Abstract
Background
Brugada Syndrome is a disorder associated with characteristic ECG precordial ST elevation and predisposes afflicted patients to ventricular fibrillation and sudden cardiac death. Despite marked achievements in outlining the organ level pathophysiology of the disorder, the understanding of human cellular phenotype has lagged due to lack of adequate human cellular models of the disorder.
Methods and Results
We recruited two patients with Type 1 Brugada Syndrome (BrS) carrying two different SCN5A variants and two healthy controls. We generated induced pluripotent stem cells (iPSCs) from their skin fibroblasts by using integration-free Sendai virus. We utilized directed differentiation to create purified populations of iPSC-derived cardiomyocytes (iPSC-CMs). BrS iPSC-CMs showed reductions in inward Na+ current density and reduced maximal upstroke velocity of action potential compared to healthy control iPSC-CMs. Furthermore, BrS iPSC-CMs showed increased burden of triggered activity, abnormal Ca2+ transients, and beating interval variation. Correction of the causative variant by genome editing was performed and resultant iPSC-CMs showed resolution of triggered activity and abnormal Ca2+ transients. Gene expression profiling of iPSC-CMs showed clustering of BrS compared to controls. Furthermore, BrS iPSC-CM gene expression correlated with gene expression from BrS human cardiac tissue gene expression.
Conclusions
Patient-specific iPSC-CMs are able to recapitulate single cell phenotype features of BrS, including blunted inward sodium current, increased triggered activity and abnormal Ca2+ handling. This novel human cellular model creates future opportunities to further elucidate cellular disease mechanism and identify novel therapeutic targets.
Keywords: Induced pluripotent stem cells, cardiomyocytes, Brugada Syndrome, arrhythmia
INTRODUCTION
Brugada Syndrome is an arrhythmic disorder that predisposes afflicted patients to ventricular fibrillation and sudden cardiac death (1). The risk of sudden death varies by clinical profile, but exceeds 25% over 2 years in some populations (2). The diagnosis of Brugada Syndrome mainly relies on a constellation of clinical characteristics: ST elevation on surface electrocardiogram (ECG), history of ventricular tachycardia or fibrillation, personal or family history of sudden cardiac death, nocturnal agonal breathing, and syncope (3). Unfortunately, early diagnosis and risk stratification is difficult as the ECG pattern of Brugada Syndrome is not consistently expressed by affected patients, and the lethality associated with some of the other diagnostic criteria makes it difficult to make the diagnosis before significant morbidity has occurred (4). Furthermore, there are limited treatments for Brugada Syndrome. A significant barrier to improving the diagnostic and therapeutic approaches to the disorder is the limited understanding of cellular disease mechanism.
Tremendous advances in understanding organ level mechanisms of Brugada Syndrome have relied on pharmacologic manipulation of canine right ventricle (RV) myocardium. These studies hypothesized that loss of human voltage-gated Na+ channel (hNav1.5) function combined with repolarization current heterogeneity across epicardium and endocardium creates a substrate for phase 2 re-entry and ventricular arrhythmias (5). More recently, a competing hypothesis stipulated that the slow depolarization across the RV creates the abnormal substrate for arrhythmia (6). A third hypothesis suggested a role for abnormal neural crest cell migration contributing to abnormal RV outflow tract development, which creates an abnormal electrical substrate (7). Nonetheless, the understanding of the single cell perturbations of BrS lagged behind owing to the lack of reliable human cellular models.
There are 12 loci possibly associated with Brugada Syndrome, the most common of which is the Na+ voltage-gated channel alpha subunit 5 (SCN5A) resulting in hNav1.5 loss of function that leads to Type 1 Brugada Syndrome (BrS). Cellular mechanistic analyses have relied on non-human cellular models to model hNav1.5 loss of function. For example, animal models are limited by both phylogenic differences from human cardiomyocytes and inability to study the mechanism of individual causative variants (8). Heterologous expression models provide information on single channel function, but fail to recapitulate the complex milieu of the human cardiomyocyte in terms of ion channel function and electrical-mechanical coupling (9). Thus, there is a strong need for a human cardiac cellular model of BrS to further scientific investigations of the disorder.
In recent years, human induced pluripotent stem cell-derived cardiomyocytes (iPSC-CMs) have been used to model multiple cardiac disorders, including long QT syndrome (10–13), catecholaminergic polymorphic ventricular tachycardia (14), arrhythmogenic right ventricular dysplasia (15,16), Barth syndrome (17), Timothy syndrome (18), LEOPARD syndrome (19), familial dilated cardiomyopathy (20–23), familial hypertrophic cardiomyopathy (24), aldehyde dehydrogenase genetic polymorphism (25), and chemotherapy-induced cardiotoxicity (26). This platform has also been used to study the mechanism of disorders such as ion channel defects associated with primary channelopathies. In addition to mirroring the electrical and mechanical features of primary cardiac disorders, iPSC-CMs recapitulated therapeutic and toxic drug responses (9–11,20).
In this study, we recruited two genetically distinct patients with BrS to generate iPSC-CMs and examine mechanism of BrS at the cellular level. We identified that reduced inward sodium current (INa), increased triggered activity, and abnormal Ca2+ transients are single cell phenotype features of BrS. Furthermore, we corrected the BrS causative SCN5A variant to wild type using genome editing and demonstrated improvement in the pro-arrhythmic phenotype, validating the association of the variant with the cellular phenotype. Collectively, these experiments better elucidate cellular mechanisms of BrS.
METHODS
Generation of iPSC lines
Somatic reprogramming was used to generate iPSC lines from skin fibroblasts of 2 patients with BrS and 2 healthy subjects using the Sendai virus reprogramming protocol described previously (27). All recruitment and consenting procedures were done under IRB approved protocol. For all analyses, both control lines were used for comparison.
Culture and maintenance of iPSCs
Control and BrS iPSCs were maintained in chemically defined medium Essential 8 (E8 medium) (Life Technologies) on Matrigel-coated (BD Bioscience, San Jose, CA) plates at 37°C with 5% (vol/vol) CO2.
Differentiation of iPSC-CMs
Control and BrS iPSCs were differentiated into iPSC-CMs using a 2D monolayer differentiation protocol and were maintained in a 5% CO2/air environment as previously published (28). Briefly, iPSC colonies were dissociated with 0.5 mM EDTA (Gibco) into single-cell suspension and re-suspended in E8 media containing 10 μM Rho-associated protein kinase inhibitor (Sigma). Approximately 100,000 cells were re-plated into Matrigel-coated 6-well plates. iPSCs were next cultured to 85% cell confluence, and then treated for 2 days with 6 μM CHIR99021 (Selleck Chemicals) in RPMI+B27 supplement without insulin to activate WNT signaling and induce mesodermal differentiation. On day 2, cells were placed on RPMI+B27 without insulin and CHIR99021 (28). On days 3–4, cells were treated with 5 μM IWR-1 (Sigma) to inhibit WNT pathway signaling and induce cardiogenesis. On days 5–6, cells were removed from IWR-1 treatment and placed on RPMI+B27 without insulin. From day 7 onwards, cells were placed on RPMI+B27 with insulin until beating was observed. At this point, cells were glucose-starved for 3 days with RPMI+B27 with insulin to purify iPSC-CMs. Following purification, cells were cultured in RPMI+B27 with insulin. When re-plating iPSC-CMs for downstream use, cells were dissociated with 0.25% trypsin-EDTA (Life Technologies) into a single-cell suspension and seeded on Matrigel-coated plates.
Genetic screening and analysis
BrS iPSCs were cultured on Matrigel-coated 6-well plates with E8 media and harvested at 80–90% confluence for subsequent analysis. Genomic DNA was extracted using a DNeasy commercial DNA isolation kit (Qiagen, Valencia, CA). Polymerase chain reaction (PCR) was carried out on S1000 Thermal Cycler (Biorad, Hercules, CA). Three single nucleotide polymorphism (SNP) loci within SCN5A gene (2053G>A/R620H, 2626G>A/R811H, 4190ΔA/1397Δ; reference sequence: NM_198056.2) were amplified and analyzed by direct sequencing, and then confirmed by sub-cloning. The primer sequences for SCN5A are listed in Supplemental Table 1 (reference sequence: NC_000003.12).
Immunofluorescence staining
Immunofluorescence was performed using appropriate primary antibodies and AlexaFluor-conjugated secondary antibodies (Invitrogen, Carlsbad, CA) as described by the manufacturer’s protocol. The primary antibodies used in this study were anti-SOX2 (Biolegend, San Diego, CA), anti-NANOG (Santa Cruz, CA), anti-cardiac Troponin T (cTnT) (Abcam, ab10214), anti-sarcomeric α-actinin (Abcam, ab90776), anti-hNav1.5 (Alomone Labs, ASC-005), and anti-SCN7A (Sigma HPA004879).
Karyotype analysis
BrS lines were trypsinized for 5 min and centrifuged at 300 g for 3 min. iPSC pellets were resuspended in 200 μl of PBS and DNA was isolated using the DNeasy blood and tissue kit (Qiagen, Valencia, CA). DNA was submitted to the Stanford Functional Genomics Facility for SNP karyotyping using the HuCytoSNP-12 chip (Illumina). CNV and SNP visualization was performed using KaryoStudio v1.4 (Illumina).
Ca2+ imaging
Dissociated control and BrS iPSC-CMs were reseeded in Matrigel-coated 8-well Lab Tek II chambers (Nalge Nuc International). Cells were recovered for 2 days and were loaded with 5 μM Fluo-4 AM with 0.02% Pluronic F-127 (Molecular Probes) in Tyrode’s solution for 15 min at 37°C, and were washed with Tyrode’s solution afterwards. Ca2+ imaging was conducted using a Zeiss LSM 510Meta confocal microscope (Carl Zeiss AG, Göttingen, Germany). Spontaneous Ca2+ transients of single beating iPSC-CMs were obtained using a time-lapse line scanning recording mode (512 pixels * 1920 lines) under 40× objective (Plan Apochromat, 0.95 NA) at 37°C, and the raw data was analyzed using customized Interactive Digital Language (IDL) script. Ca2+ signal was normalized to the intracellular basal line (F0), and transient amplitude was expressed as ΔF/F0.
RNA Sequencing
Total RNA was extracted from control or BrS iPSC-CMs using the RNeasy Mini kit (Qiagen, Valencia, CA) and DNase treated using a RNase-Free DNase kit (Qiagen). 5 ng of RNA was used as input for cDNA synthesis and amplification using the Ovation RNA-Seq System V2 (NuGEN). Illumina barcodes were ligated to cDNAs using the NEBNext® DNA Library Prep Master Mix Set for Illumina®. The ligated libraries were size selected for an average insert size of 250 bp by agarose gel excision and extraction. The ligated libraries were then amplified by PCR. Four prepared libraries were pooled and sequenced on one HiSeq2000 (Illumina) lane to obtain an average of 30 million paired-end reads per sample. Reads were mapped to the Ensembl GRCh37 reference genome using Tophat 2.1. Junction BED files generated from Tophat were used to generate an exon coordinate reference file using AltAnalyze 2.0.8 and exon counts were quantified using the reference BED files from Tophat generated BAM files. Finally, AltAnalyze was run using the default settings to calculate RPKM values per Ensembl ID. Differentially expressed genes between control and BrS samples were calculated using an empirical Bayes moderated t-test p< 0.05 with a 2-fold expression difference. These genes were imported into Ingenuity pathway analysis (Ingenuity, Redwood City, CA) and the top calls for canonical pathway analysis was reported.
Quantitative real-time PCR (qPCR)
Total mRNAs were isolated from control and BrS iPSC-CMs using the Qiagen RNeasy Mini kit. 1 μg of RNA was used to synthesize cDNA using the Superscript II First-Strand cDNA synthesis kit (Invitrogen). 0.25 μL of the reaction was used to quantify gene expression by qPCR using TaqMan Universal PCR Master Mix. Expression values were normalized to the average expression of housekeeping gene 18s.
Patch clamp recordings of iPSC-CMs
Details of patch clamp recording protocol are described in the Supplemental Methods section. In brief, whole-cell patch clamp recordings from day 40–60 iPSC-CMs were performed using an EPC-10 patch clamp amplifier (HEKA, Lambrecht, Germany) attached to a RC-26C recording chamber (Warner) and mounted onto the stage of an inverted microscope (Nikon, Tokyo, Japan). Contracting monolayer of iPSC-CMs were mechanically isolated, enzymatically dispersed into single cells, and attached to Matrigel-coated glass coverslips (Warner, USA). Current-clamp mode recordings were conducted to record action potentials (AP) from iPSC-CMs at 36 to 37°C. Voltage-clamp mode recordings were conducted to isolate Na+ current from iPSC-CMs at room temperature. Data were acquired using PatchMaster software (HEKA, Germany) and digitized at 1.0 kHz.
CRISPR/Cas9-mediated genome editing
We utilized clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 (CRISPR-associated protein 9) system to perform genome editing in BrS(p2) and modify the suspected causative SCN5A variant to a wild type sequence. Details of the genome editing methods are available in the Supplemental Methods section. We validated successful genome editing by PCR and bidirectional direct sequencing.
Statistical analysis
Statistical significance was determined by unpaired Student’s t-test (two-tail) using Prism 5. *P< 0.05 was considered statistically significant, ** indicates P< 0.01 or *** indicates P< 0.001 when compared to control iPSC-CMs. #P< 0.05 was considered statistically significant, ## indicates P< 0.01 or ### indicates P< 0.001 when compared to BrS(p2) -genome edited (BrS(p2)-GE) iPSC-CMs.
RESULTS
Clinical characteristics of BrS patients
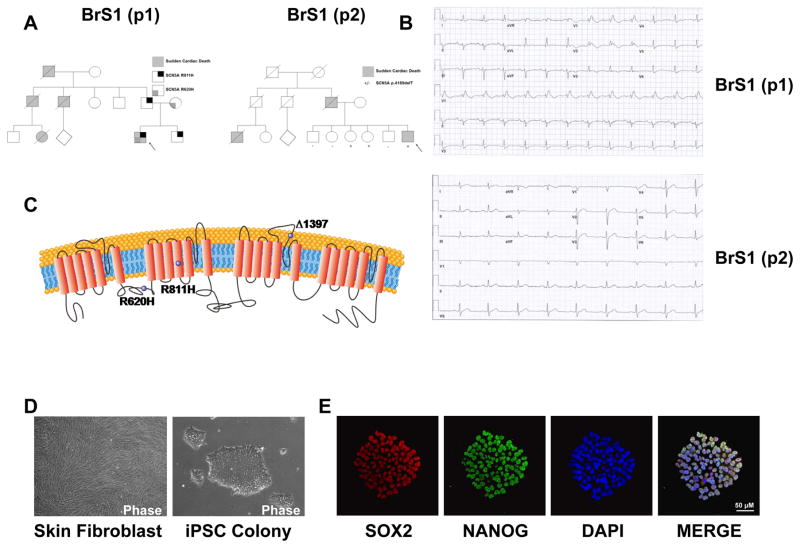
We recruited 4 patients for this study: two patients carrying a definitive diagnosis of BrS and two healthy controls (Figure 1A). Patient 1 (BrS(p1)) is a 44 year old male who presented with unstable ventricular tachycardia after multiple episodes of recurrent syncope. The patient’s resting ECG was characteristic of the Brugada ECG pattern (Figure 1B, Upper Panel). Furthermore, the patient had a family history of sudden death in two paternal uncles and a female cousin. Genetic testing revealed a double missense mutation (R620H and R811H) in SCN5A gene domain IIS4, with R811H known to be critical in voltage-dependent gating (Figure 1C, Supplemental Figure 1A, and Supplemental Table 2) (29).
Figure 1. Generation and characterization of BrS iPSC lines and iPSC-CMs.
A. The pedigrees of the 2 patients with the Type 1 Brugada Syndrome (BrS) both share a family history of sudden cardiac death. B. ECG from BrS patient 1 (BrS(p1)) shows a classic BrS ECG pattern. ECG from BrS patient 2 (BrS(p2)) shows non-specific intraventricular conduction delay. C. Schematic representation of SCN5A channel protein. Purple spheres indicate the genetic variants. D. Representative microscopy photographs of skin fibroblasts and iPSC colony derived from BrS. E. Representative immunostaining of pluripotency markers SOX2 (red) and NANOG (green) in iPSC clone derived from BrS. DAPI indicates the nuclear staining (blue). BrS(p1): Type 1 Brugada Syndrome patient 1. BrS(p2): Type 1 Brugada Syndrome patient 2
Patient 2 (BrS(p2)) is a 53 year old male with recurrent syncope presenting as seizure-like activity. The patient’s father and two paternal cousins died suddenly. Although the patient’s ECG was not typical for the Brugada ECG pattern (Figure 1B, Lower Panel), a procainamide challenge provoked classic changes of the Brugada ECG pattern (30). Subsequently, genetic testing identified one-base pair deletion mutation in the SCN5A, which causes a frame shift and truncates the hNav1.5 starting at amino acid 1397, where truncation in the domain III pore loop would be expected to produce a non-functional hNav1.5 (Figure 1C, Supplemental Figure 1B, and Supplemental Table 2). Our 2 healthy subjects were 17 (CON(p1)) and 22 (CON(p2)) year old males who had normal ECG and no personal or family history of syncope or sudden cardiac death.
Generation and characterization of control and BrS iPSC lines
Skin biopsies were collected from all subjects and fibroblasts were reprogrammed into iPSCs as described previously (27). Subsequently, typical iPSC colonies appeared and were expanded by week 4 from the reprogramming date (Figure 1D). The generated BrS iPSC lines showed normal karyotype (Supplemental Figure 2). We characterized the pluripotency of the generated iPSC lines using immunostaining of pluripotency markers such as SOX2 and NANOG (Figure 1E). At least 2 colonies were generated from each patient and healthy control subject, and were chosen for downstream characterization (Supplemental Table 2).
Differentiation of control and BrS iPSCs into cardiomyocytes
We cultured and passaged the iPSCs until at least passage 20, and then differentiated them into iPSC-CMs using a small molecule-based monolayer method as previously described (28). Two to three weeks after the induction, spontaneous beatings of the monolayer iPSC-CMs were observed in a dish. We then dissociated the monolayer iPSC-CMs into single cells, and characterized them using immunostaining for cardiac-specific markers, cardiac Troponin T (cTnT) and sarcomeric α-Actinin (Supplemental Figure 3). We detected no significant difference in cell morphology between control and BrS iPSC-CMs.
BrS iPSC-CMs show abnormal action potential profiles
Clinically, patients with BrS suffered from severe arrhythmias and exhibited abnormal ECG. To investigate if the arrhythmic predisposition is exhibited at the single-cell level, we used patch clamp recording to compare the AP of control and BrS iPSC-CMs. Ventricular-like cardiomyocytes were studied from both control and BrS iPSC-CMs, and AP parameters such as maximal diastolic potential (MDP), overshoot, action potential amplitude (APA), action potential duration at 90% repolarization (APD90), beating rate, and maximal upstroke velocity (Vmax) were quantified (Supplemental Table 3). Notably, control iPSC-CMs derived from 2 different control subjects exhibited normal AP pattern (Figure 2A). However, BrS iPSC-CMs exhibited abnormal AP profile manifesting as two distinct phenotypes of triggered activity: the more common phenotype (7.1% in control iPSC-CMs; 39.6% in BrS(p1) iPSC-CMs; 34.5% in BrS(p2) iPSC-CMs) was of a closely coupled single triggered beat (Figure 2B), and the other rare phenotype (0% in control iPSC-CMs; 5.6% in BrS(p1) iPSC-CMs; 6.8% in BrS(p2) iPSC-CMs) displayed sustained triggered activity (Figure 2C). Furthermore, BrS iPSC-CMs exhibited increased peak-peak interval variability (49.4 ± 5.5 ms, 2 control lines; 380.4 ± 96.2 ms, 2 BrS(p1) lines; 195.5 ± 60.3, 2 ms, 2 BrS(p2) lines) as well as significantly slower depolarization rate (Vmax) (15.6 ± 0.8 V/s, 2 control lines; 11.1 ± 1.0 V/s, 2 BrS(p1) lines; 7.2 ± 1.1 V/s, 2 BrS(p2) lines) compared to control cells (Figure 2D and E). Taken together, these results demonstrate that BrS iPSC-CMs exhibit distinctly abnormal AP phenotype compared to control iPSC-CMs.
Figure 2. BrS iPSC-CMs show abnormal action potential profiles.
A. Representative action potential traces of ventricular-like iPSC-CMs derived from CON(p1) (Upper Panel) and CON(p2) (Lower Panel), respectively. Dash lines indicate 0 mV. B. Representative action potential traces of ventricular-like iPSC-CMs derived from BrS(p1) (Upper Panel) and BrS(p2) (Lower Panel), showing triggered beats indicated by red arrows, respectively. Dash lines indicate 0 mV. C. Representative action potential traces of ventricular-like iPSC-CMs showing sustained triggered activity in BrS(p1) (Upper Panel) and BrS(p2) (Lower Panel), respectively, as indicated by red arrows. Dash lines indicate 0 mV. D. Bar graph comparison shows a higher incidence of peak-peak interval variability of action potentials in BrS iPSC-CMs (n = 60, 2 BrS(p1) lines; n = 22, 2 BrS(p2) lines) compared to control iPSC-CMs (n = 61, 2 control subject lines). E. Bar graph comparison of maximal upstroke velocity (Vmax) in ventricular-like iPSC-CMs between control subjects (n = 38, 2 control subject lines) and BrS patients (n = 50, 2 BrS(p1) lines; n = 7, 2 BrS(p2) lines) showing significantly reduced Vmax in BrS lines. All data were shown as mean ± s.e.m., and the statistical significance is indicated as **P< 0.01 or ***P< 0.001. CON(p1): Control subject 1. CON(p2): Control subject 2.
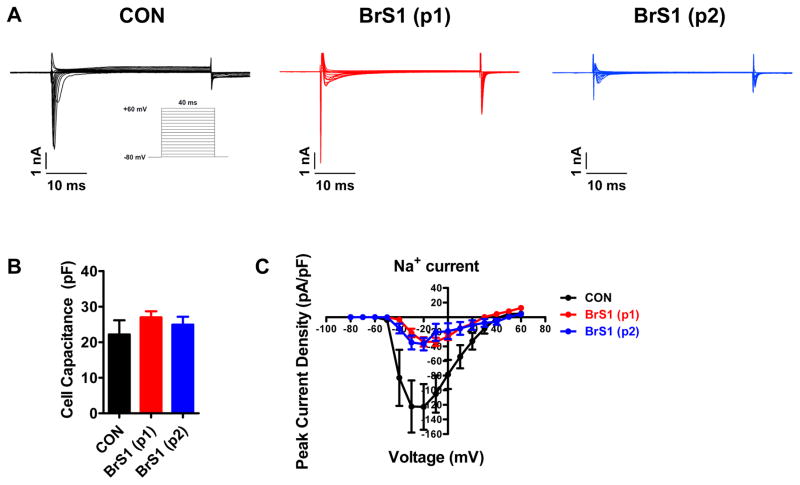
BrS iPSC-CMs show decreased INa
We next focused on measuring INa given the reported Na+ channel loss of function in BrS. We used voltage clamp recordings to measure INa. INa amplitude was dramatically reduced in BrS iPSC-CMs compared to control iPSC-CMs (Figure 3A). No significant differences in cell capacitance were observed between control and BrS iPSC-CMs, indicating that the differences in INa were not due to differences in cell size (Figure 3B). Control iPSC-CMs showed a peak INa density of −122.8 ± 31.3 pA/pF at −20 mV, whereas BrS iPSC-CMs demonstrated significantly deceased peak INa density (−33.7 ± 6.3 pA/pF and −36.8 ± 8.6 pA/pF in the 2 BrS patients, respectively), which is consistent with loss of Na+ channel function in BrS (Figure 3C and Supplemental Table 4).
Figure 3. BrS iPSC-CMs show decreased Na+ current density.
A. Representative Na+ current traces recorded from control (left panel, black), BrS(p1) (middle panel, red), and BrS(p2) (right panel, blue) iPSC-CMs. BrS iPSC-CMs show significantly reduced Na+ current. Inset is the voltage protocol used for isolating Na+ currents. B. Bar graph comparison of cell capacitance shows no significant difference in cell size between control and BrS iPSC-CMs. C. Comparison of current-voltage relationship (IV curve) between control and BrS iPSC-CMs. There is significantly decreased Na+ current density in BrS iPSC-CMs compared to control iPSC-CMs. n = 10–25 cells in 2 lines for each group.
BrS iPSC-CMs show suppressed membrane expression of hNav1.5
We subsequently sought to investigate whether the reduced INa is due to reduced membrane expression of hNav1.5. To test this notion, we stained cardiomyocytes with hNav1.5 antibody to investigate the expression level of Na+ channel protein in control and BrS iPSC-CMs. The antibody was co-stained with Caveolin 3 (Cav3), a cell membrane marker, to ensure localization to the cell membrane. BrS iPSC-CMs exhibited significantly lower membrane expression of hNav1.5 compared to control iPSC-CMs (Supplemental Figure 4). This supports the hypothesis that reduced membrane expression of hNav1.5 is a feature of BrS. To test if upregulation of hNav2.1 is as a compensatory mechanism of sodium channel loss of function, cells were stained with SCN7A antibody, which did not show a significant change in expression compared to control cells (Supplemental Figure 5).
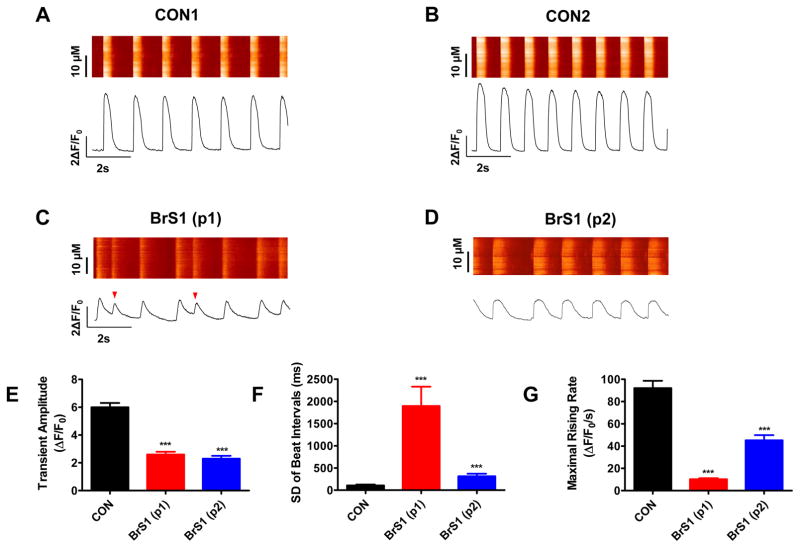
BrS iPSC-CMs show irregular Ca2+ handling
Ca2+ is an important second messenger in the heart and plays a critical role in regulating the electrical-mechanical coupling of cardiac cells. We hypothesized that the triggered activity profile observed in AP recordings will be reflected in the BrS cellular Ca2+ handling phenotype. Accordingly, we measured Ca2+ transients from control and BrS iPSC-CMs using confocal fluorescence microscopy (Figure 4A–D). Compared to control iPSC-CMs cells (5.96 ± 0.26 ΔF/F0), we observed reduced Ca2+ transient amplitudes in both BrS(p1) (2.56 ± 0.21 ΔF/F0, 57.0 ± 3.5% relative reduction) and BrS(p2) (2.12 ± 0.18 ΔF/F0, 64.4% ± 3.0% relative reduction) iPSC-CMs (Figure 4E, Supplemental Table 5). Moreover, we also observed that the variation of beating intervals were greatly increased from 108.3 ± 20.4 ms in control iPSC-CMs to 1896.7 ± 436.4 ms in BrS(p1) and 316.2 ± 58.5 ms in BrS(p2) iPSC-CMs, indicating uneven beating pattern in diseased cells (Figure 4F, Supplemental Table 5). Furthermore, maximal rising rate of [Ca]i was significantly decreased by 87.9 ± 1.1% in BrS(p1) iPSC-CMs and 50.3 ± 5.3% in BrS(p2) iPSC-CM compared to control iPSC-CMs (Figure 4G, Supplemental Table 5). Lastly, Ca2+ transient recordings displayed closely coupled beats that mirror the triggered activity seen on AP recording. These observations in Ca2+ imaging are consistent with increased beat-beat interval variability and slower depolarization of AP in the patch-clamp recording of these diseased cells. Collectively, our data demonstrates that irregular Ca2+ handling in BrS iPSC-CMs is a feature of single cardiomyocytes from BrS patients.
Figure 4. BrS iPSC-CMs show irregular Ca2+ handling.
A–D. Representative Ca2+ transient traces of iPSC-CMs derived from CON(p1), CON(p2), BrS(p1), and BrS(p2), respectively. The BrS iPSC-CMs show abnormal Ca2+ transient pattern compared to control cells (as indicated by red arrows). E. Bar graph comparison of Ca2+ transient amplitude of iPSC-CMs between control subjects and BrS patients confirm BrS iPSC-CMs indeed exhibit a lower Ca2+ transient amplitude. F. Bar graph comparison of standard deviation (SD) of beat intervals of iPSC-CMs between control subjects and BrS patients. BrS iPSC-CMs exhibit greater beat-beat interval variability, which likely contributes to the mechanism of arrhythmia seen in these cells. G. Bar graph comparison of maximal rising rate of iPSC-CMs between control subjects and BrS(p1) patients shows reduced maximal rising rate in BrS cells. n = 34–53 cells in 2 lines for each group. Unless specified, all data were shown as mean ± s.e.m and statistical significance is indicated as ***P< 0.001.
BrS iPSC-CMs show differential ion-channel gene expression profile
To investigate if there is a BrS-specific gene expression profile at the cellular level, we performed RNA sequencing to identify gene expression changes between 2 control and 2 BrS cardiomyocyte samples. Principal Components Analysis (PCA) revealed control samples clustered together while BrS iPSC-CMs samples clustered together (Supplemental Figure 6A). In using genes differentially expressed between control and BrS samples, Ingenuity Pathway Analysis (IPA) reported a marked down-regulation of genes implicated in Ca2+ signaling, cardiac hypertrophy, AMP activated kinase, and β-adrenergic signaling pathways (Supplemental Figure 6B).
A previous study reported that the expression of ion channel genes may be different in heart tissues between healthy subjects and BrS patients (31). We therefore aimed to investigate if BrS iPSC-CMs would emulate the ion channel gene expression seen in human cardiac tissue from BrS patients. We used our RNA sequencing to screen for ion channel genes reported to be affected in Brugada Syndrome patients’ tissue and used qPCR to validate differences observed in our RNA-seq dataset (Supplemental Table 6). Interestingly, BrS iPSC-CMs showed reduced gene expression of SCN5A similar to findings from human cardiac tissue (Supplemental Figure 7A). In addition, both KCND3 associated with Kv4.3 and transient outward K+ current (Ito) as well as KCNJ2 associated with Kir2.1 and inward rectifying K+ current (IK1) were markedly down-regulated compared to controls (Supplemental Figure 7B–C).
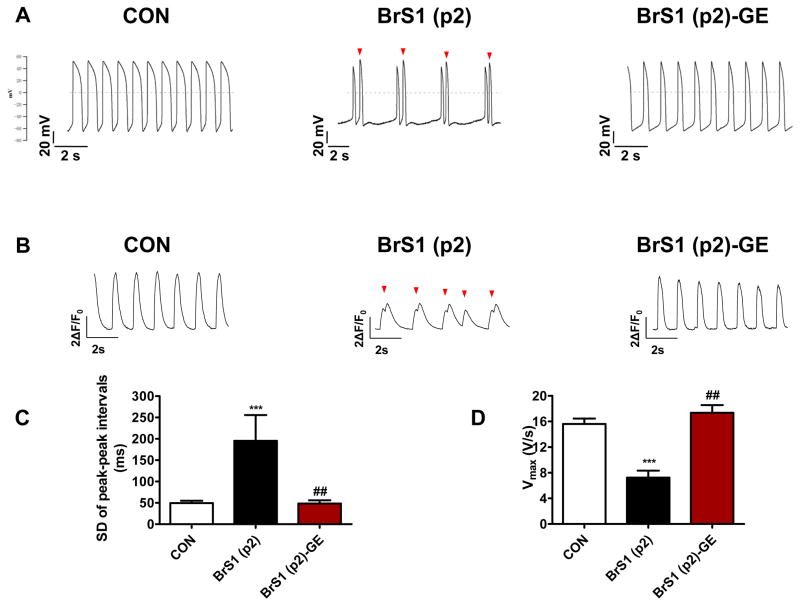
Genome editing of BrS iPSC-CMs
We next utilized CRISPR/Cas9-mediated genome editing to modify SCN5A variant implicated in BrS(p2) to validate the pathogenicity of the suspected causative variant (Supplemental Figure 8–12). The resultant BrS(p2) -genome edited (BrS(p2)-GE) iPSC-CMs had a correction of the causative SCN5A variant (rs397514446). The BrS(p2)-GE line showed marked reduction in single beat triggered activity and no sustained triggered activity, such that AP recordings resembled those from control lines (Figure 5A). Moreover, AP recording of BrS(p2)-GE demonstrated an improvement in AP maximal upstroke velocity (7.2 ± 1.1 V/s in BrS(p2) iPSC-CMs and 17.4 ± 1.2 V/s in BrS(p2)-GE iPSC-CMs) and peak-peak interval variability (313.3 ± 59.1 ms in BrS(p2) iPSC-CMs and 190.7 ± 24.6 ms in BrS(p2)-GE iPSC-CMs) parameters compared to BrS(p2) (Figure 5C–D, Supplemental Table 3). Furthermore, Ca2+ imaging validated this improved electrical profile (Figure 5B, Supplemental Table 5). Lastly, BrS(p2)-GE iPSC-CMs exhibited significantly increased hNav1.5 membrane expression (Supplemental Figure 4) compared to BrS iPSC-CMs. These findings were consistent with the SCN5A variant being associated with the abnormal electrical profile observed.
Figure 5. Genome editing of BrS iPSC-CMs.
A. Representative action potential traces of ventricular-like myocytes derived from control iPSC line (Left Panel), BrS(p2) iPSC line (Middle Panel), and BrS(p2)-GE iPSC line (Right Panel), respectively. Dash lines indicate 0 mV, red arrows indicate triggered beats. B. Representative Ca2+ transient traces of cardiomyocytes derived from control iPSC line (Left Panel), BrS(p2) iPSC line (Middle Panel), and BrS(p2)-GE iPSC line (Right Panel), respectively. Red arrow indicates triggered beat. C. Bar graph comparison of SD of peak-peak interval variability of action potentials between control, BrS(p2) and BrS(p2) -GE iPSC-CMs. The significant increase in interval variability in BrS(p2) iPSC-CMs compared to control iPSC-CMs is reduced in BrS(p2) -GE iPSC-CMs; n=22–60 cells in 2 lines for each group. D. Bar graph comparison of maximal upstroke velocity (Vmax) in control, BrS(p2) and BrS(p2)-GE iPSC-CMs demonstrating significantly reduced Vmax is BrS(p2) iPSC-CMs compared to control iPSC-CMs and improvement in Vmax after genome editing of pathogenic SCN5A variant in BrS(p2)-GE; n = 7–50 cells in 2 lines for each group. Unless specified, all data were shown as mean + s.e.m. and the statistical significance is indicated as *P< 0.05, **P< 0.01, or ***P< 0.001 when compared to control iPSC-CMs. The statistical significance is indicated as #P< 0.05, ##P< 0.01 or ###P< 0.001 when compared to BrS(p2) iPSC-CMs. BrS(p2)-GE: Type 1 Brugada Syndrome patient 2-Genome Edited.
DISCUSSION
We demonstrate that iPSC-CMs derived from BrS patients show a consistent phenotype of reduced INa, reduced AP Vmax, increased inter-beat variability, increased triggered activity, and abnormal Ca2+ transients. Using genome editing, we validate that these abnormalities are related to SCN5A variant. This model bypasses phylogenic ion channel differences of animal models and the inability of cellular transfection models to recapitulate human cardiomyocyte physiology (32). Our model provides a comprehensive analysis of BrS cellular cardiomyocyte phenotype that enhances our understanding of the mechanism of disease.
Single cell predisposition to arrhythmia
Single cell AP recordings of BrS iPSC-CMs demonstrated triggered activity manifesting as a single triggered beat or less commonly as a sustained series of triggered activity. The abnormal BrS AP profile was explained by a reduction in INa and upstroke velocity, a phenotype consistent with the loss of hNav1.5 function predicted by SCN5A variants utilizing heterologous expression models (29,33–35). The reduction in INa is similar to results by Davis et al. who studied human iPSC-CMs from subjects with an overlap syndrome with Type 3 Long QT and BrS features and demonstrated reduced INa current density and upstroke velocity (36). Reduced INa was also observed by Cerrone et al. in iPSC-CMs from BrS patients with underlying PKP2 variants (37). Additionally, we observed reduced membrane expression of hNav1.5 in BrS iPSC-CMs, which has been proposed as a cellular mechanism of BrS sodium channel loss of function (38). In summary, these findings support that BrS is a channelopathy with a single cell electrical predisposition to arrhythmia contributing to the arrhythmias observed in patients.
It is likely that there are multiple mechanisms contributing to ventricular arrhythmias seen in patients with BrS. These mechanisms include organ level such as the Ito heterogeneity across RV epicardium to endocardium, and single cell level such as triggered activity and increased inter-beat variability noted in our results. These mechanisms may be complementary, namely triggered activity may provoke arrhythmia in an abnormal cardiac substrate created by depolarization (6) or repolarization (5) heterogeneity across the myocardium. Our findings highlight the ability of iPSC-CMs to provide insight into the single cell pro-arrhythmic phenotype of SCN5A variants causing BrS.
Abnormal Ca2+Transients
Given that Ca2+ handling is closely coupled to electrical activation and correlates with AP profile, we aimed to study Ca2+ transients to validate the observed triggered activity profile in BrS iPSC-CMs. We observed reduced Ca2+ transient amplitude and reduced maximal rising rate of [Ca]i in BrS iPSC-CMs. The observed changes may be caused by the slower depolarization resulting in decreased activation of L-type Ca2+ channels, and the decreased Ca2+ influx may result in insufficient triggered Ca2+ release from the sarcoplasmic reticulum. Additionally, the cells exhibited increased inter-beat variability, a phenomenon that is known to predispose to the development of arrhythmia at the tissue or organ level. Overall, these findings define abnormal Ca2+ transients as a feature of BrS cellular phenotype and validate the triggered activity phenotype observed on AP recordings.
Genome editing of SCN5A
Genome editing takes advantage of genome nucleases to replace DNA sequences to modify a given locus of interest. This can be used in an iPSC-CM model to delineate the phenotype effect of a given cardiac variant and help identify mechanisms implicated in BrS. We performed CRISPR/Cas9 genome editing to correct the proposed causative SCN5A variant in BrS(p2) with a wild type variant to examine the precise phenotype impact of the variant. Genome edited BrS(p2) (BrS(p2)-GE) iPSC-CMs exhibited significant improvement in abnormal electrical phenotype observed in BrS(p2) iPSC-CMs. Specifically, maximal upstroke velocity and inter-beat variability both improved, which may decrease the propensity towards pro-arrhythmia. Furthermore, BrS(p2)-GE iPSC-CMs exhibited an amelioration of the pro-arrhythmic phenotype on AP recordings and Ca2+ imaging. These findings support the association between the pathogenic SCN5A variant and the pro-arrhythmic triggered activity phenotype observed. This data is in line with other investigations that support the pathogenicity of this variant (33). Our approach of using genome editing methods in iPSC-CMs can be broadly used to test genotype-phenotype associations in BrS or other channelopathies (32).
Gene expression changes in Brugada Syndrome
Changes in gene expression profile in response to changes in ion channel currents are well recognized in other channelopathies and thought to be a compensatory cellular response (39). We observed a closer homology in overall gene expression profile between the two BrS iPSC-CMs compared to the two control lines, suggesting disease specific gene expression changes. We subsequently compared our BrS gene expression profile with published reports of human cardiac tissue gene expression and noted commonalities. We observed a relative reduction in SCN5A gene expression in BrS iPSC-CMs compared to controls, which has been documented in both human and mouse tissue gene expressions studies (31,40). Furthermore, we noted a marked reduction in thlevels of KCND3, which is congruent with human tissue studies from BrS patients that demonstrated marked down regulation of the Kv4.3 associated gene (31). The reduced Kv4.3 channel expression may contribute to reduced Ito current, causing an exaggerated Ito gradient from epi- to endocardial right ventricular surface as described previously (5). The above gene expression profile correlates with BrS human cardiac tissue, validating the utility of iPSC-CMs for BrS studies.
An observation not previously described in BrS is the markedly reduced expression of KCNJ2, which encodes Kir2.1 that is largely responsible for IK1 and is important in terminal repolarization and stabilization of the resting membrane potential. Reduced IK1 would allow a smaller amount of INa to trigger an AP, which might be a compensatory adaptation to reduced Na+ channel function. However, this might also make terminal repolarization less robust, opening the door for triggered activity and dispersion of repolarization.
Potential Limitations
One limitation to our study is that iPSC-CMs lack chamber specific (right versus left) or layer specific (epicardium, myocardium, or endocardium) features, and even if present these cells remain a mixture (16,41). This is in contrast to human and murine cardiac tissue studies that utilized endocardial RV or RVOT tissue, making them more likely to identify differences in phenotype or gene expression (31,40,42). Thus, single cell phenotypes or changes that are specific to right ventricle or layer-specific cardiomyocytes may not be captured by our model. Additionally, despite the significant insight into single cell pathophysiology, it is important to note that iPSC-CMs are not a whole organ model and is not yet able to recapitulate pathophysiology of specific heart chambers or cardiac layers. Furthermore, the immature state of iPSC-CMs compared to adult ventricular cells with respect to ion channel expression and Ca2+ handling properties is a known limitation of the iPSC-CMs model (43). For example, iPSC-CMs express low levels of functional Ito current compared to adult cardiomyocytes making it difficult to measure the K+ current in our model.
CONCLUSION
In summary, we identify BrS iPSC-CM phenotypic features that include decreased INa, variability in beating intervals, abnormal Ca2+ transients, and increased triggered activity. We highlight the substantial potential of genome editing methods to examine cellular mechanisms of BrS in a model that can be applied to other channelopathies. Improved knowledge of the cellular abnormalities associated with BrS can enhance the understanding of disease mechanism and identify new therapeutic targets. This supports a role for iPSC-CMs in future studies examining the single cell physiology of inherited arrhythmic disorders with complex genetics and disease mechanism such as BrS.
Supplementary Material
PERSPECTIVES.
Competency in Clinical Knowledge: Brugada Syndrome is commonly associated with genetic variants in SCN5A gene that result in reduced inward sodium current and single cell proarrhythmic activity.
Translational outlook: Further studies using induced pluripotent stem cell-derived cardiomyocytes can provide insight into single cell mechanisms of Brugada Syndrome and accelerate drug discovery that can rescue the disease phenotype.
Acknowledgments
We thank Ms. Joan Campagna for her assistance with patient recruitment and Drs. Yongming Wang and Fangjun Jia for their assistance with experiments. We gratefully acknowledge funding support from NIH 5T32HL094274-04 (KS); American Heart Association 13EIA14420025, Steven M. Gooter Foundation (JCW), National Institutes of Health (NIH) R01 HL113006, NIH R01 HL126527, NIH R01 HL130020, and NIH R01 HL128170 (JCW).
ABBREVIATIONS
- AP
action potential
- BrS
type 1 Brugada Syndrome
- BrS(p1)
Brugada Syndrome patient 1
- BrS(p2)
Brugada Syndrome patient 2
- CON(p1)
Control subject 1
- CON(p2)
Control subject 2
- ECG
electrocardiogram
- hNav1.5
human voltage gated sodium channel
- INa
inward sodium current
- iPSC
induced pluripotent stem cells
- iPSC-CMs
induced pluripotent stem cell-derived cardiomyocytes
- Vmax
maximal upstroke velocity
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicts of interest to disclose.
References
- 1.Brugada P, Brugada J. Right bundle branch block, persistent ST segment elevation and sudden cardiac death: a distinct clinical and electrocardiographic syndrome. A multicenter report. J Am Coll Cardiol. 1992;20:1391–6. doi: 10.1016/0735-1097(92)90253-j. [DOI] [PubMed] [Google Scholar]
- 2.Brugada J, Brugada R, Brugada P. Determinants of sudden cardiac death in individuals with the electrocardiographic pattern of Brugada syndrome and no previous cardiac arrest. Circulation. 2003;108:3092–6. doi: 10.1161/01.CIR.0000104568.13957.4F. [DOI] [PubMed] [Google Scholar]
- 3.Antzelevitch C, Brugada P, Borggrefe M, et al. Brugada syndrome: report of the second consensus conference: endorsed by the Heart Rhythm Society and the European Heart Rhythm Association. Circulation. 2005;111:659–70. doi: 10.1161/01.CIR.0000152479.54298.51. [DOI] [PubMed] [Google Scholar]
- 4.Veltmann C, Schimpf R, Echternach C, et al. A prospective study on spontaneous fluctuations between diagnostic and non-diagnostic ECGs in Brugada syndrome: implications for correct phenotyping and risk stratification. Eur Heart J. 2006;27:2544–52. doi: 10.1093/eurheartj/ehl205. [DOI] [PubMed] [Google Scholar]
- 5.Yan GX, Antzelevitch C. Cellular basis for the Brugada syndrome and other mechanisms of arrhythmogenesis associated with ST-segment elevation. Circulation. 1999;100:1660–6. doi: 10.1161/01.cir.100.15.1660. [DOI] [PubMed] [Google Scholar]
- 6.Meregalli PG, Wilde AA, Tan HL. Pathophysiological mechanisms of Brugada syndrome: depolarization disorder, repolarization disorder, or more? Cardiovasc Res. 2005;67:367–78. doi: 10.1016/j.cardiores.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 7.Elizari MV, Levi R, Acunzo RS, et al. Abnormal expression of cardiac neural crest cells in heart development: a different hypothesis for the etiopathogenesis of Brugada syndrome. Heart Rhythm. 2007;4:359–65. doi: 10.1016/j.hrthm.2006.10.026. [DOI] [PubMed] [Google Scholar]
- 8.Houser SR, Margulies KB, Murphy AM, et al. Animal models of heart failure: a scientific statement from the American Heart Association. Circ Res. 2012;111:131–50. doi: 10.1161/RES.0b013e3182582523. [DOI] [PubMed] [Google Scholar]
- 9.Liang P, Lan F, Lee AS, et al. Drug screening using a library of human induced pluripotent stem cell-derived cardiomyocytes reveals disease-specific patterns of cardiotoxicity. Circulation. 2013;127:1677–91. doi: 10.1161/CIRCULATIONAHA.113.001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moretti A, Bellin M, Welling A, et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N Engl J Med. 2010;363:1397–409. doi: 10.1056/NEJMoa0908679. [DOI] [PubMed] [Google Scholar]
- 11.Matsa E, Rajamohan D, Dick E, et al. Drug evaluation in cardiomyocytes derived from human induced pluripotent stem cells carrying a long QT syndrome type 2 mutation. Eur Heart J. 2011;32:952–62. doi: 10.1093/eurheartj/ehr073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang Y, Liang P, Lan F, et al. Genome editing of isogenic human induced pluripotent stem cells recapitulates long QT phenotype for drug testing. J Am Coll Cardiol. 2014;64:451–9. doi: 10.1016/j.jacc.2014.04.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Itzhaki I, Maizels L, Huber I, et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature. 2011;471:225–9. doi: 10.1038/nature09747. [DOI] [PubMed] [Google Scholar]
- 14.Itzhaki I, Maizels L, Huber I, et al. Modeling of catecholaminergic polymorphic ventricular tachycardia with patient-specific human-induced pluripotent stem cells. J Am Coll Cardiol. 2012;60:990–1000. doi: 10.1016/j.jacc.2012.02.066. [DOI] [PubMed] [Google Scholar]
- 15.Asimaki A, Kapoor S, Plovie E, et al. Identification of a new modulator of the intercalated disc in a zebrafish model of arrhythmogenic cardiomyopathy. Sci Transl Med. 2014;6:240ra74. doi: 10.1126/scitranslmed.3008008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim C, Wong J, Wen J, et al. Studying arrhythmogenic right ventricular dysplasia with patient-specific iPSCs. Nature. 2013;494:105–10. doi: 10.1038/nature11799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang G, McCain ML, Yang L, et al. Modeling the mitochondrial cardiomyopathy of Barth syndrome with induced pluripotent stem cell and heart-on-chip technologies. Nat Med. 2014;20:616–23. doi: 10.1038/nm.3545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yazawa M, Hsueh B, Jia X, et al. Using induced pluripotent stem cells to investigate cardiac phenotypes in Timothy syndrome. Nature. 2011;471:230–4. doi: 10.1038/nature09855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carvajal-Vergara X, Sevilla A, D’Souza SL, et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature. 2010;465:808–12. doi: 10.1038/nature09005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun N, Yazawa M, Liu J, et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci Transl Med. 2012;4:130ra47. doi: 10.1126/scitranslmed.3003552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karakikes I, Stillitano F, Nonnenmacher M, et al. Correction of human phospholamban R14del mutation associated with cardiomyopathy using targeted nucleases and combination therapy. Nat Commun. 2015;6:6955. doi: 10.1038/ncomms7955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu H, Lee J, Vincent LG, et al. Epigenetic Regulation of Phosphodiesterases 2A and 3A Underlies Compromised beta-Adrenergic Signaling in an iPSC Model of Dilated Cardiomyopathy. Cell Stem Cell. 2015;17:89–100. doi: 10.1016/j.stem.2015.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinson JT, Chopra A, Nafissi N, et al. HEART DISEASE Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science. 2015;349:982–6. doi: 10.1126/science.aaa5458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lan F, Lee AS, Liang P, et al. Abnormal calcium handling properties underlie familial hypertrophic cardiomyopathy pathology in patient-specific induced pluripotent stem cells. Cell Stem Cell. 2013;12:101–13. doi: 10.1016/j.stem.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ebert AD, Kodo K, Liang P, et al. Characterization of the molecular mechanisms underlying increased ischemic damage in the aldehyde dehydrogenase 2 genetic polymorphism using a human induced pluripotent stem cell model system. Sci Transl Med. 2014;6:255ra130. doi: 10.1126/scitranslmed.3009027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Burridge PW, Li YF, Matsa E, et al. Human induced pluripotent stem cell-derived cardiomyocytes recapitulate the predilection of breast cancer patients to doxorubicin-induced cardiotoxicity. Nat Med. 2016;22:547–56. doi: 10.1038/nm.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus, an RNA virus that does not integrate into the host genome. Proc Jpn Acad Ser B Phys Biol Sci. 2009;85:348–62. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lian X, Hsiao C, Wilson G, et al. Robust cardiomyocyte differentiation from human pluripotent stem cells via temporal modulation of canonical Wnt signaling. Proc Natl Acad Sci U S A. 2012;109:E1848–57. doi: 10.1073/pnas.1200250109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calloe K, Refaat MM, Grubb S, et al. Characterization and mechanisms of action of novel NaV1. 5 channel mutations associated with Brugada syndrome. Circ Arrhythm Electrophysiol. 2013;6:177–84. doi: 10.1161/CIRCEP.112.974220. [DOI] [PubMed] [Google Scholar]
- 30.Glatter KA, Wang Q, Keating M, Chen S, Chiamvimonvat N, Scheinman MM. Effectiveness of sotalol treatment in symptomatic Brugada syndrome. Am J Cardiol. 2004;93:1320–2. doi: 10.1016/j.amjcard.2004.02.023. [DOI] [PubMed] [Google Scholar]
- 31.Gaborit N, Wichter T, Varro A, et al. Transcriptional profiling of ion channel genes in Brugada syndrome and other right ventricular arrhythmogenic diseases. Eur Heart J. 2009;30:487–96. doi: 10.1093/eurheartj/ehn520. [DOI] [PubMed] [Google Scholar]
- 32.Sallam K, Li Y, Sager PT, Houser SR, Wu JC. Finding the rhythm of sudden cardiac death: new opportunities using induced pluripotent stem cell-derived cardiomyocytes. Circ Res. 2015;116:1989–2004. doi: 10.1161/CIRCRESAHA.116.304494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen Q, Kirsch GE, Zhang D, et al. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392:293–6. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 34.Shin DJ, Kim E, Park SB, et al. A novel mutation in the SCN5A gene is associated with Brugada syndrome. Life Sci. 2007;80:716–24. doi: 10.1016/j.lfs.2006.10.025. [DOI] [PubMed] [Google Scholar]
- 35.Chiang KC, Lai LP, Shieh RC. Characterization of a novel Nav1. 5 channel mutation, A551T, associated with Brugada syndrome. J Biomed Sci. 2009;16:76. doi: 10.1186/1423-0127-16-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Davis RP, Casini S, van den Berg CW, et al. Cardiomyocytes derived from pluripotent stem cells recapitulate electrophysiological characteristics of an overlap syndrome of cardiac sodium channel disease. Circulation. 2012;125:3079–91. doi: 10.1161/CIRCULATIONAHA.111.066092. [DOI] [PubMed] [Google Scholar]
- 37.Cerrone M, Lin X, Zhang M, et al. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a Brugada syndrome phenotype. Circulation. 2014;129:1092–103. doi: 10.1161/CIRCULATIONAHA.113.003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baroudi G, Pouliot V, Denjoy I, Guicheney P, Shrier A, Chahine M. Novel mechanism for Brugada syndrome: defective surface localization of an SCN5A mutant (R1432G) Circ Res. 2001;88:E78–83. doi: 10.1161/hh1201.093270. [DOI] [PubMed] [Google Scholar]
- 39.Roden DM. Repolarization reserve: a moving target. Circulation. 2008;118:981–2. doi: 10.1161/CIRCULATIONAHA.108.798918. [DOI] [PubMed] [Google Scholar]
- 40.Martin CA, Siedlecka U, Kemmerich K, et al. Reduced Na(+) and higher K(+) channel expression and function contribute to right ventricular origin of arrhythmias in Scn5a+/− mice. Open Biol. 2012;2:120072. doi: 10.1098/rsob.120072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Witty AD, Mihic A, Tam RY, et al. Generation of the epicardial lineage from human pluripotent stem cells. Nat Biotechnol. 2014;32:1026–35. doi: 10.1038/nbt.3002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boukens BJ, Sylva M, de Gier-de Vries C, et al. Reduced sodium channel function unmasks residual embryonic slow conduction in the adult right ventricular outflow tract. Circ Res. 2013;113:137–41. doi: 10.1161/CIRCRESAHA.113.301565. [DOI] [PubMed] [Google Scholar]
- 43.Hoekstra M, Mummery CL, Wilde AA, Bezzina CR, Verkerk AO. Induced pluripotent stem cell derived cardiomyocytes as models for cardiac arrhythmias. Front Physiol. 2012;3:346. doi: 10.3389/fphys.2012.00346. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.