SUMMARY
The circadian clock controls metabolic and physiological processes through finely tuned molecular mechanisms. The clock is remarkably plastic and adapts to exogenous zeitgebers, such as light and nutrition. How a pathological condition in a given tissue influences systemic circadian homeostasis in other tissues remains an unanswered question of conceptual and biomedical importance. Here we show that lung adenocarcinoma operates as an endogenous reorganizer of circadian metabolism. High-throughput transcriptomics and metabolomics revealed unique signatures of transcripts and metabolites cycling exclusively in livers of tumor-bearing mice. Remarkably, lung cancer has no effect on the core clock, but rather reprograms hepatic metabolism through altered pro-inflammatory response via the STAT3-Socs3 pathway. This results in disruption of AKT, AMPK and SREBP signaling, leading to altered insulin, glucose and lipid metabolism. Thus, lung adenocarcinoma functions as a potent endogenous circadian organizer (ECO), which rewires the pathophysiological dimension of a distal tissue such as the liver.
INTRODUCTION
Metabolic, endocrine and behavioral functions are largely circadian and their disruption is associated with a number of disorders and pathologies, including cancer (Asher and Sassone-Corsi, 2015; Bass, 2012; Fu and Lee, 2003; Gamble et al., 2014; Masri et al., 2015; Partch et al., 2014). Circadian rhythms are governed by molecular machinery whose function is to maintain rhythmic precision within cells and synchrony between central and peripheral clocks. Importantly, circadian transcriptional circuits function in a defined tissue-specific manner by interplaying with specialized nuclear factors through poorly understood mechanisms (Masri and Sassone-Corsi, 2010; Panda et al., 2002). Under standard physiological states, the core clock machinery is coupled to the metabolic cycles with which it operates in a coherent, concerted manner. However, the clock is also able to adapt to changing metabolic fluctuations as a compensatory mechanism and it does so by utilizing alternative transcriptional strategies. For instance, restricted feeding temporally phase shifts circadian gene expression in the liver (Damiola et al., 2000; Stokkan et al., 2001; Vollmers et al., 2009) and nutritional challenge is able to reprogram circadian transcription and subsequently alter cyclic metabolism (Eckel-Mahan et al., 2013; Hatori et al., 2012; Kohsaka et al., 2007). Therefore, timing of food intake and nutritional challenge are able to uncouple the timekeeping of hepatic metabolic oscillations from the core clock machinery. Yet, aside from the consequences of nutritional challenge, the effects of other non-dietary factors that could uncouple and disrupt the hepatic clock remain largely unexplored.
Cancer cells thrive based on a heightened metabolic rate that circumvents typical physiological means for energy production through the so-called Warburg effect (Hsu and Sabatini, 2008; Vander Heiden et al., 2009). In addition, cancer cells excrete a number of factors systemically, including metabolic ‘waste’ by-products and/or inflammatory signals (Hanahan and Weinberg, 2011; Lin and Karin, 2007). For example, tumor-secreted lactate, a product of increased aerobic glycolysis of cancer cells, is associated with heightened metastatic incidence, increased angiogenesis, is responsible for metabolic reprogramming in adjacent tissues and can induce a pro-inflammatory state (Colegio et al., 2014; Doherty and Cleveland, 2013). Similarly, the cooperative effects of the inflammatory response during tumorigenesis are well documented (Gao et al., 2007; Sansone et al., 2007). Tumor-secreted cytokines, such as Interleukin-6 (IL-6), can regulate metabolism in multiple tissues (Mauer et al., 2015), suggesting a possible role in mediating tumor-induced metabolic changes systemically. Collectively, these tumor-derived metabolites and cytokines constitute the so-called tumor ‘macroenvironment’ (Al-Zoughbi et al., 2014), the systemic metabolic consequences of which remain elusive.
Importantly, the effects of a tumor on organismal homeostasis are poorly understood, and, given the unique ability of the clock in sensing metabolic discrepancies, a potential role of cancer in rewiring clock-controlled metabolism is intriguing (Sahar and Sassone-Corsi, 2009). Indeed, our results demonstrate that lung adenocarcinoma rewires the circadian hepatic transcriptome and corresponding metabolome, yet the core clock machinery remains virtually unperturbed. The tumor imposes a profound metabolic reprogramming that implicates a number of signaling pathways, which operate within the framework of the tumor macroenvironment. As a paradigm, we reveal that the inflammatory STAT3-Socs3 signaling axis is induced in the liver of lung tumor-bearing mice, resulting in inhibition of hepatic insulin signaling, glucose intolerance, and deregulated lipid metabolism. In conclusion, we illustrate a previously unappreciated role played by a distally located lung adenocarcinoma as an endogenous circadian organizer (ECO) in the rewiring of circadian homeostasis of the liver.
RESULTS
Lung cancer as an endogenous reorganizer of circadian rhythms
The KrasLSL-G12D;p53fl/fl mice are a genetic model of lung adenocarcinoma that mimics human non-small cell lung cancer (NSCLC) (Jackson et al., 2005; Jackson et al., 2001). Upon intra-tracheal delivery of equivalent adenoviral titer of Cre recombinase, which induces the genetic rearrangement of the Lox-stop-Lox cassette to activate oncogenic Kirsten rat sarcoma viral oncogene homolog (Kras) and knockout the tumor suppressor p53, mice developed defined lung adenocarcinoma (Schematic in Figure S1). This mouse model generates lung adenocarcinoma with 100% penetrance and uniform tumor burden among all mice (Jackson et al., 2001). Equivalent adenoviral titer of FlpO recombinase was administered to p53fl/fl littermates of the same pure C57BL/6J background as a control that does not induce recombination. Upon sacrifice, lung tumor-bearing (TB) mice exhibited WT expression of Kras in the liver, white adipose tissue (WAT) and muscle and no metastatic lesions were observed in the liver (Figure S2).
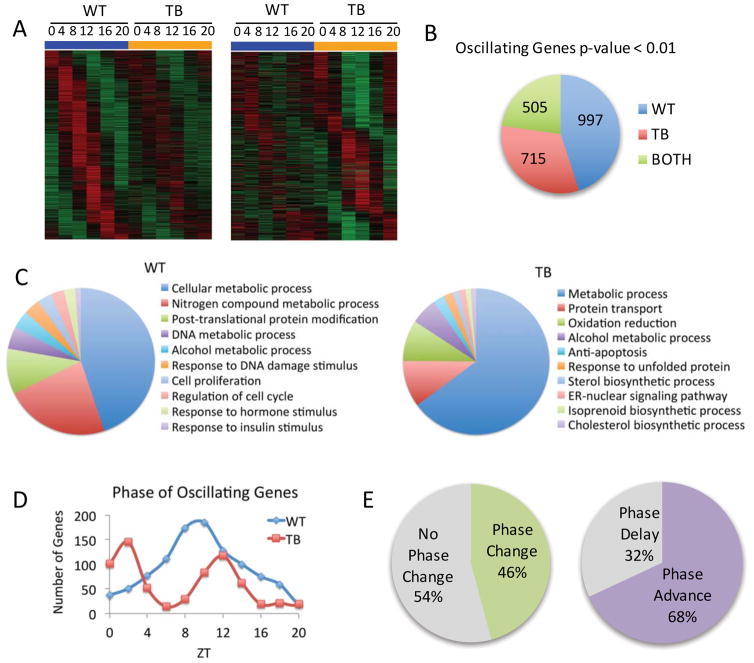
To investigate the distal effects of lung adenocarcinoma on circadian hepatic function, WT and TB mice were sacrificed every four hours over the circadian cycle (ZT 0, 4, 8, 12, 16 and 20) and livers were subjected to transcriptomics and metabolomics analyses. Heat maps for oscillating genes based on transcriptomics, as determined by JTK_cycle, display striking differences in unique sets of oscillating genes from WT (left panel) and TB (right panel) mice (Figure 1A, 1B). Gene ontology (GO) biological function was determined using DAVID pathway analysis for WT or TB oscillating genes. Pathway analysis revealed that WT-specific genes were enriched for a number of metabolic processes, including insulin response, and regulation of cell cycle and proliferation while TB-only oscillating genes were selectively enriched for endoplasmic reticulum (ER) signaling, unfolded protein response, cholesterol biosynthesis and redox state (Figure 1C and Figure S3). Phase analysis was performed for uniquely oscillating WT and TB-specific genes to determine the relative phase of circadian gene expression. The peak in phase of expression was around ZT 8 in the WT category, whereas TB oscillating genes exhibited a bi-phasic profile that peaked around ZT 0 and again at ZT 12 (Figure 1D). Using the set of 505 genes that retain oscillation in both WT and TB mice, phase analysis was performed to determine if rhythmic genes retained their peak in expression. Strikingly, 46% of circadian genes exhibited a phase change, with 68% of these genes being phase advanced and 32% were phase delayed by at least 1 hour (Figure 1E). These results demonstrate that lung adenocarcinoma significantly reprograms the circadian hepatic transcriptome.
Figure 1. Lung adenocarcinoma rewires the circadian hepatic transcriptome.
A) DNA microarray analysis was performed using mouse liver total RNA from ZT 0, 4, 8, 12, 16 and 20. Using JTK_cycle, genes selected to be circadian at a p-value <0.01 are displayed as heat maps for WT and lung tumor-bearing (TB) livers. Left panels display circadian genes exclusively in WT mice and right panels show genes with more robust oscillation in TB mice. B) Pie charts indicate actual numbers of circadian genes that oscillate exclusively in WT, TB or BOTH conditions. C) Top 10 gene ontology (GO) terms for biological process were identified by DAVID pathway analysis tool, based on a 0.01 p-value cutoff. D) Phase analysis of WT and TB-specific oscillating gene expression profiles. E) Phase analysis of ‘BOTH’ genes that remain circadian in WT and TB mice.
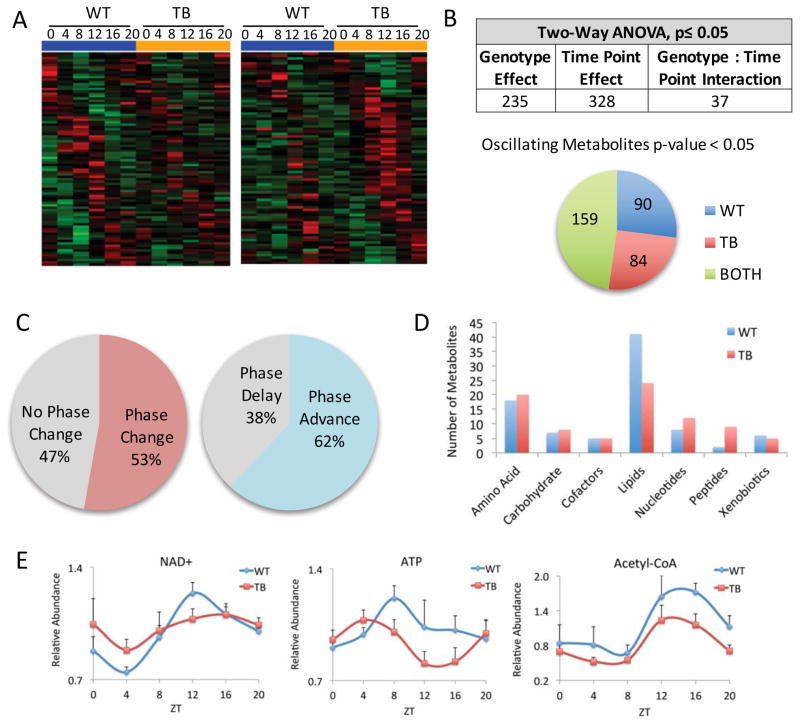
Similar to the circadian transcriptome, metabolomics analysis revealed unique sets of oscillating metabolites in the livers of WT (left panel heat maps) or TB (right panel heat maps) mice (Figure 2A). Of ~600 identified metabolites, two-way ANOVA analysis identified that 235 metabolites were differentially altered by the lung tumor and 328 metabolites were differentially expressed by time point (Figure 2B). Oscillating metabolites were further determined using JTK_cycle, and though the oscillation of 159 metabolites persisted, 90 were rhythmic exclusively in WT and 84 exclusively in TB mice (Figure 2B). Of these 159 metabolites that oscillate in WT and TB mice, 53% exhibited a change in phase, with 62% and 38% being phase advanced and delayed, respectively (Figure 2C). Classification of these metabolites into pathways demonstrated a clear reduction in oscillating lipids in TB versus WT mice (Figure 2D). In addition, a reduction in the levels of energetic metabolites, NAD+, ATP and acetyl-CoA was seen (Figure 2E). This indicates altered usage or production of these molecules resulting in disruption of liver homeostasis in TB animals. Thus, the presence of lung tumors acts to distally rewire both transcriptional and metabolic programs in the liver. As further depicted below, this circadian reorganization appears to coordinately contribute to a TB-specific hepatic metabolic profile.
Figure 2. The circadian metabolome is reorganized by lung cancer.
A) Heat maps displaying oscillating metabolites as determined by JTK_cycle (p-value <0.05) in WT and TB mice. Left panels display circadian metabolites exclusively in WT liver and right panels show metabolites with more robust oscillation in TB mice. B) Two-way ANOVA analysis using a p-value cutoff of 0.05 reveals metabolites that are responsive to lung tumors, circadian time point, or both. Numbers of oscillating metabolites using JTK_cycle are indicated from WT, TB or BOTH categories. C) Phase analysis was performed using JTK_cycle to identify the phase of peak metabolite expression. D) Oscillating metabolites are displayed based on biological sub-pathway, including amino acid, carbohydrate, cofactors, lipids, nucleotides, peptides and xenobiotics. E) Examples of energetic metabolites that are dampened in TB mice. NAD+= nicotinamide adenine dinucleotide; ATP= adenosine 5′-triphosphate.
Lung adenocarcinoma does not affect hepatic core clock components
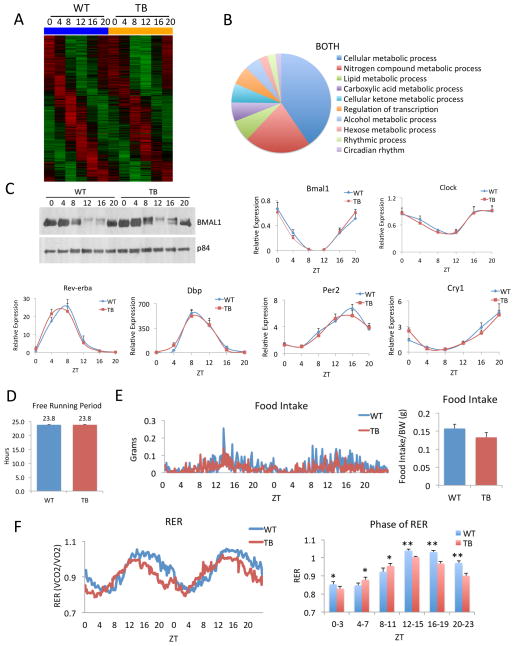
A detailed analysis of the genes that were not altered between WT and TB mice was carried out, as shown in the heat map in Figure 3A. GO pathway analysis revealed this category is enriched in select metabolic genes but also rhythmic genes pertaining to the circadian clock (Figure 3B). The phosphorylation of the aryl hydrocarbon receptor nuclear translocator-like (ARNTL or BMAL1) protein and expression of all core clock genes, including circadian locomotor output cycles kaput (Clock), Bmal1, Period (Per1-3), Cryptochrome (Cry1/2) and nuclear receptor subfamily 1, group D (Nr1d1 or Rev-Erbα), as well as the clock-controlled D site of albumin promoter binding protein (Dbp) gene, were unchanged in the livers of TB animals (Figure 3C and Figure S4). In order to better characterize the effects of lung adenocarcinoma on the clock, locomotor behavior was analyzed and no change in the free-running period was observed between WT and TB mice (Figure 3D). Similarly, behavioral actograms show that the circadian activity profile was equal during the light/dark cycles in WT and TB mice (Figure S4). Also, the feeding behavior remained rhythmic in TB mice while a non-significant decrease in food intake was observed (Figure 3E). The respiratory exchange ratio (RER) remained rhythmic but TB mice displayed an elevated RER during the light phase and a dampened RER during the dark phase (Figure 3F), in keeping with a reduction in VO2, VCO2 and heat production (Figure S5). The altered circadian metabolites (Figure 2D, 2E) in conjunction with dampened RER levels (Figure 3F and Figure S5) revealed a significant shift in the metabolic state of TB mice. Indeed, repressed energy expenditure might be a contributing factor to the uncoupling of the core clock and metabolic rhythms. Timing of food intake, which functions as a powerful zeitgeber (Damiola et al., 2000; Eckel-Mahan et al., 2013; Vollmers et al., 2009), also remains virtually unaltered in TB mice (Figure 3E).
Figure 3. The circadian clock is unaffected by lung adenocarcinoma.
A) Heatmap for ‘BOTH’ category genes that are unaltered in expression between WT and TB mice. B) GO pathway analysis using biological process for ‘BOTH’ oscillating genes. C) BMAL1 protein phosphorylation by western and circadian expression of the clock genes, Bmal1, Clock, Rev-erbα, Dbp, Per2 and Cry1, as determined by quantitative real-time PCR (RT-PCR). D) Locomotor activity analysis for WT and TB mice, as calculated by the free-running period (Tau) in dark/dark (D/D) conditions. An n=12 WT and n=12 TB mice were used for behavioral analysis. E) Food intake of WT and TB mice shown over a 48-hour period (left panel). Total food intake was normalized to body weight of each animal. An n=7 WT and n=8 TB mice were used for indirect calorimetry analysis. F) VCO2/VO2 is shown as the respiratory exchange ratio (RER) for WT and TB mice over a 48-hour period. Average RER is quantified during the light and dark phases (right panel). Error bars indicate standard error of mean (SEM). Significance was calculated using Student’s T test and * and ** indicate p-value cutoffs of 0.001 and 0.0001, respectively.
Lung adenocarcinoma rewires hepatic metabolism but not the core clock
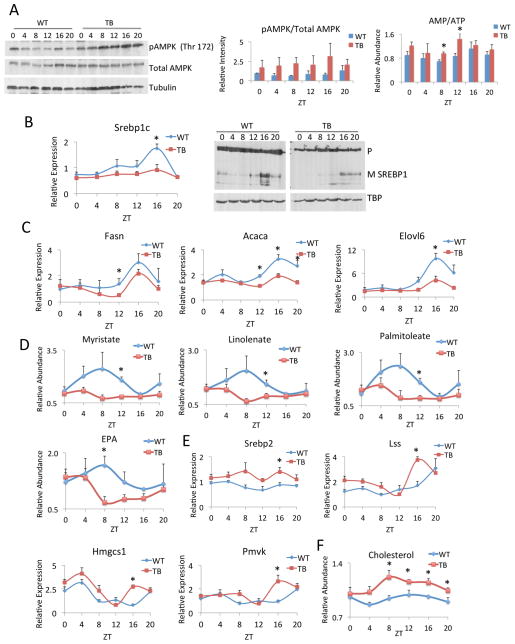
Given the changes in energy expenditure as measured by RER (Figure 3F) and the dampened lipid profiles identified by metabolomics in TB mice (Figure 2D), the effect of lung adenocarcinoma on fatty acid synthesis, breakdown by beta-oxidation and utilization for cholesterol production were further investigated. The sterol regulatory element binding protein (SREBP) pathway is known to control lipid metabolism in the liver in a circadian manner (Gilardi et al., 2014; Le Martelot et al., 2009) and its deregulation is in accordance with the observed alteration of lipid levels (Figure 2D). The SREBP pathway is known to be inhibited by the energy sensor AMP-activated protein kinase (AMPK) (Li et al., 2011; Vavvas et al., 1997). Indeed, activation of AMPKα by phosphorylation of threonine (Thr) 172 was markedly elevated in TB mice and peaked at ZT 16 (Figure 4A). Given the dampened ATP levels in TB mice (Figure 2E), these effects are aligned with the increased intracellular AMP/ATP ratios over the circadian cycle (Figure 4A). Accordingly, the SREBP1 pathway was suppressed, as both gene expression profiles and the levels of the mature form of nuclear SREBP1c protein were repressed at ZT 16 in TB mice (Figure 4B). Similarly, significant inhibition of SREBP1 target genes was observed, as seen with Fasn, Acaca and Elovl6 expression (Figure 4C). The repression of SREBP1-dependent signaling in the livers of TB mice was further substantiated by the decreased levels of long-chain fatty acids and esterified fatty acids (Figure 4D), including myristate, linolenate, palmitoleate and eicosapentaenoate (EPA). This suggests either a decrease in fatty acid biosynthesis or an increase in breakdown by beta-oxidation, the former case being most likely given the suppression of SREBP1 signaling and the unaltered peroxisome proliferator-activated receptor alpha (PPARα) and beta-oxidation gene expression profiles in livers of TB mice (Figure S6).
Figure 4. Lipid metabolism is altered in TB mice.
A) Western blot analysis for phospho-AMPK (Thr 172) and total AMPK in WT and TB mice, at the indicated circadian times. Quantification of pAMPK/total AMPK signaling is shown as a histogram. The ratio of AMP/ATP is shown over the circadian cycle and is elevated at all ZTs. B) Gene expression as determined by RT-PCR and protein expression of SREBP1c in WT and TB mice. Precursor (P) indicates uncleaved protein and mature (M) shows cleaved SREBP1 protein. C) Gene expression by RT-PCR was performed for Fasn, Acaca, and Elovl6 in WT and TB mice over the indicated ZTs. D) Levels of fatty acids and fatty acid esters as determined by metabolomics analysis for myristate, linolenate, palmitoleate and eicosapentaenoate (EPA). E) Gene expression of Srebp2 and its target genes Lss, Hmgcs1 and Pmvk as determined by RT-PCR. F) Total cholesterol levels in WT and TB mice over the circadian cycle were determined by metabolomics analysis. Error bars indicate SEM. Significance was calculated using Student’s T test and * indicates a p-value cutoff of 0.05.
In contrast to the suppression in the SREBP1 pathway, SREBP2 gene expression is not repressed in TB mice and its target genes lanosterol synthase (Lss), 3-hydroxy-3-methylglutaryl-CoA synthase (Hmgcs1) and phosphomevalonate kinase (Pmvk) showed a significant and coordinated increased peak in expression at ZT 16 (Figure 4E). As SREBP1 is primarily involved in fatty acid biosynthesis and SREBP2 is critical for cholesterol production (Brown and Goldstein, 1997; Horton et al., 2003), an increase in total cholesterol levels was observed in TB mice (Figure 4F), which paralleled SREBP2-dependent gene expression profiles (Figure 4E). Overall, TB mice displayed a deregulation of SREBP signaling, with a suppression of fatty acid synthesis and an induction of cholesterol biosynthesis. Repressed fatty acid levels and elevated cholesterol biosynthesis suggests a preferential shunting of lipids to produce cholesterol. Importantly, increased cholesterol levels are associated with a heightened inflammatory response (Ma et al., 2008; Zhao et al., 2011). These findings reveal a coordinated disruption in metabolic homeostasis in TB mice, which converge to activate AMPK in the liver and thereby differentially modulate SREBP-dependent lipid signaling. Thus, these results demonstrate that while the hepatic core clock is resilient to the distal effects of lung adenocarcinoma (Figure 3), the liver metabolic clock is altered in response to tumors.
Tumor-driven targeting of circadian inflammatory response
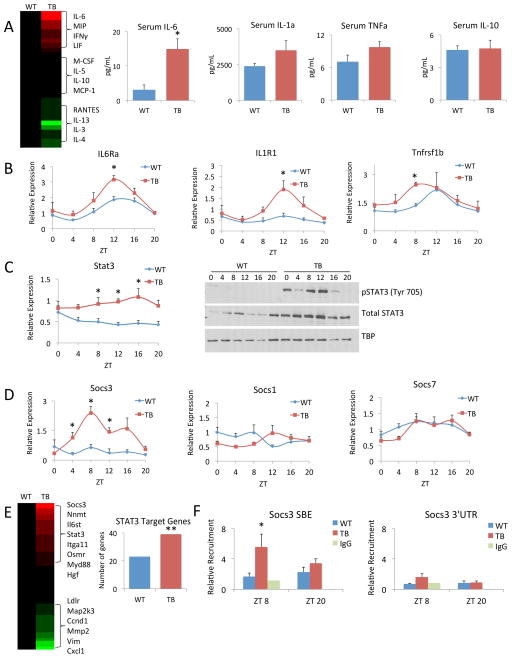
Pro-inflammatory responses mediated by tumor-secreted cytokines and chemokines are critical in cancer initiation and progression (Grivennikov et al., 2010). Specifically, the janus kinase (JAK)/signal transducer and activator of transcription (STAT) pathway has been demonstrated to play a role in multiple types of cancer (Lesina et al., 2011; Michalaki et al., 2004; Yu et al., 2014), and is known to be activated by IL-6, tumor necrosis factor alpha (TNFα), interferon gamma (IFNγ) and leukemia inhibitory factor (LIF) (Darnell et al., 1994; Fitzgerald et al., 2005; Grivennikov et al., 2009; Guo et al., 1998). Also, inflammation plays an important role in progression of lung adenocarcinoma (Gao et al., 2007; Yeh et al., 2006). Therefore, to validate the inflammatory response in TB mice, serum cytokine levels were assessed in an unbiased, multiplexed platform at ZT 12, which is the reported peak of circadian inflammatory response (Gibbs et al., 2012). Of the 31 cytokines assayed, a number were increased/decreased or unchanged in TB mouse serum versus WT and shown in a heat map (Figure 5A). Specifically, significant elevation of IL-6 was observed in TB mouse serum, along with a clear, though statistically non-significant, increase in IL-1α, TNFα, LIF and IFNγ (Figure 5A and Figure S7). In contrast, serum levels of the anti-inflammatory cytokine IL-10 and its receptor expression in the liver did not change between WT and TB mice (Figure 5A and Figure S7). Importantly, concomitant gene expression profiles of the cytokine receptors, interleukin 6 receptor (Il6rα), interleukin 1 receptor (Il1r1), tumor necrosis factor receptor subfamily (Tnfrsf1b) and interleukin 17 receptor (Il17ra) were significantly elevated in the liver and displayed circadian profiles that peaked at ZT12 in TB mice (Figure 5B and Figure S7). We focused on the effects of IL6-dependent signaling as a paradigm of how signals in TB mice lead to the expression and phosphorylation of STAT3. Gene expression of Stat3 was significantly elevated at ZT 8, ZT 12 and ZT 16, along with a corresponding increase in total protein levels in TB mice (Figure 5C), in keeping with reported IL-6-dependent STAT3 auto-regulation (Narimatsu et al., 2001). JAK-dependent phosphorylation of STAT3 at tyrosine (Tyr) 705 is known to activate STAT3 and induce its transcriptional activity by nuclear translocation (Darnell, 1997). We observed marked elevation of p-STAT3 Tyr705 in the livers of TB mice that peaked at ZT8 and ZT12 (Figure 5C). The transcriptional activation of STAT3 resulted in a significant increase in the expression of its downstream targets. Specifically, gene expression of suppressor of cytokine signaling 3 (Socs3) was drastically elevated and peaked at ZT 8 in TB mice, while the expression of Socs1 and Socs7 was unchanged (Figure 5D). Our transcriptomics data was compared to known STAT3 target genes (Bonetto et al., 2011), to determine the extent of STAT transcriptional activation in the liver. Heat maps display genes that were differentially regulated in TB normalized to WT, and of these genes, an enrichment was observed in the TB-specific gene set versus WT (Figure 5E; Supp. Table 1). Finally, there is a time-specific, significant increase of transcriptionally active p-STAT3 in TB mice at the STAT binding element (SBE), as demonstrated by chromatin immunoprecipitation (ChIP), on the Socs3 promoter (Figure 5F).
Figure 5. Lung tumor-induced inflammation in the liver.
A) Serum samples from WT and TB mice were assayed in an unbiased, multiplexed cytokine array platform and displayed as a heat map. Red indicates increased levels of cytokines and green indicates decreased cytokine levels in TB normalized to WT mouse serum. Specific profiles of pro-inflammatory cytokines (IL-6, IL-1α and TNFα) and anti-inflammatory IL-10 are shown. B) Gene expression as profiled by RT-PCR is shown for Il6rα, Il1r1 and Tnfrsf1b. C) Stat3 gene expression as shown by RT-PCR. Phospho-STAT3 (Tyr 705) and total STAT3 protein levels over the circadian cycle in WT and TB mice. D) Gene expression profiles of Socs3, Socs1 and Socs7 by RT-PCR. E) Known STAT3 target genes were compared to our transcriptomics data. Heat map displays gene expression profiles in the TB-specific group normalized to WT, with red and green representing up and down-regulated genes, respectively. Additionally, to determine enrichment of STAT3 target genes in WT and TB, Fisher’s exact test was used. The ** indicates the odds that the overlap of 39 genes in TB over random is 1.716 and the p-value is 0.00358, which satisfies a p<0.005 threshold. For WT the odds ratio is 1.345 with a p-value of 0.209. F) Recruitment of p-STAT3 to the STAT binding element (SBE) in the Socs3 promoter or to the 3′ untranslated region (UTR) as determined by chromatin immunoprecipitation (ChIP). Error bars indicate SEM. Significance was calculated using Student’s T test and * indicates a p-value cutoff of 0.05.
These results demonstrate that the pro-inflammatory response can induce transcriptional activation of STAT3 signaling in the liver, which may play a role in the hepatic metabolic rewiring observed in TB mice. Yet, this transcriptional rewiring observed in the liver is representative of a localized response, as the WAT and muscle gene expression profiles differ (Figure S6), in keeping with a tissue-specific inflammatory response. Moreover, the pro-inflammatory response is most likely not the only cause of metabolic rewiring: our preliminary results of the circadian serum metabolome from WT and TB mice show that a number of factors could be involved in tumor-dependent crosstalk with peripheral tissues (Figure S7). This data suggests that the extent of the tumor macroenvironment remains inadequately defined and that complex tissue-specific responses to these tumor-derived signaling molecules exist.
Lung cancer alters hepatic insulin signaling and glucose production
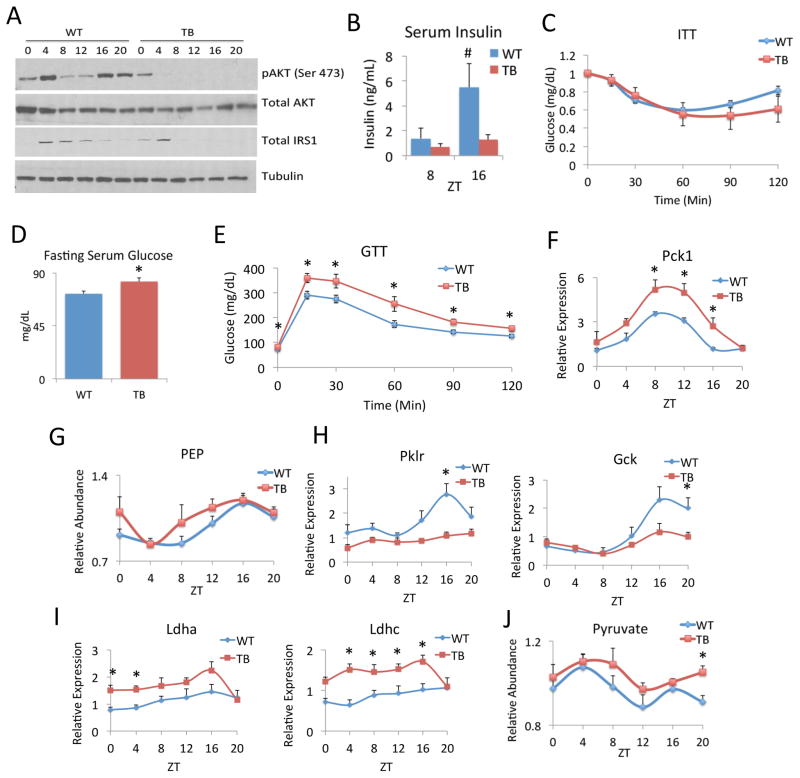
SOCS3 has been shown to play a role in modulating insulin sensitivity in adipose tissue and liver and these effects have been linked with IL-6 or TNFα-mediated inflammation (Emanuelli et al., 2001; Sachithanandan et al., 2010; Senn et al., 2003; Torisu et al., 2007). Given the induction of the STAT3 inflammatory axis and the increase in Socs3 gene expression in TB mice, hepatic insulin signaling was further investigated. Insulin-dependent phosphorylation of V-Akt murine thymoma viral oncogene homolog (AKT) at serine (Ser) 473 was dramatically inhibited in TB mice versus WT, while total levels of AKT remained unchanged (Figure 6A). Strikingly, the total levels of insulin receptor substrate 1 (IRS-1) protein were noticeably decreased in TB mice versus WT, especially from ZT8 to ZT20 (Figure 6A). Notably, these changes in IRS-1 protein levels coincided with the peak in STAT3 activation and Socs3 expression (Figure 5C, 5D), as SOCS3 is reported to target and degrade IRS-1 protein and thereby further repress hepatic insulin signaling (Rui et al., 2002). Serum insulin levels were investigated, and in line with decreased insulin signaling in the liver, systemic insulin levels were significantly low and lose their circadian oscillation in TB mice (Figure 6B). Yet, TB animals retained insulin sensitivity, as determined by insulin tolerance tests (ITT) (Figure 6C). Given these changes in insulin signaling, it would be expected that TB mice exhibit elevated levels of serum glucose. Indeed, TB mice displayed elevated fasting serum glucose levels (Figure 6D), and these mice were significantly less sensitive to exogenous glucose challenge using a glucose tolerance test (GTT) (Figure 6E). In order to elucidate the mechanism by which glucose levels were increased in TB mice, hepatic glucose production through gluconeogenesis was investigated, as this pathway is known to be clock-controlled (Zhang et al., 2010). Gene expression of phosphoenolpyruvate carboxykinase 1 (Pck1 or Pepck) remained circadian but was significantly induced at ZT 8, ZT12 and ZT16 in TB mice (Figure 6F). The level of phosphoenolpyruvate (PEP), the product of PEPCK, was elevated at ZT 8 and ZT 12 (Figure 6G). In contrast, expression of key glycolytic enzymes, such as rate-limiting glucokinase (Gck) and liver pyruvate kinase (Pklr) was significantly inhibited (Figure 6H). Though their expression was not circadian, lactate dehydrogenases (Ldha and Ldhc) that interconvert lactate and pyruvate were elevated in TB mice (Figure 6I), in keeping with the increased pyruvate levels that could be shunted into gluconeogenesis (Figure 6J). These results demonstrate that a lung tumor is responsible for the drastic change in insulin-dependent AKT signaling in the liver, leading to significant alterations in clock-controlled hepatic glucose production.
Figure 6. Lung adenocarcinoma alters hepatic insulin signaling and glucose production.
A) Western analysis of phospho-AKT (Ser 473), total AKT and total IRS1 in WT and TB mice over the circadian cycle. B) Serum insulin levels were measured by ELISA at ZT 8 and 16 in WT and TB mouse serum. Insulin levels at ZT 16 are statistically significant as indicated by # (p-value = 0.053, using Student’s T-test). C) Insulin tolerance test (ITT) in WT and TB mice. D) Overnight fasting glucose levels in WT and TB mice. E) Glucose tolerance test (GTT) in overnight fasted WT and TB mice. F) Gluconeogenic gene expression profile of Pepck (Pck1) by RT-PCR was done in livers of WT and TB mice. G) Levels of phosphoenolpyruvate (PEP) were determined by metabolomics analysis from livers of WT and TB mice. H) RT-PCR of glycolytic gene expression of L-PK (Pklr) and GK (Gck) over the circadian cycle. I) Gene expression of lactate dehydrogenases Ldha and Ldhc in WT and TB mice by RT-PCR. J) Levels of pyruvate were determined by metabolomics in livers of WT and TB mice. Error bars indicate SEM. Significance was calculated using Student’s T test and * indicates a p-value cutoff of 0.05.
DISCUSSION
Lung Tumor as an Endogenous Zeitgeber?
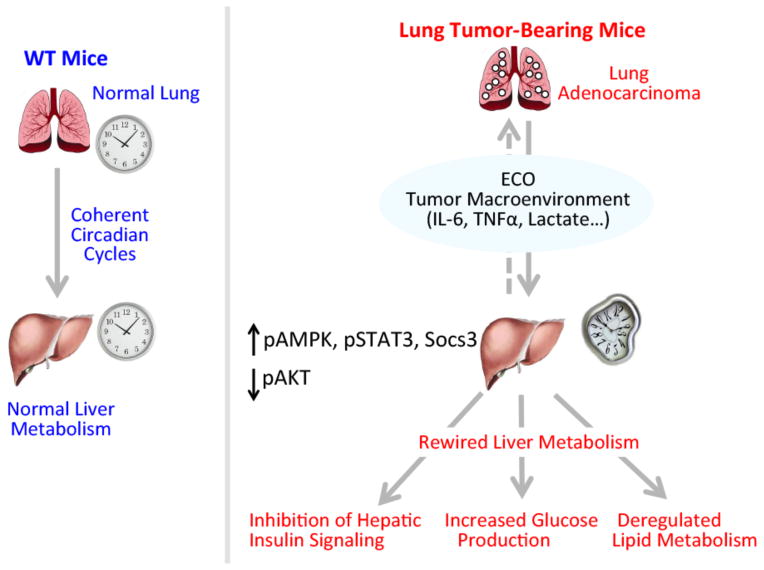
Circadian homeostasis is essential for organismal physiology and its intrinsic plasticity constitutes a highly efficient adaptation system to the changing environment. Specifically, zeitgebers such as light and nutrition are referred to as external stimuli that operate to entrain central and peripheral clocks, respectively. Here we have reported on findings that identify an endogenous circadian reorganizer that has the unique feature of rewiring circadian metabolism under unaltered light and feeding conditions. Indeed, lung adenocarcinoma contributes to the distal reprogramming of circadian hepatic gene expression and metabolic function (Figure 7). These results collectively demonstrate that lung tumors, independently of any nutritional challenge paradigm, alter circadian physiology leading to changes in cyclic energy expenditure, lipid metabolism, and hepatic insulin and glucose signaling. Importantly, since the core components of the liver molecular clock do not seem to be influenced in TB mice (Figure 3), the tumor does not appear to function as a classical zeitgeber. Yet, lung tumors act prominently on the liver by profoundly rewiring circadian metabolic control. Thus, these findings illustrate that circadian metabolism can be reprogrammed independently of exogenous inputs, such as the classical zeitgebers, light and nutrition. Indeed, lung adenocarcinoma operates as a distinctive endogenous circadian organizer (ECO) that dictates the changing pathophysiological dimension of a distal tissue such as the liver. We speculate that an ECO may function in differential manners depending on the tissue type, range of action and metabolic state.
Figure 7. Lung adenocarcinoma distally rewires circadian hepatic metabolism.
Schematic overview depicting the effects of the tumor macroenvironment on circadian hepatic metabolism. Our results show that lung tumors, acting through the inflammatory STAT3-Socs3 axis, operate to distally rewire circadian transcription and metabolism by acting as an endogenous circadian organizer (ECO). This manifests in loss of hepatic insulin signaling, glucose intolerance, and deregulated lipid metabolism through the AMPK/SREBP pathway.
Endogenous Circadian Organizer (ECO)
Given that these metabolic effects are systemic, tumor-dependent rewiring is most likely taking place in multiple organs to disrupt homeostasis. In this context it is notable that the effects of IL-6 are pleiotropic and function in a context- and tissue-specific manner to alter multiple signaling pathways (Mauer et al., 2015), and that the inflammatory response is the likely mediator of a complex web of physiological adjustments. Indeed, it does not escape our attention that the effect of a lung tumor will not be restricted to the liver alone (Figure S6), although this tissue is critical to decipher the effects of lung cancer on organismal metabolism. In further support of our results, a number of intriguing links can be made to connect tumor-derived inflammation with deregulated metabolism. Our extensive analysis demonstrates that a number of cytokines could be implicated in the rewiring observed in TB mice (Figure 5A and Figure S7). Also, our preliminary analysis of the serum metabolome indicates that the cyclic profiles of a variety of potentially critical metabolites change significantly in the TB mice (Figure S7). We have focused on IL-6 as a paradigm since the IL-6 inflammatory response has been investigated in fatty liver disease, and these effects link inflammation to altered lipid accumulation through SREBP signaling (Miller et al., 2011; Yamaguchi et al., 2010). Also, another layer of complexity exists in that IL-6 is known to activate AMPK especially in the context of exercise (Carey et al., 2006; Ruderman et al., 2006), and systemic IL-6 knockout mice are unable to stimulate AMPK signaling (Adser et al., 2011; Kelly et al., 2004). Moreover, AMPK suppresses SREBP target gene expression and attenuates hepatic steatosis (Li et al., 2011). Collectively, these notions suggest an interconnected network between inflammatory cytokines, AMPK and SREBP that could contribute to the tumor-induced liver reprogramming we observe. Moreover, the influence of cachexia on muscle and WAT is another factor that can feedback and alter liver homeostasis (Bonetto et al., 2011; Narsale et al., 2015; Tsoli et al., 2014). Our findings identify two converging pathways that might work in a coordinated manner to modulate circadian hepatic homeostasis. Altered energy expenditure can mediate a circadian metabolic rewiring that is likely compounded by the pro-inflammatory effects on the liver.
The Tumor Macroenvironment Reorganizes Homeostasis
Our results illustrate that lung adenocarcinoma has a profound effect on a variety of metabolic and signaling pathways in the liver. This tumor-derived macroenvironment is constituted by glycolytic metabolic byproducts, inflammatory cytokines, and other poorly defined circulating components (Al-Zoughbi et al., 2014). In this context, we have explored the liver pro-inflammatory response, a paradigm of the metabolic rewiring induced by the lung tumor. An additional twist to this scenario is the likely contribution of other metabolic tissues that respond to the tumor-derived macroenvironment. In turn, these tissues could provide an additional layer of signaling that would further adjust circadian homeostasis.
The pro-inflammatory response alters hepatic insulin signaling and subsequent glucose production through the STAT3-dependent activation of Socs3 (Figure 5). These findings bring to light another critical pathway that is deregulated by the distant actions of the lung tumor. The suppression of serum insulin levels suggests an important role of inflammation in the pancreas. Yet, these effects on hepatic insulin sensitivity are most likely regulated coordinately with the inflammatory response. For instance, the expression of the insulin-responsive Insig2 gene, which is known to be involved in clock-controlled SREBP function (Le Martelot et al., 2009), is unaltered in TB mice and suggests a complex signaling mechanism beyond a simple model of hypoinsulinemia. Moreover, the circadian clock controls hepatic glucose production through gluconeogenesis (Zhang et al., 2010) and our results suggest a potential crosstalk between the tumor and the liver clock. We speculate that tumor-secreted ‘waste’ such as lactate is converted to pyruvate and shunted through gluconeogenesis to produce glucose, which can further satisfy the heightened energetic demand of cancer cells. Interestingly, TB mice exhibit increased expression of lactate dehydrogenases (Ldha, Ldhc), Pepck (Pck1) and phosphoenolpyruvate (PEP), which could result in enhanced glucose intolerance (Figure 6). In conclusion, the circadian clock is highly responsive to its environment and is able to adapt to changes in energetic demand. In this context, the lung tumor macroenvironment operates as an ECO on hepatic circadian metabolism – a process that could potentially further drive tumorigenesis.
EXPERIMENTAL PROCEDURES
Animal housing and experimental procedures
KrasLSL-G12D;p53fl/fl mice have been previously described (Johnson et al., 2001). Detailed housing and infection procedures are provided as Supplementary Experimental Procedures.
DNA microarray analysis
Microarray analysis was performed as previously described (Masri et al., 2014) and further information is provided as Supplementary Experimental Procedures.
Metabolomics analysis
Metabolomics analysis was carried out by Metabolon, Inc. (Durham, NC) as previously described (Evans et al., 2009) (Masri et al., 2014). See Supplementary Experimental Procedures for further details.
Bioinformatics and pathway analysis
Bioinformatics analysis was performed using JTK_cycle and metabolomics and transcriptomics data is accessible at circadiomics.ics.uci.edu. Detailed methodology is available as Supplementary.
Metabolic Cage Analysis
Indirect calorimetry was performed using negative-flow CLAMS hardware system cages (Columbus Instruments). VO2, VCO2, RER, and food intake were measured and calculated with Oxymax software (Columbus Instruments).
Locomotor activity analysis
Animals were individually housed, using an n=12 mice/genotype for behavioral analysis. Mice were housed for 2 weeks in 12-hour standard L/D conditions and subsequently released into D/D conditions for 2 weeks. Activity was measured using optical beam motion detection (Philips Respironics) and data analyzed using Minimitter VitalView data acquisition software.
Gene expression analysis
Detailed methodology and primer sequences can be found in the Supplementary Experimental Procedures section.
Western blot analysis
Livers were homogenized in RIPA lysis buffer containing protease inhibitor cocktail, NaF and PMSF, sonicated briefly and rocked to lyse cells at 4°C. 10–30μg of protein lysate was resolved on SDS-PAGE gels. Antibodies used for western blots include: TBP, BMAL1, SREBP1 (Abcam), pAMPK, total AMPK, pSTAT3, total STAT3, pAKT, total AKT, IRS1 (Cell Signaling Technology).
Cytokine Profiling
A total of 31 mouse cytokines were profiled using a multiplex platform and data was extracted based on cytokine-specific standards by Eve Technologies (Calgary, Canada). Five independent serum samples were used from WT and TB mice. Relative change in cytokine expression between TB and WT was determined using absolute deviation values from the median and used for heat map generation.
Chromatin Immunoprecipitation (ChIP)
ChIP methodology was previously described (Masri et al., 2014). pSTAT3 antibody used for ChIP was obtained from Cell Signaling technology.
Tolerance Tests (GTT and ITT)
8 WT and 8 TB mice were fasted overnight and fasting glucose levels were measured using an ACCU-CHEK Aviva Plus glucometer (Roche). Body weight measurements were taken and insulin (0.75U/kg) or glucose (2g/kg) were IP injected and blood glucose measurements were taken 15, 30, 60, 90 and 120 minutes post injection.
Supplementary Material
Acknowledgments
We thank all members of the Sassone-Corsi laboratory for scientific discussion or technical assistance and T. Leff, G. Servillo and M. Oakes for discussions. Funding for S.M. was provided by NIH post-doctoral fellowship GM097899. K.K. was supported by a post-doctoral fellowship from the Japan Society for the Promotion of Science (JSPS). T.P. was supported by the Hope Funds for Cancer Research. Financial support for T.J. was provided by the Ludwig Center for Molecular Oncology at MIT and the Koch Institute Frontier Fund. The work of Y.L. and P.B. was supported by grants from the NSF (IIS-1321053) and the NIH (LM010235) to P.B.. Financial support for P.S-C. was provided by NIH (AG043745), Merieux Research Grant (53923) and the UC Irvine Chao Family Comprehensive Cancer Center.
Footnotes
MICROARRAY DATA
Microarray data for this study has been depositied in Gene Expression Omnibus (GEO) using the following accession number: GSE73222.
AUTHOR CONTRIBUTION
SM, TP and PSC conceptually designed the study. TP and TJ provided the mouse model. SM, TP, KK and MC performed experiments. YL and PB performed bioinformatics analysis. SM and PSC wrote the paper with input from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adser H, Wojtaszewski JF, Jakobsen AH, Kiilerich K, Hidalgo J, Pilegaard H. Interleukin-6 modifies mRNA expression in mouse skeletal muscle. Acta Physiol (Oxf) 2011;202:165–173. doi: 10.1111/j.1748-1716.2011.02269.x. [DOI] [PubMed] [Google Scholar]
- Al-Zoughbi W, Huang J, Paramasivan GS, Till H, Pichler M, Guertl-Lackner B, Hoefler G. Tumor macroenvironment and metabolism. Semin Oncol. 2014;41:281–295. doi: 10.1053/j.seminoncol.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Sassone-Corsi P. Time for food: the intimate interplay between nutrition, metabolism, and the circadian clock. Cell. 2015;161:84–92. doi: 10.1016/j.cell.2015.03.015. [DOI] [PubMed] [Google Scholar]
- Bass J. Circadian topology of metabolism. Nature. 2012;491:348–356. doi: 10.1038/nature11704. [DOI] [PubMed] [Google Scholar]
- Bonetto A, Aydogdu T, Kunzevitzky N, Guttridge DC, Khuri S, Koniaris LG, Zimmers TA. STAT3 activation in skeletal muscle links muscle wasting and the acute phase response in cancer cachexia. PLoS One. 2011;6:e22538. doi: 10.1371/journal.pone.0022538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–340. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Carey AL, Steinberg GR, Macaulay SL, Thomas WG, Holmes AG, Ramm G, Prelovsek O, Hohnen-Behrens C, Watt MJ, James DE, et al. Interleukin-6 increases insulin-stimulated glucose disposal in humans and glucose uptake and fatty acid oxidation in vitro via AMP-activated protein kinase. Diabetes. 2006;55:2688–2697. doi: 10.2337/db05-1404. [DOI] [PubMed] [Google Scholar]
- Colegio OR, Chu NQ, Szabo AL, Chu T, Rhebergen AM, Jairam V, Cyrus N, Brokowski CE, Eisenbarth SC, Phillips GM, et al. Functional polarization of tumour-associated macrophages by tumour-derived lactic acid. Nature. 2014;513:559–563. doi: 10.1038/nature13490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damiola F, Le Minh N, Preitner N, Kornmann B, Fleury-Olela F, Schibler U. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 2000;14:2950–2961. doi: 10.1101/gad.183500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JE., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- Doherty JR, Cleveland JL. Targeting lactate metabolism for cancer therapeutics. J Clin Invest. 2013;123:3685–3692. doi: 10.1172/JCI69741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckel-Mahan KL, Patel VR, de Mateo S, Orozco-Solis R, Ceglia NJ, Sahar S, Dilag-Penilla SA, Dyar KA, Baldi P, Sassone-Corsi P. Reprogramming of the Circadian Clock by Nutritional Challenge. Cell. 2013;155:1464–1478. doi: 10.1016/j.cell.2013.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelli B, Peraldi P, Filloux C, Chavey C, Freidinger K, Hilton DJ, Hotamisligil GS, Van Obberghen E. SOCS-3 inhibits insulin signaling and is up-regulated in response to tumor necrosis factor-alpha in the adipose tissue of obese mice. J Biol Chem. 2001;276:47944–47949. doi: 10.1074/jbc.M104602200. [DOI] [PubMed] [Google Scholar]
- Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem. 2009;81:6656–6667. doi: 10.1021/ac901536h. [DOI] [PubMed] [Google Scholar]
- Fitzgerald JS, Tsareva SA, Poehlmann TG, Berod L, Meissner A, Corvinus FM, Wiederanders B, Pfitzner E, Markert UR, Friedrich K. Leukemia inhibitory factor triggers activation of signal transducer and activator of transcription 3, proliferation, invasiveness, and altered protease expression in choriocarcinoma cells. Int J Biochem Cell Biol. 2005;37:2284–2296. doi: 10.1016/j.biocel.2005.02.025. [DOI] [PubMed] [Google Scholar]
- Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer. 2003;3:350–361. doi: 10.1038/nrc1072. [DOI] [PubMed] [Google Scholar]
- Gamble KL, Berry R, Frank SJ, Young ME. Circadian clock control of endocrine factors. Nat Rev Endocrinol. 2014;10:466–475. doi: 10.1038/nrendo.2014.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao SP, Mark KG, Leslie K, Pao W, Motoi N, Gerald WL, Travis WD, Bornmann W, Veach D, Clarkson B, et al. Mutations in the EGFR kinase domain mediate STAT3 activation via IL-6 production in human lung adenocarcinomas. J Clin Invest. 2007;117:3846–3856. doi: 10.1172/JCI31871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JE, Blaikley J, Beesley S, Matthews L, Simpson KD, Boyce SH, Farrow SN, Else KJ, Singh D, Ray DW, et al. The nuclear receptor REV-ERBalpha mediates circadian regulation of innate immunity through selective regulation of inflammatory cytokines. Proc Natl Acad Sci U S A. 2012;109:582–587. doi: 10.1073/pnas.1106750109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilardi F, Migliavacca E, Naldi A, Baruchet M, Canella D, Le Martelot G, Guex N, Desvergne B, Cycli XC. Genome-wide analysis of SREBP1 activity around the clock reveals its combined dependency on nutrient and circadian signals. PLoS Genet. 2014;10:e1004155. doi: 10.1371/journal.pgen.1004155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov S, Karin E, Terzic J, Mucida D, Yu GY, Vallabhapurapu S, Scheller J, Rose-John S, Cheroutre H, Eckmann L, et al. IL-6 and Stat3 are required for survival of intestinal epithelial cells and development of colitis-associated cancer. Cancer Cell. 2009;15:103–113. doi: 10.1016/j.ccr.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo D, Dunbar JD, Yang CH, Pfeffer LM, Donner DB. Induction of Jak/STAT signaling by activation of the type 1 TNF receptor. J Immunol. 1998;160:2742–2750. [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hatori M, Vollmers C, Zarrinpar A, Ditacchio L, Bushong EA, Gill S, Leblanc M, Chaix A, Joens M, Fitzpatrick JA, et al. Time-Restricted Feeding without Reducing Caloric Intake Prevents Metabolic Diseases in Mice Fed a High-Fat Diet. Cell Metab. 2012;15:848–860. doi: 10.1016/j.cmet.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton JD, Shah NA, Warrington JA, Anderson NN, Park SW, Brown MS, Goldstein JL. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci U S A. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–707. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Mercer K, Greenbaum D, Bronson RT, Crowley D, Tuveson DA, Jacks T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature. 2001;410:1111–1116. doi: 10.1038/35074129. [DOI] [PubMed] [Google Scholar]
- Kelly M, Keller C, Avilucea PR, Keller P, Luo Z, Xiang X, Giralt M, Hidalgo J, Saha AK, Pedersen BK, et al. AMPK activity is diminished in tissues of IL-6 knockout mice: the effect of exercise. Biochem Biophys Res Commun. 2004;320:449–454. doi: 10.1016/j.bbrc.2004.05.188. [DOI] [PubMed] [Google Scholar]
- Kohsaka A, Laposky AD, Ramsey KM, Estrada C, Joshu C, Kobayashi Y, Turek FW, Bass J. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metab. 2007;6:414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- Le Martelot G, Claudel T, Gatfield D, Schaad O, Kornmann B, Lo Sasso G, Moschetta A, Schibler U. REV-ERBalpha participates in circadian SREBP signaling and bile acid homeostasis. PLoS Biol. 2009;7:e1000181. doi: 10.1371/journal.pbio.1000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesina M, Kurkowski MU, Ludes K, Rose-John S, Treiber M, Kloppel G, Yoshimura A, Reindl W, Sipos B, Akira S, et al. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 2011;19:456–469. doi: 10.1016/j.ccr.2011.03.009. [DOI] [PubMed] [Google Scholar]
- Li Y, Xu S, Mihaylova MM, Zheng B, Hou X, Jiang B, Park O, Luo Z, Lefai E, Shyy JY, et al. AMPK phosphorylates and inhibits SREBP activity to attenuate hepatic steatosis and atherosclerosis in diet-induced insulin-resistant mice. Cell Metab. 2011;13:376–388. doi: 10.1016/j.cmet.2011.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin WW, Karin M. A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest. 2007;117:1175–1183. doi: 10.1172/JCI31537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma KL, Ruan XZ, Powis SH, Chen Y, Moorhead JF, Varghese Z. Inflammatory stress exacerbates lipid accumulation in hepatic cells and fatty livers of apolipoprotein E knockout mice. Hepatology. 2008;48:770–781. doi: 10.1002/hep.22423. [DOI] [PubMed] [Google Scholar]
- Masri S, Kinouchi K, Sassone-Corsi P. Circadian clocks, epigenetics, and cancer. Curr Opin Oncol. 2015;27:50–56. doi: 10.1097/CCO.0000000000000153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Rigor P, Cervantes M, Ceglia N, Sebastian C, Xiao C, Roqueta-Rivera M, Deng C, Osborne TF, Mostoslavsky R, et al. Partitioning circadian transcription by SIRT6 leads to segregated control of cellular metabolism. Cell. 2014;158:659–672. doi: 10.1016/j.cell.2014.06.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Sassone-Corsi P. Plasticity and specificity of the circadian epigenome. Nat Neurosci. 2010;13:1324–1329. doi: 10.1038/nn.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauer J, Denson JL, Bruning JC. Versatile functions for IL-6 in metabolism and cancer. Trends Immunol. 2015;36:92–101. doi: 10.1016/j.it.2014.12.008. [DOI] [PubMed] [Google Scholar]
- Michalaki V, Syrigos K, Charles P, Waxman J. Serum levels of IL-6 and TNF-alpha correlate with clinicopathological features and patient survival in patients with prostate cancer. Br J Cancer. 2004;90:2312–2316. doi: 10.1038/sj.bjc.6601814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AM, Wang H, Bertola A, Park O, Horiguchi N, Ki SH, Yin S, Lafdil F, Gao B. Inflammation-associated interleukin-6/signal transducer and activator of transcription 3 activation ameliorates alcoholic and nonalcoholic fatty liver diseases in interleukin-10-deficient mice. Hepatology. 2011;54:846–856. doi: 10.1002/hep.24517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narimatsu M, Maeda H, Itoh S, Atsumi T, Ohtani T, Nishida K, Itoh M, Kamimura D, Park SJ, Mizuno K, et al. Tissue-specific autoregulation of the stat3 gene and its role in interleukin-6-induced survival signals in T cells. Mol Cell Biol. 2001;21:6615–6625. doi: 10.1128/MCB.21.19.6615-6625.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narsale AA, Enos RT, Puppa MJ, Chatterjee S, Murphy EA, Fayad R, Pena MO, Durstine JL, Carson JA. Liver inflammation and metabolic signaling in ApcMin/+ mice: the role of cachexia progression. PLoS One. 2015;10:e0119888. doi: 10.1371/journal.pone.0119888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Antoch MP, Miller BH, Su AI, Schook AB, Straume M, Schultz PG, Kay SA, Takahashi JS, Hogenesch JB. Coordinated transcription of key pathways in the mouse by the circadian clock. Cell. 2002;109:307–320. doi: 10.1016/s0092-8674(02)00722-5. [DOI] [PubMed] [Google Scholar]
- Partch CL, Green CB, Takahashi JS. Molecular architecture of the mammalian circadian clock. Trends Cell Biol. 2014;24:90–99. doi: 10.1016/j.tcb.2013.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruderman NB, Keller C, Richard AM, Saha AK, Luo Z, Xiang X, Giralt M, Ritov VB, Menshikova EV, Kelley DE, et al. Interleukin-6 regulation of AMP-activated protein kinase. Potential role in the systemic response to exercise and prevention of the metabolic syndrome. Diabetes. 2006;55(Suppl 2):S48–54. doi: 10.2337/db06-s007. [DOI] [PubMed] [Google Scholar]
- Rui L, Yuan M, Frantz D, Shoelson S, White MF. SOCS-1 and SOCS-3 block insulin signaling by ubiquitin-mediated degradation of IRS1 and IRS2. J Biol Chem. 2002;277:42394–42398. doi: 10.1074/jbc.C200444200. [DOI] [PubMed] [Google Scholar]
- Sachithanandan N, Fam BC, Fynch S, Dzamko N, Watt MJ, Wormald S, Honeyman J, Galic S, Proietto J, Andrikopoulos S, et al. Liver-specific suppressor of cytokine signaling-3 deletion in mice enhances hepatic insulin sensitivity and lipogenesis resulting in fatty liver and obesity. Hepatology. 2010;52:1632–1642. doi: 10.1002/hep.23861. [DOI] [PubMed] [Google Scholar]
- Sahar S, Sassone-Corsi P. Metabolism and cancer: the circadian clock connection. Nat Rev Cancer. 2009;9:886–896. doi: 10.1038/nrc2747. [DOI] [PubMed] [Google Scholar]
- Sansone P, Storci G, Tavolari S, Guarnieri T, Giovannini C, Taffurelli M, Ceccarelli C, Santini D, Paterini P, Marcu KB, et al. IL-6 triggers malignant features in mammospheres from human ductal breast carcinoma and normal mammary gland. J Clin Invest. 2007;117:3988–4002. doi: 10.1172/JCI32533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senn JJ, Klover PJ, Nowak IA, Zimmers TA, Koniaris LG, Furlanetto RW, Mooney RA. Suppressor of cytokine signaling-3 (SOCS-3), a potential mediator of interleukin-6-dependent insulin resistance in hepatocytes. J Biol Chem. 2003;278:13740–13746. doi: 10.1074/jbc.M210689200. [DOI] [PubMed] [Google Scholar]
- Stokkan KA, Yamazaki S, Tei H, Sakaki Y, Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001;291:490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- Torisu T, Sato N, Yoshiga D, Kobayashi T, Yoshioka T, Mori H, Iida M, Yoshimura A. The dual function of hepatic SOCS3 in insulin resistance in vivo. Genes Cells. 2007;12:143–154. doi: 10.1111/j.1365-2443.2007.01044.x. [DOI] [PubMed] [Google Scholar]
- Tsoli M, Schweiger M, Vanniasinghe AS, Painter A, Zechner R, Clarke S, Robertson G. Depletion of white adipose tissue in cancer cachexia syndrome is associated with inflammatory signaling and disrupted circadian regulation. PLoS One. 2014;9:e92966. doi: 10.1371/journal.pone.0092966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavvas D, Apazidis A, Saha AK, Gamble J, Patel A, Kemp BE, Witters LA, Ruderman NB. Contraction-induced changes in acetyl-CoA carboxylase and 5′-AMP-activated kinase in skeletal muscle. J Biol Chem. 1997;272:13255–13261. doi: 10.1074/jbc.272.20.13255. [DOI] [PubMed] [Google Scholar]
- Vollmers C, Gill S, DiTacchio L, Pulivarthy SR, Le HD, Panda S. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc Natl Acad Sci U S A. 2009;106:21453–21458. doi: 10.1073/pnas.0909591106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi K, Itoh Y, Yokomizo C, Nishimura T, Niimi T, Fujii H, Okanoue T, Yoshikawa T. Blockade of interleukin-6 signaling enhances hepatic steatosis but improves liver injury in methionine choline-deficient diet-fed mice. Lab Invest. 2010;90:1169–1178. doi: 10.1038/labinvest.2010.75. [DOI] [PubMed] [Google Scholar]
- Yeh HH, Lai WW, Chen HH, Liu HS, Su WC. Autocrine IL-6-induced Stat3 activation contributes to the pathogenesis of lung adenocarcinoma and malignant pleural effusion. Oncogene. 2006;25:4300–4309. doi: 10.1038/sj.onc.1209464. [DOI] [PubMed] [Google Scholar]
- Yu H, Lee H, Herrmann A, Buettner R, Jove R. Revisiting STAT3 signalling in cancer: new and unexpected biological functions. Nat Rev Cancer. 2014;14:736–746. doi: 10.1038/nrc3818. [DOI] [PubMed] [Google Scholar]
- Zhang EE, Liu Y, Dentin R, Pongsawakul PY, Liu AC, Hirota T, Nusinow DA, Sun X, Landais S, Kodama Y, et al. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat Med. 2010;16:1152–1156. doi: 10.1038/nm.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Chen Y, Tang R, Chen Y, Li Q, Gong J, Huang A, Varghese Z, Moorhead JF, Ruan XZ. Inflammatory stress exacerbates hepatic cholesterol accumulation via increasing cholesterol uptake and de novo synthesis. J Gastroenterol Hepatol. 2011;26:875–883. doi: 10.1111/j.1440-1746.2010.06560.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.