Abstract
APOL1 harbors C–terminal sequence variants (G1 and G2), which account for much of the increased risk for kidney disease in sub–Saharan African ancestry populations. Expression of the risk variants has also been shown to cause injury to podocytes and other cell types, but the underlying mechanisms are not understood. We used Drosophila melanogaster and Saccharomyces cerevisiae to help clarify these mechanisms. Ubiquitous expression of the human APOL1 G1 and G2 disease risk alleles caused near-complete lethality in D. melanogaster, with no effect of the G0 nonrisk APOL1 allele, corresponding to the pattern of human disease risk. We also observed a congruent pattern of cellular damage with tissue-specific expression of APOL1. In particular, expression of APOL1 risk variants in D. melanogaster nephrocytes caused cell-autonomous accumulation of the endocytic tracer atrial natriuretic factor-red fluorescent protein at early stages and nephrocyte loss at later stages. We also observed differential toxicity of the APOL1 risk variants compared with the APOL1 nonrisk variants in S. cerevisiae, including impairment of vacuole acidification. Yeast strains defective in endosomal trafficking or organelle acidification but not those defective in autophagy displayed augmented APOL1 toxicity with all isoforms. This pattern of differential injury by the APOL1 risk alleles compared with the nonrisk alleles across evolutionarily divergent species is consistent with an impairment of conserved core intracellular endosomal trafficking processes. This finding should facilitate the identification of cell injury pathways and corresponding therapeutic targets of interest in these amenable experimental platforms.
Keywords: endocytic trafficking, Apolipoprotein L1, chronic kidney disease, nephrocyte, acidification
Two sets of DNA–derived sequence variants (termed G1 and G2 in contrast to the ancestral G0) at the human APOL1 gene are associated with a markedly increased risk of progressive CKD in sub–Saharan African ancestry populations.1–7 The G1 haplotype comprises two nonsynonymous coding variants rs73885319 (S342G) and rs60910145 (I384M), whereas the G2 (rs71785313) allele contains a 6-bp deletion (del.N388/Y389).1,2 APOL1 is one member of the APOL1–6 cluster of innate immunity genes and the only member of that cluster with a secretory signal peptide and a prominent circulating protein product.8–10 Circulating APOL1 can lyse Trypanosoma brucei species and protects humans against African sleeping sickness.11–13 The G1 and G2 risk alleles confer trypanolytic activity against the T. brucei rhodesiense subspecies that has developed resistance to the ancestral G0 allelic version of APOL1, explaining their rise to high frequency in at–risk parent populations. Genetic epidemiology studies show that kidney disease risk association is significant primarily under a recessive inheritance mode (two APOL1 risk alleles) or that there is a very marked step up in disease risk from one to two risk alleles.1–7,14–16 These studies and others also point to an important role for nongenetic second risk triggers in transforming disease risk to clinical disease.3,7,17–19 The recessive inheritance mode might suggest that the G1 and G2 mutations cause a loss of function essential to kidney functional integrity. However, APOL1 is absent and therefore, dispensable for kidney health in most species, including nonhuman primates,9,10,13,20 and the null state was not found to cause a kidney disease phenotype in at least one human individual.21 Correspondingly, transient transfection or induced expression in a variety of human cells in culture (including human podocytes and human embryonic kidney [HEK] 293 cells) and in vivo models caused transfection dose– and time–dependent cell injury, with a significantly greater effect of the G1 and G2 variants compared with G0 but still some cytotoxic effect of even the G0 allele.13,19,22–29 Taken together, these studies point rather to a gain of function of G1 and G2 compared with G0 and a very marked gene dose or expression effect, with several formulations having been proposed for the marked step up, such as the occurrence of multimeric structures.30 APOL1 protein is expressed in the circulation and Trypanosome Lytic Factor (TLF) particles (TLF1 and TLF2)31–33 as well as intracellularly in a variety of cell types, including the podocyte, endothelial cells, and other tissues, such as brain, placenta, and a variety of tumors.8,9,19,34–36 Although circulating APOL1 is responsible for trypanolysis, studies in the setting of kidney transplantation point to intracellular APOL1 as responsible for kidney disease risk, because reduced kidney transplant allograft survival tracks with the donor and not the recipient APOL1 genotype in the African ancestry population.37–40 These studies reinforce the importance of continuing to investigate gain of function mechanisms and pathways in experimental systems displaying differentially greater cell injury of G1 and G2 compared with G0 variants of APOL1.
Although human cells in culture and murine models provide the relevant complement of pathways to study mechanisms of human disease risk, simpler organisms offer the opportunity for efficient and potentially informative genetic interrogation. Accordingly, we turned to model eukaryotic organisms that are readily amenable to genetic interrogation of conserved pathways. Here, we show that the G1 and G2 allelic variants, which confer human disease risk, also confer enhanced eukaryotic cell toxicity, even in species with divergent evolutionary histories that do not naturally express APOL1 (Drosophila melanogaster and Saccharomyces cerevisiae), suggesting that APOL1 engages core pathways that are highly conserved in evolution. Genetic interrogation of the fly and yeast systems points to a local intracellular (including fly nephrocytes) rather than systemic effect of APOL1, with impairment of intracellular acidification and endosomal trafficking.
Results
APOL1 G1 and G2 Differential Toxicity Is Conserved in the Fruit Fly
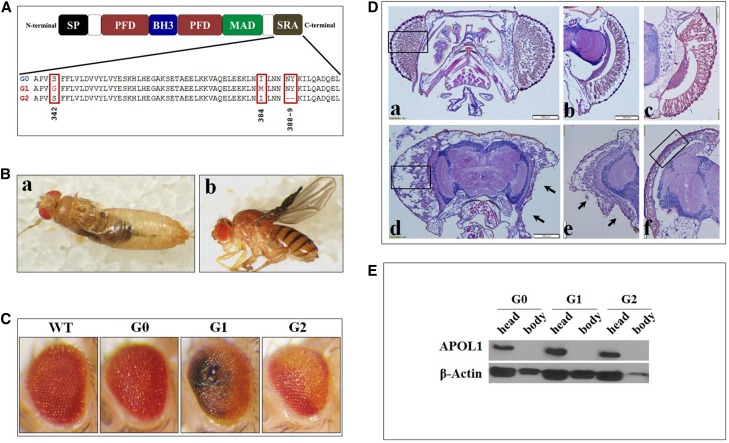
We examined the effect of expressing human APOL1 G0, G1, and G2 gene products as well as the artificial C–terminal truncated (C-tr) construct that lacks the Serum Resistance–Associated (SRA) interacting domain (Figure 1A) in D. melanogaster. The expression of APOL1 G0 under the ubiquitous driver daughterless (da)-GAL4 caused no evident phenotype compared with the wild type (WT) crossed with da-GAL4. In contrast, the G1 and G2 variants consistently led to significantly higher rates of pupal–pharate adult lethality (Figure 1B and Table 1). The C-tr construct, which lacks trypanolytic activity and toxicity in an in vivo murine model,13,41,42 was innocuous. The da-GAL4 driver induced high levels of APOL1 protein in the hemolymph (Supplemental Figure 1). To distinguish between systemic and cell-specific effects of APOL1, we used Glass multiple reporter (GMR)-GAL4 to drive APOL1 expression in the eye. As shown in Figure 1, C–E and Table 2, significantly greater percentages with a rough eye phenotype and severely disrupted ommatidia were evident in flies expressing the G1 or G2 human transgene compared with G0. As shown in Figure 1E, the observed eye phenotypes were not caused by systemic distribution of APOL1, which was restricted to the heads of adult flies when expressed under the GMR-GAL4 driver. This is consistent with an APOL1 injury mechanism originating at a local intracellular rather than systemic circulating level.
Figure 1.
APOL1 G1 and G2 are toxic to D. melanogaster. (A) Schematic representation of the protein domains in APOL1. A full–length APOL1 protein with a schematic representation of the protein domains: BH3 Domain (BH3), Membrane Addressing Domain (MAD), Pore Forming Domain (PFD), Signal Peptide (SP), and SRA-interacting domain. Lower panel shows the amino acid composition at the SRA-binding domain of APOL1 G0, G1, and G2 variants. (B) D. melanogaster expressing APOL1 kidney risk variants under the regulation of da-GAL4 shows a lethal phenotype and wing malformations. (B, a) A da-GAL4 > APOL1 G1 fly that failed to emerge from its pupal case (representing 98% of the flies of this genotype). (B, b) A da-GAL4 > APOL1 G1 fly that succeeded in eclosing but displays a severe wing phenotype. (C) Expression of APOL1 G1 and G2 gene products under the regulation of the eye–specific driver GMR-GAL4 results in an abnormal eye phenotype. Rough eye phenotypes were observed in 81% of G1, 48% of G2, and 11% of G0 flies. The rough eyes were characterized by abnormalities in architecture, often leaving a concavity. (D) Hematoxylin and eosin staining of eyes of GMR-GAL4 > APOL1 flies reveals abnormal eye histology with APOL1 G1 and G2 variants. (D, a and b) WT eye shows normal ommatidia structure. (D, c) Intact ommatidia in eye expressing APOL1 G0. (D, d) Eyes expressing APOL1 G1 and G2 (D, e) showing severely disrupted ommatidial structure (rectangle compared with D, a); black arrows show concave eye structure. (D, f) Control GMR-GAL4 eye. The ommatidial structure is abnormally condensed compared with the WT structure; however, the severe structural changes shown in the G1- and G2-expressing eyes are absent (compressed structure is shown in the rectangle). Scale bars, 100 μm in D, a, b, and d; 50 μm in D, c, e, and f. (E) Western analysis of APOL1 and β-actin in D. melanogaster head and body protein lysates from flies expressing APOL1 G0, G1, and G2 gene products under the eye driver GMR-GAL4. Protein loading is as described in Concise Methods to account for hemolymph protein in body lysates. APOL1 is expressed in the heads but not the bodies of the flies, emphasizing the role of endogenous APOL1 versus circulating APOL1.
Table 1.
APOL1 G1 and G2 differential toxicity in D. melanogaster: reduced viability of D. melanogaster expressing APOL1 kidney risk variants under da-GAL4.
| Cross | n | Survival, % |
|---|---|---|
| ♀UAS APOL1 G0 × ♂da-GAL4 | 1727 | 97a,b |
| ♀UAS APOL1 G1 × ♂da-GAL4 | 2113 | 5a,c,d |
| ♀UAS APOL1 G2 × ♂da-GAL4 | 1891 | 6b,e,f |
| ♀ WT × ♂da-GAL4 | 1028 | 98c,e |
| ♀UAS APOL1 C truncated × ♂da-GAL4 | 1795 | 98d,f |
Pairs with a statistically significant (P<0.001 for each pair) difference using one-way ANOVA. Comparison: G1-G0.
Pairs with a statistically significant (P<0.001 for each pair) difference using one-way ANOVA. Comparison: G2-G0.
Pairs with a statistically significant (P<0.001 for each pair) difference using one-way ANOVA. Comparison: WT-G1.
Pairs with a statistically significant (P<0.001 for each pair) difference using one-way ANOVA. Comparison: C truncated-G1.
Pairs with a statistically significant (P<0.001 for each pair) difference using one-way ANOVA. Comparison: WT-G2.
Pairs with a statistically significant (P<0.001 for each pair) difference using one-way ANOVA. Comparison: C truncated-G2.
Table 2.
APOL1 G1 and G2 differential toxicity in D. melanogaster: structural eye abnormality (rough eye) in D. melanogaster expressing APOL1 kidney risk variants under GMR-GAL4
| Cross | n | Rough Eye, % |
|---|---|---|
| ♀UAS APOL1 G0 × ♂GMR-GAL4 | 535 | 11 |
| ♀UAS APOL1 G1 × ♂GMR-GAL4 | 443 | 81 |
| ♀UAS APOL1 G2 × ♂GMR-GAL4 | 337 | 48 |
| ♀ WT × ♂GMR-GAL4 | 529 | 0 |
All pairwise differences are statistically significant (P<0.001 for each pair).
APOL1 G1 and G2 Differential Toxicity Is Conserved in D. melanogaster Pericardial Nephrocytes and Leads to Atrial Natriuretic Factor-Red Fluorescent Protein Accumulation
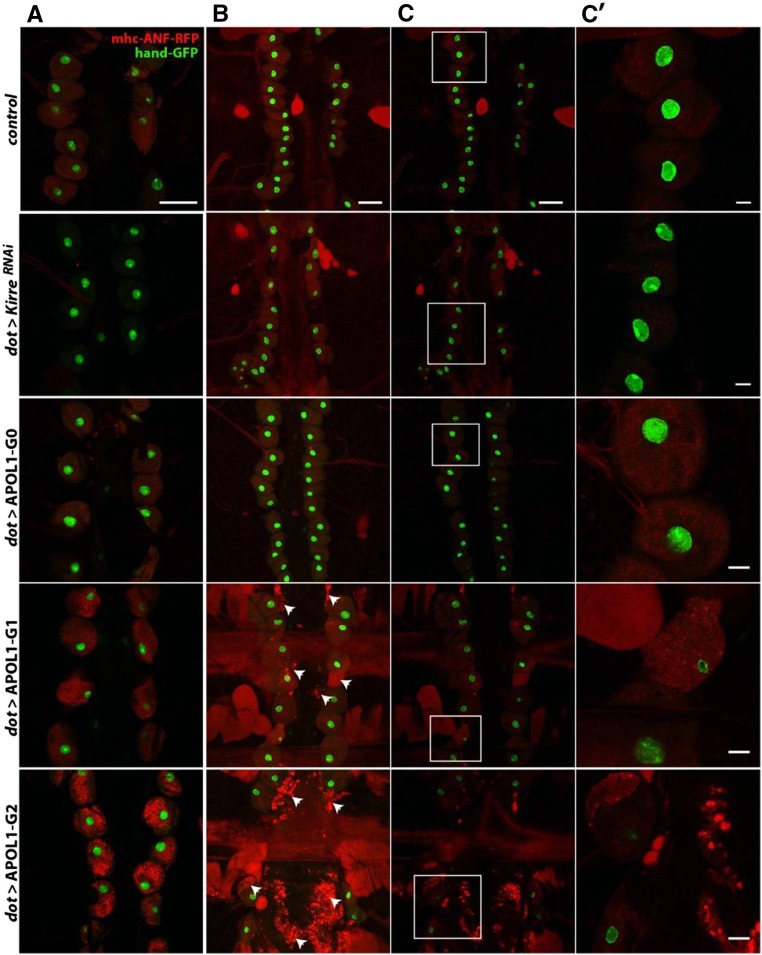
Drosophila pericardial nephrocytes share remarkable similarity with podocytes of the mammalian nephron. The fly nephrocyte diaphragm regulates filtration in a manner similar to the mammalian slit diaphragm.43–45 After passage through this filtration barrier, the main function of nephrocytes is to regulate hemolymph composition by endocytosis and metabolism or recycling of filtrate constituents. Therefore, the fly nephrocyte system enables filtration and endocytic uptake components to be monitored in a manner that is not readily recapitulated in human podocytes in cell culture. We used Dot-GAL4 to drive APOL1 expression in nephrocytes in flies expressing the Red Fluorescent Protein (RFP) reporter fused with atrial natriuretic factor (ANF) secretion peptide driven by the myosin heavy chain enhancer.44 ANF-RFP is endocytosed from the hemolymph by nephrocytes and degraded, and therefore, it serves as a marker for these nephrocyte functions.44 One-day-old APOL1 G1 and G2 but not G0 transgenic flies showed a striking increase of vesicular ANF-RFP in the pericardial nephrocytes (Figure 2). Knockdown of Kirre (fly ortholog of human NEPH1), which is known to disrupt filtration, led to a significant reduction of ANF-RFP uptake (negative control). At later time points, we observed nephrocyte toxicity, characterized by condensed or absent nuclei as well as surrounding cell fragments in G1 and G2 but not G0 transgenic flies. Taken together, the ectopic expression of G1 and G2 in nephrocytes caused nephrocyte-specific defects. The accumulation of ANF-RFP by nephrocytes followed by their death is consistent with well preserved uptake but impaired degradation, suggesting disruption of the endolysosomal processes. This led us to examine the effects of APOL1 and its risk variants on intracellular trafficking in S. cerevisiae.
Figure 2.
APOL1 G1 and G2 affect Drosophila pericardial nephrocyte function and lead to cell death during aging. Pericardial nephrocytes of adult flies were dissected, and ANF-RFP uptake and nuclear GFP were analyzed by confocal microscopy. (A) ANF-RFP uptake by pericardial nephrocytes in 1-day-old flies. (B and C) ANF-RFP uptake by pericardial nephrocytes in 15-day-old flies. In B, the maximal intensity projection of 20 focal planes was applied to visualize fragments of dead cells (arrowheads). Close-up images are shown in C′ (corresponding to the white frames in C). Genotypes used in this experiments were (from top to bottom) with myosin heavy chain (MHC)-ANF-RFP, Hand-GFP, Dot-GAL4/+ (control); with MHC-ANF-RFP, Hand-GFP; Dot-GAL4/UAS-KirreRNAi (Dot > KirreRNAi); with MHC-ANF-RFP, Hand-GFP; Dot-GAL4/+;UAS-APOL1-G0 (Dot > APOL1-G0); with MHC-ANF-RFP, Hand-GFP; Dot-GAL4/+;UAS-APOL1-G1 (dot > APOL1-G1), and with MHC-ANF-RFP, Hand-GFP; Dot-GAL4/+; UAS-APOL1-G2 (dot > APOL1-G2). Scale bars, 50 μm in A–C; 10 μm in C′.
APOL1 G1 and G2 Differential Toxicity Is Also Conserved in S. cerevisiae and Is a Manifestation of Cell Lethality
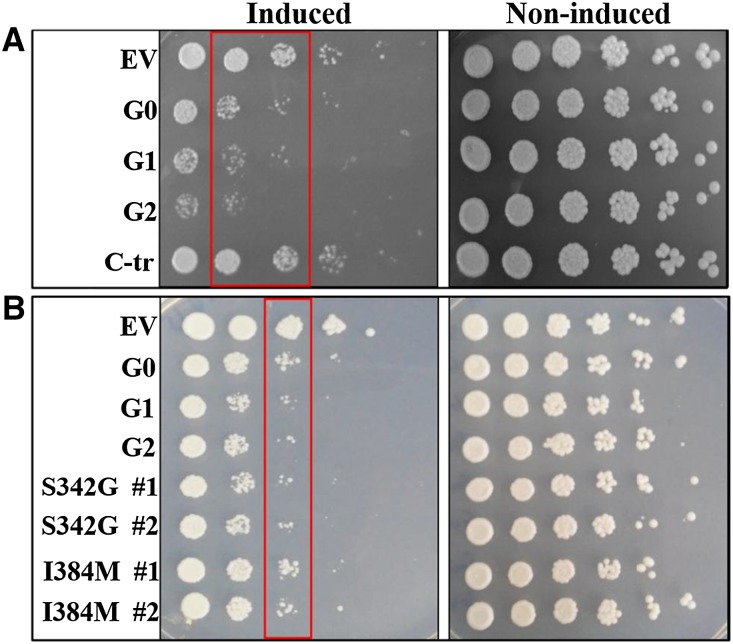
In the S. cerevisiae drop titration assay, we observed toxicity of the G0, G1, and G2 variants of APOL1 but not the C-tr version (Figure 3A). For G1, we also used constructs with S342G or I384M mutations separately and found that S342G rather than the I384M mutation suffices to recapitulate toxicity of the APOL1 G1 missense haplotype (Figure 3B). Genetic studies have also shown this to be the sequence variant in the APOL1 G1 missense haplotype that is strongly associated with kidney disease.3,46 Because in yeast, unlike D. melanogaster but similar to mammalian cells in culture,13,19,22–29 even the G0 nonrisk variant was toxic compared with vector or the C-tr construct, it was necessary to quantitatively compare the relative toxicities. To quantify and determine whether APOL1 simply inhibits yeast growth or actually increases lethality, we conducted a survival assay by counting yeast CFU after 0 or 8 hours of growth (G0, G1, G2, and C-tr) under inducing conditions for APOL1 expression followed by restoration to noninducing conditions in optimal growth media (conditions that would allow any yeast cells that had not been irreversibly injured or killed to establish countable colonies). As shown in Tables 3 and 4, expression of vector or the C-tr variant shows a similar percentage of survival. In contrast, induction of APOL1 G0 caused lethality with fewer survivors, and APOL1 G1 or G2 caused a significantly greater degree of lethality compared with G0.
Figure 3.
Expression of human APOL1 in the yeast S. cerevisiae causes reduced yeast growth. (A) Differential growth defect between APOL1 G0 versus APOL1 G1 and G2 expression determined by drop titration growth assay. (B) APOL1 S342G mutant shows reduced growth compared with the I384M APOL1 mutant. Plasmids containing the human APOL1 variants G0, G1, and G2 and the artificial APOL1 constructs C-tr, S342G, and I384M under the yeast GAL1 promoter were transformed into the WT BY4741 yeast strain. Each row represents serial fivefold dilutions (left to right) of a suspension of cells transformed with the indicated plasmid spotted on plates containing glucose (right panels; noninduced conditions) or galactose (left panels; induced conditions). The boxes highlight the dilutions where the differences are well shown. For S342G and I384M, two independent transformants were tested, and all titration assays were conducted at least twice with similar results. EV, empty vector.
Table 3.
S. cerevisiae survival assay: survival ratios at 0 and 8 hours
| APOL1 Variant | 0-h CFU Count | 8-h CFU Count | 8 h-to-0 h Ratio (95% CI) |
|---|---|---|---|
| EV | 5716 | 4935 | 0.86 (0.78 to 0.95) |
| G0 | 5220 | 3710 | 0.71 (0.64 to 0.78) |
| G1 | 5850 | 3245 | 0.55 (0.50 to 0.61) |
| G2 | 5926 | 2680 | 0.45 (0.41 to 0.50) |
| C truncated | 5160 | 4250 | 0.82 (0.74 to 0.91) |
Survival assays were conducted as described in Concise Methods, with measurement of surviving cells determined as CFU expressed as ratios at 0 and 8 hours of induction for each experimental condition. Each pair of experiments was conducted in parallel for all conditions three times, and P values for differences were determined as described in Concise Methods. 95% CI, 95% confidence interval; EV, empty vector.
Table 4.
S. cerevisiae survival assay: comparisons of survival ratios
| APOL1 Variant Comparison | P Value for Difference |
|---|---|
| G0-EV | <0.01 |
| G1-G0 | <0.001 |
| G2-G0 | <0.001 |
| G2-G1 | <0.01 |
| C truncated-EV | 0.51 |
Survival assays were conducted as described in Concise Methods, with measurement of surviving cells determined as CFU expressed as ratios at 0 and 8 hours of induction for each experimental condition. Each pair of experiments was conducted in parallel for all conditions three times, and P values for differences were determined as described in Concise Methods. EV, empty vector.
S. cerevisiae Strains Deleted in Components of the Class 3 Phosphatidylinositol 3-Kinase Complex Are Hypersensitive to APOL1 Toxicity
The recapitulation of the significantly greater APOL1 G1 and G2 toxicity in S. cerevisiae along with the suggestive endocytic trafficking perturbation leading to fly nephrocyte toxicity prompted us to explore endosomal pathways in yeast. APOL1 has been reported to be involved in autophagy regulation26,47 and induce lysosomal injury in human cells, including podocytes.22 The contributions of autophagy and endosomal trafficking to the functional integrity of podocytes have been well characterized.48–54 The class 3 phosphatidylinositol (PI) 3–kinase phosphorylates position 3 of PI, producing PI 3-phosphate, and it is a key signal for both endosomal trafficking and autophagosome formation.55–59 We monitored growth of S. cerevisiae strains deleted for class 3 PI 3–kinase complex genes with expression of the APOL1 variants. Yeast strains deleted for vacuole protein sorting 34 (VPS34), VPS30 (ATG6/human BECLIN1), or VPS15 were hypersensitive to APOL1 toxicity compared with the WT strain (Figures 3 and 4, A–C). In these strains, even the G0 version of APOL1 was toxic and indistinguishable from the G1 and G2 alleles. Notably, however, the artificial C-tr construct remained innocuous in all strains.
Figure 4.
S. cerevisiae strains deleted in components of the endocytic Vps34 complex II but not the autophagic–specific Vps34 complex I confer APOL1 hypersensitivity. Plasmids containing the human APOL1 variants under the yeast GAL1 promoter were transformed into (A) atg6Δ/Beclin, (B) vps34Δ, and (C) vps15Δ strains. (D) vps38Δ, (E) atg14Δ, and (F) atg8Δ yeast strains. Each row represents serial fivefold dilutions (left to right) of a suspension of cells transformed with the indicated plasmid spotted on plates containing glucose (right panels; noninduced conditions) or galactose (left panels; induced conditions). EV, empty vector.
Loss of the Endocytic Vps34 Complex II but Not the Autophagy–Specific Vps34 Complex I Confers APOL1 Hypersensitivity
Vps34 together with Vps30 and Vps15 form multiple complexes that are responsible for its diverse cellular functions.56,58 The autophagy–specific Vps34 complex I comprises Vps34, Vps15, Atg6/Vps30, and Atg14. The endocytic Vps34 complex II is formed by interaction with Vps38 and required for endosome to Golgi retrograde trafficking and late Golgi/TGN to endosome-VPS.55–59 To differentiate between these pathways, we investigated the viability of ATG14 or VPS38 deletion strains. Only vps38Δ showed augmented APOL1 toxicity, similar to the deletion of the VPS34, VPS30, or VPS15 component of the core complex (Figure 4), an effect that could not be attributed to elevated APOL1 protein levels (Supplemental Figure 2). Notably, the survival assay using the vps38Δ strain also confirmed irreversible injury/lethality that was significantly higher in the vps38Δ compared with the WT (Supplemental Table 1). In contrast, atg14Δ showed no such augmented APOL1–mediated toxicity and preserved the differential toxicity between the G0 and G1, G2 variants (Figure 4E). Similar results were obtained for atg8Δ (Figure 4F). These results prompted us to systematically investigate the role of anterograde and retrograde endosomal trafficking in APOL1 toxicity (Figure 5, Supplemental Table 2). Strains that enhance APOL1 toxicity have essential roles in the anterograde and retrograde Golgi–endosomal trafficking pathways,55 the endosomal sorting complex required for transport (ESCRT), the sorting of the ubiquitinated multivesicular body cargoes,60 and vacuole trafficking. This is consistent with the phenotypes of the class 3 PI 3–kinase complex II–deleted strains described above, and here, the C-tr APOL1 was also not toxic.
Figure 5.
Disruption of endocytic trafficking enhances APOL1 toxicity. Schematic representation of endosome, Golgi, multivesicular body (MVB), vacuole, and autophagosome trafficking pathways examined in relation to APOL1-mediated toxicity. Pathways marked in red are those in which deletion strains augmented APOL1–induced yeast toxicity for G0 as well as G1 and G2 risk variants. Conversely, pathways marked in blue are those in which elimination of components did not enhance APOL1 toxicity. Pathways marked in yellow were not examined. CORVET, class C core vacuole/endosome tethering factor; LE, late endosome; PI3K, phosphatidylinositol 3-kinase.
APOL1 Localization Is Altered in vps38Δ and Other Endocytic Mutants
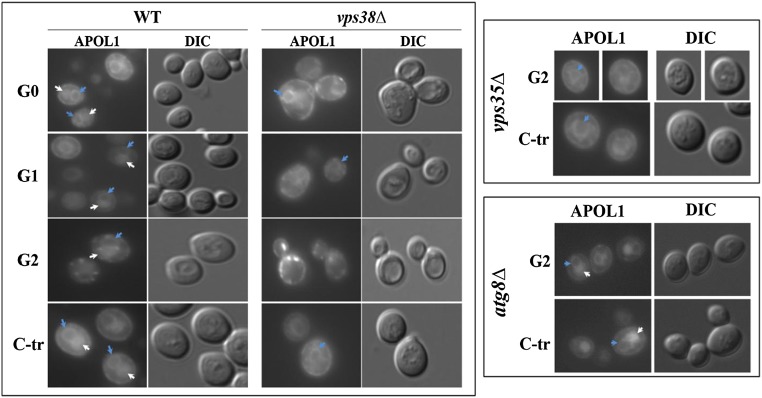
We next examined the localization of APOL1 variants in the WT versus vps38Δ. In the WT strain, expression of APOL1 was observed in the vacuole and the endoplasmic reticulum (ER) and possibly, also separately in the juxtaposed plasma membrane, with no difference evident in localization between APOL1 G0, G1, and G2 (Figure 6, Supplemental Figure 3). It should be noted that juxtaposition of the ER to the plasma membrane in yeast cells makes it difficult to determine plasma membrane localization with complete certainty. Notably, the nontoxic C-tr mutant showed the same localization pattern, more readily discerned in this case because of the absence of yeast cell damage. In contrast, in vps38Δ, APOL1 did not accumulate in the vacuole and appears most prominently in the ER. Loss of vacuole localization was also observed with deletions of the Retromer complex responsible for retrograde endosomal to Golgi trafficking (vps35Δ in Figure 6) or the ESCRT complex responsible for endosomal sorting required for transport (vps25Δ in Supplemental Figure 4), which also enhanced APOL1 toxicity. This is in contrast to strains that did not affect APOL1 toxicity, such as atg8Δ (Figure 6) and atg14Δ (Supplemental Figure 4), which were also indistinguishable from the WT strain with regard to APOL1 localization. Because there was no difference in cellular localization among APOL1 variants, including the nontoxic C-tr construct, it seems that differences in localization per se do not account for differential APOL1 toxicity. Rather, these findings point to a role for the vacuole in governing the differential toxicity of APOL1 and its risk variants in yeast and by extrapolation, endolysosomal compartments in higher eukaryotes.
Figure 6.
APOL1 localization is altered in the endocytic deleted strains. Loss of APOL1 vacuolar localization in the endocytic mutants (vps38Δ and vps35Δ) but not an autophagy-deleted strain (atg8Δ). APOL1 variants were tagged with mCherry at their N terminus (after the signal peptide) (Concise Methods) and expressed in WT and deletion (Δ) strains. APOL1 expression was induced for 3 hours with synthetic minimal media containing essential amino acids and galactose. White arrows indicate vacuoles, and blue arrows indicate perinuclear ER. The same results were observed for C-tr APOL1 variants in WT and vps38Δ (Supplemental Figure 3). DIC, differential interference contrast.
Reciprocal Interactions between APOL1 and pH
Functional integrity of the vacuole in yeast and lysosomes in higher eukaryotic cells enables their acidification, without which cells, tissues, and organs undergo cell injury and death.61 Because many of the proteins affecting APOL1 toxicity are involved in trafficking to the vacuole, we used pep4Δ, vma1Δ, and vma21Δ strains to investigate the involvement of vacuolar function in APOL1 toxicity. VMA1 encodes one of the V1 subunits of the vacuolar-ATPase, an ATP–dependent proton pump responsible for the acidification of the vacuole and other endocytic pathway components.62,63 Vma21 is needed for assembly of the V0 subunit, whereas Pep4 encodes the vacuole protein hydrolase–activating enzyme; in its absence, vacuolar proteolysis is impaired. In the vma1Δ and vma21Δ but not pep4Δ strains, the pattern of APOL1 cytotoxicity was similar to that observed for deletion of any one of the genes encoding components of endosomal trafficking (Figure 7). This suggests that it is the acidification status of the vacuole that is affecting toxicity.
Figure 7.
S. cerevisiae-deleted strains that affect vacuole acidification (VMAΔ) enhanced APOL1 toxicity, whereas deletion of vacuolar hydrolase (pep4Δ) was innocuous. Plasmids containing the human APOL1 variants under the yeast GAL1 promoter were transformed into (A) vma1Δ, (B) vma21Δ, and (C) pep4Δ yeast strains. Each row represents serial fivefold dilutions (left to right) of a suspension of cells transformed with the indicated plasmid spotted on plates containing glucose (right panels; noninduced conditions) or galactose (left panels; induced conditions). EV, empty vector.
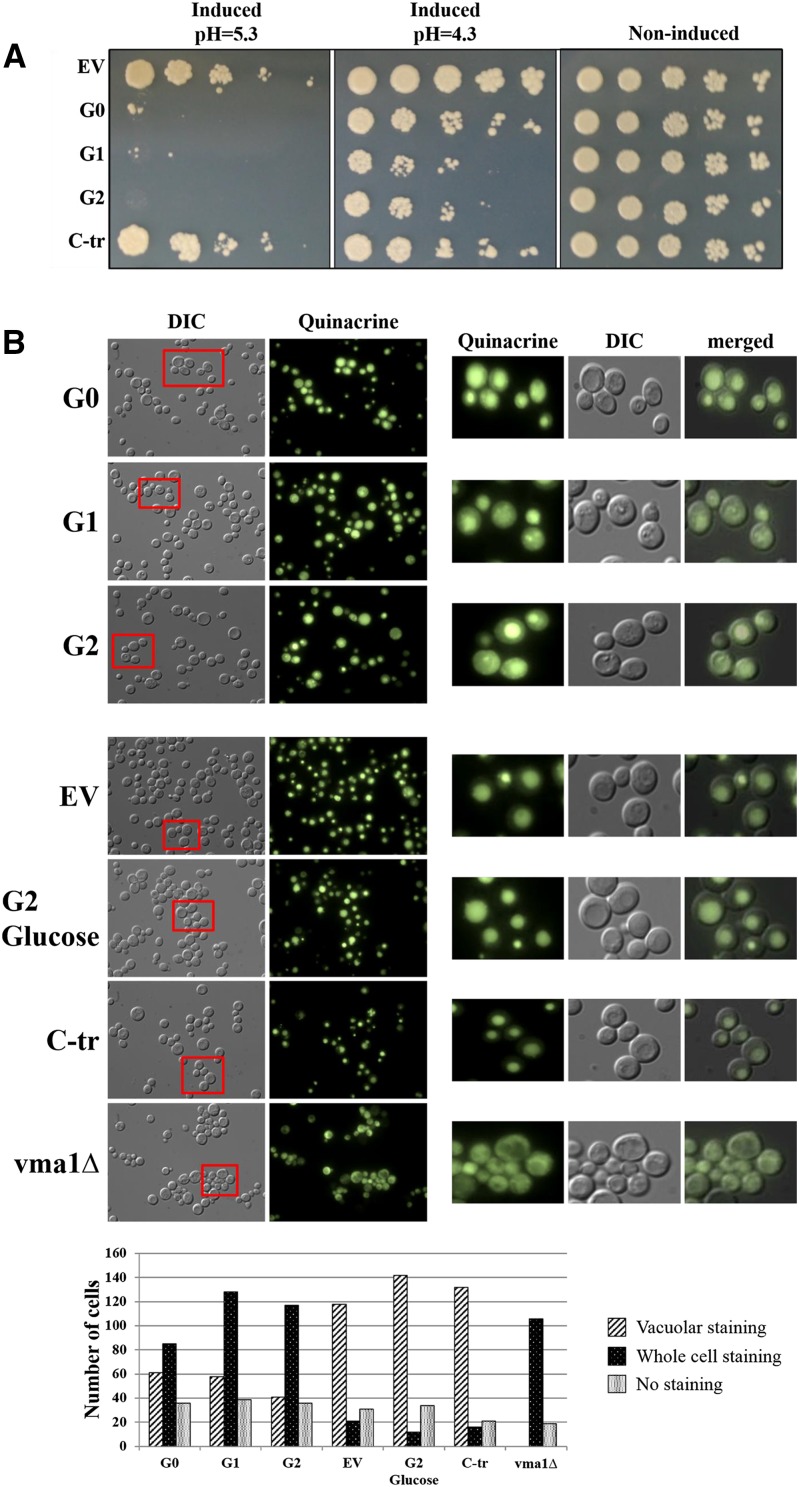
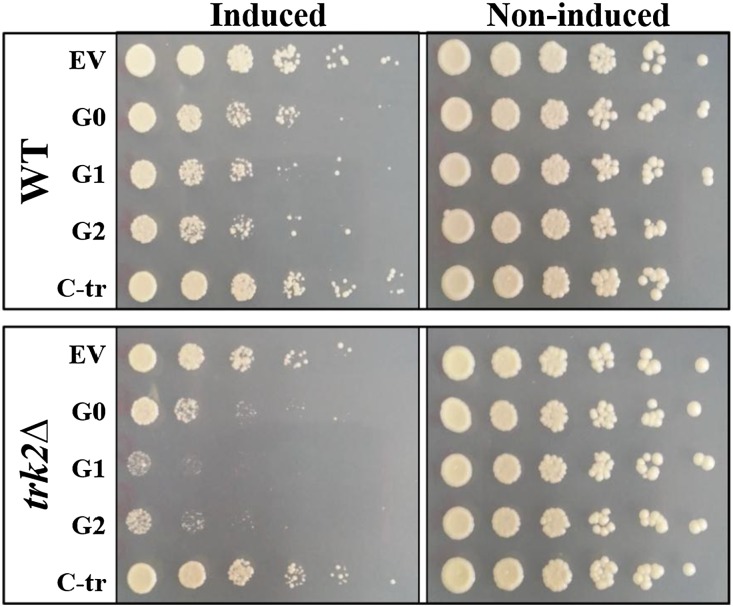
The augmented lethality in the vmaΔ strains led us to explore the relationship between vacuole acidification and APOL1 toxicity. S. cerevisiae strains lacking vacuolar ATPase activity show a conditional lethal phenotype that manifests as growth arrest at elevated pH (pH>5.0).64 Increasing pH from 4.3 to 5.3 mimicked the effect of endocytic mutants, with augmented toxicity of the G0, G1, and G2 variants of APOL1, but had no effect on the C-tr APOL1 (Figure 8A). We used quinacrine to visualize acidification of the vacuole in the absence and presence of APOL1 variants (Figure 8B). Expression of APOL1 variants resulted in an acidification defect, similar to the pattern of quinacrine staining observed in the vma1Δ strain. Although APOL1 G0 also displayed reduced vacuole acidification, this effect was more marked with the G1 and G2 and not evident at all for the nontoxic C-tr APOL1. These findings suggest that APOL1 risk variant toxicity is accompanied by impaired vacuole acidification. Because impaired vacuole acidification requires accompanying processes to balance electroneutrality, such as anion or cation influx or egress,65 we examined APOL1 toxicity in trk2Δ. The TRK2 gene product is required for low–affinity K+ influx in S. cerevisiae.66 Indeed, trk2Δ exhibited moderately enhanced sensitivity to APOL1 G0, G1, and G2 but in this case, with preservation of differential hierarchy of toxicity in the G1 and G2 variants compared with G0 and again, no toxicity of C-tr APOL1 compared with vector alone (Figure 9).
Figure 8.
Increasing medium pH augments APOL1 toxicity, whereas expression of APOL1 and risk variants results in a vacuole acidification defect. (A) Plasmids containing the human APOL1 variants G0, G1, G2, and C-tr under the yeast GAL1 promoter were transformed into the WT BY4741 strain. Each row represents serial fivefold dilutions (left to right) of a suspension of cells transformed with the indicated plasmid spotted on plates containing glucose (right panel; noninduced conditions at pH 4.3) or galactose (induced conditions) at pH 4.3 (middle panel) or pH 5.3 (left panel). (B) Quinacrine staining of S. cerevisiae transformed with APOL1 variants G0, G1, and G2 is similar to the staining of the vma1Δ strain with reduced vacuolar staining, showing a vacuole acidification defect with G1, G2 > G0. Enlarged pictures of selected areas are shown in right panels. The empty vector (EV), C-tr, and G2 in noninducing medium (glucose) show normal accumulation of the dye inside the vacuole, which indicates that the vacuolar lumen is more acidic than the cytoplasm. The corresponding graph shows the number of cells counted for each condition. DIC, differential interference contrast.
Figure 9.
Deletion of a component of the potassium transport system (trk2Δ) enhances APOL1 toxicity. Plasmids containing the human APOL1 variants under the yeast GAL1 promoter were transformed into the WT and trk2Δ strains. Each row represents serial fivefold dilutions (left to right) of a suspension of cells transformed with the indicated plasmid spotted on plates containing glucose (right panel; noninduced conditions) or galactose (left panel; induced conditions). EV, empty vector.
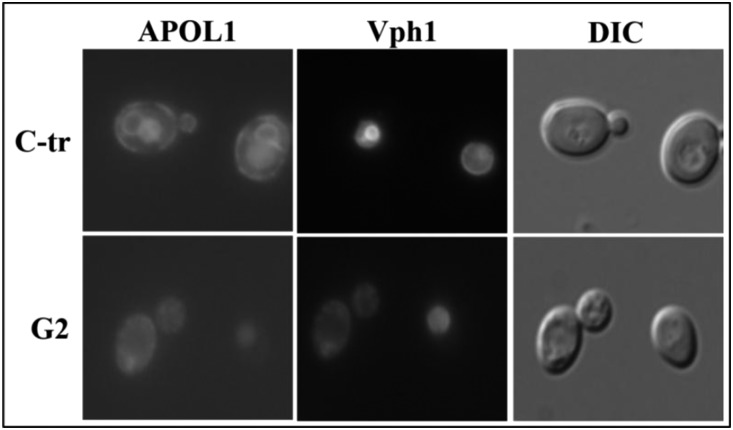
APOL1 expression induced a loss of vacuole acidification similar to that observed in the absence of the vacuolar ATPase subunit Vma1. However, growth impairment is not evident in vmaΔ strains. Therefore, we reasoned that additional mechanisms beyond disruption of VMA1–mediated vacuole acidification are involved in APOL1-mediated loss of viability. We sought to determine whether APOL1 has a broader effect on vesicle trafficking. We used Green Fluorescent Protein (GFP)–tagged Vph1, a vacuolar ATPase, as a marker for the VPS pathway.67 In the WT strain as well in the C-tr APOL1, Vph1-GFP localized normally to the vacuole, whereas APOL1 expression (G0, G1, and G2) inhibited Vph1 trafficking to the vacuole but without a differential effect of the risk versus nonrisk variants (Figure 10, Supplemental Figure 5).
Figure 10.
APOL1 inhibits the VPS pathway. Defective transport of Vph1 in yeast expressing G2 but not C-tr APOL1. Plasmids containing the human APOL1 variants under the yeast GAL1 promoter were transformed into S. cerevisiae expressing GFP-Vph1. In the C-tr APOL1, Vph1-GFP localized normally to the vacuole, whereas G2 APOL1 expression inhibited Vph1 trafficking to the vacuole, which was mis-sorted to multiple punctate compartments. DIC, differential interference contrast.
Discussion
Population genetics studies provide powerful but circumstantial evidence that the APOL1 G1 and G2 variant kidney disease risk association reflects causality,68 whereas the mechanism underlying kidney injury has not been resolved at this stage. The cardinal feature of these studies is the finding that the APOL1 toxicity displays a parallel pattern of differential toxicity between the human disease risk and nonrisk alleles in D. melanogaster and S. cerevisiae. We were able to dissect the two G1 mutations and show that it is the S342G mutation in the APOL1 G1 risk missense haplotype that is toxic to yeast, consistent with the finding that it is this amino acid change that is responsible for the association with increased risk for human kidney disease.3,46 This finding also dissociates the APOL1 requirement for APOL1 eukaryotic cell injury from the modulation of the APOL1 trypanosomal SRA interaction.13 The results in D. melanogaster indicate that cell injury does not require the presence of the gene product in the hemolymph, supporting an intracellular rather than circulating source of APOL1 in pathogenesis, consistent with human transplantation studies.37–40 Expression of APOL1 G1 and G2 but not G0 in pericardial nephrocytes led to the accumulation of constitutively secreted ANF-RFP in vesicles at earlier stages, suggesting an endolysosomal defect that may contribute to nephrocyte loss seen in older adult flies. The yeast experimental platform strengthens the hypothesis that APOL1 kidney risk variants impair endosomal trafficking, an effect mediated at least in part by impaired acidification, which may also explain the accumulation of ANF-RFP in nephrocytes.
The yeast deletion strains revealed that cell injury is especially sensitive to the integrity of PI 3-phosphate–related intracellular endosomal trafficking rather than autophagy. However, because in mammalian cells, the VPS34 regulators ATG14 and UVRAG (VPS38) are less partitioned than in yeast,69,70 a role for impairment of autophagy in human disease cannot be completely ruled out by these studies. This increased toxicity also applied to G0 (but not the C-tr construct), such that differences compared with G1 and G2 were no longer apparent. This is consistent with an action of the C-terminal domain that disrupts plasma or organellar membrane integrity, which has been recently proposed.13,41 Diversion of APOL1 from the vacuolar membrane was shown in the vps38Δ and other endocytic mutants and was not different between the G0, G1, or G2 APOL1 variant gene products or the C-tr construct. Similarly, in previous studies, attenuation of APOL1 toxicity by coexpression with MCL-1 in Xenopus oocytes has been attributed to decreased cell surface abundance.29 These yeast studies do not show a C–terminal sequence dependency of localization. Rather, diversion of APOL1 in the endocytic mutants augmented toxicity of all APOL1 variants, whereas the C-tr remained innocuous. Thus, the extent of injury induced by APOL1 depends on (1) the extent of its localization to its site of injury, which is not a function of the C terminus, and (2) the presence and protein conformation of the C terminus.71
The similar effect of perturbations in endosomal trafficking and acidification processes to increase APOL1 toxicity suggests a model wherein acidifying conditions, such as in the yeast vacuole, attenuate APOL1 activity. The pH dependence of APOL1 activity as an ion channel has been studied in trypanosomes, lipid bilayers, and indirectly, HEK cells.12,27,29,72 Although the detailed findings in these studies differ depending on the experimental system and conditions, pH dependency is observed for APOL1 insertion into the membrane or its activity as an ion channel.73 Our findings show that APOL1 inhibits both VPS (Vph1 studies) and vacuole acidification (quinacrine staining), with a greater effect of the G1 and G2 kidney disease risk variants on acidification. Enhanced APOL1 G1 and G2 toxicity compared with the G0 in the trk2Δ strain is consistent with proposed pH–dependent activity of APOL1 activity as a cationic channel,29,73 resulting in cellular potassium depletion by the kidney risk variants.27 This cation channel potassium egress is reminiscent of the activation of the inflammasome,74 which might contribute to subsequent steps of APOL1 cell injury. The findings in this study point to not only an APOL1 risk variant differential effect on acidification of intracellular membrane compartments but also, a possible role of trafficking to and acidification of such compartments in determining toxicity. This is also consistent with results previously reported for HEK cytotoxicity, which was shown to be dependent on preservation of a signal peptide sequence in exon 4 of APOL1.26 Taken together, our findings in yeast and nephrocytes are most consistent with differential APOL1 risk variant impairment of the inter–related endolysosomal trafficking and acidification processes leading to cell injury.
The D. melanogaster model shows greater fidelity with respect to recapitulating the human risk association with a greater differential toxicity of the kidney risk alleles compared with yeast (this study) and even human cell lines19,22–25,27 and has the advantage of a nephrocyte phenotype with filtration and endolysosomal functions that can be readily monitored using ANF-RFP uptake and degradation. Having shown this robust recapitulation of differential toxicity, it should be possible in the future to use such an organismal model to productively understand the relationship between a gain of function cell injury process and recessive inheritance. This could be done by coexpressing G0 with G1 or G2 to investigate the mechanistic basis for a sharp threshold on the basis of gene dosage or gene product expression or rule out a dominant protective effect of G0. Although the differential toxicity of the kidney risk alleles in the yeast platform was less robust and the G0 was more toxic than in flies, there was still a significant difference in the differential toxicity of G1/G2 that led to yeast lethality. Therefore, the yeast system, which is the most rapidly amenable system to study genetic interactions, adds a complementary piece to unraveling the puzzle of APOL1–induced cell toxicity. Indeed, the yeast studies showed endosomal trafficking effects of likely relevance to the observation of fly nephrocyte dysfunction and death. The yeast platform also facilitated investigation of the role of acidification and revealed incubation conditions and pathways that abolished the differential toxicity. These studies showed that the differential toxicity is highly sensitive to ambient pH and genetic manipulation of the endocytic trafficking pathway and vacuole acidification. As such, these findings in yeast can productively guide the subsequent relevant fly studies. The more pronounced differential toxicity between G1/G2 compared with G0 in the fly model compared with yeast and mammalian cells in culture requires additional investigation. We know from experimental systems using in vivo models and human cell lines that G0 is also toxic13,20,22–29; however, some cells express APOL1 without evidence of toxicity.34 Even in the fly platform, the differential toxicity was not uniform across drivers (G0 was more toxic under GMR compared with the da driver), suggesting cell type–related differences. Similarly, the differential toxicity of the kidney risk variants in specific cells and organs (podocytes and kidney, respectively) and the variability of the lifetime kidney disease risk (ranging from 4% to >50% in individuals with two risk alleles) are also not clear.3,7,75 Investigation of these differences should advance our understanding of environmental factors that could be mimicked in experimental systems or unique susceptibility of specific cells, organs, organisms, or even individuals to APOL1 variant–mediated deleterious effect.
Perturbations in endosomal trafficking are especially relevant to podocyte function, with functional integrity that is crucially dependent on these pathways for appropriate regulation of slit diaphragm proteins, proper functioning of the actin cytoskeletal meshwork, and protein clearance to prevent filtration barrier clogging.51,53,54 Several genes implicated in human nephrotic syndrome directly or indirectly associate with the endocytic pathway.52 The role of VPS34 as a major regulator of endocytic pathways in podocytes had been clarified,49,50 with rapid onset kidney disease observed in mice lacking podocyte Vps34 compared with late onset kidney disease occurring in Atg5 knockout mice.48
The findings of endocytic trafficking impairment by APOL1 may also be of importance with respect to mechanisms, whereby nongenetic second hits may trigger APOL1–associated kidney disease.3,6,7,17–19 HIV infection constitutes the most robust second hit that transforms APOL1 genotypic risk into a form of kidney disease designated as HIV-associated nephropathy.3,7 Multiple enveloped viruses use the ESCRT machinery for budding. Taylor et al.76 have reported the increased secretion of HIV Vif protein into microvesicles from cells transfected with APOL1. Moreover, the HIV-1–encoded Gag protein has been shown to interact with the mammalian ortholog of Vps23 (tsg101), redirecting ESCRT-1 to the plasma membrane to execute viral budding.77,78 In addition to the expectation of increased expression of APOL1 under the high IFNγ induction state that characterizes HIV infection and has been shown to induce APOL1 gene expression,19 a Gag protein–mediated redirection mechanism could also contribute to the especially powerful second hit effect of HIV infection.
The yeast and fly models described in this study have several limitations. Both of these models lack endogenous APOL1 and therefore, do not normally engage pathways that interact, transport, and degrade APOL1 protein. Our data point to well conserved endocytic trafficking and endolysosomal acidification pathways involved in APOL1-mediated toxicity; however, other pathways with specific interacting proteins developed in humans as an adaptation to APOL1 deleterious effects may have a role. These would not be discerned using nonhuman reductionist models. More complex experimental systems might show that the core conserved pathways that are differentially engaged by the different APOL1 moieties might trigger and then, be amplified or augmented by engagement of inflammatory or other mechanisms. These latter mechanisms may have a major role in human disease risk and will require moving up from simple models amenable to genetic interrogation to models with a full complement of cell injury mechanisms to understand the pathobiology of APOL1-mediated nephropathy. However, the results of this study point to the involvement of core conserved processes that distinguish the nonrisk from the risk APOL1 variants. Although fly nephrocytes share many features of mammalian podocytes,43,45 nephrocytes differ from human podocytes in several potentially relevant physiologic and molecular features. For example, nephrocytes are not exposed to high-filtrate fluxes, and nephrocytes also possess proximal tubule attributes.44,79 In mammals, the filtration system is composed of three layers: fenestrated endothelium, glomerular basement membrane, and podocyte foot processes. In Drosophila, the filtration system is composed of two: the nephrocyte basement membrane and nephrocyte diaphragm. Taken together, these limitations indicate that, although the findings in this study point to the two experimental systems that are readily amenable to genetic interrogation as valuable in uncovering core conserved mechanisms differentially engaged by the APOL1 variants found to confer risk for kidney disease, insights gleaned will require extension and validation in more complex mammalian systems.
In conclusion, we have shown that differential toxicity of APOL1 kidney disease risk and nonrisk variants is conserved across eukaryotic evolution and may be mediated by perturbation of endosomal trafficking and acidification. Because these species evolved without a known endogenous APOL1 homolog, the differential toxicity of G0 compared with the G1 and G2 risk alleles reflects an inherent difference in the mode of interaction of the respective gene products with core biologic processes without respect to evolutionary history. To the extent that the parallel pattern of differential toxicity points to mechanisms that are relevant to the increased risk of kidney disease, then this also renders the phenotypes that are readily observable in these simple experimental platforms useful for genetic and compound screens in a phenotype–based high–throughput discovery process.
Concise Methods
Fly Strains
The following strains were used in this study (described in FlyBase; http://flybase.bio.indiana.edu/80): w1118 served as WT control, da-GAL4 was used to drive transgene expression as a model for systemic APOL1 expression, and the eye–specific GMR-GAL4 driver was used as a model for tissue-specific expression. For adult pericardial nephrocyte uptake/filtration studies, virgin females myosin heavy chain-ANF-RFP, Hand-GFP; Dot-GAL4 (a gift from Zhe Han and as reported in the work by Zhang et al.44) were crossed to male UAS-APOL1 WT (G0) and UAS-APOL1 variant (G1 and G2) transgenic lines. Hand-GFP was used to label pericardial nephrocytes at all developmental stages. The Kirre RNAi line (P[mw, UAS-kirre-RNAi]GD14476) was obtained from the Vienna Drosophila RNAi Center. For the generation of APOL1 transgenic flies, APOL1 G0, G1, G2, and C-tr were cloned into the pUASTattB vector81 at NotI and BglII, and transgenic strains were generated by ΦC31 integrase–based transgenesis. All transgenes were inserted into the attP2 chromosomal landing site (Genetic Services Inc., Cambridge, MA and BestGene Inc., Chino Hills, CA). Newly eclosed flies from each cross were collected on the same day and transferred to fresh vials. Adult flies were flipped every second day, grown on standard cornmeal fly food, yeast, and molasses at 24°C and incubated at 29°C after crossing with GAL4 drivers.
In Vivo Nephrocyte Filtration Assay
Pericardial nephrocytes of 1- and 15-day-old female adult flies were dissected in cold PBS and fixed in 4% paraformaldehyde diluted in PBS for 20 minutes at room temperature. Nephrocytes were washed twice with PBS and mounted in Roti-Mount FluorCare (Roth). Images for GFP and ANF-RFP fluorescence were taken with a confocal microscope Leica TCS SP8 SMD equipped with an HC PL APO CS2 40× oil objective with a numerical aperture of 1.3, and the imaging software LAS X from Leica was used for operating the system and image acquisition. The GFP and RFP were excited sequentially (sequential scan) at 488 nm (PMT detector; 1% laser intensity) and 561 nm (HyD detector; 30% sensitivity and 2% laser intensity), respectively. Images were taken with a depth of 12 bit in the spectral ranges of the emission at 498–552 nm (for GFP) and 571–684 nm (for RFP). Images were processed with ImageJ/Fiji.82
Extraction of Larval Hemolymph
Larval hemolymph extraction was performed as described in Musselman et al.83
Fly Eye Pathology
Flies were anesthetized, and their heads were detached. The heads were fixed with 4% paraformaldehyde and embedded in paraffin. Four–micrometer paraffin sections were mounted on Super FrostPlus Microscope Slides (Menzel-Glaser, Braunschweig, Germany) and stained with hematoxylin and eosin. Digital presentations were generated using an Olympus BX51 Microscope equipped with an Olympus DP70 Camera. Pictures were processed using the analySIS 5 software (Soft Imaging Systems, Muenster, Germany).
Generation of DNA Constructs
APOL1 WT full–length (isoform B1: 414 amino acids; transcript variant 2; NM_145343), G1, and G2 cDNAs were purchased from Hy-labs. The terminology of APOL1 domains is on the basis of the APOL1 A isoform (398 amino acids). The C-tr APOL1 was cloned to remove the SRA binding domain at amino acid 354 (for isoform B1).
For expression in yeast under the Gal1 promoter, APOL1 variants were cloned to high–copy p426-GAL1 (2 μ; URA3) vector using BamHI and EcoRI restriction sites or high–copy p425-GAL1 (2 μ; LEU2) using BamHI and XhoI sites. The S342G and I384M APOL1 mutated versions were generated by site-directed mutagenesis. The N–terminal mCherry-APOL1 vectors were cloned by fusion PCR to introduce the mCherry after the signal peptide (1–47 amino acids). The C–terminal mCherry constructs were cloned by introducing the mCherry at the C terminus.
All primers are listed in Supplemental Table 3.
Yeast Strains and Media
The S. cerevisiae strains used in this study are listed in Supplemental Table 4.84 The strains were grown at 30°C in standard yeast extract/peptone/dextrose (1% yeast extract, 2% peptone, and 2% dextrose), complete yeast nitrogen base medium (1.5 g yeast nitrogen base per 1 L, 5 g ammonium sulfate per 1 L, 2% glucose or galactose, and 0.1 g/L each amino acid with the appropriate amino acids removed as required for plasmid selection), or minimal medium (1.5 g yeast nitrogen base per 1 L, 5 g ammonium sulfate per 1 L, 2% glucose or galactose, and 0.1 g/L essential amino acids).
Yeast Survival Assay
Yeast cells were grown in minimal nitrogen base media with 2% raffinose overnight. For APOL1 induction, cells were diluted to OD=0.4 and grown in minimal nitrogen base media with 2% galactose for 0 (t=0) or 8 hours (t=8). Equal aliquots of cells (normalized to OD=2∙10−3) were then plated on complete nitrogen base plates without uracil. CFU were counted after 48 hours at 30°C.
Western Blotting and Antibodies
Lysate protein samples were separated by SDS-PAGE and blotted onto WesternBright NC Nitrocellulose Membranes (Advansta). The membranes were blocked with 5% nonfat dry milk (Santa Cruz) in Tris-buffered saline with Tween 20 and incubated with primary antibodies as indicated below. The membranes were then incubated with the appropriate secondary antibodies (as indicated below). After extensive washing in Tris-buffered saline with Tween 20, the proteins were visualized using chemiluminescence reagents.
Western Blot of APOL1 and β-Actin of Fly Heads and Bodies
Because protein lysates of bodies show an extensive representation of secreted hemolymph proteins (along with cellular protein), whereas protein lysates for heads predominantly represent cellular proteins with little or no secreted hemolymph proteins, cellular proteins, such as β-actin, are less abundant in the body compared with the head. Therefore, protein loading per lane was as follows: head: 20 μg and body: 50 μg.
Antibodies Used
The antibodies used were anti-APOL1 (1:1000; HPA018885; Sigma-Aldrich) and anti–β-actin (1:5000; ab8224; Abcam or 69100 MP for flies). Anti–rabbit HRP (1:10,000; 111–035–144; Jackson) and anti–mouse HRP (1:10,000; 115–035–166; Jackson) were used as secondary antibodies.
Quinacrine Staining
The yeast cells were grown for 2 hours on YEP-HEPES (pH 7.5) with 2% galactose media; 1.5×107 cells were incubated with 200 μM Quinacrine for 10 minutes, washed twice with cold YEP-HEPES, and micrographed at ×100 magnification, and images show both bright field and corresponding software pseudocolor of intensity. For the accompanying graph, >150 cells from at least four fields were counted.
Statistical Analyses
Drosophila Studies
Differences between proportions were tested using one-way ANOVA with post hoc Tukey HSD test. P values <0.05 were considered statistically significant.
Yeast Survival Assay
Differences between 0- and 8-hour survival ratios were tested using pairwise two–sample t tests for comparisons identified as being of primary biologic interest. Ratio SEMs were obtained by running a simulation of 10,000 runs for each APOL1 group (each with three repeats) incorporating measurement error inherent to the experiment. Measurement errors (estimated at 5%) are considered to be random (no systematic trend to over- or undercounting on the basis of experimental condition) and independent of other variables. A Bonferroni correction for multiple comparisons was applied, unadjusted P values are shown, and thresholds for significance were determined using Bonferroni correction for the number of comparisons tested (P<0.01).
Disclosures
None.
Supplementary Material
Acknowledgments
The authors acknowledge the expert and thoughtful input provided by Dr. Sara Selig. We also acknowledge M. Garfa-Traoré, N. Goudin (Necker Cell Imaging Facility), and Gwenn Le Meur for technical assistance as well as Amy Galick for assistance with statistical analyses.
We acknowledge the support provided by Israel Science Foundation grant 182/15, an academic research grant from GlaxoSmithKline, and from the Ernest and Bonnie Beutler Research Grant Program in Genomic Medicine.
Part of this research work was presented as an abstract at the 11th International Podocyte Conference (Haifa, Israel, April 3–6, 2016).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Identifying the Intracellular Function of APOL1,” on pages 1008–1011.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016050546/-/DCSupplemental.
References
- 1.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR: Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, Bekele E, Bradman N, Wasser WG, Behar DM, Skorecki K: Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet 128: 345–350, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, Friedman D, Briggs W, Dart R, Korbet S, Mokrzycki MH, Kimmel PL, Limou S, Ahuja TS, Berns JS, Fryc J, Simon EE, Smith MC, Trachtman H, Michel DM, Schelling JR, Vlahov D, Pollak M, Winkler CA: APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 22: 2129–2137, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Foster MC, Coresh J, Fornage M, Astor BC, Grams M, Franceschini N, Boerwinkle E, Parekh RS, Kao WH: APOL1 variants associate with increased risk of CKD among African Americans. J Am Soc Nephrol 24: 1484–1491, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Parsa A, Kao WH, Xie D, Astor BC, Li M, Hsu CY, Feldman HI, Parekh RS, Kusek JW, Greene TH, Fink JC, Anderson AH, Choi MJ, Wright JT Jr., Lash JP, Freedman BI, Ojo A, Winkler CA, Raj DS, Kopp JB, He J, Jensvold NG, Tao K, Lipkowitz MS, Appel LJ; AASK Study Investigators; CRIC Study Investigators : APOL1 risk variants, race, and progression of chronic kidney disease. N Engl J Med 369: 2183–2196, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kruzel-Davila E, Wasser WG, Aviram S, Skorecki K: APOL1 nephropathy: From gene to mechanisms of kidney injury. Nephrol Dial Transplant 31: 349–358, 2016 [DOI] [PubMed] [Google Scholar]
- 7.Kasembeli AN, Duarte R, Ramsay M, Mosiane P, Dickens C, Dix-Peek T, Limou S, Sezgin E, Nelson GW, Fogo AB, Goetsch S, Kopp JB, Winkler CA, Naicker S: APOL1 risk variants are strongly associated with HIV-associated nephropathy in black South Africans. J Am Soc Nephrol 26: 2882–2890, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Page NM, Butlin DJ, Lomthaisong K, Lowry PJ: The human apolipoprotein L gene cluster: Identification, classification, and sites of distribution. Genomics 74: 71–78, 2001 [DOI] [PubMed] [Google Scholar]
- 9.Monajemi H, Fontijn RD, Pannekoek H, Horrevoets AJ: The apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics 79: 539–546, 2002 [DOI] [PubMed] [Google Scholar]
- 10.Smith EE, Malik HS: The apolipoprotein L family of programmed cell death and immunity genes rapidly evolved in primates at discrete sites of host-pathogen interactions. Genome Res 19: 850–858, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vanhamme L, Paturiaux-Hanocq F, Poelvoorde P, Nolan DP, Lins L, Van Den Abbeele J, Pays A, Tebabi P, Van Xong H, Jacquet A, Moguilevsky N, Dieu M, Kane JP, De Baetselier P, Brasseur R, Pays E: Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature 422: 83–87, 2003 [DOI] [PubMed] [Google Scholar]
- 12.Pérez-Morga D, Vanhollebeke B, Paturiaux-Hanocq F, Nolan DP, Lins L, Homblé F, Vanhamme L, Tebabi P, Pays A, Poelvoorde P, Jacquet A, Brasseur R, Pays E: Apolipoprotein L-I promotes trypanosome lysis by forming pores in lysosomal membranes. Science 309: 469–472, 2005 [DOI] [PubMed] [Google Scholar]
- 13.Thomson R, Genovese G, Canon C, Kovacsics D, Higgins MK, Carrington M, Winkler CA, Kopp J, Rotimi C, Adeyemo A, Doumatey A, Ayodo G, Alper SL, Pollak MR, Friedman DJ, Raper J: Evolution of the primate trypanolytic factor APOL1. Proc Natl Acad Sci USA 111: E2130–E2139, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Papeta N, Kiryluk K, Patel A, Sterken R, Kacak N, Snyder HJ, Imus PH, Mhatre AN, Lawani AK, Julian BA, Wyatt RJ, Novak J, Wyatt CM, Ross MJ, Winston JA, Klotman ME, Cohen DJ, Appel GB, D’Agati VD, Klotman PE, Gharavi AG: APOL1 variants increase risk for FSGS and HIVAN but not IgA nephropathy. J Am Soc Nephrol 22: 1991–1996, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanji Z, Powe CE, Wenger JB, Huang C, Ankers E, Sullivan DA, Collerone G, Powe NR, Tonelli M, Bhan I, Bernhardy AJ, Dibartolo S, Friedman D, Genovese G, Pollak MR, Thadhani R: Genetic variation in APOL1 associates with younger age at hemodialysis initiation. J Am Soc Nephrol 22: 2091–2097, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tzur S, Rosset S, Skorecki K, Wasser WG: APOL1 allelic variants are associated with lower age of dialysis initiation and thereby increased dialysis vintage in African and Hispanic Americans with non-diabetic end-stage kidney disease. Nephrol Dial Transplant 27: 1498–1505, 2012 [DOI] [PubMed] [Google Scholar]
- 17.Divers J, Núñez M, High KP, Murea M, Rocco MV, Ma L, Bowden DW, Hicks PJ, Spainhour M, Ornelles DA, Kleiboeker SB, Duncan K, Langefeld CD, Turner J, Freedman BI: JC polyoma virus interacts with APOL1 in African Americans with nondiabetic nephropathy. Kidney Int 84: 1207–1213, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freedman BI, Skorecki K: Gene-gene and gene-environment interactions in apolipoprotein L1 gene-associated nephropathy. Clin J Am Soc Nephrol 9: 2006–2013, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nichols B, Jog P, Lee JH, Blackler D, Wilmot M, D’Agati V, Markowitz G, Kopp JB, Alper SL, Pollak MR, Friedman DJ: Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int 87: 332–342, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capewell P, Cooper A, Clucas C, Weir W, Macleod A: A co-evolutionary arms race: Trypanosomes shaping the human genome, humans shaping the trypanosome genome. Parasitology 142[Suppl 1]: S108–S119, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnstone DB, Shegokar V, Nihalani D, Rathore YS, Mallik L, Ashish, Zare V, Ikizler HO, Powar R, Holzman LB: APOL1 null alleles from a rural village in India do not correlate with glomerulosclerosis. PLoS One 7: e51546, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lan X, Jhaveri A, Cheng K, Wen H, Saleem MA, Mathieson PW, Mikulak J, Aviram S, Malhotra A, Skorecki K, Singhal PC: APOL1 risk variants enhance podocyte necrosis through compromising lysosomal membrane permeability. Am J Physiol Renal Physiol 307: F326–F336, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lan X, Wen H, Lederman R, Malhotra A, Mikulak J, Popik W, Skorecki K, Singhal PC: Protein domains of APOL1 and its risk variants. Exp Mol Pathol 99: 139–144, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan X, Wen H, Saleem MA, Mikulak J, Malhotra A, Skorecki K, Singhal PC: Vascular smooth muscle cells contribute to APOL1-induced podocyte injury in HIV milieu. Exp Mol Pathol 98: 491–501, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng D, Weckerle A, Yu Y, Ma L, Zhu X, Murea M, Freedman BI, Parks JS, Shelness GS: Biogenesis and cytotoxicity of APOL1 renal risk variant proteins in hepatocytes and hepatoma cells. J Lipid Res 56: 1583–1593, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khatua AK, Cheatham AM, Kruzel ED, Singhal PC, Skorecki K, Popik W: Exon 4 encoded sequence is a major determinant of cytotoxicity of apolipoprotein L1. Am J Physiol Cell Physiol 309: C22–C37, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olabisi OA, Zhang JY, VerPlank L, Zahler N, DiBartolo S III, Heneghan JF, Schlöndorff JS, Suh JH, Yan P, Alper SL, Friedman DJ, Pollak MR: APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc Natl Acad Sci USA 113: 830–837, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Olabisi O, Al-Romaih K, Henderson J, Tomar R, Drummond I, MacRae C, Pollak M: From man to fish: What can Zebrafish tell us about ApoL1 nephropathy? [published online ahead of print August 10, 2016]. Clin Nephrol doi: 10.5414/CNP86S116 [DOI] [PubMed] [Google Scholar]
- 29.Heneghan JF, Vandorpe DH, Shmukler BE, Giovinazzo JA, Raper J, Friedman DJ, Pollak MR, Alper SL: BH3 domain-independent apolipoprotein L1 toxicity rescued by BCL2 prosurvival proteins. Am J Physiol Cell Physiol 309: C332–C347, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Limou S, Dummer PD, Nelson GW, Kopp JB, Winkler CA: APOL1 toxin, innate immunity, and kidney injury. Kidney Int 88: 28–34, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raper J, Nussenzweig V, Tomlinson S: The main lytic factor of Trypanosoma brucei brucei in normal human serum is not high density lipoprotein. J Exp Med 183: 1023–1029, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weckerle A, Snipes JA, Cheng D, Gebre AK, Reisz JA, Murea M, Shelness GS, Hawkins GA, Furdui CM, Freedman BI, Parks JS, Ma L: Characterization of circulating APOL1 protein complexes in African Americans. J Lipid Res 57: 120–130, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vanhollebeke B, Pays E: The trypanolytic factor of human serum: Many ways to enter the parasite, a single way to kill. Mol Microbiol 76: 806–814, 2010 [DOI] [PubMed] [Google Scholar]
- 34.Hu CA, Klopfer EI, Ray PE: Human apolipoprotein L1 (ApoL1) in cancer and chronic kidney disease. FEBS Lett 586: 947–955, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Madhavan SM, O’Toole JF, Konieczkowski M, Ganesan S, Bruggeman LA, Sedor JR: APOL1 localization in normal kidney and nondiabetic kidney disease. J Am Soc Nephrol 22: 2119–2128, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ma L, Shelness GS, Snipes JA, Murea M, Antinozzi PA, Cheng D, Saleem MA, Satchell SC, Banas B, Mathieson PW, Kretzler M, Hemal AK, Rudel LL, Petrovic S, Weckerle A, Pollak MR, Ross MD, Parks JS, Freedman BI: Localization of APOL1 protein and mRNA in the human kidney: Nondiseased tissue, primary cells, and immortalized cell lines. J Am Soc Nephrol 26: 339–348, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reeves-Daniel AM, DePalma JA, Bleyer AJ, Rocco MV, Murea M, Adams PL, Langefeld CD, Bowden DW, Hicks PJ, Stratta RJ, Lin JJ, Kiger DF, Gautreaux MD, Divers J, Freedman BI: The APOL1 gene and allograft survival after kidney transplantation. Am J Transplant 11: 1025–1030, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee BT, Kumar V, Williams TA, Abdi R, Bernhardy A, Dyer C, Conte S, Genovese G, Ross MD, Friedman DJ, Gaston R, Milford E, Pollak MR, Chandraker A: The APOL1 genotype of African American kidney transplant recipients does not impact 5-year allograft survival. Am J Transplant 12: 1924–1928, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Freedman BI, Julian BA, Pastan SO, Israni AK, Schladt D, Gautreaux MD, Hauptfeld V, Bray RA, Gebel HM, Kirk AD, Gaston RS, Rogers J, Farney AC, Orlando G, Stratta RJ, Mohan S, Ma L, Langefeld CD, Hicks PJ, Palmer ND, Adams PL, Palanisamy A, Reeves-Daniel AM, Divers J: Apolipoprotein L1 gene variants in deceased organ donors are associated with renal allograft failure. Am J Transplant 15: 1615–1622, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freedman BI, Pastan SO, Israni AK, Schladt D, Julian BA, Gautreaux MD, Hauptfeld V, Bray RA, Gebel HM, Kirk AD, Gaston RS, Rogers J, Farney AC, Orlando G, Stratta RJ, Mohan S, Ma L, Langefeld CD, Bowden DW, Hicks PJ, Palmer ND, Palanisamy A, Reeves-Daniel AM, Brown WM, Divers J: APOL1 genotype and kidney transplantation outcomes from deceased African American donors. Transplantation 100: 194–202, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Molina-Portela MP, Samanovic M, Raper J: Distinct roles of apolipoprotein components within the trypanosome lytic factor complex revealed in a novel transgenic mouse model. J Exp Med 205: 1721–1728, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lecordier L, Vanhollebeke B, Poelvoorde P, Tebabi P, Paturiaux-Hanocq F, Andris F, Lins L, Pays E: C-terminal mutants of apolipoprotein L-I efficiently kill both Trypanosoma brucei brucei and Trypanosoma brucei rhodesiense. PLoS Pathog 5: e1000685, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weavers H, Prieto-Sánchez S, Grawe F, Garcia-López A, Artero R, Wilsch-Bräuninger M, Ruiz-Gómez M, Skaer H, Denholm B: The insect nephrocyte is a podocyte-like cell with a filtration slit diaphragm. Nature 457: 322–326, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang F, Zhao Y, Han Z: An in vivo functional analysis system for renal gene discovery in Drosophila pericardial nephrocytes. J Am Soc Nephrol 24: 191–197, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhuang S, Shao H, Guo F, Trimble R, Pearce E, Abmayr SM: Sns and Kirre, the Drosophila orthologs of Nephrin and Neph1, direct adhesion, fusion and formation of a slit diaphragm-like structure in insect nephrocytes. Development 136: 2335–2344, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ito K, Bick AG, Flannick J, Friedman DJ, Genovese G, Parfenov MG, Depalma SR, Gupta N, Gabriel SB, Taylor HA Jr., Fox ER, Newton-Cheh C, Kathiresan S, Hirschhorn JN, Altshuler DM, Pollak MR, Wilson JG, Seidman JG, Seidman C: Increased burden of cardiovascular disease in carriers of APOL1 genetic variants. Circ Res 114: 845–850, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wan G, Zhaorigetu S, Liu Z, Kaini R, Jiang Z, Hu CA: Apolipoprotein L1, a novel Bcl-2 homology domain 3-only lipid-binding protein, induces autophagic cell death. J Biol Chem 283: 21540–21549, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hartleben B, Gödel M, Meyer-Schwesinger C, Liu S, Ulrich T, Köbler S, Wiech T, Grahammer F, Arnold SJ, Lindenmeyer MT, Cohen CD, Pavenstädt H, Kerjaschki D, Mizushima N, Shaw AS, Walz G, Huber TB: Autophagy influences glomerular disease susceptibility and maintains podocyte homeostasis in aging mice. J Clin Invest 120: 1084–1096, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen J, Chen MX, Fogo AB, Harris RC, Chen JK: mVps34 deletion in podocytes causes glomerulosclerosis by disrupting intracellular vesicle trafficking. J Am Soc Nephrol 24: 198–207, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bechtel W, Helmstädter M, Balica J, Hartleben B, Kiefer B, Hrnjic F, Schell C, Kretz O, Liu S, Geist F, Kerjaschki D, Walz G, Huber TB: Vps34 deficiency reveals the importance of endocytosis for podocyte homeostasis. J Am Soc Nephrol 24: 727–743, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akilesh S, Huber TB, Wu H, Wang G, Hartleben B, Kopp JB, Miner JH, Roopenian DC, Unanue ER, Shaw AS: Podocytes use FcRn to clear IgG from the glomerular basement membrane. Proc Natl Acad Sci USA 105: 967–972, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Soda K, Ishibe S: The function of endocytosis in podocytes. Curr Opin Nephrol Hypertens 22: 432–438, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sampogna RV, Al-Awqati Q: Taking a bite: Endocytosis in the maintenance of the slit diaphragm. J Clin Invest 122: 4330–4333, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Soda K, Balkin DM, Ferguson SM, Paradise S, Milosevic I, Giovedi S, Volpicelli-Daley L, Tian X, Wu Y, Ma H, Son SH, Zheng R, Moeckel G, Cremona O, Holzman LB, De Camilli P, Ishibe S: Role of dynamin, synaptojanin, and endophilin in podocyte foot processes. J Clin Invest 122: 4401–4411, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Backer JM: The regulation and function of Class III PI3Ks: Novel roles for Vps34. Biochem J 410: 1–17, 2008 [DOI] [PubMed] [Google Scholar]
- 56.Kihara A, Noda T, Ishihara N, Ohsumi Y: Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol 152: 519–530, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim J, Kim YC, Fang C, Russell RC, Kim JH, Fan W, Liu R, Zhong Q, Guan KL: Differential regulation of distinct Vps34 complexes by AMPK in nutrient stress and autophagy. Cell 152: 290–303, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nakatogawa H, Suzuki K, Kamada Y, Ohsumi Y: Dynamics and diversity in autophagy mechanisms: Lessons from yeast. Nat Rev Mol Cell Biol 10: 458–467, 2009 [DOI] [PubMed] [Google Scholar]
- 59.Obara K, Sekito T, Ohsumi Y: Assortment of phosphatidylinositol 3-kinase complexes--Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell 17: 1527–1539, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Katzmann DJ, Babst M, Emr SD: Ubiquitin-dependent sorting into the multivesicular body pathway requires the function of a conserved endosomal protein sorting complex, ESCRT-I. Cell 106: 145–155, 2001 [DOI] [PubMed] [Google Scholar]
- 61.Eskelinen EL, Tanaka Y, Saftig P: At the acidic edge: Emerging functions for lysosomal membrane proteins. Trends Cell Biol 13: 137–145, 2003 [DOI] [PubMed] [Google Scholar]
- 62.Corbacho I, Teixidó F, Olivero I, Hernández LM: Dependence of Saccharomyces cerevisiae Golgi functions on V-ATPase activity. FEMS Yeast Res 12: 341–350, 2012 [DOI] [PubMed] [Google Scholar]
- 63.Smardon AM, Diab HI, Tarsio M, Diakov TT, Nasab ND, West RW, Kane PM: The RAVE complex is an isoform-specific V-ATPase assembly factor in yeast. Mol Biol Cell 25: 356–367, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kane PM: The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol Mol Biol Rev 70: 177–191, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huotari J, Helenius A: Endosome maturation. EMBO J 30: 3481–3500, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ko CH, Gaber RF: TRK1 and TRK2 encode structurally related K+ transporters in Saccharomyces cerevisiae. Mol Cell Biol 11: 4266–4273, 1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Toshima JY, Nishinoaki S, Sato Y, Yamamoto W, Furukawa D, Siekhaus DE, Sawaguchi A, Toshima J: Bifurcation of the endocytic pathway into Rab5-dependent and -independent transport to the vacuole. Nat Commun 5: 3498, 2014 [DOI] [PubMed] [Google Scholar]
- 68.Genovese G, Friedman DJ, Pollak MR: APOL1 variants and kidney disease in people of recent African ancestry. Nat Rev Nephrol 9: 240–244, 2013 [DOI] [PubMed] [Google Scholar]
- 69.Kim HJ, Zhong Q, Sheng ZH, Yoshimori T, Liang C, Jung JU: Beclin-1-interacting autophagy protein Atg14L targets the SNARE-associated protein Snapin to coordinate endocytic trafficking. J Cell Sci 125: 4740–4750, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Liang C, Lee JS, Inn KS, Gack MU, Li Q, Roberts EA, Vergne I, Deretic V, Feng P, Akazawa C, Jung JU: Beclin1-binding UVRAG targets the class C Vps complex to coordinate autophagosome maturation and endocytic trafficking. Nat Cell Biol 10: 776–787, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sharma AK, Friedman DJ, Pollak MR, Alper SL: Structural characterization of the C-terminal coiled-coil domains of wild-type and kidney disease-associated mutants of apolipoprotein L1. FEBS J 283: 1846–1862, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vanwalleghem G, Fontaine F, Lecordier L, Tebabi P, Klewe K, Nolan DP, Yamaryo-Botté Y, Botté C, Kremer A, Burkard GS, Rassow J, Roditi I, Pérez-Morga D, Pays E: Coupling of lysosomal and mitochondrial membrane permeabilization in trypanolysis by APOL1. Nat Commun 6: 8078, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Thomson R, Finkelstein A: Human trypanolytic factor APOL1 forms pH-gated cation-selective channels in planar lipid bilayers: Relevance to trypanosome lysis. Proc Natl Acad Sci USA 112: 2894–2899, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Stutz A, Golenbock DT, Latz E: Inflammasomes: Too big to miss. J Clin Invest 119: 3502–3511, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dummer PD, Limou S, Rosenberg AZ, Heymann J, Nelson G, Winkler CA, Kopp JB: APOL1 kidney disease risk variants: An evolving landscape. Semin Nephrol 35: 222–236, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Taylor HE, Khatua AK, Popik W: The innate immune factor apolipoprotein L1 restricts HIV-1 infection. J Virol 88: 592–603, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Garrus JE, von Schwedler UK, Pornillos OW, Morham SG, Zavitz KH, Wang HE, Wettstein DA, Stray KM, Côté M, Rich RL, Myszka DG, Sundquist WI: Tsg101 and the vacuolar protein sorting pathway are essential for HIV-1 budding. Cell 107: 55–65, 2001 [DOI] [PubMed] [Google Scholar]
- 78.Martin-Serrano J, Zang T, Bieniasz PD: HIV-1 and Ebola virus encode small peptide motifs that recruit Tsg101 to sites of particle assembly to facilitate egress. Nat Med 7: 1313–1319, 2001 [DOI] [PubMed] [Google Scholar]
- 79.Zhang F, Zhao Y, Chao Y, Muir K, Han Z: Cubilin and amnionless mediate protein reabsorption in Drosophila nephrocytes. J Am Soc Nephrol 24: 209–216, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.St Pierre SE, Ponting L, Stefancsik R, McQuilton P; FlyBase Consortium : FlyBase 102--advanced approaches to interrogating FlyBase. Nucleic Acids Res 42: D780–D788, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bischof J, Maeda RK, Hediger M, Karch F, Basler K: An optimized transgenesis system for Drosophila using germ-line-specific phiC31 integrases. Proc Natl Acad Sci USA 104: 3312–3317, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A: Fiji: An open-source platform for biological-image analysis. Nat Methods 9: 676–682, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Musselman LP, Fink JL, Narzinski K, Ramachandran PV, Hathiramani SS, Cagan RL, Baranski TJ: A high-sugar diet produces obesity and insulin resistance in wild-type Drosophila. Dis Model Mech 4: 842–849, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Spokoini R, Moldavski O, Nahmias Y, England JL, Schuldiner M, Kaganovich D: Confinement to organelle-associated inclusion structures mediates asymmetric inheritance of aggregated protein in budding yeast. Cell Rep 2: 738–747, 2012 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.