Abstract
APOL1 G1 and G2 variants facilitate kidney disease in blacks. To elucidate the pathways whereby these variants contribute to disease pathogenesis, we established HEK293 cell lines stably expressing doxycycline-inducible (Tet-on) reference APOL1 G0 or the G1 and G2 renal-risk variants, and used Illumina human HT-12 v4 arrays and Affymetrix HTA 2.0 arrays to generate global gene expression data with doxycycline induction. Significantly altered pathways identified through bioinformatics analyses involved mitochondrial function; results from immunoblotting, immunofluorescence, and functional assays validated these findings. Overexpression of APOL1 by doxycycline induction in HEK293 Tet-on G1 and G2 cells led to impaired mitochondrial function, with markedly reduced maximum respiration rate, reserve respiration capacity, and mitochondrial membrane potential. Impaired mitochondrial function occurred before intracellular potassium depletion or reduced cell viability occurred. Analysis of global gene expression profiles in nondiseased primary proximal tubule cells from black patients revealed that the nicotinate phosphoribosyltransferase gene, responsible for NAD biosynthesis, was among the top downregulated transcripts in cells with two APOL1 renal-risk variants compared with those without renal-risk variants; nicotinate phosphoribosyltransferase also displayed gene expression patterns linked to mitochondrial dysfunction in HEK293 Tet-on APOL1 cell pathway analyses. These results suggest a pivotal role for mitochondrial dysfunction in APOL1-associated kidney disease.
Keywords: chronic kidney disease, APOL1, mitochondria
The association between APOL1 G1 and G2 renal-risk variants and nondiabetic CKD has transformed our understanding of glomerulosclerosis. HIV-associated nephropathy, FSGS, hypertension-attributed ESRD, severe lupus nephritis, and sickle cell nephropathy reside in the APOL1-associated nephropathy spectrum and contribute to approximately 70% of nondiabetic ESRD in blacks.1–6
The frequency of APOL1 renal-risk variants has increased likely because of the protection they provide from the parasite causing African sleeping sickness during natural selection.1 Intrinsic APOL1 gene expression in kidney cells appears critical for development of nephropathy because circulating APOL1 protein concentrations are independent of the APOL1 genotype,7,8 and donor (but not recipient) APOL1 variation contributes to poorer allograft survival after kidney transplantation.9–11 APOL1 protein and mRNA are present in human podocytes, renal tubule cells, and glomerular endothelial cells.12,13 Hence, kidney synthesized APOL1 risk variants likely contribute to glomerulosclerosis.
To detect APOL1-associated pathways potentially involved in nephropathy, global gene expression profiles were determined using a cell model transfected with APOL1 G0, G1, and G2 in tetracycline (Tet)-inducible HEK Tet-on 3G cell lines derived from HEK293 cells. These cells stably expressed APOL1 G0, G1, and G2 protein at similar levels when induced by low dose doxycycline (Dox), a derivative of Tet, whereas cell viability was minimally effected. Functional assays were used to validate bioinformatics findings. Additional experiments were performed using nondiseased primary renal proximal tubule cells (PTCs) from black patients to determine whether the most differentially expressed genes attributed to APOL1 renal-risk variants replicated findings from the HEK293 Tet-on cell model.
Results
Cell Model and Global Gene Expression
As summarized in the study design (Supplemental Figure 1), global gene expression profiles of HEK293 Tet-on APOL1 (G0, G1, and G2) cells were analyzed in paired and pattern-based analyses to identify affected pathways through enrichment of key transcripts.
Cell Viability of HEK293 Cells Stably Expressing APOL1 G0, G1, and G2
Before induction, HEK293 Tet-on APOL1 G0, G1, and G2 variant cells (including those stably transfected with empty pTRE2hyg vector) had similar viability. With Dox-induction, empty vector (EV), G0, G1, and G2 cells remained viable until 16 hours, when G1 and G2 cells showed <10% reduction in viability (Supplemental Figure 2).
APOL1 mRNA and Protein Levels in HEK293 Cells Stably Expressing G0, G1, and G2
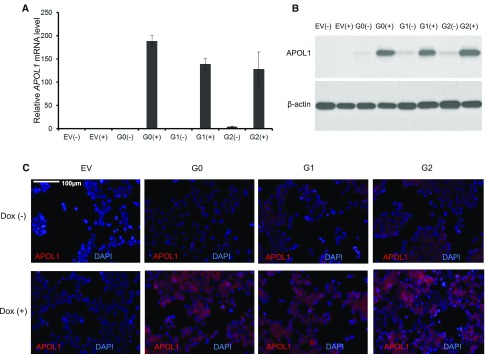
Comparison of global gene expression pattern changes across HEK293 Tet-on APOL1 (G0, G1, and G2) cell lines relied on similar APOL1 expression among cell lines with Dox induction. By adjusting final concentrations of Dox in the culture media, the APOL1 G0 mRNA level, determined by RT-PCR, was intentionally set slightly higher than G1 and G2 (Figure 1A) to ensure the gene expression profile changes attributed to G1 and G2 renal-risk variants could be convincingly detected relative to G0 at 16 hours postinduction. APOL1 protein levels, estimated by immunoblot and immunofluorescence (Figure 1, B and C, respectively), were comparable among the three APOL1 genotypes. As anticipated, APOL1 expression was low in the absence of Dox induction.
Figure 1.
APOL1 expression levels with Dox induction in HEK293 Tet-on G0, G1, and G2 cells are comparable. (A) Relative APOL1 mRNA levels in HEK293 Tet-on cells stably expressing G0, G1, or G2 APOL1 variants. HEK293 Tet-on cells with (+) or without (−) Dox induction were grown in complete DMEM growth media for 16 hours. Without Dox induction, APOL1 mRNA levels in HEK293 Tet-on cells were negligible compared with Dox induction. The ΔΔCT method was used to quantify relative APOL1 mRNA expression. Fold changes were normalized to mRNA levels of G0 cells without Dox induction. Data are expressed as mean±SD. APOL1 mRNA levels in G0 cells without Dox induction set at 1 (reference). (B) Relative APOL1 levels from total protein lysates in HEK293 Tet-on cells stably expressing G0, G1, or G2 APOL1 variants. HEK293 Tet-on cells with (+) or without (−) Dox induction were grown in complete DMEM growth media for 16 hours. Four micrograms of total cell lysate protein was loaded onto a 4%–20% SDS-PAGE gel and probed with APOL1 antibody. HEK293 Tet-on empty pTRE2hyg vector cells did not express APOL1 with or without Dox induction. HEK293 Tet-on APOL1 G0, G1, and G2 cells expressed similar amounts of APOL1 with Dox induction, whereas trace amounts of APOL1 were present in HEK293 Tet-on cells without Dox induction. Data are representative of three trials of immunoblotting with similar results. (C) APOL1 immunofluorescence in HEK293 Tet-on cells stably expressing G0, G1, or G2 APOL1 variants with and without Dox induction. HEK293 Tet-on cells grown with (+) or without (−) Dox for 16 hours. After washing with PBS and fixing with 4% paraformaldehyde, cells were stained for APOL1 (red) with Epitomics anti-APOL1 antibody and counterstained with DAPI (blue). Elevated APOL1 signal intensities were comparable in HEK293 Tet-on G0, G1, and G2 cells with Dox induction, whereas APOL1 was absent in HEK293 Tet-on empty pTRE2hyg vector cells. EV, HEK293 Tet-on cell line with empty pTRE2hyg plasmid; DAPI, 4′,6-diamidino-2-phenylindole.
Pathway Analysis of HEK293 Cells Stably Expressing G0, G1, and G2
To maximize the effect of G1 and G2 renal-risk variants to differential gene expression versus G0, Dox induction was terminated at 16 hours, along with the noninduced controls. RNAs were collected to compare gene expression. Illumina human HT-12 v4 array-based paired analyses were performed as outlined in Supplemental Figure 1. Descriptions of paired analyses (methods and purpose) are summarized in Table 1. Gene expression profile changes were negligible in paired analysis for HEK293 Tet-on EV cells with and without Dox induction (data not shown). Absent Dox induction, pair-wise analyses among HEK293 Tet-on G0, G1, and G2 cells did not reveal significant pathways attributed to APOL1 renal-risk variants (data not shown).
Table 1.
Description of paired analysis for HEK293 Tet-on APOL1 cells using Illumina HT-12 v4 arrays
| Paired Analysis | Purpose |
|---|---|
| EV(+) versus EV(−) | Clarify the effect of Dox on global gene expression |
| G1(−) versus G0(−) | |
| G2(−) versus G0(−) | Test differential gene expression by APOL1 genotype at baseline (Dox absent) |
| G1(−) versus G2(−) | |
| G0(+) versus G0(−) | Examine the effect of reference APOL1 (G0) when overexpressed with Dox induction |
| G1(+) versus G1(−) | Test differentially expressed genes attributed to overexpression of G1 |
| G2(+) versus G2(−) | Test differentially expressed genes attributed to overexpression of G2 |
| G1(+) versus G0(+) | Explore APOL1 G1-attributed gene expression (compared with G0) with Dox induction |
| G2(+) versus G0(+) | Explore APOL1 G2-attributed gene expression (compared with G0) with Dox induction |
| G0(−) versus EV(−) | |
| G1(−) versus EV(−) | Narrow down the corresponding pathway alterations of interest under Dox induction |
| G2(−) versus EV(−) |
Relative global gene expression levels were determined for stabilized HEK293 Tet-on APOL1 cells grown with (+) or without (−) Dox induction. EV, HEK293 Tet-on cell line with empty pTRE2hyg plasmid.
HEK293 Tet-on APOL1 G0 cells with and without Dox induction revealed 1056 differentially expressed genes. These were analyzed using Cytoscape-based BiNGO software to identify associated pathways (Supplemental Table 1), and Ingenuity Pathway Analysis (IPA) software to highlight top canonical pathways and upstream regulators (Supplemental Tables 2 and 3). With Dox induction, key components of endoplasmic reticulum (ER) stress were significantly suppressed (Supplemental Figure 3), and genes involved in inhibition of N-linked glycosylation (inhibited by tunicamycin) were downregulated (Supplemental Table 3). Unfolded protein response (ER stress response) inhibition appeared to play an important role in the biologic function of APOL1 G0. Unlike in HEK293 G0 cells, the ER stress pathway was not inhibited by Dox-induced APOL1 G1 and G2 variants in HEK293 Tet-on cells (compared with corresponding baseline cells without Dox induction).
Differentially expressed genes by Dox-induced APOL1 overexpression (versus without Dox-induction) in the HEK293 G1 Tet-on cell line revealed the top associated pathways involved the nucleus and membrane-bound organelles, including mitochondria (Supplemental Table 4). Differentially expressed genes by Dox-induced APOL1 overexpression in the HEK293 G2 Tet-on cell line (versus without Dox-induction) indicated similar pathways (Supplemental Table 5) as HEK293 G1 cells. Paired comparisons for Dox-induced HEK293 Tet-on APOL1 G1 versus G0 and G2 versus G0 cells were performed to assess potential biologic functions of APOL1 renal-risk variants. On the basis of the differentially expressed genes, Cytoscape-based BiNGO identified common pathways related to the function of intracellular membrane-bound organelles, including mitochondria (Supplemental Tables 6 and 7).
To narrow down the differentially expressed genes attributed to overexpression of APOL1 and its G1 and G2 risk variants, additional comparisons were performed for baseline HEK293 G0 versus EV, G1 versus EV, and G2 versus EV cells in the absence of Dox. ER components were effected for G0, G1, and G2 cells, in contrast to EV cells where APOL1 was virtually absent (data not shown). This is not unexpected because APOL1 protein is secreted; however, in contrast to Dox-induced HEK293 G1 and G2 cells (Supplemental Tables 4 and 5), mitochondrial or nucleus pathways were not involved (data not shown). These data, along with pair-wise analyses among baseline HEK293 Tet-on G0, G1, and G2 cells, suggest the need for higher APOL1 expression to distinguish specific pathways attributed to APOL1 renal-risk variants.
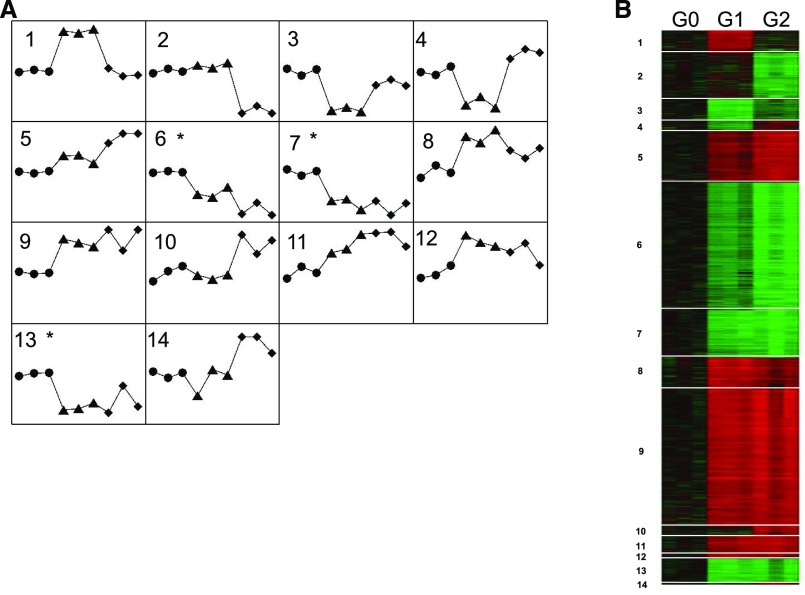
To increase power for identifying pathways attributable to APOL1 renal-risk variants, transcript pattern analyses were performed using the java-based gene expression profile clustering method, Extracting microarray gene expression Patterns and Identifying coexpressed Genes (EPIG; http://www.niehs.nih.gov/research/resources/software/biostatistics/epig/index.cfm) to extract gene expression profiles of HEK293 Tet-on APOL1 G0, G1, and G2 cells with Dox induction on Illumina human HT-12 v4 arrays. EPIG identified 1699 significantly effected genes and 14 patterns (Figure 2A for line patterns; Figure 2B for heat map) in Illumina analysis mode. The top three pathways for optimized patterns are summarized in Table 2 on the basis of Cytoscape BiNGO; additional information is provided in Supplemental Table 8. For Illumina analysis mode, patterns 6, 7, and 13 were enriched with mitochondrial pathways (false discovery rate [FDR] P=1.33×10−27). Patterns 5 and 9 appear to represent top pathways involved in gene regulation by nuclear transcription factor(s) (FDR P=1.01×10−14).
Figure 2.
Illumina human HT-12 v4 arrays identify significantly clustered gene expression patterns for HEK293 Tet-on APOL1 G0, G1, and G2 cells. From top to bottom are 1699 transcripts, in order from pattern 1 to pattern 14. (A) Gene expression patterns most altered in HEK293 Tet-on G0 (circle), G1 (triangle), and G2 (diamond) cells with Dox induction. Asterisk indicates that mitochondrial pathway enrichment is present (patterns 6, 7, and 13). (B) Heat map of clustered gene expression patterns: transcript expression in G0 shown in black; red and green correspond to up and downregulation, respectively, by G1 or G2. Lighter colors denote less differential expression.
Table 2.
Pattern-based pathway analysis on Illumina human HT-12 v4 arrays and Affymetrix HTA 2.0 arrays using Cytoscape based BiNGO
| Analysis Mode | Pattern | GO ID | Description | x | n | X | N | P Value | FDR P Value |
|---|---|---|---|---|---|---|---|---|---|
| Illumina | 5 | 5634 | Nucleusa | 74 | 5183 | 130 | 17,783 | 3.30×10−11 | 6.90×10−08 |
| 50794 | Regulation of cellular process | 80 | 6221 | 130 | 17,783 | 5.50×10−10 | 5.76×10−07 | ||
| 50789 | Regulation of biologic process | 81 | 6551 | 130 | 17,783 | 3.02×10−09 | 1.35×10−06 | ||
| 6 | 5739 | Mitochondriona | 87 | 1273 | 342 | 17,780 | 2.80×10−26 | 7.93×10−23 | |
| 44444 | Cytoplasmic part | 169 | 5147 | 342 | 17,780 | 7.02×10−16 | 9.92×10−13 | ||
| 44429 | Mitochondrial parta | 45 | 612 | 342 | 17,780 | 9.48×10−15 | 8.94×10−12 | ||
| 9 | 5634 | Nucleusa | 143 | 5181 | 317 | 17,767 | 9.40×10−10 | 2.19×10−06 | |
| 44451 | Nucleoplasm part | 34 | 585 | 317 | 17,767 | 1.49×10−09 | 2.19×10−06 | ||
| 5515 | Protein binding | 195 | 8110 | 317 | 17,767 | 7.50×10−09 | 5.48×10−06 | ||
| 7 and 13 | 5739 | Mitochondriona | 34 | 1272 | 191 | 17,785 | 6.66×10−07 | 1.34×10−03 | |
| 5 and 9 | 5634 | Nucleusa | 215 | 5180 | 443 | 17,761 | 2.83×10−18 | 1.01×10−14 | |
| 50794 | Regulation of cellular process | 239 | 6210 | 443 | 17,761 | 1.04×10−16 | 1.85×10−13 | ||
| 50789 | Regulation of biologic process | 246 | 6540 | 443 | 17,761 | 4.26×10−16 | 5.08×10−13 | ||
| 6, 7, and 13 | 5739 | Mitochondriona | 121 | 1273 | 533 | 17,774 | 3.82×10−31 | 1.33×10−27 | |
| 44444 | Cytoplasmic part | 248 | 5147 | 533 | 17,774 | 2.30×10−18 | 4.01×10−15 | ||
| 44429 | Mitochondrial parta | 62 | 612 | 533 | 17,774 | 3.06×10−17 | 3.55×10−14 | ||
| Affymetrix | 2 | 5739 | Mitochondriona | 274 | 1272 | 1357 | 17,736 | 9.68×10−61 | 5.19×10−57 |
| 43231 | Intracellular membrane-bounded organellea | 899 | 8330 | 1357 | 17,736 | 3.57×10−50 | 9.58×10−47 | ||
| 43227 | Membrane-bounded organellea | 899 | 8337 | 1357 | 17,736 | 5.61×10−50 | 1.00×10−46 | ||
| 4 | 5634 | Nucleusa | 543 | 5167 | 1020 | 17,727 | 6.00×10−62 | 3.06×10−58 | |
| 10468 | Regulation of gene expression | 364 | 2920 | 1020 | 17,727 | 1.49×10−53 | 3.79×10−50 | ||
| 60255 | Regulation of macromolecule metabolic process | 398 | 3369 | 1020 | 17,727 | 2.30×10−53 | 3.92×10−50 | ||
| 1 and 2 | 5739 | Mitochondriona | 288 | 1272 | 1468 | 17,733 | 3.07×10−61 | 1.70×10−57 | |
| 43231 | Intracellular membrane-bounded organelle | 976 | 8330 | 1468 | 17,733 | 9.56×10−56 | 2.65×10−52 | ||
| 43227 | Membrane-bounded organelle | 976 | 8337 | 1468 | 17,733 | 1.57×10−55 | 2.90×10−52 | ||
| 3 and 4 | 50794 | Regulation of cellular process | 728 | 6201 | 1346 | 17,713 | 2.90×10−50 | 1.69×10−46 | |
| 5634 | Nucleusa | 640 | 5167 | 1346 | 17,713 | 8.47×10−50 | 2.47×10−46 | ||
| 50789 | Regulation of biologic process | 750 | 6530 | 1346 | 17,713 | 1.55×10−48 | 3.01×10−45 |
Top three pathways for each pattern with FDR P<0.01; detail in Supplemental Tables 8 and 9. GO, gene ontology; x, number of gene IDs identified in the designated pathway among input gene IDs; n, number of known gene IDs in the designated pathway; X, number of input gene IDs selected by BiNGO; N, number of background gene IDs selected from the Gene Ontology pool by BiNGO.
Highly replicated pathway.
To verify pathways detected with Illumina HT-12 v4 arrays, pattern-based comparisons were repeated on Dox-induced HEK293 APOL1 Tet-on G0, G1, and G2 cell lines using the Affymetrix HTA 2.0 array gene expression platform, which probes all exons of known transcripts. Pattern-based analysis identified ten patterns based upon 3620 significantly effected genes, using EPIG (Supplemental Figure 4 for gene expression line patterns and heat map) in Affymetrix analysis mode. Table 2 summarizes the most significant pathways detected with Cytoscape BiNGO; additional information is provided in Supplemental Table 9. For Affymetrix analysis mode, pattern 2 highlighted mitochondrial pathways as top hits (FDR P=5.19×10−57). Pattern 4 contained nuclear regulation pathways via transcription factors (top FDR P=3.06×10−58). On the basis of similar trends for pattern 1 with 2, and pattern 3 with 4, integrated analyses were performed and results are summarized in Table 2 (detailed in Supplemental Table 9). Patterns 1, 2, 3, and 4 were verified using IPA software; this demonstrated that patterns 1 and 2 were enriched with mitochondrial dysfunction pathways (Figure 3, Supplemental Figure 5, Supplemental Table 10). In the IPA-based analysis, transfer RNA (tRNA) charging surpassed the mitochondrial dysfunction rating (Supplemental Table 10); all captured genes in the tRNA charging pathway encoded different aminoacyl tRNA synthetases and approximately 50% were mitochondria specific. This strengthened the hypothesis that APOL1 renal-risk variant-mediated mitochondrial dysfunction is a key pathway altering cellular metabolism. Patterns 3 and 4 represent top genes enriched in activated TGF-β signaling (Supplemental Figure 6, Supplemental Table 10) in the nuclear regulation pathway in paired and pattern-based analyses (Cytoscape-based BiNGO; Supplemental Table 10, Table 2). Mitochondrial dysfunction pathways topped common lists that were based on different analytic approaches.
Figure 3.
APOL1 G1 and G2 renal-risk variants contribute to mitochondrial dysfunction. Affymetrix patterns 1 and 2 were analyzed using IPA for HEK293 Tet-on APOL1 G1 and G2 versus G0 cells. Results indicate marked downregulation of enzymes in mitochondrial complexes I–V.
All relevant gene expression data performed on Illumina HT-12 v4 and Affymetrix HTA 2.0 arrays are available at http://www.ncbi.nlm.nih.gov/geo (accession ID: GSE85921).
Intracellular Potassium Concentrations in HEK293 Tet-on G0, G1, and G2 Cells
Intracellular potassium concentration [K+] was measured using the potassium-sensitive fluorescence indicator Asante Potassium Green-2 AM in HEK293 Tet-on G0, G1, and G2 cells, with and without Dox induction for 8 hours and 16 hours. Intracellular [K+] was comparable in Tet-on G0, G1, and G2 cells with or without 8-hour Dox induction. After 16 hours of Dox induction, intracellular [K+] differed between G0, G1, and G2 cells (ANOVA P=0.02); relative to G0, G1 cells had lower [K+] (P=0.01) and G2 cells trended toward lower [K+] (P=0.07) (Supplemental Figure 7).
APOL1 G1 and G2 Renal-Risk Variant Expression Results in Mitochondrial Dysfunction
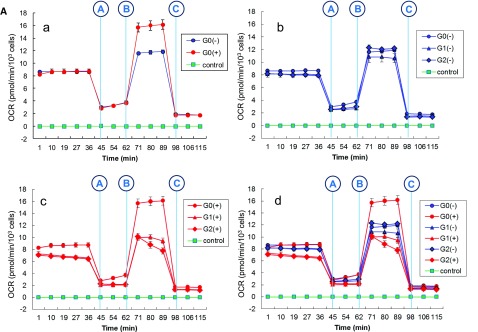
To assess whether mitochondrial dysfunction was attributable to APOL1 renal-risk variants in HEK293 Tet-on APOL1 cells, the 8-hour time point with Dox induction was chosen to perform mitochondrial respirometry. At 8 hours, cell viability and intracellular [K+] were similar for G0, G1, and G2 cells (Supplemental Figures 2 and 7); APOL1 expression levels were also comparable (Supplemental Figure 8). Immunofluorescence revealed that APOL1 colocalized with mitochondria in HEK293 Tet-on APOL1 cells with Dox induction by dual staining of ATP synthase 5A1 (ATP5A1, a mitochondrial marker) and APOL1 antibody (Figure 4, Supplemental Figure 9, Supplemental Video 1). Time effects on APOL1 expression are shown in Supplemental Figure 10 in noninduced controls, and after 4-hour and 8-hour Dox induction. With Dox induction, respirometric assessments indicated that the maximum oxygen consumption rate (ANOVA P<0.0001) and spare respiratory capacity (ANOVA P=0.0007) were lower in HEK293 Tet-on cells overexpressing APOL1 G1 and G2 variants, relative to G0 (Figure 5). The fluorescence signal intensity of mitochondrial membrane potential marker tetramethyl rhodamine ethyl ester (TMRE) was markedly reduced in Dox-induced HEK293 Tet-on G1 and G2 cells (Supplemental Figure 11, A and B). However, mitochondrial content on the basis of immunoblot analysis of cytochrome c oxidase subunit 4 (COXIV) and voltage-dependent anion-selective channel protein 1(VDAC1) in HEK293 Tet-on APOL1 cells was not different across the G0, G1, and G2 variants (Supplemental Figure 11C), as confirmed by comparable mRNA levels of VDAC1 and COXIV across G0, G1, and G2 cells on the Illumina and Affymetrix arrays (data not shown). This supports that APOL1 G1- or G2-mediated mitochondrial dysfunction resulted from impaired mitochondrial respiration and membrane potential, not reductions in mitochondrial mass. Carbonyl cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP) depolarization controls were provided for mitochondrial membrane potential in Dox-induced HEK293 Tet-on APOL1 cells (Supplemental Figure 12).
Figure 4.
APOL1 colocalizes with mitochondria in HEK293 Tet-on APOL1 cells with Dox induction. HEK293 Tet-on APOL1 cells with 8-hour Dox induction (stably expressing G0, G1, or G2 APOL1 variants) were washed with PBS and fixed with 4% paraformaldehyde. Cells were stained for APOL1 (red) with Epitomics anti-APOL1 antibody, mitochondria (green) with ATP synthase 5 A1 (ATP5A1) antibody, and counterstained with nuclear dye 4′,6-diamidino-2-phenylindole (DAPI; blue).
Figure 5.
APOL1 G1 and G2 renal-risk variants decrease mitochondrial respiration. (A) a-d: Mitochondrial stress tests in HEK293 Tet-on APOL1 cells. Cells were seeded on a V7 cell culture plate (Seahorse Bioscience) with final density 100,000 cells/well before assay. Effects of G0, G1, and G2 with (+) and without (−) Dox induction were compared using a Seahorse XF-24 extracellular flux analyzer to measure oxygen consumption rate (OCR), a measure of oxidative phosphorylation, in the presence of a series of metabolic inhibitors and uncoupling agents. The first injection (shown as vertical blue line A) was oligomycin, an inhibitor of ATP synthesis via blockade of the proton channel of ATP synthase (complex V). Oligomycin distinguishes the OCR from ATP synthesis by blocking the oxygen consumption required to overcome proton leakage across the inner mitochondrial membrane, and basal respiration rate, by blocking nonmitochondrial respiration. The second injection (shown as vertical blue line B) was FCCP, an uncoupling agent that disrupts ATP synthesis by transporting hydrogen ions across the mitochondrial membrane instead of the ATP synthase proton channel (complex V). Collapse of the mitochondrial membrane potential leads to rapid energy and oxygen consumption without generation of ATP. FCCP is used to calculate the spare respiration capacity of cells, defined as the quantitative difference between maximal and basal respiration rates. The third injection (shown as vertical blue line C) was a combination of rotenone, a complex I inhibitor, and antimycin A, a complex III inhibitor; this combination blocks mitochondrial respiration and enables calculation of mitochondrial and nonmitochondrial cellular respiration. At least three wells were assigned for each type of HEK293 Tet-on cells with (+) or without (−) Dox induction on the same 24-well V7 cell culture plate. Data were expressed as mean±SEM of each time point, grouped by HEK293 Tet-on G0, G1, and G2 cells with or without Dox induction. (B) Bioenergetic profiles for HEK293 Tet-on APOL1 G0, G1, and G2 cells. APOL1 stably transfected HEK293 cells were incubated with (+) or without (−) Dox for 8 hours before assay on a Seahorse XF-24 extracellular flux analyzer to measure the OCR. After a 15 minute equilibration period, three measures of OCR were made for each stage, i.e., preoligomycin, between oligomycin and FCCP, between FCCP and rotenone/antimycin, and postrotenone (injections denoted as vertical blue lines, see A). The average of three measures was used to represent the OCR of the corresponding stage for each well. Data were grouped by APOL1 genotype and Dox treatment (+/−). Basal respiration rate, maximum respiration rate, and spare respiration capacity were expressed as mean±SD; n refers to replicated well number for each type of HEK293 Tet-on cells on the 24-well V7 cell culture plate. (a) For G0 cells, induction of APOL1 did not alter the basal respiration rate (t test P=0.68), but improved maximum respiration rate and spare respiration capacity (t test P values 0.005 and 0.005, respectively). (b) For G1 cells, induction of APOL1 did not alter the basal respiration rate (t test P=0.08), maximum respiration rate, and spare respiration capacity (t test P values 0.48 and 0.68, respectively). (c) For G2 cells, induction of APOL1 diminished the basal respiration rate, maximum respiration rate, and spare respiration capacity (t test P values 0.002, 0.003, and 0.01, respectively). (d) Without Dox induction, G0, G1, and G2 cells appeared to have similar basal respiration rate, maximum respiration rate, and spare respiration capacity (ANOVA P=0.78, 0.22, and 0.13, respectively). (e) With Dox induction, the basal respiration rate, maximum respiration rate, and spare respiration capacity for G1 and G2 were significantly reduced (ANOVA P=0.004, <0.0001, and 0.0007, respectively) compared with G0. (f) Overview of bioenergetic profiles for HEK293 Tet-on APOL1 G0, G1, and G2 cells.
Differential Gene Expression by APOL1 Renal-Risk (Two Risk Alleles) versus Nonrisk (G0G0) Genotypes in Primary PTCs
Global gene expression (mRNA) levels were examined on Affymetrix HTA 2.0 arrays in primary PTCs cultured from nondiseased kidneys in 53 black patients without CKD who underwent nephrectomy for localized renal cell carcinoma. In this group, 25 had zero, 23 had one, and five had two APOL1 renal-risk alleles. To detect differentially expressed gene profiles attributable to APOL1 renal-risk genotypes, only PTCs with two renal-risk alleles (n=5) and PTCs lacking renal-risk alleles (n=25) were included in comparisons of global gene expression. Supplemental Table 11 lists the most differentially expressed genes; the fibrinogen β chain gene (FGB) topped the list. In pattern-based analysis, nicotinate phosphoribosyltransferase (NAPRT/NAPRT1) and zinc finger E-box binding homeobox 2 (ZEB2)/SMAD-interacting protein 1 (SMADIP1) appeared in pattern 7 and pattern 9, respectively (Figure 2, Supplemental Table 8), representing mitochondria- and nucleus-related pathways, respectively.
Discussion
Little is known regarding underlying mechanisms whereby APOL1 G1 and G2 renal-risk variants contribute to kidney disease. To assess differences in gene expression profiles relating to APOL1 variants, an HEK293 Tet-on cell model stably expressing Dox-inducible APOL1 G0, G1, and G2 was established. Relative to G0, modest overexpression of APOL1 G1 and G2 renal-risk variants led to compromised mitochondrial function. APOL1 expression was absent in parental HEK293 cells12 and EV HEK293 Tet-on cells (Figure 1), excluding interference from endogenous APOL1. Functional studies, including mitochondrial stress testing (respirometry), confirmed mitochondrial dysfunction in HEK293 cells stably overexpressing APOL1 G1 or G2 before reductions in cell viability or intracellular [K+]. Sixteen hours was chosen as the optimal time point in order to maximize the effect of G1 and G2 renal-risk variants on differential gene expression (relative to G0), because cell viability remained largely intact at that time point. At 24 hours, the viability of G0 cells began to decline, whereas that of G1 and G2 cells declined to a greater extent. Together with reduced NAPRT/NAPRT1 in primary PTCs from black patients with two APOL1 renal-risk variants (Supplemental Table 11), these findings support mitochondrial dysfunction in Dox-induced HEK293 G1 and G2 cells (versus G0) (Figure 2, Supplemental Table 8: pattern 7). Results strongly suggest a pivotal role for mitochondrial dysfunction in APOL1-associated kidney disease. The reduced NAPRT may lead to insufficient biosynthesis of NAD14 and result in mitochondrial dysfunction15 and CKD.16
Using a biotinylated membrane protein assay, Olabisi et al. reported that APOL1 was present on plasma membranes and that expression of APOL1 G1 and G2 renal-risk variants affected ion channels, decreased intracellular [K+], and resulted in cellular injury.17 We and others have shown that APOL1 resides mainly in internal organelles,12,13 typically in the ER18 and mitochondria (Figure 4, Supplemental Figure 9). We note the absence of a plasma membrane pattern for APOL1 localization compared with the report by Olabisi et al. We used monoclonal anti-APOL1 antibody (Epitomics) to localize cellular APOL1. The conformation of APOL1 in complexes on the cell membrane and in organelles may differ because of diverse protein and lipid compositions, potentially resulting in different exposed epitopes. Therefore, antibodies that successfully recognize intracellular APOL1 may not be able to capture plasma membrane-bound APOL1 as part of protein complexes. Importantly, our study found that compromised mitochondrial function occurred 8 hours after Dox induction, before G1- and G2-induced cell death or decreases in intracellular [K+] (measured with a fluorescence indicator in live cells; Supplemental Figure 7). This suggested that mitochondrial dysfunction was a cause, rather than a result of cell injury relating to intracellular [K+] depletion. As in Olabisi et al., we observed decreased intracellular [K+] but at a later time point (16 hours). This would be an expected finding in the presence of mitochondrial dysfunction. Although VDAC1 is also expressed in the plasma membrane, it mainly displays a cytoplasmic pattern in immunofluorescence ;studies. To assess mitochondrial mass, both outer and inner mitochondrial membrane markers were chosen (VDAC1 and COXIV, respectively; Supplemental Figure 11C) to quantify the corresponding mitochondrial marker proteins.19
Mitochondrial dysfunction negatively effects the activity of plasma membrane sodium-potassium ATPase. Subsequent reductions in intracellular ATP can result in failure to maintain transmembrane Na+/K+ gradients, leading to loss of the membrane potential, secondary Cl− and water influx, intracellular swelling, and accumulation of intracellular Ca2+, triggering downstream enzymatic cascades that lead to cell injury.20 The presence of APOL1 protein in mitochondria and other membrane-bound organelles, including ER, supports potential roles in the regulation of cellular metabolism and the stress response,18 although the precise role of mitochondrial APOL1 remains to be elucidated.
An unbiased gene expression pattern identification method (EPIG) was used to detect pathways induced by APOL1 renal-risk variants by grouping transcript patterns; this obviated arbitrary logFC thresholds. Without Dox induction, few pathways were distinguishable between HEK293 APOL1 G0, G1, and G2 cells. Therefore, only Dox-induced HEK293 Tet-on APOL1 cells were included in pattern-based pathway analyses. Unbiased global gene expression profiles with Dox induction were assessed in HEK293 Tet-on cells on both Illumina HT-12 v4 arrays and Affymetrix HTA 2.0 arrays, the latter covers all exons of transcripts in known UCSC (University of California Santa Cruz) genes, UCSC lincRNA transcripts, Ensembl, RefSeq, and Broad Institute Human Body Map lincRNAs and TUCP (transcripts of uncertain coding potential) catalog (www.affymetrix.com). Mitochondrial pathways appeared at the top of pattern-based analyses on both arrays.
Reductions in mitochondrial superoxide dismutase (mtSOD)/superoxide dismutase 2 (SOD2) and catalase (CAT) transcripts were also detected in APOL1 G1 and G2 expressing cells (Supplemental Figure 5; blue arrows). This could exacerbate reactive oxygen species (ROS)-mediated mitochondrial dysfunction. Decreases in SOD2/mtSOD may enhance mitochondrial oxidative damage.21,22 CAT is an enzyme that degrades ROS. It is possible that decreased SOD2 and CAT (pattern analysis shown in Supplemental Figure 5) worsen mitochondrial oxidative stress, resulting in inhibition of cellular respiration as seen in the mitochondrial stress test for APOL1 G1 and G2 variants (Figure 5).
The most significant differences in Dox-induced HEK293 G1 and G2 cells, evident after addition of the respiratory chain uncoupler FCCP, were the reduced maximal respiratory rate and spare respiratory capacity. Because uncouplers act as protonophores across all membranes, acidifying the cytosolic compartment and disrupting the function of endosomes and other compartments, it is possible that the reduced maximal respiratory rate observed after addition of FCCP in G1 and G2 resulted from endosomal disruption and reduced substrate delivery to the mitochondria (as APOL1 is not restricted to mitochondria). It is currently unclear how APOL1 renal-risk variants effect mitochondrial membrane proton permeability in human cells. However, the link between APOL1-induced trypanolysis and apoptosis-like mitochondrial membrane permeabilization23 may shed light on the potential mechanism of APOL1 renal-risk variant-mediated mitochondrial dysfunction in renal cells.
In addition, an upregulated TGF-β signaling pathway was detected with IPA (Supplemental Figure 6, Supplemental Table 10). ZEB2/SMADIP1, one of the top upregulated genes by APOL1 renal-risk variants in primary PTCs, is an essential component of the SMAD signaling pathway critical in transmitting the TGF-β signal to the nucleus and for regulation of transcriptional responses.24 An activated TGF-β pathway has long been recognized as a cause of FSGS and renal fibrosis.25 Mitochondrial and TGF-β pathways are also known to interact. Mitochondrial ROS regulate TGF-β signaling. Increased oxidative stress amplifies activation of the canonical TGF-β pathway, a major contributor to cellular fibrosis.26 Genetically disrupting mitochondrial complex III-generated ROS production attenuates TGF-β –induced profibrotic gene expression.27 TGF-β1 induces prolonged mitochondrial ROS generation through decreased complex IV activity with senescent arrest.28 Such crosstalk may be evident in our bioinformatics data.
Because the primary PTCs we analyzed were derived from nondiseased kidneys and APOL1 expression levels were lower than HEK293 Tet-on cells with Dox induction, effects of APOL1 risk alleles were more subtle. However, two of the most differentially expressed genes in primary PTCs corresponded to patterns in pathway analyses of HEK293 Tet-on APOL1 cells; these represented mitochondrial dysfunction and activated TGF-β signaling. ANOVA quantifies both “between” and “within” group variation. It is especially useful when sample sizes are small and unequal variances are present. We used both fold-change and P values to define differential gene expression. The P values in Supplemental Table 11 were not adjusted for multiple testing. However, the gene expression fold changes were marked (log2FC>1.5) and fewer than 25 top over- or underexpressed genes were included, on the basis of the presence of two APOL1 renal-risk variants. The pathways detected in our study are unlikely to be the sole mechanisms leading to APOL1-associated nephropathy. Other stressors may amplify pathologic effects in the form of second hits,29 including ischemia,30 viral infections,31–33 interferons,34 and lupus.5,35
Although no significantly altered pathways were present in the tubulointerstitial compartment of black patients with nephrotic syndrome or FSGS in the Nephrotic Syndrome Study Network study,36 the 200 most significantly underexpressed genes (<1% of all expressed genes) on the basis of APOL1 high risk genotypes (two renal-risk variants) in glomeruli of patients with FSGS were nominally enriched for the mitochondrial pathway (FDR P=0.04; Cytoscape-based BiNGO analysis).
A limitation of this study is that transcript-based screening may miss functional changes that relate to post-transcriptional and post-translational modification of proteins. However, to the best of our knowledge, this is the first study that links APOL1 renal-risk variants with mitochondrial dysfunction via an unbiased systems biology-based approach. Findings were confirmed by mitochondrial respirometry. Increasing evidence supports mitochondrial dysfunction and the generation of ROS in the pathogenesis of CKD.16 The mechanisms of APOL1-associated kidney disease identified in this study may provide valuable insights toward developing novel therapeutic targets for nondiabetic kidney disease in black patients.
Concise Methods
Detailed methods and information on pathway analysis are available in Supplemental Material.
Establishment of HEK293 Tet-on G0, G1, G2, and EV Cells
HEK293 Tet-on APOL1 cell lines were established to stably express G0, G1, G2, and empty pTRE2hyg vector, on the basis of previously reported methods.18 Individual clones were verified by direct sequencing (ABI 3730XL, Applied Biosystems, Foster City, CA). RT-PCR and immunoblotting were used to determine APOL1 expression levels and APOL1 protein size.
Global Gene Expression and Pathway Analyses
Based on the study design (Supplemental Figure 1), global gene expression profiles were performed on HEK293 Tet-on G0, G1, G2, and EV cells with and without Dox induction using Illumina human HT-12 v4 arrays. Another independent gene expression array system, Affymetrix HTA 2.0, was used to verify the results of Illumina arrays. Paired and pattern-based analyses were applied to detect the mostly effected pathways because of overexpression and by APOL1 genotypes.
Mitochondrial Respirometry
Twenty thousand HEK293 Tet-on APOL1 cells were seeded on a V7 cell culture plate (Seahorse Bioscience, Billerica, MA) for 24 hours in complete DMEM media. Dox was used to induce APOL1 expression (concentrations provided in Supplemental Material) on the basis of the G0, G1, and G2 genotype, 8 hours before assessing cellular respiration. The final preassay density of HEK293 Tet-on APOL1 cells reached approximately 100,000/well. Cellular respiration was measured using a Seahorse Bioscience XF-24–3 analyzer. To measure oxygen consumption rate, cell culture media was replaced with assay media (3 mM glucose, 1 mM sodium pyruvate, and 1.5 mM glutamine without FBS). For bioenergetic profiling, final concentrations of compounds after port injections were 1.0 μM oligomycin, 0.25 μM FCCP, 1 μM antimycin A, and 1 μM rotenone. After respirometric assessments, cell numbers were recounted for normalization of oxygen consumption rate.
Human Subjects and Primary PTC Lines
Black patients undergoing nephrectomy for localized renal cell carcinoma with preoperative eGFR>60 ml/min per 1.73 m2 (without proteinuria) were recruited to provide nondiseased kidney tissue. Fifty three individuals (men/women, 31:22; mean±SD age, 58.3±10.7 years; and serum creatinine 1.08±0.33 mg/dl) were enrolled between March 2010 and May 2015. The study was approved by the Wake Forest School of Medicine (WFSM) Institutional Review Board and all participants provided written informed consent. Primary PTC lines were established using an established protocol.12 DNA was isolated from peripheral blood of participants using a Promega DNA isolation wizard kit (Promega, Madison, WI) and two single nucleotide polymorphisms in the APOL1 G1 renal-risk allele (rs73885319; rs60910145) and an insertion/deletion for the G2 renal-risk allele (rs71785313) were genotyped using a custom assay designed at WFSM on the Sequenom platform (Sequenom Laboratories, San Diego, California).37 G1 and G2 genotype calls were visually inspected for quality control. Genotyping efficiency was 100% and 41 blind duplicates resulted in a 100% concordance rate.
Disclosures
Wake Forest University Health Sciences and B.I.F. have filed for a patent related to APOL1 genetic testing. B.I.F. receives research funding from Novartis (Basel, Switzerland) and B.I.F. and J.S.P. are consultants for Ionis Pharmaceuticals (Carlsbad, CA). All other authors have nothing to disclose.
Supplementary Material
Acknowledgments
We thank Dr. Martin R. Pollak for sharing APOL1 vectors.
This work was supported by National Institutes of Health grant R01DK070941 (to B.I.F.).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorial, “Identifying the Intracellular Function of APOL1,” on pages 1008–1011.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2016050567/-/DCSupplemental.
References
- 1.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR: Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, Bekele E, Bradman N, Wasser WG, Behar DM, Skorecki K: Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Hum Genet 128: 345–350, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Freedman BI, Kopp JB, Langefeld CD, Genovese G, Friedman DJ, Nelson GW, Winkler CA, Bowden DW, Pollak MR: The apolipoprotein L1 (APOL1) gene and nondiabetic nephropathy in African Americans. J Am Soc Nephrol 21: 1422–1426, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larsen CP, Beggs ML, Saeed M, Walker PD: Apolipoprotein L1 risk variants associate with systemic lupus erythematosus-associated collapsing glomerulopathy. J Am Soc Nephrol 24: 722–725, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Freedman BI, Langefeld CD, Andringa KK, Croker JA, Williams AH, Garner NE, Birmingham DJ, Hebert LA, Hicks PJ, Segal MS, Edberg JC, Brown EE, Alarcón GS, Costenbader KH, Comeau ME, Criswell LA, Harley JB, James JA, Kamen DL, Lim SS, Merrill JT, Sivils KL, Niewold TB, Patel NM, Petri M, Ramsey-Goldman R, Reveille JD, Salmon JE, Tsao BP, Gibson KL, Byers JR, Vinnikova AK, Lea JP, Julian BA, Kimberly RP; Lupus Nephritis–End‐Stage Renal Disease Consortium : End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis Rheumatol 66: 390–396, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ashley-Koch AE, Okocha EC, Garrett ME, Soldano K, De Castro LM, Jonassaint JC, Orringer EP, Eckman JR, Telen MJ: MYH9 and APOL1 are both associated with sickle cell disease nephropathy. Br J Haematol 155: 386–394, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Weckerle A, Snipes JA, Cheng D, Gebre AK, Reisz JA, Murea M, Shelness GS, Hawkins GA, Furdui CM, Freedman BI, Parks JS, Ma L: Characterization of circulating APOL1 protein complexes in African Americans. J Lipid Res 57: 120–130, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bruggeman LA, O’Toole JF, Ross MD, Madhavan SM, Smurzynski M, Wu K, Bosch RJ, Gupta S, Pollak MR, Sedor JR, Kalayjian RC: Plasma apolipoprotein L1 levels do not correlate with CKD. J Am Soc Nephrol 25: 634–644, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reeves-Daniel AM, DePalma JA, Bleyer AJ, Rocco MV, Murea M, Adams PL, Langefeld CD, Bowden DW, Hicks PJ, Stratta RJ, Lin J-J, Kiger DF, Gautreaux MD, Divers J, Freedman BI: The APOL1 gene and allograft survival after kidney transplantation. Am J Transplant 11: 1025–1030, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freedman BI, Julian BA, Pastan SO, Israni AK, Schladt D, Gautreaux MD, Hauptfeld V, Bray RA, Gebel HM, Kirk AD, Gaston RS, Rogers J, Farney AC, Orlando G, Stratta RJ, Mohan S, Ma L, Langefeld CD, Hicks PJ, Palmer ND, Adams PL, Palanisamy A, Reeves-Daniel AM, Divers J: Apolipoprotein L1 gene variants in deceased organ donors are associated with renal allograft failure. Am J Transplant 15: 1615–1622, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freedman BI, Pastan SO, Israni AK, Schladt D, Julian BA, Gautreaux MD, Hauptfeld V, Bray RA, Gebel HM, Kirk AD, Gaston RS, Rogers J, Farney AC, Orlando G, Stratta RJ, Mohan S, Ma L, Langefeld CD, Bowden DW, Hicks PJ, Palmer ND, Palanisamy A, Reeves-Daniel AM, Brown WM, Divers J: APOL1 Genotype and Kidney Transplantation Outcomes From Deceased African American Donors. Transplantation 100: 194–202, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma L, Shelness GS, Snipes JA, Murea M, Antinozzi PA, Cheng D, Saleem MA, Satchell SC, Banas B, Mathieson PW, Kretzler M, Hemal AK, Rudel LL, Petrovic S, Weckerle A, Pollak MR, Ross MD, Parks JS, Freedman BI: Localization of APOL1 protein and mRNA in the human kidney: nondiseased tissue, primary cells, and immortalized cell lines. J Am Soc Nephrol 26: 339–348, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madhavan SM, O’Toole JF, Konieczkowski M, Ganesan S, Bruggeman LA, Sedor JR: APOL1 localization in normal kidney and nondiabetic kidney disease. J Am Soc Nephrol 22: 2119–2128, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hara N, Yamada K, Shibata T, Osago H, Hashimoto T, Tsuchiya M: Elevation of cellular NAD levels by nicotinic acid and involvement of nicotinic acid phosphoribosyltransferase in human cells. J Biol Chem 282: 24574–24582, 2007 [DOI] [PubMed] [Google Scholar]
- 15.Hipkiss AR: Aging, Proteotoxicity, Mitochondria, Glycation, NAD and Carnosine: Possible Inter-Relationships and Resolution of the Oxygen Paradox. Front Aging Neurosci 2: 10, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kawakami T, Gomez IG, Ren S, Hudkins K, Roach A, Alpers CE, Shankland SJ, D’Agati VD, Duffield JS: Deficient Autophagy Results in Mitochondrial Dysfunction and FSGS. J Am Soc Nephrol 26: 1040–1052, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Olabisi OA, Zhang J-Y, VerPlank L, Zahler N, DiBartolo S 3rd, Heneghan JF, Schlöndorff JS, Suh JH, Yan P, Alper SL, Friedman DJ, Pollak MR: APOL1 kidney disease risk variants cause cytotoxicity by depleting cellular potassium and inducing stress-activated protein kinases. Proc Natl Acad Sci U S A 113: 830–837, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng D, Weckerle A, Yu Y, Ma L, Zhu X, Murea M, Freedman BI, Parks JS, Shelness GS: Biogenesis and cytotoxicity of APOL1 renal risk variant proteins in hepatocytes and hepatoma cells. J Lipid Res 56: 1583–1593, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schweikert E-M, Devarajan A, Witte I, Wilgenbus P, Amort J, Förstermann U, Shabazian A, Grijalva V, Shih DM, Farias-Eisner R, Teiber JF, Reddy ST, Horke S: PON3 is upregulated in cancer tissues and protects against mitochondrial superoxide-mediated cell death. Cell Death Differ 19: 1549–1560, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benarroch EE: Na+, K+-ATPase: functions in the nervous system and involvement in neurologic disease. Neurology 76: 287–293, 2011 [DOI] [PubMed] [Google Scholar]
- 21.Van Remmen H, Williams MD, Guo Z, Estlack L, Yang H, Carlson EJ, Epstein CJ, Huang TT, Richardson A: Knockout mice heterozygous for Sod2 show alterations in cardiac mitochondrial function and apoptosis. Am J Physiol Heart Circ Physiol 281: H1422–H1432, 2001 [DOI] [PubMed] [Google Scholar]
- 22.Kokoszka JE, Coskun P, Esposito LA, Wallace DC: Increased mitochondrial oxidative stress in the Sod2 (+/-) mouse results in the age-related decline of mitochondrial function culminating in increased apoptosis. Proc Natl Acad Sci U S A 98: 2278–2283, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vanwalleghem G, Fontaine F, Lecordier L, Tebabi P, Klewe K, Nolan DP, Yamaryo-Botté Y, Botté C, Kremer A, Burkard GS, Rassow J, Roditi I, Pérez-Morga D, Pays E: Coupling of lysosomal and mitochondrial membrane permeabilization in trypanolysis by APOL1. Nat Commun 6: 8078, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Espinosa-Parrilla Y, Amiel J, Augé J, Encha-Razavi F, Munnich A, Lyonnet S, Vekemans M, Attié-Bitach T: Expression of the SMADIP1 gene during early human development. Mech Dev 114: 187–191, 2002 [DOI] [PubMed] [Google Scholar]
- 25.Böttinger EP: TGF-beta in renal injury and disease. Semin Nephrol 27: 309–320, 2007 [DOI] [PubMed] [Google Scholar]
- 26.Hagler MA, Hadley TM, Zhang H, Mehra K, Roos CM, Schaff HV, Suri RM, Miller JD: TGF-β signalling and reactive oxygen species drive fibrosis and matrix remodelling in myxomatous mitral valves. Cardiovasc Res 99: 175–184, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jain M, Rivera S, Monclus EA, Synenki L, Zirk A, Eisenbart J, Feghali-Bostwick C, Mutlu GM, Budinger GRS, Chandel NS: Mitochondrial reactive oxygen species regulate transforming growth factor-β signaling. J Biol Chem 288: 770–777, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon Y-S, Lee J-H, Hwang S-C, Choi KS, Yoon G: TGF beta1 induces prolonged mitochondrial ROS generation through decreased complex IV activity with senescent arrest in Mv1Lu cells. Oncogene 24: 1895–1903, 2005 [DOI] [PubMed] [Google Scholar]
- 29.Freedman BI, Skorecki K: Gene-gene and gene-environment interactions in apolipoprotein L1 gene-associated nephropathy. Clin J Am Soc Nephrol 9: 2006–2013, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Anderson BR, Howell DN, Soldano K, Garrett ME, Katsanis N, Telen MJ, Davis EE, Ashley-Koch AE: In vivo Modeling Implicates APOL1 in Nephropathy: Evidence for Dominant Negative Effects and Epistasis under Anemic Stress. PLoS Genet 11: e1005349, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, Friedman D, Briggs W, Dart R, Korbet S, Mokrzycki MH, Kimmel PL, Limou S, Ahuja TS, Berns JS, Fryc J, Simon EE, Smith MC, Trachtman H, Michel DM, Schelling JR, Vlahov D, Pollak M, Winkler CA: APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. J Am Soc Nephrol 22: 2129–2137, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Divers J, Núñez M, High KP, Murea M, Rocco MV, Ma L, Bowden DW, Hicks PJ, Spainhour M, Ornelles DA, Kleiboeker SB, Duncan K, Langefeld CD, Turner J, Freedman BI: JC polyoma virus interacts with APOL1 in African Americans with nondiabetic nephropathy. Kidney Int 84: 1207–1213, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ohta A, Nishiyama Y: Mitochondria and viruses. Mitochondrion 11: 1–12, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nichols B, Jog P, Lee JH, Blackler D, Wilmot M, D’Agati V, Markowitz G, Kopp JB, Alper SL, Pollak MR, Friedman DJ: Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney Int 87: 332–342, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leishangthem BD, Sharma A, Bhatnagar A: Role of altered mitochondria functions in the pathogenesis of systemic lupus erythematosus. Lupus 25: 272–281, 2016 [DOI] [PubMed] [Google Scholar]
- 36.Sampson MG, Robertson CC, Martini S, Mariani LH, Lemley KV, Gillies CE, Otto EA, Kopp JB, Randolph A, Vega-Warner V, Eichinger F, Nair V, Gipson DS, Cattran DC, Johnstone DB, O’Toole JF, Bagnasco SM, Song PX, Barisoni L, Troost JP, Kretzler M, Sedor JR; Nephrotic Syndrome Study Network : Integrative Genomics Identifies Novel Associations with APOL1 Risk Genotypes in Black NEPTUNE Subjects. J Am Soc Nephrol 27: 814–823, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Freedman BI, Langefeld CD, Turner J, Núñez M, High KP, Spainhour M, Hicks PJ, Bowden DW, Reeves-Daniel AM, Murea M, Rocco MV, Divers J: Association of APOL1 variants with mild kidney disease in the first-degree relatives of African American patients with non-diabetic end-stage renal disease. Kidney Int 82: 805–811, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.