Abstract
Wnt signaling through β-catenin plays a crucial role in skin development and homeostasis. Disruption or hyperactivation of this pathway results in skin defects and diseases (Lim and Nusse, Cold Spring Harb Perspect Biol 5(2), 2013). Monitoring Wnt signaling in skin under normal and abnormal conditions is therefore critical to understand the role of this pathway in development and homeostasis.
In this chapter, we provide methods to detect Wnt/β-catenin (canonical) signaling in the skin. We present a comprehensive list of Wnt reporter mice and detail the processing of skin tissue to detect reporter genes. From this list, we focus on the three most recent lines that, according to reports, are the most sensitive in skin. Additionally, we describe a protocol to detect nuclear β-catenin, a hallmark of active Wnt signaling, although this technique should be used with caution due to its limited sensitivity. The techniques outlined below will be useful for detecting active Wnt signaling in skin.
Keywords: Skin, Epidermis, Hair follicles, Wnt reporter mice
1 Introduction
The skin is a self-renewing tissue made up of the stratified epidermis, its appendages and the underlying dermis [1]. The stratified epidermis, which provides protective barrier function of the skin, continually sheds its outermost layer. Cells in the basal layer proliferate, move upward and differentiate to replenish the outermost layer. Whereas the stratified epidermis self-renews continuously, hair follicles undergo periodic degeneration and regeneration, depending on signals from a specific dermal niche, the dermal papilla [2]. In response to wounding, stem cells from both the stratified epidermis and hair follicles contribute to repair of the epidermis [3–9].
Canonical Wnt signaling plays a critical role in skin epidermal development and homeostasis [10]. During morphogenesis, Wnt/β-catenin signaling is required in both dermal and epidermal cells to specify formation of the hair placode and the dermal papilla precursor, the dermal condensate [11]. Subsequently, Wnt/β-catenin signaling is essential in both hair follicular progenitor cells and the dermal papilla for hair regeneration to occur [12–15]. Under homeostatic conditions, Wnt/β-catenin controls normal proliferation of the basal cells of the stratified epidermis [16, 17].
Given the crucial role of Wnt/β-catenin signaling in skin biology, monitoring Wnt signaling in the skin is of vital interest to skin researchers. Because canonical Wnt signaling results in the translocation of β-catenin into the nucleus, detection of nuclear β-catenin has been used to demonstrate active Wnt signaling. However, due to the low sensitivity of the assay, nuclear β-catenin may reflect only very high Wnt activity. A more sensitive method is based on the detection of Wnt target genes. Wnt target genes are transcribed when nuclear β-catenin binds to TCF/LEF, which bind to TCF/LEF binding sites of the promoters of Wnt target genes, and transactivate their transcription. In order to detect Wnt activity, a series of Wnt reporter mice have been developed. These mice can be categorized into two groups according to the types of promoter. One group expresses reporter gene lacZ or green fluorescent protein (GFP) or its variants under the promoter containing multimerized TCF /LEF binding sites [18–25]. The other group contains the reporter gene knock-in under the control of the endogenous promoter of the Wnt target gene Axin2, or inducible cre-recombinase (creERT2) knocked into the Axin2 locus and used with Rosa26 reporter lines [17, 26–30].
In this chapter, we summarize the current Wnt reporter strains used to monitor canonical Wnt signaling and provide methods for detecting different reporter genes in the skin. We also provide an immunostaining protocol for detecting nuclear β-catenin.
2 Materials
2.1 Wnt Reporter Mice
See Table 1. We used wild-type C57BL/6 mice, TCF/LEF:H2BGFP mice [25], BAT-GAL mice [20], and Axin2lacZ mice [27] as specific examples in this chapter. Low Wnt activity is detected in basal cells of the epidermis only in TCF/LEF:H2B-GFP mice and Wnt reporter mice with Axin2 promoter [16, 17].
Table 1.
Wnt reporter mice
| Transgenic reporter mice with promoter containing multimerized TCF/LEF binding sites | ||
|---|---|---|
| Promoter | References | |
| TOP-GAL | 3 TCF/LEF binding sites with c-fos minimal promoter |
DasGupta and Fuchs, 1999 [18] |
| TOP-lacZ | 3 TCF/LEF binding sites with TK minimal promoter |
Staal et al., 2001 [19] |
| BAT-GAL | 7 TCF/LEF binding sites with siamois minimal promoter |
Maretto et al., 2003 [20] |
| TCF/LEF-lacZ | 6 TCF/LEF binding sites with hsp68 minimal promoter |
Mohamed et al., 2004 [21] |
| BAT-lacZ | 8 TCF/LEF binding sites with minimal siamois promoter |
Nakaya et al., 2005 [22] |
| Ins-TOPGAL | 6 TCF/LEF binding sites with TK minimal promoter |
Moriyama et al., 2007 [23] |
| Ins-TOPEGFP | 6 TCF/LEF binding sites with TK minimal promoter |
Moriyama et al., 2007 [23] |
| LEF-EGFP | 7 TCF/LEF binding sites with c-fos minimal promoter |
Currier et al., 2010 [24] |
| TCF/LEF:H2B-GFP | 6 TCF/LEF binding sites with hsp68 minimal promoter |
Ferrer-Vaquer et al., 2010 [25] |
| Reporter mice with Wnt target gene promoter (Axin2) | ||
| Axin2-EGFP | 5.6 kb genomic fragment containing promoter and first intron of Axin2 |
Jho et al., 2002 [26] |
| Axin2lacZ | Endogenous Axin2 promoter | Lustig et al., 2002 [27] |
| Axin2creERT2;Rosa26RmTmG | Endogenous Axin2 promoter | van Amerongen et al., 2012; Muzumdar et al., 2007 [28, 29] |
| Axin2creERT2;Rosa26RlacZ | Endogenous Axin2 promoter | van Amerongen et al., 2012; Soriano 1999 [28, 30] |
| Axin2creERT/tdT;Rosa26RmTmG | Endogenous Axin2 promoter | Choi et al., 2013; Muzumdar et al., 2007 [17, 29] |
2.2 Tissue Embedding Components
Electric clipper.
4-in. scissors.
4-in. dissecting forceps.
Razor blade.
Brown paper towels.
Cryomolds 25 × 20 mm.
O.C.T. compound.
Dry ice block.
2.3 Tissue Fixation Reagents and Components
30 % sucrose solution. Filtrate with 0.45 um filter. Store at 4 °C.
10 % sucrose solution. Filtrate with 0.45 um filter. Store at 4 °C.
Phosphate-buffered saline (PBS).
EMS 16 % paraformaldehyde (PFA) in 10 mL glass ampules.
Falcon 5 mL 12 × 75 mm snap-cap polypropylene tubes.
Aluminum foil.
Shaker.
2.4 Tissue Sectioning and Staining Components
Cryostat.
Adhesion Micro Slides.
Microscope cover glass.
Hematoxylin 2.
Glass staining jar.
10 % Triton X-100 solution.
20 % bovine serum albumin (BSA). Filtrate with 0.22 um filter, aliquot, and store at −20 °C.
Gelatin from cold water fish skin (Sigma).
10 % normal donkey serum (NDS).
PBSGT buffer: 2 % fish gelatin, 0.2 % Triton, in PBS.
Blocking buffer: PBSGT buffer with 2 % BSA with 10 % NDS.
Staining buffer: PBSGT buffer with 1 % BSA with 5 % NDS.
Hoechst 33342.
ProLong Gold Antifade (Molecular Probes, Thermofisher).
Nail polish.
Additional components for immunohistochemistry staining of β-catenin using M.O.M kit
Citrus clearing solvent as xylene substitute.
Ethanol series: 100, 95, 85, 70, 50 %.
10 mM citric acid buffer, pH 6.0.
0.3 % Hydrogen peroxidase (H2O2). Prepare fresh from 30 % hydrogen peroxide.
Mouse on Mouse (M.O.M. ™) basic kit (Vector).
Avidin/Biotin blocking kit.
M.O.M blocking buffer with Avidin: Add 45 µL of IgG reagent (from M.O.M. kit), four drops of avidin solution (from Avidin/Biotin Blocking kit) to 1 mL PBSGT buffer.
M.O.M. Diluent Buffer with Biotin: Add 100 µL of Protein Concentrate reagent (from M.O.M. kit), four drops of biotin solution (from Avidin/Biotin Blocking kit) to 1 mL PBSGT buffer.
M.O.M. Diluent Buffer without Biotin: Add 80 µL of Protein Concentrate reagent (from M.O.M. kit) to 1 mL PBSGT buffer.
Vectastain Elite ABC reagent: Prepare fresh. Add one drop of Reagent A to 1.25 mL PBS, mixing well, then adding one drop of Reagent B, then mixing well again. Allow ABC mixture to stand for 30 min at room temperature before use.
ImmPACT DAB Peroxidase (HRP) substrate: Prepare fresh. Add one drop of concentrated DAB to 1 mL of the provided diluent buffer.
Clarifier 2.
Bluing reagent.
Permount Mounting Medium.
Coplin staining jar.
Hybridization oven.
Microwave oven.
2.5 Primary and Secondary Antibodies
Rabbit α GFP (Molecular Probes, G10362, dilution 1:250).
Rabbit α mouse keratin 5 primary antibody.
Rat α β-4 integrin (BD, 553745, dilution 1:200).
Mouse α β-catenin monoclonal antibody (Sigma, 15B8, dilution 1:500).
AF488-conjugated donkey α rabbit secondary antibody.
RRX-conjugated donkey α rabbit secondary antibody (2° Abs: affinity-purified whole IgG, cross-adsorbed).
RRX-conjugated donkey α rat secondary antibody (2° Abs: affinity-purified whole IgG, cross-adsorbed).
2.6 X-gal Staining
25 % glutaraldehyde solution.
VECTOR Nuclear Fast Red solution (Vector Lab).
20 mg/mL X gal stock: Dissolve X-gal in N′N′ dimethylformamide (DMF). Store at −20 °C protect from light.
1 M Na-phosphate solution.
1 M MgCl2.
50 mM K3Fe(CN)6. Store at 4 °C, protect from light.
50 mM K4Fe(CN)6. Store at 4 °C, protect from light.
10 % Na deoxycholate.
10 % NP40.
80 % Glycerol.
X-gal staining solution: 100 mM Na-phosphate (pH 7.3), 1.3 mM MgCl2, 3 mM K3Fe(CN)6, 3 mM K4Fe(CN)6, 0.01 % Na deoxycholate, 0.2 % NP40, 2 mg/mL X-Gal.
2.7 Microscopy and Imaging Setup
Transmitted light microscope.
Fluorescence microscope.
3 Methods
We describe here the use of two types of Wnt reporter mice to monitor the activity of Wnt/β-catenin signaling: TCF /LEF:H2B- GFP [25] and BAT-GAL [20] or Axin2lacZ [27]. The reporter genes are either GFP- or β-galactosidase-based. TCF/LEF:H2B-GFP is a transgenic line expressing histone H2B fused with GFP under a promoter containing 6 TCF/LEF binding sites [25]. BAT- GAL is a transgenic line expressing lacZ reporter gene under a promoter containing 6 TCF /LEF binding sites [20]. Axin2lacZ line contains the lacZ gene inserted into the Axin2 locus [27]. Immunofluorescent detection of GFP and immunohistochemical detection of β-galactosidase activity are both performed on fresh cryosections. To visualize GFP signal in situ, pre-fixation (fixation before tissue embedding) is recommended to preserve the GFP signal.
3.1 Harvesting Backskin Tissue
Sacrifice the mice according to institutional guidelines (see Note 1) and closely shave all hair on the back with an electric shaver.
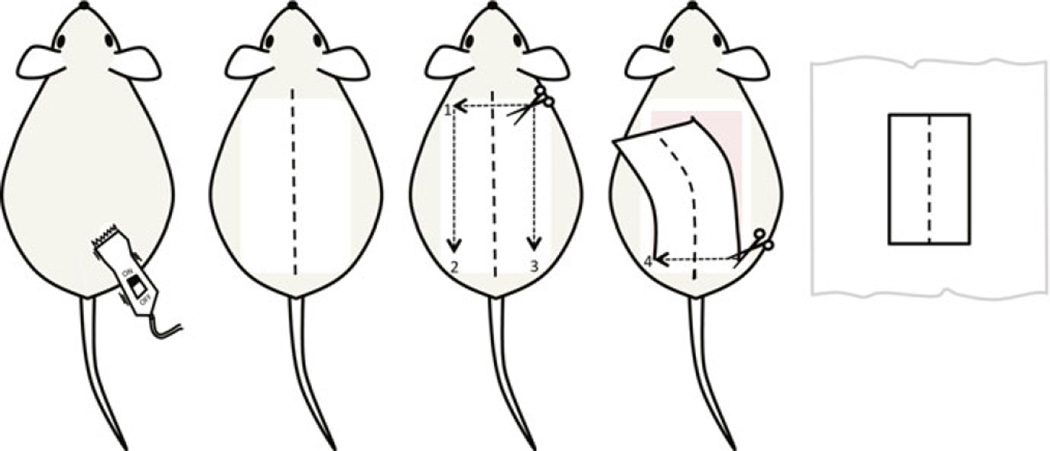
Mark the midline from neck to tail with a marker. Lift the backskin at the back of the neck with the fine forceps and make an incision. Insert the scissors underneath the skin and cut a rectangle shape from upper back to the lower back. Gently peel the entire backskin away from the body (Fig. 1).
Place the skin onto a paper towel with the epidermis side up and the dermis on top of the towel. Use forceps to spread the skin so that midline is straight. When working with multiple mice, align all pieces of skin in the same head-to-tail orientation (see Note 2).
Use a razor blade to cut the skin attached to the paper towel at the midline and on two sides parallel to the midline to yield two strips of skin 0.5 cm × 2.5 cm (see Note 3).
With GFP reporter mice proceed to the tissue processing with prefixation Subheading 3.2. When using lacZ reporter mice proceed to the embedding Subheading 3.3.
Fig. 1.
Stepwise illustration of backskin tissue harvesting. The mouse backskin is closely shaved before being marked at the midline from the neck to the base of the tail. An incision is made across the upper back and then along the two sides parallel to the midline. The skin is gently peeled off with the final cut across the back above the hind legs. The skin is spread on a brown paper towel with the midline aligned straight. The skin then can be cut into strips for processing
3.2 Prefixation of Tissue for GFP Preservation
For GFP reporter mice, prefixation of tissue prevents GFP signal loss. However, in the TCF /LEF: H2B-GFP [25] the GFP is fused with histone H2B, which renders the GFP much more stable, and therefore with this mouse line the prefixation step is optional. Prefixation is required for all other GFP reporter lines.
Transfer the skin strips to an aluminum foil-wrapped Falcon 5 mL tube to protect it from light throughout the whole procedure. All incubation steps below are performed with gentle rocking on the shaker.
Fix with fresh 4 % PFA for 2 h. Ensure the strip is fully immersed (see Note 4). Discard PFA as hazardous waste.
Wash with PBS 2–3 times, 5 min each wash.
Soak tissues in 10 % sucrose and incubate for 2 h. Aspirate.
Soak tissues in 30 % sucrose and incubate overnight at 4 °C in the dark.
The next day, equilibrate with 1:1 O.C.T. slurry (30 % sucrose: O.C.T. = 1:1) for 6 h. O.C.T. slurry can be made by damping out half of the sucrose and adding O.C.T. for 1:1 mixture. Proceed to embedding backskin in O.C.T. for cryosectioning (Subheading 3.3).
3.3 Embedding Backskin in OCT Block
Fill the cryomold with O.C.T.
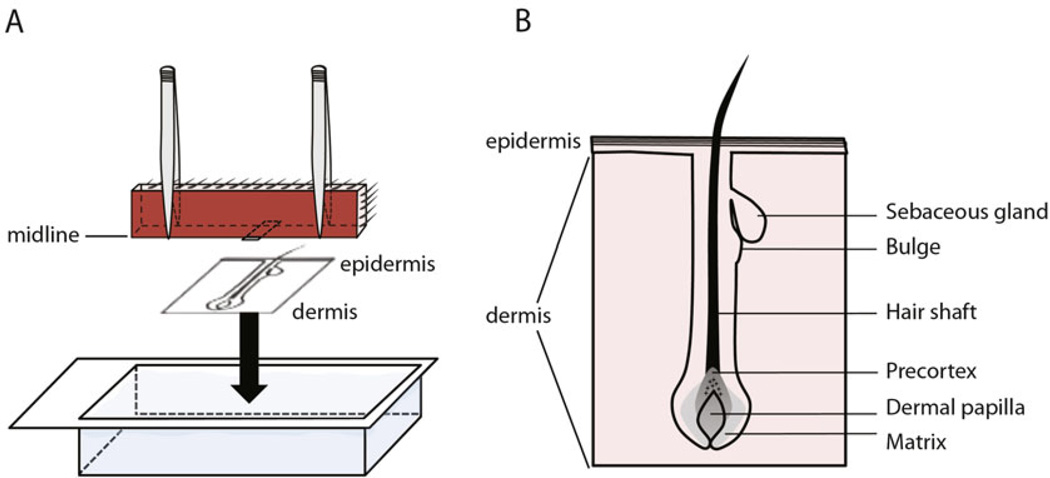
Hold both ends of the skin strip with fine forceps, and place the skin strip with the midline touching the bottom of O.C.T.-filled cryomold (Fig 2a). Use fine forceps to hold the skin straight and to make sure the skin freezes exactly perpendicular to the bottom of the block (see Note 5). Hold the skin straight while placing the O.C.T. block on the dry ice to freeze (see Note 6).
Transfer the frozen O.C.T. block to prechilled cryostat for sectioning or store at −80 °C.
Fig. 2.
Illustration of skin embedding to acquire longitudinal sections of hair follicles. (a) Skin strip is placed into O.C.T. filled cryomold with the midline facing and touching the bottom of the cryomold. (b) Cartoon depicting a good day-28 skin section with an intact hair follicle
3.4 Cryosectioning
Follow cryostat instruction (see Note 7) to section tissue in 8 µm (range: 6–12 µm), two sections per slide.
Air-dry the first collected section on the slide and counterstain with hematoxylin solution for 1 min.
Gently wash off excess staining solution with tap water. Visualize hair follicles under a light microscope and adjust block angle until intact hair follicles can be detected (Fig. 2b, see Note 8).
Collect sections, air dry for 20 min (range: 15–30 min), and freeze at −80 °C for long-term storage.
3.5 Visualizing GFP Signal in GFP Reporter Mouse Skin
GFP can be visualized directly under a fluorescence microscope (see Note 9). If stored at −80 °C, thaw at room temperature for 10 min before use. To colocalize GFP signal with epidermal markers (or other markers), proceed to the next step (see Note 10). If GFP signal is weak, proceed to next step and immunostain for GFP expression (Fig. 3a).
Use a pap pen to draw a circle around the skin tissue to create a hydrophobic barrier.
Transfer the slide to an aluminum foil-wrapped, humidified staining chamber (see Note 11) and incubate with blocking buffer (150–200 µL/section in general) for 1 h.
Add primary antibody (α β-4 integrin 1:200 and optional α GFP 1:250 diluted in staining buffer) and incubate for 2 h at room temperature or overnight (16–24 h) at 4 °C in humidified chamber.
The next day, wash with PBS in the staining jar, three times, 5 min each time.
Add fluorophore-conjugated secondary antibody (AF488 1:150 and RRX 1:200 diluted in staining buffer) for 45 min in the humidified chamber.
Wash in PBS briefly.
Counterstain in Hoechst 33342 (1:2000 dilution in PBS) at room temperature for 2 min.
Wash with PBS three times, 10 min total.
Aspirate excess buffer and mount the section with ProLong Gold antifade and cure overnight at room temperature in the dark.
Seal the slide with nail polish and air-dry for 30 min before imaging with a fluorescence microscope. If immediate imaging is not possible, slides can be stored at −20 °C (see Note 9).
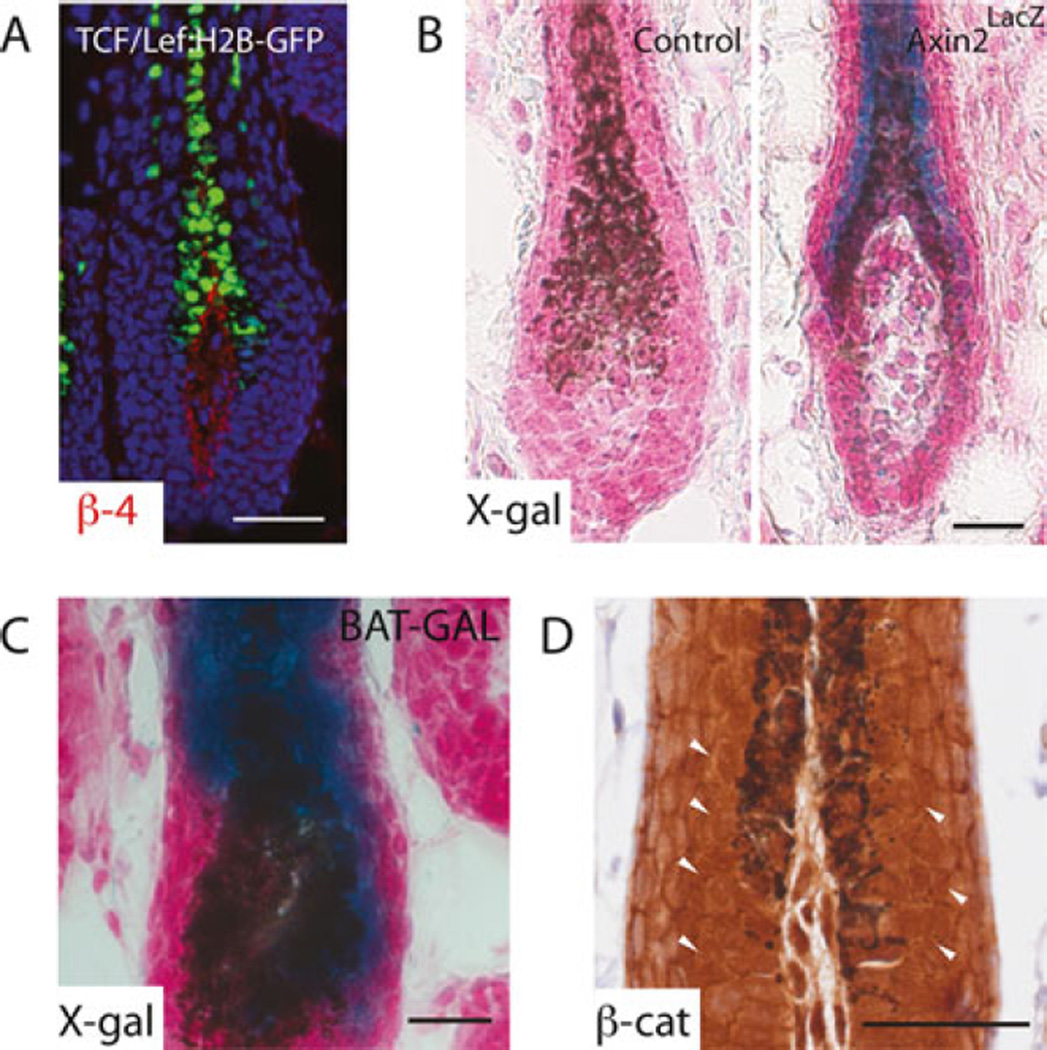
Fig. 3.
Detecting Wnt/β-catenin signaling. (a) Fluorescence image of endogenous GFP signal in the hair follicle of 28-day-old TCF /LEF:H2B-GFP mouse, with nuclei counterstained with Hoechst. β4-integrin co-staining in red labels the basement membrane, delineating the interface between epithelial cells and the dermal papilla. X-Gal staining of 28-day-old (b) wild-type or Axin2lacZ and (c) BAT-GAL mice. Skins were counterstained with Nuclear Fast Red to mark nuclei. Blue signal in the precortex of hair follicle indicates Wnt activity. (d) Immunostaining of β-catenin in 28-day-old wild-type mouse skin. Arrowheads point to nuclear β-catenin. Scale bar denotes 50 µm
3.6 X-gal Staining of lacZ Reporter Mouse Skin
Thaw slides from −80 °C for 10 min at room temperature (see Note 12).
Fix in freshly prepared 0.1 % glutaraldehyde in PBS for 2 min.
Wash with PBS in the staining jar at least five times, 5 min per wash.
Prepare X-Gal staining solution fresh by adding X-Gal (2 mg/mL) to pre-warmed base solution.
Incubate the slides at 37 °C in the hybridization oven. Cover the jar with slides with aluminum foil. Active β-galactosidase digests X-Gal as a substrate and converts it into a blue precipitate. Check the color development from 2 to 3 h of staining and let it go overnight if necessary.
Wash with PBS three times, 5 min each.
Rinse with distilled water.
Counterstain with Nuclear Fast Red for 5 min.
Wash with running water for 10 min.
Mount slides with 80 % glycerol and seal with nail polish.
Acquire images with a light microscope (Fig. 3b, c).
3.7 Immunostaining for Nuclear β-catenin
3.7.1 Dehydration and Antigen Retrieval of FFPE Sample
Use 5 µm skin sections from a formaldehyde-fixed paraffinembedded (FFPE) block (see Note 13).
Dewax paraffin sections by placing slides in a 60 °C prewarmed citrus solvent and incubate in the hybridization oven at 60 °C for 10 min.
Rehydrate the slides by sequential incubation in an alcohol series, 3 min each step: 100 % ethanol, 100 % ethanol, 95 % ethanol, 85 % ethanol, 70 % ethanol, 50 % ethanol.
Rinse the slides twice briefly in distilled water and transfer to the Coplin jar.
- Antigen unmasking is achieved using the heat-induced epitope retrieval (HIER) method:
- Fill the jar to the top with citric acid buffer (pH 6.0).
- Boil in a microwave at 500 W (50 % power in regular microwave) for three rounds, 5 min each (15 min in total).
- Between each round, refill citric buffer to the top to prevent the sections from drying. Set the jar on the benchtop and chill for 20 min until it reaches room temperature.
3.7.2 Blocking and Antibody Staining with M.O.M Kit
Briefly wash slides with distilled water.
Block endogenous peroxidase activity by incubating slides in 0.3 % H2O2 solution for 15 min in the glass staining jar (see Note 14).
Wash with PBS three times, 5 min each.
Aspirate residual PBS without drying the tissue.
Draw a circle around the skin tissue with a Pap pen to create a hydrophobic border.
To block nonspecific antibody binding and endogenous biotin, add 100–200 µL M.O.M blocking buffer with Avidin per section and incubate for 1 h in the humidified staining chamber (see Note 15).
Remove the blocking buffer and rinse with PBS.
Add 100–200 µL M.O.M diluent buffer per section and incubate for 5 min.
Aspirate the diluent.
Add 100–200 µL of primary antibody (Mouse α β-catenin, 1:500) in M.O.M diluent buffer with biotin onto the skin section and incubate overnight in the humidified staining chamber at 4 °C.
The next day, wash with PBS three times, 5 min each.
3.7.3 Signal Amplification and Development
Aspirate excess PBS buffer around the tissue after the last wash.
Prepare secondary antibody by diluting α Mouse IgG Biotinylated antibody (1:200, from the M.O.M. kit) in M.O.M. diluent buffer without biotin.
Add 100–200 µL of secondary antibody solution per section and incubate for 30 min (see Note 16). At the same time, prepare Vectastain Elite ABC reagent.
Wash with PBS three times, 5 min each.
Add 100–200 µL ABC reagent mixture onto the skin section and let incubate for 30 min.
Wash with PBS two times, 5 min each.
For color development, prepare DAB solution and add 100–200 µL DAB substrate to the section. Monitor DAB colorization (brown) under the light microscope until the intensity of the color is satisfactory (between 3 and 5 min, see Note 17) and wash with tap water for 5 min (Fig. 3d).
3.7.4 Hematoxylin Counterstain and Imaging
Hematoxylin counterstain marks the cell’s nucleus to reveal histology and also enhances the contrast from DAB development.
Stain the slides in hematoxylin for 30 s.
Wash with tap water for 1 min.
Incubate in clarifier for 1 min.
Wash with tap water for 1 min.
Incubate in bluing reagent for 1 min.
Wash with tap water for 1 min.
Wash four times total with increasing concentrations of Ethanol: 70 % ethanol, 95 % ethanol, 100 % ethanol, 100 % ethanol, then two times with citrus solvent, 1 min each time.
Allow evaporation of citrus solvent in the chemical hood for 2–5 min.
Mount the slide with a drop (~50 µL) of Permount solution and let solidify overnight.
The next day, image slides or store slides at room temperature for future imaging.
Footnotes
CO2 euthanasia of mice is performed according to the American Veterinary Medical Association (AVMA) guidelines (CO2: 3 min on and 2 min off at a defined flow rate in closed chamber).
When harvesting skins of multiple mice, keep the skins moist with PBS until all the skins are processed. Maximum five mice per batch are recommended.
The phases of the hair cycle may differ in different regions of the backskin. Therefore, always collect skin from the same region of the back.
During tissue harvesting and fixation, ensure the skin strip is fully immersed in PFA without air bubbles. Since the skin will become more rigid after fixation, avoid tissue distortion or bending during PFA incubation.
If multiple skin pieces are embedded in one cryomold, stack them on top of each other with O.C.T. compound squeezed between them. Make sure that the midlines of all the skin strips are facing and touching the bottom of the cryomold. A maximum of three skin strips in one cryomold is strongly recommended.
Use dry ice blocks with a flat surface to ensure direct contact and immediate freezing of the O.C.T. compound.
The cryostat setting for skin sectioning is CT = −24 °C, OT = −27 °C (CT = chamber temperature, OT = object temperature).
A good embedding minimizes angle adjustment during cryostat sectioning. A good section should include a couple of hair follicles that are contiguous to the basal layer of the epidermis at the top and has contact with the dermal papilla at the bottom.
The GFP signal in TCF/LEF:H2B-GFP reporter mice is relatively stable compared with other GFP-based Wnt reporter mice. Unfixed TCF/LEF:H2B-GFP skin in O.C.T. retains GFP signal for at least 2 years.
Co-immunostaining of structural protein such as β4-integrin or Keratin 5 can help define the structure of hair follicles. β4-integrin antibody labels the basement membrane, which marks the interface between epidermal and dermal compartments. Keratin 5 antibody marks keratinocytes in the basal layer of the interfollicular epidermis and the outer root sheath of the hair follicle.
A humidified chamber is assembled with a homemade slide rack and PBS-soaked paper towels underneath to minimize reagent evaporation during the incubation step. To create a homemade slide rack, we align two serological pipettes (10 mL) that have been shortened to fit into a BioAssay Dish. The pipettes are attached by tape to the base of the dish to form two parallel tubes where the slides can be placed on top. To minimize light exposure during staining, the lid and bottom dish of BioAssay Dish are wrapped with aluminum foil.
The best X-gal staining result is obtained with freshly embedded/sectioned skin of Axin2lacZ reporter mice.
Detection of nuclear β-catenin in skin is best achieved by immunohistochemistry with paraffin embedded skin.
H2O2 in 0.3 % solution should be prepared fresh from a 30 % stock with tap water in a glass jar.
The M.O.M kit is used in conjunction with mouse-raised primary antibody in mouse tissue.
The ABC reagent doesn’t spread because it has no serum or protein to reduce surface tension. Ensure the complete coverage of specimen by gently adding reagent directly onto the specimen.
A satisfactory level for DAB development is defined by the balance between the intensity of the brown signal in the nucleus and the overall background signal.
References
- 1.Blanpain C, Fuchs E. Epidermal stem cells of the skin. Annu Rev Cell Dev Biol. 2006;22:339–373. doi: 10.1146/annurev.cellbio.22.010305.104357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schneider MR, Schmidt-Ullrich R, Paus R. The hair follicle as a dynamic miniorgan. Curr Biol. 2009;19(3):R132–R142. doi: 10.1016/j.cub.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 3.Tumbar T, et al. Defining the epithelial stem cell niche in skin. Science. 2004;303(5656):359–363. doi: 10.1126/science.1092436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito M, et al. Stem cells in the hair follicle bulge contribute to wound repair but not to homeostasis of the epidermis. Nat Med. 2005;11(12):1351–1354. doi: 10.1038/nm1328. [DOI] [PubMed] [Google Scholar]
- 5.Levy V, et al. Epidermal stem cells arise from the hair follicle after wounding. FASEB J. 2007;21:1–9. doi: 10.1096/fj.06-6926com. [DOI] [PubMed] [Google Scholar]
- 6.Snippert HJ, et al. Lgr6 marks stem cells in the hair follicle that generate all cell lineages of the skin. Science. 2010;327(5971):1385–1389. doi: 10.1126/science.1184733. [DOI] [PubMed] [Google Scholar]
- 7.Mascre G, et al. Distinct contribution of stem and progenitor cells to epidermal maintenance. Nature. 2012;489(7415):257–262. doi: 10.1038/nature11393. [DOI] [PubMed] [Google Scholar]
- 8.Page ME, et al. The epidermis comprises autonomous compartments maintained by distinct stem cell populations. Cell Stem Cell. 2013;13(4):471–482. doi: 10.1016/j.stem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Howard JM, et al. Tcf3 expression marks both stem and progenitor cells in multiple epithelia. Development. 2014;141(16):3143–3152. doi: 10.1242/dev.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lim X, Nusse R. Wnt signaling in skin development, homeostasis, and disease. Cold Spring Harb Perspect Biol. 2013;5(2) doi: 10.1101/cshperspect.a008029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D, et al. Dermal beta-catenin activity in response to epidermal Wnt ligands is required for fibroblast proliferation and hair follicle initiation. Development. 2012;139(8):1522–1533. doi: 10.1242/dev.076463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huelsken J, et al. beta-Catenin controls hair follicle morphogenesis and stem cell differentiation in the skin. Cell. 2001;105(4):533–545. doi: 10.1016/s0092-8674(01)00336-1. [DOI] [PubMed] [Google Scholar]
- 13.Lowry WE, et al. Defining the impact of beta-catenin/Tcf transactivation on epithelial stem cells. Genes Dev. 2005;19(13):1596–1611. doi: 10.1101/gad.1324905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enshell-Seijffers D, et al. Beta-catenin activity in the dermal papilla of the hair follicle regulates pigment-type switching. Proc Natl Acad Sci U S A. 2010;107(50):21564–21569. doi: 10.1073/pnas.1007326107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsai SY, et al. Wnt/beta-catenin signaling in dermal condensates is required for hair follicle formation. Dev Biol. 2014;385(2):179–188. doi: 10.1016/j.ydbio.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lim X, et al. Interfollicular epidermal stem cells self-renew via autocrine Wnt signaling. Science. 2013;342(6163):1226–1230. doi: 10.1126/science.1239730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi YS, et al. Distinct functions for Wnt/beta-catenin in hair follicle stem cell proliferation and survival and interfollicular epidermal homeostasis. Cell Stem Cell. 2013;13(6):720–733. doi: 10.1016/j.stem.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.DasGupta R, Fuchs E. Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development. 1999;126(20):4557–4568. doi: 10.1242/dev.126.20.4557. [DOI] [PubMed] [Google Scholar]
- 19.Staal FJ, et al. Wnt signaling is required for thymocyte development and activates Tcf-1 mediated transcription. Eur J Immunol. 2001;31(1):285–293. doi: 10.1002/1521-4141(200101)31:1<285::AID-IMMU285>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 20.Maretto S, et al. Mapping Wnt/beta-catenin signaling during mouse development and in colorectal tumors. Proc Natl Acad Sci U S A. 2003;100(6):3299–3304. doi: 10.1073/pnas.0434590100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mohamed OA, Clarke HJ, Dufort D. Beta-catenin signaling marks the prospective site of primitive streak formation in the mouse embryo. Dev Dyn. 2004;231(2):416–424. doi: 10.1002/dvdy.20135. [DOI] [PubMed] [Google Scholar]
- 22.Nakaya MA, et al. Wnt3a links left-right determination with segmentation and anteroposterior axis elongation. Development. 2005;132(24):5425–5436. doi: 10.1242/dev.02149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Moriyama A, et al. GFP transgenic mice reveal active canonical Wnt signal in neonatal brain and in adult liver and spleen. Genesis. 2007;45(2):90–100. doi: 10.1002/dvg.20268. [DOI] [PubMed] [Google Scholar]
- 24.Currier N, et al. Dynamic expression of a LEF-EGFP Wnt reporter in mouse development and cancer. Genesis. 2010;48(3):183–194. doi: 10.1002/dvg.20604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ferrer-Vaquer A, et al. A sensitive and bright single-cell resolution live imaging reporter of Wnt/ss-catenin signaling in the mouse. BMC Dev Biol. 2010;10:121. doi: 10.1186/1471-213X-10-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jho EH, et al. Wnt/beta-catenin/Tcf signaling induces the transcription of Axin2, a negative regulator of the signaling pathway. Mol Cell Biol. 2002;22(4):1172–1183. doi: 10.1128/MCB.22.4.1172-1183.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lustig B, et al. Negative feedback loop of Wnt signaling through upregulation of conductin/axin2 in colorectal and liver tumors. Mol Cell Biol. 2002;22(4):1184–1193. doi: 10.1128/MCB.22.4.1184-1193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Amerongen R, Bowman AN, Nusse R. Developmental stage and time dictate the fate of Wnt/beta-catenin-responsive stem cells in the mammary gland. Cell Stem Cell. 2012;11(3):387–400. doi: 10.1016/j.stem.2012.05.023. [DOI] [PubMed] [Google Scholar]
- 29.Muzumdar MD, et al. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 30.Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21(1):70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]