Abstract
Objective
We hypothesized that a low glycemic diet combined with exercise would increase expression of nuclear regulators of fat transport and oxidation in insulin-resistant skeletal muscle.
Method
Nineteen subjects (64±1 yrs; 34±1 kg/m2) were randomized to receive isocaloric high- (HiGIX; 80±0.6 units, N=10) or low-glycemic index (LoGIX; 40±0.3 units, N=9) diets combined with supervised exercise (1 h/d, 5 d/wk at ~85% HRmax) for 12 weeks. Insulin sensitivity was determined by hyperinsulinemic-euglycemic clamp. Skeletal muscle biopsies were obtained before and after the intervention to assess fasting gene and protein expression.
Results
Weight loss was similar for both groups (9.5±1.3 kg). Likewise, improvements in insulin sensitivity (P<0.002), and PPARγ (P<0.002), PGC-1α (P=0.003), CD36 (P=0.003), FABP3 [mRNA, P=0.01 and protein, P=0.02], and CPT1B [mRNA, P=0.03 and protein, P=0.008] expression were similar for both interventions. Increased insulin sensitivity correlated with increased PGC-1α expression (P=0.04), and increased fasting fat oxidation correlated with increased FABP3 (P=0.04) and CPT1B (P=0.05) expression.
Conclusions
An exercise/diet program resulting in an 8–10% weight loss improved insulin sensitivity and key molecular mechanisms in skeletal muscle that are controlled by PGC-1α. These effects were independent of the glycemic index of the diets.
Keywords: diabetes, obesity, exercise, glycemic index and gene regulation
Introduction
Lifestyle modifications, including diet management and increased physical activity are effective interventions to combat obesity and its comorbidities. Endurance exercise, with or without weight loss, reduces insulin resistance (1–4). However, the role of diet composition on insulin sensitivity, particularly the effect of the glycemic index (GI) of the diet is somewhat controversial. Recent data on dietary GI concluded that a low glycemic diet did not improve insulin sensitivity (5). In contrast, we previously reported that when combined with exercise, both high and low-glycemic diets induce similar improvement in peripheral and hepatic insulin sensitivity (6).
Skeletal muscle is a key site in the regulation of metabolism and insulin-regulated glucose uptake, and is exquisitely responsive to environmental changes, such as nutritional status and physical activity. The adaptations are largely caused by changes in muscle mitochondrial size, content and number, and are associated with changes in fat transport and oxidative enzymes, and oxidative respiratory components that can contribute to ATP production. Peroxisome proliferator-activated receptor γ coactivator-1α (PGC-1α) is a transcriptional coactivator that plays a central role in regulating mitochondrial function and oxidative capacity (7) by augmenting the expression of proteins that promote transcription of both nuclear-encoded mitochondrial genes and mitochondrial DNA (8). Suppression of skeletal muscle PGC-1α is associated with increased energy intake (9, 10), and the development of obesity, diabetes and metabolic diseases (11). On the other hand, exercise is associated with increased expression of PGC-1α and has been shown to facilitate skeletal muscle mitochondrial adaptations, including enhanced mitochondrial biogenesis, bioenergetics, dynamics and oxidative capacity (12, 13).
Fat transport and utilization in skeletal muscle also plays an integral role in metabolic homeostasis. Long-chain fatty acid transport across the plasma membrane is controlled by protein-mediated mechanisms that include CD36, fatty acid binding protein (FABP), and carnitine palmitoyltransferase 1B (CPT1B). Recently, Kawaguchi et al. (14) reported that differences in muscle FABP expression may affect insulin sensitivity, while Kim et al. (15) reported that deletion of one allele of CPT1B in mice caused impairment of muscle insulin signaling that led to severe insulin resistance upon high fat feeding.
Exercise is a widely prescribed physiological intervention for treating and preventing insulin resistance (2, 3, 6). Some of the improvement in glucose homeostasis that is seen following exercise training is attributed to mitochondrial adaptations in skeletal muscle, including enhanced mitochondrial biogenesis, bioenergetics, dynamics and oxidative capacity (13, 16, 17). However, whether dietary carbohydrate quality has an impact on exercise training induced expression of PGC-1α in relation to insulin sensitivity is not known. Carbohydrate quality may be important since high glycemic diets promote hyperinsulinemia and down-regulate genes related to fat metabolism (18), while low glycemic diets have been shown to increase fat oxidation, thus improving glycemic control (19, 20) . Previous experiments in lean healthy individuals suggest that when low glycemic meals are consumed prior to exercise there is a shift in substrate utilization that favors fat as the main fuel during exercise (21, 22). Thus, it appears that exercise, independent of diet composition, may induce changes within skeletal muscle that restore metabolic flexibility thereby correcting muscle metabolism. However, to date this hypothesis has not been tested.
The objective of this study was to determine whether an intervention program that included low or high glycemic diet plus exercise would induce differential change in expression of genes and proteins related to mitochondrial function and lipid metabolism, and whether these adaptations would be associated with improved insulin sensitivity and/or fat metabolism. We hypothesized that participants provided a low glycemic diet would have greater improvements in the expression of fatty acid transport (FAT/CD36 and FABP3) and oxidative enzymes (CPT1B and β-HAD), as well as the master regulator of mitochondrial biogenesis (PGC-1α) and whole body glucose and lipid homeostasis (PPARγ), and this improvement would correlate with increased whole body fat oxidation and insulin sensitivity.
Experiment Design and Method
Subjects
Nineteen older (64±1 year), previously sedentary humans who were obese (BMI: 34±1 kg/m2) (Table 1) were recruited to participate in a 12-week diet and exercise intervention. The study was conducted using a randomized, parallel-group, repeated measures design. All volunteers were non-smokers, and underwent comprehensive medical screening prior to participation to rule out overt disease (type 2 diabetes, cardiovascular, renal, etc.). Female subjects were postmenopausal and not using hormone replacement therapy. Subjects taking medications or dietary supplements known to affect the primary outcomes were excluded from the study. All subjects were verbally briefed and signed informed consent documents in accordance with the Cleveland Clinic Institutional Review Board.
Table 1.
Effect of the HiGIX and LoGIX interventions on body composition, plasma lipids, insulin resistance, and aerobic fitness
| HiGIX (n=10) | LoGIX (n=9) | ANOVA (P value) | ||||
|---|---|---|---|---|---|---|
| pre | post | pre | Post | time | Group × time | |
| Sex (F/M) | 4/6 | 4/6 | 5/4 | 5/4 | ||
| Age, yr | 64 ± 1 | - | 67 ± 1 | - | ||
| Weight, kg | 104 ± 5.2 | 93 ± 4.4 | 93 ± 4.9 | 85 ± 3.8 | 0.05 | 0.78 |
| BMI, kg/m2 | 35.1 ± 1.1 | 31.6 ± 1.3 | 32.6 ± 1.0 | 29.8 ± 0.7 | 0.005 | 0.74 |
| Fat mass, kg | 44.1 ± 2.7 | 34.5 ± 3.6 | 40.6 ± 2.5 | 34.7 ± 2.0 | 0.013 | 0.52 |
| Fat free mass, kg | 57.9 ± 5.0 | 56.7 ± 4.7 | 50.7 ± 3.7 | 49.7 ± 3.4 | 0.81 | 0.97 |
| Fasting plasma glucose, mg/dl | 96.5 ± 2.7 | 92.1 ± 2.0 | 102.8 ± 5.5 | 98.8 ± 2.4 | 0.22 | 0.95 |
| Fasting plasma insulin, µU/ml | 11.5 ± 1.3 | 9.8 ± 1.3 | 14.9 ± 1.2 | 13.1 ± 1.0 | 0.17 | 0.94 |
| HOMA-IR | 2.8 ± 0.3 | 2.2 ± 0.3 | 3.8 ± 0.4 | 3.2 ± 0.3 | 0.10 | 0.93 |
| Fasting fat oxidation rate (g/min) | 0.031 ± 0.01 | 0.045 ± 0.01 | 0.037 ± 0.01 | 0.053 ± 0.01 | 0.006 | 0.82 |
| Energy utilization as fat (%) | 48.3 ± 6.2 | 57.7 ± 5.0 | 48.4 ± 3.3 | 60.0 ± 5.1 | 0.05 | 0.84 |
| Fasting CHO oxidation rate (g/min) | 0.12 ± 0.02 | 0.09 ± 0.01 | 0.13 ± 0.02 | 0.08 ± 0.02 | 0.05 | 0.66 |
| Energy utilization as CHO (%) | 51.7 ± 6.2 | 42.2 ± 5.0 | 51.6 ± 3.3 | 40.0 ± 5.1 | 0.05 | 0.84 |
| REE (kcal/kg FFM/min) | 0.019 ± 0.001 | 0.019 ± 0.001 | 0.019 ± 0.001 | 0.020 ± 0.001 | 0.34 | 0.14 |
| Insulin sensitivity (mg/kgFFM/min/µU.ml) | 0.06 ± 0.01 | 0.10 ± 0.01 | 0.04 ± 0.01 | 0.06 ± 0.01 | 0.002 | 0.52 |
| Glucose disposal rate (mg/kgFFM/min) | 5.6 ± 0.8 | 8.4 ± 0.7 | 4.0 ± 0.4 | 6.7 ± 0.6 | 0.0002 | 0.94 |
| VO2 max, ml/kg/min | 21.7 ± 1.4 | 29.7 ± 2.5 | 21.2 ± 0.9 | 30.0 ± 4.2 | 0.002 | 0.88 |
Data represent mean ± SE. HiGIX, high-glycemic index diet with exercise; LoGIX, low-glycemic index diet with exercise; BMI, body mass index; CHO, carbohydrate; REE, resting energy expenditure, VO2max, Maximal oxygen uptake.
Lifestyle Intervention
Participants were randomly assigned to receive either a high- (HiGIX, n=10) or low- (LoGIX, n=9) GI diet. This was a fully provisioned diet study; all meals were prepared in our metabolic kitchen and provided on a daily basis throughout the intervention as previously described (6). Caloric intake of each individual subject’s diet was calculated from their resting metabolic rate multiplied by a sedentary factor of 1.25, and were formulated by the study dietitian (HB), using GI data tables from Foster-Powell et al. (23). The dietary macronutrient composition (including fiber) was matched between groups, however the glycemic index content of the diet was designed so that the LoGIX diet had a GI of 40 arbitrary units (au) and the HiGIX had a GI of 80-au (6, 24, 25). Dietary adherence was ensured via food container weigh-backs and weekly dietary counseling. Dietary analysis was performed with Nutritionist Pro software (Axxya Systems, Stafford, TX).
The intervention also included a supervised aerobic exercise program consisting of treadmill walking and cycle ergometry performed at approximately 85% of maximum heart rate (HRmax), 5d/wk for 60-min/d. Exercise compliance was calculated from the number of sessions attended divided by the number of exercise sessions in the intervention (i.e., 60), and is expressed as percent. Pre- and post-intervention, the participants were admitted for a 3-day inpatient stay in our Clinical Research Unit to facilitate control of diet and activity prior to all metabolic testing. During the post-study inpatient stay subjects continued their corresponding diet, and metabolic testing (clamp, calorimetry and muscle biopsy) was performed approximately 16 hours after the last exercise bout.
Anthropometric and Fitness Testing
Body composition (i.e. total body fat and fat-free mass) via dual-energy x-ray absorptiometry, aerobic fitness (VO2max), basal fat and carbohydrate oxidation rate, and percentage energy source as fat and carbohydrate were determined as routinely performed (26).
OGTT
The plasma glucose response to a 75-g oral glucose solution was assessed after an overnight fast at pre- and post-intervention. Following a baseline blood draw, glucose was ingested and blood samples were drawn every 30 minutes up to 180 minutes. Plasma glucose was measured immediately on a YSI glucose analyzer (YSI 2300 STAT Plus, Yellow Springs, OH). The incremental area under the blood glucose response curve was calculated as previously described (26).
Insulin Sensitivity
After an overnight fast, a hyperinsulinemic-euglycemic clamp (40 mU•m−2•min−1) was performed to determine insulin-stimulated glucose uptake, as previously described (1). Glucose infusion rates were corrected for glucose distribution space to derive glucose disposal rates (GDR). Whole-body insulin sensitivity was defined as GDR divided by plasma insulin concentrations. A secondary measure of insulin sensitivity was assessed using the homeostatic model assessment of insulin resistance (HOMA-IR) approach (27).
Biochemical Blood Analysis
Plasma glucose was determined using a glucose oxidase assay and plasma insulin was measured by radioimmunoassay (Millipore, Billerica, MA).
Skeletal Muscle Biopsy
A muscle sample was obtained from the vastus lateralis after an overnight fast. A biopsy needle with suction was used to obtain approximately 250 mg of tissue as described (28). The muscle was dissected free from visible fat and connective tissue, and quickly frozen and stored in liquid nitrogen until analysis.
RNA Extraction
Muscle RNA was extracted with TRI Reagent (Sigma, St Louis, MO). Briefly, 10–20 mg of tissue was homogenized in TRI Reagent with repetitive short bursts, and incubated at room temperature for 5–10 minutes, followed by centrifugation at 12,000xg for 10 minutes at 4°C. The supernatant was collected and RNA was separated into an aqueous phase using 1-bromo-3-chloropropane, and precipitated with isopropanol. Isolated RNA was washed with 75% ethanol and dissolved in nuclease-free water. The RNA concentration and purity was determined by measuring the absorbance at 230, 260 and 280 nm using a NanoDrop ND-1000 Spectrophotometer (Thermo Scientific, Wilmington, DE). RNA integrity was randomly assessed using an Agilent bioanalyzer (Agilent, Santa Clara, CA). Isolated RNA was aliquoted and stored at −80°C.
cDNA Synthesis
Prior to cDNA synthesis, RNA samples were treated with DNaseI (Invitrogen, Carlsbad, CA). One microgram of total RNA was reverse transcribed into cDNA (iScript cDNA synthesis kit, Biorad, Hercules, CA) using a PX2 Thermal Cycler (Thermo Scientific). The synthesis was performed following the manufacturer’s instructions. cDNA were stored at −20°C.
qRT-PCR Primer Pairs
Primer pairs for target genes were obtained from PrimerBank (pga.mgh.harvard.edu/primerbank/; Table 2). All primers were checked for specificity to the genes of interest by conducting a Blast analysis.
Table 2.
Primer List for Quantitative Real Time PCR analysis
| Gene | Forward primer | Reverse primer | GenBank Accession # |
Amplicon size (nt) |
|---|---|---|---|---|
| GAPDH | CAC CAA CTG CTT AGC ACC CC | TGG TCA TGA GTC CTT CCA CG | NM_002046 | 70 |
| PGC-1α | TCC TCA CAG AGA CAC TAG ACA G | GGC AAT CCG TCT TCA TCC ACA | NM_013261 | 49 |
| PPARγ | ACC AAA GTG CAA TCA AAG TGG A | AGG CTT ATT GTA GAG CTG AGT CT | NM_138711 | 74 |
| PPARα | AGC TTT GGC TTT ACG GAA TAC CA | CCA CAG GAT AAG TCA CCG AGG A | NM_005036 | 114 |
| PPARβ/δ | ACT GAG TTC GCC AAG AGC ATC | GAA GGG TAA CCT GGT CGT TGA | NM_177435 | 64 |
| FAT/CD36 | GTA CAG AGT TCG TTT TCT AGC CA | GCA GGA AAG AGA CTG TGT C | NM_000072 | 71 |
| FABP3 | GTC ACT CGG TGT GGG TTT TG | TTC GAT TGT GGT AGG CTT G | NM_004102 | 67 |
| CPT1B | CAT GTA TCG CCG TAA ACT GGA C | GCC ATC ACA GGC TTG ATT TCT | NM_004377 | 46 |
| ACC-β | CAA GCC GAT CAC CAA GAG TAA A | CCC TGA GTT ATC AGA GGC TGG | NM_001093 | 79 |
| ACADL | GAT TAA AAG CCC AGG ATA CCG C | GCT GGC AAC CGT ATA TCT TCA A | NM_001608 | 55 |
| HADH | ACC AGG CAG TTC ATG CGT T | TGC TTG ACG ATT ATC TTC TTG GC | NM_001184705 | 74 |
GAPDH – glyceraldehyde-3-phosphate dehydrogenase; PGC-1α – peroxisome proliferative activated receptor (PPAR) γ coactivator-1α; FAT/CD36 – fatty acid translocase, CD36; FABP3 –fatty acid binding protein 3; CPT1B – carnitine palmitoyltransferase-1B; ACC-β – acetyl-CoA carboxylase-β; ACADL – acyl CoA dehydrogenase, long chain; HADH – hydroxyacyl-CoA dehydrogenase.
Semi-Quantitative rtPCR Analysis
Determination of relative mRNA expression was performed in duplicate as previously described (13). Human GAPDH was used as an internal standard for sample normalization (29). The relative changes in mRNA abundance were calculated using the comparative ΔΔCt method (30).
Protein Analysis
Muscle homogenates were prepared by grinding tissue with ice-cold lysis buffer cell extraction buffer (Invitrogen) as previously described (13). Protein concentrations were measured using a BCA protein assay (Thermo Scientific). Proteins were separated by 4–20% Novex Tris-Glycine SDS-PAGE (Invitrogen) and transferred to a PVDF membrane (Biorad). Membranes were incubated overnight with anti-FABP3 (Lifespan Biosciences, Seattle, WA; LS-C138955), anti-CPT1B (Abcam, Cambridge, MA; ab15703) or anti-actin (Santa Cruz, Dallas, TX; sc-1616), followed by incubation with appropriate secondary horseradish peroxidase-conjugated antibodies, anti-rabbit (GE Healthcare, Piscataway, NJ; NA931) and anti-goat (Santa Cruz, sc-2020). Immunoreactive proteins were visualized by chemiluminescence reagent (ECL Prime; GE Healthcare) and quantified by densitometric analysis using ImageQuantTL (GE Healthcare).
Statistical Analysis
Data were analyzed using GraphPad Prism 5 (GraphPad, San Diego, CA). Values were tested for normality using the D’Agostino and Pearson omnibus normality test. An independent t-test was used to assess baseline group differences. There were no statistical differences at baseline for any variable between groups (HiGIX vs LoGIX). A two-way repeated measures analysis of variance (ANOVA) was used to detect group by test interactions. The non-parametric Mann-Whitney t-test and Wilcoxon matched test were used for comparison of gene expression between groups and before and after lifestyle intervention, respectively. Pearson correlation analysis was used to assess associations between variables and to identify the best predictors of the investigated variables. Data represent the mean ± SEM. Statistical significance was accepted as P<0.05.
Results
Diet and Exercise Adherence
The HiGIX and LoGIX groups were provided diets that were based on their pre-intervention resting metabolic rate, adjusted for a sedentary activity factor, and were formulated by the study dietitian (HB), with the only difference being the GI of the diet (6, 24). Compliance with training was 95%, and there were no differences in exercise training intensity between the groups (HiGIX: 83.3±1.5% vs. LoGIX: 84.3±1.0% HRmax; P=0.60).
Body Weight and Aerobic Fitness
Both HiGIX and LoGIX groups reduced body weight (−10.7±2.0 vs. −8.2±1.7 kg, P=0.36, respectively); BMI [Δ= −3.5±0.6 vs. −2.8±0.5 kg/m2, P=0.40]; and fat mass [Δ= −9.6±1.7 vs −5.8±1.6 kg, P=0.14], but differences in weight loss and changes in body composition were not significantly different between the groups. Fat-free mass did not change after either intervention. Aerobic fitness, i.e. VO2max, was significantly increased in each group (P<0.05, Table 1), and the increase was significant when expressed as ml/kg/min and ml/kg FFM/min.
Insulin Sensitivity
Both HiGIX and LoGIX programs led to improved insulin-stimulated glucose disposal rates (74.3±23.2% vs. 78.1±20.4%, respectively; effect of time; P<0.0001). Likewise, both interventions increased whole-body insulin sensitivity 80.1±26.0% vs. 77.4±18.5%, respectively; time effect, P<0.0001, (Table 1). Fasting plasma glucose was unchanged after both interventions. HOMA-IR was reduced after both interventions, although this was not statistically significant (Table 1).
Basal Fat and Carbohydrate Oxidation, PGC-1α, and Lipid Transport Gene Expression
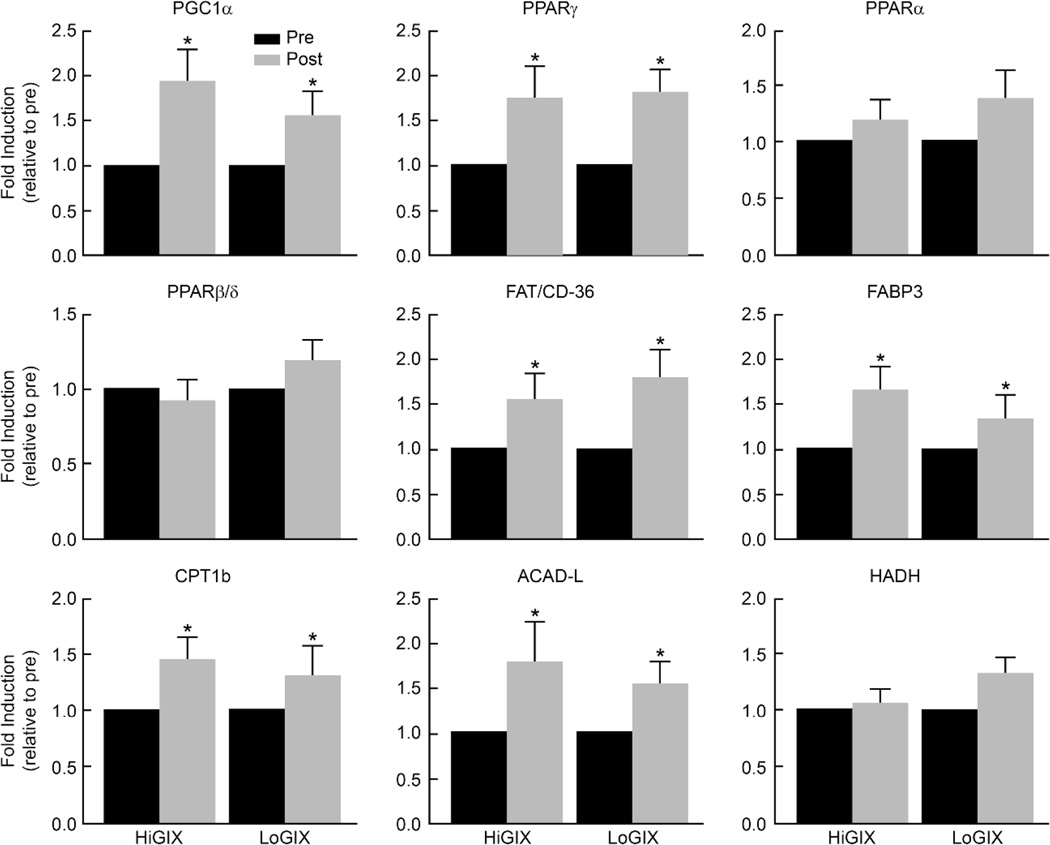
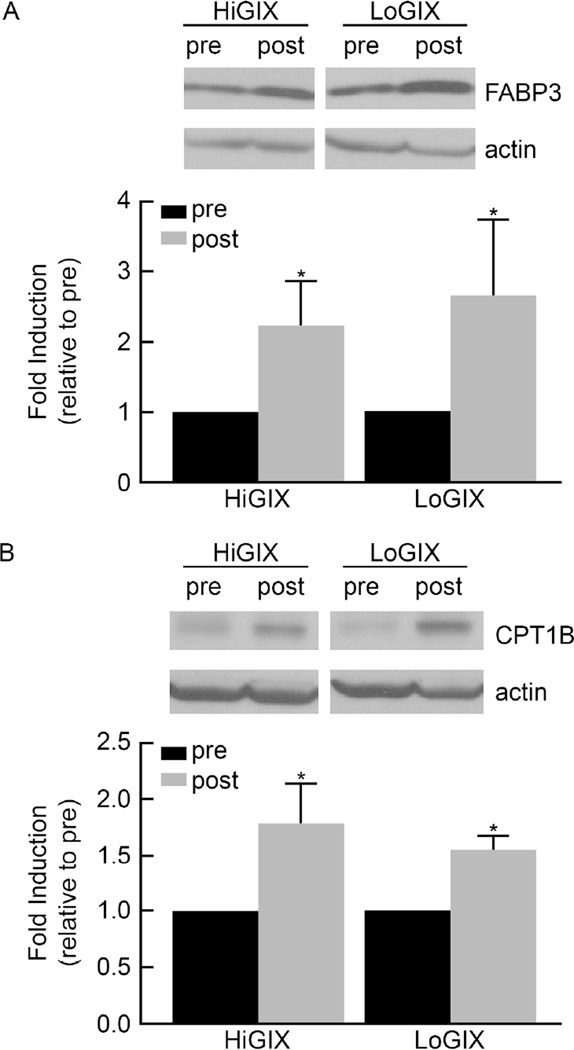
Resting whole body fat oxidation increased in both groups after the intervention (time effect, P=0.02, Table 1) while the carbohydrate oxidation rate decreased (time effect, P=0.05). The increase in fat oxidation was significant whether expressed as g/min (P=0.006), mg/kg/min (P=0.02) or mg/kg FFM/min (P=0.04). However, the decrease in carbohydrate oxidation was not significant when expressed as mg/kg/min (P=0.25) or mg/kg FFM/min (P=0.12). The change in substrate oxidation was similar for both arms of the trial (Table 1). Resting energy expenditure did not change significantly (Table 1). Both lifestyle programs increased the expression of genes involved in skeletal muscle mitochondrial biogenesis (PGC-1α, PPARγ), fatty acid transport (FABP3 and FAT/CD36), and fat oxidation (CPT1B and long-chain acyl-CoA dehydrogenase) (Figure 1). Due to a limited amount of muscle tissue, only FABP3 and CPT1B protein expression levels were determined to confirm findings at the molecular level. The data show that at post-intervention, FABP3 and CPT1B protein abundance were increased in both groups (Figure 2).
Figure 1.
The effect of exercise combined with high and low glycemic diet interventions on the expression of genes involved in fat transport and oxidation. A biopsy of the vastus lateralis muscle was performed after an overnight fast before and after the lifestyle intervention and RNA was extracted from 10–20 mg of the sample using the TRI-method. The expression of genes was analyzed by quantitative–real time PCR analysis, calculated by the ΔΔCt method, and expressed as fold induction relative to pre-exercise. Data are mean ± SE. *Significant difference pre- vs. post-intervention (P<0.05).
Figure 2.
Change in FABP3 and CPT1B protein expression in skeletal muscle. Muscle homogenates were prepared as described and protein was separated by SDS-PAGE electrophoresis, and probed with FABP3 (A) and CPT1B (B) antibodies, respectively. Actin was used as internal loading control. Band intensity were quantified and expressed as fold induction relative to pre-intervention. Data are mean ± SE. *Significant difference pre- vs. post-intervention (P<0.05)
Correlations
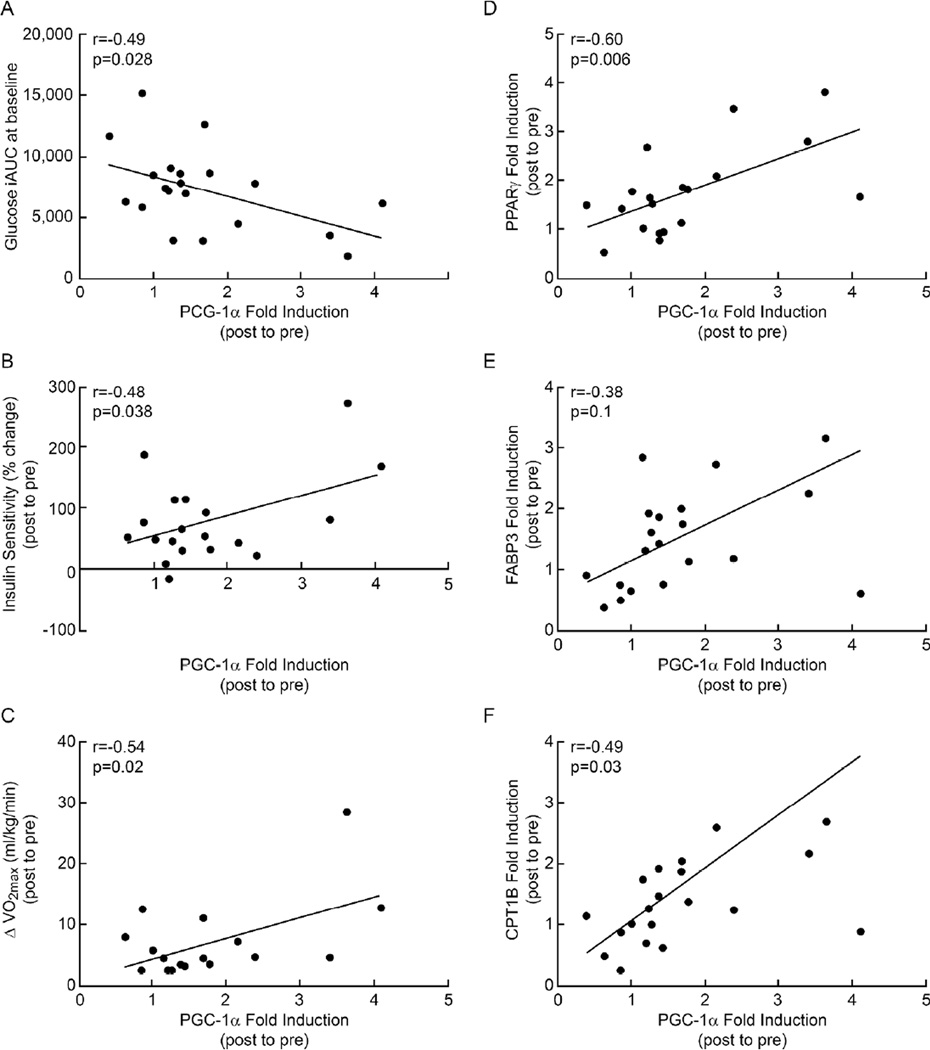
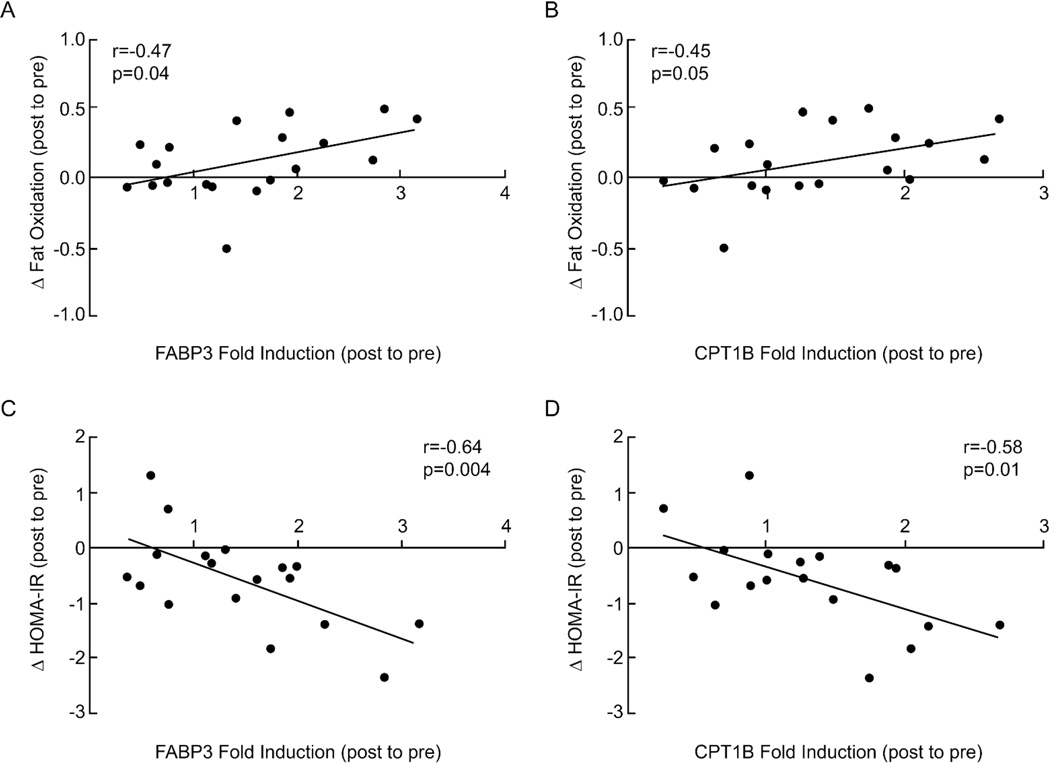
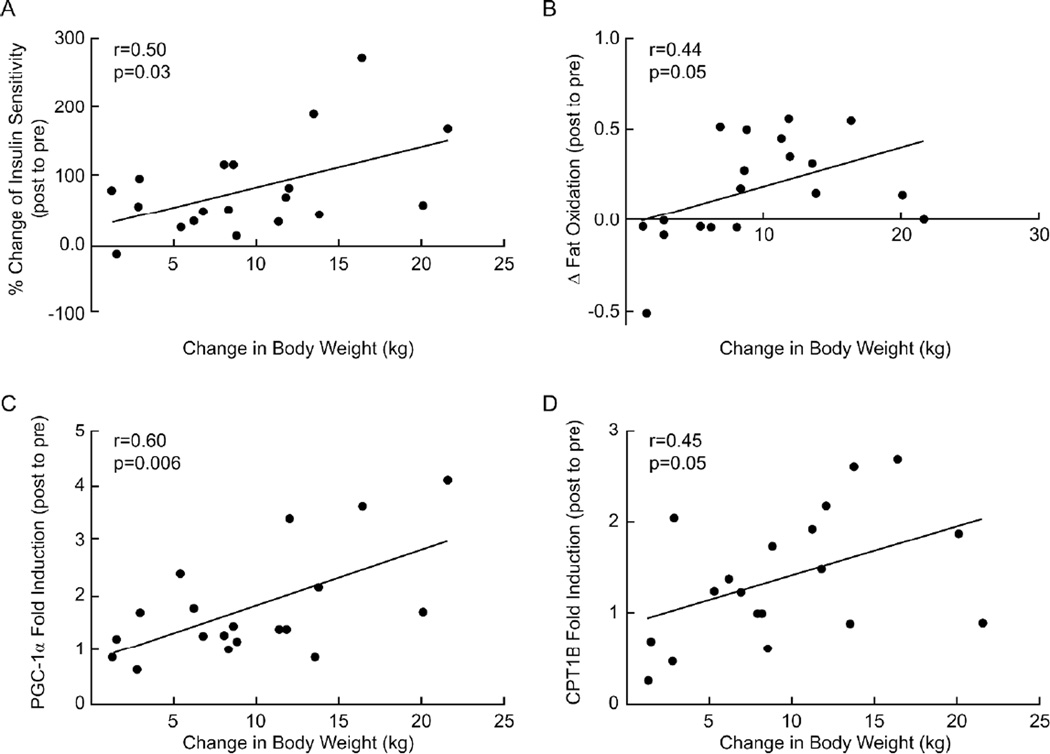
Glucose incremental area under the glucose tolerance curve (iAUC) before the intervention was negatively correlated with changes in PGC-1α mRNA expression after the intervention (r=−0.49, P=0.03; Figure 3A. The increase in PGC-1α mRNA expression was positively correlated with increased insulin sensitivity (r=0.48, P=0.04; Figure 3B), VO2max (r=0.54, P=0.02; Figure 3C), PPARγ (r=0.60, P=0.006; Figure 3D), FABP3 (r=0.38, P=0.1; Figure 3E) and CPT1B (r=0.49, P=0.03; Figure 3F) mRNA expression. Increased VO2max after the intervention was also positively correlated with elevated FAT/CD36 mRNA (r=0.47, P=0.06) and fat oxidation (r=0.46, P<0.05) (data not shown). Enhanced fat oxidation was positively correlated with increased FABP3 and CPT1B mRNA expression (r=0.47, P=0.04; r=0.45, P=0.05, respectively, Figure 4A and 4B). Increases in FABP3 and CPT1B mRNA were negatively associated with reductions in HOMA-IR (r=−0.64, P=0.004; r=−0.58, P=0.01, respectively, Figure 4C and 4D), but not with the clamp measures of insulin sensitivity (data not shown). Weight loss was positively associated with increased insulin sensitivity (r=0.5, P=0.03; Figure 5A), changes in fat oxidation (r=0.44, P=0.05, Figure 5B), and PGC-1α and CPT1B mRNA levels (r=0.6, P=0.006; r=0.45, P=0.05, respectively, Figure 5C, 5D).
Figure 3.
Comparison between fold induction of the PGC-1α gene and baseline incremental area under the glucose response curves (A), changes in lifestyle intervention-induced insulin sensitivity (% changes post relative to pre-intervention) (B), delta VO2max (C), PPARΔ (D), CPT1B (E) and FABP3 (F) fold induction in gene expression.
Figure 4.
Correlation analyses between fold induction of FABP3 (A, C) and CPT1B (B, D) genes with change in fat oxidation (delta post to pre) and change in HOMA-IR (delta post to pre), respectively.
Figure 5.
Correlation analyses between decreased body weight and increased insulin sensitivity (A), increased fat oxidation (post to pre) (B) and fold induction of PGC-1α (C) and CPT1B (D) mRNA levels.
Discussion
To our knowledge, this is the first report on the combined influence of both dietary GI and exercise training on skeletal muscle PGC-1α and fatty acid transport/metabolism gene and/or protein expression level in humans who were older, obese, and insulin resistant. Our data show that a lifestyle intervention that included supervised exercise and either a fully provisioned high- or low-GI diet increased PGC-1α expression, downstream fat transport and oxidation, as well as peripheral insulin sensitivity. The improvement in skeletal muscle fat transport/oxidation gene expression was positively correlated with increased insulin sensitivity and fat oxidation, and importantly, weight loss. The amount of weight loss was consistent with energy expenditure during the supervised exercise program. These data highlight the effect of lifestyle intervention featuring both exercise and controlled dietary intake on cellular and molecular changes in skeletal muscle that lead to important mitochondrial adaptations that are known to regulate insulin sensitivity. These outcomes are more influenced by exercise and weight loss than the GI of the diet.
Although several investigators report positive effects of a low glycemic diet on substrate oxidation and insulin sensitivity (20, 31), these observations have not been universal (32). Some of the inconsistencies may be explained by differences in experimental design, subject population, weight loss, and control of confounding variables. In the current study, improvements in metabolic parameters were followed by changes at the molecular level in skeletal muscle, and all of these responses were independent of dietary glycemic content. However, there was significant weight loss in both groups. The magnitude of weight loss was remarkably consistent with energy expenditure from exercise. The duration and intensity of the exercise program was designed such that the expected energy expenditure was 500 kcal per exercise session. Thus, subjects were expected to expend approximately 30,000 kcals during the 12-week program. From an energy balance perspective, the 8–10 kg weight loss would require a 24–30,000 kcal deficit, and tracks closely with the prescribed energy expenditure from the exercise program. It should be noted that energy intake in this study was fixed and was based on measured resting metabolic rate adjusted for a sedentary activity level. Thus, these data support the view that exercise causes significant weight loss in humans when they do not engage in compensatory eating practices. The data also support the view that when humans lose significant amounts of weight, metabolic outcomes are improved regardless of the GI of the meals that are being consumed. These observations have important implications for dietary counseling and clinical management of obesity and type 2 diabetes.
The underlying mechanisms that contribute to the improvement in insulin sensitivity, substrate utilization, and fat oxidation in our study are partly evident from skeletal muscle adaptations at the molecular level, and in the main pathways that control mitochondrial biogenesis and function. We focused specifically on the coordinated actions of PGC-1α, which is known to control gene programs that optimize cellular bioenergetics, and genes involved in mitochondrial biogenesis, oxidative metabolism, and substrate metabolism (33). It is known that one way to enhance skeletal muscle capacity to handle fatty acids is by increasing proteins and enzymes involved in fatty acid transport (FAT/CD36 and FABP3) and oxidation (CPT1B) (34). Using genetically engineered mice, Summermatter et al. (35) have shown that overexpression of muscle PGC-1α specifically induced expression of FAT/CD36, FABP3, and CPT1B. Exercise also robustly activates PGC-1α expression in skeletal muscle, and this is associated with improvement in metabolic parameters and weight loss (36). Exercise also increases total muscle FABP3 and FAT/CD36 in mitochondria and sarcolemma as well as CPT1B and these adaptations lead to increased mitochondrial content (37, 38). We observed a significant increase in nuclear factors: PPARγ and PGC-1α, fat transport genes: FAT/CD36 and FABP3, and lipid oxidation gene: CPT1B and long chain acyl-CoA dehydrogenase after both interventions. Notably, these responses appear to be more influenced by exercise and weight loss than the GI of the diet. The correlation between increased PGC-1α and PPARγ, FABP3, and CPT1B in our study is consistent with previous reports showing that exercise training increases lipid utilization by improving transport and entry of fat into the mitochondria (38). We also observed that the increase in fat transport (i.e. FABP3 and CPT1B) was correlated with increased whole body basal fat oxidation. Combined these observations point to the likelihood that increased insulin sensitivity after these interventions is a result of better coupling between β-oxidation and the Krebs cycle, leading to more complete lipid oxidation (39), and/or reduced lipid intermediates in skeletal muscle (2, 3). In either case, our findings suggest a strong link between oxidative gene expression and insulin sensitivity after exercise induced weight loss.
One of the major strengths of this study was implementation of a controlled diet and supervised exercise training intervention with monitored energy intake for 12-weeks in individuals who were obese, insulin resistant. Compliance with the program was excellent. In addition, insulin sensitivity was assessed using the gold-standard hyperinsulinemic-euglycemic clamp. However, this study has some limitations that may impact interpretation. First, we cannot exclude the possibility that at least some of the lifestyle-induced improvement in insulin sensitivity is not due to a PGC-1α related mechanism (40). However, the consistent association between PGC-1α, fat transporters and lipid utilization strongly suggests that enhanced mitochondrial function contributes in some manner to the increase in insulin-stimulated glucose uptake after the interventions. Second, we did not directly measure fat oxidation in skeletal muscle. While our whole-body calorimetry measures include skeletal muscle fat oxidation this is only a fraction of the total oxidation and increased oxidation in other tissues such as adipose tissue cannot be discounted. Third, we did not measure parameters related to mitochondrial function in skeletal muscle, therefore we cannot establish a causal link between the observed molecular changes and increased insulin sensitivity. Fourth, due to a limited amount of available tissue, we could only measure gene and total protein expression, and could not determine protein concentrations at the sarcolemma and mitochondrial level as described by others (37). Lastly, we cannot conclude whether the increases in gene expression are the net result of increased mRNA synthesis or mRNA stability. In either case, the directionality of the changes in gene expression coupled with increased insulin sensitivity and fat metabolism suggests that these participants who were obese had effectively lowered their diabetes risk.
In conclusion, this study demonstrated that a combination of exercise training and controlled diet reduced body weight and induced molecular changes in the PGC-1α pathway that upregulates fat transport and oxidation in skeletal muscle of individuals who were older, obese and insulin-resistant. The metabolic effects were not influenced by the GI of the diet. These observations highlight the importance of weight loss for improved insulin sensitivity regardless of the GI of the meals provided during the intervention.
What is already known about this subject?
PGC1-α is a transcriptional coactivator that serves as a master regulator of mitochondrial biogenesis, respiration, fat metabolism and many metabolic processes.
PGC-1α is downregulated in obesity and insulin resistance.
Exercise training upregulates the expression of PGC-1α and its downstream fat transport/oxidation genes.
What does your study add?
Demonstrates that the improvement in skeletal muscle PGC-1α, PPAR-γ and fat transport/oxidation gene and protein expression after lifestyle intervention is independent of dietary glycemic index in human obesity.
Provides data from humans with obesity to show that the association between exercise training induced changes in skeletal muscle at the molecular and cellular level and improved insulin sensitivity are independent of glycemic index of the diet.
Demonstrates that exercise induced changes in resting fat metabolism correlated with increased expression of genes responsible for fat transport/oxidation, and these associations are independent of the glycemic index of the diet.
Acknowledgments
We thank the CRU staff, the participants for their dedication and contributions to the study, and the exceptional organizational assistance of Julianne Filion, R.N., B.S.N.
Funding Agencies: This research was supported by NIH Grants RO1 AG-12834 (JPK) and NIH National Center for Research Resources, 1UL1RR024989, Cleveland OH. JMH was supported by T32 HL-007887, and KRK and SKM were supported by T32 DK007319.
Footnotes
Disclosure: The authors declared no conflict of interest.
Authors Contributions: JPK and HB developed the primary study hypotheses, AM and JPK developed the PGC-1α hypothesis. AM was primarily responsible for sample and data analysis. JMH, TPJS. KRK, MR, HB, and JPK performed data collection. SKM assisted with statistical analysis. AM and JPK wrote the manuscript and remaining authors edited the manuscript. JPK takes responsibility for data integrity.
References
- 1.Kirwan JP, Solomon TP, Wojta DM, Staten MA, Holloszy JO. Effects of 7 days of exercise training on insulin sensitivity and responsiveness in type 2 diabetes mellitus. Am J Physiol Endocrinol Metab. 2009;297(1):E151–E156. doi: 10.1152/ajpendo.00210.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dube JJ, Amati F, Stefanovic-Racic M, Toledo FG, Sauers SE, Goodpaster BH. Exercise-induced alterations in intramyocellular lipids and insulin resistance: the athlete's paradox revisited. Am J Physiol Endocrinol Metab. 2008;294(5):E882–E888. doi: 10.1152/ajpendo.00769.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bruce CR, Thrush AB, Mertz VA, Bezaire V, Chabowski A, Heigenhauser GJ, et al. Endurance training in obese humans improves glucose tolerance and mitochondrial fatty acid oxidation and alters muscle lipid content. Am J Physiol Endocrinol Metab. 2006;291(1):E99–E107. doi: 10.1152/ajpendo.00587.2005. [DOI] [PubMed] [Google Scholar]
- 4.Tuomilehto J, Lindstrom J, Eriksson JG, Valle TT, Hamalainen H, Ilanne-Parikka P, et al. Prevention of type 2 diabetes mellitus by changes in lifestyle among subjects with impaired glucose tolerance. N Engl J Med. 2001;344(18):1343–1350. doi: 10.1056/NEJM200105033441801. [DOI] [PubMed] [Google Scholar]
- 5.Sacks FM, Carey VJ, Anderson CA, Miller ER, 3rd, Copeland T, Charleston J, et al. Effects of high vs low glycemic index of dietary carbohydrate on cardiovascular disease risk factors and insulin sensitivity: the OmniCarb randomized clinical trial. JAMA. 2014;312(23):2531–2541. doi: 10.1001/jama.2014.16658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Solomon TP, Haus JM, Kelly KR, Cook MD, Filion J, Rocco M, et al. A low-glycemic index diet combined with exercise reduces insulin resistance, postprandial hyperinsulinemia, and glucose-dependent insulinotropic polypeptide responses in obese, prediabetic humans. Am J Clin Nutr. 2010;92(6):1359–1368. doi: 10.3945/ajcn.2010.29771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ventura-Clapier R, Garnier A, Veksler V. Transcriptional control of mitochondrial biogenesis: the central role of PGC-1alpha. Cardiovasc Res. 2008;79(2):208–217. doi: 10.1093/cvr/cvn098. [DOI] [PubMed] [Google Scholar]
- 8.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1(6):361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Morino K, Petersen KF, Dufour S, Befroy D, Frattini J, Shatzkes N, et al. Reduced mitochondrial density and increased IRS-1 serine phosphorylation in muscle of insulin-resistant offspring of type 2 diabetic parents. J Clin Invest. 2005;115(12):3587–3593. doi: 10.1172/JCI25151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Morino K, Petersen KF, Shulman GI. Molecular mechanisms of insulin resistance in humans and their potential links with mitochondrial dysfunction. Diabetes. 2006;55(Suppl 2):S9–S15. doi: 10.2337/db06-S002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Summermatter S, Handschin C. PGC-1alpha and exercise in the control of body weight. Int J Obes (Lond) 2012;36(11):1428–1435. doi: 10.1038/ijo.2012.12. [DOI] [PubMed] [Google Scholar]
- 12.Baar K, Wende AR, Jones TE, Marison M, Nolte LA, Chen M, et al. Adaptations of skeletal muscle to exercise: rapid increase in the transcriptional coactivator PGC-1. FASEB J. 2002;16(14):1879–1886. doi: 10.1096/fj.02-0367com. [DOI] [PubMed] [Google Scholar]
- 13.Fealy CE, Mulya A, Lai N, Kirwan JP. Exercise training decreases activation of the mitochondrial fission protein dynamin-related protein-1 in insulin-resistant human skeletal muscle. J Appl Physiol (1985) 2014;117(3):239–245. doi: 10.1152/japplphysiol.01064.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kawaguchi M, Tamura Y, Kakehi S, Takeno K, Sakurai Y, Watanabe T, et al. Association between expression of FABPpm in skeletal muscle and insulin sensitivity in intramyocellular lipid-accumulated nonobese men. J Clin Endocrinol Metab. 2014;99(9):3343–3352. doi: 10.1210/jc.2014-1896. [DOI] [PubMed] [Google Scholar]
- 15.Kim T, He L, Johnson MS, Li Y, Zeng L, Ding Y, et al. Carnitine Palmitoyltransferase 1b Deficiency Protects Mice from Diet-Induced Insulin Resistance. J Diabetes Metab. 2014;5(4):361. doi: 10.4172/2155-6156.1000361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holloszy JO. Regulation by exercise of skeletal muscle content of mitochondria and GLUT4. J Physiol Pharmacol. 2008;59(Suppl 7):5–18. [PubMed] [Google Scholar]
- 17.Horowitz JF, Leone TC, Feng W, Kelly DP, Klein S. Effect of endurance training on lipid metabolism in women: a potential role for PPARalpha in the metabolic response to training. Am J Physiol Endocrinol Metab. 2000;279(2):E348–E355. doi: 10.1152/ajpendo.2000.279.2.E348. [DOI] [PubMed] [Google Scholar]
- 18.Isken F, Klaus S, Petzke KJ, Loddenkemper C, Pfeiffer AF, Weickert MO. Impairment of fat oxidation under high- vs. low-glycemic index diet occurs before the development of an obese phenotype. Am J Physiol Endocrinol Metab. 2010;298(2):E287–E295. doi: 10.1152/ajpendo.00515.2009. [DOI] [PubMed] [Google Scholar]
- 19.Lopes da Silva MV, de Cassia Goncalves Alfenas R. Effect of the glycemic index on lipid oxidation and body composition. Nutr Hosp. 2011;26(1):48–55. [PubMed] [Google Scholar]
- 20.Kahlhofer J, Lagerpusch M, Enderle J, Eggeling B, Braun W, Pape D, et al. Carbohydrate intake and glycemic index affect substrate oxidation during a controlled weight cycle in healthy men. Eur J Clin Nutr. 2014;68(9):1060–1066. doi: 10.1038/ejcn.2014.132. [DOI] [PubMed] [Google Scholar]
- 21.Kirwan JP, Cyr-Campbell D, Campbell WW, Scheiber J, Evans WJ. Effects of moderate and high glycemic index meals on metabolism and exercise performance. Metabolism. 2001;50(7):849–855. doi: 10.1053/meta.2001.24191. [DOI] [PubMed] [Google Scholar]
- 22.Stevenson EJ, Williams C, Mash LE, Phillips B, Nute ML. Influence of high-carbohydrate mixed meals with different glycemic indexes on substrate utilization during subsequent exercise in women. Am J Clin Nutr. 2006;84(2):354–360. doi: 10.1093/ajcn/84.1.354. [DOI] [PubMed] [Google Scholar]
- 23.Foster-Powell K, Holt SH, Brand-Miller JC. International table of glycemic index and glycemic load values: 2002. Am J Clin Nutr. 2002;76(1):5–56. doi: 10.1093/ajcn/76.1.5. [DOI] [PubMed] [Google Scholar]
- 24.Kelly KR, Haus JM, Solomon TP, Patrick-Melin AJ, Cook M, Rocco M, et al. A low-glycemic index diet and exercise intervention reduces TNF(alpha) in isolated mononuclear cells of older, obese adults. J Nutr. 2011;141(6):1089–1094. doi: 10.3945/jn.111.139964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solomon TP, Haus JM, Kelly KR, Cook MD, Riccardi M, Rocco M, et al. Randomized trial on the effects of a 7-d low-glycemic diet and exercise intervention on insulin resistance in older obese humans. Am J Clin Nutr. 2009;90(5):1222–1229. doi: 10.3945/ajcn.2009.28293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Leary VB, Jorett AE, Marchetti CM, Gonzalez F, Phillips SA, Ciaraldi TP, et al. Enhanced adiponectin multimer ratio and skeletal muscle adiponectin receptor expression following exercise training and diet in older insulin-resistant adults. Am J Physiol Endocrinol Metab. 2007;293(1):E421–E427. doi: 10.1152/ajpendo.00123.2007. [DOI] [PubMed] [Google Scholar]
- 27.Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, Turner RC. Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia. 1985;28(7):412–419. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 28.Kirwan JP, del Aguila LF, Hernandez JM, Williamson DL, O'Gorman DJ, Lewis R, et al. Regular exercise enhances insulin activation of IRS-1-associated PI3-kinase in human skeletal muscle. J Appl Physiol (1985) 2000;88(2):797–803. doi: 10.1152/jappl.2000.88.2.797. [DOI] [PubMed] [Google Scholar]
- 29.Mahoney DJ, Carey K, Fu MH, Snow R, Cameron-Smith D, Parise G, et al. Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise. Physiol Genomics. 2004;18(2):226–231. doi: 10.1152/physiolgenomics.00067.2004. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Clapp JF, Lopez B. Low-versus high-glycemic index diets in women: effects on caloric requirement, substrate utilization and insulin sensitivity. Metab Syndr Relat Disord. 2007;5(3):231–242. doi: 10.1089/met.2006.0040. [DOI] [PubMed] [Google Scholar]
- 32.Diaz EO, Galgani JE, Aguirre CA, Atwater IJ, Burrows R. Effect of glycemic index on whole-body substrate oxidation in obese women. Int J Obes (Lond) 2005;29(1):108–114. doi: 10.1038/sj.ijo.0802592. [DOI] [PubMed] [Google Scholar]
- 33.Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell. 1999;98(1):115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 34.McFarlan JT, Yoshida Y, Jain SS, Han XX, Snook LA, Lally J, et al. In Vivo, Fatty Acid Translocase (CD36) Critically Regulates Skeletal Muscle Fuel Selection, Exercise Performance, and Training-induced Adaptation of Fatty Acid Oxidation. J Biol Chem. 2012;287(28):23502–23516. doi: 10.1074/jbc.M111.315358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Summermatter S, Baum O, Santos G, Hoppeler H, Handschin C. Peroxisome proliferator-activated receptor {gamma} coactivator 1{alpha} (PGC-1{alpha}) promotes skeletal muscle lipid refueling in vivo by activating de novo lipogenesis and the pentose phosphate pathway. J Biol Chem. 2010;285(43):32793–32800. doi: 10.1074/jbc.M110.145995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Correia JC, Ferreira DM, Ruas JL. Intercellular: local and systemic actions of skeletal muscle PGC-1s. Trends Endocrinol Metab. 2015;26(6):305–314. doi: 10.1016/j.tem.2015.03.010. [DOI] [PubMed] [Google Scholar]
- 37.Talanian JL, Holloway GP, Snook LA, Heigenhauser GJ, Bonen A, Spriet LL. Exercise training increases sarcolemmal and mitochondrial fatty acid transport proteins in human skeletal muscle. Am J Physiol Endocrinol Metab. 2010;299(2):E180–E188. doi: 10.1152/ajpendo.00073.2010. [DOI] [PubMed] [Google Scholar]
- 38.Tunstall RJ, Mehan KA, Wadley GD, Collier GR, Bonen A, Hargreaves M, et al. Exercise training increases lipid metabolism gene expression in human skeletal muscle. Am J Physiol Endocrinol Metab. 2002;283(1):E66–E72. doi: 10.1152/ajpendo.00475.2001. [DOI] [PubMed] [Google Scholar]
- 39.Muoio DM, Koves TR. Skeletal muscle adaptation to fatty acid depends on coordinated actions of the PPARs and PGC1 alpha: implications for metabolic disease. Appl Physiol Nutr Metab. 2007;32(5):874–883. doi: 10.1139/H07-083. [DOI] [PubMed] [Google Scholar]
- 40.Leick L, Wojtaszewski JF, Johansen ST, Kiilerich K, Comes G, Hellsten Y, et al. PGC-1alpha is not mandatory for exercise- and training-induced adaptive gene responses in mouse skeletal muscle. Am J Physiol Endocrinol Metab. 2008;294(2):E463–E474. doi: 10.1152/ajpendo.00666.2007. [DOI] [PubMed] [Google Scholar]