Abstract
Immunoglobulin heavy chain (IgH) class switch recombination (CSR) occurs through the deliberate introduction of AID-instigated DNA double strand breaks (DSBs) into the IgH loci. Since DSBs are generally highly toxic, mechanisms that regulate AID expression are of much relevance to both CSR and genomic integrity; however, effectors of such regulatory processes are still poorly understood. Here we show that the transcription factor Sox2 is expressed in activated B cells but almost exclusively in those that have undergone CSR. We demonstrate that enforced expression of Sox2 in splenic B cells severely inhibits AID expression and CSR, while deletion of Sox2 increases the frequency of IgH;c-Myc translocations. These results suggest that Sox2 may regulate AID expression in class-switched B cells to suppress genomic instability associated with CSR.
Introduction
Mature IgM-expressing B cells in peripheral lymphoid organs such as the spleen, lymph nodes and Peyer’s patches are activated upon antigen encounter and undergo class switch recombination (CSR), a deletional-recombination reaction that replaces the Cμ constant region segment for a set of downstream CH genes (Cγ, Cε, or Cα) (1, 2). The B cell thereby changes from expressing IgM to one expressing IgG, IgE or IgA, with each secondary isotype having a distinct effector function during an immune response. CSR occurs between repetitive DNA elements termed “switch” (S) regions that precede each CH segment and requires “germline” transcription through the CH genes and activation-induced cytidine deaminase (AID) (1, 2). Current models posit that AID-mediated deamination of transcribed S regions instigates generation of DNA double strand breaks (DSBs) and end-joining of DSBs between donor (usually Sμ) and downstream acceptor S regions (Sγ, Sε, or Sα) completes CSR (1, 2). While S regions are primary targets of AID during CSR, off-target activity of AID within B cells can generate translocations commonly seen in the development of lymphomas (1, 3). AID expression is therefore tightly regulated at multiple levels and its transcription alone is controlled by at least a dozen different transcription factors (4). However, the mechanism that turns-off AID in B cells that have already initiated and/or completed CSR is poorly understood.
The transcription factor Sox2 (Sex Determining Region Y HMG-Box 2) is primarily known for its role in early development, where it can be found in embryonic stem cells, the differentiating eye and gut, and in primordial germ cells (5–7). It has also been detected in some adult stem cell populations and skin cells (8, 9). More recently, Sox2 expression was demonstrated in neutrophils, where it was shown to play a non-canonical role as a pathogen sensor in the cytoplasm (10). Finally, Sox2 is also one of the four transcription factors (with Klf4, Oct4, and c-Myc) vital for reprogramming somatic cells into induced pluripotent stem cells (11).
Here we report that Sox2 is expressed in class-switched B cells and that its enforced expression led to a drastic reduction in levels of AID and frequency of CSR. Conversely, deletion of Sox2 in mature B cells increased both AID expression and frequency of IgH:c-Myc translocations. Thus Sox2 appears to be a novel factor that turns off critical aspects of the CSR program in class-switched B cells to maintain genomic integrity.
Materials and Methods
Mice, B cell cultures and retroviral transduction
Wild-type C57Bl/6J mice were obtained from the Jackson Laboratory and AID−/− mice were a gift from T Honjo (12). Primary B cells were isolated from the spleen, stimulated with anti-CD40 (1μg/ml; HM40-3; eBioscience) and IL-4 (12.5μlg/ml; 404-ML; R&D Systems), LPS (10μg/ml; Sigma) or LPS+IL-4 and analyzed for CSR as described previously (13). Sox2 conditional mice (Sox2flox) were generated by Shaham et al (14) and purchased from the Jackson Laboratory (http://www.informatics.jax.org/reference/J:152850). Splenic B cells were transduced with retroviral vectors pMIR or pMIR-Sox2 using spinfection as described earlier (13). Immunization with NP-CGG and analysis of germinal center B cells and NP-specific response by flow-cytometry and ELISA were performed as described earlier (15). To purify germinal center B cells, splenocytes from immunized mice were stained with B220, GL7 and Fas, and B220+GL7+Fas+ (GC+) or B220+GL7- Fas- (GC-) cells were collected by passing through a cell sorter. IgM+ and IgG1+ B cells were purified by cell sorting following stimulation of splenic B cells with anti-CD40 +IL-4 for 4 days.
Western blotting and cell division assays
Whole-cell extracts were prepared in RIPA lysis buffer (13). Primary antibodies were as follows: anti-AID (16), anti-Sox2 (R&D Systems), anti-Gapdh (6C5; Millipore), anti-tubulin (T9026; Sigma), anti-Batf (D7C5; Cell Signaling Technology), and anti-p21 (B-2), anti-cyclinD2 (M-20), anti-Irf4 (M-17), all from Santa Cruz Biotechnology. Uninfected and retrovirally infected cells were stained with the CellTrace CFSE Cell Proliferation Kit (Molecular Probes) immediately following the first spin-infection (or at 24h of stimulation for control cells) according to the manufacturer’s instructions. Staining was detected by flow cytometry at 0h and 48h.
Quantitative real-time PCR
Total RNA was extracted using TRIzol reagent according to the manufacturer’s instructions, followed by cDNA synthesis of 1μg total RNA with Quanta qScript™ cDNA Synthesis Kit (Quanta Biosciences). iQ SYBR Green Supermix (Bio-Rad) was used for quantification of mRNA, which was performed on a Bio-Rad CFX96 Real-Time PCR Detection system. CFX Manager software was used to determine the crossing points for incorporation of the SYBR Green. Relative expression was determined with β-actin as a reference gene. Primers are as follows: Batf 5′-AGCTTCAGCCGCTCTCCT-3′ and 5′-GGTGTCGGCTTTCTGTGTCT-3′, Irf4 5′-ACGCTGCCCTCTTCAAGG-3′ and 5′-GCTCCTCTCGACCAATTCCT-3′, p21 5′-TCCACAGCGATATCCAGACA-3′ and 5′-GCGCAACTGCTCACTGTC-3′, CyclinD2 5′-GCTAGGAACATGCACACTGC-3′ and 5′-CTGGGGCTTCACAGAGTTGT-3′, γ1-GLT: 5′-GGCCCTTCCAGATCTTTGAG-3′ and 5′-GGATCCAGAGTTCCAGGTCACT-3′, γ3-GLT: 5′-AACTACTGCTACCACCACCACCAG and 5′-ACCAAGGGATAGACAGATGGGG-3′, μ-GLT: 5′-CTCTGGCCCTGCTTATTGTTG-3′ and 5′-GAAGACATTTGGGAAGGACTGACT-3. Primers for AID, β-actin, μ-chain, γ1-chain and γ3 have been previously described (17).
RNA-Seq
A paired end RNA-Seq library was prepared as described and sequencing was performed on an Illumina HiSeq2000 with 30×10ˆ6 reads per sample. Determination of differential expression was based on a minimum of a 2-fold change in expression and at least 200 average reads in the higher sample. Gene ontology analysis was carried out using the Database for Annotation, Visualization and Integrated Discovery (DAVID). The heat maps were generated using R software packages. The RNAseq data has being submitted to NCBI (GEO submission GSE93131).
Translocation assay
Translocations between IgH and c-Myc were assessed by PCR using primers and PCR conditions described previously (18). Briefly, genomic DNA from approximately 1×105 cells was subjected to 2 rounds of amplification using nested primers using Expand Long Template PCR system (Roche), products resolved on agarose gel and probed by Southern blotting using IgH and c-Myc probes as previously described (18).
Statistical Analysis
Statistical significance was assessed using a Student’s t-test. The values for significance are as follows: * p < 0.05, ** p < 0.01, *** p <0.001.
Results and Discussion
Enforced Sox2 expression severely impairs CSR
While CSR is essential for B cell-mediated immunity, it proceeds through the deliberate generation of DSBs into the B cell genome (2). It is therefore critical that the process is finely regulated, not only for proper control of CSR, but also for the maintenance of genomic integrity. However, factors that regulate AID expression once CSR has been initiated are poorly defined. In our effort to understand the process by which the combination of the four transcription factors Sox2, c-Myc, Klf4 and Oct4 converts a B cell into an induced pluripotent stem cell, we discovered that Sox2 was expressed in splenic B cells activated to undergo CSR to IgG1 ex vivo with anti-CD40 and IL-4 (Fig. 1A). Interestingly, when activated B cells were sorted into IgM+ and IgG1+ populations (Supplemental Fig. 1A), Sox2 protein was found to be more abundant in the class-switched IgG1+ fraction (Fig. 1B). Additionally, Sox2 expression in AID-deficient activated B cells was markedly lower than in wild type B cells (Fig. 1C). The delayed kinetics of Sox2 expression and its abundance in class-switched B cells prompted us to query the effect of enforced Sox2 expression on CSR. Mouse splenic B cells were retrovirally infected with either empty vector pMIR (expressing mCherry marker) or pMIR-Sox2 and CSR to IgG1 was assessed in transduced B cells. Overexpression of Sox2 led to a reproducible and marked reduction in CSR to IgG1 (Fig. 1D, Fig 1E, Supplemental Fig. 1B).
FIGURE 1.
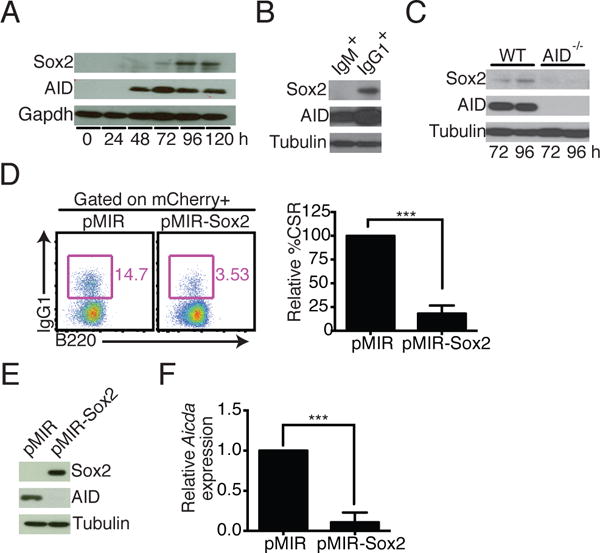
Enforced Sox2 expression alters CSR and AID levels. (A) Immunoblot of protein extracts from wild type splenic B cells stimulated with anti-CD40 and IL-4 for the indicated time and probed for expression of Sox2, AID or Gapdh (loading control) (n = 4). (B) Representative immunoblot of purified IgM+ or IgG1+ splenic B cells obtained from anti-CD40 and IL-4 cultures; tubulin, loading control (n=3). (C) Representative immunoblot of activated splenic B cells from wild type or AID−/− mice (n=3). (D) Representative flow cytometry of B cells infected with pMIR-Sox2 or pMIR control vector. pMIR expresses mCherry from an IRES element downstream of the test gene. Cells were gated first on the mCherry+ population as a marker for infection and then on B220+ IgG1+ cells. Bar graph shows CSR of 15 independent experiments; CSR in the pMIR control population was set at 100%. (E) Immunoblot shows expression of Sox2, AID and tubulin (loading control). (F) mCherry+ B cells were sorted 72h after infection and analyzed for Aicda expression by qPCR; Aicda level in pMIR sample was set to 1 (n = 3).
Consistent with the role of Sox2 in promoting cellular proliferation (19), enforced expression of Sox2 in B cells led to decreased levels of the cell cycle inhibitor p21, and increased levels of Cyclin D2, the major cyclin involved in B cell proliferation (Supplemental Fig. 1C) (20, 21). Furthermore, dilution of CFSE, a surface staining dye, showed that Sox2-expressing cells proliferated slightly better than control cells (Supplemental Fig. 1D); however, CSR was markedly impaired regardless of the extent of proliferation (Supplemental Fig. 1D). Thus, impaired CSR in Sox2-expresing cells was not due to a defect in cell proliferation.
AID is induced by CSR stimuli in B cells and is absolutely required for CSR (12, 22, 23). We therefore assessed AID levels in B cells transduced with pMIR-Sox2. Sox2 overexpression led to a drastic reduction in AID protein (Fig. 1E) and Aicda (encoding AID) mRNA (Fig. 1F). The effect of Sox2 expression on AID levels was not limited to cells stimulated to undergo CSR to IgG1. Overexpression of Sox2 in splenic B cells stimulated with LPS also impaired CSR and AID expression (Supplemental Fig. 1E, 1F). Thus, reduction in AID levels could account for the impaired CSR observed in B cells over-expressing Sox2.
Sox2 targets known regulators of AID expression
To examine if restoring AID levels could rescue CSR, mouse splenic B cells were co-infected with pMIR-Sox2 and pMIG-AID, or control vectors. Despite comparable levels of AID in pMIR/pMIG-AID and pMIR-Sox2/pMIG-AID co-infected samples, CSR to IgG1 was not significantly restored (Fig. 2A–C), suggesting that Sox2 could be impacting multiple parameters effecting CSR. Indeed, germline transcription from Iγ1 was markedly impaired while that through Iμ, which is constitutively transcribed, was unaltered (Fig. 2D). Similar reduction in germline transcription from Iγ3 was observed in Sox2 overexpressing cells stimulated with LPS (Supplemental Fig. 1G). As expected, exogenous AID expression did not restore levels of germline transcripts (Supplemental Fig. 1H). It appears therefore that Sox2 regulates transcription of induced promoters in activated B cells and that the CSR defect observed in Sox2-expressing B cells is likely due to a defect in both induced germline transcription and AID expression.
FIGURE 2.
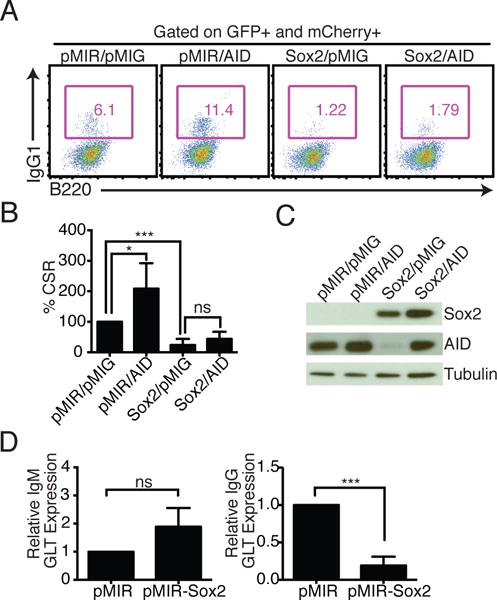
Exogenous AID expression cannot restore CSR in Sox2 overexpressing cells. (A) Representative flow cytometry of splenic B cells co-infected with pMIR, pMIR-Sox2 (Sox2), pMIG, and pMIG-AID (AID). mCherry from pMIR and GFP from pMIG-based vectors served as surrogates for infection. (B) Average CSR in co-infected cells with CSR in pMIR/pMIG co-infected cells set at 100%; n=3. (C) Representative immunoblot showing expression of exogenous Sox2 and AID in the experiment from (A). (D) IgM and IgG1 germline transcripts (GLTs) were analyzed at 72h post-infection following sorting of infected cells based on mCherry expression. For qPCRs, the pMIR sample was set to 1 (n = 3).
To examine the global effects of Sox2 overexpression, we purified pMIR and pMIR-Sox2 transduced B cells based on mCherry expression and subjected them to RNA-seq analysis. Sox2 overexpression led to a significant increase in 702 genes and a decrease in 388 genes (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE93131). These genes were grouped into putative functional categories using the DAVID database. Importantly, while Sox2 overexpression did lead to an increase in the expression of many genes involved in developmental pathways, the master regulators of B cell identity (including Pax5, E2A, and Bcl6) (24–26) were unaltered (Fig. 3A). Consistent with results described above, amongst the most severely decreased genes were Aicda, and several IgH genes (Fig. 3A). Interestingly, Batf and Irf4, two genes known to be involved in regulating AID expression (27, 28), were also significantly downregulated (Fig. 3A), an observation that was independently confirmed at both the mRNA and protein levels (Fig. 3B, 3C). Several genes involved in B cell activation and terminal differentiation were also differentially expressed, although no clear pattern emerged to suggest differentiation into either memory or plasma cells (Fig. 3A). Overall, the RNA-seq analysis suggested that Sox2 could regulate multiple aspects of CSR, including transcription of Aicda.
FIGURE 3.
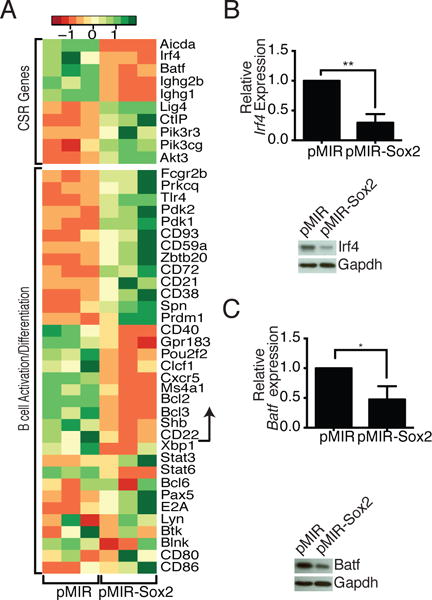
Sox2 dampens expression of the AID regulators Batf and Irf4. Cells were sorted based on mCherry expression as a marker of infection. (A) RNA-Seq was performed on samples from three independent infections. Differential expression of genes involved in CSR and in regulating AID expression (top) or genes expressed during B cell activation and differentiation (bottom) are shown. Genes shown below the arrow were not significantly different in the Sox2 expressing cells. (B, C) qPCR and immunoblot analysis of Irf4 and Batf expression levels in infected cells (n = 3).
Deletion of Sox2 increases IgH:c-Myc translocations
To determine if Sox2 is expressed in B cells in vivo, mice were immunized with the model T-dependent antigen NP-CGG and germinal center B cells were purified (Supplemental Fig. 2A). Sox2 and AID were detectable in the germinal center B cell fraction, but not in the non-germinal center B cell population (Supplemental Fig. 2B). To assess the effect of Sox2 deletion during an immune response, Sox2flox mice in which the coding sequence of Sox2 was flanked by loxP sites (14), were bred to AID-cre transgenic mice in which the Aicda transgene drives expression of the Cre-recombinase in activated B cells (29). Germinal center B cells from Sox2flox/flox:AID-cre mice exhibited similar frequency of CSR to IgG1 and NP-responsiveness relative to control mice (Supplemental Fig. 2C). Additionally, serum immunoglobulin levels in Sox2flox/flox:AID-cre mice were indistinguishable from controls (Supplemental Fig. 2D).
We next assessed the effect of Sox2 deletion on AID levels. For these mechanistic studies, we deleted Sox2 from splenic B cells derived from Sox2flox/flox mice via retroviral transduction of the Cre-recombinase (Fig. 4A). Sox2flox/flox B cells infected with retroviral Cre underwent similar frequency of CSR to IgG1 as uninfected cells (Fig. 4B), even though Sox2 expression was undetectable (Fig. 4C, 4D). There was however, a clear increase in AID protein and Aicda mRNA in Sox2 deleted B cells (Fig. 4D, E).
FIGURE 4.
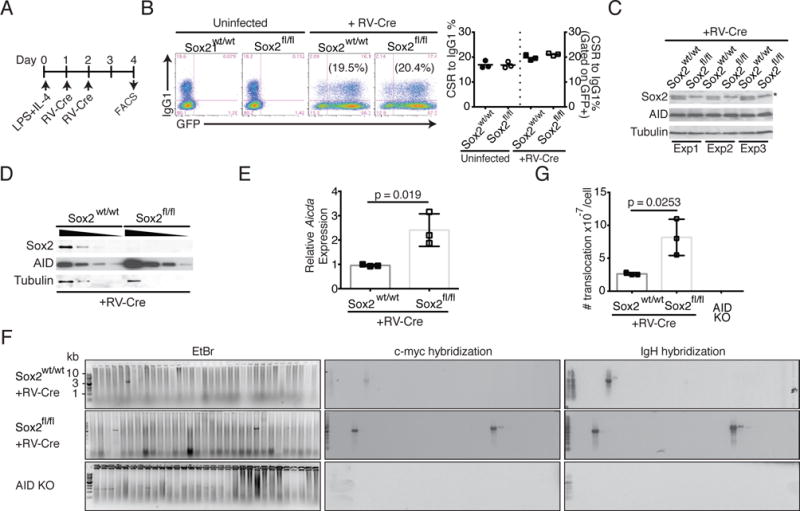
Loss of Sox2 increases IgH:c-Myc translocations. (A) Strategy to delete Sox2 in ex vivo stimulated splenic B cells. B cells were harvested from wild type (Sox2wt/wt) or Sox2flox/flox (Sox2fl/fl) mice, activated with LPS+IL-4 and retrovirally infected with virus expressing Cre-recombinase and a GFP marker. (B) CSR to IgG1 was assayed by flow cytometry and quantified (n=3). (C) Cells were harvested and analyzed by immunoblotting for expression of Sox2, AID and tubulin (loading control). * represents a background polypeptide that would sometimes react with Sox2 antibodies. (D) Immunoblot with 2-fold dilution of protein extracts to assess AID levels. The 4 lanes for each genotype represent 10, 5, 2.5 and 1.25 μg of extract. (E) RNA from infected cells was analyzed for aicda levels by qPCR relative to β-actin; expression of wild type B cells was normalized to 1. (F) Genomic DNA from activated splenic B cells was subjected to two rounds of PCR, resolved on agarose gel, stained with Ethidium bromide (EtBr panel) and followed by Southern blot analysis with IgH and c-Myc probes to assess translocation frequency. (G) Translocation frequency; n=3.
To assess Sox2 deletion induces genomic instability, we determined the frequency of IgH:c-Myc translocation, an established parameter of AID-dependent genome destabilization in B cells (18, 30). There was a significant increase in the frequency of IgH:c-Myc translocations upon loss of Sox2 (Fig. 4F, G). These results provide strong evidence that Sox2 represses AID expression once B cells have undergone CSR to promote genomic integrity.
Collectively, our data show that Sox2 is expressed in activated B cells and negatively regulates expression of AID following completion of CSR. The kinetics of Sox2 expression in B cells also suggests that it could be relevant in the setting of reprogramming. Numerous studies have demonstrated that AID is involved in active demethylation processes, both in artificial settings such as heterokaryon formation and transcription factor-mediated reprogramming, as well as in the natural processes occurring in fertilized oocytes and primordial germ cells (31–34). AID has also been implicated in the demethylation events that occur as part of the epithelial-mesenchymal transition during development and in tumorigenesis (35). For a potent mutator like AID to be involved in these natural processes, it is logical to think that the cell would want a system in place to prevent unwanted AID activity once its role in demethylation is completed. Our recent data showing that AID expression is only needed in the early phase of transcription factor-mediated reprogramming, further suggests a need to limit its expression (33). The data presented in this work therefore have implications for understanding termination of CSR as well as in the field of reprogramming.
Supplementary Material
Acknowledgments
The authors wish to thank T. Honjo for AID−/− mice and members of the Chaudhuri laboratory for their comments and discussion.
Grant Support: This work was supported by grants from NIH/NIAID (1RO1AI072194 and 1RO1AI124186), NIH/NCI Cancer Center Support Grant (P30 CA008748) and the Starr Cancer Research Foundation (I7-A767) to JC.
Abbreviations
- CSR
Class switch recombination
- AID
activation-induced cytidine deaminase
- GLT
germline transcription
- GC
germinal center
- sox2
Sex Determining Region Y HMG-Box 2
- DSBs
Double strand breaks
References
- 1.Hwang JK, Alt FW, Yeap LS. Related Mechanisms of Antibody Somatic Hypermutation and Class Switch Recombination. Microbiology spectrum. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0037-2014. MDNA3-0037-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stavnezer J, Guikema JE, Schrader CE. Mechanism and regulation of class switch recombination. Annual review of immunology. 2008;26:261–292. doi: 10.1146/annurev.immunol.26.021607.090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nussenzweig A, Nussenzweig MC. Origin of chromosomal translocations in lymphoid cancer. Cell. 2010;141:27–38. doi: 10.1016/j.cell.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matthews AJ, Zheng S, DiMenna LJ, Chaudhuri J. Regulation of immunoglobulin class-switch recombination: choreography of noncoding transcription, targeted DNA deamination, and long-range DNA repair. Advances in immunology. 2014;122:1–57. doi: 10.1016/B978-0-12-800267-4.00001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Campolo F, Gori M, Favaro R, Nicolis S, Pellegrini M, Botti F, Rossi P, Jannini EA, Dolci S. Essential role of Sox2 for the establishment and maintenance of the germ cell line. Stem cells. 2013;31:1408–1421. doi: 10.1002/stem.1392. [DOI] [PubMed] [Google Scholar]
- 6.Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes & development. 2001;15:1272–1286. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Que J, Okubo T, Goldenring JR, Nam KT, Kurotani R, Morrisey EE, Taranova O, Pevny LH, Hogan BL. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lesko MH, Driskell RR, Kretzschmar K, Goldie SJ, Watt FM. Sox2 modulates the function of two distinct cell lineages in mouse skin. Developmental biology. 2013;382:15–26. doi: 10.1016/j.ydbio.2013.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pevny LH, Nicolis SK. Sox2 roles in neural stem cells. The international journal of biochemistry & cell biology. 2010;42:421–424. doi: 10.1016/j.biocel.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 10.Xia P, Wang S, Ye B, Du Y, Huang G, Zhu P, Fan Z. Sox2 functions as a sequence-specific DNA sensor in neutrophils to initiate innate immunity against microbial infection. Nature immunology. 2015;16:366–375. doi: 10.1038/ni.3117. [DOI] [PubMed] [Google Scholar]
- 11.Yamanaka S, Blau HM. Nuclear reprogramming to a pluripotent state by three approaches. Nature. 2010;465:704–712. doi: 10.1038/nature09229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 13.Vuong BQ, Lee M, Kabir S, Irimia C, Macchiarulo S, McKnight GS, Chaudhuri J. Specific recruitment of protein kinase A to the immunoglobulin locus regulates class-switch recombination. Nature immunology. 2009;10:420–426. doi: 10.1038/ni.1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaham O, Smith AN, Robinson ML, Taketo MM, Lang RA, Ashery-Padan R. Pax6 is essential for lens fiber cell differentiation. Development. 2009;136:2567–2578. doi: 10.1242/dev.032888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pucella JN, Yen WF, Kim MV, van der Veeken J, Luo CT, Socci ND, Naito Y, Li MO, Iwai N, Chaudhuri J. miR-182 is largely dispensable for adaptive immunity: lack of correlation between expression and function. Journal of immunology. 2015;194:2635–2642. doi: 10.4049/jimmunol.1402261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaudhuri J, Tian M, Khuong C, Chua K, Pinaud E, Alt FW. Transcription-targeted DNA deamination by the AID antibody diversification enzyme. Nature. 2003;422:726–730. doi: 10.1038/nature01574. [DOI] [PubMed] [Google Scholar]
- 17.Nowak U, Matthews AJ, Zheng S, Chaudhuri J. The splicing regulator PTBP2 interacts with the cytidine deaminase AID and promotes binding of AID to switch-region DNA. Nature immunology. 2011;12:160–166. doi: 10.1038/ni.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramiro AR, Jankovic M, Eisenreich T, Difilippantonio S, Chen-Kiang S, Muramatsu M, Honjo T, Nussenzweig A, Nussenzweig MC. AID is required for c-myc/IgH chromosome translocations in vivo. Cell. 2004;118:431–438. doi: 10.1016/j.cell.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Chou MY, Hu FW, Yu CH, Yu CC. Sox2 expression involvement in the oncogenicity and radiochemoresistance of oral cancer stem cells. Oral oncology. 2015;51:31–39. doi: 10.1016/j.oraloncology.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 20.Chiles TC. Regulation and function of cyclin D2 in B lymphocyte subsets. Journal of immunology. 2004;173:2901–2907. doi: 10.4049/jimmunol.173.5.2901. [DOI] [PubMed] [Google Scholar]
- 21.Solvason N, Wu WW, Parry D, Mahony D, Lam EW, Glassford J, Klaus GG, Sicinski P, Weinberg R, Liu YJ, Howard M, Lees E. Cyclin D2 is essential for BCR-mediated proliferation and CD5 B cell development. International immunology. 2000;12:631–638. doi: 10.1093/intimm/12.5.631. [DOI] [PubMed] [Google Scholar]
- 22.Muramatsu M, Sankaranand VS, Anant S, Sugai M, Kinoshita K, Davidson NO, Honjo T. Specific expression of activation-induced cytidine deaminase (AID), a novel member of the RNA-editing deaminase family in germinal center B cells. The Journal of biological chemistry. 1999;274:18470–18476. doi: 10.1074/jbc.274.26.18470. [DOI] [PubMed] [Google Scholar]
- 23.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2) Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 24.Basso K, Dalla-Favera R. BCL6: master regulator of the germinal center reaction and key oncogene in B cell lymphomagenesis. Advances in immunology. 2010;105:193–210. doi: 10.1016/S0065-2776(10)05007-8. [DOI] [PubMed] [Google Scholar]
- 25.Medvedovic J, Ebert A, Tagoh H, Busslinger M. Pax5: a master regulator of B cell development and leukemogenesis. Advances in immunology. 2011;111:179–206. doi: 10.1016/B978-0-12-385991-4.00005-2. [DOI] [PubMed] [Google Scholar]
- 26.Singh H, Medina KL, Pongubala JM. Contingent gene regulatory networks and B cell fate specification. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:4949–4953. doi: 10.1073/pnas.0500480102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fillatreau S, Radbruch A. IRF4 - a factor for class switching and antibody secretion. Nature immunology. 2006;7:704–706. doi: 10.1038/ni0706-704. [DOI] [PubMed] [Google Scholar]
- 28.Ise W, Kohyama M, Schraml BU, Zhang T, Schwer B, Basu U, Alt FW, Tang J, Oltz EM, Murphy TL, Murphy KM. The transcription factor BATF controls the global regulators of class-switch recombination in both B cells and T cells. Nature immunology. 2011;12:536–543. doi: 10.1038/ni.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Robbiani DF, Bothmer A, Callen E, Reina-San-Martin B, Dorsett Y, Difilippantonio S, Bolland DJ, Chen HT, Corcoran AE, Nussenzweig A, Nussenzweig MC. AID is required for the chromosomal breaks in c-myc that lead to c-myc/IgH translocations. Cell. 2008;135:1028–1038. doi: 10.1016/j.cell.2008.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramiro AR, Jankovic M, Callen E, Difilippantonio S, Chen HT, McBride KM, Eisenreich TR, Chen J, Dickins RA, Lowe SW, Nussenzweig A, Nussenzweig MC. Role of genomic instability and p53 in AID-induced c-myc-Igh translocations. Nature. 2006;440:105–109. doi: 10.1038/nature04495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010;463:1042–1047. doi: 10.1038/nature08752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bombardieri M, Barone F, Humby F, Kelly S, McGurk M, Morgan P, Challacombe S, De Vita S, Valesini G, Spencer J, Pitzalis C. Activation-induced cytidine deaminase expression in follicular dendritic cell networks and interfollicular large B cells supports functionality of ectopic lymphoid neogenesis in autoimmune sialoadenitis and MALT lymphoma in Sjogren’s syndrome. Journal of immunology. 2007;179:4929–4938. doi: 10.4049/jimmunol.179.7.4929. [DOI] [PubMed] [Google Scholar]
- 33.Kumar R, DiMenna L, Schrode N, Liu TC, Franck P, Munoz-Descalzo S, Hadjantonakis AK, Zarrin AA, Chaudhuri J, Elemento O, Evans T. AID stabilizes stem-cell phenotype by removing epigenetic memory of pluripotency genes. Nature. 2013;500:89–92. doi: 10.1038/nature12299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Popp C, Dean W, Feng S, Cokus SJ, Andrews S, Pellegrini M, Jacobsen SE, Reik W. Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency. Nature. 2010;463:1101–1105. doi: 10.1038/nature08829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munoz DP, Lee EL, Takayama S, Coppe JP, Heo SJ, Boffelli D, Di Noia JM, Martin DI. Activation-induced cytidine deaminase (AID) is necessary for the epithelial-mesenchymal transition in mammary epithelial cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2977–2986. doi: 10.1073/pnas.1301021110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


