Abstract
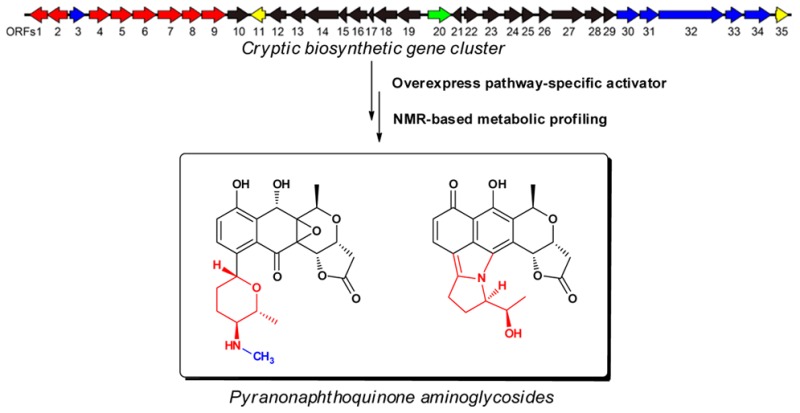
Mining of microbial genomes has revealed that actinomycetes harbor far more biosynthetic potential for bioactive natural products than anticipated. Activation of (cryptic) biosynthetic gene clusters and identification of the corresponding metabolites has become a focal point for drug discovery. Here, we applied NMR-based metabolomics combined with bioinformatics to identify novel C-glycosylpyranonaphthoquinones in Streptomyces sp. MBT76 and to elucidate the biosynthetic pathway. Following activation of the cryptic qin gene cluster for a type II polyketide synthase (PKS) by constitutive expression of its pathway-specific activator, bioinformatics coupled to NMR profiling facilitated the chromatographic isolation and structural elucidation of qinimycins A–C (1–3). The intriguing structural features of the qinimycins, including 8-C-glycosylation, 5,14-epoxidation, and 13-hydroxylation, distinguished these molecules from the model pyranonaphthoquinones actinorhodin, medermycin, and granaticin. Another novelty lies in the unusual fusion of a deoxyaminosugar to the pyranonaphthoquinone backbone during biosynthesis of the antibiotics BE-54238 A and B (4, 5). Qinimycins showed weak antimicrobial activity against Gram-positive bacteria. Our work shows the utility of combining bioinformatics, targeted activation of cryptic gene clusters, and NMR-based metabolic profiling as an effective pipeline for the discovery of microbial natural products with distinctive skeletons.
Actinomycetes are prolific sources of bioactive natural products (NPs). From the roughly 18 000 known bioactive bacterial compounds, more than 10 000 were discovered from the actinomycete genus Streptomyces.1,2 Even so, their biosynthetic potential is far from exhausted, though the enthusiasm to discover new molecules produced by these bacteria is dampened by chemical redundancy and the consequential poor return of investment of high-throughput screening campaigns.3−5 On a more positive note, whole genome sequencing revealed that actinomycetes harbor numerous silent and hence likely untapped biosynthetic gene clusters that may not be associated with known metabolites; indeed, even the very extensively studied model organism Streptomyces coelicolor was shown to possess a far greater producing potential than anticipated.6 Furthermore, bioinformatics tools developed specifically for mining genome sequences for the identification of biosynthetic gene clusters allow the prediction of the chemical output on the basis of accumulated biosynthetic knowledge.7,8 These developments in genome mining mark the start of a new era of genomics-based drug discovery, with the potential of greatly expanding the chemical space of bioactive natural products.
A bottleneck is that many of the biosynthetic pathways uncovered by genome sequencing are in a dormant state under routine laboratory conditions, generally referred to as cryptic or silent gene clusters, and specific approaches to activate their expression are required.9,10 The transcription of genes encoding the biosynthetic machinery for secondary metabolites in actinomycetes involves multiple regulatory cascades and networks.11 The regulatory signals are transmitted through global regulatory networks and ultimately transmitted to the pathway-specific regulatory genes that control the expression of the biosynthetic genes.12
A promising approach is via a drug-discovery pipeline that is based on combining bioinformatics-driven genome mining with approaches to elicit the production of (cryptic) antibiotics, followed by the metabolic profiling-based identification of the bioactivity of interest. Strategies to circumvent the regulatory networks that silence cryptic biosynthetic gene clusters and trigger the biosynthesis of the corresponding natural product(s) include manipulating the pleiotropic regulatory networks as well as targeting pathway-specific regulators.10,13 NMR-based metabolic profiling allows the efficient identification of the relevant NPs within the context of the complex metabolic matrix. The available bioinformatic information allows prediction of the metabolite produced by the biosynthetic gene clusters, and this knowledge can be directly related to the 1H NMR spectrum by examining the expected chemical shift and/or splitting pattern of typical protons in the predicted molecular motifs. In turn, as one compound usually contains multiple 1H resonances, any unique NMR feature(s) in its structure can be used to check for the expression of the corresponding biosynthetic gene cluster and subsequently for NMR-guided chromatographic separation. This should allow scientists to bridge the gap between bioinformatics-driven gene cluster analysis and experimental NPs discovery.14
In this study, we applied a workflow of NMR-based metabolomics and bioinformatics to identify novel pyranonaphtoquinones. Constitutive expression of the pathway-specific activator of a cryptic type II PKS gene cluster (designated qin) in Streptomyces sp. MBT76 activated the biosynthesis of a family of pyranonaphtoquinones with intriguing chemical architecture. Genomics and structural analysis identified a family of 8-C-glycosyl-pyranonaphthoquinones (1–5). The qinimycins A–C (1–3) encompass structural features of a rare 5,14-epoxidation and unprecedented 13-hydroxylation in pyranonaphthoquinones. In addition, an unusual fusion of a deoxyaminosugar into a pyranonaphthoquinone backbone was seen for the previously described antibiotics BE-54238A (4) and BE-54238B (5).
Results and Discussion
Identification and Activation of the Cryptic Type II PKS Gene Cluster (qin) for Glycosylated Pyranonaphthoquinones
Streptomyces sp. MBT76, which originates from the Qinling mountains in China, was previously identified as a prolific producer of antibiotics, including those with efficacy against multiple Gram-positive and Gram-negative multidrug resistant pathogens.15 Further detailed metabolic characterization of the strain identified many natural products often with interesting chemistry, including isocoumarins, prodiginines, acetyltryptamine, and fervenulin, among others.16Streptomyces sp. MBT76 was subjected to Illumina/Solexa whole genomic sequencing, and the genome was assembled in 13 contigs, with a total genome size of 8.64 Mb. In total 7974 coding sequences (CDS) were predicted using the GeneMark algorithm.17 Analysis of the contigs by AntiSMASH18 presented a possible 55 putative biosynthetic gene clusters specifying secondary metabolites, 22 of which encoding polyketide synthases (PKS).
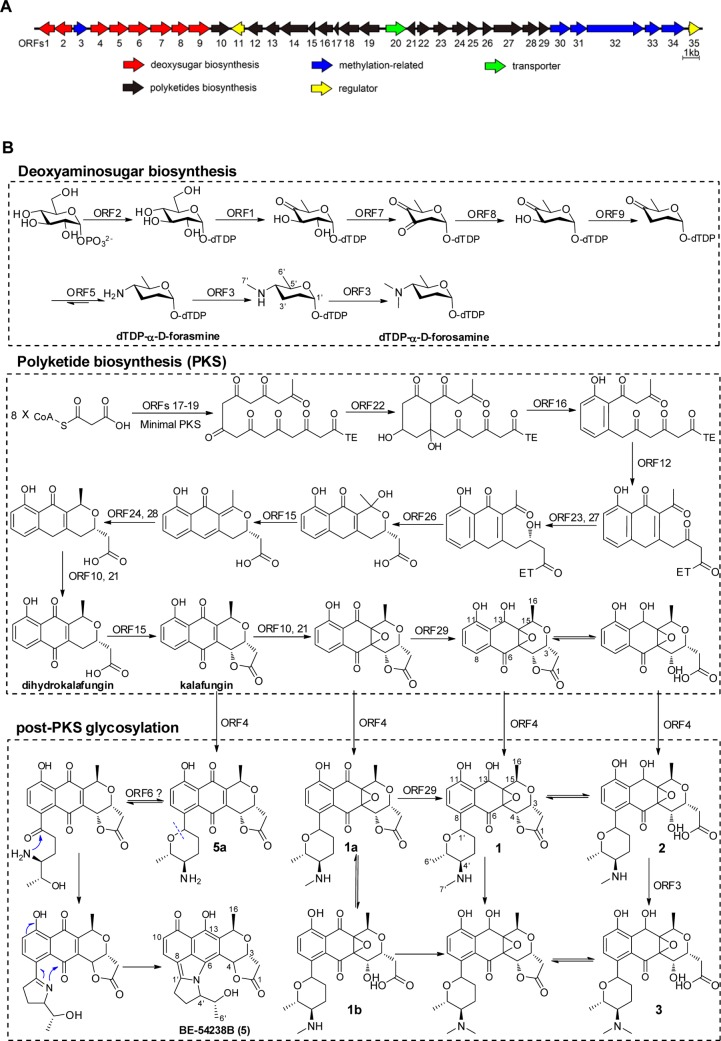
One 41 kb biosynthetic gene cluster for a type II PKS, designated qin (Table 1) was of particular interest considering its potential to specify pyranonaphthoquinones, a well-studied family of aromatic polyketides with highly complex chemical architecture and pronounced bioactivities,19,20 including the representative members actinorhodin,21 medermycin,22 and granaticin.23 In the qin gene cluster, besides the central PKS genes that are responsible for the biosynthesis of the pyranonaphthoquinone backbone, the presence of genes for the deoxyaminosugar d-forosamine strongly suggested that the end product should be glycosylated. This genetic organization was similar to the clusters for the synthesis of the glycosylated pyranonaphthoquinone medermycin in Streptomyces sp. AM-7161 (med)22 and granaticin in Streptomyces violaceoruber Tü22 (gra).23 Coclustering of glycosylation-associated biosynthetic genes with those for the aglycones is typical of microbial genomes, which facilitates matching the biosynthetic gene cluster to the corresponding NPs.24 In comparison, qin-ORF29 encoding an NADPH-dependent FMN reductase was absent in either the med or gra biosynthetic gene clusters, and glycosylation with a d-forosamine is unprecedented in the pyranonaphthoquinone family. This promoted an investigation into the potentially novel product(s) of the qin gene cluster. Despite our previous detailed chemical investigations of Streptomyces sp. MBT76,15,16 the corresponding molecules had not been identified, suggesting that the gene cluster may be cryptic under the many different growth conditions that had been tested.
Table 1. Predicted Functions of the Gene Products of the Qinimycin Biosynthetic Gene Cluster (qin) of Streptomyces sp. MBT76.
| CDS | length | putative function | nearest orthologue found in | aa identity | accession number |
|---|---|---|---|---|---|
| 1 | 322 | dTDP-glucose 4,6-dehydratase | Streptomyces sp. NRRL S-623 | 79% | WP_031121809.1 |
| 2 | 359 | dTDP-1-glucose synthase | Streptomyces vietnamensis | 69% | ADO32770.1 |
| 3 | 280 | N-methyltransferase | Streptomyces clavuligerus | 39% | WP_003953258.1 |
| 4 | 365 | C-glycosyltransferase | Streptomyces sp. SCC 2136 | 48% | CAF31363.2 |
| 5 | 385 | NDP-hexose aminotransferase | Streptomyces ambofaciens | 72% | CAM96587.1 |
| 6 | 421 | hypothetical protein | Streptomyces violaceoruber | 45% | CAA09645.1 |
| 7 | 436 | NDP-hexose 2,3-dehydratase | Streptomyces sp. NRRL S-623 | 67% | WP_031121795.1 |
| 8 | 351 | NDP-hexose 3-ketoreductase | Streptomyces sp. NRRL S-623 | 64% | WP_032774990.1 |
| 9 | 432 | NDP-hexose-3,4-dehydratase | Streptomyces lydicus | 79% | CBA11561.1 |
| 10 | 378 | flavin-dependent monooxygenase | Streptomyces sp. AM-7161 | 59% | BAC79043.1 |
| 11 | 267 | SARP family transcriptional regulator | Streptomyces cyaneofuscatus | 70% | WP_030562180.1 |
| 12 | 312 | polyketide cyclase | Streptomyces aureofaciens | 73% | YP_009060633.1 |
| 13 | 287 | 4-phosphopantetheinyl transferase | Streptomyces albus | 45% | WP_031175178.1 |
| 14 | 571 | type I polyketide synthase component | Streptomyces scabiei | 44% | YP_003491848.1 |
| 15 | 142 | hydroxylacyl-CoA dehydrogenase | Streptomyces sp. NRRL F-5135 | 69% | WP_030744938.1 |
| 16 | 334 | bifunctional cyclase/dehydratase | Streptomyces lavenduligriseus | 70% | WP_030792829.1 |
| 17 | 87 | acyl carrier protein | Streptomyces sp. R1128 | 69% | AAG30201.1 |
| 18 | 410 | chain-length factor (polyketide beta-ketoacyl synthase beta subunit) | Streptomyces lividans | 73% | WP_003973890.1 |
| 19 | 418 | actinorhodin polyketide beta-ketoacyl synthase alpha subunit | Streptomyces lavenduligriseus | 85% | WP_030792818.1 |
| 20 | 420 | putative transporter | Streptomyces viridochromogenes | 50% | WP_003988487.1 |
| 21 | 166 | flavin reductase | Streptomyces fulvoviolaceus | 57% | WP_052425229.1 |
| 22 | 261 | ketoacyl reductase | Streptomyces chartreusis | 72% | WP_01003439 |
| 23 | 309 | hydroxylacyl-CoA dehydrogenase | Streptomyces sp. NRRL S-31 | 67% | WP_030735501.1 |
| 24 | 303 | quinone oxidoreductase | Nocardia sp. CNY236 | 72% | WP_028476080.1 |
| 25 | 217 | DSBA-type oxidoreductase | Streptosporangium roseum | 63% | WP_012891327.1 |
| 26 | 191 | dehydratase | Streptomyces sp. AM-7161 | 55% | BAC79041.1 |
| 27 | 576 | stereospecific keto reductase | Streptomyces sp. AM-7161 | 64% | BAC79036.1 |
| 28 | 328 | quinone oxidoreductase | Streptomyces sp. AM-7161 | 71% | BAC79039.1 |
| 29 | 201 | NADPH-dependent FMN reductase | Streptomyces sp. NRRL S-623 | 64% | WP_031121793.1 |
| 30 | 402 | S-adenosylmethionine synthetase | Streptomyces roseoverticillatus | 95% | WP_030365339.1 |
| 31 | 339 | adenosine kinase | Streptomyces albulus | 72% | AIA08031.1 |
| 32 | 1167 | methionine synthase | Streptomyces roseoverticillatus | 93% | WP_030367617.1 |
| 33 | 312 | 5,10-methylenetetrahydrofolate reductase | Streptomyces sp. NRRL F-5135 | 81% | WP_030742694.1 |
| 34 | 473 | adenosylhomocysteinase | Streptomyces roseoverticillatus | 90% | WP_030365247.1 |
| 35 | 227 | TetR family transcriptional regulator | Streptomyces lividans | 49% | WP_003972639.1 |
The qin gene cluster contains two putative regulatory genes, namely, the SARP-family transcriptional regulatory gene qin-ORF11 and the TetR-family regulatory gene qin-ORF35 (Table 1). Many pathway-specific activators for secondary metabolite production in streptomycetes belong to the SARP family,12 while TetR-family regulators often act as repressors.25 As an example, in the type II PKS gene cluster aur1 of Streptomyces aureofaciens CCM 3239, which specifies the angucycline-family auricin,26aur1P and aur1PR3 encode SARP-family activators,27 while the tetR-type aur1R encodes a negative regulator.28 To activate the qin gene cluster, we opted to overexpress the likely pathway-specific activator gene qin-ORF11. For this, the gene was amplified by PCR and inserted behind the ermE* promoter29 in the conjugative and integrative vector pSET152. The resulting plasmid was then integrated into the attB site of the chromosome of MBT76 to create the recombinant derivative MBT76-1.
NMR-Based Metabolic Profiling Expedited Characterization of Novel Glycosylated Pyranonaphthoquinones
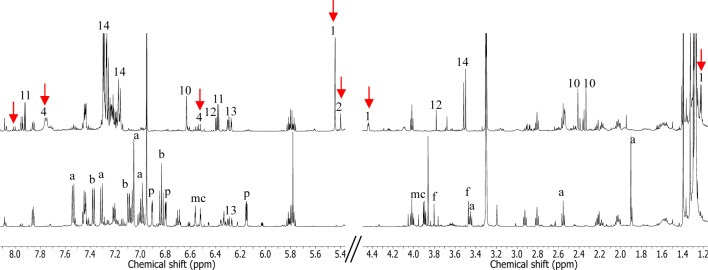
Ex-conjugant MBT76-1 and its parent MBT76 were fermented in parallel in liquid modified NMMP media.16 After 5 days of growth, the cultures were harvested by centrifugation and extracted with EtOAc. The metabolites obtained were subjected to 1H NMR profiling.30 As shown in Figure 1, MBT76-1 presented a very different metabolic profile in comparison to the parental strain. The metabolites usually produced by the wild-type strain, such as 1H-pyrrole-2-carboxamide, acetyltryptamine, fervenulin, and 2-hydroxy-3-methoxy-benzamide,16 were aborted in MBT76-1. While the production of 6,8-dihydroxy-3-methyl-isocoumarin (9) was not affected by ORF11 overexpression, its post-PKS methoxylation was hampered as shown by the disappearance of methoxyl groups in the region δ 3.45–4.25.16 Conversely, the 1H NMR spectra of MBT76-1 exhibited proton resonances absent in the parental strain, indicating that different compounds were produced. These included molecules with characteristic NMR signals such as δ 4.45 (dd, J = 4.2, 1.8 Hz) for the methine in a substituted pyran ring and the methyl doublet at δ 1.29 (d, J = 6.6 Hz), which were in agreement with kalafungin (16-CH3) or 6′-deoxy-CH3 of a sugar moiety predicted by bioinformatics. In the aromatic region, coupling doublets at δ 8.01 (d, J = 8.4 Hz), and 6.56 (d, J = 8.4 Hz) were indicative of an α,β-unsaturated ketone aryl moiety typical of pyranonaphthoquinones. All these NMR features indicated that indeed the qin gene cluster was actively expressed in MBT76-1. In light of the substantial change in the metabolome of MBT76 and MBT76-1, we hypothesized that qin-ORF11 may control multiple biosynthetic gene clusters for specialized metabolites.
Figure 1.
Comparison of 1H NMR spectra. 1H NMR (600 MHz, in CD3OD) was obtained on the crude extracts from wild-type strain Streptomyces sp. MBT76 (bottom) and its ex-conjugant MBT76-1 (top). The presented chemical shifts are for aromatic region δ 5.4–8.1 and aliphatic region δ 1.2–4.5. The signals attributable to compounds produced in MBT76 wild type were indicated as the following: a, acetyltryptamine; b, 2-hydroxy-3-methoxy-benzamide; p, 1H-pyrrole-2-carboxamide; f, fervenulin; mc, methoxylated isocoumarins. The signals attributable to compounds produced in MBT76-1 were labeled according to the numbering in Figure 2, and the detailed signals assignments were summarized in Table S1. The highlighted signals by red arrows are attributable to glycosylated pyranonaphthoquinone molecules.
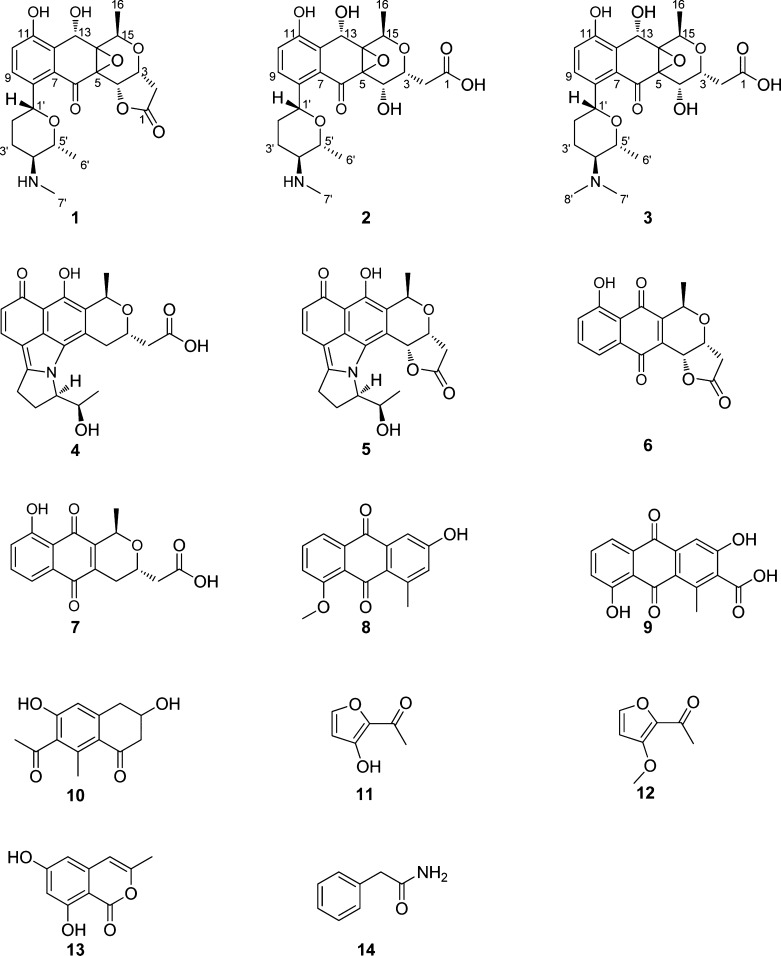
To identify the metabolic product(s) of the qin gene cluster, the crude extract of MBT76-1 was separated by semipreparative HPLC-UV chromatography. The resulting fractions (Fr1–16) were analyzed by 1H NMR spectroscopy, which showed that Fr1 (2.1 mg) contained the sought-after deoxysugar-pyranonaphthoquinones. Further UHPLC-UV-ToF-HRMS analysis confirmed Fr1 contained mainly three structurally related compounds, with molecular formulas C23H27NO8 (1), C23H29NO9 (2), and C24H31NO9 (3) with a relative abundance of 6:2:1 (Figure S1). Elucidation of the final structure of major compound 1 was done on the basis of extensive NMR analysis, including 1H NMR, HSQC, HMBC, COSY, and APT. The 1H NMR data for compound 1 (Table 2) presented characteristic resonances for the bioinformatics-predicted deoxyaminosugar forosamine, including a 6′-deoxy-CH3 doublet at δH 1.29 (d, J = 6.6 Hz), a 7′-N-CH3 singlet at δH 2.89 (s), and highly overlapping signals for 2′,3′-deoxy-CH2 at δH 2.20–2.40. Aided by COSY and HSQC experiments (Figure S2), the backbone of the aglycone was confirmed as a kalafungin-type molecule, as established by the spin system H2-2/H-3/H-4 for a γ-lactone ring, together with two coupling protons at δH 1.41 (d, J = 6.6 Hz, H-16) and 4.93 (q, J = 6.6 Hz, H-15) for a pyran ring. The additional two aromatic coupling doublets δH 7.65 (d, J = 8.4 Hz, H-9) and 7.25 (d, J = 8.4 Hz, H-10) indicated that the deoxyaminosugar was linked to the aglycone through the C-8 residue of the benzene ring, which was confirmed by the HMBC correlations (Figure S2) from H2-2′ (δH 2.30, 2.24) to C-8 (δC 124.2) and from H-9 (δH 7.65) to C-1′ (δC 68.7). More importantly, the key HMBC correlations from the H-13 (δH 5.48) to C-11 (δC 158.1), C-12 (δC 129.2), C-7 (δC 131.1), C-5 (δC 60.6), C-14 (δC 68.1), and C-15 (δC 65.9) demonstrated that the pyranonaphthoquinone skeleton of kalafungin (6) acquired two variations, via the epoxidation of the Δ5,14 double bond and hydroxylation of the ketone at C-13. The stereochemistry at C-13 was established as R, based on the NOESY correlation between H-13/H3-16, but no correlation was observed for H-13/H-15. However, the configuration of 5,14-epoxy remained unclear. Consequently, the final structure of major compound 1 (Figure 2) in Fr1 was elucidated as a novel glycosylated pyranonaphthoquinone, and named qinimycin A. Similar NMR spectral analyses revealed that compound 2 was a hydration product of 1 in the γ-lactone ring (Table 2), and accordingly named qinimycin B. Though compound 3 (qinimycin C) was not resolved by NMR spectra due to its low abundance, the additional mass of 14.0153 (Figure S1) corresponding exactly to a CH2 unit suggested that the imide in 2 was further methylated into a tertiary amine, because the N-methyltransferase qin-ORF3 is able to di-methylate the nitrogen atom of dTDP-D-forosamine in spinosyn pathway.31 The known compounds 4–14 (Figure 2) were identified in Fr2–Fr16 by 1H NMR and/or UHPLC-UV-ToF-HRMS, and the results were compared with spectroscopic data from the literature (Table S1).
Table 2. 1H and 13C NMR Data for Compounds 1 and 2a.
|
1 |
2 |
|||
|---|---|---|---|---|
| no. | δC, type | δH (J in Hz) | δC, type | δH (J in Hz) |
| 1 | 177.4, C | 174.4, C | ||
| 2 | 37.3, CH2 | 2.91, dd (15.6, 4.2); | 36.5, CH2 | 2.58, d (7.2) |
| 2.40, dd (15.6, 1.8) | ||||
| 3 | 68.0, CH | 4.45, dd (4.2, 1.8) | 67.1, CH | 4.16, m |
| 4 | 74.0, CH | 4.98, d (1.2) | 64.0, CH | 4.32, d (12.6) |
| 5 | 60.6, C | 63.0, C | ||
| 6 | 194.6, C | 197.1, C | ||
| 7 | 131.1, C | 131.5, C | ||
| 8 | 124.2, C | 124.1, C | ||
| 9 | 132.2, CH | 7.65, d (8.4) | 132.1, CH | 7.62, d (8.4) |
| 10 | 121.4, CH | 7.25, d (8.4) | 121.3, CH | 7.23, d (8.4) |
| 11 | 158.1, C | 158.1, C | ||
| 12 | 129.2, C | 129.3, C | ||
| 13 | 61.0, CH | 5.48, s | 61.2, CH | 5.43, s |
| 14 | 68.1, C | 67.7, C | ||
| 15 | 65.9, CH | 4.93, q (6.6) | 65.7, CH | 4.90, q (6.6) |
| 16 | 14.1, CH3 | 1.41, d (6.6) | 14.4, CH3 | 1.40, d (6.6) |
| 1′ | 68.7, CH | 5.36, m | 68.7, CH | 5.36, m |
| 2′ | 32.1, CH2 | 2.30, m; 2.24, m | 32.1, CH2 | 2.30, m; 2.24, m |
| 3′ | 22.5, CH2 | 2.38, m; 2.21, m | 22.5, CH2 | 2.38, m; 2.21, m |
| 4′ | 75.0, CH | 3.63, td (7.2, 1.8) | 75.0, CH | 3.63, td (7.2, 1.8) |
| 5′ | 63.8, CH | 4.27, m | 63.8, CH | 4.27, m |
| 6′ | 19.9, CH3 | 1.29, d (6.6) | 19.9, CH3 | 1.29, d (6.6) |
| 7′ | 39.0, CH3 | 2.89, s | 39.0, CH3 | 2.89, s |
1 and 2 were recorded in CD3OD. Proton coupling constants (J) in Hz are given in parentheses. All chemical shift assignments were done on the basis of 1D and 2D NMR techniques.
Figure 2.
Secondary metabolites produced by Streptomyces sp. MBT76-1. Compounds identification was done on the basis of NMR and HRMS techniques, and their spectroscopy data were summarized in Table 2 and Table S1.
The qinimycins have intriguing chemical features, representing a new branch within the extensively studied pyranonaphthoquinone family of natural products. The 5,14-epoxidation was previously described in pyranonaphthoquinone aglycones,32,33 but hydroxylation of the pyranonaphthoquinone backbone at C-13 is unprecedented. The 8-C-glycosylation of pyranonaphthoquinone is rare, though previously seen in the antibiotic Sch 38519,34,35 and xiakemycin A.36 Within the pyranonaphthoquinones, glycosylation with the deoxyaminosugar forosamine is uniquely seen in the qinimycins. With this chemical architecture, the qinimycins rival the well-known pyranonaphthoquinone glycosides medermycin22 or granaticin23 in terms of structural complexity. To establish if the qinimycins might function as antimicrobials, their bioactivity was tested using agar diffusion assays. Growth inhibition was seen for the Gram-positive bacteria Bacillus subtilis 168 and Staphylococcus aureus CECT976, but not for Gram-negative Escherichia coli JM109 or Pseudomonas aeruginosa PAO1, thus establishing bioactivity against Gram-positive but not Gram-negative bacteria (Table 3). However, assessment of the MIC of the mixture of molecules revealed minimal inhibition concentrations (MICs) against B. subtilis and S. aureus of 50 and 100 μg/mL, respectively. These high values are indicative of very limited bioactivity of the qinimycins under the conditions tested, and no further bioactivity analysis was performed.
Table 3. Antimicrobial Activity of Qinimycinsa.
| inhibition
zone (mm) |
||||
|---|---|---|---|---|
| compound no. | Bacillus subtilis | Escherichia coli | Staphylococcus aureus | Pseudomonas aeruginosa |
| Fr1 | 15 | 0 | 20 | 0 |
| AMP | 23 | 20 | 10 | 7 |
| APRA | 10 | 7 | 7 | 15 |
| NC | 0 | 0 | 0 | 0 |
For Fr1, 25 μL was spotted of a 2 mg/mL solution in methanol. For AMP and APRA, 5 μL was spotted of a 1 mg/mL solution in miliQ water. AMP, ampicillin; APRA, apramycin; NC, negative control (methanol).
Proposed Biosynthetic Pathway for Qinimycins Based on Bioinformatics Analysis
A biosynthetic model for qinimycins is proposed on the basis of the functional assignments from sequence analysis and the known metabolites that are produced (Figure 3). The assembly line includes three steps: (i) biosynthesis of an activated form of dTDP-D-forosamine, similar to that found in the spinosyn pathway;31 (ii) assembly of the kalafungin aglycone by a minimal PKS similar to that found in the biosynthetic gene clusters for medermycin22 and granaticin;23 and (iii) regioselective installing of a dTDP-D-forosamine moiety at the C-8 position of kalafungin by a C-glycosyltransferase (most likely encoded by qin-ORF4).
Figure 3.
Biosynthetic pathway of qinimycins and BE-54238A/B. (A) Organization of the qin locus in Streptomyces sp. MBT76. For annotation of the respective gene products see Table 1. Genes for the minimal PKS (presented in black) are similar to those for biosynthesis of the pyranonaphthoquinone kalafungin, while the eight genes in red encode enzymes for production of deoxyaminosugar d-forosamine. (B) Proposed biosynthetic route to qinimycins. The exact function of each gene was assigned in every specific biosynthesis step. The intriguing feature for qinimycins biosynthesis is reduction of the C-13 ketone probably catalyzed by qin-ORF29, while the pyrrole ring in the antibiotics BE-54328A/B originated from the six-membered deoxyaminosugar forosamine.
In a pilot study of actinorhodin biosynthesis, a two-component flavin-dependent monooxygenase (FMO), encoded by actVA-ORF5 (oxygenase) and actVB (flavin reductase), was demonstrated to perform the two consecutive oxygenations at C-6 and C-8.37 The FMO genes are commonly present in the med and gra biosynthetic gene clusters for medermycin22 and granaticin,23 respectively. The ActVA homologue Gra-21 was shown to be bifunctional at C-6 and C-8, whereas Med-7 monofunctionally oxygenizes the C-6 position. Interestingly, the gene product of qin-ORF10 shows higher similarity to Med-7 (59% aa identity) than to ActVA-ORF5 (53% aa identity), suggesting possible monofunctionality at C-6. The single oxygenation at C-6 leading to a pyranonaphthoquinone skeleton is in good agreement with the subsequent C-glycosylation for qinimycins at C-8 and medermycin at C-10, where the selectivity of the responsible glycosyl transferase qin-ORF4 is of great interest. Further studies33 on ActVA-ORF5/ActVB revealed its additional in vitro epoxidation activity of kalafungin. The co-occurrence of three qinimycins (1–3) is a clear example of in vivo activity of a two-component FMO encoded by qin-ORF10/qin-ORF21. Compared with the act,6med,22 and gra(23) gene clusters, qin-ORF29 encoding an NADPH-dependent FMN reductase is unique in the qin gene cluster. We propose that this enzyme may execute the unprecedented reduction of the C-13 ketone.
It is noteworthy that, on the basis of the biosynthetic pathway (Figure 3), we predict that qinimycins in addition to compounds (1–3) are produced by Streptomyces sp. MBT76, which may have been missed due to lower yields. For instance, UHPLC-UV-ToF-HRMS analysis of MBT76-1 crude extract showed molecules with the molecular formulas C23H27NO9 and C23H25NO8 for the putative compounds 1a and 1b, respectively. The post-PKS glycosylation of 8-C-glycosylpyranonaphthoquinones BE-54238A (4) and BE-54238B (5)38 involved rearrangement of the deoxyaminosugar moiety. After glycosylation of kalafungin into 5a, the six-membered amino pyran ring was first rearranged into a five-membered 2H-3,4-dihydropyrrole ring through a Mannich reaction, whereby the gene product of qin-ORF6 might mediate the ring-opening of the appended forosamine to a linear ketone. The newly formed imide was then further cyclized with the aglycone quinone at C-6 to give an intermediary iminium ion, which subsequently underwent proton tautomerization to produce antibiotic BE-54238A/B. Interestingly, this naturally occurring biosynthetic pathway mimics the total synthesis of BE-54238B from rhamnose.39,40 The anthraquinones 11-O-methyl-aloesaponarin II (8) and 3,8-dihydroxy-1-methyl-anthraquinone-carboxylic acid (9) were shunt products of the kalafungin biosynthetic pathway,41 but it remains unclear whether polyketide molecules 10–12 were also derived from the qin gene cluster.
Conclusions
The replication issues that have frustrated high-throughput screening regimes necessitate new approaches to expand the chemical diversity of natural products and keep filling the discovery pipelines with new lead compounds. Our work shows that the combination of NMR-based metabolomics and genome mining is an efficient way to streamline the discovery of novel molecules from actinomycetes. Prior knowledge of the types of NPs that can be expected based on bioinformatics thereby simplifies the chromatographic isolation process and structure determination of target compound(s), while NMR profiling of molecules in highly complex matrices allows rapid linkage of chemotype to genotype. We here provide proof of concept for this principle, whereby activation of the cryptic qin type II PKS gene cluster by constitutive expression of its pathway-specific activator gene (qin-ORF11) followed by NMR-based metabolic profiling, identified novel glycosylated pyranonaphthoquinones (1–5). The elucidated biosynthetic pathway for the qinimycins in the genetically tractable Streptomyces sp. MBT76 offers new insights into the biosynthesis of this family of natural products. The best studied pyranonaphtoquinone is undoubtedly actinorhodin produced by Streptomyces coelicolor A3(2), which is the archetype of this family of polyketides. The qinimycins described in this work have intriguing structural features that make them stand out, namely 8-C-glycosylation by the deoxyaminosugar d-forosamine, as well as 5,14-epoxidation and 13-hydroxylation. Another novelty lies in the unusual fusion of the deoxyaminosugar to the pyranonaphthoquinone backbone during biosynthesis of the antibiotics BE-54238 A and B. Thus, the qinimycins form a new branch of the family of pyranonaphthoquinone-type antibiotics and provides important insights into the biosynthesis of this class of natural products.
Experimental Section
General Experimental Procedures
FT-IR spectra were recorded on a Shimadzu FT-IR 83000 spectrometer (Shimadzu Corporation, Japan). UV measurements were performed using a Shimadzu UV mini-1240 (Shimadzu Corporation, Japan). NMR spectra were recorded in CD3OD on a Bruker 600 MHz calibrated to a residual CD3OD (3.30 ppm). The UHPLC-TOF-MS analyses were performed on an Ultimate 3000 UHPLC system (Thermoscientific, Pittsburgh, PA, U.S.A.) coupled to a micro-ToF-2Q mass spectrometer from Bruker Daltonics (Bremen, Germany) with an electrospray (ESI) interface. HPLC analysis was performed with an Agilent 1200 series HPLC apparatus (Agilent technologies Inc., Santa Clara, CA, U.S.A.), using a 150 × 4.6 mm Luna 5 μm C18 (2) 100 Å column equipped with a guard column containing C18 4 × 3 mm cartridges (Phenomenex Inc., Torrance, CA, U.S.A.). Semipreparative HPLC separation was performed on reversed-phase column (Phenomenex Luna 5 μm C18 (2) 100 Å column, 250 × 10 mm). Silica gel 60 F254 (Merck, Darmstadt, Germany) was used for TLC analysis, migrated with CHCl3/MeOH (10:1), and visualized with anisaldehyde/sulfuric acid reagent. Polymerase chain reactions (PCR) were performed on a T100 Thermal Cycler (Bio-Rad, Hercules, CA, U.S.A.). The ZR Plasmid Miniprep-Classic kit (Zymo Research, Irvine, CA, U.S.A.) was used for plasmid extraction. All organic solvents and chemicals were of analytical or HPLC grade, depending on the experiment.
Bacterial Strains and Culturing Conditions
Streptomyces sp. MBT76 was obtained from the culture collection of Molecular Biotechnology, IBL, Leiden University. Escherichia coli JM10942 was used for routine cloning. Escherichia coli ET1256743 containing pUZ800244 was used for introducing nonmethylated DNA into Streptomyces by conjugation. The basal medium for Streptomyces sp. MBT76 growth was modified minimal liquid medium NMMP44 without PEG6000 and containing 1% (w/v) glycerol and 0.5% (w/v) mannitol as the carbon sources, which was further supplemented with 0.8% (w/v) Bacto peptone.16 Tryptone soy broth with 10% (w/v) sucrose (TSBS) was used to grow MBT76 mycelia as receptor for conjugation experiments. Soy flour mannitol (SFM) agar plates44 were used to grow overexpression conjugants of MBT76. For culturing of MBT76 for chemical analysis, 50 mL modified NMMP was inoculated with 106 spores of Streptomyces sp. MBT76 in 250 mL Erlenmeyer flasks equipped with a spring, and grown at 30 °C with constant shaking at 220 rpm. The incubation lasted for 120 h.
Genome Sequencing, Assembly, and Annotation
DNA was extracted from Streptomyces sp. MBT76 as described previously.44 Genome sequencing and annotation was done essentially as described previously.45 Illumina/Solexa sequencing on Genome Analyzer IIx and sequencing on PacBio RS were outsourced to BaseClear BV (Leiden, The Netherlands). In general, 100-nt paired-end reads were obtained, and the quality of the short reads were verified using FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/). Depending on the quality, reads were trimmed to various lengths at both ends. Processed raw reads were subsequently used as input for the Velvet assembly algorithm. Genomes were annotated using the RAST server with default options. Contigs were also annotated using GeneMark.hmm17 for ORF prediction, BLASTP for putative function prediction, and HMMER for protein-domain prediction, manually inspected for some and visualized using Artemis. The genome has been deposited at GenBank under the accession LNBE00000000.
Overexpression of the Pathway-Specific SARP Regulator qin-ORF11
qin-ORF11 was amplified by PCR from Streptomyces sp. MBT76 genomic DNA as described46 using primers SC06_0044_F_EcoRI 5′-CGATGAATTCCGTCCGGCGCTTCGTGTG and SC06_0044_R_EcoRI_NdeI 5′-CGATGAATTCGCACCCGGTACAGGAGTGTGTCATATGCGATTCAACCTCATC. The PCR product was subcloned as an EcoRI fragment and subsequently ligated as an NdeI-HindIII fragment behind the constitutive ermE promoter in pSET152,29 which integrates at the φC31 attachment site in the Streptomyces chromosome. The final recombinant plasmid pCSW01 (pSET152/ermE/qin-ORF11) was subsequently transformed into Escherichia coli ET12567/pUZ8002, and the positive transformant was selected by 100 μg/mL apramycin. This strain was then used to conjugate two-day old mycelia of Streptomyces sp. MBT76. The ex-conjugant was confirmed by PCR using primers SC06_0044_SF 5′-TTTCCCAGTCACGACGTTG and SC06_0044_SR 5′-GGATAACAATTTCACACAGG.
Metabolomics and Compound Purification
Initially, 50 mL cultures of Streptomyces sp. MBT76 or MBT76-1 were harvested by centrifugation at 4000 rpm for 10 min, and the supernatant was extracted twice with 20 mL of ethyl acetate (EtOAc). The organic phase was washed with 30 mL of water and subsequently dried with 5 g of anhydrous Na2SO4. EtOAc was removed under vacuum at 38 °C, and the residue was dissolved in 2.0 mL of EtOAc in a microtube (Eppendorf type-5415C, Hamburg, Germany). The solvent was then evaporated at room temperature under nitrogen gas and subsequently dipped into liquid nitrogen and lyophilized using a freeze-dryer (Edwards Ltd., Crawley, England). The NMR sample preparation and measurements were performed according to our previously published protocol.30 For details, see ref (47).
For separation of metabolites, 15 replicates of Streptomyces sp. MBT76-1 culture (750 mL in total) were pooled. After EtOAc extraction, 0.26 g of crude extract was partitioned between methanol and n-hexane to remove the lipids. The resolved methanol fraction was subsequently separated by semipreparative reversed-phase HPLC (Phenomenex Luna 5 μm C18 (2) 100 Å column, 250 × 10 mm) on an Agilent 1200 series HPLC (Agilent technologies Inc., Santa Clara, CA, U.S.A.), eluting with a gradient of ACN in H2O adjusted with 0.1% TFA from 20% to 60% at a flow rate of 2 mL/min in 40 min. Sixteen fractions (Fr1–Fr16) were manually collected by peak detection at 254 nm, which were numbered in ascending order of retention time. After rotary evaporation at 42 °C under vacuum, these 16 factions were further subjected to 1H NMR profiling and UHPLC-UV-ToF-MS analysis. Qinimycins were contained in Fr1 (2.1 mg) at retention time 8.12 min.
Qinimycins (1–3)
The products had the consistency of brown gum; UV (MeOH) λmax (log ε) 222 (3.87), 270 (3.28), 326 (3.15) nm; IR νmax 3400, 3299, 1792, 1674, 1585, 1304, 1202, 1148, 837, 800, 719 cm–1; 1H NMR (600 MHz, methanol-d4) and 13C NMR (150 MHz, methanol-d4) data, see Table 2; HRMS (positive mode) m/z 446.1854 [M + H]+ for qinimycin A (calcd for C23H28NO8, 446.1809), 464.1944 [M + H]+ for qinimycin B (calcd for C23H30NO9, 464.1915), 478.2097 [M + H]+ for qinimycin C (calcd for C24H32NO9, 478.2072).
UHPLC-UV-ToF-MS Analysis
UHPLC-ToF-MS analyses were performed on an UHPLC system (Ultimate 3000, ThermoScientific, Germany) coupled to an ESI-llQ-TOF spectrometer (micrOTOF-QII, Bruker Daltonics, Germany) in the positive mode.48 The chromatographic separation was done using a Kinetex C18 UHPLC 2.6 μm particle size column 150 × 2.0 mm (Phenomenex) at a flow rate of 0.3 mL/min and a column temperature of 30 °C. Samples (3 μL) were eluted using a gradient of solvent A (water) and B (acetonitrile), both with 0.1% formic acid (v/v). The initial percentage of B was 5%, which was linearly increased to 90% in 19.5 min, followed by a 2 min isocratic period, and then re-equilibrated with original conditions in 2 min. Nitrogen was used as drying and nebulizing gas. The gas flow was set at 10.0 L/min at 250 °C, and the nebulizer pressure was 2.0 bar. The MS data were acquired over m/z range of 100–1000. The capillary voltage was 3.5 kV. For internal calibration, a 10 mM solution of sodium formate (Fluka, Steinheim, Germany) was infused. Formic acid, water, and acetonitrile were LCMS grade (Optima, Fisher Scientific, NJ, U.S.A.).
Antimicrobial Activity Assays
Antimicrobial activity of qinimycins was determined according to a disc diffusion method as described.49,50 Initially, 25 μL of Fr1 (2 mg/mL in methanol) was spotted onto paper discs (6 mm diameter) placed on agar plates containing a soft agar overlay with indicator bacteria. Indicator bacteria were Bacillus subtilis 168, Escherichia coli ASD19, Staphylococcus aureus CECT976, or Pseudomonas aeruginosa PAO1. Ampicillin and apramycin were used as positive controls, whereby 5 μL was spotted of a 1 mg/mL solution in miliQ water. The solvent methanol was used as the negative control. After incubation at 37 °C for 18 h, growth inhibition zones (in mm) were recorded as antimicrobial activity.
The MIC assay against B. subtilis and S. aureus was carried out in 96-well plate by serial double dilution method, as previously described.15 All MIC determinations were performed in duplicate.
Acknowledgments
C.S.W. was supported by a grant from the China Scholarship Council.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.jnatprod.6b00478.
Detailed spectral data of qinimycins, including MS, UV, IR, 1H NMR, APT, COSY, HSQC, HMBC, and NOESY NMR (PDF)
The authors declare no competing financial interest.
Supplementary Material
References
- Bérdy J. J. Antibiot. 2012, 65, 385–395. 10.1038/ja.2012.27. [DOI] [PubMed] [Google Scholar]
- Barka E. A.; Vatsa P.; Sanchez L.; Gaveau-vaillant N.; Jacquard C.; Klenk H.; Clément C.; Ouhdouch Y.; van Wezel G. P. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. 10.1128/MMBR.00019-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper M. A.; Shlaes D. Nature 2011, 472, 32. 10.1038/472032a. [DOI] [PubMed] [Google Scholar]
- Kolter R.; van Wezel G. P. Nat. Microbiol. 2016, 1, 15020. 10.1038/nmicrobiol.2015.20. [DOI] [PubMed] [Google Scholar]
- Payne D. J.; Gwynn M. N.; Holmes D. J.; Pompliano D. L. Nat. Rev. Drug Discovery 2007, 6, 29–40. 10.1038/nrd2201. [DOI] [PubMed] [Google Scholar]
- Bentley S. D.; Chater K. F.; Cerdeno-Tarraga A.-M.; Challis G. L.; Thomson N. R.; James K. D.; Harris D. E.; Quail M. A.; Kieser H.; Harper D.; Bateman A.; Brown S.; Chandra G.; Chen C. W.; Collins M.; Cronin A.; Fraser A.; Goble A.; Hidalgo J.; Hornsby T.; Howarth S.; Huang C.-H.; Kieser T.; Larke L.; Murphy L.; Oliver K.; O'Neil S.; Rabbinowitsch E.; Rajandream M.-A.; Rutherford K.; Rutter S.; Seeger K.; Saunders D.; Sharp S.; Squares R.; Squares S.; Taylor K.; Warren T.; Wietzorrek A.; Woodward J.; Barrell B. G.; Parkhill J.; Hopwood D. A. Nature 2002, 417, 141–147. 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Medema M. H.; Kottmann R.; Yilmaz P.; Cummings M.; Biggins J. B.; Blin K.; de Bruijn I.; Chooi Y. H.; Claesen J.; Coates R. C.; Cruz-Morales P.; Duddela S.; Düsterhus S.; Edwards D. J.; Fewer D. P.; Garg N.; Geiger C.; Gomez-Escribano J. P.; Greule A.; Hadjithomas M.; Haines A. S.; Helfrich E. J. N.; Hillwig M. L.; Ishida K.; Jones A. C.; Jones C. S.; Jungmann K.; Kegler C.; Kim H. U.; Kötter P.; Krug D.; Masschelein J.; Melnik A. V.; Mantovani S. M.; Monroe E. a; Moore M.; Moss N.; Nützmann H.-W.; Pan G.; Pati A.; Petras D.; Reen F. J.; Rosconi F.; Rui Z.; Tian Z.; Tobias N. J.; Tsunematsu Y.; Wiemann P.; Wyckoff E.; Yan X.; Yim G.; Yu F.; Xie Y.; Aigle B.; Apel A. K.; Balibar C. J.; Balskus E. P.; Barona-Gómez F.; Bechthold A.; Bode H. B.; Borriss R.; Brady S. F.; Brakhage A. a; Caffrey P.; Cheng Y.-Q.; Clardy J.; Cox R. J.; De Mot R.; Donadio S.; Donia M. S.; van der Donk W. a; Dorrestein P. C.; Doyle S.; Driessen A. J. M.; Ehling-Schulz M.; Entian K.-D.; Fischbach M. a; Gerwick L.; Gerwick W. H.; Gross H.; Gust B.; Hertweck C.; Höfte M.; Jensen S. E.; Ju J.; Katz L.; Kaysser L.; Klassen J. L.; Keller N. P.; Kormanec J.; Kuipers O. P.; Kuzuyama T.; Kyrpides N. C.; Kwon H.-J.; Lautru S.; Lavigne R.; Lee C. Y.; Linquan B.; Liu X.; Liu W.; Luzhetskyy A.; Mahmud T.; Mast Y.; Méndez C.; Metsä-Ketelä M.; Micklefield J.; Mitchell D. a; Moore B. S.; Moreira L. M.; Müller R.; Neilan B. a; Nett M.; Nielsen J.; O’Gara F.; Oikawa H.; Osbourn A.; Osburne M. S.; Ostash B.; Payne S. M.; Pernodet J.-L.; Petricek M.; Piel J.; Ploux O.; Raaijmakers J. M.; Salas J. a; Schmitt E. K.; Scott B.; Seipke R. F.; Shen B.; Sherman D. H.; Sivonen K.; Smanski M. J.; Sosio M.; Stegmann E.; Süssmuth R. D.; Tahlan K.; Thomas C. M.; Tang Y.; Truman A. W.; Viaud M.; Walton J. D.; Walsh C. T.; Weber T.; van Wezel G. P.; Wilkinson B.; Willey J. M.; Wohlleben W.; Wright G. D.; Ziemert N.; Zhang C.; Zotchev S. B.; Breitling R.; Takano E.; Glöckner F. O. Nat. Chem. Biol. 2015, 11, 625–631. 10.1038/nchembio.1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedorova N. D.; Moktali V.; Medema M. H. Methods Mol. Biol. 2012, 944, 23–45. 10.1007/978-1-62703-122-6_2. [DOI] [PubMed] [Google Scholar]
- Zhu H.; Sandiford S. K.; van Wezel G. P. J. Ind. Microbiol. Biotechnol. 2014, 41, 371–386. 10.1007/s10295-013-1309-z. [DOI] [PubMed] [Google Scholar]
- Rutledge P. J.; Challis G. L. Nat. Rev. Microbiol. 2015, 13, 509–523. 10.1038/nrmicro3496. [DOI] [PubMed] [Google Scholar]
- van Wezel G. P.; McDowall K. J. Nat. Prod. Rep. 2011, 28, 1311–1333. 10.1039/c1np00003a. [DOI] [PubMed] [Google Scholar]
- Bibb M. J. Curr. Opin. Microbiol. 2005, 8, 208–215. 10.1016/j.mib.2005.02.016. [DOI] [PubMed] [Google Scholar]
- Abdelmohsen U. R.; Grkovic T.; Balasubramanian S.; Kamel M. S.; Quinn R. J.; Hentschel U. Biotechnol. Adv. 2015, 33, 798–811. 10.1016/j.biotechadv.2015.06.003. [DOI] [PubMed] [Google Scholar]
- Wu C.; Choi Y. H.; van Wezel G. P. J. Ind. Microbiol. Biotechnol. 2016, 43, 299–312. 10.1007/s10295-015-1666-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H.; Swierstra J.; Wu C.; Girard G.; Choi Y. H.; van Wamel W.; Sandiford S. K.; van Wezel G. P. Microbiology 2014, 160, 1714–1725. 10.1099/mic.0.078295-0. [DOI] [PubMed] [Google Scholar]
- Wu C.; Zhu H.; van Wezel G. P.; Choi Y. H. Metabolomics 2016, 12, 90. 10.1007/s11306-016-1025-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukashin A. V.; Borodovsky M. Nucleic Acids Res. 1998, 26, 1107–1115. 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K.; Medema M. H.; Kazempour D.; Fischbach M. A.; Breitling R.; Takano E.; Weber T. Nucleic Acids Res. 2013, 41, W204–W212. 10.1093/nar/gkt449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metsä-Ketelä M.; Oja T.; Taguchi T.; Okamoto S.; Ichinose K. Curr. Opin. Chem. Biol. 2013, 17, 562–570. 10.1016/j.cbpa.2013.06.032. [DOI] [PubMed] [Google Scholar]
- Oja T.; Galindo P. S. M.; Taguchi T.; Manner S.; Vuorela P. M.; Ichinose K.; Metsä-Ketelä M.; Fallarero A. Antimicrob. Agents Chemother. 2015, 59, 6046–6052. 10.1128/AAC.00991-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto S.; Taguchi T.; Ochi K.; Ichinose K. Chem. Biol. 2009, 16, 226–236. 10.1016/j.chembiol.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Ichinose K.; Ozawa M.; Itou K.; Kunieda K.; Ebizuka Y. Microbiology 2003, 149, 1633–1645. 10.1099/mic.0.26310-0. [DOI] [PubMed] [Google Scholar]
- Ichinose K.; Bedford D. J.; Tornus D.; Bechthold A.; Bibb M. J.; Revill W. P.; Floss H. G.; Hopwood D. A. Chem. Biol. 1998, 5, 647–659. 10.1016/S1074-5521(98)90292-7. [DOI] [PubMed] [Google Scholar]
- Kersten R. D.; Ziemert N.; Gonzalez D. J.; Duggan B. M.; Nizet V.; Dorrestein P. C.; Moore B. S. Proc. Natl. Acad. Sci. U. S. A. 2013, 110, E4407–E4416. 10.1073/pnas.1315492110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos J. L.; Martinez-Bueno M.; Molina-henares A. J.; Teran W.; Watanabe K.; Zhang X.; Gallegos M. T.; Brennan R.; Tobes R. Microbiol. Mol. Biol. Rev. 2005, 69, 326–356. 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novakova R.; Homerova D.; Feckova L.; Kormanec J. Microbiology 2005, 151, 2693–2706. 10.1099/mic.0.28019-0. [DOI] [PubMed] [Google Scholar]
- Novakova R.; Rehakova A.; Kutas P.; Feckova L.; Kormanec J. Microbiology 2011, 157, 1629–1639. 10.1099/mic.0.047795-0. [DOI] [PubMed] [Google Scholar]
- Novakova R.; Kutas P.; Feckova L.; Kormanec J. Microbiology 2010, 156, 2374–2383. 10.1099/mic.0.037895-0. [DOI] [PubMed] [Google Scholar]
- Bierman M.; Logan R.; O’Brien K.; Seno E. T.; Rao R. N.; Schoner B. E. Gene 1992, 116, 43–49. 10.1016/0378-1119(92)90627-2. [DOI] [PubMed] [Google Scholar]
- Kim H. K.; Choi Y. H.; Verpoorte R. Nat. Protoc. 2010, 5, 536–549. 10.1038/nprot.2009.237. [DOI] [PubMed] [Google Scholar]
- Hong L.; Zhao Z.; Melancon C. E.; Zhang H.; Liu H.-w. J. Am. Chem. Soc. 2008, 130, 4954–4967. 10.1021/ja0771383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kara M.; Soga S.; Shono K.; Eishima J.; Mizukami T. J. Antibiot. 2001, 54, 182–186. 10.7164/antibiotics.54.182. [DOI] [PubMed] [Google Scholar]
- Taguchi T.; Okamoto S.; Hasegawa K.; Ichinose K. ChemBioChem 2011, 12, 2767–2773. 10.1002/cbic.201100571. [DOI] [PubMed] [Google Scholar]
- Hegde V. R.; King A. H.; Patel M. G.; Puar M. S.; McPhail A. T. Tetrahedron Lett. 1987, 28, 4485–4488. 10.1016/S0040-4039(00)96543-2. [DOI] [Google Scholar]
- Patel M.; Hegde V.; Horan A.; Barrett T.; Bishop R.; King A.; Marquez J.; Hare R.; Gullo V. J. Antibiot. 1989, 42, 1063–1069. 10.7164/antibiotics.42.1063. [DOI] [PubMed] [Google Scholar]
- Jiang Z.; Guo L.; Chen C.; Liu S.; Zhang L.; Dai S.; He Q.; You X.; Hu X.; Tuo L.; Jiang W.; Sun C. J. Antibiot. 2015, 68, 771–774. 10.1038/ja.2015.70. [DOI] [PubMed] [Google Scholar]
- Taguchi T.; Yabe M.; Odaki H.; Shinozaki M.; Metsä-Ketelä M.; Arai T.; Okamoto S.; Ichinose K. Chem. Biol. 2013, 20, 510–520. 10.1016/j.chembiol.2013.03.007. [DOI] [PubMed] [Google Scholar]
- Tsukamoto M.; Nakajima S.; Murooka K.; Hirayama M.; Hirano K.; Yoshida S.; Kojiri K.; Suda H. J. Antibiot. 2000, 53, 26–32. 10.7164/antibiotics.53.26. [DOI] [PubMed] [Google Scholar]
- Tatsuta K.; Hirabayashi T.; Kojima M.; Suzuki Y.; Ogura T. J. Antibiot. 2004, 57, 291–297. 10.7164/antibiotics.57.291. [DOI] [PubMed] [Google Scholar]
- Tatsuta K.; Hosokawa S. Sci. Technol. Adv. Mater. 2006, 7, 397–410. 10.1016/j.stam.2006.05.001. [DOI] [Google Scholar]
- Taguchi T.; Itou K.; Ebizuka Y.; Malpartida F.; Hopwood D. A.; Surti C. M.; Booker-Milburn K. I.; Stephenson G. R.; Ichinose K. J. Antibiot. 2000, 53, 144–152. 10.7164/antibiotics.53.144. [DOI] [PubMed] [Google Scholar]
- Sambrook J.; Fritsch E. F.; Maniatis T.. Molecular Cloning: A Laboratory Manual; Cold Spring Harbor Laboratory Press: New York, 1989. [Google Scholar]
- MacNeil D. J.; Gewain K. M.; Ruby C. L.; Dezeny G.; Gibbons P. H.; MacNeil T. Gene 1992, 111, 61–68. 10.1016/0378-1119(92)90603-M. [DOI] [PubMed] [Google Scholar]
- Kieser T, Bibb M, Buttner M, Chater K H. D.. Practical Streptomyces Genetics; John Innes Foundation: Norwich, United Kingdom, 2000. [Google Scholar]
- Girard G.; Willemse J.; Zhu H.; Claessen D.; Bukarasam K.; Goodfellow M.; van Wezel G. P. Antonie van Leeuwenhoek 2014, 106, 365–380. 10.1007/s10482-014-0209-1. [DOI] [PubMed] [Google Scholar]
- Colson S.; Stephan J.; Hertrich T.; Saito A.; van Wezel G. P.; Titgemeyer F.; Rigali S. J. Mol. Microbiol. Biotechnol. 2006, 12, 60–66. 10.1159/000096460. [DOI] [PubMed] [Google Scholar]
- Wu C.; Zacchetti B.; Ram A. F. J.; van Wezel G. P.; Claessen D.; Choi Y. H. Sci. Rep. 2015, 5, 10868. 10.1038/srep10868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N.; Kuo C.-L.; Paniagua G.; van den Elst H.; Verdoes M.; Willems L. I.; van der Linden W. a; Ruben M.; van Genderen E.; Gubbens J.; van Wezel G. P.; Overkleeft H. S.; Florea B. I. Nat. Protoc. 2013, 8, 1155–1168. 10.1038/nprot.2013.065. [DOI] [PubMed] [Google Scholar]
- Wu C.; Medema M. H.; Läkamp R. M.; Zhang L.; Dorrestein P. C.; Choi Y. H.; van Wezel G. P. ACS Chem. Biol. 2016, 11, 478–490. 10.1021/acschembio.5b00801. [DOI] [PubMed] [Google Scholar]
- Wu C.; van Wezel G. P.; Choi Y. H. J. Antibiot. 2015, 68, 445–452. 10.1038/ja.2015.14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.