Abstract
Proteins and RNA are often found in ribonucleoprotein particles (RNPs), where they function in cellular processes to synthesize proteins (the ribosome), chemically modify RNAs (small nucleolar RNPs), splice pre-mRNAs (the spliceosome), and, on a larger scale, sequester RNAs, degrade them, or process them (P bodies, Cajal bodies, and nucleoli). Each RNA–protein interaction is a story in itself, as both molecules can change conformation, compete for binding sites, and regulate cellular functions. Recent studies of Xist long non-coding RNP, the U4/5/6 tri-small nuclear RNP complex, and an activated state of a spliceosome reveal new features of RNA interactions with proteins, and, although their stories are incomplete, they are already fascinating.
Keywords: Ribonucleoprotein Particles, RNPs, RNA-protein interaction, Xist, lncRNA, U4/5/6 tri-snRNP complex
Introduction
RNA molecules in the cell are rarely naked. Rather, proteins are bound to them in some arrangement consistent with their regulation, protection from nucleases, transport, or formation of ribonucleoprotein particles (RNPs). A 2014 compendium of RNA-binding proteins in humans 1 concluded that 7.5% of 20,500 known protein-coding genes are found in RNPs or bound to mRNAs, where they regulate RNA metabolism. This is likely to be an underestimate, since their structural heterogeneity makes them difficult to identify de novo.
The recent discovery of a plethora of non-coding RNAs 2 in cells has invigorated investigation of proteins that bind to RNA. New methods of probing the proteins in a transcriptome have allowed simultaneous identification of a protein and its RNA-binding site. Typically, these are crosslinking-immunoprecipitation (CLIP) experiments 3– 9. Intact cells can be irradiated with ultraviolet (UV) light or treated with formaldehyde to crosslink proteins to RNA, then the complexes are purified from the milieu by immunoprecipitation. To identify proteins bound to mRNAs, cellular UV RNA–protein crosslinking is followed by isolation of all poly(A)-RNA 7. Alternatively, proteins bound to a specific RNA could be recovered by annealing biotin-oligonucleotides complementary to the RNA and selective purification by streptavidin 9. Proteins bound to RNAs could then be identified by mass spectrometry. Several groups applied this method to identify mRNA-binding proteins in human cell lines, mouse embryonic stem cells (ESCs), and Saccharomyces cerevisiae yeast cells (reviewed in Gerstberger et al. 1).
Assuming that there are indeed more than 1,500 RNA-binding proteins in human cells, books will be written about them and their roles in RNA biology. Here, I focus on recent advances that reveal the variety and mystery of RNPs.
Xist, the RNA that inactivates an X chromosome
Xist is a long non-coding RNA (lncRNA) that is responsible for transcriptional silencing of one of two X chromosomes in female cells 10– 13. There are approximately 200 Xist molecules bound to a single X chromosome, and each 18 kb of Xist is bound by proteins ( Figure 1). Proteins could participate in any aspect of its biology: Xist has to associate with the X chromosome, then spread along it, and finally inhibit RNA polymerase II (Pol II) transcription. After more than twenty years of efforts to identify those proteins, the power of mass spectroscopy has been applied to proteins crosslinked in cellulo to Xist.
Figure 1. Xist wraps around nucleosomes in the X chromosome.
Approximately 200 Xist molecules bind to an X chromosome, spread along it, and inhibit RNA polymerase II from transcribing the DNA. Xist is bound by many proteins at unknown sites and with unknown stoichiometry, which subsequently interact with each other through disordered regions or structured domains. RNA is shown as a yellow/orange strand and protein linkers as blue strands. RRM, RNA recognition motif.
Two research groups have recently published compendia of Xist-bound proteins. Each group first crosslinked RNA to protein in cellulo, selected Xist through oligonucleotide-directed annealing, then used quantitative mass spectrometry to identify bound proteins. An overall comparison of their results shows great similarity but also some curious and intriguing differences. Table 1 and Table 2 list the most abundant proteins recovered from each study.
Table 1. Top 15 Xist-binding proteins from Chl-MS recovery in mouse cells 14.
| Crosslinked
proteins |
In order of
abundance |
Protein structural
motifs |
Length
*
(number of amino acids) |
|---|---|---|---|
| hnRNP M | 1 | 3 RRM | 728 |
| hnRNP U (Saf-A) | 2 | RGG, KH, acidic
region, DNA binding |
793 |
| hnRNP K | 3 | 3 KH, proline-rich | 463 |
| hnRNP A2/B1 | 4 | 2 RRM, RGG,
glycine-rich |
353 |
| MYEF2 | 5 | 2 non-canonical
RRMs, homology to hnRNP M4 |
591 |
| hnRNP A1 | 6 | 2 RRM, glycine-rich,
RGG |
320 |
| DDX5 | 7 | DEAD box protein | |
| Spen (SHARP) | 8 | 3 RRM, SPOC | 3,640 |
| RBM XL1 | 9 | RRM | |
| hnRNP AB | 10 | 2 RRM | |
| hnRNP D (AUF1) | 11 | 2 RRM | 355 |
| hnRNP L | 12 | 4 RRM, glycine-rich | 589 |
| hnRNP A3 | 13 | 2 RRM, glycine-rich | 379 |
| hnRNP C | 14 | 1 RRM, acid rich | 293 |
| TARDBP (TDP-43) | 15 | 2 RRM, glycine-rich,
DNA-binding protein |
414 |
*Many proteins have isoforms with varying lengths; the longest variant in Homo sapiens is listed.
Table 2. Mouse embryonic stem cells: top 10 Xist-binding proteins from RAP-MS 23.
| Crosslinked
proteins |
In order of
abundance |
Protein structural
motifs |
Length
(number of amino acids) |
|---|---|---|---|
| SHARP
(SPEN) |
1 | 3 RRM, SPOC | 3,640 |
| RBM15 | 2 | 3 RRM, SPOC | 969 |
| MYEF2
(hnRNP M) |
3 | 3 RRMs, homology to
hnRNP M4 |
591 |
| CELF1 | 4 | 3 RRMs | 486 |
| hnRNP C | 5 | 1 RRM | 313 |
| LBR | 6 | Chromatin-interaction
domain, transmembrane region, lamin-interacting domain |
626 |
| SAF-A
(hnRNP U) |
7 | RGG, SPRY domain,
ATPase domain |
793 |
| RALY
(hnRNP C) |
8 | 1 RRM | 312 |
| hnRNP M | 9 | 3 RRM | 729 |
| PTBP1
(hnRNP I) |
10 | 4 RRM | 555 |
hnRNP, heterogeneous nuclear ribonucleoprotein particle; RAP-MS, RNA antisense purification-mass spectrometry; RRM, RNA recognition motif; SILAC, stable isotope labeling by amino acids in culture; SPOC, Spen paralog and ortholog C-terminal domain.
The groups of Heard and Chang 14 identified 81 proteins in toto bound to Xist. Using formaldehyde, they crosslinked proteins to Xist in three different mouse cell types: a male ESC line containing an inducible Xist gene, an epiblast stem cell line, and trophoblast stem cells. Each cell type represents one stage of Xist expression. Combining all datasets, three proteins were identified as being most abundant: heterogeneous nuclear RNP (hnRNP) K, hnRNP U, and hnRNP M. In addition, a detailed examination of Xist 5′ 0.9 kb sequence revealed several localized proteins. In particular, SPEN (aka SHARP) was found to be necessary for transcriptional silencing.
There is a preponderance of hnRNP proteins. These heterogeneous nuclear ribonucleoproteins are abundant in metazoan cells, where they are mostly found in the nucleus 15, 16. A recent review of them traced their ancestry 17, concluding that there are 13 families, each with isoforms or variants. For example, hnRNP A has four homologues in humans (A0, A1, A2, and A3), while hnRNP M has two (MYEF2 and hnRNP M). These proteins typically use RNA recognition motifs (RRMs) to bind RNA, while their other domains engage in protein–protein interactions. Several are involved in pre-mRNA splicing, where they repress splice site selection (hnRNP A 18) or regulate exon inclusion (hnRNP I 19). hnRNP I (aka polypyrimidine tract binding protein 1 [PTB1]) also facilitates translation from internal ribosome entry sites (IRES) 20, 21. hnRNP functions in Xist are unknown, with the exception of hnRNP U (aka Saf-A), which facilitates Xist localization on chromatin 22.
In contrast, a group of investigators headed by Guttman 23 took a different approach to finding Xist proteins during transcriptional silencing. After Xist induction in mouse ESCs, cells were UV-crosslinked, Xist RNP was recovered with long antisense oligonucleotides, and Xist proteins were identified by mass spectrometry. Two batches of mouse ESCs were cultured, one in 15N- and one in 14N-media to allow quantification by mass spectrometry (SILAC). Among their ten most abundant proteins, they found SHARP (SPEN) and RMD15, two proteins related in their architecture (they are SPEN family proteins). They also recovered six hnRNP proteins ( Table 2).These are exciting findings. In a curious coincidence, SHARP has another life in a nuclear RNP with the steroid receptor RNA activator (SRA) 24. SRA is a lncRNA that co-regulates the transcription of nuclear receptors 24. Bound to SRA, SHARP represses SRA transactivation when it recruits histone deacetylate 25. Does it carry out a similar task in Xist 2, 10?
In fact, McHugh et al. found that SHARP was required for the inhibition of Pol II transcription at sites where Xist was bound 23. The mechanism of inhibition could lie in the recruitment of SMRT and/or HDAC3 25. HDAC3 is a histone deacetylase 26 that is thought to be responsible for transcriptional repression by changing chromatin structure 27. Loss of SHARP, LBR, or hnRNP U in knockdown experiments was sufficient to eliminate silencing 23, but each protein appears to have unique contributions. The role of the other seven proteins was not tested directly, but since each binds directly to Xist, they could have functions in localization, recruitment of other enzymes, stabilization, etc. (for example, binding to Polycomb repressive complex 2 [PRC2]).
The identification of LBR bound to Xist explains localization of the Xist-X chromosome to the nuclear lamina 12. Transmembrane helices anchor LBR to the lamina, while its tail contacts Xist. Positioning of Xist-X on the lamina changes the structure of the DNA and facilitates protein-mediated spreading of the Xist molecules along the length of the chromatin.
Rather than discovering unknown proteins, these investigations have re-discovered known proteins. They present a new challenge: to understand why they are particularly useful in the Xist context and how their use, and corresponding abundance, is modulated according to developmental stage or cell lineage. The general challenge is not only to understand how proteins use their RNA-binding domains and intervening sequences and disordered tails to control formation of RNPs but must also account for their temporal exchange.
RNA recognition motifs
A striking feature of proteins bound to Xist is the recurring use of tandem RRM domains. There are certainly advantages to this scheme, since affinity and specificity can be modulated by increasing the number of contacts between RNA and protein. However, neither Xist-binding sites for its associated proteins nor their binding stoichiometry are known. These biochemical characterizations are important to understand how they select their target sites on the RNA, how they bind to Xist in the milieu of other RNAs in the cell, and how they hang onto the RNA while they also bind to other cellular compartments or recruit other proteins.
RRMs 1 are the most common structural motif used in eukaryotes to bind RNA ( Figure 2) and are estimated to be found in 225 human genes. When RRMs are present in multiples, deciphering the contributions of each RRM to the whole can be quite difficult 28– 31. A recent biophysical study of two tandem RRMs revealed how they partition function.
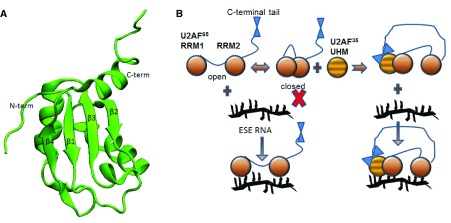
Figure 2. Regulation of RNA recognition motif (RRM) binding to RNA.
A. An RRM has a four-stranded anti-parallel β-sheet, with two α-helices on one side 92– 95. RNA often sits on the surface of the β-sheet. B. The two RRMs of U2 auxiliary factor (U2AF) exhibit closed/open transitions at equilibrium, but only in the open state can RNA bind. Binding of the U2AF homology motif (UHM) from U2AF35 to RRM1 shifts the equilibrium to favor the open state, which facilitates RNA binding. The C-terminal tail of U2AF65 contacts the U2AF35 UHM. ESE, exonic splicing enhancer.
U2 auxiliary factory (U2AF) is a heterodimer of U2AF65 and U2AF35 32, 33, which in pre-mRNA splicing aids in the recognition of a 3′ splice site 34– 38. U2AF65 has two RRMs (RRM1 and RRM2) that bind polypyrimidine tracts, but U2AF35 has a single UHM, a “U2AF homology motif”, that is structurally homologous to an RRM 39, 40. RRM1 and RRM2 are tethered by a short linker (~20 amino acids) that allows them to undergo relative motion and orientation 36. Since they bind to polypyrimidine tracts of variable length and sequence, they must be able to expand or contract to span the site 41.
The Sattler and Lamb laboratories collaborated on a comprehensive study of the spatiotemporal disposition of U2AF65 RRM1 and RRM2 and their role in RNA binding. von Voithenberg et al. 35 showed that RRM1 and RRM2 undergo dynamic exchange between a closed or open orientation at equilibrium ( Figure 2). In the closed state, RRM1 and RRM2 do not bind RNA, but when the conformation is open, a polypyrimidine tract can bind. If binding is weak (i.e. the polypyrimidine tract is too short or contains multiple purine nucleotides), the exchange between open and closed states is relatively unperturbed. If RNA binding is tight, RRM1 and RRM2 will be trapped in an open state. Thus, the RNA shifts the equilibrium of U2AF RRM1 and RRM2 between open and closed states in an example of conformational selection.
These experiments were conducted using single pair Förster resonance energy transfer (spFRET) that observed single molecules, each containing a donor and acceptor fluorophore. One fluorophore was attached to either RRM, such that the open and closed orientations were distinguished by the FRET efficiency. Combining measurements of fluorophore lifetimes with spFRET facilitated temporal characterization of exchange between open and closed states. In experimental conditions, free RRM1 and RRM2 occupied an open state ~67% of the time. Addition of RNA trapped RRM1 and RRM2 in the open conformation 90% of the time.
U2AF65 and U2AF35 have been the subject of many biochemical and structural investigations, since they are essential proteins for pre-mRNA splicing. In particular, experimental studies of protein–protein interactions between U2AF and other proteins have identified sites where interactions occur 42– 45. These latest experiments revealed a mechanism of protein–protein interaction involving the UHM of U2AF35 and U2AF65 RRM1 and RRM2. A combination of nuclear magnetic resonance (NMR) structure and dynamics experiments identified the binding site of U2AF35 UHM to be a surface of U2AF65 RRM1. Binding of the UHM to RRM1 shifts the RRM1 and RRM2 conformational equilibrium to the open state, thereby favoring RNA binding. The authors suggest that allostery drives the RRM1 and RRM2 conformational switch. Allosteric modulation of binding is a powerful mechanism to provide discrimination and affinity 46– 49, but, by its nature, it is almost impossible to anticipate and cannot be gleaned from static structures.
Many RNA-binding proteins are modular, with an RNA-binding domain, intervening sequences, and disordered tails. Here, U2AF uses two proteins to regulate splicing; other examples include the Sxl-Unr heterodimer that regulates translation via interactions between Sxl RRM and a Unr cold-shock domain 50, while the SR protein (serine-arginine) SRSF1 is regulated by phosphorylation of its RS tail that blocks intramolecular interaction with its RRMs 51, 52. Regulation by intermolecular and intramolecular interactions adds another level of complexity to RNA-binding proteins.
The spliceosome and its small nuclear ribonucleoprotein particles
It is estimated that 94% of all human genes contain introns 53– 55, thereby providing protein isoform diversity. The process of removing introns and joining exons is carried out by the spliceosome, a multi-component and dynamic assembly of RNPs 56. A great challenge in the field of pre-mRNA splicing has been to understand how the spliceosome is physically able to carry out the concerted transesterification reactions of the splicing chemistry to yield mRNAs.
The spliceosome consists of five small nuclear RNPs (snRNPs) that dynamically associate with each other and with pre-mRNA. The major spliceosome uses U1, U2, U4, U5, and U6 snRNPs in the process of splicing 57. Each snRNP contains a single RNA (snRNA) and multiple proteins, but while U1 and U2 snRNPs are independent, U4 and U6 form a di-snRNP that goes on to become a U4/U5/U6 tri-snRNP 58. The tri-snRNP is recruited to a bona-fide intron and is then remodeled, losing U4 snRNP and leaving U5 and U6 snRNPs to form the active spliceosome.
The goal of snRNP rearrangement is to allow and facilitate snRNA conformational rearrangements in the spliceosome to produce the active site for catalysis 59– 61. Rearrangements of pre-mRNA and snRNAs to prepare and position them for catalysis are mainly accomplished by protein helicases 62. There are eight such type SF2 helicases that associate with the spliceosome along the reaction pathway 63, 64. These ATP-dependent RNA helicases are not sequence specific; they can unwind any RNA duplex. Rather, their specific targets appear to be defined by where and when they associate with the spliceosome. The Brr2 helicase is particularly critical in the transformation of pre-spliceosome intermediates 64– 67. Brr2 is unusual: it has two helicase domains (only one is active) and a long (450-amino-acid) N-terminal domain 64, 65, 68, 69.
Brr2, a unique RNA helicase
Brr2 enters the nucleus independently and associates with the U5 snRNP. U5 snRNP then joins the U4/U6 di-snRNP to become the U4/U5/U6 tri-snRNP 68. The tri-snRNP is recruited by U1 and U2 snRNPs to form a pre-spliceosome.
To form the active spliceosome, two snRNPs must be displaced. U1 snRNP is released from the 5′ splice site, and U4 snRNP is removed from the tri-snRNP. It is the latter remodeling that requires Brr2, as U4 and U6 snRNAs are joined by 22 perfect base pairs and Brr2 is the helicase that separates them. Only when U6 snRNA is free of U4 snRNA can it rearrange to base pair with U2 snRNA and pre-mRNA and so form the catalytic center of the spliceosome. Clearly, Brr2 activity must be regulated such that it is inactive in the tri-snRNP but active in the pre-spliceosome. How is it regulated?
Several recent studies have delved into the details of Brr2 regulation. In a series of papers from the Wahl lab 70– 74, Brr2 structure and function were addressed by crystallography and biochemistry. The goal of Brr2 in the tri-snRNP is to maintain stasis. As biochemistry experiments of Brr2 show 64, there is a plug domain at the N-terminus of Brr2’s long N-terminal region (NTR). This plug folds back over the entrance of the helicase to block access of the U4/U6 snRNA duplex to the active site of Brr2. This is a unique intramolecular regulatory device, and more experiments are required to understand how it is directed to this position (and how it is displaced).
The tri-snRNP is an intermediate in the pathway to spliceosome formation. Years of enormous efforts to map intermediates 42, 63, 75– 77 have now been coupled with technological advances in cryo-electron microscopy (cryo-EM) to visualize select transitional complexes 70– 72, 78, 79. Those efforts have produced a cryo-EM structure of human tri-snRNP that captures Brr2 in its plugged conformation 72 (PDB ID 3jcr). This state of the tri-snRNP, illustrated in Figure 3, might represent its structure as an autonomous particle before it joins the pre-spliceosome, where U4 and U6 snRNAs are still base-paired to each other. If so, then proteins and RNAs in the tri-snRNP must rearrange to present U4 and/or U6 tails to the helicase active site.
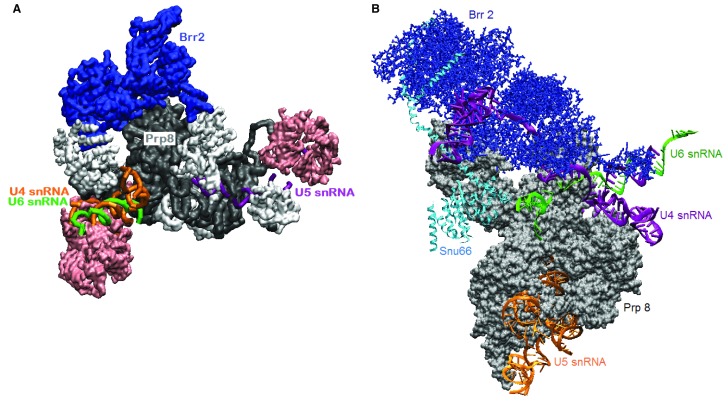
Figure 3. Two tri-small nuclear ribonucleoprotein particle (snRNP) structures trap different states of Brr2.
A. Human tri-snRNP cryo-electron microscopy (cryo-EM) at 7 Å resolution 72 shows Brr2 sitting on Prp8 (PDB ID 3jcr). A U4/U6 snRNA duplex is visible. Sm and Lsm rings are pink; other proteins are white. B. In a yeast tri-snRNP complex 70, (PDB ID 5GAN), U4 snRNA is threaded through Brr2 in the RNA-binding tunnel. These structures might correspond to the tri-snRNP in the nucleus ( A) and the tri-snRNP poised for activation by Brr2 as it joins the pre-spliceosome ( B). Visualized with visual molecular dynamics (VMD).
In the tri-snRNP, Brr2 sits on the Jab1 domain of Prp8, but its orientation and contacts change during activation of the particle. In contrast to the structure of the human tri-snRNP, in a structure of yeast tri-snRNP, a single-stranded region of U4 snRNA occupies the RNA-binding tunnel of Brr2 73, 80, 81 (illustrated in Figure 3). Is Brr2 now poised to completely separate U4 snRNA from U6 snRNA? Does this separation occur before the tri-snRNP is recruited to the pre-spliceosome, or is this a paused state that requires further activation?
There is another competitive inhibitor of Brr2. Prp8’s Jab1 domain has a C-terminal disordered tail that sneaks into the RNA tunnel of Brr2 to compete with U4 82. The intramolecular plug interaction and Prp8 Jab1 cooperate to inhibit unwinding. Removing the Jab1 tail activates Brr2 helicase activity; Brr2 without its intramolecular plug also has enhanced activity 75. Do both inhibitors operate in the isolated tri-snRNP?
Brr2 remains in the spliceosome after U4 snRNP has been expelled from the spliceosome. It is seen in a structure of yeast-activated spliceosome, which is defined by the loss of U1 and U4 snRNP and rearrangements of the remaining snRNAs to interact with each other and pre-mRNA. A cryo-EM structure of activated yeast spliceosomes (B act) shows Brr2 perched on Prp8’s Jab1 domain 79, with its helicase activity blocked by both inhibitor interactions (PDB ID 5lqw). In an illustration from this structure, U2, U5, and U6 snRNAs are remote from Brr2 ( Figure 4). Although not clear from the perspective of Figure 4, Prp8 is entwined with other proteins and the snRNAs in this complex, even as it binds Brr2.
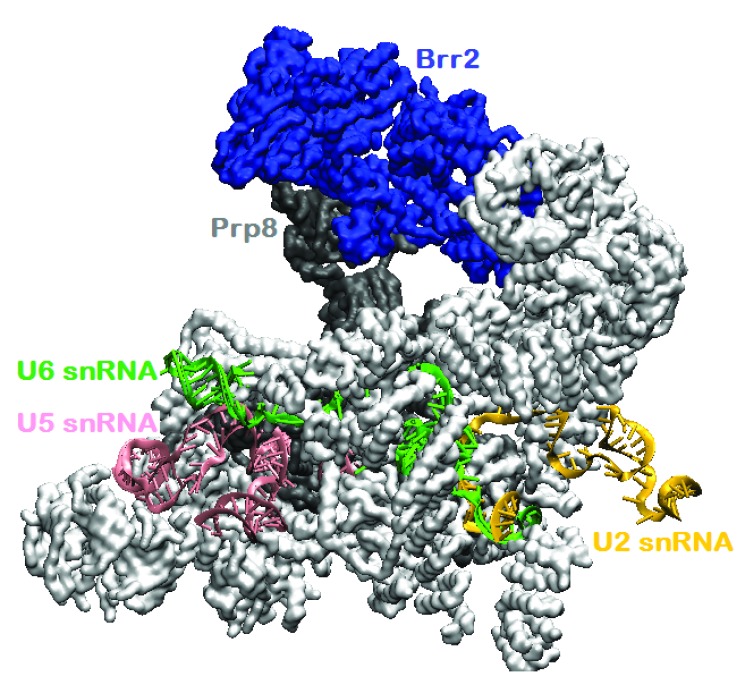
Figure 4. Yeast-activated (B act) spliceosome 79 (PDB ID 5LQW; cryo-electron microscopy [cryo-EM] 5.8Å).
Brr2 has separated U4 and U6 small nuclear RNAs (snRNAs), and U4 small nuclear ribonucleoprotein particle (snRNP) has been expelled from the spliceosome. Brr2 is bound to the Jab1 domain of Prp8. All 27 proteins are shown in surface representation; most are colored white. Visualized with visual molecular dynamics (VMD).
As the spliceosome progresses through its cycle, there are many short RNA duplexes that need to be unwound. The other seven SF2 RNA helicases are recruited to the spliceosome when they are needed, and then they dissociate. Brr2 remains with the spliceosome until it has completed a splicing cycle, but there are no data suggesting that it is active at any time other than in the conversion from pre-spliceosome to B act. If it is not required for its helicase activity, perhaps its long NTR contributes something to splicing. Brr2 is reported to contribute to catalysis 74, 83, to stabilize U5 and U6 in the spliceosome 68, and to assist in the final disruption of the spliceosome and release of ligated exons 84. If these states of the spliceosome could be trapped for structural studies, Brr2 might be captured in action.
The spliceosome is composed of hundreds of proteins 56, many of which simply bind RNA, but others actively remodel it. In the past year, spliceosome structures have revealed connections between RNA and proteins that explain previous observations but also raise new questions. This year, structures of the spliceosome C/C* complex show another helicase, prp16, at work on remodelling 85– 87. Slowly, this RNA enzyme is giving up its secrets.
Conclusions
There is a need to not only understand specific RNPs but also define general rules of engagement, since RNA–protein interactions dominate RNA biology. Indeed, the most mysterious are the membrane-less organelles that contain RNAs and proteins 88, 89. These conglomerates of RNAs bound by RNA-binding proteins are variously thought to be centers of RNA processing, degradation, transcription, and exchange: P bodies and stress granules in the cytoplasm and nucleoli, Cajal bodies, speckles, and PML bodies in the nucleus. A current model is that disordered domains of the proteins form a fluid matrix that allows a flux of molecules through these liquid droplets 90, 91. It is a sure bet that these droplets will be objects of intense scrutiny for years to come.
Acknowledgements
I thank my reviewers for their thoughtful reading and perceptive comments.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Shinichi Nakagawa, RNA Biology Laboratory, Faculty of Pharmaceutical Sciences, Hokkaido University, Sapporo, Japan
Markus Wahl, Department of Biology, Chemistry, Pharmacy, Freie Universität Berlin, Berlin, Germany
Xiang-Dong Fu, Department of Cellular & Molecular Medicine, University of California San Diego, San Diego, CA, USA
Funding Statement
The author(s) declared that no grants were involved in supporting this work.
[version 1; referees: 3 approved]
References
- 1. Gerstberger S, Hafner M, Tuschl T: A census of human RNA-binding proteins. Nat Rev Genet. 2014;15(12):829–45. 10.1038/nrg3813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Melé M, Mattioli K, Mallard W, et al. : Chromatin environment, transcriptional regulation, and splicing distinguish lincRNAs and mRNAs. Genome Res. 2017;27(1):27–37. 10.1101/gr.214205.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jensen KB, Darnell RB: CLIP: crosslinking and immunoprecipitation of in vivo RNA targets of RNA-binding proteins. Methods Mol Biol. 2008;488:85–98. 10.1007/978-1-60327-475-3_6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ascano M, Hafner M, Cekan P, et al. : Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip Rev RNA. 2012;3(2):159–77. 10.1002/wrna.1103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Scheibe M, Butter F, Hafner M, et al. : Quantitative mass spectrometry and PAR-CLIP to identify RNA-protein interactions. Nucleic Acids Res. 2012;40(19):9897–902. 10.1093/nar/gks746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Garzia A, Meyer C, Morozov P, et al. : Optimization of PAR-CLIP for transcriptome-wide identification of binding sites of RNA-binding proteins. Methods. 2016. pii: S1046-2023(16)30384-X. 10.1016/j.ymeth.2016.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Munschauer M, Schueler M, Dieterich C, et al. : High-resolution profiling of protein occupancy on polyadenylated RNA transcripts. Methods. 2014;65(3):302–9. 10.1016/j.ymeth.2013.09.017 [DOI] [PubMed] [Google Scholar]
- 8. Huppertz I, Attig J, D'Ambrogio A, et al. : iCLIP: protein-RNA interactions at nucleotide resolution. Methods. 2014;65(3):274–87. 10.1016/j.ymeth.2013.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. McHugh CA, Russell P, Guttman M: Methods for comprehensive experimental identification of RNA-protein interactions. Genome Biol. 2014;15(1):203. 10.1186/gb4152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Minajigi A, Froberg JE, Wei C, et al. : Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349(6245): pii: aab2276. 10.1126/science.aab2276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Galupa R, Heard E: X-chromosome inactivation: new insights into cis and trans regulation. Curr Opin Genet Dev. 2015;31:57–66. 10.1016/j.gde.2015.04.002 [DOI] [PubMed] [Google Scholar]
- 12. Chen CK, Blanco M, Jackson C, et al. : Xist recruits the X chromosome to the nuclear lamina to enable chromosome-wide silencing. Science. 2016;354(6311):468–72. 10.1126/science.aae0047 [DOI] [PubMed] [Google Scholar]
- 13. Engreitz JM, Pandya-Jones A, McDonel P, et al. : The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science. 2013;341(6147):1237973. 10.1126/science.1237973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chu C, Zhang QC, da Rocha ST, et al. : Systematic discovery of Xist RNA binding proteins. Cell. 2015;161(2):404–16. 10.1016/j.cell.2015.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Görlach M, Burd CG, Portman DS, et al. : The hnRNP proteins. Mol Biol Rep. 1993;18(2):73–8. 10.1007/BF00986759 [DOI] [PubMed] [Google Scholar]
- 16. Geuens T, Bouhy D, Timmerman V: The hnRNP family: insights into their role in health and disease. Hum Genet. 2016;135(8):851–67. 10.1007/s00439-016-1683-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Busch A, Hertel KJ: Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA. 2012;3(1):1–12. 10.1002/wrna.100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jean-Philippe J, Paz S, Caputi M: hnRNP A1: the Swiss army knife of gene expression. Int J Mol Sci. 2013;14(9):18999–9024. 10.3390/ijms140918999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keppetipola N, Sharma S, Li Q, et al. : Neuronal regulation of pre-mRNA splicing by polypyrimidine tract binding proteins, PTBP1 and PTBP2. Crit Rev Biochem Mol Biol. 2012;47(4):360–78. 10.3109/10409238.2012.691456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kafasla P, Morgner N, Robinson CV, et al. : Polypyrimidine tract-binding protein stimulates the poliovirus IRES by modulating eIF4G binding. EMBO J. 2010;29(21):3710–22. 10.1038/emboj.2010.231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sawicka K, Bushell M, Spriggs KA, et al. : Polypyrimidine-tract-binding protein: a multifunctional RNA-binding protein. Biochem Soc Trans. 2008;36(Pt 4):641–7. 10.1042/BST0360641 [DOI] [PubMed] [Google Scholar]
- 22. Sakaguchi T, Hasegawa Y, Brockdorff N, et al. : Control of Chromosomal Localization of Xist by hnRNP U Family Molecules. Dev Cell. 2016;39(1):11–2. 10.1016/j.devcel.2016.09.022 [DOI] [PubMed] [Google Scholar]
- 23. McHugh CA, Chen CK, Chow A, et al. : The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015;521(7551):232–6. 10.1038/nature14443 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 24. Colley SM, Leedman PJ: SRA and its binding partners: an expanding role for RNA-binding coregulators in nuclear receptor-mediated gene regulation. Crit Rev Biochem Mol Biol. 2009;44(1):25–33. 10.1080/10409230802661719 [DOI] [PubMed] [Google Scholar]
- 25. Shi Y, Downes M, Xie W, et al. : Sharp, an inducible cofactor that integrates nuclear receptor repression and activation. Genes Dev. 2001;15(9):1140–51. 10.1101/gad.871201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gregoretti IV, Lee YM, Goodson HV: Molecular evolution of the histone deacetylase family: functional implications of phylogenetic analysis. J Mol Biol. 2004;338(1):17–31. 10.1016/j.jmb.2004.02.006 [DOI] [PubMed] [Google Scholar]
- 27. Tessarz P, Kouzarides T: Histone core modifications regulating nucleosome structure and dynamics. Nat Rev Mol Cell Biol. 2014;15(11):703–8. 10.1038/nrm3890 [DOI] [PubMed] [Google Scholar]
- 28. Mackereth CD, Sattler M: Dynamics in multi-domain protein recognition of RNA. Curr Opin Struct Biol. 2012;22(3):287–96. 10.1016/j.sbi.2012.03.013 [DOI] [PubMed] [Google Scholar]
- 29. Safaee N, Kozlov G, Noronha AM, et al. : Interdomain allostery promotes assembly of the poly(A) mRNA complex with PABP and eIF4G. Mol Cell. 2012;48(3):375–86. 10.1016/j.molcel.2012.09.001 [DOI] [PubMed] [Google Scholar]
- 30. Maynard CM, Hall KB: Interactions between PTB RRMs induce slow motions and increase RNA binding affinity. J Mol Biol. 2010;397(1):260–77. 10.1016/j.jmb.2009.12.051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clerte C, Hall KB: The domains of polypyrimidine tract binding protein have distinct RNA structural preferences. Biochemistry. 2009;48(10):2063–74. 10.1021/bi8016872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ruskin B, Zamore PD, Green MR: A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52(2):207–19. 10.1016/0092-8674(88)90509-0 [DOI] [PubMed] [Google Scholar]
- 33. Zamore PD, Green MR: Identification, purification, and biochemical characterization of U2 small nuclear ribonucleoprotein auxiliary factor. Proc Natl Acad Sci U S A. 1989;86(23):9243–7. 10.1073/pnas.86.23.9243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shao C, Yang B, Wu T, et al. : Mechanisms for U2AF to define 3' splice sites and regulate alternative splicing in the human genome. Nat Struct Mol Biol. 2014;21(11):997–1005. 10.1038/nsmb.2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Voith von Voithenberg L, Sánchez-Rico C, Kang HS, et al. : Recognition of the 3' splice site RNA by the U2AF heterodimer involves a dynamic population shift. Proc Natl Acad Sci U S A. 2016;113(46):E7169–E7175. 10.1073/pnas.1605873113 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 36. Agrawal AA, Salsi E, Chatrikhi R, et al. : An extended U2AF(65)-RNA-binding domain recognizes the 3' splice site signal. Nat Commun. 2016;7:10950. 10.1038/ncomms10950 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 37. Zorio DA, Blumenthal T: Both subunits of U2AF recognize the 3' splice site in Caenorhabditis elegans. Nature. 1999;402(6763):835–8. 10.1038/45597 [DOI] [PubMed] [Google Scholar]
- 38. Wu S, Romfo CM, Nilsen TW, et al. : Functional recognition of the 3' splice site AG by the splicing factor U2AF35. Nature. 1999;402(6763):832–5. 10.1038/45590 [DOI] [PubMed] [Google Scholar]
- 39. Loerch S, Kielkopf CL: Unmasking the U2AF homology motif family: a bona fide protein-protein interaction motif in disguise. RNA. 2016;22(12):1795–807. 10.1261/rna.057950.116 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 40. Kielkopf CL, Lucke S, Green MR: U2AF homology motifs: protein recognition in the RRM world. Genes Dev. 2004;18(13):1513–26. 10.1101/gad.1206204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Jenkins JL, Laird KM, Kielkopf CL: A Broad range of conformations contribute to the solution ensemble of the essential splicing factor U2AF(65). Biochemistry. 2012;51(26):5223–5. 10.1021/bi300277t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Chen L, Weinmeister R, Kralovicova J, et al. : Stoichiometries of U2AF35, U2AF65 and U2 snRNP reveal new early spliceosome assembly pathways. Nucleic Acids Res. 2017;45(4):2051–2067. 10.1093/nar/gkw860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Selenko P, Gregorovic G, Sprangers R, et al. : Structural basis for the molecular recognition between human splicing factors U2AF65 and SF1/mBBP. Mol Cell. 2003;11(4):965–76. 10.1016/S1097-2765(03)00115-1 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 44. Zhang Y, Madl T, Bagdiul I, et al. : Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition. Nucleic Acids Res. 2013;41(2):1343–54. 10.1093/nar/gks1097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Corsini L, Hothorn M, Stier G, et al. : Dimerization and protein binding specificity of the U2AF homology motif of the splicing factor Puf60. J Biol Chem. 2009;284(1):630–9. 10.1074/jbc.M805395200 [DOI] [PubMed] [Google Scholar]
- 46. Koshland DE, Jr, Nemethy G, Filmer D: Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5(1):365–85. 10.1021/bi00865a047 [DOI] [PubMed] [Google Scholar]
- 47. Cooper A, Dryden DT: Allostery without conformational change. A plausible model. Eur Biophys J. 1984;11(2):103–9. 10.1007/BF00276625 [DOI] [PubMed] [Google Scholar]
- 48. Motlagh HN, Wrabl JO, Li J, et al. : The ensemble nature of allostery. Nature. 2014;508(7496):331–9. 10.1038/nature13001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Williams SG, Hall KB: Linkage and allostery in snRNP protein/RNA complexes. Biochemistry. 2014;53(22):3529–39. 10.1021/bi500192a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hennig J, Militti C, Popowicz GM, et al. : Structural basis for the assembly of the Sxl-Unr translation regulatory complex. Nature. 2014;515(7526):287–90. 10.1038/nature13693 [DOI] [PubMed] [Google Scholar]
- 51. Serrano P, Aubol BE, Keshwani MM, et al. : Directional Phosphorylation and Nuclear Transport of the Splicing Factor SRSF1 Is Regulated by an RNA Recognition Motif. J Mol Biol. 2016;428(11):2430–45. 10.1016/j.jmb.2016.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 52. Cléry A, Sinha R, Anczuków O, et al. : Isolated pseudo-RNA-recognition motifs of SR proteins can regulate splicing using a noncanonical mode of RNA recognition. Proc Natl Acad Sci U S A. 2013;110(30):E2802–11. 10.1073/pnas.1303445110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pan Q, Shai O, Lee LJ, et al. : Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–5. 10.1038/ng.259 [DOI] [PubMed] [Google Scholar]
- 54. Wang ET, Sandberg R, Luo S, et al. : Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–6. 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Nilsen TW, Graveley BR: Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463(7280):457–63. 10.1038/nature08909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Will CL, Lührmann R: Spliceosome structure and function. Cold Spring Harb Perspect Biol. 2011;3(7): pii: a003707. 10.1101/cshperspect.a003707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wahl MC, Lührmann R: SnapShot: Spliceosome Dynamics III. Cell. 2015;162(3):690–690.e1. 10.1016/j.cell.2015.07.033 [DOI] [PubMed] [Google Scholar]
- 58. Wahl MC, Lührmann R: SnapShot: Spliceosome Dynamics I. Cell. 2015;161(6):1474–e1. 10.1016/j.cell.2015.05.050 [DOI] [PubMed] [Google Scholar]
- 59. Nguyen TH, Galej WP, Fica SM, et al. : CryoEM structures of two spliceosomal complexes: starter and dessert at the spliceosome feast. Curr Opin Struct Biol. 2016;36:48–57. 10.1016/j.sbi.2015.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wahl MC, Will CL, Lührmann R: The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–18. 10.1016/j.cell.2009.02.009 [DOI] [PubMed] [Google Scholar]
- 61. Raghunathan PL, Guthrie C: RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol. 1998;8(15):847–55. 10.1016/S0960-9822(07)00345-4 [DOI] [PubMed] [Google Scholar]
- 62. De I, Schmitzova J, Pena V: The organization and contribution of helicases to RNA splicing. Wiley Interdiscip Rev RNA. 2016;7(2):259–74. 10.1002/wrna.1331 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 63. Semlow DR, Blanco MR, Walter NG, et al. : Spliceosomal DEAH-Box ATPases Remodel Pre-mRNA to Activate Alternative Splice Sites. Cell. 2016;164(5):985–98. 10.1016/j.cell.2016.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Absmeier E, Santos KF, Wahl MC: Functions and regulation of the Brr2 RNA helicase during splicing. Cell Cycle. 2016;15(24):3362–77. 10.1080/15384101.2016.1249549 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 65. Absmeier E, Wollenhaupt J, Mozaffari-Jovin S, et al. : The large N-terminal region of the Brr2 RNA helicase guides productive spliceosome activation. Genes Dev. 2015;29(24):2576–87. 10.1101/gad.271528.115 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 66. Zhang L, Li X, Hill RC, et al. : Brr2 plays a role in spliceosomal activation in addition to U4/U6 unwinding. Nucleic Acids Res. 2015;43(6):3286–97. 10.1093/nar/gkv062 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 67. Theuser M, Hobartner C, Wahl MC, et al. : Substrate-assisted mechanism of RNP disruption by the spliceosomal Brr2 RNA helicase. Proc Natl Acad Sci U S A. 2016;113(28):7798–803. 10.1073/pnas.1524616113 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 68. Nguyen TH, Li J, Galej WP, et al. : Structural basis of Brr2-Prp8 interactions and implications for U5 snRNP biogenesis and the spliceosome active site. Structure. 2013;21(6):910–9. 10.1016/j.str.2013.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Santos KF, Jovin SM, Weber G, et al. : Structural basis for functional cooperation between tandem helicase cassettes in Brr2-mediated remodeling of the spliceosome. Proc Natl Acad Sci U S A. 2012;109(43):17418–23. 10.1073/pnas.1208098109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Nguyen TH, Galej WP, Bai XC, et al. : Cryo-EM structure of the yeast U4/U6.U5 tri-snRNP at 3.7 Å resolution. Nature. 2016;530(7590):298–302. 10.1038/nature16940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Nguyen TH, Galej WP, Bai XC, et al. : The architecture of the spliceosomal U4/U6.U5 tri-snRNP. Nature. 2015;523(7558):47–52. 10.1038/nature14548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Agafonov DE, Kastner B, Dybkov O, et al. : Molecular architecture of the human U4/U6.U5 tri-snRNP. Science. 2016;351(6280):1416–20. 10.1126/science.aad2085 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 73. Cornilescu G, Didychuk AL, Rodgers ML, et al. : Structural Analysis of Multi-Helical RNAs by NMR-SAXS/WAXS: Application to the U4/U6 di-snRNA. J Mol Biol. 2016;428(5 Pt A):777–89. 10.1016/j.jmb.2015.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 74. Hahn D, Kudla G, Tollervey D, et al. : Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev. 2012;26(21):2408–21. 10.1101/gad.199307.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Absmeier E, Becke C, Wollenhaupt J, et al. : Interplay of cis- and trans-regulatory mechanisms in the spliceosomal RNA helicase Brr2. Cell Cycle. 2017;16(1):100–12. 10.1080/15384101.2016.1255384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Weber G, Trowitzsch S, Kastner B, et al. : Functional organization of the Sm core in the crystal structure of human U1 snRNP. EMBO J. 2010;29(24):4172–84. 10.1038/emboj.2010.295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Pomeranz Krummel DA, Oubridge C, Leung AK, et al. : Crystal structure of human spliceosomal U1 snRNP at 5.5 Å resolution. Nature. 2009;458(7237):475–80. 10.1038/nature07851 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 78. Galej WP, Wilkinson ME, Fica SM, et al. : Cryo-EM structure of the spliceosome immediately after branching. Nature. 2016;537(7619):197–201. 10.1038/nature19316 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 79. Rauhut R, Fabrizio P, Dybkov O, et al. : Molecular architecture of the Saccharomyces cerevisiae activated spliceosome. Science. 2016;353(6306):1399–405. 10.1126/science.aag1906 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 80. Mozaffari-Jovin S, Santos KF, Hsiao HH, et al. : The Prp8 RNase H-like domain inhibits Brr2-mediated U4/U6 snRNA unwinding by blocking Brr2 loading onto the U4 snRNA. Genes Dev. 2012;26(21):2422–34. 10.1101/gad.200949.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Boesler C, Rigo N, Agafonov DE, et al. : Stable tri-snRNP integration is accompanied by a major structural rearrangement of the spliceosome that is dependent on Prp8 interaction with the 5' splice site. RNA. 2015;21(11):1993–2005. 10.1261/rna.053991.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Mozaffari-Jovin S, Wandersleben T, Santos KF, et al. : Inhibition of RNA helicase Brr2 by the C-terminal tail of the spliceosomal protein Prp8. Science. 2013;341(6141):80–4. 10.1126/science.1237515 [DOI] [PubMed] [Google Scholar]
- 83. Cordin O, Hahn D, Alexander R, et al. : Brr2p carboxy-terminal Sec63 domain modulates Prp16 splicing RNA helicase. Nucleic Acids Res. 2014;42(22):13897–910. 10.1093/nar/gku1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Small EC, Leggett SR, Winans AA, et al. : The EF-G-like GTPase Snu114p regulates spliceosome dynamics mediated by Brr2p, a DExD/H box ATPase. Mol Cell. 2006;23(3):389–99. 10.1016/j.molcel.2006.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yan C, Wan R, Bai R, et al. : Structure of a yeast step II catalytically activated spliceosome. Science. 2017;355(6321):149–155. 10.1126/science.aak9979 [DOI] [PubMed] [Google Scholar]
- 86. Fica SM, Oubridge C, Galej WP, et al. : Structure of a spliceosome remodelled for exon ligation. Nature. 2017;542(7641):377–380. 10.1038/nature21078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bertram K, Agafonov DE, Liu WT, et al. : Cryo-EM structure of a human spliceosome activated for step 2 of splicing. Nature. 2017;542(7641):318–323. 10.1038/nature21079 [DOI] [PubMed] [Google Scholar]
- 88. Banani SF, Rice AM, Peeples WB, et al. : Compositional Control of Phase-Separated Cellular Bodies. Cell. 2016;166(3):651–63. 10.1016/j.cell.2016.06.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Zhu L, Brangwynne CP: Nuclear bodies: the emerging biophysics of nucleoplasmic phases. Curr Opin Cell Biol. 2015;34:23–30. 10.1016/j.ceb.2015.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Nott TJ, Petsalaki E, Farber P, et al. : Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol Cell. 2015;57(5):936–47. 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Zhang H, Elbaum-Garfinkle S, Langdon EM, et al. : RNA Controls PolyQ Protein Phase Transitions. Mol Cell. 2015;60(2):220–30. 10.1016/j.molcel.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Nagai K, Oubridge C, Jessen TH, et al. : Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348(6301):515–20. 10.1038/348515a0 [DOI] [PubMed] [Google Scholar]
- 93. Oubridge C, Ito N, Evans PR, et al. : Crystal structure at 1.92 Å resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372(6505):432–8. 10.1038/372432a0 [DOI] [PubMed] [Google Scholar]
- 94. Hoffman DW, Query CC, Golden BL, et al. : RNA-binding domain of the A protein component of the U1 small nuclear ribonucleoprotein analyzed by NMR spectroscopy is structurally similar to ribosomal proteins. Proc Natl Acad Sci U S A. 1991;88(6):2495–9. 10.1073/pnas.88.6.2495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Birney E, Kumar S, Krainer AR: Analysis of the RNA-recognition motif and RS and RGG domains: conservation in metazoan pre-mRNA splicing factors. Nucleic Acids Res. 1993;21(25):5803–16. 10.1093/nar/21.25.5803 [DOI] [PMC free article] [PubMed] [Google Scholar]