Abstract
Background
When left untreated, age-related macular degeneration (AMD) is the leading cause of vision loss in people over fifty in the US. Currently it is estimated that about eight million US individuals have the intermediate stage of AMD that is often asymptomatic with regard to visual deficit. These individuals are at high risk for progressing to the advanced stage where the often treatable choroidal neovascular form of AMD can occur. Careful monitoring to detect the onset and prompt treatment of the neovascular form as well as dietary supplementation can reduce the risk of vision loss from AMD, therefore, preferred practice patterns recommend identifying individuals with the intermediate stage in a timely manner.
Methods
Past automated retinal image analysis (ARIA) methods applied on fundus imagery have relied on engineered and hand-designed visual features. We instead detail the novel application of a machine learning approach using deep learning for the problem of ARIA and AMD analysis. We use transfer learning and universal features derived from deep convolutional neural networks (DCNN). We address clinically relevant 4-class, 3-class, and 2-class AMD severity classification problems.
Results
Using 5664 color fundus images from the NIH AREDS dataset and DCNN universal features, we obtain values for accuracy for the (4-,3-,2-) class classification problem of (79.4%, 81.5%, 93.4%) for machine vs. (75.8%, 85.0%, 95.2%) for physician grading.
Discussion
This study demonstrates the efficacy of machine grading based on deep universal features/transfer learning when applied to ARIA and is a promising step in providing a pre-screener to identify individuals with intermediate AMD and also as a tool that can facilitate identifying such individuals for clinical studies aimed at developing improved therapies. It also demonstrates comparable performance between computer and physician grading.
Keywords: Deep learning, Deep Convolutional Neural Networks, DCNNs, universal features, retinal image analysis, Age-related macular degeneration, AMD, transfer learning
1. Introduction
Age-related macular degeneration (AMD) is the leading cause of blindness for people over 50 in the United States. ([1],[2],[3],[7]). The intermediate stage of AMD is typically asymptomatic, but it often proceeds to the advanced stage during which substantial central vision loss may set in, affecting basic tasks including reading, driving, or face recognition ([7]).
AMD is caused by degeneration of the central portion of the retina known as the macula. The intermediate stage of AMD is characterized by the presence of extensive medium-sized (63μ to 125μ) drusen or at least one large (>125μ) druse or geographic atrophy not involving the fovea. Drusen are made up of long-spacing collagen and phospholipid vesicles between Bruch's membrane (the basement membrane of the choriocapillaris) and the basement membrane of the retinal pigment epithelium (RPE).
Advanced AMD is characterized by damage to the macula through either the choroidal neovascular (“wet”) form or geographic atrophy (GA) of the retinal pigment epithelium involving the center of the macula (“dry”) form of AMD. Both forms of advanced AMD can lead to rapid or gradual loss of visual acuity caused by a loss of photoreceptors that, in the case of wet form, are replaced by scar tissue or, in the case of the dry geographic atrophy form, degenerate in roughly circular regions (diameters > ∼175 μm) of hypopigmentation, depigmentation, or the apparent absence of the RPE [5]. It is estimated that there are currently between 1.75 and 3 million people in the United States with some form of advanced stage AMD [7].
While there is currently no definite cure for AMD, the Age Related Eye Disease Study (AREDS) has suggested benefits of certain dietary supplements for slowing the progression of the disease from the intermediate stage to the advanced stage [35]. Additionally, the worsening of vision loss due to CNV can be slowed substantially by intravitreous injections of anti-VEGF agents, which reduce the chance of vision loss compared to no treatment [8] or when compared with alternative treatments such as photodynamic therapy with verteporfin [6] or laser photocoagulation [1]. Early detection is therefore key: the sooner a patient with wet AMD begins anti-VEGF injections, the less the risk of vision loss, resulting in improved quality of visual acuity for a longer period of time. Thus, it is imperative that people with the intermediate stage of AMD be identified as soon as possible and referred to a health care provider for education on monitoring for the development of wet AMD and possible use of dietary supplements to reduce the risk of developing wet AMD. No treatment comparable to anti-VEGF injections currently exists for GA and, consequently, numerous clinical studies are being conducted with the goal of finding a treatment for slowing or eliminating GA growth rate [17],[20],[30],[31].
Developing automated diagnostic techniques that detect AMD and assess its severity are a key endeavor for several reasons: (1) Cost: Fundus image grading can be a tedious and time consuming process requiring the expertise of an extensively trained health care professional. (2) Access: access to ophthalmology healthcare at least every two years to detect intermediate AMD after age fifty -- so that individuals with intermediate AMD can be identified and referred to a physician -- can be difficult in many health care environments. Because of the large at risk population of people over fifty (∼110 million in the United States [33]), the logistics for screening individuals is prohibitive due to numbers alone. (3) Monitoring treatment: Another key role of such automated computer techniques is in helping assess the efficacy of treatments for geographic atrophy, where it is critical to monitor quantitatively and objectively the disease progression under therapy.
2. Deep Features
This paper proposes the usage of deep convolutional neural networks (DCNN)-derived universal features on the specific automated retinal image analysis (ARIA) problem of AMD categorization. For the past decades, visual recognition algorithms have leveraged various methods with hand-designed feature-based approaches becoming widely used techniques [12],[21],[22]. Traditional ARIA methods have primarily followed an algorithmic pattern consisting of two steps: i) an image analysis step whereby the image is pre-processed and some visual features are computed and ii) a classification step using one of the many machine learning methods of record (e.g. SVMs, random forests, logistic regression, boosting, etc.) [9],[10],[15],[32],[37]. This type of approach can be used either locally, to find and delineate specific lesions, or globally, to diagnose disease from an entire image.
In contrast, the past few years have seen a shift towards methods employing deep learning (DL) approaches. In particular, recent progress in DCNN have exploited deeper networks, better DCNN parameter optimization, and expanded computational power afforded by GPUs, which in turn have led to substantial performance improvement in visual recognition tasks [16],[18],[23],[24],[25],[26],[27],[28],[29]. It is therefore natural for the focus of ARIA algorithms to leverage these emerging techniques. This paper demonstrates the use of one such techniques: using DCNN universal features, to replace hand-designed features, for global retinal image analysis and multi-class AMD classification.
When applied to image classification, an important difference between DCNNs and conventional machine vision and machine learning methods referred to above is the process of feature selection. In particular, prior to classification, conventional methods have included a step in which specific visual features are computed such as wavelets or SIFT features [9],[14],[15]. This manual selection (or feature design) can result in a set of features that are suboptimal and overly specialized to a specific data set resulting in poor generalization to larger or unknown data sets.
As a departure from the manual approach described above, DCNN's features are not ‘hand-crafted’ or specially designed but are found by training a DCNN ([10],[27]). However, this optimization frequently requires a very large number of images to reliably learn DCNN weights. In contrast, typical ARIA datasets often consists of only a few thousand images. One idea instead is to use universal features and transfer learning: one can tap into fully connected layers of a DCNN because these provide very compact and pooled spatial feature representations. This can be accomplished by using a network that has already been pre-trained on millions of general purpose images (ImageNet [26]) without any additional retraining needed for the DCNN on our specific dataset. When neural networks trained on large general-purpose datasets are repurposed, in a transfer learning sense, to address problems applied to image domains different from those that the networks were trained on, the features are referred to as universal features. The features are then used as input to a classifier like an SVM or Random Forrest that is then trained on the specific ARIA data. This is the approach we use for ARIA and AMD classification in this study.
3. Problem Structure
We now discuss how this study addresses several clinically relevant N-class AMD classification problems including 4-class, 3-class, and 2-class problems. In particular, important objectives are to develop techniques that can address AMD severity classification that discriminates among individuals with no AMD vs. early stage AMD vs. intermediate stage AMD vs. advanced AMD (4-class) as well as among individuals with no or early stage AMD vs. intermediate stage AMD vs. advanced AMD (3-class). Such algorithms would be beneficial in clinical studies designed to assess new or alternative therapies where it is critical to differentiate between intermediate and advanced AMD. In addition, these algorithms can be used to automatically and remotely monitor the change in an individual level of AMD as it progresses from, say, early stage AMD to intermediate stage AMD. Also, because of the importance of identifying individuals in the at risk population with the asymptomatic intermediate stage we also consider the important 2-class problem that discriminates among individuals with no or early stage AMD vs. intermediate stage AMD or advanced AMD. This particular 2-class problem is important in that it identifies individuals that need to be referred to a physician immediately and could be implemented in public settings especially in under resourced areas where there is limited access to health care.
4. Fundus image Data
The Age-Related Eye Disease Study (AREDS) was a 12-year longitudinal study designed to increase understanding of disease progression and risk factors for both AMD and age-related cataracts [2,4,30]. During the study, color fundus photographs were taken of each patient at the initial baseline visit as well as during follow-up visits. One of the notable features of the AREDS study is that these images were quantitatively graded for AMD severity by experts at various grading centers [4]. Each image was assigned a category from 1 to 4 with category 1 denoting no evidence of AMD, category 2 denoting early stage AMD, category 3 denoting intermediate stage AMD, and category 4 denoting one of the advanced forms of AMD. Figure 1 shows examples of fundus images from each category. We remark here that this set of graded images provides a unique “gold standard” data set that can be used to assess the efficacy of machine learning algorithms. It is this set of images that we used for characterizing the performance of our N-class classification problem.
Figure 1.
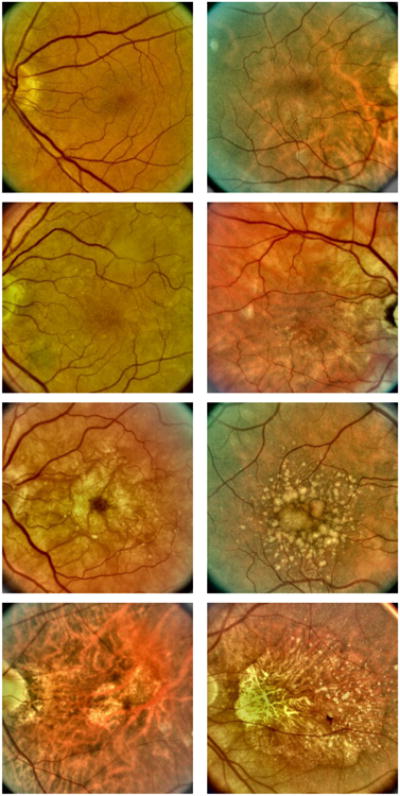
Fundus images showing two examples for each of the four AMD categories. Top row: AMD category 1 (healthy retina). Second row: AMD category 2 (early stage AMD). Third row: AMD category 3 (intermediate stage AMD). Bottom row: AMD category 4 (advanced AMD).
5. Methods
2.1 Overview of Approach
The algorithm first preprocesses the image, and then computes universal features, including computation over concentric grids inscribed within the retinal fundus image. Thereafter, universal features computed in each grid are concatenated into a feature vector that is passed on to a linear support vector machine (LSVM) classifier for testing and training. The main steps of the method are detailed below. A summary pseudo-code of the entire algorithmic pipeline is also shown here:
Pseudo-code
|
2.2 Implementation Details
(1) Preprocessing
Pre-processing the fundus images involves (a) detecting the outer boundaries of the retina to crop the images to the square inscribed within the retinal boundary (Figure 1). This is done by thresholding out the black background, fitting a circle and computing an inscribed square. (b) resizing them to size 231×231 to conform to the expected OverFeat input network (see below). As detailed below, additional cropping to smaller inscribed squares is also performed.
(2) Universal Features computation
We use OverFeat (OF) features. These use the OverFeat DCNN that was pre-trained on an ImageNet data set consisting of over a million ImageNet general purpose (non-medical) images including one thousand image classes consisting of various objects (e.g. animals, food, household objects, and so forth). The implementation code is provided in [36]. Features are calculated by tapping into the second to last network layer output. This yields a 4096 long feature vector. This vector is normalized (as a unit vector) for illumination invariance.
(3) Linear Support Vector Machine Classification
The normalized feature vectors are used to train and test on a LSVM classifier [13]. For training and testing we used the standard machine learning approach of n-fold cross-validation [34]. This consists of subdividing the data into n equally sized subsets (i.e. folds) and using n-1 folds for training and the remaining nth fold for testing. This process is repeated n times, leaving a different fold out of the training step each time in order to use it for testing. For this study we used n=10.
(4) Multi-grid processing
As illustrated in Figure 2, we use a multi-grid approach to generate the feature vector. Although the details of this approach have been described previously [10], we nevertheless provide a brief summary of the method here. In particular, the solid light blue line in Figure 2 is the inscribed square that includes the largest usable area of the fundus image. The dashed light blue lines show two successive square regions centered on the macula. The final feature vector that is ultimately used is obtained by concatenating feature vectors generated from each of these three regions. That is, we first generate a feature vector of OF features obtained from the fully inscribed square (solid light blue line in Figure 2). Next, we generate a separate OF feature vector using only the area of the image within the inner most dashed square. Then, a third OF feature vector is generated using the area of the image within the outer most dashed square (the area of inner dashed square is included). Finally, the three feature vectors are concatenated together to form the feature vector used for classification. We note here that this approach is motivated by the fact that, for color fundus images, features near the center of the macula are of greater significance for classifying the AMD severity category[10][14].
Figure 2.
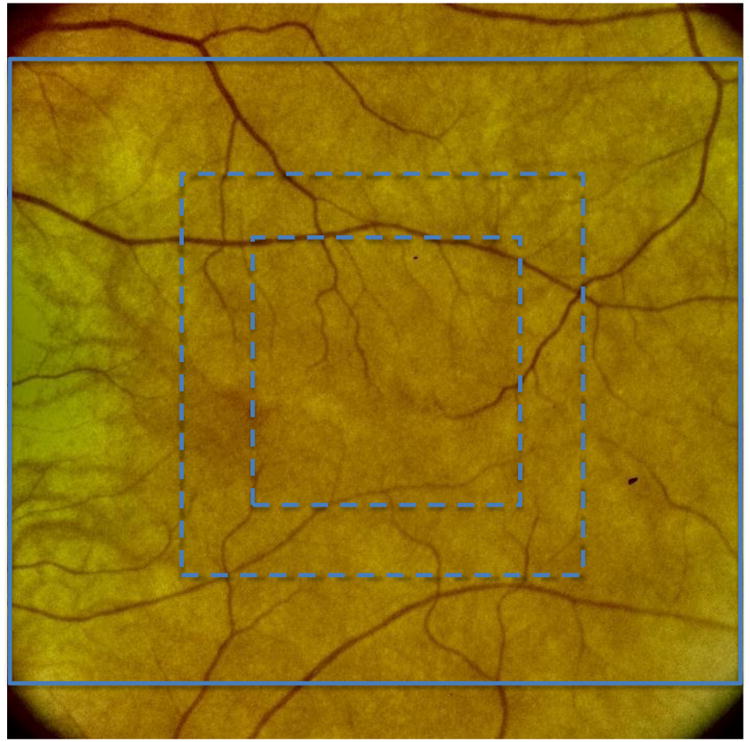
Grid areas used to compute the Overfeat feature vectors.
6. Experiments
6.1 AREDS Dataset for testing
For training and testing, we used color fundus images from the NIH AREDS database and subsequently characterized the performance of the universal feature-based algorithm. Since they are centered on the macula, only field 2 fundus images were used and, to avoid using virtually the same image twice, one of the images in cases with stereo pairs was removed. This resulted in a total of 5664 available images. Table 1 shows the number of images available in each class. Note that these images are a combination of all available baseline and follow-up images. As seen in the table below, all classes are well represented with the exception of category 2 for which there is significant imbalance and under-representation.
Table 1.
Number of AREDS images in each AMD category after eliminating redundant data in cases in which two stereo images of the same eye were taken on a given visit.
| Total Number of Images | Images in Category 1 | Images in Category 2 | Images in Category 3 | Images in Category 4 |
|---|---|---|---|---|
| 5664 | 1566 | 198 | 1853 | 2047 |
6.2 Performance metrics
To assess the results, we provide both counting and confusion matrices. In the counting matrix the columns represent the numbers of samples in the true class and the rows provide the classifier prediction breakdown. For example, for the 4-class classification problem, the first column corresponds to the breakdown for actual AMD category 1 (class 1), the second column correspond to actual AMD category 2 (class 2), the third column corresponds to actual AMD category 3 (class 3), and the fourth column corresponds to actual AMD category 4 (class 4). Each diagonal element of the counting matrix gives the number of images classified correctly for the corresponding class (i.e. column). Off diagonal elements show the number of images misclassified and how they were was misclassified. For example, the matrix element in the first row and second column shows the number of images whose true AMD score is category 1, but were misclassified as AMD category 2. The overall accuracy is determined by dividing the total number of correctly classified images (obtained by adding the diagonal elements in the counting matrix) and dividing by the total number of images (i.e. 5664). Finally, Along with the counting matrix we report normalized values (i.e. percentages) which are obtained by dividing the counts in each of the matrix cell by the total number of samples in the specific class, this results in what is usually referred to as the confusion matrix.
The algorithm's performance was further assessed by comparing to results obtained by a physician. Specifically, an ophthalmologist independently graded all 5664 fundus images and the results were compared to both the AREDS AMD scores and the computer's. Finally, paired gradings were compared for computer vs. AREDS and the physician vs. AREDS using kappa statistics.
6.3 Results for N class classification
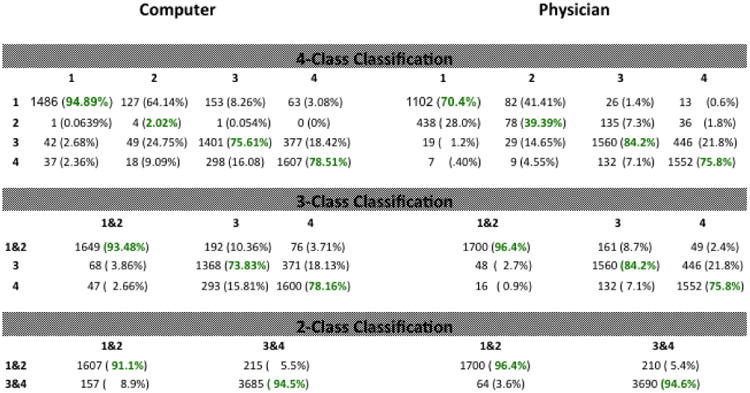
Table 2 is the table of counting/confusion matrices comparing the results of the machine with those of the physician. In particular, the left column gives the counting matrices for the 4-class, 3-class, and 2-class classification problems for the results obtained by the machine, while the right column gives the corresponding results for the physician. Notice that each of the six entries in table 2 is a matrix (i.e., a table in its own right) and that each of these matrices has its own rows and columns associated with it. Specifically, the rows and columns for each of the six matrices in table 2 correspond to the AMD severity category associated with each type of classification problem. As described above, the numbers in each matrix indicate the accuracy as well as the types of errors made by either the computer or the physician. We note here that, the physician only performed 4-class classification. The physicians' results for the 3-class and 2-class classification problem were then derived by appropriately combining the classes from the 4-class classification results. On the other hand, the computer performed each of the N-class problems separately, starting from scratch (so to speak) each time with absolutely no knowledge of results and analysis (i.e. training) from a previous classification problem.
Table 2. Counting/confusion matrices comparing the computer with the physician.
|
In the 4-class problem, it is seen that the computer and physician' results are roughly similar with the computer correctly classifying AMD category 1 and 4 more often while the physician correctly classified AMD category 2 and 3 more often. One of the salient features of the 4-class results is the difficulty both the computer and the physician had distinguishing AMD category 1 from AMD category 2. The computer only classified a total of four AMD category 2 images correctly and misclassified 127 (64.1%) as AMD category 1. This poor performance is to be expected and is due to the relatively very small number (198) of AMD category 2 images available to train the classifier. Additionally, because of the much larger number of category 1 images present in the training set the classifier is biased to choose category 1 over 2. On the other hand, the physician classified 78 AMD category 2 images correctly for only a 39.4% correct rate. Furthermore, the physician misclassified 28% of AMD category 1 images as category 2 and misclassified 41.4% of AMD category 2 images as AMD category 1. The 28% and 41.4% represent the two largest misclassification rates in the physicians' 4-class classification counting matrix. These misclassification challenges affecting both machine and human are also likely due to the fact that categories 1 and 2 present very similarly, as category 2 entails the presence of very small number of drusen present outside the macula. As in the 4-class classification problem, the computer and physician' results are also similar in performance for the 3-class and 2-class problems, with the physicians' results being slightly better due to the combining of AMD category 1 AMD category 2 into a single class. For the 3-class problem, the physician does better for class 3 while the machine does better for class 4. For the two-class problem, the physician does slightly better for classes 1&2 while 3&4 are nearly identical for physician and computer.
Table 3 provides a succinct comparison between the computer and physician. Specifically, the table compares the overall accuracy between the two, showing the computer accuracy is 3.6% better in the 4-class problem and the physicians' is 3.5% and 1.8% better in the 3-class problem and 2-class problem respectively.
Table 3. Comparison of overall accuracy and kappa values between the computer and physician.
| Computer | Physician | |||||
|---|---|---|---|---|---|---|
|
| ||||||
| Classification Problem | Accuracy | Kappa | Weighted kappa | Accuracy | Kappa | Weighted kappa |
| 4-class | 79.4% | 0.6962 | 0.7875 | 75.8% | 0.6583 | 0.7889 |
| 3-class | 81.5% | 0.7226 | 0.7693 | 85.0% | 0.7748 | 0.8167 |
| 2-class | 93.4% | 0.8482 | NA | 95.2% | 0.8897 | NA |
The table also shows both un-weighted and linear weighted kappa scores for the computer and the physician. The kappa score is generally considered to be more robust than simple percent agreement because it accounts for agreement occurring by chance [4],[11],[19]. Following Landis and Koch, the kappa scores for both the computer and the physician show substantial agreement with the AREDS grading for the 4-class and 3-class problem and values that are usually considered almost perfect agreement for the 2-class problem [4],[19].
The evaluation of our proposed automated algorithm performance and the physician grading is made in comparison with the AREDS severity grading that was performed by Reading Center Experts at national AREDS grading centers and which was used in this study as a gold standard.
As a final comparison between the computer and physician, we note that once the OF features were computed, which took about 0.5 to 1s per image to carry out on a non-GPU processor, the computer took about 68 minutes to complete all three classification problems on a MacBook Pro with a 2.8 GHz Intel Core i7 processor, while the physician took several days of actual time dedicated exclusively (i.e., not counting time possibly spent performing other duties or sleeping) to completing the task.
In summary, from the accuracy values and the kappa scores it appears that both the machine grading and the physician grading perform comparably. This is a very promising outcome for the deployment of such algorithms in clinical environments and their use for assessing the evolution of disease under treatment in longitudinal studies. In this study, the reduced performance of the machine for category 2 AMD can be attributed to the relative imbalance and lack of representation of that class that impairs training and introduces bias in the prediction. It is expected that with an even larger dataset it would be of interest, as future work, to assess if a machine could possibly perform better and possibly have fewer errors than a human operator.
Of note, the current study was performed on all of the available AREDS images which are commonly referred to as AREDS1. We did not exclude images based on any kind of quality criteria because we intended to use a dataset that would be as representative as possible of the type of problems that would be seen in actual practice. Although some AREDS1 images have artifacts, these images are in general well behaved. Should we have excluded some images based on quality this would have undoubtedly raised the performance numbers reported above. One mitigating factor in this dataset is that it is made up of digitized images taken from analog photographs. This is also a factor that may lower the resulting performance.
7. Conclusion
We used a large publicly available dataset of AMD images, the NIH AREDS dataset, to characterize the performance of deep learning universal feature-based multi-class AMD severity categorization. The results show that -- with virtually no additional processing -- universal deep features combined with support vector machines provide attractive performance characteristics and transfer well to the domain of retinal imagery. Considering that universal features were used here and that no further DCNN optimization was attempted for this problem, the results are very encouraging. The universal features were taken from a fully connected layer of the OF DCNN, and the results show that they encapsulate well, and in a compact fashion, low level features and spatial characteristics that are also well adapted for analyzing retinal fundus images. Clinically, the approach is important since it allows for fine categorization of clinically relevant features of AMD as well as the isolation of cases of individuals with the often asymptomatic intermediate stage who require immediate referral to an ophthalmologist.
Future work may be dedicated to finding techniques for direct DCNN fine tuning and retraining for ARIA classification problems in general and AMD in particular. We believe that additional ARIA problems other than AMD classification can benefit from the same basic approach, this includes for example diabetic retinopathy classification, but it also includes tasks such as vessel and optical disk segmentation. More importantly, this approach could be beneficial for not only classifying severity within a particular retinal disease, but could also assist in diagnosis. That is, distinguishing among various retinal pathologies and subsequently classifying the severity level within the identified pathology.
Acknowledgments
Research reported in this publication was supported primarily by the National Eye Institute of the National Institutes of Health under award number R21EY024310. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. Supported in part by James P. Gills Professorship and unrestricted research funds to the Retina Division for macular degeneration and related diseases research. Additional support from JHU Whiting School of Engineering SPUR program and the Retina Division research fund of the Brazilian Center of Vision Eye Hospital is acknowledged.
Footnotes
Conflict of interest: The authors have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Subfoveal neovascular lesions in age-related macular degeneration: Guidelines for evaluation and treatment in the macular photocoagulation study. Archives of Ophthalmology. 1991;1099:1242–1257. URL + http://dx.doi.org/10.1001/archopht.1991.01080090066027. [PubMed] [Google Scholar]
- 2.Potential public health impact of age-related eye disease study results: AREDS report no. 11. Archives of Ophthalmology. 2003;12111:1621–1624. doi: 10.1001/archopht.121.11.1621. URL+ http://dx.doi.org/10.1001/archopht.121.11.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Abramoff M, Garvin M, Sonka M. Retinal imaging and image analysis. Biomedical Engineering, IEEE Reviews in. 2010;3:169–208. doi: 10.1109/RBME.2010.2084567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Age-Related Eye Disease Study Research Group and others. The age-related eye disease study system for classifying age-related macular degeneration from stereoscopic color fundus photographs: the age-related eye disease study report number 6. American Journal of Ophthalmology. 2001;1325:668. doi: 10.1016/s0002-9394(01)01218-1. [DOI] [PubMed] [Google Scholar]
- 5.Bird A, Bressler N, Bressler S, Chisholm I, Coscas G, Davis M, de Jong P, Klaver C, Klein B, Klein R, Mitchell P, Sarks J, Sarks S, Soubrane G, Taylor H, Vingerling J. An international classification and grading system for age-related maculopathy and age-related macular degeneration. Survey of Ophthalmology. 1995;395:367–374. doi: 10.1016/s0039-6257(05)80092-x. URL http://www.sciencedirect.com/science/article/pii/S003962570580092X. [DOI] [PubMed] [Google Scholar]
- 6.Blinder KJ, Bradley S, Bressler NM, Bressler SB, Donati G, Hao Y, Ma C, Menchini U, Miller J, Potter MJ, Pournaras C, Reaves A, Rosenfeld PJ, Strong HA, Stur M, Su XY, Virgili G Treatment of Age-related Macular Degeneration with Photodynamic Therapy study group, Verteporfin in Photodynamic Therapy study group. Effect of lesion size, visual acuity, and lesion composition on visual acuity change with and without verteporfin therapy for choroidal neovascularization secondary to age-related macular degeneration: Tap and vip report no. 1. American journal of ophthalmology. 2003;1363:407–418. doi: 10.1016/s0002-9394(03)00223-x. URL http://dx.doi.org/10.1016/S0002-9394(03)00223-X. [DOI] [PubMed] [Google Scholar]
- 7.Bressler N. Age-related macular degeneration is the leading cause of blindness. JAMA. 2004;29115:1900–1901. doi: 10.1001/jama.291.15.1900. URL + http://dx.doi.org/10.1001/jama.291.15.1900. [DOI] [PubMed] [Google Scholar]
- 8.Bressler NM, Chang TS, Suñer IJ, Fine JT, Dolan CM, Ward J, Ianchulev T. Vision- related function after ranibizumab treatment by better- or worse-seeing eye. Ophthalmology. 2010;1174:747–756.e4. doi: 10.1016/j.ophtha.2009.09.002. URL http://www.aaojournal.org/article/S0161-6420(09)00981-6/abstract. [DOI] [PubMed] [Google Scholar]
- 9.Burlina P, Freund DE, Dupas B, Bressler N. Automatic screening of age-related macular degeneration and retinal abnormalities. Engineering in Medicine and Biology IEEE. 2011:3962–3966. doi: 10.1109/IEMBS.2011.6090984. [DOI] [PubMed] [Google Scholar]
- 10.Burlina P, Freund DE, Joshi N, Wolfson Y, Bressler N. Detection of Age-Related Macular Degeneration via Deep Learning. Proceedings - International Symposium on Biomedical Imaging. 2016 Jun;:184–188. 7493240. [Google Scholar]
- 11.Cardillo G. Cohens kappa: compute the Cohen's kappa ratio on a 2×2 matrix. 2007 http://www.mathworks.com/matlabcentral/fileexchange/15365.
- 12.Dalal N, Triggs B. Histograms of oriented gradients for human detection. IEEE Computer Vision and Pattern Recognition. 2005;2005:886–893. [Google Scholar]
- 13.Fan RE, Chang KJ, Hsieh CJ, Wang XR, Lin CJ. LIBLINEAR: A Library for Large Linear Classification. Journal of Machine Learning Research. 2008:1871–1874. [Google Scholar]
- 14.Feeny AK, Tadarati M, Freund DE, Bressler NM, Burlina P. Automated segmentation of geographic atrophy of the retinal epithelium via random forests in AREDS color fundus images. Comput Biol Med. 2015;65:124–136. doi: 10.1016/j.compbiomed.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Freund DE, Bressler N, Burlina P. Automated detection of drusen in the macula. IEEE International Symposium on Biomedical Imaging. 2009:61–64. [Google Scholar]
- 16.Girshick R, Donahue J, Darrell T, Malik J. Rich feature hierarchies for accurate object detection and semantic segmentation. IEEE conference on computer vision and pattern recognition. 2014 [Google Scholar]
- 17.Holz FG, Strauss EC, Schmitz-Valckenberg S, van Lookeren Campagne M. Geographic atrophy: Clinical features and potential therapeutic approaches. Ophthalmology. 2014;1215:1079–1091. doi: 10.1016/j.ophtha.2013.11.023. URL http://www.sciencedirect.com/science/article/pii/S0161642013011020. [DOI] [PubMed] [Google Scholar]
- 18.Krizhevsky A, Sutskever I, Hinton G. Imagenet classification with deep convolutional neural networks. Advances in neural information processing systems. 2012 [Google Scholar]
- 19.Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–174. [PubMed] [Google Scholar]
- 20.Lim LS, Mitchell P, Seddon JM, Holz FG, Wong TY. Age-related macular degeneration. The Lancet. 2012;3799827:1728–1738. doi: 10.1016/S0140-6736(12)60282-7. URL http://www.sciencedirect.com/science/article/pii/S0140673612602827. [DOI] [PubMed] [Google Scholar]
- 21.DGLowe Distinctive Image Features from scale invariants keypoints. International Journal of Computer Vision. 2004;60(2):91–110. [Google Scholar]
- 22.Rajagopalas AN, Burlina P, Chellappa R. Detection of people in images. International Joint Conference on Neural Networks. 1999;4:2747–2752. [Google Scholar]
- 23.Razavian A, et al. CNN features off-the-shelf: an astounding baseline for recognition. IEEE Conference on Computer Vision and Pattern Recognition Workshops. 2014 [Google Scholar]
- 24.Redmon J, et al. You only look once: Unified, real-time object detection. arXiv preprint arXiv, 2015;1506.02640 [Google Scholar]
- 25.Ren S, He K, Girshick R, Sun J. Faster R-CNN: Towards real-time object detection with region proposal networks. Advances in Neural Information Processing. 2015:91–99. doi: 10.1109/TPAMI.2016.2577031. [DOI] [PubMed] [Google Scholar]
- 26.Russakovsky O, et al. Imagenet large scale visual recognition challenge. International Journal of Computer Vision. 2015;115:211–252. [Google Scholar]
- 27.Sermanet P, Eigen D, Zhang X, Mathieu M, Fergus R, LeCun Y. Overfeat: Integrated recognition, localization and detection using convolutional networks. arXiv preprint arXiv, 2013, 1312.6229 [Google Scholar]
- 28.Simonyan K, Zisserman Z. Very deep convolutional networks for large-scale image recognition. arXiv preprint, 2014, 1409.1556 [Google Scholar]
- 29.Szegedy C, et al. Going deeper with convolutions. IEEE Conference on Computer Vision and Pattern Recognition. 2015 [Google Scholar]
- 30.The AREDS Research Group. Change in area of geographic atrophy in the age-related eye disease study: AREDS report number 26. Archives of Ophthalmology. 2009;1279:1168–1174. doi: 10.1001/archophthalmol.2009.198. URL + http://dx.doi.org/10.1001/archophthalmol.2009.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tolentino MJ, Dennrick A, John E, Tolentino MS. Drugs in phase II clinical trials for the treatment of age-related macular degeneration. Expert Opinion on Investigational Drugs. 2015;242:183–199. doi: 10.1517/13543784.2015.961601. URL http://dx.doi.org/10.1517/13543784.2015.961601. [DOI] [PubMed] [Google Scholar]
- 32.Trucco E, Ruggeri A, Karnowski T, Giancardo L, Chaum E, Hubschman JP, al Diri B, Cheung CY, Wong D, Abràmoff M, et al. Validating retinal fundus image analysis algorithms: issues and a proposal. Investigative ophthalmology & visual science. 2013;545:3546–3559. doi: 10.1167/iovs.12-10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.U.S. Department of Commerce USCB. The 2012 Statistical Abstract. The National Data Book. 2012 [Google Scholar]
- 34.Vapnik . Statistical Learning Theory. Wiley; New York: 1998. pp. 416–417. [Google Scholar]
- 35.Age-Related Eye Disease Study (AREDS) Research Group. A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119:1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.https://github.com/sermanet/OverFeat
- 37.Pacheco K, et al. Evaluation of automated drusen detection system for fundus photographs of patients with age-related macular degeneration. Investigative Ophthalmology & Visual Science. 2016:1611–1611. [Google Scholar]


