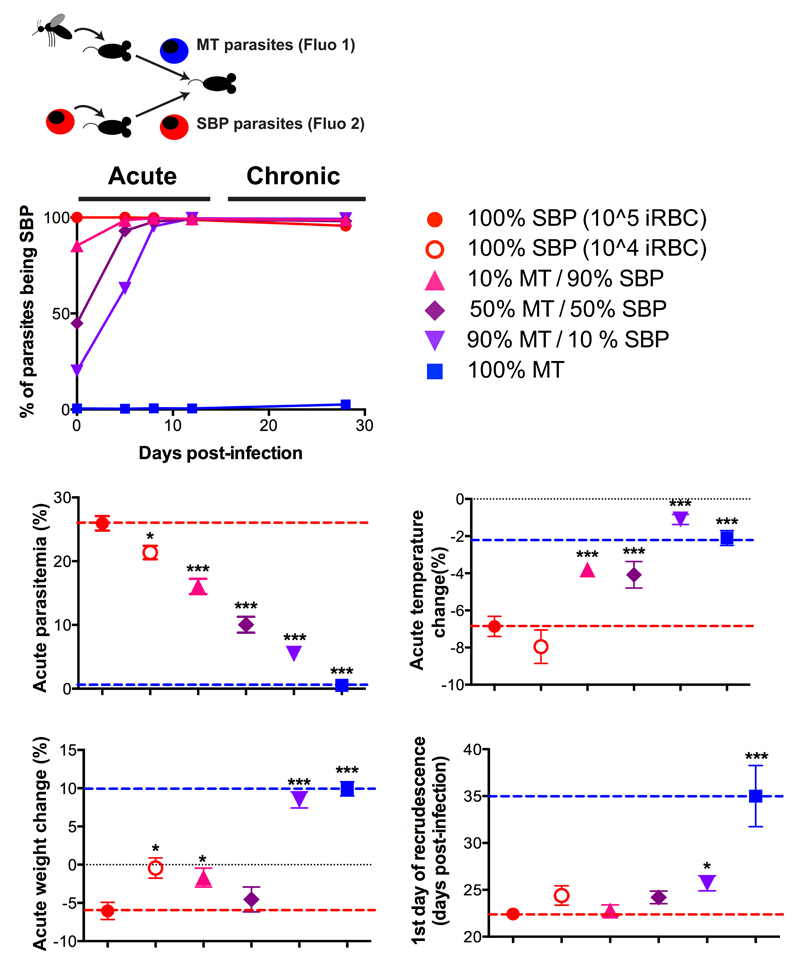
Figure 4. Chronicity and virulence of infection are dictated by the initial composition of P. chabaudi population.
Parasites tagged with mCherry (Fluo 1) were Mosquito-Transmitted (MT), and mNeonGreen (Fluo 2) tagged parasites were Serially Blood-Passaged (SBP). Different proportions of the two were used to infect recipient mice by i.p. injection of 105 parasites (n= 10 mice per group). As a control, a group of 10 recipient mice was infected by i.p. injection of 104 SBP parasites. The mean percentage of parasites being SBP (as determined by flow cytometry) is indicated (upper panel). We show maximum parasitaemia, temperature change, weight change (middle panel) and time of recrudescence (lower panel) for each group compared to mice infected with 100% MT parasites (blue dotted line) or 100% SBP parasites (red dotted line). Each dot represents the mean ± standard error of the mean (SEM) for each group of 10 mice. Each group has been compared to mice infected with 105 SBP parasites (*P<0,05; **P<0,01; ***P<0,001, two-sided Mann Whitney Test).
When a similar infection mixing SBP and MT parasites was carried out in RAG1-/- mice (lacking both T and B cells) and RAG-/- mice treated with chlodronate liposomes to deplete macrophages (Supplementary Figure 12A), virulent parasites similarly rapidly outgrew AAPL-expressing parasites. Thus, this early selection takes place even in the absence of B cells, T cells, and macrophages indicating that other mechanisms are operating (Supplementary Figure 12B).
Unexpectedly, when mice were infected with 104 virulent parasites plus 105 avirulent parasites, the acute parasitaemia and pathology were lower than in mice infected with 104 virulent parasites alone (Figure 4B, Supplementary Figure 11). This suggests that AAPL-expressing parasites act to lower the virulence of parasites expressing largely virulence-associated L1 pirs and delay the onset of chronicity. Therefore, the initial composition of parasites expressing different pir genes regulates parasite recrudescence.
To determine whether our findings apply more generally, beyond P. chabaudi AS infections of laboratory mice, we established chronic infections with alternative host/parasite combinations. We used the more virulent P. chabaudi CB strain in C57BL/6 mice and P. chabaudi AS in the African thicket rat Grammomys surdaster. This is a natural host of rodent Plasmodium (Supplementary Figures 13A & 13B). Reduced AAPL and increased ChAPL gene expression during chronic P. chabaudi infection were observed in both cases, suggesting that our findings are a general feature of P. chabaudi chronic infections irrespective of strain of parasite or host (Supplementary Figures 13C & 13D). We then investigated chronic P. berghei infections in Brown Norway rats (Supplementary Figure 14A). The subtelomeric regions and pir repertoire of P. berghei are quite different from those of P. chabaudi 14. Loci resembling the AAPLs and ChAPLs of P. chabaudi are absent. Nevertheless, we observed a general decrease in pir expression during the chronic phase with an increase in expression of L2 pir genes (Supplementary Table 9; Supplementary Figure 14B). The L2 pir genes of P. berghei are similar to the P. chabaudi L1 genes we found in ChAPLs. They are present in recently copied loci (Supplementary Figure 14C), contain long variable central domains and have a longer N-terminal domain that may contain a second transmembrane helix (Supplementary Figure 15). This suggests our findings are a general feature of malaria parasites infecting rodents.
Our results show that mosquito transmission of P. chabaudi gives rise to a clonally variant population of parasites, the majority expressing AAPLs and a minority ChAPLs. During the acute phase of infection, AAPL-expressing parasites are controlled by the host response, while ChAPL-expressing parasites survive and establish a chronic infection. The more ChAPL-expressing parasites that are present early in infection, the more rapidly the chronic infection is established.
While it is thought that maintenance of chronic P. falciparum infection depends on the differential expression of var genes7,8, how similar infections are established and maintained by P. vivax, P. malariae and P. ovale, that lack these variant antigens, is unknown. Here, we show that the establishment of chronic Plasmodium infection in rodent models is reproducibly associated with phenotypically distinct parasites, expressing particular pir gene clusters. Importantly, contrary to what is understood about maintenance of chronic infection in P. falciparum9, this change in the parasite population is independent of adaptive immunity. Antigenic variation may still operate in maintaining chronicity, but our results clearly show that other mechanisms allow for the establishment of chronic infection in parasites that infect rodents.
Our finding that the initial population of parasites comprises individuals, each expressing different pir genes, suggests that the chronic infection is established by a small number of parasites which evade the early immune response. The selective advantage during infection of ChAPL-expressing over AAPL-expressing parasites could be of multiple origins. Although our previous results showed that growth rates of avirulent MT parasites and virulent SBP parasite were similar in RAG1-/- mice12, red cell invasion efficiency might still play a role in selection of virulent clones for establishment of chronic infection. We are also investigating rosette formation21,22, red cell deformability23, sequestration24, and the capacity to escape a macrophage-independent innate immune response25 as possibilities. It has been shown that the expression of subsets of P. falciparum rif genes (which may be relatives of pir genes10) and P. vivax pir genes are associated with cytoadherence of iRBCs to human endothelial cells26,27 and red blood cells21,28,29, two processes implicated in parasite survival and pathogenicity30. Furthermore, it has previously been suggested that the same molecules are involved in cytoadherence and immune evasion in P. chabaudi9,24. Differences in cytoadhesive properties could therefore promote the expansion of more virulent parasites. Characterizing this process and determining the function of pir genes in vivo may identify mechanisms that universally regulate immune evasion by Plasmodium and lead to novel approaches to reduce their survival and transmission.
Methods
Mouse and parasite lines used
All experiments were performed in accordance with the UK Animals (Scientific Procedures) Act 1986 (PPL 80/2538) and the guidelines provided by the National Institutes of Health (NIH) Animal Care and Use Committee (ACUC) and approved by the Francis Crick Institute Ethical Committee and by the Institutional Animal Care and Use Committee (IACUC). T cell receptor-α chain knockout (TCRα-/-)31, mice homozygous for a targeted mutation of the transmembrane exon of the IgM μ chain, μMT32, and V(D)J recombination activation gene RAG-1 knockout (RAG-1-/-) 33 on a C57Bl/6 background, and C57 Bl/6 Wildtype (WT) mice were obtained from the specific-pathogen free (SPF) unit and subsequently conventionally housed with autoclaved cages, bedding and food at the BRF, of the Francis Crick Institute. The Grammomys surdaster rats used in this study were captured in the wild at Fungurume and Lumata in the Katanga province, south of DR Congo, were bred and maintained in a conventional unit in sterile cages, with autoclaved water and bedding at NIH/NIAID/LMIV 34. Inbred Brown Norway rats were obtained from Charles River Laboratories and housed at the Animal Holding Unit of the Wellcome Trust Sanger Institute. Experiments were performed with 6-8 week old female mice and Grammomys surdaster rats housed under reverse light conditions (light 19:00-07:00, dark 07:00-19:00) and 6-week old female Brown Norway rats housed under normal light conditions, at 20-22°C. Measurements of clinical pathology were taken at 11:00. Core body temperature was measured with a rectal thermometer and erythrocyte density was determined with a VetScan HMII haematology system (Abaxis). Changes in body temperature, body weight and erythrocyte density were calculated relative to a baseline measurement performed 1 day before infection. An infection was considered virulent when it induced a statistically significant decrease in the mouse temperature, weight and/or red blood cells compared with uninfected control mice.
Cloned lines of Plasmodium chabaudi chabaudi AS and CB parasites were used. To initiate infection, mice were either injected intraperitoneally (IP) or intravenously (IV) with 1, 104 or 105 infected erythrocytes or submitted to the bite of 20 infected mosquitoes as previously described12. Grammomys surdaster rats were infected by I.V. injection of 2500 sporozoites of Plasmodium chabaudi chabaudi AS. Brown Norway rats were infected by I.V. injection of 7200 sporozoites of Plasmodium berghei ANKA. Parasitaemia was monitored by Giemsa-stained thin blood films on blinded samples. The limit of detection for patent parasitaemia was 0.001% infected erythrocytes.
Chronic infections in wild type and genetically altered rodents
Out of 10 wild-type C57Bl/6 mice, 10 μMT mice and 10 TCRα-/- mice infected with P. chabaudi AS via mosquito bite, 9 WT C57Bl/6 mice, 5 μMT mice and 3 TCRα-/- mice were randomly selected for transcriptomic analysis. 3 C57Bl/6 mice infected with P. chabaudi CB, 3 Grammomys surdaster rats infected with P. chabaudi AS and 3 Brown Norway rats infected with P. berghei ANKA (all via mosquito bites or injection of sporozoites) were also used for transcriptomic analysis. Our aim was to use a minimum of three biological replicates per condition for all experiments, as this is considered to be sufficient for general surveys of differential expression35. Blood was extracted from the same rodents at two time points, therefore analysing the changes occurring over time in the same parasite populations. During the acute phase of infection, 100μL of blood was collected from the tail after the completion of seven cycles of schizogony. During the chronic phase, exsanguination was performed between 30 and 40 days post-infection in μMT mice, at 30 days post-infection in TCRα-/- mice, at day 15 post-infection in Brown Norway rats or when the parasitaemia reached 0.1% between 27 and 40 days post-infection in wild type C57Bl/6 mice and G. surdaster rats. After data collection, the acute sample from wild-type mouse 454 and both acute and chronic samples from μMT mouse 433 were excluded, as they were outliers in a multi-dimensional scaling analysis of the gene expression data.
Phenotypic analysis of acute and chronic parasites
Donor mice were infected via mosquito bites. Parasites from donor mice (n= 3) were blood-passaged into recipient mice at 1, 3, 4 and 6 weeks post-mosquito transmission. Recipient mice (n=5 per donor mouse, the minimum number required for an 80% chance of significant differences (p<=0.05) in parasitemia, weight and erythrocyte counts) were infected by IP injection of 105 infected erythrocytes. Parasitaemia and signs of pathology were monitored in recipient mice on blinded samples.
Cloning of single parasites from acute and chronic phases
One mouse was infected via mosquito bite. Infected blood was then collected from the tail at two time points: 1 week post-infection (acute phase) and six weeks post-infection (chronic phase). Both times, parasites were cloned by dilution and single parasites were injected IV in wild type mice. After expansion, each cloned line was blood passaged into eight experimental mice by IP injection of 105 infected erythrocytes. Five mice were used to monitor parasitaemia and signs of pathology and three mice were sacrificed and exsanguinated after the completion of seven cycles of schizogony for RNA extraction.
RNA extraction
During all RNA extractions experiments, blood-stage parasites were isolated at 11:00, when >90% of the parasites were at the late trophozoite stage of development (Table S8). Parasite RNA was isolated as previously described12. Briefly, whole blood was depleted of leukocytes by filtration (Plasmodipur, EuroProxima) and from globin RNA and red cell debris by saponin lysis (Sigma) and centrifugation. Purified parasites were resuspended in TRIzol, snap-frozen on dry ice and kept at -80°C until use. RNA was then extracted, resuspended in water and its quantity and quality were determined on an Agilent 2100 Bionalayzer RNA 6000 Nano or Pico chip.
RNA-seq library prep and sequencing
The majority of RNA was used to make 200-450bp fragment libraries using Illumina's TruSeq RNA Sample Prep v2 kit, with 10 cycles of PCR amplification using KAPA Hifi Polymerase rather than the kit-supplied Illumina PCR Polymerase. Two samples required 14 cycles of PCR (T1_46-4 and T1_47-4). Mosquito-transmitted P. chabaudi CB samples (MTCB) were subjected to 15 cycles of PCR.
The libraries were sequenced using an Illumina HiSeq 2000 v3 with 100bp paired-end reads (samples beginning ‘T’), an Illumina HiSeq 4000 with 75bp paired-end reads (MTCB samples) or an Illumina HiSeq 2500 with 75bp paired-end reads for the rest. RNA from cloned parasites was used to make 100-300bp fragment libraries produced using Illumina’s TruSeq Stranded mRNA Sample Prep Kit with 10 cycles of PCR amplification using KAPA Hifi Polymerase. These libraries were sequenced using an Illumina HiSeq 2500 with 75bp paired-end reads. A description of the libraries can be found in Supplementary Table 10.
Analysis of gene expression
Tag sequences were removed from sequenced reads. Where individual samples were run over multiple lanes, the reads from these were merged before mapping. Reads were then mapped against spliced gene sequences (exons, but not UTRs) from the v3 P. c. chabaudi AS (this work) or the v3 P. berghei ANKA (Fougère et al., under review) reference genomes using Bowtie2 v2.1.036 (-a -X 800 –x). Read counts per transcript were estimated using eXpress v1.3.037, with default parameters. Genes with an effective length cutoff below 10 in any sample were removed. Summing over transcripts generated read counts per gene.
Differential expression analysis was performed using edgeR v3.8.638 on genes with ≥ 3 counts per million. The Fisher’s exact test was used with cutoffs FDR < 0.01 and fold change ≥ 2, except for the P.berghei analysis where FDR < 0.1 and fold change ≥ 1 was used. Functional categories of genes were identified by orthology using GeneDB 39 from several different P. falciparum datasets: invasion genes40, sexual genes41 and subtelomeric (by manual inspection of chromosomes).
Gene expression clustering was used to identify which genes within ChAPLs and AAPLs were co-expressed. This was done by generating a toroidal 5x5 Self-Organising Map (SOM) using the Kohonen package in R42. FPKM values for each gene were logged (base 2) and mean-normalised per gene. The SOM was trained in 100 iterations.
Sequencing and assembly of P. c. chabaudi AS v3 genome
High molecular weight DNA was prepared as follows. Heparinised blood from two mice infected with P. chabaudi AS (blood passaged parasites, days seven & eight post-infection) was pooled, diluted 1:3 in PBS, and passed through a Plasmodipur (Euro-Diagnostica) filter. Erythrocyte membranes were removed by saponin lysis (0.15% in ice cold PBS). Parasitised erythrocytes were recovered by centrifugation (2000g 10 min 4 °C) and washed twice in ice cold PBS. The cell pellet was taken up in 50mM Tris HCl pH 7.5, 50mM EDTA pH8.0, 100mM NaCl, 0.5% SDS and digested with RNase A (1mg/ml, Life Technologies) for 30 min at 37°C. Proteinase K (Roche) was added to a final concentration of 1mg/ml, incubated at 45°C overnight, followed by extraction with phenol chloroform and ethanol precipitation.
For preparation of long-read sequencing libraries, 5μg of P. c. chabaudi AS genomic DNA was sheared to 20-25kb by passing through a 25mm blunt ended needle. SMRT bell template libraries were generated using the Pacific Biosciences issued protocol (20 kb Template Preparation Using BluePippin™ Size-Selection System). After 7kb-20kb size-selection using the BluePippin™ Size-Selection System (Sage Science, Beverly, MA) the library was sequenced using P6 polymerase and chemistry version 4 (P6C4) on 5 single-molecule real-time (SMRT) cells, each with a 240 minutes movie length. The five SMRT cells were processed using a Pacific Biosciences RSII. Reads were filtered using SMRT portal v2.2 with default parameters (minimum subread length 50, minimum polymerase read quality 75, minimum polymerase read length 50). The yield was 258214 reads totaling 1.51Gb, with read N50 9003bp and read quality 0.833. The reads were assembled using HGAP v2.2.043 with expected coverage 75 and other parameters as default.
The 28 unitigs from the HGAP assembly were BLASTed against the P. chabaudi AS v2 assembly14 to identify sequences relating to nuclear, mitochondrial and apicoplast genomes. Four unitigs were found to be mouse contamination. Fourteen unitigs represented the fourteen chromosomes from the v2 assembly including most of the unplaced contigs from v2 and some additional new sequence. All fourteen had telomeric repeats at both ends. The only major rearrangement between the assemblies was between the ends of chromosomes 6 and 13. We mapped Quiver-corrected PacBio reads against v2 and v3 assemblies and confirmed that the v3 assembly was correct. However, we noticed base errors and in particular many indels in the v3 assembly due to the relatively high error rate of PacBio sequencing. The v2 assembly had had the benefit of Sanger and Illumina technologies such that at the base level it was more accurate than the PacBio assembly. Therefore we generated 2x250bp paired-end pseudoreads from the v2 assembly with a fragment size of 600bp to a depth of 50x and used these to correct the v3 assembly using iCORN244. We transferred gene models from the v2 assembly to the corrected v3 assembly using RATT44. We then trained Augustus v2.5.545 using the v2 gene models with default parameters and predicted a new set of models for the corrected v3 assembly. We kept only those, which did not overlap with the RATT-transferred gene models. We then manually assessed the transferred models, annotated the new Augustus models and renamed the locus tags. In general we added a zero to the end of the v2 locus tags, but where a gene did not exist in v2 or had moved we added an odd number at the end. We maintained numerical ordering of locus tags across each chromosome so that they reflect relative chromosomal location.
Sequence analysis of pir genes
Pir gene sub clade assignments were initially made using Hidden Markov Models (HMMs) built from the rodent malaria pir subclades published in Otto et al.14. There were some discrepancies between the two sets, e.g. where the clade in the published tree did not agree with the top HMM hit. In particular we identified three members of the S3 subclade, which was not previously thought to be present in P. chabaudi. We used hmmsearch from HMMer i1.1rc346 with an E-value cutoff of 1e-10 and took the best hit to a subclade. We thus identified 207 pir genes which we could assign to a rodent malaria parasite (RMP) pir subclade, while a further five genes could not be classified into existing subfamilies (PCHAS_0401400, PCHAS_0602000 and PCHAS_1247000). HMM assignments of L2 pirs in P. berghei were consistent with the original tree-based classification presented in Otto et al. (2014).
We used FIRE v1.1a47 to identify putative promoter sequences upstream of genes in the AAPLs and ChAPLs. Firstly we used RNA-seq data from the study to determine 5’ unstranslated regions (UTRs) of pir genes and then manually identified untranscribed regions upstream of each UTR, reaching until the proximal UTR of the next upstream gene. Where there was insufficient RNA-seq data or other ambiguity, we did not identify a region. This was successful for 27/32 ChAPL genes and 22/28 AAPL genes. We then defined AAPL and ChAPL genes as two distinct expression clusters and ran FIRE with -- exptype=discrete and --nodups=1 options.
Sequence alignments were performed using Muscle48 with default parameters and visualized using Jalview49.
Regression analysis of pir genes against parasitological parameters
DESeq2 was used to determine whether variation in gene expression between parasite populations in different mice was correlated with parasitological parameters such as parasitaemia and time of recrudescence50. In particular we found that the expression level during the acute phase of 152 genes was correlated with the time to recrudescence across nine mice (p-value < 0.01; fold change >= 2). We discretized the time in days at recrudescence as follows: mice 214 (32 days), 421 (31 days), 427 (30 days) were short, 2014 (34 days), 2114 (36 days), 452 (33 days) and 456 (34 days) were medium, 211 (43 days) and 212 (39 days) were long. We then used this single discrete factor in the DESeq2 design formula. Those genes where expression level during the acute phase was negatively correlated with time at recrudescence were, with a single exception, pir genes from 3L, 6L and 6R subtelomeres (Table S6). These are the three ChAPLs and another L1-rich locus (4R). Interestingly, the strongest effect was from an exported gene of unknown function (PCHAS_1201300). The strongest positive effects were from pir genes in AAPLs.
Mixed infection
Transgenic lines of Plasmodium chabaudi AS expressing mCherry or mNeonGreen under the control of the constitutive promoter EF1α were generated by transfection with the plasmid PcEFp230p_mCherry and PcEFp230p_mNeonGreen, respectively. These plasmids targeted the silent 230p genomic locus, and were generated by replacement of the ssu rRNA locus of pPc-mCHCAM, described in51, with the P. chabaudi p230p locus (PCHAS_03_v3 276,451-283,284). For PcEFp230p_mNeonGreen, the mCherry tag was replaced with mNeonGreen52. Parasites were cycled ON and OFF acid water containing pyrimethamine to select for stably transfected parasites, and cloned by serial dilution. The insertion was verified by PCR amplification and Southern blot analysis and fluorochrome expression by Flow cytometry analysis (Figure S9). mNeonGreen-expressing parasites were then serially blood passaged (SBP) 10 times while mCherry-expressing parasites were mosquito transmitted (MT) into donor mice. Both parasites were then mixed at different ratios and 105 infected erythrocytes were injected i.p. into ten recipient mice per group. Competition between parasite lines as well as parasitaemia and pathology were then monitored in recipient mice. To determine the ratio of mNeonGreen- and mCherry-expressing parasites during infection, tail blood was collected and stained with Hoechst 33342 (New England Biolab). Samples were acquired on a BD LSRII flow cytometer and data were analysed with FlowJo software (TreeStar).
Clodronate liposomes treatment
B6.RAG-1-/- mice were treated with 200uL i.v. of either saline (Sigma; negative control) or clodronate liposomes (clodronateliposomes.org). Flow cytometric analysis was carried out to assess the depletion of phagocytic cells. Single-cell suspensions of splenocytes were prepared, , and cells enumerated 24h after in vivo treatment. After staining with Zombie Aqua (Biolegend) for live/dead discrimination, cells were stained with monoclonal antibodies using appropriate combinations of fluorochromes (CD11c-BV786, CD169-BV605, F4/80-BV421, Ly6G-PerCPCy5.5, MHCII-FITC, Ter119-PE-Cy7, CD68-PE, Ly6c-APC-Cy7, CD11b-AF647, all from Biolegend). The samples were acquired on a BD Fortessa/X20 (BD Biosciences) using Diva acquisition software (BD Biosciences). FlowJo (Tree Star) software was used to analyze the data.
To determine the role of T-, B- and phagocytic cells in the selection of virulent parasites, B6.RAG-1-/- mice were treated with saline or clodronate liposomes as described above at days -1 and 4 after i.p. infection with 105 iRBCs containing a mix of Neon-Green-expressing SBP parasites and mCherry-expressing MT parasites (see “Mixed infections” section).
Data availability
RNA-seq datasets used in this study have been submitted to the European Nucleotide Archive (ENA) under study accession ERP017479. These datasets are described further in Supplementary Table 10. The Pacific Biosciences RSII genomic sequencing reads, used to generate the Plasmodium chabaudi chabaudi AS v3 genome sequence, have been submitted to the ENA under experiment accession ERX613966. The assembled genome sequence and annotation can be accessed from GeneDB (http://www.genedb.org/Homepage/Pchabaudi) and PlasmoDB (http://plasmodb.org/plasmo/). The authors declare that no competing interests exist.
Supplementary Material
Acknowledgements
The Francis Crick Institute receives its core funding from the UK Medical Research Council (U117584248), Cancer Research UK, and the Wellcome Trust (WT104777MA). The Wellcome Trust Sanger Institute is funded by the Wellcome Trust (grant WT098051). CN is supported by the Wellcome Trust (WT104792), and S.C. and P.E.D. are supported by the Intramural Research Program of the U.S. National Institute of Allergy and Infectious Diseases.
The authors would like to thank Biological Research Facility and Flow Cytometry facility at the Francis Crick, institute for their skilled technical assistance, the staff of the Illumina Bespoke Sequencing team at the Wellcome Trust Sanger Institute for their contribution, Edward Smith for assistance in making constructs for fluorescence tagging of the parasites, and M. Blackman, C. van Ooij, G. Kassiotis and J. Rayner for critical reading of the manuscript.
Footnotes
Author contributions
T.B., A.J.R., C.N., M.B. and J.L. (last author) designed the study. T.B. designed and performed the mouse experiments with the help of I.T., G.K., S.M. and P.S.. A.J.R. designed the sequencing experiments and performed the bioinformatic analyses. M.S. coordinated sequencing experiments. U.B. manually annotated the Plasmodium chabaudi AS genome sequence. D.C. and J.L. (7th author) created transgenic parasites. P.E.D. and S.C performed thicket rat experiments, T.M, E.B. and O.B carried out the experiments in the Brown Norway rats. T.B., A.J.R., C.N., M.B. and J.L. (last author) wrote the manuscript.
Competing financial Interests
The authors declare no competing financial interests.
References
- 1.Ashley EA, White NJ. The duration of Plasmodium falciparum infections. Malar J. 2014;13:500. doi: 10.1186/1475-2875-13-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Assennato SM, et al. Plasmodium genome in blood donors at risk for malaria after several years of residence in Italy. Transfusion. 2014;54:2419–2424. doi: 10.1111/trf.12650. [DOI] [PubMed] [Google Scholar]
- 3.Brown KN, Brown IN. Immunity to malaria: antigenic variation in chronic infections of Plasmodium knowlesi. Nature. 1965;208:1286–1288. doi: 10.1038/2081286a0. [DOI] [PubMed] [Google Scholar]
- 4.Bruce MC, et al. Genetic diversity and dynamics of plasmodium falciparum and P. vivax populations in multiply infected children with asymptomatic malaria infections in Papua New Guinea. Parasitology. 2000;121(Pt 3):257–272. doi: 10.1017/s0031182099006356. [DOI] [PubMed] [Google Scholar]
- 5.Vinetz JM, Li J, McCutchan TF, Kaslow DC. Plasmodium malariae infection in an asymptomatic 74-year-old Greek woman with splenomegaly. The New England journal of medicine. 1998;338:367–371. doi: 10.1056/NEJM199802053380605. [DOI] [PubMed] [Google Scholar]
- 6.Bousema T, Okell L, Felger I, Drakeley C. Asymptomatic malaria infections: detectability, transmissibility and public health relevance. Nature reviews Microbiology. 2014;12:833–840. doi: 10.1038/nrmicro3364. [DOI] [PubMed] [Google Scholar]
- 7.Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol. 2008;62:445–470. doi: 10.1146/annurev.micro.61.080706.093134. [DOI] [PubMed] [Google Scholar]
- 8.Kyes S, Horrocks P, Newbold C. Antigenic variation at the infected red cell surface in malaria. Annual review of microbiology. 2001;55:673–707. doi: 10.1146/annurev.micro.55.1.673. [DOI] [PubMed] [Google Scholar]
- 9.Warimwe GM, et al. Plasmodium falciparum var gene expression is modified by host immunity. Proc Natl Acad Sci U S A. 2009;106:21801–21806. doi: 10.1073/pnas.0907590106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janssen CS, Phillips RS, Turner CM, Barrett MP. Plasmodium interspersed repeats: the major multigene superfamily of malaria parasites. Nucleic Acids Res. 2004;32:5712–5720. doi: 10.1093/nar/gkh907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham D, Lawton J, Jarra W, Preiser P, Langhorne J. The pir multigene family of Plasmodium: antigenic variation and beyond. Mol Biochem Parasitol. 2010;170:65–73. doi: 10.1016/j.molbiopara.2009.12.010. [DOI] [PubMed] [Google Scholar]
- 12.Spence PJ, et al. Vector transmission regulates immune control of Plasmodium virulence. Nature. 2013;498:228–231. doi: 10.1038/nature12231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ndungu FM, et al. Functional memory B cells and long-lived plasma cells are generated after a single Plasmodium chabaudi infection in mice. PLoS Pathog. 2009;5:e1000690. doi: 10.1371/journal.ppat.1000690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otto TD, et al. A comprehensive evaluation of rodent malaria parasite genomes and gene expression. BMC Biol. 2014;12:86. doi: 10.1186/s12915-014-0086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rovira-Graells N, et al. Transcriptional variation in the malaria parasite Plasmodium falciparum. Genome Res. 2012;22:925–938. doi: 10.1101/gr.129692.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Roode JC, et al. Virulence and competitive ability in genetically diverse malaria infections. Proc Natl Acad Sci U S A. 2005;102:7624–7628. doi: 10.1073/pnas.0500078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bell AS, de Roode JC, Sim D, Read AF. Within-host competition in genetically diverse malaria infections: parasite virulence and competitive success. Evolution. 2006;60:1358–1371. [PubMed] [Google Scholar]
- 18.De Roode JC, Read AF, Chan BH, Mackinnon MJ. Rodent malaria parasites suffer from the presence of conspecific clones in three-clone Plasmodium chabaudi infections. Parasitology. 2003;127:411–418. doi: 10.1017/s0031182003004001. [DOI] [PubMed] [Google Scholar]
- 19.Schneider P, et al. Virulence, drug sensitivity and transmission success in the rodent malaria, Plasmodium chabaudi. Proc Biol Sci. 2012;279:4677–4685. doi: 10.1098/rspb.2012.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wargo AR, de Roode JC, Huijben S, Drew DR, Read AF. Transmission stage investment of malaria parasites in response to in-host competition. Proceedings. Biological sciences / The Royal Society. 2007;274:2629–2638. doi: 10.1098/rspb.2007.0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goel S, et al. RIFINs are adhesins implicated in severe Plasmodium falciparum malaria. Nat Med. 2015;21:314–317. doi: 10.1038/nm.3812. [DOI] [PubMed] [Google Scholar]
- 22.Moll K, Palmkvist M, Ch'ng J, Kiwuwa MS, Wahlgren M. Evasion of Immunity to Plasmodium falciparum: Rosettes of Blood Group A Impair Recognition of PfEMP1. PLoS One. 2015;10:e0145120. doi: 10.1371/journal.pone.0145120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang X, et al. Differential Spleen Remodeling Associated with Different Levels of Parasite Virulence Controls Disease Outcome in Malaria Parasite Infections. mSphere. 2016;1 doi: 10.1128/mSphere.00018-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gilks CF, Walliker D, Newbold CI. Relationships between sequestration, antigenic variation and chronic parasitism in Plasmodium chabaudi chabaudi--a rodent malaria model. Parasite Immunol. 1990;12:45–64. doi: 10.1111/j.1365-3024.1990.tb00935.x. [DOI] [PubMed] [Google Scholar]
- 25.Stevenson MM, Riley EM. Innate immunity to malaria. Nat Rev Immunol. 2004;4:169–180. doi: 10.1038/nri1311. [DOI] [PubMed] [Google Scholar]
- 26.Carvalho BO, et al. On the cytoadhesion of Plasmodium vivax-infected erythrocytes. J Infect Dis. 2010;202:638–647. doi: 10.1086/654815. [DOI] [PubMed] [Google Scholar]
- 27.Claessens A, et al. A subset of group A-like var genes encodes the malaria parasite ligands for binding to human brain endothelial cells. Proc Natl Acad Sci U S A. 2012;109:E1772–1781. doi: 10.1073/pnas.1120461109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Niang M, et al. STEVOR is a Plasmodium falciparum erythrocyte binding protein that mediates merozoite invasion and rosetting. Cell Host Microbe. 2014;16:81–93. doi: 10.1016/j.chom.2014.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yam XY, et al. Characterization of the Plasmodium Interspersed Repeats (PIR) proteins of Plasmodium chabaudi indicates functional diversity. Sci Rep. 2016;6:23449. doi: 10.1038/srep23449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith JD, Rowe JA, Higgins MK, Lavstsen T. Malaria's deadly grip: cytoadhesion of Plasmodium falciparum-infected erythrocytes. Cell Microbiol. 2013;15:1976–1983. doi: 10.1111/cmi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Philpott KL, et al. Lymphoid development in mice congenitally lacking T cell receptor alpha beta-expressing cells. Science. 1992;256:1448–1452. doi: 10.1126/science.1604321. [DOI] [PubMed] [Google Scholar]
- 32.Kitamura D, Roes J, Kuhn R, Rajewsky K. A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature. 1991;350:423–426. doi: 10.1038/350423a0. [DOI] [PubMed] [Google Scholar]
- 33.Mombaerts P, et al. RAG-1-deficient mice have no mature B and T lymphocytes. Cell. 1992;68:869–877. doi: 10.1016/0092-8674(92)90030-g. [DOI] [PubMed] [Google Scholar]
- 34.Conteh S, et al. Grammomys surdaster, the natural host for Plasmodium berghei parasites, as a model to study whole organism vaccines against malaria. American Journal of Tropical Medicine and Hygiene. doi: 10.4269/ajtmh.16-0745. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conesa A, et al. A survey of best practices for RNA-seq data analysis. Genome Biol. 2016;17:13. doi: 10.1186/s13059-016-0881-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roberts A, Pachter L. Streaming fragment assignment for real-time analysis of sequencing experiments. Nat Methods. 2013;10:71–73. doi: 10.1038/nmeth.2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Logan-Klumpler FJ, et al. GeneDB--an annotation database for pathogens. Nucleic Acids Res. 2012;40:D98–108. doi: 10.1093/nar/gkr1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hu G, et al. Transcriptional profiling of growth perturbations of the human malaria parasite Plasmodium falciparum. Nat Biotechnol. 2010;28:91–98. doi: 10.1038/nbt.1597. [DOI] [PubMed] [Google Scholar]
- 41.Young JA, et al. The Plasmodium falciparum sexual development transcriptome: a microarray analysis using ontology-based pattern identification. Mol Biochem Parasitol. 2005;143:67–79. doi: 10.1016/j.molbiopara.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 42.Wehrens R, Buydens LMC. Self- and super-organizing maps in R: The kohonen package. J Stat Softw. 2007;21:1–19. [Google Scholar]
- 43.Chin CS, et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nature Methods. 2013;10:563–+. doi: 10.1038/Nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 44.Swain MT, et al. A post-assembly genome-improvement toolkit (PAGIT) to obtain annotated genomes from contigs. Nature Protocols. 2012;7:1260–1284. doi: 10.1038/nprot.2012.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stanke M, Schoffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. Bmc Bioinformatics. 2006;7 doi: 10.1186/1471-2105-7-62. Artn 62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Eddy SR. Accelerated Profile HMM Searches. Plos Comput Biol. 2011;7 doi: 10.1371/journal.pcbi.1002195. ARTN e1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Elemento O, Slonim N, Tavazoie S. A universal framework for regulatory element discovery across all genomes and data types. Molecular cell. 2007;28:337–350. doi: 10.1016/j.molcel.2007.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh34032/5/1792. [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15 doi: 10.1186/s13059-014-0550-8. ARTN 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Spence PJ, et al. Transformation of the rodent malaria parasite Plasmodium chabaudi. Nat Protoc. 2011;6:553–561. doi: 10.1038/nprot.2011.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shaner NC, et al. A bright monomeric green fluorescent protein derived from Branchiostoma lanceolatum. Nature Methods. 2013;10:407–+. doi: 10.1038/Nmeth.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq datasets used in this study have been submitted to the European Nucleotide Archive (ENA) under study accession ERP017479. These datasets are described further in Supplementary Table 10. The Pacific Biosciences RSII genomic sequencing reads, used to generate the Plasmodium chabaudi chabaudi AS v3 genome sequence, have been submitted to the ENA under experiment accession ERX613966. The assembled genome sequence and annotation can be accessed from GeneDB (http://www.genedb.org/Homepage/Pchabaudi) and PlasmoDB (http://plasmodb.org/plasmo/). The authors declare that no competing interests exist.