Abstract
Excessive subconjunctival scarring is the main reason of failure of glaucoma filtration surgery. We analyzed conjunctival and systemic gene expression patterns after non penetrating deep sclerectomy (NPDS). To find expression patterns related to surgical failure and their correlation with the clinical outcomes. This study consisted of two consecutive stages. The first was a prospective analysis of wound-healing gene expression profile of six patients after NPDS. Conjunctival samples and peripheral blood samples were collected before and 15, 90,180, and 360 days after surgery. In the second stage, we conducted a retrospective analysis correlating the late conjunctival gene expression and the outcome of the NPDS for 11 patients. We developed a RT-qPCR Array for 88 key genes associated to wound healing. RT-qPCR Array analysis of conjunctiva samples showed statistically significant differences in 29/88 genes in the early stages after surgery, 20/88 genes between 90 and 180 days after surgery, and only 2/88 genes one year after surgery. In the blood samples, the most important changes occurred in 12/88 genes in the first 15 days after surgery. Correspondence analyses (COA) revealed significant differences between the expression of 20/88 genes in patients with surgical success and failure one year after surgery. Different expression patterns of mediators of the bleb wound healing were identified. Examination of such patterns might be used in surgery prognosis. RT-qPCR Array provides a powerful tool for investigation of differential gene expression wound healing after glaucoma surgery.
Introduction
Glaucoma is characterized by progressive deterioration of retinal nerve fiber layer and optic nerve, leading to defects in the visual field and optic atrophy. In most cases, the condition is associated with increased intraocular pressure (IOP) caused by obstruction of drainage of aqueous humor from the eye. Currently, the main treatment for glaucoma is reduction of IOP using drugs and laser or filtration surgery.
Glaucoma Filtering Surgery (GFS) success depends primarily on the creation and maintenance of a communication channel between the anterior chamber and subconjunctival space. The conjunctival and episcleral healing is the main limiting factor in this surgery; postoperative IOP depends on the balance between scarring and tissue regeneration [1]. Thus, complete healing would cause failure of filtration surgery. In contrast, lack of healing would result in ocular hypotony.
Numerous pharmacological strategies have been employed to modulate conjunctival scarring. These include corticosteroids [2], antimitotics (5-fluorouracil, 5-FU and mitomycin C, MMC) [3–5], beta-radiation [6,7], inhibitors of growth factors (lederlimumab, bevacizumab) [8,9], and metalloproteinase inhibitors (ilomastat) [10]. However, some of these drugs are relatively nonspecific and lead to complications such as hypotony, endophthalmitis, etc.
Healing after glaucoma surgery consists of several stages: vascular response, coagulation, inflammation, proliferation and tissue remodeling [11]. Knowledge of the molecular mechanisms regulating this process should help in the development of new treatments for improved healing control. Likewise, categorization of patients based on gene expression profiles of the conjunctiva and Tenon’s capsule could help on identifying cases with high risk of failure, and thus improve the therapeutic options.
Reverse Transcription Real-Time Quantitative PCR Array (RT-qPCR Array) is the most reliable, specific and sensitive technology for analyzing the expression of a specific panel of genes. We present a customized RT-qPCR Array, used to analyze the expression of 88 genes implicated in the wound-healing process. These genes encode extracellular matrix (ECM) remodeling factors, inflammatory cytokines, as well as growth factors and main signaling molecules.
The purpose of this study was to investigate the conjunctival and systemic gene expression patterns at several different time points before and after non penetrating deep sclerctomy (NPDS), to correlate them with the clinical outcomes of the surgery, and to search for gene expression patterns related to surgical success and failure.
Materials and methods
Patients
The study used 11 eyes from 11 caucasian patients (four men and seven women, with a mean age ± standard deviation, SD of 67.16 ± 5.94 years) with uncontrolled primary open-angle glaucoma (POAG). The mean number (± SD) of preoperative topical anti-glaucoma drugs was 2.16 ± 0.75. Uncontrolled glaucoma was defined as an IOP over 21 mmHg after taking the maximum tolerated medication, with characteristic visual field and optic disc changes. Patients were recruited from the Glaucoma Unit of the Instituto Clínico Quirúrgico de Oftalmología (Bilbao, Spain), and Mendaro Hospital (Mendaro, Spain), after obtaining their signed informed consent.
The exclusion criteria were: concomitant administration of steroids or antimetabolites, previous anti-glaucoma surgery or conjunctival incisional surgery, cataract surgery during the three months before NPDS, diabetic retinopathy and other causes of ocular neovascularization. We also excluded cases of glaucoma with high risk of failure such as neovascular, aphakic, inflammatory, juvenile, traumatic, and postoperative glaucoma.
Ethics statement
The study followed the principles established in the Declaration of Helsinki and was approved by the Euskadi Review Board (ethics committee for clinical research) of Cruces Hospital. Written informed consent was obtained from all the study participants and all samples were anonymized to preserve patient confidentiality.
Study design
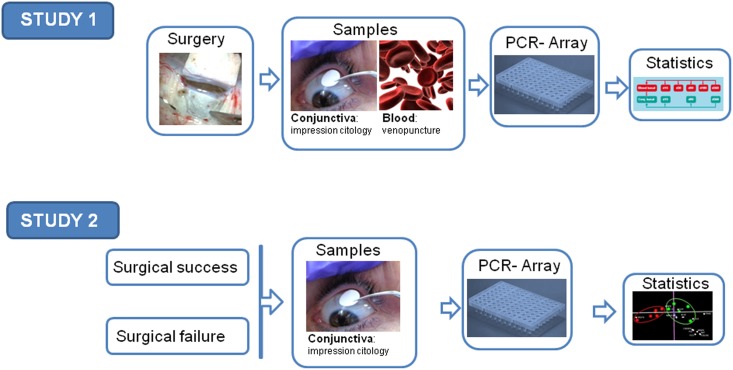
The work was divided into two consecutive studies. In the first study, we performed a prospective analysis of the wound-healing gene expression profiles of six patients after NPDS. Conjunctival samples (obtained by impression cytology, IC) and peripheral blood samples were collected before the operation and 15, 90,180, and 360 days after surgery. We examined gene expression patterns at these time points. In the second study, we conducted a retrospective analysis correlating the late conjunctival gene expression (a year or more after the intervention), and the success or failure of the NPDS. From each patient, a single conjunctival sample was collected using IC. (Fig 1)
Fig 1. Workflow for PCR-Array analysis of glaucoma filtering surgery.
Surgical failure was defined as an IOP ≥21 mmHg in the absence of ocular hypotensive treatment. In this study, we enrolled 11 patients (five patients with surgical failure and six patients from the first study).
Surgical technique and follow-up
NPDS was performed under topical anesthesia by the same surgeon (IRA). After excision of the deep scleral flap, a non-degradable HEMA implant (Esnoper, AJL Ophthalmic, Miñano, Álava, Spain) was sutured to the scleral bed with one 10–0 nylon suture. No antimetabolites were employed during the surgical procedure or in the follow-up. All patients received tobramycin and dexamethasone drops during one month after the surgery, in a decreasing pattern (Tobradex, AlconCusi, El Masnou, Barcelona, Spain). Follow-up consisted of biomicroscopic examination of anterior segment, checking bleb characteristics (morphology and presence of corkscrew vessels and epithelial microcysts), and IOP using Goldmann applanation tonometry.
Sample collection
For IC, cellulose acetate membrane (HAWP304, Millipore, Bedford, MA, USA) was applied onto the upper bulbar conjunctiva after instillation of topical anesthetic (Colircusi anestésico doble, Alcon Cusi, El Masnou, Barcelona, Spain). IC samples were used to study preoperative ocular surface (PAS-hematoxylin staining) and for RNA purification to analyze gene expression profiles before and after surgery.
IC samples for PAS-hematoxylin staining were obtained on 5×5 mm strips of cellulose acetate and immediately fixed in 96% ethanol. IC samples for gene expression analysis were obtained from the upper bulbar conjunctiva at the surgical site, by applying both sides of an 8 mm-diameter disc. These discs were immediately placed in an RNA conserving buffer (RNAprotect Cell Reagent, Qiagen, Germantown, MD, USA) and stored at 4°C until use.
At the same time, peripheral blood samples were collected with standard methods using PAX gene blood collection tubes (Qiagen) for RNA stabilization.
PAS-hematoxylin staining
PAS-hematoxylin staining was performed according to the Locquin and Langeron protocol modified by Rivas [12]. These samples were analyzed microscopically to establish the degree of metaplasia of the non-secreting cells, the presence of cytoplasmic or nuclear alterations, and cytoplasm-nucleus ratio.
RT-qPCR array design
The search for genes related to the healing process was conducted in three stages. In the first stage, a subset of genes was selected through a literature review (PubMed). In the second stage, another subset of genes was obtained from the bioinformatic analysis of functional networks focused on wound healing and related scarring processes (http://geneontology.org/, DAVID, EGAN). In the third stage, 88 genes were selected after processing both subsets and discarding redundancies. Subsequently, a custom 96-well plate RT-qPCR Array containing the 88 selected genes was designed and produced specifically for this study (SA Biosciences, Frederick, MD, USA) (Table 1). The remaining wells corresponded to the five housekeeping genes, a negative genomic DNA control, and positive reverse transcription and PCR controls.
Table 1. List of genes selected and analyzed by RT-q PCR Array.
a Genomic DNA Control (GDC) primer set that specifically detects nontranscribed genomic DNA contamination with a high level of sensitivity.
b Reverse Transcription Controls (RTC) to test the efficiency of the RT PCR Array.
c Positive PCR Controls (PPC) to test the efficiency of the polymerase chain reaction itself using a pre-dispensed artificial DNA sequence and the primer set that detects it.
RNA purification and RT-qPCR array processing
Total RNA was isolated from blood and conjunctiva. Whole peripheral blood was collected in PAXgene tubes (PreAnalytics) to grant RNA stabilization. After centrifugation, the supernatant was discarded and the pellet was treated with a series of buffers and silica-column purified following the manufacturer’s instructions (PAXgene Blood RNA kit, PreAnalytics, Qiagen). Thus, total blood RNA was purified for the study.
Conjunctival IC discs were processed with RNeasy Plus Micro Kit (Qiagen), with slight modifications of the manufacturer's protocol to perform an on-membrane lysis [13]. These modifications include a thorough vortexing during cell lysis to ensure maximum RNA recovery from both sides of the disc. Additionally, after transferring the cell lysate to the spin columns provided with the kit, the IC membrane alone was spun again to recover as much lysate as possible before this processing. Finally, after having added the elution buffer to the column, 10 minutes incubation was performed prior to RNA elution. After elution, the eluate loaded again into the column, allowing incubation for additional 10 minutes to maximize RNA concentration. These steps considerably increased RNA yield.
The quantification and evaluation of RNA quality were performed using an Agilent 2100 Bioanalyzer and RNA 6000 Pico Kit for low sample amounts (Agilent Technologies, Santa Clara, CA, USA). This method provides data on the integrity of isolated RNA (RNA Integrity Number; RIN), from which it extrapolates the RNA concentration. Conjunctiva and blood samples with RNA concentrations above 4 and 25 ng/μl, respectively, were processed; those with RIN less than 6 were discarded in both cases. cDNA was synthesized by reverse transcription of RNA using the RT2 First Strand Kit (SA Biosciences), following the manufacturer’s instructions. PCR was performed in iQ5 Thermal Cycler (Bio-Rad, Munich, Germany), using diluted cDNA as template (10 μl of cDNA, 10 μl of Genomic DNA Elimination Mixture and 91 μl of RNAse free water) and the RT2 SYBR Green qPCR Master Mix (SA Biosciences), according to the manufacturer's guidelines. All samples were analyzed in duplicate for each gene set.
Generation of expression values
Quantification results were normalized against housekeeping genes (ACTB, GAPDH, HPRT1, B2M and RPL13A), whose expression is constitutive under our experimental conditions. To do that, we normalized the primary data, represented in threshold cycle (Ct), using the geometric mean of the Cts of the constitutive genes included in each plate. Then expression changes were calculated using the ΔCt method, followed by the calculation of up or down regulation fold.
Statistical analysis
According to the small sample size, nonparametric tests were used for statistical comparisons. The Wilcoxon test was used to compare both types of samples (blood and conjunctiva) with their baseline levels.
Correlation between gene expression and IOP was examined using Spearman’s correlation test. Gene expression patterns related to surgical success and failure were identified using Correspondence Analysis (COA). Statistical analysis was performed using SPSS 19.0 (SPSS Sciences, Chicago, IL, USA). The level of statistical significance was P <0.05.
Results
None of the patients had intraoperative or postoperative complications. Regarding to the existence of systemic diseases, two patients were hypertensive patients treated with angiotensin-converting-enzyme inhibitors (ACE inhibitors), one patient suffered hypothyroidism treated by hormone replacement therapy and two patients were taking gastric protector. Finally, one patient was non-insulin dependent diabetic and was being treated with oral antidiabetic drugs.
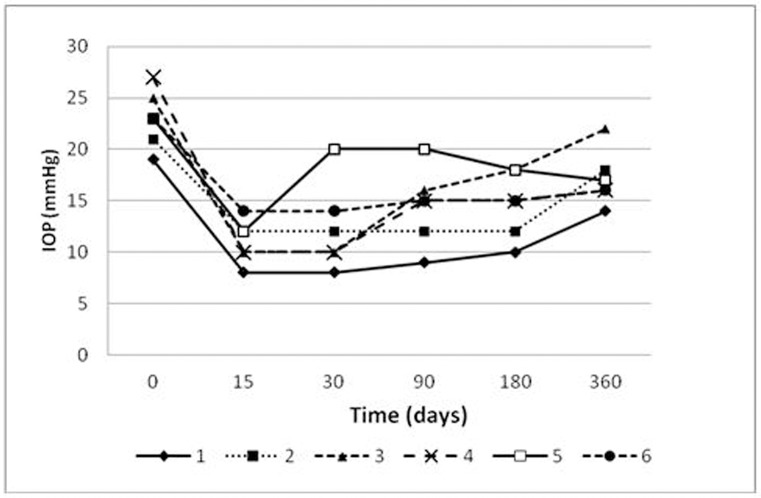
IOP values decreased significantly in comparison with baseline values, without specifying pharmacological treatment in any of the cases (Fig 2). Surgery failed in one case, one year after the NPDS (patient 3 IOP: 22 mmHg), requiring reintroduction of drug therapy.
Fig 2. Schematic representation of the fluctuations in Intraocular Pressure (IOP) as a result of surgical treatment, at different stages of the study.
In the group of patients analyzed in the first study, there was only one case of failure, a year after the surgery (IOP of 21 mmHg). For the remaining patients in this group, the intervention was successful, with IOPs maintained below 21 mmHg, which is the established criterion for determining the success of the surgery. The surgical failure case shows a scarring area where there it had been a channel allowing the drainage of aqueous humor.
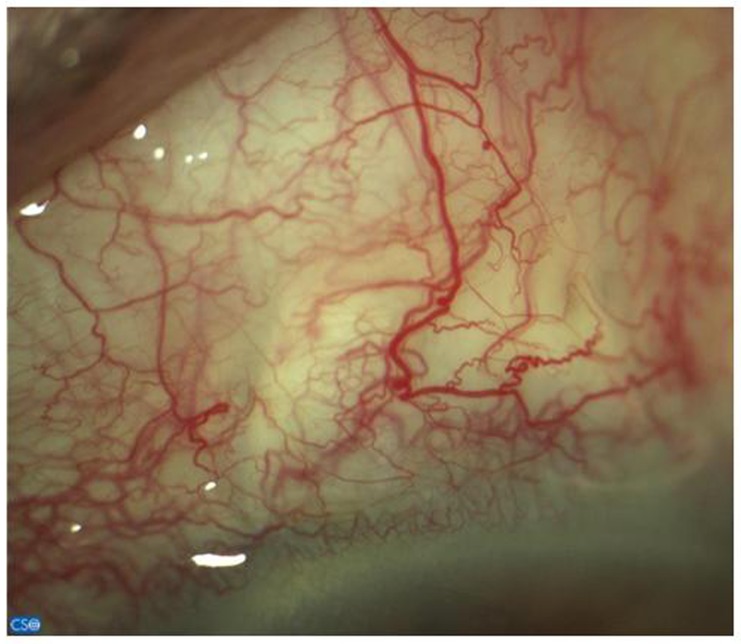
Bleb morphology was predominantly diffuse (100% of patients after 15 days, decreasing to 60% of cases after one year). The remaining cases had flat blebs, except for patient 3 who presented a cystic bleb 180 days after the intervention. Corkscrew vessels were detected throughout the study, reaching its maximum 30 days after surgery (83% of patients) (Fig 3). In all cases, epithelial microcysts were found during the first 30 days after surgery, which later disappeared.
Fig 3. Bleb characteristics.
Corkscrew vessels and microcysts.
PAS-hematoxylin staining of the conjunctiva
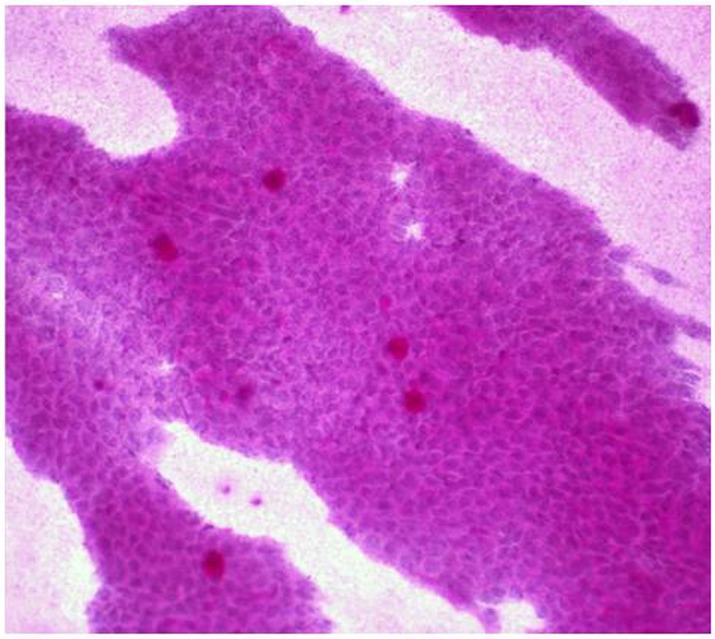
All patients included in the study had grade-0 squamous metaplasia in the preoperative conjunctiva (Fig 4). The epithelial cells were small and round, with eosinophilic cytoplasm. The nuclei were large, basophilic, with a nucleo-cytoplasmic ratio of 1:2. Goblet cells were abundant, plump, and oval and had an intensely PAS-positive cytoplasm. No deterioration of the ocular surface due to drug treatment was observed before the operation.
Fig 4. Microphotograph of preoperative conjunctival Impression Cytology (IC) with periodic Acid-Schiff-hematoxylin staining, obtained from a glaucoma patient.
The nonsecretory epithelial cells are small and round and retain the intercellular junctions. The nuclei are large and N:C ratio is 1:2. The goblet cells are abundant, plump, and oval. Magnification 40x.
Gene expression analyses
In the first study, where we compared conjunctival gene expression with baseline expression at each time point (prospective study) (S1 Table), we observed statistically significant changes for 29 of the 88 genes studied (Table 2). In the postoperative phase (15 days), were detected the most significant changes in gene expression (upregulation of CDKN1A and CDKN2A, IL8, TGFA, and VEGFA genes) were detected. In the second phase (90 days), we found expression changes in 20 genes (all of these were downregulated with the exception of TGFA). After one year, only two genes showed expression changes (downregulation of TGFB1 and ITGB3).
Table 2. List of conjunctival genes showing expression differences at 15, 90, and 360 days after surgery.
| Gene | Name | P | Fold | Days |
|---|---|---|---|---|
| IL8 | Interleukin 8 | 0.017 | 140.57 | 0 to 15 |
| VEGF-A | Vascular Endothelial Growth Factor alpha | 0.033 | 7.01 | |
| CDKN 2A | Cyclin-dependent kinase inhibitor 2A | 0.017 | 5.96 | |
| CDKN 1A | Cyclin-dependent kinase inhibitor 1A | 0.017 | 4.08 | |
| TGF-A | Transforming Growth Factor alpha | 0.017 | 3.08 | |
| IL18 | Interleukin 18 | 0.017 | -4.00 | |
| MYC | Myelocytomatosis oncogene | 0.017 | -9.09 | |
| TGF-A | Transforming Growth Factor alpha | 0.042 | 2.17 | 0 to 90 |
| JUNB | Jun B proto-oncogene | 0.042 | -2.00 | |
| MIF | Macrophage migration Inhibitory Factor | 0.024 | -3.12 | |
| TGF B1 | Transforming Growth Factor beta 1 | 0.012 | -3.33 | |
| HIF 1A | Hypoxia-Inducible Factor 1 alpha | 0.012 | -3.57 | |
| SMARC A4 | SWI/SNF related, Matrix associated, Actin dependent Regulator of Chromatin, subfamily A, member 4 | 0.012 | -3.57 | |
| CLU | Clusterin | 0.042 | -3.84 | |
| IL 18 | Interleukin 18 | 0.006 | -3.84 | |
| CD44 | Glycoprotein CD44 | 0.042 | -4.00 | |
| TFAP 2α | Transcription Factor AP-2 alpha | 0.012 | -4.54 | |
| TGF B3 | Transforming Growth Factor beta 3 | 0.024 | -4.76 | |
| HTT | Huntingtin | 0.012 | -5.55 | |
| EGFR | Epidermal Growth Factor Receptor | 0.006 | -7.14 | |
| PDGF A | Platelet Derived Growth Factor A | 0.006 | -8.33 | |
| BCL2 | B-Cell Lymphoma 2 | 0.024 | -10.00 | |
| MLL2 | Myeloid Lymphoid Leukemia 2 | 0.012 | -11.11 | |
| CTGF | Conective Tissue Growth Factor | 0.012 | -14.28 | |
| MYC | Myelocytomatosis oncogene | 0.006 | -16.66 | |
| TGF B2 | Transforming Growth Factor beta 2 | 0.006 | -20.00 | |
| MMP2 | Matrix Metalloproteinase 2 | 0.024 | -25.00 | |
| ITG B3 | Integrin beta 3 | 0.033 | -1.06 | 0 to 360 |
| TGF B1 | Transforming Growth Factor beta 1 | 0.017 | -4.16 |
The analysis of gene expression changes in peripheral blood, relative to baseline (S2 Table), showed expression changes of 12 genes among the 88 genes studied (Table 3).
Table 3. List of genes in blood samples showing expression differences at 15, 90, and 360 days after surgery.
| Gene | Name | P | Fold | Days |
|---|---|---|---|---|
| IL 1β | Interleukin 1 | 0.03 | 2.05 | 0 to 15 |
| CEBPD | CCAAT / Enhancer Binding Protein Delta | 0.009 | 1.7 | |
| HIF 1A | Hypoxia-Inducible Factor 1 alpha | 0.009 | 1.69 | |
| ITG β2 | Integrin beta 2 | 0.009 | 1.49 | |
| TNFRSF 1A | Tumor Necrosis Factor Receptor Superfamily member 1A | 0.017 | 1.29 | |
| CASP 1 | Caspase 1 | 0.003 | 1.12 | |
| MET | Met proto-oncogene (hepatocyte growth factor | 0.026 | -1,96 | 0 to 90 |
| TGF B2 | Transforming Growth Factor beta 2 | 0.026 | -2,7 | |
| HIF 1A | Hypoxia-Inducible Factor 1 | 0.038 | 1.94 | 0 to 180 |
| EREG | Epiregulin | 0.038 | 1.97 | |
| IL 1β | Interleukin 1 | 0.038 | 2.27 |
These changes were more pronounced in the first 15 days (upregulation of IL1B, CEBPD, HIF1A, ITGB2, TNFRSF1A and CASP1). The remaining expression changes happened between 90, and 180 days after surgery. One year after the intervention (as in the conjunctiva), the baseline expression levels of most genes were restored, with the exception of HIF1A, EREG and IL1B.
Correlation between gene expression and IOP
We found a significant correlation between gene expression and the values of IOP for 15 genes in the conjunctiva and only a single gene in the peripheral blood (Table 4) samples. Conjunctival TGFA and IL18 showed the strongest correlation values.
Table 4. Correlation IOP-gene expression in blood and conjunctiva samples.
| Spearman coefficient rs | P | |
|---|---|---|
| Blood genes | ||
| IL 1β | -0.387 | 0.038 |
| Conjunctival genes | ||
| IL 18 | 0.604 | 0.008 |
| BCL2 | 0.576 | 0.012 |
| PDGF A | 0.570 | 0.014 |
| TGF B3 | 0.560 | 0.016 |
| CTGF | 0.547 | 0.019 |
| MMP2 | 0.547 | 0.019 |
| MYC | 0.547 | 0.019 |
| COL3A1 | 0.529 | 0.024 |
| CLU | 0.504 | 0.033 |
| TGF B2 | 0.481 | 0.043 |
| MIF | 0.479 | 0.044 |
| TGF-A | -0.668 | 0.002 |
| CDKN 2A | -0.509 | 0.031 |
| CDKN 1A | -0.499 | 0.035 |
| CASP 3 | -0.485 | 0.041 |
Clustering gene expression patterns related to surgical success and failure
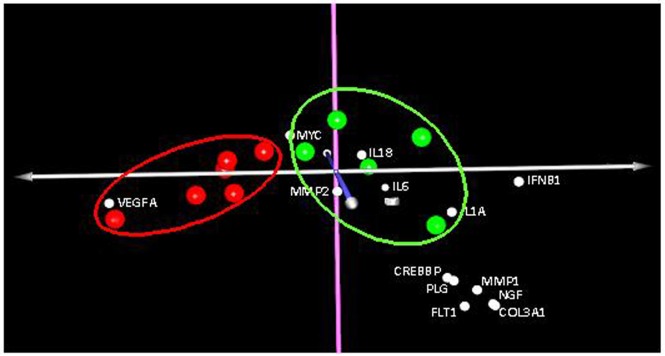
Correspondence Analysis (COA) of gene expression of conjunctival samples in study 2 showed a clear clustering of patients with surgical success and failure (Fig 5). The statistical analysis revealed 20 genes whose expression differed significantly between these two groups (Table 5). From this analysis a clear over expression of VEGFA gene was related with failure of surgery whereas a notable decrease in IFNB1 gene was also related.
Fig 5. Correspondence Analysis (COA).
Distribution of genes that separate the group of patients with successful surgery (green dots) from the patients with surgical failure (red dots).
Table 5. Genes with significant expression changes in patients with surgical failure group in comparison with those of the success group.
| Gene | Fold | P |
|---|---|---|
| VEGF-A | 3.00 | 0.0003 |
| HIF 1A | -1.52 | 0.035 |
| MIF | -1.62 | 0.025 |
| ITG B3 | -2.30 | 0.037 |
| MYC | -2.55 | 0.017 |
| IL18 | -2.70 | 0.009 |
| CTGF | -3.08 | 0.043 |
| MMP2 | -3.24 | 0.012 |
| BTG2 | -3.35 | 0.045 |
| CREBBP | -3.65 | 0.044 |
| MMP3 | -4.90 | 0.040 |
| PLG | -6.16 | 0.038 |
| FLT1 | -6.32 | 0.044 |
| MMP1 | -7.14 | 0.044 |
| NGF | -7.65 | 0.049 |
| IL1A | -7.69 | 0.015 |
| COL3A1 | -8.37 | 0.048 |
| F2 | -8.37 | 0.048 |
| IL6 | -8.84 | 0.038 |
| IFN β1 | -10.72 | 0.043 |
Discussion
Despite the downward trend in glaucoma surgery since the nineteen-nineties [14], it is still a procedure with major advantages; it is inexpensive [15] and allows an IOP reduction more drastic [16] and less fluctuating than pharmacological treatment [17]. Although the most common glaucoma surgical technique worldwide is the trabeculectomy, some alternative interventions have been proposed to reduce the IOP. The NPDS could be considered as a variation of the trabeculectomy which does not require iridectomy and allows the aqueous humor to drain gradually, reducing IOP in a less abrupt manner. Long-term results seem to be not worse than those of trabeculectomy [18]. However, subconjunctival and episcleral scarring remain the main impediment to the long-term success of any filtering surgery [19], irrespective of whether it is perforating or non-perforating procedure [20].
Analysis of biochemical mechanisms involved in wound healing and failure of filtering surgery requires sampling of the conjunctiva and Tenon’s capsule. IC allows the collection of cells from the conjunctiva in a nearly painless and non-invasive way, for the purpose of analyses (cytology, flow cytometry, PCR…) and diagnosis. There are other different methods to obtain conjunctival cells such as brushing or biopsy, however these techniques are more invasive than IC and are not indicated in studies requiring repeated conjunctival sampling. Although collecting conjunctival cells from IC is technically challenging, the IC has been standardized and widely used in many studies for years [21–27]. Recently, Lopez-Miguel A et al., have published a paper which shows that the impression cytology is a robust and reproducible technique to obtain acceptable quantities of RNA [21]. In their work they conclude that there are no statistical differences between two methods employed for obtaining the sample: the conventional IC and the EyePrim device (OPIA Technologies, Paris, France). Previous studies from our research group were used to optimize the storage and processing conditions of the conjunctival IC samples and to provide reliable amounts of medium-high RNA quality, adequate for gene expression analysis [13,28,29]. In summary, IC is the technique that assures us the highest reproducibility and lower discomfort for the patient.
It could be argued that, using our technique, we only analyze the conjunctival epithelium, neglecting deeper strata such as conjunctival stroma and Tenon's capsule, associated with surgical failure [1]. However, firstly given the close relationship between the conjunctiva and anterior Tenon's capsule [30], the postoperative gene expression in these tissues should not differ substantially. Secondly, the low cellularity of Tenon’s capsule makes it difficult to extract its genetic material; in practice, it is usually extracted together with the conjunctiva. Thirdly, the typical location of the fibrosis under the conjunctival flap border in the case of surgical bleb revision could indicate the contribution of the conjunctiva to the fibrosis in a sort of epithelial mesenchymal transition. Unfortunately, this fact has not been demonstrated yet in glaucoma surgery. Moreover, not only conjunctival cells but also inflammatory cells could contribute to the surgical failure. In this respect it is worth mentioning that some changes could be due to the effect of the topical antiglaucoma drugs in the ocular surface. In the current clinical practice, we mainly operate patients with uncontrolled IOP in spite of the maximum tolerated medication. So, although these changes could be different among patients, they are unavoidable and should not interfere in the study; at last, they are expected to cause the same gene expression variations throughout different patients and sample collections, which would not affect our differential expression study.
To obtain a homogenous set of samples, we only enrolled patients with POAG. We excluded patients with glaucoma with high risk of surgical failure and those using supplementary antimetabolites, to rule out interference in the healing process. Furthermore, although the patients had received multiple topical treatments, which have been previously associated with surgical failure [31], they did not show significant changes in the ocular surface (as confirmed by the results of conjunctival impression cytology stained with PAS-hematoxylin).
In our knowledge, this is the first study in gene expression in humans after NPDS. Recently, Mahale A et al. have studied the fibrosis related gene expression after Ahmed glaucoma valve [32]. As in our study they have employed a fibrosis targeted PCR Array, however they have focused their study in a single-time point (median time about 25 months) which could be considered as a late capsule excision time.
Our results showed that, in the first stage, the main changes in conjunctival gene expression occurred on day 15 after the surgical trauma. In this phase, we observed three patterns of gene expression: upregulation (CDKN1A and CDKN2A genes, IL8, TGFA, and VEGFA), downregulation (IL18 and MYC) and no change (other genes included in the study). The upregulated genes encode factors favoring chemotaxis of inflammatory cells (IL8) [33,34], angiogenesis (VEGFA, IL8) [35], and proliferation and migration of keratinocytes (IL8 and TGFA) [36, 37], all of which are associated to processes typical of the first stage of wound healing [11]. Elevated expression of IL8 was found in three patients and could be explained as a typical chemokine response in the acute phase of healing [37]. There are some doubts about the role of CDKN genes, which slow down the cell cycle. However, the overall effect of these genes on the cell cycle also depends on their effectors, which were not analyzed in this work. A decrease in the expression of IL18 might be mediated by overexpression of TGFA, as it has been previously described [38]. It has been postulated that IL18 exerts its proinflammatory effect in early stages of healing and shows expression changes at this stage [38]. The downregulation of MYC would have pro-healing effect, given the inhibitory effect of this protein on the process of wound healing [39].
In the intermediate phase of the healing process (90 days), we detected downregulation of some genes involved in crucial biological processes associated with healing. These genes have been implicated in the positive regulation of cell proliferation (HIF1A, EGFR, MYC, MLL2, and TFAP2A) [40, 41], response to wound healing (CLU, CTGF, TGFB1, TGFB2, BCL2, PDGFA and MIF) [42–44], fibroblast migration (TGFB1, TGFB2, TGFB3, MMP2 and CTGF) [45–48] and angiogenesis (PDGF) [49–51].
In the final stage of the first study, one year after the surgery, virtually no changes in conjunctival gene expression were observed and basal expression levels were recovered. Only TGFB1 and ITGB3, involved in the activation of healing showed a decrease in their expression [52].
Gene expression analysis in blood samples found the most significant changes during the first 15 days after surgery. During this phase, genes associated with pro-inflammatory processes (CASP1, IL1B) [53], pro-apoptotic genes (HIF1A and ITGB2) [54] and in addition, genes associated with anti-apoptotic or anti-inflammatory processes such as CEBPD [55]. During the next stages, expression changes were minimal; some cell migration- and inflammation-related genes showed expression changes. It is clear that, despite detectable changes in overall gene expression, they are less pronounced in the blood and differ from those found in the conjunctiva. This finding, also reported by other authors [56], reflects the primarily local nature of the healing process. Unfortunately, we could not perform paired comparisons of gene expression between blood and conjunctiva throughout the survey, due to the small number of adequate conjunctival RNA samples.
Regarding the analysis of the correlation between IOP and gene expression, we found that the most significant changes occurred in two genes expressed in conjunctiva (IL18 and TGFA). Whereas IL18 gene showed a positive correlation, TGFA gene showed a negative correlation with IOP. This data together with the gene expression analysis suggest that local factors play an important role in the surgical outcome.
In our second study, we decided to analyze only conjunctival samples from patients undergoing glaucoma filtering surgery once the wound healing process had concluded. At this time, the first group (designated "success”) maintained IOP below 21 mmHg as a result of good filtration of aqueous humor. However, in the second group (called "failure"), the surgery had failed, thus being IOP levels over 21 mmHg. The comparison of gene expression levels performed using COA revealed on the one hand, a trend of association among patients with successful surgery and, on the other hand, among patients with failed surgery (Fig 5). This indicates that the expression of genes related to wound healing in patients with failed surgery, differs from that of patients for whom the treatment succeeded. As in the first study, we showed that the expression levels one year after intervention were comparable with the basal levels. Ultimately this analysis could theoretically differentiate between success and failure. From the genes with different expression levels in success group in comparison with failure group, VEGFA was the furthest gene from the origin of the graph and the closest to the failure group, thus contributing significantly to the differentiation of the latter group (Fig 5). This was consistent with the data shown in Table 5, which showed 3-fold higher expression of this gene in the failure group than in the success group. In the latter group, genes that contributed most to the separation were IFNB1, IL1A, IL18, IL6, F2, COL3A1, NGF, MMP1, FTL1, PLG, MMP3 and CREBBP, whose decreased expression hindered the success of glaucoma surgery.
On the assumption that gene expression levels one year after the NPDS would not differ from the preoperative status, our analysis could provide a predictive classifying tool for forecasting the success or failure of glaucoma operation. Prediction of the risk of scarring based on individual gene expression profiles has a great potential in the development of personalized and stratified therapies to prevent ocular fibrosis. The combination of therapeutic targets in different biological pathways could offer a more effective treatment than conventional monotherapies. In addition, patient management before, during and after the intervention (use of antimitotics, dosage, etc.) could be substantially improved with the use of such tools.
To our knowledge, this study is the first in which expression of a broad spectrum of genes after NPDS in patients with POAG has been examined. This RT-qPCR Array analysis allows to determine not only the roles and mechanisms of various mediators of the conjunctival wound healing process, but also to identify the mediators implicated in the prognosis of the surgery.
However, there are limitations in our study. First, due to the low number of patients, our results should be interpreted with caution until further studies with larger sample sizes will be carried out. Second, a subsequent validation study performed at a protein level would be desirable in order to confirm the gene expression levels determined. Unfortunately, due to the small number of adequate conjunctival RNA samples, we could not perform this kind of study. Regardless of these limitations, our findings demonstrate clear alterations in fibrosis related gene pathways in NPDS and these preliminary findings could help form the basis for systematic future investigations.
In conclusion, this study identified an expression profile of genes related to inflammation, angiogenesis and cell proliferation, whose expression levels change after glaucoma filtering surgery. This type of analysis might become a valuable predictive tool for forecasting the success or failure of glaucoma surgery.
Supporting information
(XLSX)
(XLSX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work has been partially supported by the Ministry of Science and innovation through PROFIT program, grant dex-580000-2008-41, and the Ministry of Science and innovation through Apply Research, grant PPT-010000-2009-30. The funding organizations had no role in the design or conduct of this research.
References
- 1.Hitchings RA, Grierson I. Clinicopathological correlation in eyes with failed fistulizing surgery. Trans Ophthalmol Soc UK 1983; 103: 84–8. [PubMed] [Google Scholar]
- 2.Araujo SV, Spaeth GL, Roth SM, Starita RJ. A ten-year follow-up on a prospective, randomized trial of postoperative corticosteroids after trabeculectomy. Ophthalmology 1995; 102: 1753–9. [DOI] [PubMed] [Google Scholar]
- 3.Heuer DK, Parrish RK 2nd, Gressel MG, Hodapp E, Palmberg PF, Anderson DR. 5-fluorouracil and glaucoma filtering surgery. II. A pilot study. Ophthalmology 1984; 91: 384–94. [DOI] [PubMed] [Google Scholar]
- 4.Smith MF, Sherwood MB, Doyle JW, Khaw PT. Results of intraoperative 5-fluorouracil supplementation on trabeculectomy for open-angle glaucoma. Am J Ophthalmol 1992; 114: 737–41. [DOI] [PubMed] [Google Scholar]
- 5.Chen CW, Huang HT, Bair JS, Lee CC. Trabeculectomy with simultaneous topical application of mitomycin-C in refractory glaucoma. J Ocul Pharmacol 1990; 6: 175–82. [DOI] [PubMed] [Google Scholar]
- 6.Cohen LB, Graham TF, Fry WE. Beta radiation; as an adjunct to glaucoma surgery in the Negro. Am J Ophthalmol 1959; 47:54–61. [PubMed] [Google Scholar]
- 7.Kirwan JF, Rennie C, Evans JR. Beta radiation for glaucoma surgery. Cochrane Database Syst Rev. 2009;2:CD003433. [DOI] [PubMed] [Google Scholar]
- 8.Cordeiro MF, Gay JA, Khaw PT. Human anti-transforming growth factor-beta2 antibody: a new glaucoma anti-scarring agent. Invest Ophthalmol Vis Sci 1999; 40: 2225–34. [PubMed] [Google Scholar]
- 9.Grewal DS, Jain R, Kumar H, Grewal SP. Evaluation of subconjunctival bevacizumab as an adjunct to trabeculectomy a pilot study. Ophthalmology 2008; 115: 2141–45. 10.1016/j.ophtha.2008.06.009 [DOI] [PubMed] [Google Scholar]
- 10.Daniels JT, Cambrey AD, Occleston NL, Garrett Q, Tarnuzzer RW, Schultz GS, et al. Matrix metalloproteinase inhibition modulates fibroblast-mediated matrix contraction and collagen production in vitro. Invest Ophthalmol Vis Sci 2003; 44: 1104–10. [DOI] [PubMed] [Google Scholar]
- 11.Seibold LK, Sherwood MB, Kahook MY. Wound Modulation After Filtration Surgery. Survey of Ophthalmology 2012; 57: 530–50. 10.1016/j.survophthal.2012.01.008 [DOI] [PubMed] [Google Scholar]
- 12.Rivas L, Oroza MA, Perez-Esteban A, Murube-del-Castillo J. Topographical distribution of ocular surface cells by the use of impression cytology. Acta Ophthalmol (Copenh) 1991; 69: 371–76. [DOI] [PubMed] [Google Scholar]
- 13.Garcia I, Etxebarria J, Boto-de-Los-Bueis A, Díaz-Valle D, Rivas L, Martínez-Soroa I, et al. Comparative study of limbal stem cell deficiency diagnosis methods: detection of MUC5AC mRNA and goblet cells in corneal epithelium. Ophthalmology 2012; 119: 923–9. 10.1016/j.ophtha.2011.10.031 [DOI] [PubMed] [Google Scholar]
- 14.Whittaker KW, Gillow JT, Cunliffe IA. Is the role of trabeculectomy in glaucoma management changing?. Eye 2001; 15: 449–52. 10.1038/eye.2001.153 [DOI] [PubMed] [Google Scholar]
- 15.Lindblom B, Nordmann JP, Sellem E, Chen E, Gold R, Polland W, et al. A multicentre, retrospective study of resource utilization and costs associated with glaucoma management in France and Sweden. Acta Ophthalmol Scand 2006; 84: 74–83. 10.1111/j.1600-0420.2005.00560.x [DOI] [PubMed] [Google Scholar]
- 16.Lichter PR, Musch DC, Gillespie BW, Guire KE, Janz NK, Wren PA, Mills RP; CIGTS Study Group. Interim clinical outcomes in the Collaborative Initial Glaucoma Treatment Study comparing initial treatment randomized to medications or surgery. Ophthalmology 2001; 108: 1943–53. [DOI] [PubMed] [Google Scholar]
- 17.Migdal C, Gregory W, Hitchings R. Long-term functional outcome after early surgery compared with laser and medicine in open-angle glaucoma. Ophthalmology 1994; 101: 1651–6. [DOI] [PubMed] [Google Scholar]
- 18.Mermoud A, Schnyder CC, Sickenberg M, Chiou AG, Hédiguer SE, Faggioni R. Comparison of deep sclerectomy with collagen implant and trabeculectomy in open-angle glaucoma. J Cataract Refract Surg 1999; 25: 323–31. [DOI] [PubMed] [Google Scholar]
- 19.Khaw PT, Occleston NL, Schultz G, Grierson I, Sherwood MB, Larkin G. Activation and suppression of fibroblast function. Eye1994; 8:188–95. [DOI] [PubMed] [Google Scholar]
- 20.Rulli E, Biagioli E, Riva I, Gambirasio G, De Simone I, Floriani I, et al. Efficacy and safety of trabeculectomy vs nonpenetrating surgical procedures: a systematic review and meta-analysis. JAMA Ophthalmol 2013; 131: 1573–82. 10.1001/jamaophthalmol.2013.5059 [DOI] [PubMed] [Google Scholar]
- 21.López-Miguel A, Gutiérrez-Gutiérrez S, García-Vázquez C, Enríquez-de-Salamanca A. RNA Collection From Human Conjunctival Epithelial Cells Obtained With a New Device for Impression Cytology. Cornea. 2017; 36: 59–63. 10.1097/ICO.0000000000000977 [DOI] [PubMed] [Google Scholar]
- 22.Pflugfelder SC, De Paiva CS, Moore QL, Volpe EA, Li DQ, Gumus K, et al. Aqueous Tear Deficiency Increases Conjunctival Interferon-γ (IFN-γ) Expression and Goblet cells loss. Invest Ophthalmol Vis Sci 2015; 56: 7545–50. 10.1167/iovs.15-17627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang L, Su Z, Zhang Z, Lin J, Li DQ, Pflugfelder SC. Effects of azithromycin on gene expression profiles of proinflammatory and anti-inflammaroty mediators in the eyelid margin and conjunctiva of patients with meibomian gland disease. JAMA Ophthalmol 2015; 133: 1117–23. 10.1001/jamaophthalmol.2015.2326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cocho L, Fernández I, Calonge M, Martínez V, González-García MJ, Caballero D, et al. Gene Expression-based Predicive Models on Graft Versus Host Disese-Associate Dry Eye. Invest Ophthalmol Vis Sci 2015; 56: 4570–81. 10.1167/iovs.15-16736 [DOI] [PubMed] [Google Scholar]
- 25.Niu L, Zhang S, Wu J, Chen L, Wang Y. Upregulation of NLRP3 inflmammasome in the tears and ocular surface of dry eye patients. PloS One 2015. 10: e0126277 10.1371/journal.pone.0126277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Moore QL, De Paiva CS, Pflugfelder SC. Effects of Dry eye Therapies on Enviromentally induced ocular surface disease. Am J Ophthalmol 2015; 160: 135–42. 10.1016/j.ajo.2015.04.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corrales RM, Narayanan S, Fernández I, Mayo A, Galarreta DJ, Fuentes- Páez G, et al. Ocular mucin geen expression levels as biomarkers for the diagnosis’s of dry eye syndrome. Invest Ophthalmol Vis Sci 2011; 52: 8363–9. 10.1167/iovs.11-7655 [DOI] [PubMed] [Google Scholar]
- 28.García I, Etxebarria J, Merayo-Lloves J, Torras J, Boto-de-Los-Bueis A, Díaz-Valle D, et al. Novel molecular diagnostic system of limbal stem cell deficiency based on MUC5AC transcript detection in corneal epithelium by PCR-reverse dot blot. Invest Ophthalmol Vis Sci 2013; 54: 5643–52. 10.1167/iovs.13-11933 [DOI] [PubMed] [Google Scholar]
- 29.López-García JS, García-Lozano I, Rivas L, Giménez C, Acera A, Suárez-Cortés T. Effects of Autologous Serum Eye Drops on Conjunctival Expression of MUC5AC in Patients With Ocular Surface Disorders. Cornea 2016; 35: 336–41. 10.1097/ICO.0000000000000726 [DOI] [PubMed] [Google Scholar]
- 30.Lockwood CB. The anatomy of the muscles, ligaments, and fasciae of the orbit, including an account of the capsule of tenon, the check ligaments of the recti, and the suspensory ligaments of the eye. J Anat Physiol 1885; 20: 2–25. [PMC free article] [PubMed] [Google Scholar]
- 31.Broadway DC, Grierson I, O'Brien C, Hitchings RA. Adverse effects of topical antiglaucoma medication. II. The outcome of filtration surgery. Arch Ophthalmol 1994; 112: 1446–54. [DOI] [PubMed] [Google Scholar]
- 32.Mahale A, Othman MW, Al Shahwan S, Al Jadaan I, Owaydha O, Khan Z, Altered Expression of fibrosis genes in capsules of failed Ahmed Glaucoma valve implants. PloS One 2015; 10: e0122409 10.1371/journal.pone.0122409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Z, Yang Y, Yang H, Capó-Aponte JE, Tachado SD, Wolosin JM, Reinach PS. NF-êB feedback control of JNK1 activation modulates TRPV1-induced increases in IL-6 and IL-8 release by human corneal epithelial cells. Mol Vis. 2011; 17:3137–46. [PMC free article] [PubMed] [Google Scholar]
- 34.Reidy T, Rittenberg A, Dwyer M, D'Ortona S, Pier G, Gadjeva M. Homotrimeric macrophage migration inhibitory factor (MIF) drives inflammatory responses in the corneal epithelium by promoting caveolin-rich platform assembly in response to infection. J Biol Chem 2013; 288: 8269–78. 10.1074/jbc.M112.351064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoshida S, Ono M, Shono T, Izumi H, Ishibashi T, Suzuki H, et al. Involvement of interleukin-8, vascular endothelial growth factor, and basic fibroblast growth factor in tumor necrosis factor alpha-dependent angiogenesis. Mol Cell Biol 1997; 17: 4015–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Z, Van Bergen T, Van de Veire S, Van de Vel I, Moreau H, Dewerchin M, et al. Inhibition of vascular endothelial growth factor reduces scar formation after glaucoma filtration surgery. Invest Ophthalmol Vis Sci 2009; 50: 5217–25. 10.1167/iovs.08-2662 [DOI] [PubMed] [Google Scholar]
- 37.Barrientos S, Stojadinovic O, Golinko MS, Brem H, Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Rep Reg 2008; 16: 585–601. [DOI] [PubMed] [Google Scholar]
- 38.Kämpfer H, Mühl H, Manderscheid M, Kalina U, Kauschat D, Pfeilschifter J, et al. Regulation of interleukin-18 (IL-18) expression in keratinocytes (HaCaT): implications for early wound healing. Eur Cytokine Netw 2000; 11: 626–33. [PubMed] [Google Scholar]
- 39.Stojadinovic O, Brem H, Vouthounis C, Lee B, Fallon J, Stallcup M, et al. Molecular pathogenesis of chronic wounds: the role of beta-catenin and c-myc in the inhibition of epithelialization and wound healing. Am J Pathol 2005; 167: 59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lang SA, Klein D, Moser C, Gaumann A, Glockzin G, Dahlke MH, et al. Inhibition of heat shock protein 90 impairs epidermal growth factor-mediated signaling in gastric cancer cells and reduces tumor growth and vascularization in vivo. Mol Cancer Ther 2007; 6: 1123–32. 10.1158/1535-7163.MCT-06-0628 [DOI] [PubMed] [Google Scholar]
- 41.Rao GN, Broeckx JL, Church RL. Expression of c-myc protooncogene in primary cultures of human corneal stromal cells. Curr Eye Res 1989; 8: 405–13. [DOI] [PubMed] [Google Scholar]
- 42.Chang L, Crowston JG, Cordeiro MF, Akbar AN, Khaw PT. The role of the immune system in conjunctival wound healing after glaucoma surgery. Surv Ophthalmol 2000; 45: 49–68 [DOI] [PubMed] [Google Scholar]
- 43.Sriram S, Gibson DJ, Robinson P, Pi L, Tuli S, Lewin AS, et al. Assessment of anti-scarring therapies in ex-vivo organ cultured rabbit corneas. Exp Eye Res 2014; 125: 173–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zheng X, Gai X, Han S, Moser CD, Hu C, Shire AM, et al. The human sulfatase 2 inhibitor 2,4-disulfonylphenyl-tert-butylnitrone (OKN-007) has an antitumor effect in hepatocellular carcinoma mediated via suppression of TGFB1/SMAD2 and Hedgehog/GLI1 signaling. Genes Chromosomes Cancer 2013; 52: 225–36. 10.1002/gcc.22022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pan H, Chen J, Xu J, Chen M, Ma R. Antifibrotic effect by activation of peroxisome proliferator-activated receptor-gamma in corneal fibroblasts. Mol Vis 2009; 15: 2279–86. [PMC free article] [PubMed] [Google Scholar]
- 46.Saika S, Yamanaka O, Okada Y, Miyamoto T, Kitano A, Flanders KC, et al. Effect of overexpression of PPARgamma on the healing process of corneal alkali burn in mice. Am J Physiol Cell Physiol 2007; 293: C75–86. 10.1152/ajpcell.00332.2006 [DOI] [PubMed] [Google Scholar]
- 47.Servold SA. Growth factor impact on wound healing. Clin Podiatr Med Surg 1991; 8: 937–53 [PubMed] [Google Scholar]
- 48.Neufeld G, Tessler S, Gitay-Goren H, Cohen T, Levi BZ. Vascular endothelial growth factor and its receptors. Prog Growth Factor Res 1994; 5: 89–97. [DOI] [PubMed] [Google Scholar]
- 49.Dell S, Peters S, Müther P, Kociok N, Joussen AM. The role of PDGF receptor inhibitors and PI3-kinase signaling in the pathogenesis of corneal neovascularization. Invest Ophthalmol Vis Sci 2006; 47: 1928–37 10.1167/iovs.05-1071 [DOI] [PubMed] [Google Scholar]
- 50.Jo N, Mailhos C, Ju M, Cheung E, Bradley J, Nishijima K, et al. Inhibition of platelet-derived growth factor B signaling enhances the efficacy of anti-vascular endothelial growth factor therapy in multiple models of ocularneovascularization. Am J Pathol 2006; 168: 2036–53. 10.2353/ajpath.2006.050588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bo Q, Zhang Y, Wu Y, Wang F. Research advances of platelet derived growth factor family and its significance in neovascular eye diseases. Zhonghua Yan Ke Za Zhi 2014; 50: 471–5. [PubMed] [Google Scholar]
- 52.Li DQ, Lee SB, Tseng SC. Differential expression and regulation of TGF-beta1, TGF-beta2, TGF-beta3, TGF-betaRI, TGF-betaRII and TGF-betaRIII in cultured human corneal, limbal, and conjunctival fibroblasts. Curr Eye Res. 1999; 19: 154–61. [DOI] [PubMed] [Google Scholar]
- 53.Wree A, McGeough MD, Peña CA, Schlattjan M, Li H, Inzaugarat ME, et al. NLRP3 inflammasome activation is required for fibrosis development in NAFLD. J Mol Med (Berl) 2014; 92: 1069–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kong T, Eltzschig HK, Karhausen J, Colgan SP, Shelley CS. Leukocyte adhesion during hypoxia is mediated by HIF-1-dependent induction of beta2 integrin gene expression. Proc Natl Acad Sci USA 2004; 101: 10440–5. 10.1073/pnas.0401339101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moore F, Santin I, Nogueira TC, Gurzov EN, Marselli L, Marchetti P, et al. The transcription factor C/EBP delta has anti-apoptotic and anti-inflammatory roles in pancreatic beta cells. PLoS One 2012; 7: e31062 10.1371/journal.pone.0031062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seet LF, Su R, Barathi VA, Lee WS, Poh R, Heng YM, et al. SPARC deficiency results in improved surgical survival in a novel mouse model of glaucoma filtration surgery. PLoS One 2010; 5: e9415 10.1371/journal.pone.0009415 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.