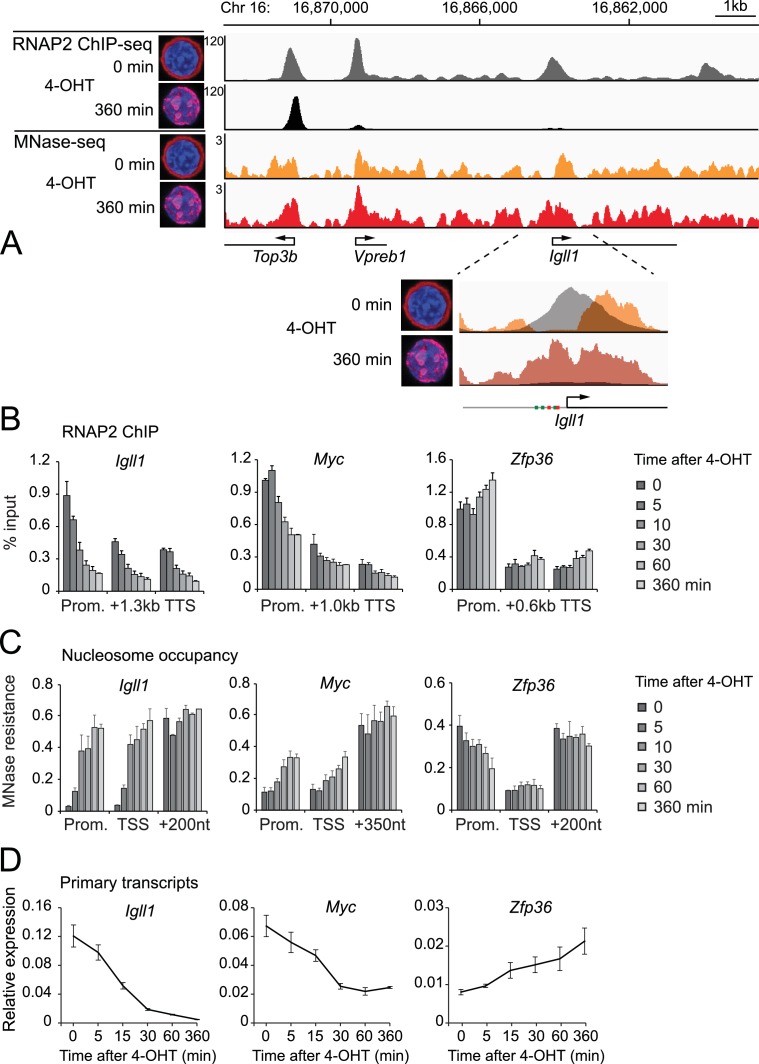
Figure 2. Kinetics of Ikaros-mediated changes in RNAP2 occupancy, locus accessibility and transcription.
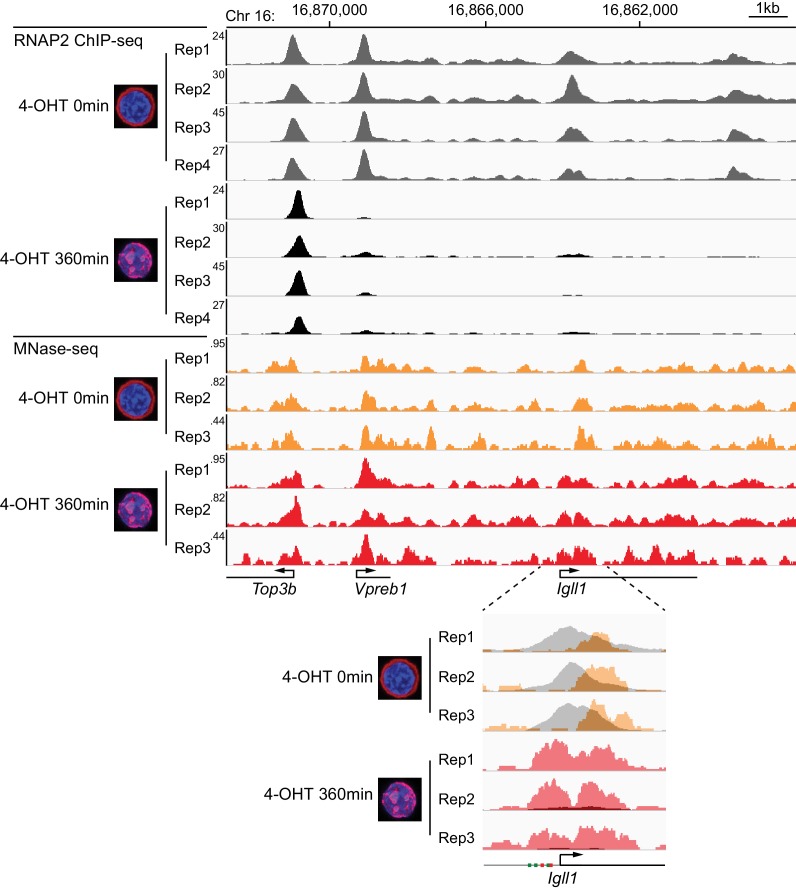
(A) Ikaros triggers RNAP2 eviction and nucleosome recruitment at the Igll1 promoter. RNAP2 ChIP-seq: pooled data from 4 independent biological replicates. MNase-seq: pooled data from 3 independent biological replicates. Figure 2—figure supplement 1 shows the data for each individual replicate. (B) Kinetics of RNAP2 binding at Igll1, Myc, and Zfp36 after 4-OHT by ChIP-PCR. Mean ± SE, n = 3. RNAP2 binding was significantly reduced at the Igll1 promoter, gene body and TTS from 15 min and the Myc promoter from 30 min. (C) Kinetics of nucleosome occupancy at Igll1, Myc, and Zfp36 after 4-OHT by MNase-PCR of 80–120 bp amplicons. 3 independent biological replicates. Nucleosome occupancy was significantly increased at the Igll1 promoter and TSS from 5 min and the Myc promoter and TSS from 15 min. (D) Kinetics of primary Igll1, Myc, and Zfp36 transcripts after 4-OHT by RT-PCR with primer pairs designed to span an intron-exon boundary (Igll1 and Myc) or located within an intron (Zfp36). Mean ± SE, 4 independent biological replicates. Primary Igll1 and Myc transcripts were significantly reduced from 15 and 5 min, respectively.
DOI: http://dx.doi.org/10.7554/eLife.22767.005