Abstract
Mutations in the isocitrate dehydrogenase genes IDH1 and IDH2 are among the first genetic alterations observed during the development of lower-grade glioma (LGG). LGG-associated IDH mutations confer gain-of-function activity by converting α-ketoglutarate to the oncometabolite R-2-hydroxyglutarate (2HG). Clinical samples and gene expression data from The Cancer Genome Atlas (TCGA) demonstrate reduced expression of cytotoxic T lymphocyte–associated genes and IFN-γ–inducible chemokines, including CXCL10, in IDH-mutated (IDH-MUT) tumors compared with IDH-WT tumors. Given these findings, we have investigated the impact of IDH mutations on the immunological milieu in LGG. In immortalized normal human astrocytes (NHAs) and syngeneic mouse glioma models, the introduction of mutant IDH1 or treatment with 2HG reduced levels of CXCL10, which was associated with decreased production of STAT1, a regulator of CXCL10. Expression of mutant IDH1 also suppressed the accumulation of T cells in tumor sites. Reductions in CXCL10 and T cell accumulation were reversed by IDH-C35, a specific inhibitor of mutant IDH1. Furthermore, IDH-C35 enhanced the efficacy of vaccine immunotherapy in mice bearing IDH-MUT gliomas. Our findings demonstrate a mechanism of immune evasion in IDH-MUT gliomas and suggest that specific inhibitors of mutant IDH may improve the efficacy of immunotherapy in patients with IDH-MUT gliomas.
Introduction
Gliomas are the most common primary brain tumors and are typically classified on the basis of WHO criteria as grades I–IV, in order of increasing anaplasia (1). Grade IV glioblastomas (GBMs) are the most aggressive, with a median survival time of less than 15 months (2). While the majority of WHO grade I gliomas are curable, lower-grade (WHO grade II or III) diffuse gliomas (1) are considered malignant because of their invasive growth, resistance to therapy, and high risk of transforming into higher-grade gliomas (3).
Recent genomic studies, including those of The Cancer Genome Atlas (TCGA), have guided us toward a better understanding of the molecular characterizations of lower-grade gliomas (LGGs) (4). Among the earliest signature molecular alterations, mutations in the isocitrate dehydrogenase IDH1 and IDH2 genes are of particular interest. These mutations have been found to be early and frequent (70%–80%) genetic alterations in LGG patients (5), as well as in a small fraction of GBM patients, especially those with secondary GBM who progress from LGG (6). IDH mutations persist throughout multiple recurrences, chemotherapy, and resections (7). All IDH1 and IDH2 mutations identified to date affect a single amino acid located within the isocitrate-binding site (IDH1 [R132] or IDH2 [R140 and R172]) and confer a novel gain-of-function activity by converting α-ketoglutarate (αKG) to its (R)-enantiomer of 2-hydroxyglutarate (2HG). Reduced availability of αKG and accumulation of 2HG by mutant IDH (8) may coordinate genome-wide epigenetic changes, such as the glioma–CpG island methylator phenotype, that predispose the cells toward malignant transformation (reviewed in ref. 5).
Immunotherapy remains an unproven but promising approach for treating brain tumors (reviewed in refs. 9, 10). We have demonstrated that tumor-specific type 1 CD8+ T cells, which predominantly secrete IFN-γ, but not type 2 CD8+ T cells, can efficiently traffic into brain tumor sites and mediate effective tumor cell killing (11) via the type 1 chemokine CXCL10 (11–14). Despite the importance of the type 1 T cell response, gliomas and other cancers secrete numerous type 2 cytokines (15–17) that promote tumor proliferation (18, 19) and immune escape (20–22).
Therefore, we asked whether IDH mutations could directly suppress genes required for a type 1 CD8+ T cell response, allowing for glioma growth. Our analyses of clinical samples and TCGA RNA-sequencing (RNA-seq) data demonstrated reduced expression of CD8, type 1–associated effector molecules, and chemokines. Using orthotopic syngeneic glioma models, we demonstrated that IDH1 mutations and 2HG recapitulate this phenotype. As intrinsic events in an IDH-mutated (IDH-MUT) tumor cell, we found that either the IDH1 R132H mutation or 2HG can suppress the protein-level expression of STAT1, leading to the observed decrease in type 1–associated chemokines, such as CXCL10. Finally, use of IDH-C35, the specific inhibitor of mutant IDH1, improved antitumor immunity and enhanced the efficacy of a peptide vaccine. Our findings demonstrate what we believe to be a novel, IDH-MUT–mediated mechanism of immune evasion and further suggest that mutant IDH inhibitors can be used to enhance glioma immunotherapy in patients with IDH-MUT gliomas.
Results
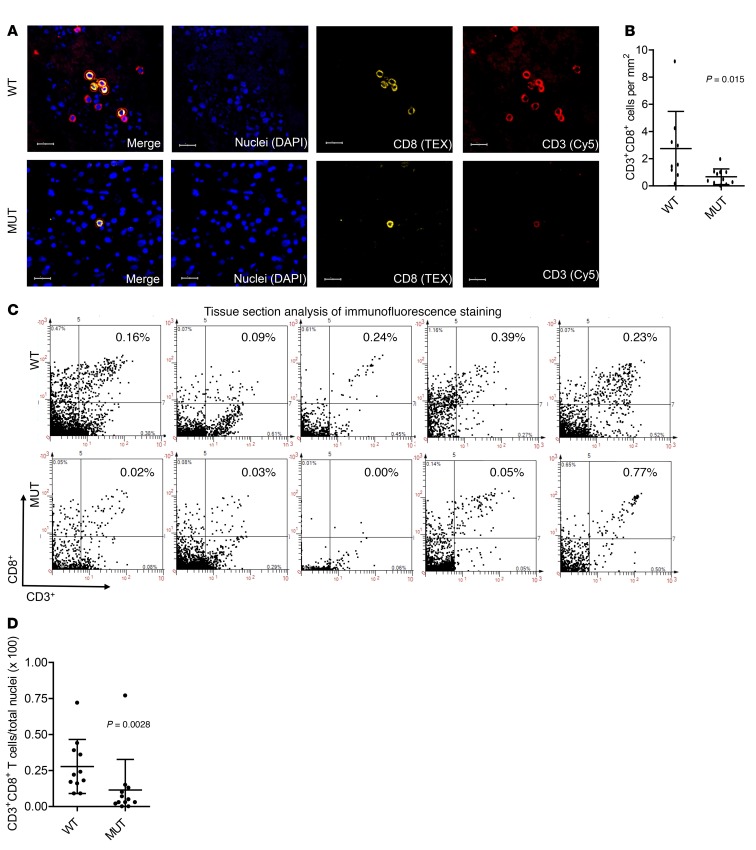
Immunofluorescence evaluation confirms reduced CD3+CD8+ T cell infiltration levels in IDH-MUT WHO grade III gliomas compared with levels in IDH-WT gliomas.
To understand how IDH mutations impact the immunological environment of LGGs, we first performed immunofluorescence analysis on formalin-fixed, paraffin-embedded (FFPE) tissue sections derived from IDH-MUT (n = 11) and IDH-WT (n = 9) WHO grade III gliomas. We confirmed IDH1 and IDH2 status by sequencing, using the method previously described (ref. 23 and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI90644DS1.org). Using TissueFAXs and StrataQuest tissue analysis software, CD3+CD8+ double-positive (dp) cells were identified (indicated by red circles) on the IDH-WT and IDH-MUT tissue sections (Figure 1A and Supplemental Figure 2). We observed greater numbers of CD3+CD8+ cells per tumor area in IDH-WT tumors than in IDH-MUT tumors (Figure 1B). To ensure that the higher numbers of T cells were not merely due to differences in overall cell density between the 2 groups, we confirmed the greater numbers of CD3+CD8+ cells per total DAPI+ nuclei, which represent all cells, in IDH-WT compared with IDH-MUT tumors (Figure 1, C and D). On the other hand, we observed no difference in CD3+CD8– cells between IDH-WT and IDH-MUT cases (Supplemental Figure 3). The CD3+CD8– cell population included CD3+CD4+ T cells, but we were unable to detect any γδ+ T cells in IDH-WT or IDH-MUT cases (Supplemental Figure 3). Also, we were unable to reliably enumerate CD4+ T cells. Overall, these data demonstrate a significant reduction of CD3+CD8+ T cell accumulation in IDH-MUT tumors compared with accumulation in IDH-WT tumors.
Figure 1. Reduced CD8+ T cell numbers in WHO grade III IDH-MUT gliomas compared with IDH-WT gliomas.
FFPE sections from IDH-WT (n = 9) and IDH-MUT (n = 11) WHO grade III gliomas were stained for CD3 (Cy5) and CD8 (Texas Red [TEX]) and analyzed using StrataQuest software. (A) Representative staining for CD3 (red), CD8 (yellow), and nuclei (blue) either alone or merged in both IDH-WT (WT) and IDH-MUT (MUT) cases. Red circles represent automated cell masks on CD3 and CD8 dp cells. Scale bars: 20 μm. (B) Number of CD3+CD8+ dp cells per area (mm2) of tumor calculated for each case. The average tumor area per tissue section was 111.21 mm2 for IDH-WT and 165.35 mm2 for IDH-MUT cases. (C) Scatter diagrams illustrating the staining intensity of CD3 and CD8 on the x and y axes, respectively. The percentage of cells in each quadrant was based on the total number of DAPI+ (blue) nuclei. Each dot represents an individual case. (D) CD3+CD8+ dp cells as a percentage of total DAPI+ cells (nuclei) calculated for each case. In B and D, each dot represents a value from a single patient, black lines represent the mean, and error bars indicate the SD of samples in a group. P values were obtained using a 2-sided, unpaired Mann-Whitney U test.
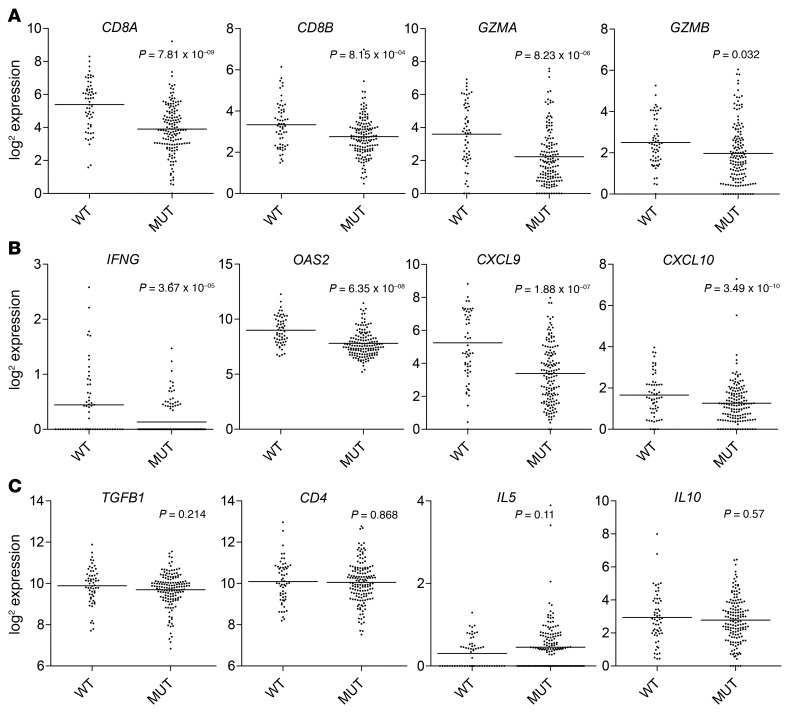
IDH mutations are associated with lower levels of CD8+ cytotoxic T cell infiltration and IFN-γ–induced chemokine gene expression in patients with LGG.
Using level 3 gene expression data from TCGA database, we compared gene expression profiles of IDH-MUT (n = 149) and IDH-WT (n = 58) cases. As oligodendroglioma has a different biology and prognosis than does astrocytoma, we excluded cases with chromosome 1p/19q deletion, which is a marker for oligodendroglioma (1). Genes related to CD8+ CTLs (CD8A, CD8B, GZMA, and GZMB) (Figure 2A), IFN-γ and IFN-γ–inducible genes (IFNG and OAS2), as well as the chemokines CXCL9 and CXCL10 (Figure 2B) were significantly lower in IDH-MUT cases compared with IDH-WT counterparts. Conversely, we observed no differences in the Th2- or Treg-associated cytokines TGFB1, IL5, IL10, or CD4 (Figure 2C).
Figure 2. Lower expression levels of CD8+ T cell–associated genes in IDH-MUT versus IDH-WT patients.
TCGA level 3 gene expression data were downloaded from the University of California Santa Cruz Cancer Genome Browser for 1p/19q intact, WHO grade II or III glioma cases (n = 207) (http://www.cbioportal.org/). IDH-MUT (n = 149) and IDH-WT (n = 58) cases were compared for immune cell and immune effector gene expression levels. Differentially expressed genes associated with (A) CD8+ CTLs and (B) IFNG and IFN-γ–inducible genes. (C) Gene expression associated with immunosuppression (TGFB1), CD4 T cells (CD4), and type 2 immunity (IL5 and IL10). Graphs depict the log2 expression value of each gene: each dot represents 1 patient; black bars show the average value. P values were determined using 2-sided, unpaired t tests, with Benjamini-Hochberg adjustment for multiple testing.
To evaluate genes that are upregulated or downregulated in IDH-MUT cases in an unbiased and comprehensive manner, we evaluated Gene Ontology (GO) terms, each containing genes annotated by each term (24), in these cases. We found that 465 GO terms are upregulated (Supplemental Table 1) and 704 GO terms are downregulated in IDH-MUT (Supplemental Table 2) cases compared with IDH-WT cases. Among them, only 2 immune-related GO terms are upregulated in IDH-MUT cases (Supplemental Table 1), while 74 immune-related GO terms are downregulated in IDH-MUT cases (Supplemental Table 2).
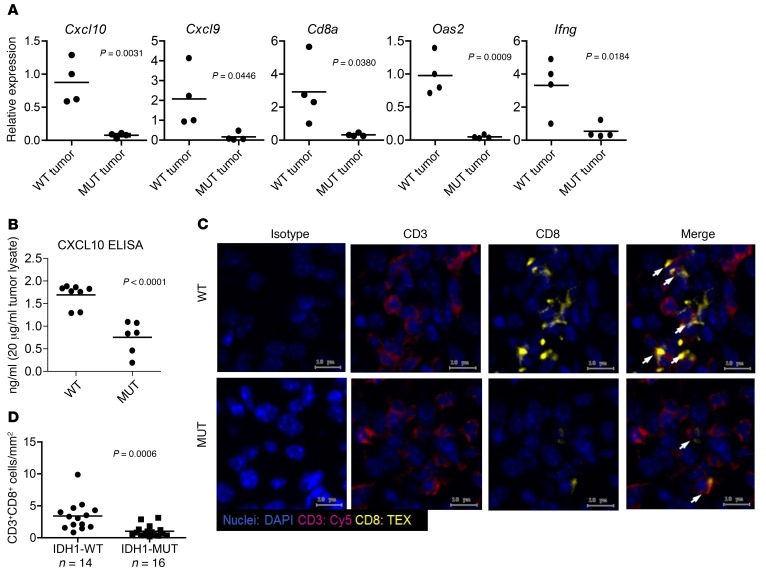
Syngeneic mouse gliomas with the IDH1 R132H mutation recapitulate the immune phenotype of human IDH-MUT gliomas in vivo.
To determine what mechanisms underlie the observed suppression of CD8+ CTL accumulation in IDH-MUT gliomas, we stably transfected mouse GL261 and SB28 glioma cell lines as well as normal human astrocytes (NHAs) with cDNA encoding either WT-IDH1(WT) or IDH1 containing the R132H mutation (R132H), the most common mutant IDH mutation (25). Liquid chromatography–mass spectrometry (LC-MS) analysis verified the functional expression of the mutant IDH1 through detection of oncometabolite 2HG production in GL261R132H, NHAR132H, and SB28R132H cells (Supplemental Figure 4).
We harvested glioma tissues from C57BL/6 mice implanted with IDH1WT or IDH1R132H gliomas in the brain and then evaluated the expression of immune response–related genes by real-time PCR (RT-PCR). Consistent with TCGA data, both SB28R132H (Figure 3A) and GL261R132H (Supplemental Figure 5) tumors had reduced levels of CTL-associated and IFN-γ–inducible chemokine-related genes compared with WT counterpart tumors. Additionally, CXCL10 ELISA revealed significantly reduced levels of CXCL10 in GL261R132H tumors compared with levels in GL261WT tumors (Figure 3B). Furthermore, immunofluorescence staining showed reduced CTL accumulation in intracranial GL261R132H gliomas compared with accumulation in GL261WT gliomas (Figure 3, C and D). These data demonstrate the direct role of IDH1R132H in the suppression of T cell accumulation in gliomas.
Figure 3. Syngeneic IDH-MUT murine gliomas demonstrate reduced chemokine- and IFN-γ pathway–related genes and CTL infiltration.
C57BL/6 mice received intracranial injections of 1 × 105 GL261 or SB28 glioma cells stably transfected with cDNA for either WT or the R132H form of IDH1. Day-21 tumors were removed from mice and further assessed. (A) RT-PCR analysis of SB28 tumor–derived RNA showing decreased expression of CTL response–related genes in SB28-MUT (MUT tumor; n = 4) versus SB28-WT (WT tumor; n = 4) tumors. (B) CXCL10 ELISA was performed on tumor-derived protein extracts from mice bearing GL261-WT or GL261-MUT tumors. (C) Representative fluorescence microscopy images from sections stained with DAPI (blue), CD3 (magenta), and CD8 (yellow) on frozen tumor sections from mice bearing GL261-WT or GL261-MUT tumors. Cells were imaged as described in Figure 2. Scale bars: 10 μm. (D) Quantification of CD3+CD8+ dp cells from GL261-WT (n = 14) and GL261-MUT (n = 16) tumors using StrataQuest software. P values were obtained using a 2-sided, unpaired t test.
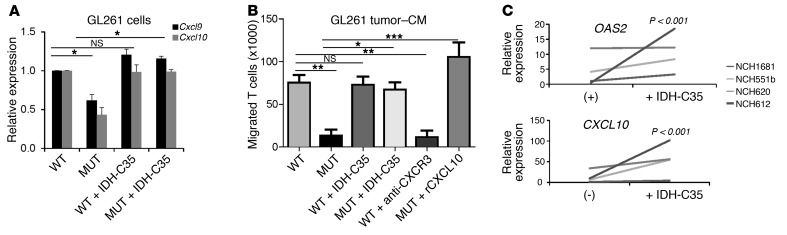
IDH-MUT glioma cells produce reduced levels of CTL-attracting chemokines.
The observed expression of immune-related genes in LGG and murine glioma tissues led us to examine whether these genes are expressed by glioma cells themselves. RT-PCR evaluation of in vitro–cultured cells revealed that the NHA cell line and mouse glioma cell lines GL261 and SB28 expressed the CTL-attracting chemokines CXCL9 and CXCL10 (Figure 4A and Supplemental Figure 6), but not CD8A, CD8B, GZMB, or IFNG (Supplemental Figure 7). GL261R132H cells expressed reduced levels of the CTL-attracting chemokines Cxcl9 and Cxcl10 in comparison with GL261WT cells (Figure 4A). We observed similar results by RT-PCR in both SB28 and NHA models and by CXCL10-specific ELISA in the NHA model (Supplemental Figure 6). Treatment of GL261R132H cells with a specific inhibitor of mutant IDH1 (IDH-C35) reversed the levels of Cxcl9 and Cxcl10 to those observed with GL261-WT cells. However, IDH-C35 did not affect the Cxcl9 or Cxcl10 levels in GL261-WT cells (Figure 4A). These findings indicate that mutant IDH1 leads to decreased expression levels of CTL-attracting chemokines within glioma cells.
Figure 4. Decreased chemokines and chemotaxis of T cells to IDH-MUT–derived CM compared with media derived from IDH-WT cells.
(A and B) GL261-WT or GL261- MUT cells treated with either DMSO (control) or 1 μM IDH-C35 for 3 days were stimulated with 10 ng/ml recombinant murine IFN-γ for 6 hours prior to analysis. (A) RT-PCR analysis of Cxcl9 and Cxcl10 from GL261-WT (WT) and GL261-MUT (MUT) cells treated with or without IDH-C35. Gene expression levels were normalized to Gapdh and are shown relative to levels in WT samples. (B) Chemotaxis of activated CD8+ T cells toward GL261 glioma cell–CM by Transwell assay. CM were diluted 1:1 with fresh media. For CXCL10 conditions, 100 ng/ml recombinant murine CXCL10 (rCXCL10) was added to GL261-MUT–CM. For blockade of CXCR3, T cells were preincubated in 10 μg/ml anti-CXCR3 for 30 minutes prior to use. Results are representative of 3 independent experiments, each performed in triplicate with highly similar results. (C) RT-PCR analysis of OAS2 and CXCL10 gene expression from 4 unique glioma patient–derived neurospheres with IDH1 mutations either untreated (–) or treated with IDH-C35 for 3 days. Each line (and sample name) corresponds to a neurosphere generated from a different patient. Gene expression levels were normalized to GAPDH and are shown relative to expression levels in untreated neurosphere NCH1681. P values were obtained using 1-way ANOVA with a Newman-Keuls multiple comparisons test (A and B) and a 2-tailed, paired t test (C). *P < 0.05, **P < 0.01, ***P < 0.001; NS, P > 0.05.
We next performed a Transwell assay to assess the migration of murine splenocyte-derived CD8+ T cells toward glioma cell line–conditioned media (CM) derived from either IDH1WT or IDHR132H cell lines. Migration of CD8+ T cells toward CM of IDHR132H tumors was reduced compared with that observed in IDH1WT CM using both SB28 and GL261 models (Figure 4B and Supplemental Figure 8). Importantly, antibody-mediated blockade of CXCR3, the receptor for CXCL9 and CXCL10, reduced T cell migration toward GL261WT CM. Furthermore, the addition of recombinant murine CXCL10 to CM from IDHR132H cultures increased T cell migration to levels observed when using IDHWT CM (Figure 4B). Remarkably, when CM was derived from GL261R132H or SB28R132H cells treated with IDH-C35, migration of T cells recovered to the levels observed with IDH1WT CM (Figure 4B and Supplemental Figure 8, respectively). Taken together, these observations demonstrate that IDH-MUT glioma cells produce reduced levels of CTL-attracting chemokines, which can directly lead to the decreased migration of CD8+ T cells.
Furthermore, treatment of neurosphere cultures derived from 4 IDH-MUT glioma patients (NCH612, grade III oligodendroglioma; NCH1681, grade III astrocytoma; NCH551b and NCH620, secondary GBM) with IDH-C35 resulted in enhanced OAS2 and CXCL10 expression (Figure 4C). Of note, these neurosphere cultures expressed cancer stem cell–related genes including CD133, SOX2, CD44, CSPG4, CD90, and nestin (ref. 26 and Supplemental Figure 9), suggesting that the observed mechanism may also be relevant to glioma stem cells.
Decreased STAT1 in IDH-MUT and 2HG-treated tumor cell lines.
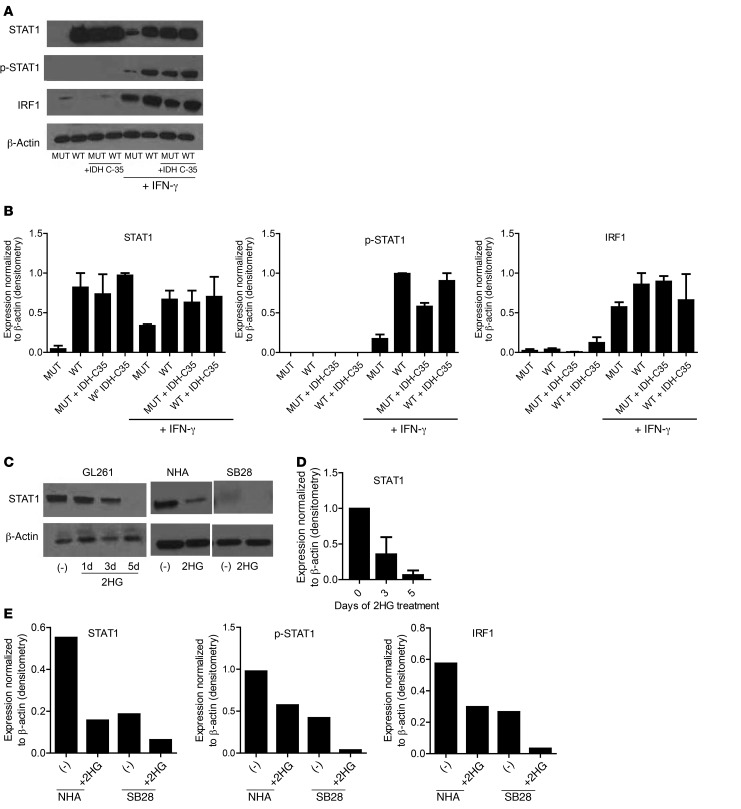
To understand the mechanism by which IDH-MUT cells produce reduced levels of CTL-attracting chemokines, we evaluated the regulators of CXCL9 and CXCL10, IFN regulatory factor 1 (IRF1), STAT1, and phosphorylated STAT1 (p-STAT1) by Western blotting (ref. 27 and Figure 5, A and B). Surprisingly, in addition to decreased levels of IRF1 and p-STAT1, the total amount of STAT1 protein was markedly decreased in GL261R132H cells compared with that in GL261WT cells, despite STAT1 activity being commonly regulated through its phosphorylation (ref. 28 and Figure 5, A and B). We observed reduced STAT1 signaling even in the presence of IFN-γ, which is a primary activator of STAT1 signaling (28). Additionally, IDH-C35 treatment of GL261R132H restored STAT1 levels to those of GL261WT cells (Figure 5, A and B). Since mutant IDH leads to production of the oncometabolite 2HG, we next examined the ability of 2HG to directly suppress STAT1 protein. Treatment of parental GL261 cells with 3 mM 2HG for 1, 3, or 5 days resulted in the gradual loss of STAT1 protein (Figure 5, C and D). Similarly, NHA and SB28 cells treated with 3 mM 2HG for 5 days showed strongly reduced STAT1, p-STAT1, and IRF1 protein levels (Figure 5, C and E).
Figure 5. Decreased STAT1 protein levels in IDH-MUT and 2HG-treated tumor cell lines.
(A) Western blotting was performed on GL261-WT and GL261-MUT cell lysates in the presence or absence of 1 μM IDH-C35 and 100 ng/ml recombinant murine IFN-γ. (B) Quantification of Western blot bands by ImageJ. Data represent the mean ± SD of band density/β-actin of 2 to 4 experiments. (C) Western blotting was performed on cell lines treated with 3 mM 2HG. Data shown represent GL261 cells treated with 2HG for 1, 3, or 5 days and NHA and SB28 cells treated with 2HG for 5 days. (D) ImageJ quantification of Western blot bands from C. Data represent the mean ± SD of band density/β-actin band density from 3 independent experiments. (E) ImageJ quantification of Western blot band densities of STAT1, p-STAT1, and IRF1, normalized to β-actin levels for NHA and SB28 cells treated with 2HG. Data are representative of 2 independent experiments with similar results.
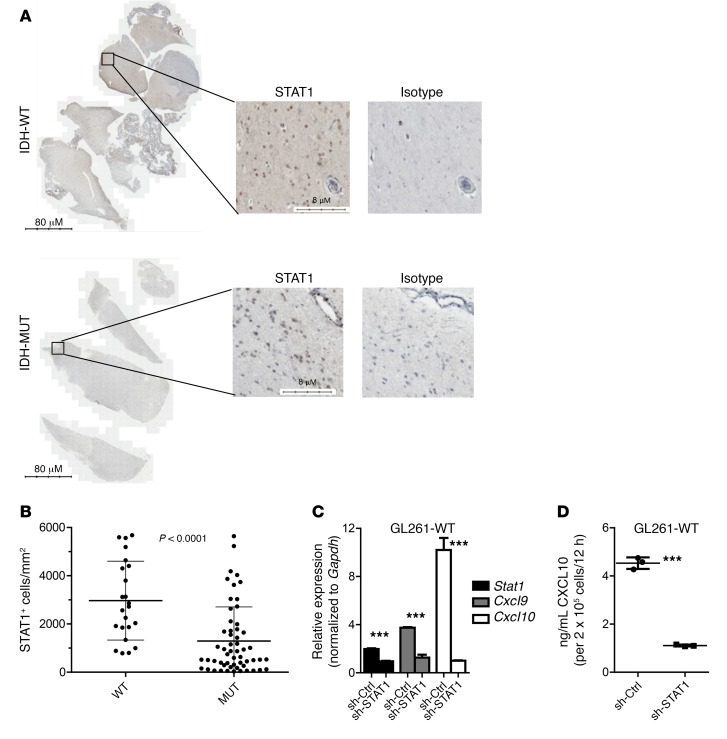
We further examined STAT1 expression on IDH-WT and IDH-MUT LGG tissue sections by immunohistochemical staining (Figure 6A). IDH-MUT tumors (23 sections from 11 cases) demonstrated decreased STAT1+ cells per area of tumor (mm2) compared with IDH-WT tumors (56 sections from 9 cases) (Figure 6B).
Figure 6. Decreased STAT1 positive cells in IDH-MUT compared with IDH-WT LGG tumor sections.
(A) Representative images showing STAT1 staining on IDH-WT and IDH-MUT WHO grade III LGG samples. Scale bars: 80 μm and 8 μm. (B) Quantification of STAT1+ cells per area of tumor (mm2) on IDH-WT (56 sections from 9 cases) and IDH-MUT (23 sections from 11 cases) tumors. Data represent the mean ± SD. Data obtained by 2-tailed unpaired t test. (C) RT-PCR analysis of Stat1, Cxcl9, and Cxcl10 expression in GL261WT cells stimulated with 10 ng/ml recombinant murine IFN-γ for 12 hours, 36 hours after transfection with 1 μg plasmid encoding either scrambled shRNA (OriGene Technologies; TR30013) or 4 shRNAs targeting STAT1 (OriGene Technologies; TG502153). (D) CXCL10 ELISA was performed using supernatant from the cells described in C. Data shown in C and D represent the mean ± SD of 3 biologic replicate samples and are representative of 2 independent experiments; ***P < 0.0001. sh-Ctrl, sh-control.
To demonstrate whether STAT1 directly regulates CXCL9 and CXCL10 in glioma cells, GL261WT cells were transfected with retroviral vectors encoding either scrambled shRNA or 4 unique shRNAs targeting STAT1 and then stimulated for 12 hours with recombinant murine IFN-γ. GL261WT cells transfected with STAT1 shRNA demonstrated decreased Stat1, Cxcl9, and Cxcl10 levels by RT-PCR (Figure 6C) and decreased CXCL10 as measured by ELISA (Figure 6D) compared with cells transfected with scrambled shRNA.
IDH-C35 enhanced the efficacy of a peptide-based vaccine in mice challenged with GL261.
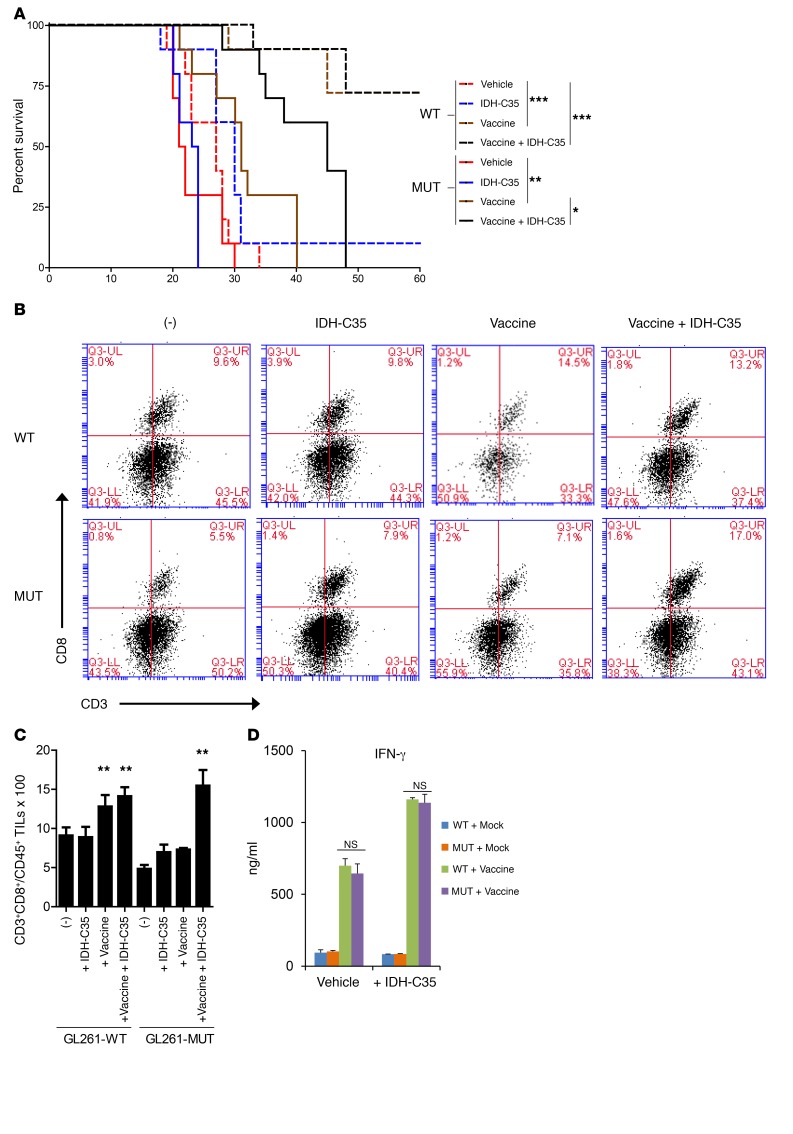
To determine whether IDH mutation status impacts the efficacy of peptide-based vaccine therapy, C57BL/6 mice received 3 doses of prophylactic vaccinations with synthetic peptides derived from glioma-associated antigens (GAAs) (EPHA2671–679, EPHA2682–689, TRP2180–188, GARC1177–185, and HBV core128–140) (29, 30) emulsified in incomplete Freund’s adjuvant (IFA) with polyinosinic-polycytidylic acid with polylysine and carboxymethylcellulose (poly-ICLC) as an adjuvant. Vaccinated mice were then challenged with GL261WT or GL261R132H tumors. While more than 80% of mice challenged with GL261WT tumors rejected the tumor, the vaccine treatment prolonged the survival of mice challenged with GL261R132H tumors only marginally (median survival of 21.5 vs. 23.5 days; P = 0.0016) compared with mice treated with vehicles alone and was unable to lead to long-term survival beyond 40 days after tumor inoculation (Figure 7A).
Figure 7. Treatment with IDH-C35 improves the efficacy of peptide vaccines in mice bearing GL261-MUT tumors.
C57BL/6 mice were vaccinated 3 times with synthetic peptides encoding GAAs presented by GL261 cells (EPHA2671–679, EPHA2682–689, TRP2180–188, GARC1177–185, and HBV core128–140) emulsified in IFA with 20 μg poly-ICLC as an adjuvant. Control mock vaccines consisted of 100 μg HBV core128–140, but without GAA peptides emulsified in IFA with 20 μg Poly-ICLC. Vaccinated mice received intracranial injections of either 1 × 105 GL261-WT or 1 × 105 GL261-MUT cells and received daily treatment with vehicle or 450 mg/kg/day IDH-C35. (A) Kaplan-Meier curves demonstrating survival of mice bearing GL261-WT or GL261-MUT gliomas in the brain (n = 10/group). (B and C) Mice that received treatments and intracranial tumor challenge as above were sacrificed on day 21 after tumor inoculation for evaluation of immune responses. Data are representative of 2 independent experiments with similar results. (B) Representative flow cytometric analysis of tumor-infiltrating CD3+CD8+ T cells gated on CD45+ live lymphocytes. Q3, gating on CD45+ live lymphocytes; UL, upper left; UR, upper right; LL, lower left; LR, lower right. (C) Mean percentage of tumor-infiltrating CD3+CD8+ cells from each treatment group (n = 3 tumors/group). (D) IFN-γ ELISA evaluation of antigen-specific T cell responses in peripheral blood samples derived from treated mice on day 10 after tumor inoculation. Data are expressed as the mean ± SD. Results were analyzed by log-rank (Mantel-Cox) and 1-way ANOVA with Dunnett’s multiple comparisons test for data sets in A and C, respectively. *P < 0.05, **P < 0.01, ***P < 0.0001, and NS, P > 0.05.
We then asked whether IDH-C35 could improve vaccine efficacy in IDH-MUT tumor–bearing mice. Vaccinated mice challenged with GL261WT or GL261R132H tumors received daily treatment by oral gavage with IDH-C35 (450 mg/kg/day; Figure 7A). The treatment had no affect on the survival of mice bearing GL261WT gliomas, with or without prior peptide vaccination (Figure 7A), or of mice bearing GL261R132H gliomas without prior vaccination (Figure 7A). On the other hand, IDH-C35 significantly improved the survival of GAA-vaccinated mice bearing GL261R132H gliomas compared with GL261R132H glioma–bearing mice that received peptide vaccine but no IDH-C35 (median survival of 31 vs. 45 days; P = 0.0177; Figure 7A). Furthermore, IDH-C35 treatment of vaccinated mice enhanced CD3+CD8+ T cell infiltration (Figure 7, B and C) and Cxcl10 expression (Supplemental Figure 10) in the glioma cells compared with mice that received vaccine treatment alone. Consistent with our immunofluorescence analyses of WHO grade III cases (Supplemental Figure 3), there were no differences in the level of CD3+CD8– T cell infiltration (Supplemental Figure 11) between GL261-WT and GL261-MUT tumors for either treatment condition. Of note, peripheral blood–derived T cells from prevaccinated mice with or without IDH-C35 treatment showed equivalent levels of IFN-γ response to peptides in the vaccine, indicating that the IDH mutation status in the intracranial glioma did not affect systemic responses to the peptide-based vaccine (Figure 7D).
Discussion
To understand how IDH mutations influence the immunological microenvironment of LGGs, we compared immune responses in IDH-MUT and IDH-WT LGGs using clinical specimens, the TCGA database, and newly created syngeneic murine models of IDH-WT and IDH-MUT gliomas. We found that IDH-MUT LGGs exhibited a markedly reduced CD8+ CTL signature compared with IDH-WT LGGs. Using the syngeneic SB28 and GL261 mouse models, we found that the IDH1 R132H mutation in murine gliomas led to a reduction of CD8+ T cells and CTL-attracting chemokines. Our in vitro experiments, designed to investigate glioma cell–intrinsic mechanisms, revealed that IDH-MUT tumors had reduced expression of the T cell–attracting chemokines CXCL9 and CXCL10, leading to reduced migration of T cells toward CM from IDH-MUT cells compared with media from IDH-WT cells. Furthermore, compared with IDH-WT cell lines, IDH-MUT cells expressed lower levels of STAT1 protein, an important regulator of CXCL 9 and CXCL10. We also found that these changes were mediated through 2HG. Finally, IDH-C35, a selective inhibitor that blocks the ability of IDH1 mutations to produce 2HG, reversed the inhibition of CXCL10 and T cell migration and enhanced the efficacy of GAA vaccines in mice bearing GL261R132H gliomas.
Our findings highlight what we believe to be a novel mechanism of immune evasion and demonstrate that mutations in IDH suppress chemokines critical for the accumulation of CD8+ T cells in the tumor. We have previously reported the importance of CXCL10 in the recruitment of CD8+ T cells to murine gliomas (11, 13, 31) and the preliminary clinical activity of vaccinations with α-type 1–polarized DCs, which induced CXCL10 production in patients with GBM (32). Our data in the current study corroborated these previous observations in both clinical and preclinical LGG models and suggest that the development of T cell–based immunotherapies for LGGs should take into consideration the altered chemokine profiles of IDH-MUT tumors.
IDH mutations are believed to be one of the initial mutations to occur in gliomas (7). Thus, all currently available models are inherently limited, in that these gliomas lack an IDH mutation in early development. Nonetheless, in the current study, we used multiple cell models, including the NHA model, which has been extensively characterized as a model for tumorigenic transformation of astrocytes, especially in the presence of an IDH1 mutation (33, 34). Additionally, we used the C57BL/6 mouse syngeneic SB28 model, which was driven by transfection of the human oncogenes PDGFB , NRAS, and short hairpin targeting P53 (35), thereby genetically resembling proneural GBMs, which commonly harbor IDH mutations (36). Although GL261 is a chemically induced glioma cell line (37), this model was critical for the current study, as it is one of the most extensively studied syngeneic mouse glioma models with established T cell epitopes for vaccines (29, 30). Although none of these models perfectly resembles human IDH-MUT LGGs, our data from these models consistently demonstrated the impact of IDH mutations on STAT1 and the subsequent immunological milieu. Furthermore, the use of primary spheroid cultures derived from IDH-MUT glioma patients confirmed our observations related to the reversal of CXCL10 production following IDH-C35 treatment (Figure 4C).
Patients with IDH-MUT LGGs have better survival rates than do patients with IDH-WT LGGs (38). This may seem inconsistent with our syngeneic mouse models, in which we observed no difference in survival between mice with GL261-WT and those with GL261-MUT tumors (Figure 7). Furthermore, our observations that IDH-MUT gliomas have reduced numbers of infiltrating T cells may seem contradictory to those of previous studies demonstrating that tumor infiltration by CD8+ T cells is predictive of better patient survival (39–41). However, it is important to note that human IDH-WT LGGs may be driven by other oncogenic signals that are not present in IDH-MUT tumors, and poor prognosis may indicate that the biology of IDH-WT LGGs more closely resembles that of GBM than of LGGs (42). In our models, genetic backgrounds for IDH-MUT versus IDH-WT cells (with SB28, GL261, and NHA) were the same except for the status of IDH, which is not the case in human LGGs. Future studies are warranted to better understand the tumor properties that account for the differential survival of patients with IDH-WT LGGs and those with IDH-MUT LGGs.
The biological activities of STAT1 are known to be regulated by phosphorylation of STAT1 at tyrosine 701 and serine 727 sites and involve homodimerization of STAT1 (43). However, regulation of STAT1 signaling may also occur at the expression level of STAT1 protein itself (44–46). As 2HG is known to cause CpG site hypermethylation (47), we hypothesize that STAT1 expression might be regulated by the methylation of its promoter. However, our analysis of STAT1 methylation site β values from TCGA 450k methylation data (Supplemental Figure 12) and Illumina 850k array on NHA cells revealed no significant differences in STAT1 promoter methylation between IDH-WT and IDH-MUT samples (48). Further studies are warranted to understand the mechanism by which mutant IDH and 2HG suppress STAT1 protein levels.
In the current study, we showed the efficacy of IDH-C35 in recovering STAT1, CXCL10, and T cell chemotaxis. Recently, Johannessen et al. (33) demonstrated that IDH mutations irreversibly drive anchorage-independent growth of NHA cells, with the role of mutant IDH then changing from a driver to a passenger mutation. The fact that IDH-C35 did not reverse histone methylation and the phenotype of tumor cells (33) creates a challenge and may raise a question about the use of IDH inhibitors as a monotherapy in patients. This is consistent with our finding that IDH-C35 monotherapy did not delay GL261-MUT tumor growth in vivo compared with vehicle controls. IDH-C35 improved symptom-free survival when used in combination with vaccine, thereby supporting the use of IDH inhibitors in combination with immunotherapy.
Several reports have demonstrated that CD8+ T cells can be tolerized following chronic antigen exposure by downregulating the CD8 coreceptor (49, 50). However, our GO term analysis of TCGA data (Supplemental Table 2) demonstrated that many CD8+ T cell–related genes, and not just the CD8 gene, are downregulated in IDH-MUT compared with IDH-WT cases. In particular, the GO term “lymphocyte aggregation” revealed that 47 genes are enriched in WT cases when compared with MUT cases (Supplemental Table 2), suggesting that there is an actual decrease in CD8+ T cell numbers rather than a downregulation of the CD8 coreceptor. Further studies are warranted to examine T cell tolerance in IDH-MUT gliomas. Other GO terms comparing IDH-WT and IDH-MUT cases supported decreased STAT1 signaling. For example, the term “response to interferon-gamma” was enriched with 22 genes in IDH-WT compared with IDH-MUT cases. Furthermore, terms such as “lymphocyte chemotaxis” (7 genes) and “T cell migration” (8 genes) were enriched in IDH-WT compared with IDH-MUT cases.
We believe the CD8+ T cells that we found to be decreased in IDH-MUT cases were αβ T cells rather than γδ T cells, as we did not observe any γδ T cells in WHO grade III glioma cases by immunofluorescence (Supplemental Figure 3). Furthermore, it has previously been reported in syngeneic GL261 tumors that the γδ T cell response occurs on day 10 and then diminishes in developing tumors (51). Last, our GO term analysis demonstrated enrichment of αβ T cell activation genes in IDH-WT compared with IDH-MUT cases. We acknowledge that IDH mutations may also regulate T cells through additional mechanisms, as the GO terms “lymphocyte activation” and “lymphocyte differentiation” were enriched in IDH-WT compared with IDH-MUT cases, although this may also be due to increased CD8+ T cell accumulation at the tumor site.
In our analysis of TCGA data, staining of grade III glioma tissue, and flow cytometric analysis of GL261 tumor-infiltrating lymphocytes (TILs), in contrast to CD8+ T cells, we did not observe any difference in glioma infiltration of CD8– T cells between IDH-WT and IDH-MUT samples. One possible explanation for this discrepancy would be that CD4+ T cells include Tregs, which are known to migrate toward chemokines such as CCL2 (52) and CCL22 (53). CCL2 and CCL22 gene expression levels were not differentially regulated between IDH-WT and IDH-MUT cases in TCGA data. Moreover, Schumacher et al. (54) demonstrated that CD4+ T cells can respond to a class II–restricted neoantigen epitope encompassing the R132H IDH mutation. Hence, it is speculated that IDH1-MUT gliomas may attract mutant IDH1–specific CD4+ T cells. Ultimately, effective vaccine strategies will likely require the cooperation of CD4+ and CD8+ T cells. The current study provides a rationale for the combination of an IDH inhibitor with peptide-based vaccine strategies, such as one targeting the IDH1 R132H–derived neoantigen epitope.
Recent studies have demonstrated that mutation load levels within tumors such as GBMs are associated with response to immunotherapy (55, 56). As the decreased cytotoxic T lymphocyte (CTL) signature in IDH-MUT compared with IDH-WT cases could be due to differences in mutation load, we compared the mutation load levels in TCGA data. Interestingly, IDH-MUT LGGs have more mutations than do IDH-WT LGGs (Supplemental Figure 13), indicating that the differential mutation load is not likely to be the reason for our results in the current study. Furthermore, the increased mutation load in IDH-MUT cases may be due to decreased levels of immune surveillance and subsequent immune-selective pressure.
The prophylactic vaccine setting of the current study is relevant, as one of the goals in the treatment of patients with LGG is prevention of recurrence. Furthermore, IDH-C35 was given therapeutically to glioma-bearing mice, in a manner similar to how it would be administered to patients. We have previously reported the induction of robust CTL responses in LGG patients treated with synthetic peptides for high-grade glioma-derived GAAs (57).
In the current study, we evaluated the role of IDH mutations and 2HG in tumor-intrinsic pathways that subsequently reduce T cell accumulation in the tumors. It is known that 2HG can be present at millimolar levels in IDH-MUT tumors (58). Therefore, there is a strong possibility that 2HG may also impact nontumor cells such as microglia, macrophages, and T cells in the tumor microenvironment. Interestingly, in our preliminary experiments, treatment of the BV2 microglia cell line with 2HG resulted in reduced STAT1 and CXCL10 levels (Supplemental Figure 14). Further studies are warranted to examine the role of 2HG in cells in the tumor stroma.
In summary, we report for the first time to our knowledge that IDH mutations in glioma cells lead to decreased STAT1 protein levels, which regulate T cell–attracting chemokines and impact CD8+ T cell accumulation. Strategies to improve CD8+ T cell accumulation in the tumor, such as the use of mutant IDH inhibitors, should be considered for integration into T cell–based therapies for patients with IDH-MUT gliomas.
Methods
Experimental design.
Vehicle-treated controls were used, and mice were randomized into groups when appropriate. Mechanistic studies on cells lines were performed without blinding. Sample sizes were chosen on the basis of power calculations based on previous studies to estimate the population mean and SD. All data were included in the analysis, and the criteria for interpretation were established prospectively. Experiments were performed a minimum of 2 times (as indicated in the figure legends).
Reagents.
The following reagents were purchased: RPMI 1640, DMEM, L-glutamine, sodium pyruvate (NA-Pyr), β-mercaptoethanol (2ME), nonessential amino acids (NEAAs), penicillin/streptomycin (p/s), and anti-CD3/anti-CD28 Dynabeads (Thermo Fisher Scientific); heat-inactivated FBS (Gemini Bio-Products); and recombinant human IL-2 (rhIL-2) (PeproTech). Poly-ICLC was provided by Oncovir Inc. The following peptides were synthesized by the automated solid-phase peptide synthesizer at the University of Pittsburgh Peptide and Peptoid Synthesis Core: H-2Db-binding mEPHA2671–679 (FSHHNIIRL); H-2Db–binding mGARC-177–85 (AALLNKLYA) (49); H-2Db–binding human gp100 (hgp100)25–33 (KVPRNQDWL); H-2Kb–binding mEPHA2682–689 (VVSKYKPM); and H-2Kb–binding mTRP2180–188 (SVYDFFVWL). I-Ab–binding HBV core128–140 (TPPAYRPPNAPIL) was purchased from A&A Labs.
Cell culture.
Murine GL261, provided by Robert Prins (UCLA, Los Angeles, California, USA) and SB28 (35) glioma cell lines were maintained in RPMI 1640 supplemented with 10% FBS, NEAA, NA-Pyr, 2ME, and p/s. Immortalized, untransformed NHA cells stably expressing either WT or R132H-mutant IDH (59) were cultured in DMEM containing 10% FBS, NEAA, NA-Pyr, 2ME, and p/s. Primary glioma spheres derived from patients with IDH1-MUT WHO grade III glioma were cultured in DMEM/Ham’s F-12 with p/s, L-glutamine (100 U/ml), and 20% BIT 9500 Serum Substitute (STEMCELL Technologies) and supplemented with 0.02 ng/ml basic FGF (bFGF) and EGF (ReliaTech GmbH). All cell lines were maintained in a humidified incubator in 5% CO2 at 37°C.
Generation of cell lines transduced with IDH cDNA.
Aliquots of 5 × 105 GL261 and SB28 cells were plated overnight in antibiotic-free media onto 6-well plates. Cells were then transfected with 10 μg plasmids encoding cDNAs for either the IDH-WT (OriGene Technologies; RC210582) or the R132H IDH-MUT (OriGene Technologies; RC400096) with Lipofectamine 2000 and Opti-MEM (Thermo Fisher Scientific), according to the manufacturer’s protocol. Forty-eight hours after transfection, G418 selective media were added for one week to select for stably transfected cells. Stable transfection was confirmed using Western blotting for the DDK tag.
TCGA analysis.
TCGA level 3 gene expression (~20,000 genes) data and microRNA data (~1,000 microRNAs) were downloaded from the University of California Santa Cruz (UCSC) cancer genome browser for WHO grades II and III glioma patients (started with approximately 500 cases). 450k methylation data were downloaded directly from TCGA database. As oligodendroglioma has a different biology and prognosis than does astrocytoma, we excluded cases with chromosome 1p/19q deletion, which is a marker for oligodendroglioma (1). Data on the IDH mutation status were then obtained from the remaining cases using cBioportal, and cases were stratified into 2 groups on the basis of nonmutated WT (58 cases) or mutated (MUT; 150 cases) status in IDH1 or IDH2. Log2 expression of genes between the IDH-WT and IDH-MUT groups was used to compare expression levels of 310 genes related to immune responses in WT versus MUT cases. A 2-sided, unpaired t test was performed, with a Benjamini-Hochberg adjustment for multiple testing using Microsoft Excel. For GO term analysis, the clusterProfiler R package (60) was used to perform GO enrichment analyses. Genes upregulated in IDH-WT and IDH-MUT tumors were defined as genes with a Benjamini-Hochberg–corrected P value of less than 0.01 and a difference in mean log2 expression of greater than 1. We used a background gene list of all genes with expression data.
Intracranial injection of GL261 glioma cells.
C57BL/6j mice used for the study were purchased from The Jackson Laboratory. The procedure used in the current study has been described previously (13, 61). Briefly, using a Hamilton syringe (Hamilton Company), 1 × 105 GL261-WT or GL261-MUT cells in 2 μl PBS were stereotactically injected through an entry site from the bregma, 3 mm to the right of sagittal suture, and 3.0 mm below the surface of the skull of anesthetized mice by using a stereotactic frame (Kopf). Mice were monitored daily for symptom-free survival.
Prophylactic vaccination with GAAs.
Prophylactic vaccinations were administered as described previously (31). Briefly, mice received s.c. vaccinations with 100 μg HBV core128–140 and GAA peptides, 100 μg each of hgp10025–33, mEPHA2682–689, mEPHA2671–679, mTRP2180–188, and mGARC-177–85, emulsified in IFA (Difco Laboratories), and 20 μg poly-ICLC on days –25, –15 and –5 relative to the date of tumor inoculation. Control mice received poly-ICLC and HBV core128–140 emulsified in IFA. Some mice received 450 mg/kg/day IDH-C35 (MedKoo Biosciences) by oral gavage.
Transwell migration assay.
CD8+ lymphocytes were selected from C57BL/6j mouse–derived splenocytes by positive selection using magnetic beads (Miltenyi Biotec). CD8+ cells were stimulated with anti-CD3/anti-CD28 Dynabeads at a 1:1 bead/cell ratio with 30 units/ml hIL-2 for 72 hours, and 5 × 105 cells in 100 μl complete media were loaded into the top chamber of Transwell inserts (5.0-μm pore size; Costar). The bottom well was filled with RPMI medium with recombinant murine CXCL10 (R&D Systems), glioma cell–CM (diluted 1:1 with fresh media), or media with anti-CXCR3–neutralizing antibody (BioLegend; 126517). Glioma cell–CM were derived from media with confluent GL261 and SB28 glioma cell lines. Plates were incubated at 37°C overnight, the contents of the lower chamber were collected, and the percentage of viable CD8+ cells present in the bottom chamber was determined using the Countess Cell Counter System (Thermo Fisher Scientific) and the WST-1 cell viability assay (Roche).
RT-PCR.
Tumor samples or cells were placed in Buffer RLT (QIAGEN; 79216), lysed using QIAshredder columns (QIAGEN; 79654), and total RNA isolated using the RNAeasy Kit (QIAGEN). Total RNA (10 ng) was further converted to cDNA using an XLT cDNA Synthesis Kit (Quanta Bioscience; 95161), and RT-PCR was done with 2 μl cDNA using Taqman Gene Expression Assays (Thermo Fisher Scientific; 4331182) and a StepOne Plus System (Thermo Fisher Scientific). GAPDH was used as a housekeeping gene in all gene expression experiments.
ELISA.
Tumor tissue cell extracts in RIPA buffer were used for ELISA. CXCL10 protein levels were determined using either the human or mouse CXCL10/IP-10 Quantikine ELISA Kit (R&D Systems; DIP100 and MCX100, respectively), according to the manufacturer’s protocol. Absorbance was measured using an Epoch Spectrophotometer (BioTek).
Western blot analysis.
Total protein was isolated from cells or tissue by lysis with ice-cold RIPA buffer containing protease and phosphatase inhibitors. Protein concentration was measured with the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific; 24225). The proteins were denatured at 95°C for 5 minutes using Blue Loading Gel Dye and DTT at 1× concentration from the Blue Loading Buffer Pack (Cell Signaling Technology; 7722). Western blotting was performed using primary rabbit anti-IRF1 (Cell Signaling Technology; 8478); rabbit anti-STAT1 (Cell Signaling Technology; 9172); rabbit anti–p-STAT1 (Cell Signaling Technology; 9167); and rabbit anti–β actin (Cell Signaling Technology; 4967) according to the manufacturer’s recommended protocols. Quantification of band densities on Western blot films was performed using ImageJ (NIH).
Immunofluorescence staining of mouse samples.
Euthanized mice received transcardial perfusion with 10 ml PBS. Brains were removed and fixed in 4% paraformaldehyde, followed by 30% sucrose solution and embedded in O.C.T. Compound (Tissue-Tek; 4583; Sakura Finetek). Sections (10-μm-thick) were fixed in acetone solution at –20°C for 10 minutes and then placed in 10% normal goat serum (Abcam; ab7481) for blocking. The following antibodies were used: anti-CD3 (1:100; Abcam; ab11089); anti-CD8 (1:50; Biorbyt; orb323288); goat anti-rat (1:500; Abcam; ab150167); and goat anti-rabbit (1:500; Abcam; ab150086). Samples were dehydrated in ethanol (95%–100%) and mounted with DAPI (ProLong Gold Antifade Reagent with DAPI; Life Technologies, Thermo Fisher Scientific).
Image acquisition and analysis.
Images were acquired using a Zeiss Axio Imager 2 microscope (×20 magnification) and TissueFAXS scanning software (TissueGnostics). Identical exposure times and threshold settings were used for each channel on all sections of similar experiments. Quantification of dp cells was performed using StrataQuest Analysis Software (TissueGnostics), with detection engines set for our purposes. In brief, the algorithm detected nuclei on the basis of the signal from the DAPI channel, then expanded and built a mask over the cytoplasm. On the generated mask, the algorithm searched for colocalization of TEX and Cy5 signals. Results were plotted onto scattergrams or histograms, and events were manually verified for all quadrants. The number of cells per area (mm2) and total number of cells (as a percentage of total DAPI+ nuclei) were calculated.
LC-MS detection of 2HG.
Metabolite extraction and LC-MS were performed as described previously (8). Fresh media were added to 1 × 106 cells for 24 hours, and media were then collected for analysis. Cells were washed twice with cold PBS and then resuspended and vortexed in a solution of 80% methanol and 20% water. Cell extractions were then immediately stored at –80°C until analysis. Analysis was performed at the UCSF Lipid Mass Spectroscopy Core.
Statistics.
Statistical analysis was performed using Excel (Microsoft) or Prism V (GraphPad Software). Data are expressed as the mean ± SD. P values of less than 0.05 were considered statistically significant. For TCGA data analysis (Figure 2), P values were obtained using a 2-sided, unpaired t test, with the Benjamini-Hochberg adjustment for multiple testing. For animal survival studies (Figure 7), P values were determined by log-rank (Mantel-Cox) test and 1-way ANOVA.
Study approval.
Clinical samples were obtained from the Brain Tumor Research Center Tissue Core at UCSF under a protocol approved by the UCSF Committee on Human Research. All mouse studies were predefined and performed under a UCSF IACUC–approved protocol.
Author contributions
HO, GK, BJA, and NAA conceived the study. GK, HO, NAA, TM, CHM, and JFC designed research studies. GK, DAC, SS, BJA, NJ, ZDC, KMD, CB, and PBW conducted experiments. GK, HO, DAC, NJ, CB, RW, and TM analyzed data. GK and HO wrote the manuscript. All authors reviewed and proofed the manuscript.
Supplementary Material
Acknowledgments
The authors would like to thank Anny Shai and Joanna Philips at the UCSF BTRC Tissue Core for providing tissue analysis and Yong Huang at the UCSF Lipid Mass Spectroscopy Core for analysis of 2HG. NHA cells with WT-IDH1 or IDH1 containing the R132H mutation (R132H) were provided by Russel Pieper (UCSF). Poly-ICLC was provided by Andres Salazar, Oncovir Inc. The current study was supported by the National Institute of Neurological Disorders and Stroke (NINDS), NIH (2R01NS055140-06, to HO; R21NS093654, to HO and JC and R21NS083171, to HO); the National Cancer Institute, NIH (P50CA097257, to HO and R01CA169316, to JC); the Cancer Research Institute (to HO); Will Power Research Fund (to HO); loglio funds from the Dabierre Foundation (to HO and JC); the Parker Institute for Cancer Immunotherapy (to HO); the NIH (1T32CA151022, to GK and JC); and The Walter L. Copeland Fund of The Pittsburgh Foundation (to GK).
Footnotes
BJA’s present address is: Department of Medicine, University of Pittsburgh Medical Center School of Medicine, Pittsburgh, Pennsylvania, USA.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(4):1425–1437.https://doi.org/10.1172/JCI90644.
See the related Commentary at Resisting fatal attraction: a glioma oncometabolite prevents CD8+ T cell recruitment.
Contributor Information
Gary Kohanbash, Email: gary.kohanbash@ucsf.edu.
Shruti Shrivastav, Email: shrutishrivastav92@gmail.com.
Naznin Jahan, Email: naznin.jahan@ucsf.edu.
Tali Mazor, Email: tali.mazor@ucsf.edu.
Casey Beppler, Email: casey.beppler@ucsf.edu.
Rolf Warta, Email: rolf.warta@med.uni-heidelberg.de.
Christel Herold-Mende, Email: christel.herold-mende@med.uni-heidelberg.de.
Hideho Okada, Email: okadah@neurosurg.ucsf.edu.
References
- 1.Perry A, Wesseling P. Histologic classification of gliomas. Handb Clin Neurol. 2016;134:71–95. doi: 10.1016/B978-0-12-802997-8.00005-0. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 3.Sanai N, Chang S, Berger MS. Low-grade gliomas in adults. J Neurosurg. 2011;115(5):948–965. doi: 10.3171/2011.7.JNS101238. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. et al. Comprehensive, integrative genomic analysis of diffuse lower-grade gliomas. N Engl J Med. 2015;372(26):2481–2498. doi: 10.1056/NEJMoa1402121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ichimura K. Molecular pathogenesis of IDH mutations in gliomas. Brain Tumor Pathol. 2012;29(3):131–139. doi: 10.1007/s10014-012-0090-4. [DOI] [PubMed] [Google Scholar]
- 6.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson BE, et al. Mutational analysis reveals the origin and therapy-driven evolution of recurrent glioma. Science. 2014;343(6167):189–193. doi: 10.1126/science.1239947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dang L, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin Y, Okada H. Cellular immunotherapy for malignant gliomas. Expert Opin Biol Ther. 2016;16(10):1265–1275. doi: 10.1080/14712598.2016.1214266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reardon DA, et al. Mitchell DA, Fecci PE, Sampson JH, et al. An update on vaccine therapy and other immunotherapeutic approaches for glioblastoma. Expert Rev Vaccines. 2013;12(6):597–615. doi: 10.1586/erv.13.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nishimura F, et al. Adoptive transfer of type 1 CTL mediates effective anti-central nervous system tumor response: critical roles of IFN-inducible protein-10. Cancer Res. 2006;66(8):4478–4487. doi: 10.1158/0008-5472.CAN-05-3825. [DOI] [PubMed] [Google Scholar]
- 12.Fujita M, et al. Inhibition of STAT3 promotes the efficacy of adoptive transfer therapy using type-1 CTLs by modulation of the immunological microenvironment in a murine intracranial glioma. J Immunol. 2008;180(4):2089–2098. doi: 10.4049/jimmunol.180.4.2089. [DOI] [PubMed] [Google Scholar]
- 13.Fujita M, et al. Effective immunotherapy against murine gliomas using type 1 polarizing dendritic cells — significant roles of CXCL10. Cancer Res. 2009;69(4):1587–1595. doi: 10.1158/0008-5472.CAN-08-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujita M, et al. COX-2 blockade suppresses gliomagenesis by inhibiting myeloid-derived suppressor cells. Cancer Res. 2011;71(7):2664–2674. doi: 10.1158/0008-5472.CAN-10-3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roussel E, Gingras MC, Grimm EA, Bruner JM, Moser RP. Predominance of a type 2 intratumoural immune response in fresh tumour-infiltrating lymphocytes from human gliomas. Clin Exp Immunol. 1996;105(2):344–352. doi: 10.1046/j.1365-2249.1996.d01-753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weller M, Fontana A. The failure of current immunotherapy for malignant glioma. Tumor-derived TGF-beta, T-cell apoptosis, and the immune privilege of the brain. Brain Res Brain Res Rev. 1995;21(2):128–151. doi: 10.1016/0165-0173(95)00010-0. [DOI] [PubMed] [Google Scholar]
- 17.Nitta T, Hishii M, Sato K, Okumura K. Selective expression of interleukin-10 gene within glioblastoma multiforme. Brain Res. 1994;649(1–2):122–128. doi: 10.1016/0006-8993(94)91055-3. [DOI] [PubMed] [Google Scholar]
- 18.Jarnicki AG, Lysaght J, Todryk S, Mills KH. Suppression of antitumor immunity by IL-10 and TGF-β-producing T cells infiltrating the growing tumor: influence of tumor environment on the induction of CD4+ and CD8+ regulatory T cells. J Immunol. 2006;177(2):896–904. doi: 10.4049/jimmunol.177.2.896. [DOI] [PubMed] [Google Scholar]
- 19.Prokopchuk O, Liu Y, Henne-Bruns D, Kornmann M. Interleukin-4 enhances proliferation of human pancreatic cancer cells: evidence for autocrine and paracrine actions. Br J Cancer. 2005;92(5):921–928. doi: 10.1038/sj.bjc.6602416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn BJ, Pollack IF, Okada H. Immune-checkpoint blockade and active immunotherapy for glioma. Cancers (Basel) 2013;5(4):1379–1412. doi: 10.3390/cancers5041379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohanbash G, Okada H. Myeloid-derived suppressor cells in gliomas and glioma-development. Immunol Invest. 2012;41(6–7):658–679. doi: 10.3109/08820139.2012.689591. [DOI] [PubMed] [Google Scholar]
- 22.Nduom EK, Weller M, Heimberger AB. Immunosuppressive mechanisms in glioblastoma. Neuro Oncol. 2015;17(suppl 7):vii9–vii14. doi: 10.1093/neuonc/nov151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elkhaled A, et al. Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4(116):116ra5. doi: 10.1126/scitranslmed.3002796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashburner M, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Watanabe T, Vital A, Nobusawa S, Kleihues P, Ohgaki H. Selective acquisition of IDH1 R132C mutations in astrocytomas associated with Li-Fraumeni syndrome. Acta Neuropathol. 2009;117(6):653–656. doi: 10.1007/s00401-009-0528-x. [DOI] [PubMed] [Google Scholar]
- 26.Pavon LF, et al. In vitro analysis of neurospheres derived from glioblastoma primary culture: a novel methodology paradigm. Front Neurol. 2014;4:214. doi: 10.3389/fneur.2013.00214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jaruga B, Hong F, Kim WH, Gao B. IFN-γ/STAT1 acts as a proinflammatory signal in T cell-mediated hepatitis via induction of multiple chemokines and adhesion molecules: a critical role of IRF-1. Am J Physiol Gastrointest Liver Physiol. 2004;287(5):G1044–G1052. doi: 10.1152/ajpgi.00184.2004. [DOI] [PubMed] [Google Scholar]
- 28.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264(5164):1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 29.Ueda R, et al. Systemic inhibition of transforming growth factor-beta in glioma-bearing mice improves the therapeutic efficacy of glioma-associated antigen peptide vaccines. Clin Cancer Res. 2009;15(21):6551–6559. doi: 10.1158/1078-0432.CCR-09-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hatano M, et al. Vaccination with EphA2-derived T cell-epitopes promotes immunity against both EphA2-expressing and EphA2-negative tumors. J Transl Med. 2004;2(1):40. doi: 10.1186/1479-5876-2-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhu X, et al. Poly-ICLC promotes the infiltration of effector T cells into intracranial gliomas via induction of CXCL10 in IFN-α and IFN-γ dependent manners. Cancer Immunol Immunother. 2010;59(9):1401–1409. doi: 10.1007/s00262-010-0876-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okada H, et al. Induction of CD8+ T-cell responses against novel glioma-associated antigen peptides and clinical activity by vaccinations with {alpha}-type 1 polarized dendritic cells and polyinosinic-polycytidylic acid stabilized by lysine and carboxymethylcellulose in patients with recurrent malignant glioma. J Clin Oncol. 2011;29(3):330–336. doi: 10.1200/JCO.2010.30.7744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johannessen TA, et al. Rapid conversion of mutant IDH1 from driver to passenger in a model of human gliomagenesis. Mol Cancer Res. 2016;14(10):976–983. doi: 10.1158/1541-7786.MCR-16-0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu C, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kosaka A, Ohkuri T, Okada H. Combination of an agonistic anti-CD40 monoclonal antibody and the COX-2 inhibitor celecoxib induces anti-glioma effects by promotion of type-1 immunity in myeloid cells and T-cells. Cancer Immunol Immunother. 2014;63(8):847–857. doi: 10.1007/s00262-014-1561-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ausman JI, Shapiro WR, Rall DP. Studies on the chemotherapy of experimental brain tumors: development of an experimental model. Cancer Res. 1970;30(9):2394–2400. [PubMed] [Google Scholar]
- 38.Eckel-Passow JE, et al. Glioma groups based on 1p/19q, IDH, and TERT promoter mutations in tumors. N Engl J Med. 2015;372(26):2499–2508. doi: 10.1056/NEJMoa1407279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma P, et al. CD8 tumor-infiltrating lymphocytes are predictive of survival in muscle-invasive urothelial carcinoma. Proc Natl Acad Sci U S A. 2007;104(10):3967–3972. doi: 10.1073/pnas.0611618104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kmiecik J, et al. Elevated CD3+ and CD8+ tumor-infiltrating immune cells correlate with prolonged survival in glioblastoma patients despite integrated immunosuppressive mechanisms in the tumor microenvironment and at the systemic level. J Neuroimmunol. 2013;264(1–2):71–83. doi: 10.1016/j.jneuroim.2013.08.013. [DOI] [PubMed] [Google Scholar]
- 41.Lohr J, et al. Effector T-cell infiltration positively impacts survival of glioblastoma patients and is impaired by tumor-derived TGF-β. Clin Cancer Res. 2011;17(13):4296–4308. doi: 10.1158/1078-0432.CCR-10-2557. [DOI] [PubMed] [Google Scholar]
- 42.Arita H, et al. A combination of TERT promoter mutation and MGMT methylation status predicts clinically relevant subgroups of newly diagnosed glioblastomas. Acta Neuropathol Commun. 2016;4(1):79. doi: 10.1186/s40478-016-0351-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wen Z, Zhong Z, Darnell JE. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell. 1995;82(2):241–250. doi: 10.1016/0092-8674(95)90311-9. [DOI] [PubMed] [Google Scholar]
- 44.Lehtonen A, Matikainen S, Julkunen I. Interferons up-regulate STAT1, STAT2, and IRF family transcription factor gene expression in human peripheral blood mononuclear cells and macrophages. J Immunol. 1997;159(2):794–803. [PubMed] [Google Scholar]
- 45.Gil MP, Salomon R, Louten J, Biron CA. Modulation of STAT1 protein levels: a mechanism shaping CD8 T-cell responses in vivo. Blood. 2006;107(3):987–993. doi: 10.1182/blood-2005-07-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci U S A. 1998;95(26):15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ohba S, et al. Mutant IDH1 expression drives TERT promoter reactivation as part of the cellular transformation process. Cancer Res. 2016;76(22):6680–6689. doi: 10.1158/0008-5472.CAN-16-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prins RM, Graf MR, Merchant RE. Cytotoxic T cells infiltrating a glioma express an aberrant phenotype that is associated with decreased function and apoptosis. Cancer Immunol Immunother. 2001;50(6):285–292. doi: 10.1007/s002620100202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Xiao Z, Mescher MF, Jameson SC. Detuning CD8 T cells: down-regulation of CD8 expression, tetramer binding, and response during CTL activation. J Exp Med. 2007;204(11):2667–2677. doi: 10.1084/jem.20062376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beck BH, et al. Dynamics of circulating γδ T cell activity in an immunocompetent mouse model of high-grade glioma. PLoS One. 2015;10(5):e0122387. doi: 10.1371/journal.pone.0122387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang AL, et al. CCL2 produced by the glioma microenvironment is essential for the recruitment of regulatory t cells and myeloid-derived suppressor cells. Cancer Res. 2016;76(19):5671–5682. doi: 10.1158/0008-5472.CAN-16-0144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Curiel TJ, et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med. 2004;10(9):942–949. doi: 10.1038/nm1093. [DOI] [PubMed] [Google Scholar]
- 54.Schumacher T, et al. A vaccine targeting mutant IDH1 induces antitumour immunity. Nature. 2014;512(7514):324–327. doi: 10.1038/nature13387. [DOI] [PubMed] [Google Scholar]
- 55.Giannakis M, et al. Genomic correlates of immune-cell infiltrates in colorectal carcinoma. Cell Rep. 2016;17(4):1206. doi: 10.1016/j.celrep.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bouffet E, et al. Immune checkpoint inhibition for hypermutant glioblastoma multiforme resulting from germline biallelic mismatch repair deficiency. J Clin Oncol. 2016;34(19):2206–2211. doi: 10.1200/JCO.2016.66.6552. [DOI] [PubMed] [Google Scholar]
- 57.Okada H, et al. Induction of robust type-I CD8+ T-cell responses in WHO grade 2 low-grade glioma patients receiving peptide-based vaccines in combination with poly-ICLC. Clin Cancer Res. 2015;21(2):286–294. doi: 10.1158/1078-0432.CCR-14-1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalinina J, et al. Detection of “oncometabolite” 2-hydroxyglutarate by magnetic resonance analysis as a biomarker of IDH1/2 mutations in glioma. J Mol Med. 2012;90(10):1161–1171. doi: 10.1007/s00109-012-0888-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ohba S, Mukherjee J, See WL, Pieper RO. Mutant IDH1-driven cellular transformation increases RAD51-mediated homologous recombination and temozolomide resistance. Cancer Res. 2014;74(17):4836–4844. doi: 10.1158/0008-5472.CAN-14-0924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu G, Wang LG, Han Y, He QY. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kohanbash G, et al. GM-CSF promotes the immunosuppressive activity of glioma-infiltrating myeloid cells through interleukin-4 receptor-α. Cancer Res. 2013;73(21):6413–6423. doi: 10.1158/0008-5472.CAN-12-4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.