Abstract
Background
Injury to the airways after smoke inhalation is a major mortality risk factor in victims of burn injuries, resulting in a 15–45% increase in patient deaths. Damage to the airways by smoke may induce acute respiratory distress syndrome (ARDS), which is partly characterized by hypoxemia in the airways. While ARDS has been associated with bacterial infection, the impact of hypoxemia on airway microbiota is unknown. Our objective was to identify differences in microbiota within the airways of burn patients who develop hypoxemia early after inhalation injury and those that do not using next-generation sequencing of bacterial 16S rRNA genes.
Results
DNA was extracted from therapeutic bronchial washings of 48 patients performed within 72 hours of hospitalization for burn and inhalation injury at the North Carolina Jaycee Burn Center. DNA was prepared for sequencing using a novel molecule tagging method and sequenced on the Illumina MiSeq platform. Bacterial species were identified using the MTToolbox pipeline. Patients with hypoxemia, as indicated by a PaO2/FiO2 ratio ≤ 300, had a 30% increase in abundance of Streptococcaceae and Enterobacteriaceae and 84% increase in Staphylococcaceae as compared to patients with a PaO2/FiO2 ratio > 300. Wilcoxon rank-sum test identified significant enrichment in abundance of OTUs identified as Prevotella melaninogenica (p = 0.042), Corynebacterium (p = 0.037) and Mogibacterium (p = 0.048). Linear discriminant effect size analysis (LefSe) confirmed significant enrichment of Prevotella melaninognica among patients with a PaO2/FiO2 ratio ≤ 300 (p<0.05). These results could not be explained by differences in antibiotic treatment.
Conclusions
The airway microbiota following burn and inhalation injury is altered in patients with a PaO2/FiO2 ratio ≤ 300 early after injury. Enrichment of specific taxa in patients with a PaO2/FiO2 ratio ≤ 300 may indicate airway environment and patient changes that favor these microbes. Longitudinal studies are necessary to identify stably colonizing taxa that play roles in hypoxemia and ARDS pathogenesis.
Introduction
Smoke-induced inhalation injury occurs in up to 43% of burn victims, increasing death rates by up to 20% as compared to patients with burn injury alone [1]. Inhalation injury predisposes these patients to respiratory failure, acute respiratory distress syndrome (ARDS), and pneumonia. Pneumonia, in combination with burn and inhalation injury, further increases patient mortality to 60% and is a contributing risk factor to development of ARDS [2,3]. ARDS is a life-threatening condition resulting from either direct or indirect injury to the lung, and is diagnosed clinically by the presence of bilateral opacities on chest imaging and airway hypoxemia [3,4]. Hypoxemia is determined by the ratio of the partial pressure of arterial oxygen (PaO2) to the fraction of inspired oxygen (FiO2). To meet the Berlin definition of ARDS, this ratio must be less than or equal to 300 mm Hg, with a minimum positive end expiratory pressure (PEEP) of 5 cm H2O [4]. Although bacterial infection is frequently the first step towards pneumonia and sepsis, and can induce direct injury to the lung and contribute to the pathogenesis of ARDS, its relationship with the disease is complex and not well understood [5].
Early antibiotic therapy is critical to improved patient outcomes once infection and pneumonia occur, but identification of the organisms can be challenging [6]. Current methodologies rely on culture or polymerase chain reaction (PCR) techniques to identify the causative agent [7]; however, these methods require specific knowledge of the organism’s growth and metabolic requirements and a period of 1–2 days for identification and susceptibility testing, which are prone to false positive results [7]. These limitations often result in broad-spectrum antibiotic treatment that may have little impact on the target organism, promote the development of antibiotic resistance, and ultimately increase mortality [6,7].
To address the limitation of organism identification, we utilized next-generation sequencing of bacterial 16S rRNA genes to characterize the bacterial communities (collectively known as microbiota) in the airways of burn patients following smoke inhalation with or without a PaO2/FiO2 ratio ≤ 300, regardless of the presence of ARDS. Study of the microbiota has revealed the key roles they play in the development and function of the host immune system, and how dysbiosis, or perturbation of the communities, contributes to disease [8–10]. Although host-microbiota interactions are complex and poorly understood, recent studies underscore the importance of low-abundance species in dysbiosis and disease progression, particularly in the airways [10,11]. We hypothesized that inhalation injury and a low P/F ratio (≤ 300) would create conditions within the airways that favor distinct communities of bacteria. We show that facultative anaerobic taxa are enriched among all burn patients, and that specific, low-abundance bacterial taxa are associated with low P/F ratios within the first 24 to 72 hours after injury.
Methods
Patients and sample collection
Therapeutic bronchial washings from patients hospitalized for burn and inhalation injury at the North Carolina Jaycee Burn Center were collected as previously described [12]. Briefly, patients with suspected inhalation injury underwent clinically indicated bronchoscopy within 24 hours of admission. All patients were intubated, bronchial washes performed, and inhalation injury severity scored on the basis of examination. Only those patients for whom inhalation injury was confirmed by bronchoscopy were included in the present study. Only samples taken within three days of injury were utilized. Clinical cultures were grown to detect bacteria within these bronchoscopy samples. Organisms detected per patient and antibiotic treatment are listed in S3 Table in the additional data. Differential cell counts were not done for the bronchial washings. According to the Berlin definition of ARDS, hypoxemia was defined as the ratio of the partial pressure of arterial oxygen (PaO2) to the fraction of inspired oxygen (FiO2) ≤300 [4,12]. Ratios >300 were defined as normal oxygenation levels [3]. The P/F ratio for each patient in this study was measured the same day the bronchial washing was done. Other clinical information, such as patient demographics and total body surface area burned, were collected upon admission. The study protocol was approved by the Institutional Review Board at the University of North Carolina School of Medicine in Chapel Hill (IRB# 10–0959 and #12–2475). All patients or their legally authorized representative gave written informed consent for collection of their bronchial washings for inclusion in a repository as previously described [12]. Analysis of the microbiota in bronchial washings was not an original part of the study and was added after completion of sample collection (IRB #12–2475).
DNA extraction and sequencing
Bronchial washes were transported on ice and processed within 24–48 hours. DNA was extracted from the cellular portion of the wash and quantified (online data supplement). Positive Staphylococcus aureus and negative reagent and human DNA controls were extracted simultaneously and prepared in parallel with the patient samples for sequencing. Sequencing of all DNA was performed in duplicate by a molecule tagging method recently described by Lundberg et al., [13]. This approach allows us to confidently identify operational taxonomic units (OTU) that diverge at the 3% threshold. Briefly, a short round of polymerase chain reaction (PCR) was performed to attach molecule tags to each DNA molecule, followed by a round of full PCR to label each individual sample with a barcode and attach the adapters necessary for sequencing. The primers targeted the V4 region of the bacterial 16S rRNA gene with forward sequence GTGCCAGCMGCCGCGGTAA (515F) and reverse sequence TAATCTWTGGGVHCATCAGG (806R) [13]. Sequencing was performed on the Illumina MiSeq platform at the High Throughput Sequencing Facility at the University of North Carolina at Chapel Hill.
Sequencing data and statistical analysis
We used the MTToolbox [14] pipeline to minimize sequencing errors and match reads to the GreenGenes 16S rRNA database [15]. Sequences that did not match a 16S GreenGenes sequence were removed from the OTU table and those remaining were corrected for variation among 16S rRNA operon number. An R-squared read number correlation was performed on technical replicates in order to determine an appropriate threshold of low read count OTUs to remove from the data [16]. This method improves reproducibility of the results while minimizing loss of data. Total raw counts per OTU of duplicate patient samples were averaged and count thresholds were set for the OTU tables using an R-squared correlation analysis as detailed previously [16]. The samples varied according to the date of sequencing and thresholds were set separately for each group (S1 and S2 Figs). Samples from patients with and without hypoxemia were distributed among the sequencing plates as shown in S1 Table. OTU tables with appropriate thresholds were imported into the program Explicet [17] for normalization and subsequent diversity analyses and the Wilcoxon rank-sum and two-proportions tests. Rarefaction was performed on sample counts within Explicet before bootstrapping to calculate Chao1 diversity indices. The Wilcoxon test is a non-parametric, continuity-corrected test appropriate for analysis of differential OTU abundances [17]. The two-proportions test performs a continuity-adjusted chi-square test to determine differences in detection among OTUs [17]. One-way analysis of variance (ANOVA) was used to identify differences among the abundance of aerobic and anaerobic bacterial taxa present in patients with and without a P/F ratio ≤ 300 (performed in R as anova = lm(Taxa_per_seq_count~Group, data = ALI)) [18]. The phyloseq package within R was used to create a principle components analysis (PCA) plot to compare beta diversity across patients [19]. The linear discriminant analysis (LDA) effect size (LEfSe) method [20] was used to determine the significance of differences in taxa abundance by biologically relevant classes, which included patient P/F ratio and antibiotic treatment. LEfSe first performs a factorial Kruskal-Wallis test to determine differential distribution of OTUs among the biological classes. If subclasses are present, a pairwise Wilcoxon test is done on those with p values greater than 0.05. OTUs with significant differences are then used to build a linear discriminant analysis model, which uses the relative differences of OTUs among classes to rank those that are most discriminative. To determine the influence of antibiotic treatment and sequencing batch effect on these results, we performed a non-parametric differential abundance analysis adjusted for antibiotic treatment and batch effects [21].
Results
Patients
Of the 48 patients included in this study, 50% had P/F ratios ≤ 300 (Table 1). Of the 24 patients with P/F ratios > 300, 7 subsequently had a P/F ratio < 300. All patients who did not survive had initial P/F ratios < 300. The rate of positive bacterial cultures in both patients with (21%) and without (25%) a P/F ratio ≤ 300 was similar to the overall rate (23%). However, the rate of antibiotic treatment within the first 72 hours of injury in patients with a P/F ratio ≤ 300 was lower (29%) than either patients with a higher P/F ratio (46%) or the entire group (40%). Antibiotic treatment was not associated with P/F ratio ≤ 300 (chi-square test, p = 0.4).
Table 1. Patient clinical characteristics.
| Clinical Variable | Total | PaO2/FiO2 ≤ 300 | PaO2/FiO2 > 300 | T Test p Value |
|---|---|---|---|---|
| Patients | 48 | 24 | 24 | NA |
| Males | 36 (75%) | 18 (75%) | 18 (75%) | NA |
| Females | 12 (25%) | 6 (25%) | 6 (25%) | NA |
| BMI | 27 (14–51) | 30 (17–51) | 25 (14–42) | 0.1107 |
| Age | 41 (1–75) | 42 (8–76) | 41 (1–75) | 0.8143 |
| %TBSA | 19 (0–85) | 27 (0–85) | 10 (0–40) | 0.002 |
| Antibiotic Treated | 18 (40%) | 7 (29%) | 11 (46%) | 0.3711¶ |
| Baux Score | 60 (1–115) | 71 (31–115) | 50 (1–96) | 0.0109 |
| Endotracheal Tube | 29 (60%) | 17 (71%) | 12 (50%) | 0.3099¶ |
| Days on Ventilator | 35 (0–105) | 45 (0–105) | 25 (0–79) | 0.0136 |
| Positive Cultures | 11 (23%) | 5 (21%) | 6 (25%) | 0.894¶ |
| Survived | 41 (87%) | 17 (71%)* | 24 (100%) | 0.02497¶ |
Patient clinical characteristics were grouped by total population and subdivided by P/F ratio. The data are represented as mean (range) or number (percent). PaO2/FiO2 > 300 and PaO2/FiO2 ≤ 300 group percentages are calculated per group total.
*Cause of death was either or both cardiac and pulmonary failure.
¶Indicates the p value from Pearson’s chi-squared test.
%TBSA = percent total body surface area burn.
The airway microbiota among all patients
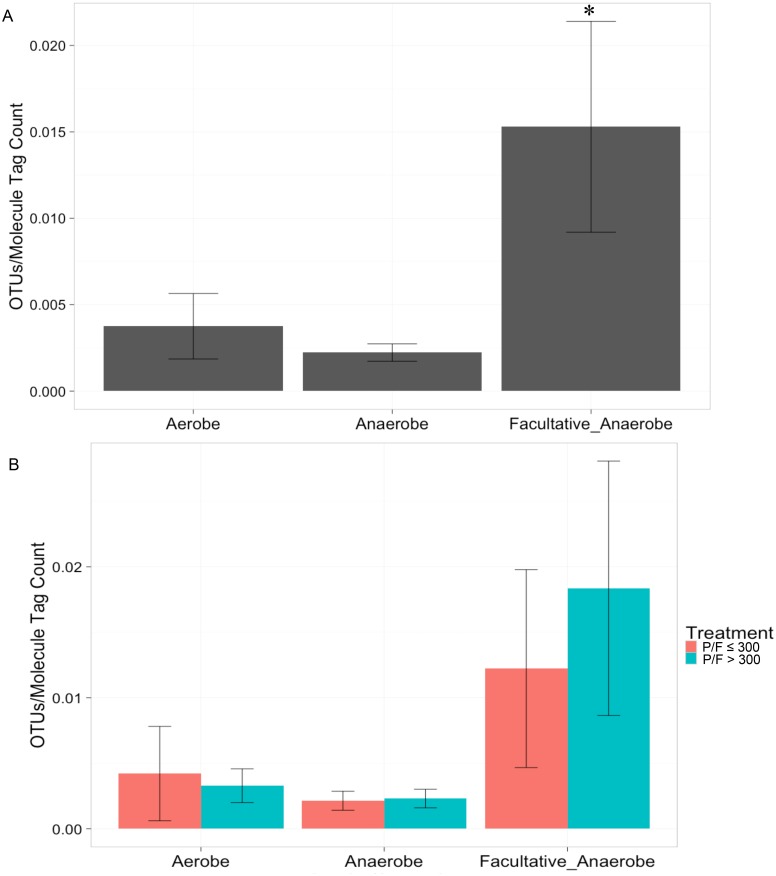
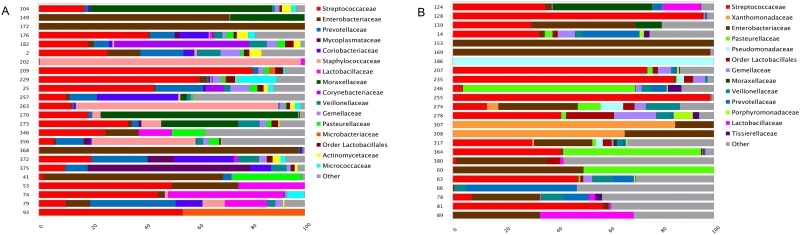
Among all patient samples, OTUs identified as facultative anaerobic taxa were detected at a significantly higher rate than OTUs identified as either obligate anaerobic or obligate aerobic taxa (Fig 1A; ANOVA p = 0.029). When we split the data by PaO2/FiO2 ratio, patients with PaO2/FiO2 >300 contained more unique facultative anaerobic OTUs than those with PaO2/FiO2 ≤ 300, but this difference was not significant (Fig 1B). The most abundant OTUs among all patient samples at the family level were Streptococcaceae and Enterobacteriaceae, which accounted for 26% and 18% of total family-level OTUs, respectively. The remaining 56% of OTUs consisted of 45 additional families, each present at 7% of the total family-level OTUs or less. Fig 2 shows the composition of the microbiota per patient for those with (Fig 2A) and without (Fig 2B) hypoxemia. Positive and negative control samples were sequenced with the patient samples and S2 Table quantifies the total reads and molecule tags produced and S3 Fig shows the composition of these reads.
Fig 1. Increased detection of unique facultative anaerobic taxa.
(A) Unique facultative anaerobic OTUs were detected significantly more frequently than obligate aerobes or anaerobes among all patients. (B) No significant difference was found between number of unique facultative anaerobic OTUs when the data was split by patient PaO2/FiO2 ratio. OTUs were identified as facultative anaerobes, obligate anaerobes, or obligate aerobes among all patients. OTUs were quantified and normalized to the molecule tag count and averaged by bacterial aerobic or anaerobic capability. One-way ANOVA detected a significant difference among the mean taxa of facultative anaerobes (p = 0.029). (n = 48)
Fig 2. Patient airway microbiota composition.
OTUs identified as the families Streptococcaceae and Enterobacteriaceae dominate the airway microbiota within 72 hours following burn and inhalation injury. (A) The microbiota composition among patients with hypoxemia. (B) The microbiota composition among patients without hypoxemia. Each horizontal bar represents an individual patient microbiota normalized to 100%. Different colored sections within each bar indicate abundance of specific families. The category ‘Other’ includes bacterial taxa present at less than 1% of the total community. (n = 24)
Enrichment of low-abundance OTUs among patients with PaO2/FiO2 ≤ 300
The Streptococcaceae and Enterobacteriaceae family-level OTU abundances were not significantly different between patients with and without PaO2/FiO2 ≤ 300 (Wilcoxon test, p >0.05). At the lowest level of OTU identification, Enterobacteriaceae family-level OTUs, Streptococcus genus-level OTUs, and Staphylococcus genus-level OTUs were detected in 80% of patients both with and without PaO2/FiO2 ≤ 300 (Table 2; the average abundance refers to the percent of each OTU within each group). However, when compared to patients with PaO2/FiO2 > 300, patients with PaO2/FiO2 ≤ 300 had a 27% increase in OTUs identified as Streptococcus spp., a 32% increase in Enterobacteriaceae, and an 83% increase in Staphylococcus spp, calculated as the percent change. An additional six OTUs were detected in 80% of patients with PaO2/FiO2 ≤ 300 at 3.1% or less of the total OTUs in this group (Table 2). All OTUs detected in 80% of patients were either facultative or obligate anaerobes. Figs 2 and 3 display OTU abundances at the family level that account for greater than 1% of the total OTUs among individual patients without and with PaO2/FiO2 ≤ 300, respectively.
Table 2. Average abundance of taxa detected among 80% of patients.
| Bacteria | Aerobe/Anaerobe | Average Abundance Among Patients with PaO2/FiO2 > 300* | Average Abundance Among Patients with PaO2/FiO2 ≤ 300§ |
|---|---|---|---|
| OTU10: Enterobacteriaceae | Facultative anaerobe | 20.0 | 17.0 |
| OTU75: Streptococcus spp. | Facultative anaerobe | 27.8 | 22.1 |
| OTU58: Staphylococcus spp | Facultative anaerobe | 2.7 | 9.2 |
| OTU35: Atopobium spp. | Facultative anaerobe | NA | 3.1 |
| OTU62: Gemellaceae | Facultative anaerobe | NA | 1.8 |
| OTU108: Veillonella dispar | Obligate anaerobe | NA | 1.2 |
| OTU65: Lactobacillales | Facultative anaerobe | NA | 0.7 |
| OTU45: Prevotella spp. | Obligate anaerobe | NA | 2.4 |
| OTU47: Prevotella melaninogenica | Obligate anaerobe | NA | 2.5 |
Taxa detected in 80% of patients with and without PaO2/FiO2 ≤ 300. Taxa names represent the lowest level of identification of the corresponding OTU.
*Percent of total OTUs among 24 patients with PaO2/FiO2 > 300.
§Percent of total OTUs among 24 patients with PaO2/FiO2 ≤ 300.
NA indicates that these OTUs were not present among 80% of patients with PaO2/FiO2 > 300.
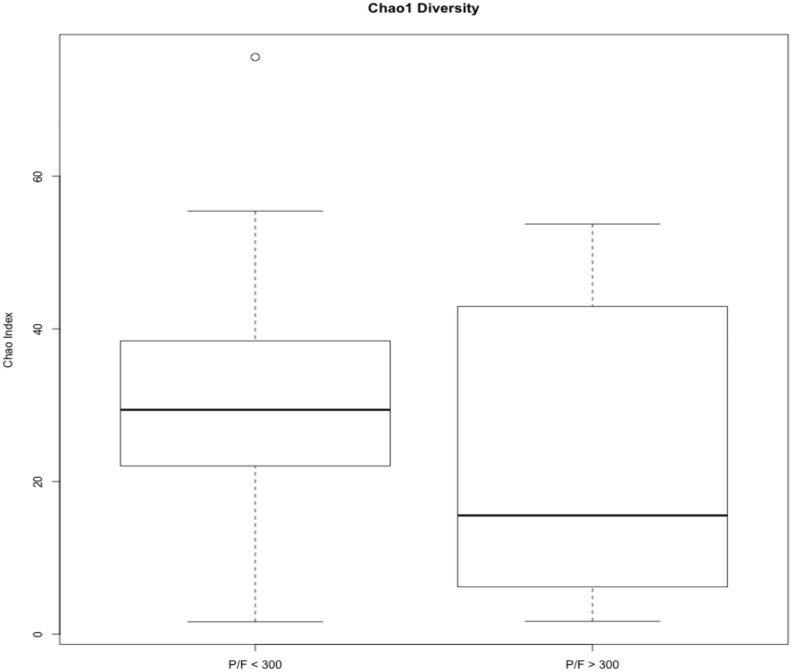
Fig 3. Chao1 diversity among patients with and without hypoxemia.
Samples were rarefied prior to calculation of the Chao1 diversity index and averaged based on PaO2/FiO2 ratio. Student’s T test did not show significant differences between the patient groups. (n = 48)
Alpha diversity among patients with and without PaO2/FiO2 ≤ 300
The Chao1 diversity index, which is a non-parametric species richness estimator [22], did not show significant differences in number of different OTUs between patients with and without PaO2/FiO2 ≤ 300 (Fig 3). Though the median Chao1 index in patients with PaO2/FiO2 > 300 is less than that of patients with PaO2/FiO2 ≤ 300, it shows a much broader range in patients with PaO2/FiO2 > 300.
Beta diversity among patients with and without PaO2/FiO2 ≤ 300
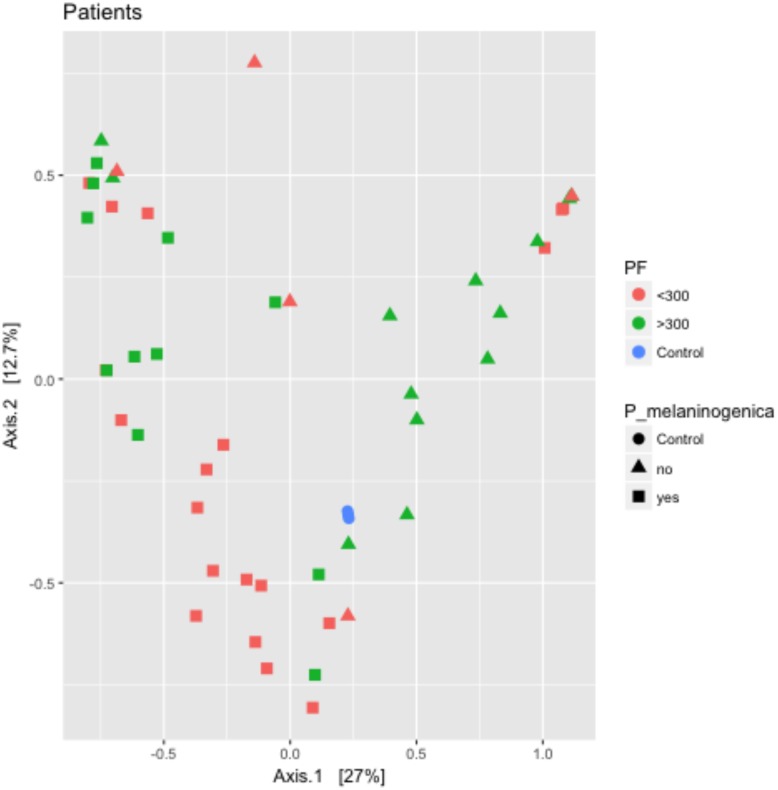
The PCA plot in Fig 4 displays similarity of microbiota among burn patients with inhalation injury. Data points represent individual patients, which are colored by PaO2/FiO2 ratio. The shape of each data point represents whether P. melaninogenica was detected in the community. Patients with PaO2/FiO2 ratios > 300 with P. melaninogenica present cluster to the left above the horizontal zero axis on the graph, while those with PaO2/FiO2 ratios < 300 with P. melaninogenica cluster to the left below it. Patients with PaO2/FiO2 ratios > 300 without P. melaninogenica are more spread out along the right side of the graph while those with PaO2/FiO2 ratios < 300 and without P. melaninogenica are scattered throughout. This clustering pattern suggests that the presence of P. melaninogenica is associated with microbiota similarity regardless of PaO2/FiO2 ratio, while in the absence of this microbe a PaO2/FiO2 ratio higher than 300 may influence similarity. Low PaO2/FiO2 ratio without P. melaninogenica does not appear to be associated with microbiota similarity.
Fig 4. Clustering by principle components analysis suggests microbiota similarity in the presence of P. melaninogenica.
The R package phyloseq was used to generate a PCA plot of patient microbiota abundance. Patient samples containing P. melaninogenica cluster to the left side of the graph and are split based on PaO2/FiO2 ratio. Manhattan distance was used to perform the ordination prior to plotting the PCA graph. Data points are colored by patient PaO2/FiO2 ratio (Red: P/F < 300; Green: P/F > 300) and data point shape indicates presence (square) or absence (triangle) of P. melaninogenica. Control samples are included for comparison (blue circles). (n = 48).
Significant enrichment of specific OTUs among patients with PaO2/FiO2 ≤ 300
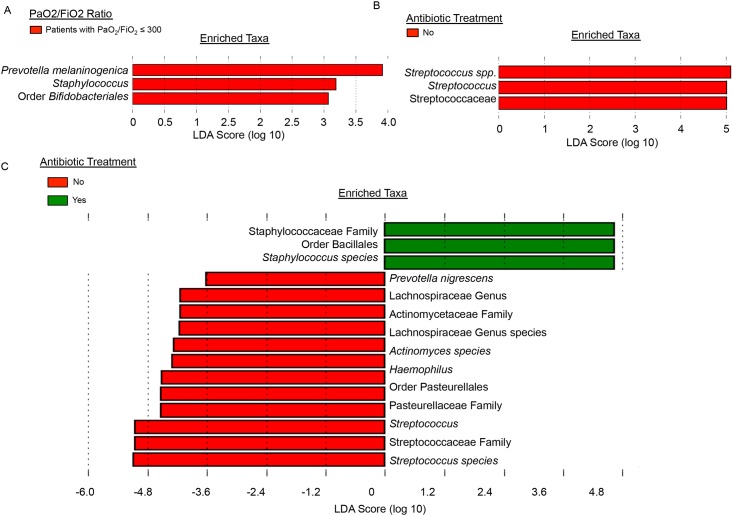
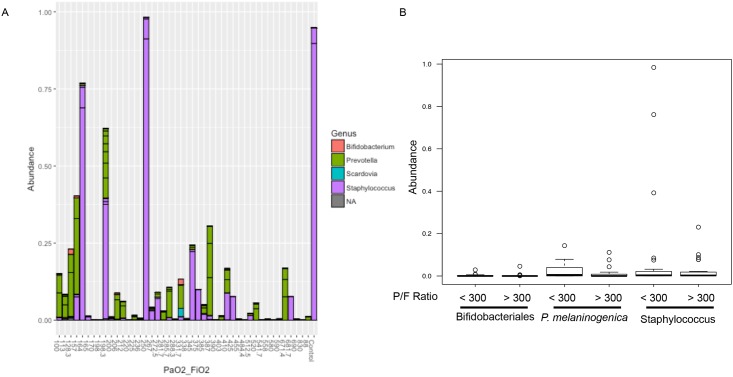
Four OTUs were identified as significantly different in abundance and detection between patients with and without PaO2/FiO2 ≤ 300. OTUs identified as Prevotella melaninogenica, Mogibacterium spp., and Corynebacterium spp. were significantly increased in abundance among patients with PaO2/FiO2 ≤ 300 (Table 3). Patients with PaO2/FiO2 ≤ 300 had 72% more of the OTU represented by Prevotella melaninogenica than patients with ratios > 300, 79% more Corynebacterium genus-level OTU, and 86% more of the Mogibacterium genus-level OTU. Prevotella melaninogenica OTUs were also detected significantly more frequently among patients with PaO2/FiO2 ≤ 300, while Corynebacterium OTUs were significantly more frequent in patients with ratios > 300 (Table 4). OTUs identified as Fusobacterium spp. were also detected significantly more frequently among patients with PaO2/FiO2 ≤ 300. LEfSe [20] was used to confirm these results. LEfSe identified the Prevotella melaninogenica OTU as most discriminative among patients with PaO2/FiO2 ≤ 300 as compared to those with ratios > 300, followed by Staphylococcus genus-level and then Bifidobacteriales order-level OTUs (Fig 5A). Though Staphylococcus OTUs are more abundant among the patients (Fig 6A), their average abundance within patients with hypoxemia is less than that of P. melaninogenica (Fig 6B). Prevotella is consistently present among the patients (Fig 6A) and its average abundance in patients with hypoxemia is greater than that for the other two significantly enriched taxa (Fig 6B). Additional analysis with LEfSe indicated significant enrichment of Staphylococcus spp. OTUs in the presence of antibiotics, while enrichment of Prevotella melaninogenica OTUs was not affected (Fig 5B and 5C). Among all patients in the study, LEfSe identified significant enrichment of taxa only in patients with hypoxemia (Fig 5A). Among only patients with hypoxemia, LEfSe found significant enrichment of taxa only in patients treated with antibiotics (Fig 5B). LEfSe identified significant enrichment of taxa in patients that did and did not receive antibiotics when all patients were included in the analysis (Fig 5C). Among patients without hypoxemia, several bacterial taxa were enriched with and without antibiotic treatment (S5 Fig). Non-parametric differential abundance analysis adjusted for antibiotic treatment and sequencing batch effect confirmed that enrichment of P. melaninogenica was not affected by antibiotic treatment or batch effect.
Table 3. Percent abundance OTU level differences.
| Taxa | Percent Abundance Among Patients with PaO2/FiO2 ≤ 300 | Percent Abundance Among Patients with PaO2/FiO2 > 300 | P-Value |
|---|---|---|---|
| OTU47: Prevotella melaninogenica | 1.56 (0.04) | 0.44 (7.7x10-3) | 0.042 |
| OTU18: Corynebacterium spp. | 1.53 (0) | 0.32 (1.8x10-3) | 0.037 |
| OTU82: Mogibacterium spp. | 0.07 (1.0x10-3) | 0.01 (1.3x10-4) | 0.048 |
OTU level significant differences in abundance as determined by Wilcoxon rank-sum test. Taxa names represent the lowest level of identification of the corresponding OTU. Values are percent abundance (interquartile range).
Table 4. OTU level detection differences.
| Taxa | Detection Rate Among Patients with PaO2/FiO2 ≤ 300 (# of patients) | Detection Rate Among Patients with PaO2/FiO2 > 300 (# of patients) | P-Value |
|---|---|---|---|
| OTU47: Prevotella melaninogenica | 19 | 11 | 0.037 |
| OTU18: Corynebacterium spp. | 6 | 14 | 0.040 |
| OTU115: Fusobacterium spp. | 17 | 9 | 0.043 |
OTU level significant differences in detection as determined by the two-proportions test. Taxa names represent the lowest level of identification of the corresponding OTU.
Fig 5. Significant enrichment of specific taxa.
Specific bacterial taxa are enriched among patients with PaO2/FiO2 ≤ 300. (A) LEfSe analysis detected significant enrichment of OTUs identified as Prevotella melaninogenica, Staphylococcus spp., and the order Bifidobacteriales among patients with hypoxemia. No significant enrichment was detected among patients without hypoxemia. (B) Among only those patients with hypoxemia, only those not treated with antibiotics contained significantly enriched taxa, all of which were in the Streptococcaceae family. No significant enrichment of taxa was detected among patients not treated with antibiotics in this comparison. (C) Antibiotic treatment alters the microbiota among all patients, with a specific increase in Staphylococcus among patients treated with antibiotics, but does not impact association of the Prevotella melaninogenica OTU with hypoxemia. LEfSe uses a Kruskal-Wallis rank-sum test, Wilcoxon rank-sum test, and linear discriminant analysis to determine the biological relevance of significant enrichment of taxa and ranks them by effect size. LDA score indicates the magnitude of the effect size.
Fig 6. Percent abundance of OTUs with significant enrichment detected by LEfSe.
Of the three taxa LEfSe identified as significantly enriched in patients with hypoxemia, Staphylococcus had the highest percent increase in abundance. However, Prevotella melaninogenica was more consistently present among patients with hypoxemia, resulting in its higher ranking over Staphylococcus spp. (A) Bacterial abundances for taxa identified as significantly enriched by LEfSe are displayed per patient. The X axis is labeled with each patient’s PaO2/FiO2 ratio and the Y axis displays relative abundance of the taxa. (B) The range of abundances of the three significantly enriched taxa are split by patient hypoxemia status along the X axis. (n = 48).
Discussion
Our work details differences in the airway microbiota in patients with PaO2/ FiO2 ratios ≤ 300 and > 300, following burn and inhalation injury. A cut-off of 300 was chosen based on the Berlin definition of airway hypoxemia in ARDS [4]. We identify several low-abundance OTUs with significant enrichment in patients with PaO2/FiO2 ≤ 300, of which the OTU identified as Prevotella melaninogenica was the most significant. In addition, we show that while antibiotic treatment alters the airway microbiota, it does not explain the enrichment of a specific OTU among patients with PaO2/FiO2 ≤ 300.
Patients with a PaO2/ FiO2 ratio that was less than or equal to 300 within 72 hours of burn and inhalation injury had consistently worse indicators of poor prognosis. Table 1 shows the average values for patients with and without PaO2/FiO2 ≤ 300 for several clinical variables that are predictive of injury severity. In patients with inhalation injury, several studies have demonstrated that age, percent TBSA and PaO2/ FiO2 ratio predict mortality [2]. The PaO2/ FiO2 ratio itself has been shown to be more predictive of patient outcomes on the day after patients meet the Berlin definition of ARDS rather than the day of [23]. In our study, patients with PaO2/FiO2 ≤ 300 within 72 hours of injury had, on average, a higher Baux score (age + %TBSA), spent longer on the ventilator, were intubated more frequently, and had lower survival rates. Only percent TBSA and the PaO2/ FiO2 ratio were significantly different among the patient groups (Student’s t test, p = 0.002 and 5.293e-11, respectively). Patients were 41 years old on average, but ranged from 1 to 75 years. Though not statistically more prevalent in this cohort, patients at the ends of this spectrum are more susceptible to infection, pneumonia, and poor outcomes [24]. Over a lifetime, a patient will range from increased susceptibility to infection after burn and inhalation injury, to decreased susceptibility in mid-life, to increased in old age. While fewer patients with PaO2/FiO2 ≤ 300 received antibiotic treatment than those with ratios > 300, rates of positive clinical bacterial cultures were similar between the two groups. This discrepancy may be partly due to the challenges in predicting bacterial infection and development of pneumonia in this patient population. Pneumonia is the primary complication of inhalation injury [25] and early, adequate antibiotic treatment has been shown to improve outcomes in these patients [6]. Criteria to predict pneumonia early after injury have been developed and include age > 60 years, TBSA > 20%, and initial PaO2/ FiO2 ratio of ≤ 300 [26]. The patients with PaO2/FiO2 ≤ 300 in our study meet the TBSA and initial PaO2/ FiO2 ratio criteria, but not the age criteria, which may explain why they did not receive as many antibiotics. A major limitation of this scoring system is its failure to take into account bacteria within the airways, emphasizing the need for one that does, perhaps through a combination of clinical cultures and next-generation sequencing of bacterial 16S rRNA genes.
Though we have focused on the PaO2/FiO2 ratio in alterations of the airway microbiota, TBSA may also contribute to the differences we detected. Increasing TBSA is a known predictor of patient mortality [25], which is compounded in the presence of inhalation injury. Burns greater than 20% TBSA induce systemic changes similar to those seen in trauma and surgical patients [27]. The injury induces a systemic inflammatory response, but compromises global immune function, increasing susceptibility to bacterial, viral, and fungal infections. Patients with PaO2/FiO2 ≤ 300 in our study had, on average, 27% TBSA, indicating immune dysfunction that could predispose them to airway bacterial colonization and infection. Though we cannot determine whether the burn injury itself induces PaO2/FiO2 ≤ 300 through systemic changes or if this is a direct result of inhalation injury, it is clear that TBSA may be contributing indirectly to alterations in the airway microbiota in our patient population. A mouse model of burn and inhalation injury is necessary to determine the extent to which TBSA influences changes in the airway microbiota.
Among all patients in the study, there were significantly more unique OTUs identified as facultative anaerobes than either strict anaerobes or aerobes (Fig 1A). Anaerobic taxa are normally associated with mucosal surfaces, but may lead to infection following disruption by trauma and surgery [28]. All patients within this study, regardless of PaO2/FiO2 ratio, presumably experienced disruption of their mucosa through the double trauma of burn and inhalation injury. Recent work has demonstrated that the mouth serves as the primary source community for the airway microbiota [29]. Inhalation injury may have increased microbial immigration through disruption of the mouth and upper airways’ mucosal surface, dislodging taxa that subsequently traveled down the airways to the bronchi. Alteration of airway conditions by inhalation injury may have selected for enrichment of facultative anaerobic taxa among all patients, which was significantly different from strict aerobic and anaerobic taxa (ANOVA, p = 0.029). When we subdivided the data by PaO2/FiO2 ratio, we did not see significant differences in strict aerobes, anaerobes or facultative anaerobes between the two patient groups (Fig 1B, p > 0.05). These results suggest that PaO2/FiO2 ≤ 300 early after burn and inhalation injury does not select for overall taxa in the airways based on their aerobic or anaerobic capabilities, but that burn and inhalation injury do. Development of PaO2/FiO2 ≤ 300 within 72 hours of burn and inhalation injury may not be enough time to observe significant change in the abundances of overall taxa between the two groups.
We detected OTUs identified as Enterobacteriaceae, Streptococcus spp., and Staphylococcus spp. in 80% of patients with and without PaO2/FiO2 ≤ 300 (Table 2). All three of these OTUs are facultative anaerobes and their dominance across patients implies similarity in the mechanism of injury to the airways selecting for these taxa and their related functions. Inhalation injury may induce fluctuations in oxygen availability in the airways, perhaps creating both aerobic and anaerobic microenvironments that favor taxa that can withstand these changes. Our finding of overall significant enrichment of facultative anaerobic taxa supports this idea. Patients with PaO2/FiO2 ≤ 300 demonstrated a 32%, 27%, and 83% increase in Enterobacteriaceae, Streptococcus spp., and Staphylococcus spp. OTUs, respectively, when compared to those with PaO2/FiO2 > 300 (Table 2). Additionally, 80% of patients with PaO2/FiO2 ≤ 300 contained six more OTUs that represented 3.1% and less of the total community among these patients (Table 2). This suggests that, although facultative anaerobes are enriched over strict anaerobes and aerobes among all patients, there are differences in enrichment of specific, low-abundance OTUs depending on PaO2/FiO2 ratio.
Enterobacteriaceae, Streptococcus spp., and Staphylococcus spp. have all been consistently detected in previous airway microbiome studies in both healthy and diseased airways [30]. Members of the Enterobacteriaceae family have been implicated in inflammation-driven colorectal cancer in the gut microbiome [31,32], are enriched in patients with COPD and asthma, but can also be detected in healthy airways [33–35]. Similarly, Streptococcus is consistently found in healthy airways but is enriched in COPD [35], idiopathic pulmonary fibrosis (IPF) [36], and pneumonia [37]. Staphylococcus, while a normal commensal in the nasal microbiome [38,39], is largely associated with disease in the lung, such as IPF [36], and cystic fibrosis, in which it is correlated with increased inflammation [40,41]. Given the inconsistency with which these three taxa are associated with health or disease, it is difficult to interpret the importance of their detection across patients with and without PaO2/FiO2 ≤ 300. They may indicate an underlying core airway microbiota among all burn and inhalation injury patients but it is not clear whether their presence is beneficial or detrimental. Given that sampling was done within 72 hours of injury, some of the taxa we have detected could be acquired nosocomial pathogens rather than commensal organisms, implying a detrimental impact. A longitudinal study of patients with burn and inhalation injury could clarify the role of these taxa.
Due to its association with health outcomes, overall diversity has long been a focus in many microbiome studies; however, we observed no difference in alpha diversity between patients with PaO2/FiO2 ≤ 300 and those with PaO2/FiO2 > 300 (Fig 3). Despite this, the PCA plot in Fig 4 suggests similarity in the microbiota between patients depending first on whether P. melaninogenica is present and then on patient PaO2/FiO2 ratio. Patients with P. melaninogenica clustered to the left side of the graph and were largely split by PaO2/FiO2 ratio. Patients without P. melaninogenica and PaO2/FiO2 ratio > 300 tended to cluster together, but those without this microbe and PaO2/FiO2 ratio < 300 were scattered throughout the graph. This agrees with recent studies demonstrating that diversity (especially in the airways) is a complex, multifactorial trait that encompasses more than an indication of positive or negative outcomes [34,41]. Many of these studies have emphasized the critical roles of specific taxa during disease and their interactions with other taxa [10,11]. They suggest that rare and less abundant taxa, which are overlooked by traditional culture methods, may play significant roles in the development of disease. Dysbiosis of the microbiota is followed by enrichment of a specific bacterial taxa that is either rarely found or present at very low abundance [10,37]. Changes in the balance of bacterial taxa alters how the microbes interact with each other along with their associated functions, allowing species that may have been suppressed by the presence of other bacteria to overgrow [10]. What was considered a harmless commensal in a healthy individual may become a harmful pathogen under dysbiosis-inducing conditions [42]. Accordingly, in our study, we observed significant differences not in the species dominating the overall community, but in less abundant taxa. While these taxa do not differ in microbial diversity between patient groups, they may differ by functional diversity, which ultimately plays a greater role in patient outcomes [30]. Most significantly, we identified enrichment of the OTU Prevotella melaninogenica among patients with PaO2/FiO2 ≤ 300 within 72 hours of burn and inhalation injury (Fig 5A).
Prevotella melaninogenica is a gram-negative obligate anaerobe that is part of the normal microbiota but is also a significant source of infection [43]. The specifics of Prevotella melaninogenica’s function in the microbiome remain unclear. In the gut, it has been identified as a normal commensal family, but within dental plaque it is a potential pathogen [44]. In the upper airways, the presence of Prevotella melaninogenica is associated with health while lactobacilli, Rothia spp., and Streptococcus pneumoniae dominate bacterial profiles in patients with pneumonia [37]. Prevotella melaninogenica’s positive role in the airways is supported by its ability to decrease production of T cell-activating IL-12p70 by dendritic cells exposed to Haemophilus influenzae [45]. This highlights the ability of bacteria within microbial communities to regulate each other’s functions as well as that of the host immune system. Several studies indicate that Prevotella melaninogenica could also play a non-beneficial or harmful role in the airways under certain conditions. Prevotella melaninogenica was a dominant bacterial species isolated from the airways of intubated patients [46] as well as cystic fibrosis (CF) patients, where characterized species varied phenotypically over time [47]. Prevotella species in CF airways have been shown to be virulent and contribute to the pathophysiology of the disease [48]. Prevotella melaninogenica specifically produces short-chain fatty acids that generate IL-8 production by host epithelial cells, presumably drawing neutrophils to the airways that contribute to the inflammatory status of the patient [49]. Though present at low abundance, we identified a consistent and significant enrichment of the Prevotella melaninogenica OTU among patients with PaO2/FiO2 ≤ 300 within 72 hours of burn and inhalation injury. While facultative anaerobic taxa were enriched among all patients in the study, Prevotella melaninogenica was enriched specifically in patients whose airways have the lowest PaO2/FiO2 ratio, which may select for growth of this obligate anaerobe. Without pre-injury samples from the patients, it is not possible to determine whether enrichment of Prevotella melaninogenica precedes PaO2/FiO2 ≤ 300 or if a low PaO2/FiO2 ratio precedes this enrichment. If confirmed in a longitudinal study, the consistent presence of this OTU throughout the hospital stay of patients with PaO2/FiO2 ≤ 300 would suggest that it is in some way altering the airway environment to favor Prevotella melaninogenica. This could be achieved through elimination of other OTUs that Prevotella melaninogenica interacts with that cannot thrive in hypoxic conditions or outgrowth of those that can. Early changes in both oxygen availability and other OTUs may impact Prevotella melaninogenica’s ability to act as a pathogen depending on whether species it interacts with are increased or eliminated or airway conditions alter its growth and pathogenicity. Given that Prevotella melaninogenica is an obligate anaerobe, hypoxic conditions may favor its growth, but it is impossible to predict its pathogenicity without further study. While determining a causal link between PaO2/FiO2 ≤ 300 and Prevotella melaninogenica is beyond the scope of the current study, future studies will examine its pathogenicity from patients with and without PaO2/FiO2 ≤ 300 as well as its role in either preceding or following hypoxemia.
Infection is a serious concern in these immunocompromised patients, for whom mortality rates increase to 20% with inhalation injury alone and triple to 60% when present with pneumonia [50]. Prophylactic antibiotic treatment is a common strategy to prevent infection, but results in as many as 25% of patients without infections receiving antibiotics [51], which may alter the microbiota in deleterious ways and encourage outgrowth of resistant bacteria [52]. Antibiotic treatment has been shown to perturb the gut microbiome and immune cell response by eliminating commensal species and allowing drug-resistant bacteria to take over [53,54]. In the airways, antibiotic treatment in asthma shows a similar response, in which elimination of certain species provides a niche for establishment of other infectious species [55]. In COPD, three months of varying types of antibiotic treatment in patients did not reduce overall bacterial load but instead increased antibiotic resistance across all groups [56]. While a powerful tool for controlling bacterial growth, antibiotic treatment is a double-edged sword that can create communities of bacteria resistant to treatment. Our poor understanding of bacterial interactions within the microbiota and their roles in patient outcomes combined with antibiotics’ lack of specificity results in overkilling of beneficial organisms that could aid in improving patient outcomes. In our study, 18 total patients were treated with antibiotics; 9 of these had negative culture results and for 2, cultures were not done (S3 Table). If negative culture results indicate absence of infection in these patients, antibiotic treatment is unnecessarily altering the airway microbiota, possibly contributing to development of resistance and poor outcomes. Among all patients treated with antibiotics, analysis with LEfSe indicated significant enrichment of bacteria in the Staphylococcaceae family and the order Bacillales (Fig 5C). These bacteria may be resistant to the drugs or not targeted by them, leading to overgrowth of these particular species. Methicillin-resistant Staphylococcus aureus is a known problematic infection in hospitals, including the Jaycee Burn Center, but its role within burn patient microbiota is unknown and requires further study. Despite alteration of other taxa by antibiotic treatment, enrichment with the Prevotella melaninogenica OTU among patients with PaO2/FiO2 ≤ 300 was not affected, implying that its association with hypoxemia is independent of antibiotic treatment, at least within 72 hours of injury (Fig 5B and 5C). Further study is necessary to determine the role of this OTU in early development of hypoxemia and whether targeted antibiotic treatment may be beneficial.
There are several limitations to the study. Although unique in its examination of a heterogeneous group of burn patients, our work is also limited by this variability. The heterogeneity of clinical diagnoses in this group makes interpretation of results challenging. We were unable to extract significantly more DNA from healthy human bronchial washings than we did from our negative reagent controls, and therefore were unable to include healthy control samples for comparison (S4 Fig). However, the number of patients studied is comparable to or larger than previous studies of microbiota in airway disease [10,35]. Longitudinal samples were taken throughout the course of each patient’s hospital stay, but the variation in drug treatments (including antibiotics) and therapies precluded achievement of statistical significance among the microbiota detected in these samples. Despite these limitations, this study is pioneering in its examination of the injured airway microbiota among burn patients and its association with patient outcomes.
In conclusion, we have demonstrated differences in the airway microbiota of patients with and without PaO2/FiO2 ≤ 300 within 72 hours of burn and inhalation injury. We detected overall enrichment of facultative anaerobes among all patients with differences in specific OTUs among patients with and without PaO2/FiO2 ≤ 300. Significant differences between these patients reside among the less abundant OTUs, specifically the Prevotella melaninogenica OTU, an obligate anaerobe whose role in the microbiome is unclear. Hypoxic conditions indicative of ARDS development may favor Prevotella melaninogenica enrichment and alter its pathogenicity. Alternatively, hypoxemia may develop due to increased abundance of this OTU following inhalation injury. A mouse model of inhalation injury is needed to determine whether development of hypoxemia drives enrichment of Prevotella melaninogenica or enrichment of this OTU induces hypoxemia. Given the cross-sectional nature of this study, more work is necessary to determine the long-term impact of Prevotella melaninogenica and its role in the airway microbiome of burn patients with inhalation injury who develop PaO2/FiO2 ≤ 300 within 72 hours of injury. Importantly, antibiotic treatment did not alter this association, supporting a link between this OTU and PaO2/FiO2 ≤ 300 early after burn and inhalation injury.
Supporting information
Variability was minimized by grouping samples by the plate on which they were sequenced. The Y axis displays log10 transformed raw read counts per OTU for the first replicate and the X axis displays these values for the second replicate. (A) Log10 transformed raw counts per OTU from Plate 2, samples sequenced January 2015. (B) Log10 transformed raw counts per OTU from Plate 1, samples sequenced January 2015. (C) Log10 transformed raw counts per OTU from Plate 3, samples sequenced December 2014.
(TIF)
The threshold for log10 transformed raw counts per OTU per sequencing batch was set where the regression began to plateau (red line), indicating acceptable levels of read count correlation between the replicate samples. (A) R2 values for raw counts for samples on Plate 2, samples sequenced January 2015. (B) R2 values for raw counts for samples on Plate 1, samples sequenced January 2015. (C) R2 values for raw counts for samples on Plate 3, samples sequenced December 2014.
(TIF)
Each bar represents OTUs detected among human (16HBE), Staphylococcus aureus (SAUR), and reagent (CNTRL) DNA controls normalized to 100%. n = 6
(TIF)
Bronchoscopy was performed on healthy volunteers and DNA was extracted from airway washings in the same manner as the burn patient samples. Extracted DNA was quantified using the universal primer set developed by Maeda et. al. DNA extracted from Staphylococcus aureus and Klebsiella pneumoniae were used as positive controls and DNA from the human cell line 16HBE was used as a negative control. A water-only reagent control was included as well. DNA extracted from six healthy volunteers did not contain significantly more DNA than the negative control. (n = 6).
(TIF)
Specific taxa are enriched among non-hypoxemic patients who did and did not receive antibiotics. Analysis with LEfSe detects significant enrichment of bacteria in the Enterobacteriales order with antibiotic treatment, while several other taxa were enriched without treatment.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Acknowledgments
The authors would like to thank Paula Murphy for initial processing of the bronchoalveolar lavage samples, Dr. Juliette J. Kahle for input on development of DNA extraction methods, Sur Herrera for assistance in statistical data analysis, and Dr. Marianne Muhlebach for reviewing the manuscript. The research described in this article has been reviewed by the Environmental Protection Agency and approved for publication. The contents of this article do not necessarily represent Agency policy, nor does mention of trade names or commercial products constitute endorsement or recommendations for use.
Abbreviations
- ANOVA
analysis of variance
- ARDS
acute respiratory distress syndrome
- CF
cystic fibrosis
- DNA
deoxyribonucleic acid
- FiO2
fraction of inspired oxygen
- IPF
idiopathic pulmonary fibrosis
- LEfSe
linear discriminant analysis effect size
- OTU
operational taxonomic unit
- PaO2
partial pressure of arterial oxygen
- PCA
principle components analysis
- PCR
polymerase chain reaction
- rDNA
ribosomal deoxyribonucleic acid
Data Availability
The dataset generated during the current study is available in the Figshare data repository (A replicate FASTQ files: https://dx.doi.org/10.6084/m9.figshare.3496412, B replicate FASTQ files: https://dx.doi.org/10.6084/m9.figshare.3496538, Metadata: https://dx.doi.org/10.6084/m9.figshare.3485201, R Code: https://dx.doi.org/10.6084/m9.figshare.3474923).
Funding Statement
This work was supported by the following: I.J. received funding from the United States Environmental Protection Agency (www.epa.gov) Cooperative Agreement CR83578501 “Human Health Effects of Environmental Pollutants,” which was used to purchase all necessary supplies and to pay for sequencing and publication of this manuscript; D.M.W.’s stipend came from the National Institutes of Health (www.nih.gov) Toxicology Training grant T32 ES007126; S.W.J. received funding from the National Institutes of Health North Carolina TraCS Institute UL1RR02574, National Institutes of Health UNC Center for Environmental Health and Susceptibility P30ES010126, and National Institutes of Health 5K08GM109106, each of which paid for supplies and personnel to collect the patient samples and establish the repository as detailed in Jones et. al. [12]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.El-Helbawy RH, Ghareeb FM. Inhalation injury as a prognostic factor for mortality in burn patients. Ann Burns Fire Disasters. 2011;24: 82–8. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=3230152&tool=pmcentrez&rendertype=abstract [PMC free article] [PubMed] [Google Scholar]
- 2.Dries DJ, Endorf FW. Inhalation injury: epidemiology, pathology, treatment strategies. Scand J Trauma Resusc Emerg Med. 2013;21: 31 10.1186/1757-7241-21-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bastarache JA, Ware LB, Bernard GR. Acute Lung Injury and Respiratory Distress Syndrome Textbook of Critical Care. 2011. pp. 388–397. [Google Scholar]
- 4.The ARDS Task Force. Acute Respiratory Distress Syndrome. Jama. 2012;307: 2526–2533. 10.1001/jama.2012.5669 [DOI] [PubMed] [Google Scholar]
- 5.Dreyfuss D, Ricard J-D. Acute Lung Injury and Bacterial Infection. Clin Chest Med. 2005;26: 105–112. 10.1016/j.ccm.2004.10.014 [DOI] [PubMed] [Google Scholar]
- 6.Brusselaers N, Logie D, Vogelaers D, Monstrey S, Blot S. Burns, inhalation injury and ventilator-associated pneumonia: value of routine surveillance cultures. Burns. 2012;38: 364–70. 10.1016/j.burns.2011.09.005 [DOI] [PubMed] [Google Scholar]
- 7.Zumla A, Al-Tawfiq JA, Enne VI, Kidd M, Drosten C, Breuer J, et al. Rapid point of care diagnostic tests for viral and bacterial respiratory tract infections—needs, advances, and future prospects. Lancet Infect Dis. 2014;14: 1123–35. 10.1016/S1473-3099(14)70827-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marsland BJ, Yadava K, Nicod LP. The airway microbiome and disease. Chest. 2013;144: 632–7. 10.1378/chest.12-2854 [DOI] [PubMed] [Google Scholar]
- 9.Honda K, Littman DR. The microbiome in infectious disease and inflammation. Annu Rev Immunol. 2012;30: 759–95. 10.1146/annurev-immunol-020711-074937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abreu N a, Nagalingam N a, Song Y, Roediger FC, Pletcher SD, Goldberg AN, et al. Sinus microbiome diversity depletion and Corynebacterium tuberculostearicum enrichment mediates rhinosinusitis. Sci Transl Med. 2012;4: 151ra124 10.1126/scitranslmed.3003783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10: 497–506. 10.1016/j.chom.2011.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones SW, Zhou H, Ortiz-Pujols SM, Maile R, Herbst M, Joyner BL, et al. Bronchoscopy-derived correlates of lung injury following inhalational injuries: a prospective observational study. PLoS One. 2013;8: e64250 10.1371/journal.pone.0064250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lundberg DS, Yourstone S, Mieczkowski P, Jones CD, Dangl JL. Practical innovations for high-throughput amplicon sequencing. Nat Methods. 2013;10: 999–1002. 10.1038/nmeth.2634 [DOI] [PubMed] [Google Scholar]
- 14.Yourstone SM, Lundberg DS, Dangl JL, Jones CD. MT-Toolbox: improved amplicon sequencing using molecule tags. BMC Bioinformatics. 2014;15: 284 10.1186/1471-2105-15-284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a Chimera-Checked 16S rRNA Gene Database and Workbench Compatible with ARB. Appl Environ Microbiol. 2006;72: 5069–5072. 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S, et al. Defining the core Arabidopsis thaliana root microbiome. Nature. 2012;488: 86–90. 10.1038/nature11237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robertson CE, Harris JK, Wagner BD, Granger D, Browne K, Tatem B, et al. Explicet: graphical user interface software for metadata-driven management, analysis and visualization of microbiome data. Bioinformatics. 2013;29: 3100–3101. 10.1093/bioinformatics/btt526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.R Core Team. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2013; http://www.r-project.org/ [Google Scholar]
- 19.McMurdie PJ, Holmes S. Phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data. PLoS One. 2013;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. BioMed Central Ltd; 2011;12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hale VL, Chen J, Johnson S, Harrington SC, Yab TC, Smyrk TC, et al. Shifts in the fecal microbiota associated with adenomatous polyps. Cancer Epidemiol Biomarkers Prev. 2016; 85–95. 10.1158/1055-9965.EPI-16-0337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haegeman B, Hamelin J, Moriarty J, Neal P, Dushoff J, Weitz JS. Robust estimation of microbial diversity in theory and in practice. ISME J. 2013; 1092–1101. 10.1038/ismej.2013.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai C-C, Sung M-I, Liu H-H, Chen C-M, Chiang S-R, Liu W-L, et al. The Ratio of Partial Pressure Arterial Oxygen and Fraction of Inspired Oxygen 1 Day After Acute Respiratory Distress Syndrome Onset Can Predict the Outcomes of Involving Patients. Medicine (Baltimore). 2016;95: e3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.American Burn Association. National Burn Repository 2015 Report version 11.0. 2015.
- 25.Edelman D a, Khan N, Kempf K, White MT. Pneumonia after inhalation injury. J Burn Care Res. 2007;28: 241–6. 10.1097/BCR.0B013E318031D049 [DOI] [PubMed] [Google Scholar]
- 26.Lin C-C, Liem a a, Wu C-K, Wu Y-F, Yang J-Y, Feng C-H. Severity score for predicting pneumonia in inhalation injury patients. Burns. Elsevier Ltd and International Society of Burns Injuries; 2012;38: 203–7. [DOI] [PubMed] [Google Scholar]
- 27.Jeschke MG, Herndon DN. Burns Sabiston Textbook of Surgery. Twentieth 2017. pp. 505–531. [Google Scholar]
- 28.Cohen-Poradosu R, Kasper DL. Anaerobic Infections In: Bennett JE, Dolin R, Blaser MJ, editors. Mandell, Douglas, and Bennett’s Principles and Practice of Infectious Diseases. Eight 2015. p. 2736 0 2743. [Google Scholar]
- 29.Dickson RP, Erb-Downward JR, Huffnagle GB. Homeostasis and its Disruption in the Lung Microbiome. Am J Physiol—Lung Cell Mol Physiol. 2015; ajplung.00279.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dickson RP, Erb-Downward JR, Martinez FJ, Huffnagle GB. The Microbiome and the Respiratory Tract. Annu Rev Physiol. 2015; 1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arthur JC, Gharaibeh RZ, Mühlbauer M, Perez-Chanona E, Uronis JM, McCafferty J, et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat Commun. 2014;5: 4724 10.1038/ncomms5724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mira-Pascual L, Cabrera-Rubio R, Ocon S, Costales P, Parra A, Suarez A, et al. Microbial mucosal colonic shifts associated with the development of colorectal cancer reveal the presence of different bacterial and archaeal biomarkers. J Gastroenterol. 2014;50: 167–179. 10.1007/s00535-014-0963-x [DOI] [PubMed] [Google Scholar]
- 33.Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. Comparison of the Respiratory Microbiome in Healthy Nonsmokers and Smokers. Am J Respir Crit Care Med. 2013;187: 1067–1075. 10.1164/rccm.201210-1913OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang YJ, Nelson CE, Brodie EL, Desantis TZ, Baek MS, Liu J, et al. Airway microbiota and bronchial hyperresponsiveness in patients with suboptimally controlled asthma. J Allergy Clin Immunol. Elsevier Ltd; 2011;127: 372–381– 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pragman AA, Kim HB, Reilly CS, Wendt C, Isaacson RE. The lung microbiome in moderate and severe chronic obstructive pulmonary disease. PLoS One. 2012;7: e47305 10.1371/journal.pone.0047305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Han MK, Zhou Y, Murray S, Tayob N, Noth I, Lama VN, et al. Lung microbiome and disease progression in idiopathic pulmonary fibrosis: An analysis of the COMET study. Lancet Respir Med. 2014;2: 448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Steenhuijsen Piters W a a, Huijskens EGW, Wyllie AL, Biesbroek G, van den Bergh MR, Veenhoven RH, et al. Dysbiosis of upper respiratory tract microbiota in elderly pneumonia patients. ISME J. 2015; 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lemon KP, Klepac-ceraj V, Schiffer HK, Brodie EL, Lynch S V, Kolter R. Comparative Analyses of the Bacterial Microbiota of the Human Nostril and Oropharynx. MBio. 2010;1: 4–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frank DN, Feazel LM, Bessesen MT, Price CS, Janoff EN, Pace NR. The human nasal microbiota and Staphylococcus aureus carriage. PLoS One. 2010;5: e10598 10.1371/journal.pone.0010598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zemanick ET, Wagner BD, Robertson CE, Stevens MJ, Szefler SJ, Accurso FJ, et al. Assessment of Airway Microbiota and Inflammation in Cystic Fibrosis Using Multiple Sampling Methods. Ann Am Thorac Soc. 2015;12: 221–229. 10.1513/AnnalsATS.201407-310OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dickson RP, Erb-Downward JR, Huffnagle GB. The role of the bacterial microbiome in lung disease. Expert Rev Respir Med. 2013;7: 245–257. 10.1586/ers.13.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Abt MC, Artis D. The dynamic influence of commensal bacteria on the immune response to pathogens. Curr Opin Microbiol. Elsevier Ltd; 2013;16: 4–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buckingham SC. Bacteroides, Fusobacterium, and Prevotella In: Cherry JD, Harrison GJ, Kaplan SL, Steinbach WJ, Hotez PJ, editors. Feigin and Cherry’s Textbook of Pediatric Infectious Diseases. Seventh 2014. pp. 1825–1834. [Google Scholar]
- 44.Faust K, Sathirapongsasuti JF, Izard J, Segata N, Gevers D, Raes J, et al. Microbial co-occurrence relationships in the human microbiome. PLoS Comput Biol. 2012;8: e1002606 10.1371/journal.pcbi.1002606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Larsen JM, Steen-Jensen DB, Laursen JM, Søndergaard JN, Musavian HS, Butt TM, et al. Divergent pro-inflammatory profile of human dendritic cells in response to commensal and pathogenic bacteria associated with the airway microbiota. PLoS One. 2012;7: e31976 10.1371/journal.pone.0031976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Agvald-Ohman C, Wernerman J, Nord CE, Edlund C. Anaerobic bacteria commonly colonize the lower airways of intubated ICU patients. Clin Microbiol Infect. 2003;9: 397–405. [DOI] [PubMed] [Google Scholar]
- 47.Field TR, Sibley CD, Parkins MD, Rabin HR, Surette MG. The genus Prevotella in cystic fibrosis airways. Anaerobe. 2010;16: 337–344. 10.1016/j.anaerobe.2010.04.002 [DOI] [PubMed] [Google Scholar]
- 48.Ulrich M, Beer I, Braitmaier P, Dierkes M, Kummer F, Krismer B, et al. Relative contribution of Prevotella intermedia and Pseudomonas aeruginosa to lung pathology in airways of patients with cystic fibrosis. 2010; 978–984. [DOI] [PubMed] [Google Scholar]
- 49.Mirković B, Murray MA, Lavelle GM, Molloy K, Azim AA, Gunaratnam C, et al. The Role of Short-Chain Fatty Acids, Produced by Anaerobic Bacteria, in the Cystic Fibrosis Airway. Am J Respir Crit Care Med. 2015;192: 1314–1324. 10.1164/rccm.201505-0943OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shirani KZ, Pruitt B a, Mason a D. The influence of inhalation injury and pneumonia on burn mortality. Ann Surg. 1987;205: 82–7. Available: http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1492872&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schultz L, Walker S a N, Elligsen M, Walker SE, Simor A, Mubareka S, et al. Identification of predictors of early infection in acute burn patients. Burns. Elsevier Ltd and International Society of Burns Injuries; 2013;39: 1355–66. [DOI] [PubMed] [Google Scholar]
- 52.Sommer MO a, Dantas G. Antibiotics and the resistant microbiome. Curr Opin Microbiol. Elsevier Ltd; 2011;14: 556–63. [DOI] [PubMed] [Google Scholar]
- 53.Ubeda C, Pamer EG. Antibiotics, microbiota, and immune defense. Trends Immunol. Elsevier Ltd; 2012;33: 459–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hill D a, Hoffmann C, Abt MC, Du Y, Kobuley D, Kirn TJ, et al. Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol. 2010;3: 148–58. 10.1038/mi.2009.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Collie D, Glendinning L, Govan J, Wright S, Thornton E, Tennant P, et al. Lung Microbiota Changes Associated with Chronic Pseudomonas aeruginosa Lung Infection and the Impact of Intravenous Colistimethate Sodium. PLoS One. 2015;10: e0142097 10.1371/journal.pone.0142097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Brill SE, Law M, El-Emir E, Allinson JP, James P, Maddox V, et al. Effects of different antibiotic classes on airway bacteria in stable COPD using culture and molecular techniques: a randomised controlled trial. Thorax. 2015;70: 930–8. 10.1136/thoraxjnl-2015-207194 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Variability was minimized by grouping samples by the plate on which they were sequenced. The Y axis displays log10 transformed raw read counts per OTU for the first replicate and the X axis displays these values for the second replicate. (A) Log10 transformed raw counts per OTU from Plate 2, samples sequenced January 2015. (B) Log10 transformed raw counts per OTU from Plate 1, samples sequenced January 2015. (C) Log10 transformed raw counts per OTU from Plate 3, samples sequenced December 2014.
(TIF)
The threshold for log10 transformed raw counts per OTU per sequencing batch was set where the regression began to plateau (red line), indicating acceptable levels of read count correlation between the replicate samples. (A) R2 values for raw counts for samples on Plate 2, samples sequenced January 2015. (B) R2 values for raw counts for samples on Plate 1, samples sequenced January 2015. (C) R2 values for raw counts for samples on Plate 3, samples sequenced December 2014.
(TIF)
Each bar represents OTUs detected among human (16HBE), Staphylococcus aureus (SAUR), and reagent (CNTRL) DNA controls normalized to 100%. n = 6
(TIF)
Bronchoscopy was performed on healthy volunteers and DNA was extracted from airway washings in the same manner as the burn patient samples. Extracted DNA was quantified using the universal primer set developed by Maeda et. al. DNA extracted from Staphylococcus aureus and Klebsiella pneumoniae were used as positive controls and DNA from the human cell line 16HBE was used as a negative control. A water-only reagent control was included as well. DNA extracted from six healthy volunteers did not contain significantly more DNA than the negative control. (n = 6).
(TIF)
Specific taxa are enriched among non-hypoxemic patients who did and did not receive antibiotics. Analysis with LEfSe detects significant enrichment of bacteria in the Enterobacteriales order with antibiotic treatment, while several other taxa were enriched without treatment.
(TIF)
(DOCX)
(DOCX)
(DOCX)
(DOCX)
Data Availability Statement
The dataset generated during the current study is available in the Figshare data repository (A replicate FASTQ files: https://dx.doi.org/10.6084/m9.figshare.3496412, B replicate FASTQ files: https://dx.doi.org/10.6084/m9.figshare.3496538, Metadata: https://dx.doi.org/10.6084/m9.figshare.3485201, R Code: https://dx.doi.org/10.6084/m9.figshare.3474923).