Abstract
Shohat-type spondyloepimetaphyseal dysplasia (SEMD) is a skeletal dysplasia that affects cartilage development. Similar skeletal disorders, such as spondyloepiphyseal dysplasias, are linked to mutations in type II collagen (COL2A1), but the causative gene in SEMD is not known. Here, we have performed whole-exome sequencing to identify a recurrent homozygous c.408+1G>A donor splice site loss-of-function mutation in DDRGK domain containing 1 (DDRGK1) in 4 families affected by SEMD. In zebrafish, ddrgk1 deficiency disrupted craniofacial cartilage development and led to decreased levels of the chondrogenic master transcription factor sox9 and its downstream target, col2a1. Overexpression of sox9 rescued the zebrafish chondrogenic and craniofacial phenotype generated by ddrgk1 knockdown, thus identifying DDRGK1 as a regulator of SOX9. Consistent with these results, Ddrgk1–/– mice displayed delayed limb bud chondrogenic condensation, decreased SOX9 protein expression and Col2a1 transcript levels, and increased apoptosis. Furthermore, we determined that DDRGK1 can directly bind to SOX9 to inhibit its ubiquitination and proteasomal degradation. Taken together, these data indicate that loss of DDRGK1 decreases SOX9 expression and causes a human skeletal dysplasia, identifying a mechanism that regulates chondrogenesis via modulation of SOX9 ubiquitination.
Introduction
Chondrodysplasias are a clinically and genetically diverse group of skeletal dysplasias that primarily affect cartilage and present with disproportionate short stature and premature osteoarthritis (OA) (1–3). Shohat type spondyloepimetaphyseal dysplasia (SEMD) is a chondrodysplasia characterized by vertebral, epiphyseal, and metaphyseal abnormalities. These abnormalities include scoliosis with vertebral compression factures, flattened vertebral bodies, and hypomineralization of the long bones. Additionally, affected individuals may have a small trunk, short neck, small limbs, joint laxity, bowlegs, and/or abdominal distension with hepatosplenomegaly (4–6).
Shohat-type SEMD shares some clinical features with the spondyloepiphyseal dysplasia (SED) spectrum, including long-bone and vertebral defects. Disorders of the SED spectrum, which include SED congenita and X-linked SED tarda, are usually caused by mutations altering the expression and/or processing of type II collagen (COL2A1), the most abundant extracellular matrix protein in cartilage (1, 2, 7, 8). Specifically, mutations in COL2A1 were the first reported cause of a human chondrodysplasia, SED congenita. Shohat-type SEMD is not caused by any known mutations in genes that affect the structure or trafficking of COL2A1. However, its phenotypic overlap with the SED spectrum led us to hypothesize that its causative gene may control COL2A1 processing and/or cartilage formation.
To identify the genetic cause of Shohat-type SEMD, we performed whole-exome sequence (WES) on a proband and identified a homozygous c.408+1G>A donor splice site mutation in the DDRGK domain containing 1 (DDRGK1) gene. DDRGK1, also known as UFM1-binding protein 1 containing a PCI domain (UFBP1), has been linked to protein stability and activation. For instance, in vitro knockdown of Ddrgk1 indirectly inhibits IκBα ubiquitination by increasing phosphorylation of IκBα, a prerequisite for its ubiquitination (9). DDRGK1 is also required for estrogen receptor α (ERα) transactivation, as it mediates activating signal cointegrator 1 (ASC1) ufmylation, an ubiquitin-like posttranslational modification (10). While the cellular functions of Ddrgk1 have been investigated in other systems (9–13), its role in skeletal development remains unknown.
Here, we report that Ddrgk1 is essential for cartilage development and the differentiation of chondrocytes, the cells that produce and maintain cartilage. Furthermore, we demonstrate that DDRGK1 loss of function causes Shohat-type SEMD pathogenesis by increasing ubiquitin-dependent proteasomal degradation of SOX9.
Results
Shohat-type SEMD is caused by a loss-of- function mutation within the DDRGK1 gene.
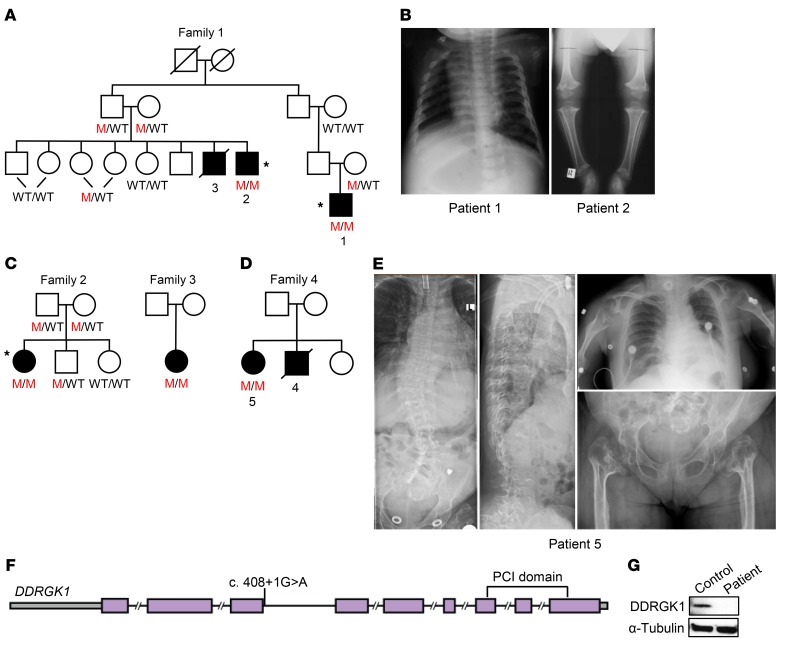
Family 1, of Jewish Iraqi descent, was first reported and diagnosed in 1993 with Shohat-type SEMD (Figure 1A). As previously described, these patients have a disproportionate short stature, short neck, small chest, mild abdominal distension with hepatosplenomegaly, lordosis, short limbs, genu varum, and joint hypermobility (6). Radiographically, they have delayed epiphyseal ossification, platyspondyly with central notches in the vertebral end plates, radiolucency of the femoral metaphyses, and relative fibular overgrowth (6), as seen in the radiographs of patients 1 and 2 at ages 9 months and 5 years, respectively (Figure 1B). At age 18, because of his narrowed trachea, patient 1 required a lifesaving tracheostomy during an aborted orthopedic surgery. Unfortunately, this surgical complication was the cause of death for patients 3 and 4 from families 1 and 4, respectively. In addition, now at age 26, patient 1 has joint pain and hoarseness due to his severely narrowed laryngx and bronchi.
Figure 1. Homozygous DDRGK1 loss-of-function mutation identified in Iraqi Jewish families with Shohat-type SEMD.
(A) Pedigree of family 1. M represents mutant allele. Asterisks indicate individuals who were selected for WES. (B) Radiographs from family 1 reveal platyspondyly and hypomineralized epiphyses and metaphyses in patients 1 and 2, respectively. (C and D) Pedigrees of families 2, 3, and 4. (E) Radiographs show severe scoliosis, vertebral compression factures, platyspondyly, broaden hypomineralized metaphyses, and smaller than average hypomineralized epiphyses in patient 5 from family 4. (F) Schematic of the c.408+1G>A mutation in the DDRGK1 gene. (G) Western analysis of whole cell lysates reveals absence of DDRGK1 expression in patient LCLs.
The affected individuals in families 2 through 4, of Jewish Iraqi descent, were diagnosed with very short limbs in utero, and at birth, had vertebral and long-bone defects similar to those of the patients in family 1 (Figure 1, C and D). In addition to severe scoliosis, vertebral compression factures, platyspondyly, broadened hypomineralized metaphyses, and hypomineralized epiphyses (Figure 1E), patient 5 had a narrow trachea and severe upper airway obstruction requiring a tracheostomy (Figure 1E) with continuous positive airway pressure (CPAP) therapy while sleeping.
To identify the genetic cause of Shohat-type SEMD, we performed WES on 3 affected individuals from the first 2 Jewish families of Iraqi descent (Figure 1, A and C). By applying WES with an established method and variant filtering (14), we identified 1,951 genes with rare coding variants, 53 of which were shared by all 3 individuals. Out of the 53 genes, 13 had homozygous variants in all 3 patients; among these, SLC4A11 and DDRGK1 had rare variants having minor allele frequencies of less than 0.05 and were located at the same region of chromosome 20. The SLC4A11 variant found in our patients is a known variant corresponding to dbSNP rs141705330, for which some individuals in the general population in the Exome Aggregation Consortium (ExAC; http://exac.broadinstitute.org/) database were homozygous. SLC4A11 defects cause corneal endothelial dystrophy and sensorineural hearing loss in humans and mice (15, 16), while DDRGK1 has been linked to protein stability and activation (9, 10). Therefore, based on the known variant and function of SLC4A11, DDRGK1 appeared to be the best candidate gene for Shohat-type SEMD.
The 3 affected individuals had a homozygous c.408+1G>A donor splice site mutation in the third intron of the DDRGK1 gene, while none of the unaffected family members were homozygous for this mutation (Figure 1F). Additionally, by conventional Sanger sequencing, we identified 2 more patients from families 3 and 4 who were homozygous for the same mutation (Figure 1, C and D). This variant was not found in the ExAC database, but there were other potential DDRGK1 deleterious variants in the ExAC database, all heterozygous with minor allele frequencies of less than 0.001. To assess whether the c.408+1G>A mutation affected DDRGK1 transcription and translation, we performed quantitative real-time PCR (qRT-PCR) and Western blot analysis on RNA and whole-cell lysates from lymphoblastoid cells (LCLs) of patient 1. While control LCLs expressed WT DDRGK1 RNA, patient LCLs expressed a mix of 2 aberrant DDRGK1 RNA species (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/JCI90193DS1). The first aberrant RNA was a read-through of the third intron and the second one was the result of use of a cryptic splice site, both resulting in a premature stop codon (Supplemental Figure 1B). When we examined protein expression, LCLs from the patient did not express DDRGK1 (Figure 1G). These results suggest that the c.408+1G>A homozygous donor splice site mutation in DDRGK1 leads to nonsense-mediated decay and is a founder, disease-causing variant of Shohat-type SEMD.
Knockdown of ddrgk1 disrupts cartilage development in zebrafish embryos.
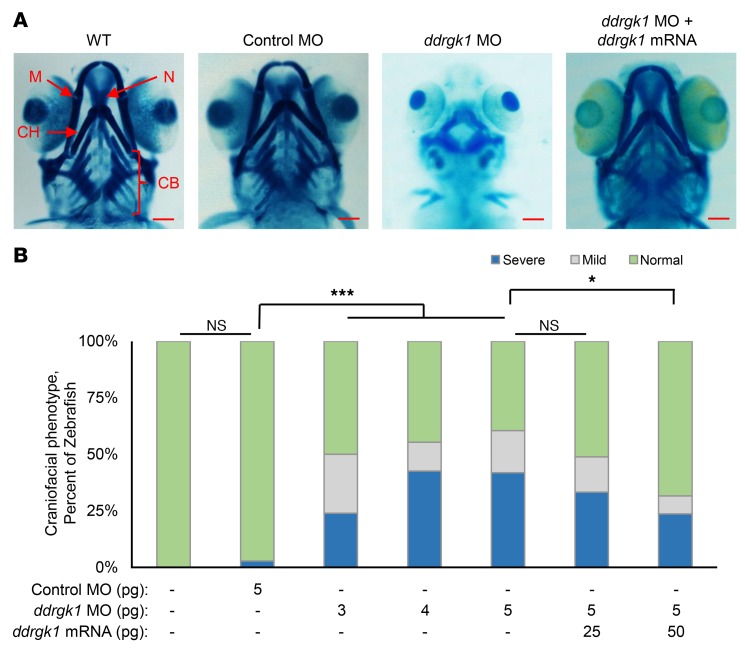
In zebrafish, ddrgk1 is highly expressed in the head at 60 hours post fertilization (h.p.f.) and the sequence of its Ddrgk1 protein is 53% identical to its human protein ortholog (17). Therefore, zebrafish were used to evaluate the functional consequences of ddrgk1 deficiency in cartilage development. During zebrafish embryogenesis, chondrogenesis starts as early as 52 h.p.f., and well-developed craniofacial cartilage structures can be visualized by Alcian blue staining by 120 h.p.f. (18–20). We transiently knocked down the ddrgk1 in zebrafish embryos with morpholino oligonucleotide (MO) injections. Similarly to WT, approximately 97% of embryos injected with control MO (control morphants) had well-developed Meckel’s cartilage, ceratohyal cartilage, ceratobranchial cartilage, and neurocranial cartilage by 120 h.p.f. (Figure 2, A and B). Interestingly, 50% to 61% of embryos injected with ddrgk1 MO (ddrgk1 morphants) showed craniofacial defects, which increased in severity in a ddrgk1 MO dosage–dependent manner (Figure 2, A and B). These defects included shortened and dysmorphic Meckel’s cartilage and ceratohyal cartilage and the absence of ceratobranchial cartilage and neurocranial cartilage. The craniofacial defects in ddrgk1 morphants thus suggest that ddrgk1 is required for cartilage development.
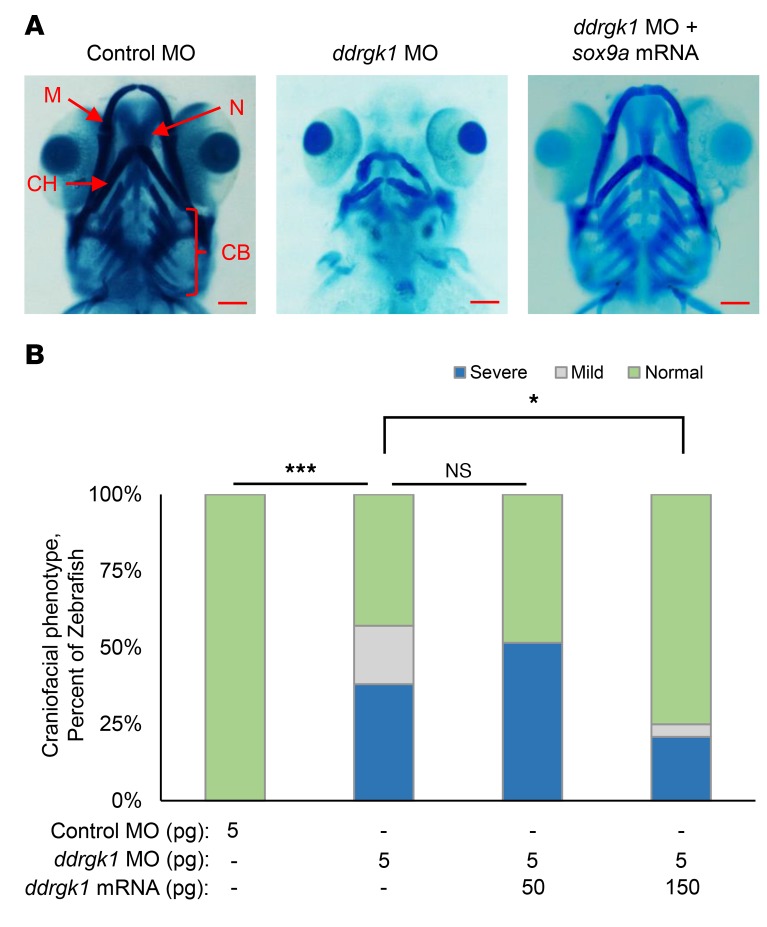
Figure 2. Overexpression of ddrgk1 mRNA rescues the ddrgk1 knockdown cartilage phenotype in zebrafish.
(A) Ventral view at 120 h.p.f. of Alcian blue–stained WT embryos and embryos injected with either control MO or ddrgk1 MO with or without exogenous ddrgk1 mRNA. M, Meckel’s cartilage; CH, ceratohyal cartilage; N, neurocranial cartilage; CB, ceratobranchial cartilage. Scale bars: 100 μM. (B) Quantification of the craniofacial phenotype in the embryos. Embryos with normal craniofacial features had properly formed Meckel’s cartilage, ceratohyal cartilage, neurocranial cartilage, and ceratobranchial cartilage. Embryos lacking ceratobranchial cartilage showed mild craniofacial defects, while embryos with several craniofacial defects had no neurocranial or ceratobranchial cartilage and poorly developed Meckel’s cartilage and ceratohyal cartilage. WT, n = 44; 5 pg control MO, n = 36; 3 pg ddrgk1 MO, n = 50; 4 pg ddrgk1 MO, n = 47; 5 pg ddrgk1 MO, n = 43; 5 pg ddrgk1 MO + 25 pg ddrgk1 mRNA, n = 45; 5 pg ddrgk1 MO + 50 pg ddrgk1 mRNA, n = 38. *P < 0.05; ***P < 0.001; Kruskal-Wallis rank-sum test followed by Wilcoxon’s rank-sum test with continuity correction.
To confirm that the craniofacial defects in ddrgk1 morphants were caused by ddrgk1 knockdown, we performed rescue experiments by coinjecting ddrgk1 MO with ddrgk1 mRNA resistant to MO targeting. Overexpression of ddrgk1 mRNA rescued the cartilage malformations in ddrgk1 morphants in a dosage-dependent manner, with the majority of ddrgk1 morphants achieving normal craniofacial features. As the ddrgk1 mRNA dose increased in ddrgk1 morphants from 25 pg to 50 pg, the proportion of embryos with normal craniofacial structures increased from 51% to 68% (Figure 2, A and B). This rescue of the cartilage phenotype in ddrgk1 morphants by overexpression of ddrgk1 mRNA confirms that ddrgk1 is essential for cartilage development in zebrafish and supports the hypothesis that the c.408+1G>A DDRGK1 loss-of-function mutation causes Shohat-type SEMD.
Ddrgk1–/– mice show delayed chondrogenic mesenchymal condensation in the limb buds.
To further characterize the in vivo function of Ddrgk1 in a mammalian context, we generated Ddrgk1 knockout mice using CRISPR/Cas9. After injecting Cas9 mRNA and Ddrgk1 guide RNA into mouse embryos, we identified a 310-bp c.-249_61del mutation in the Ddrgk1 gene in a founder mouse. This mutation deletes a fragment of the 5′ UTR and most of the first exon, including the start codon (Supplemental Figure 2, A and B). RT-PCR and Western blot analysis confirmed that Ddrgk12BL/2BL (Ddrgk1–/–) mice do not express Ddrgk1 mRNA or protein (Supplemental Figure 2, C and D). As previously reported, we confirmed that Ddrgk1 loss of function results in embryonic lethality between E11.5 and E12.5 (ref. 13, Supplemental Figure 2E and Supplemental Table 1) because of an erythropoiesis deficiency (13). Deletion of Ddrgk1 decreases the number of erythrocytes and their progenitor cells in peripheral blood, while increasing the number of abnormal multinucleated erythrocytes in E11.5 embryos (13). However, there are no reports of Shohat-type SEMD patients with any erythropoiesis deficiency or phenotype (5, 6). These results suggest that erythropoiesis deficiency is potentially a mouse-specific phenotype, which would explain why embryonic lethality occurs in Ddrgk1–/– mice and not in Shohat-type SEMD patients.
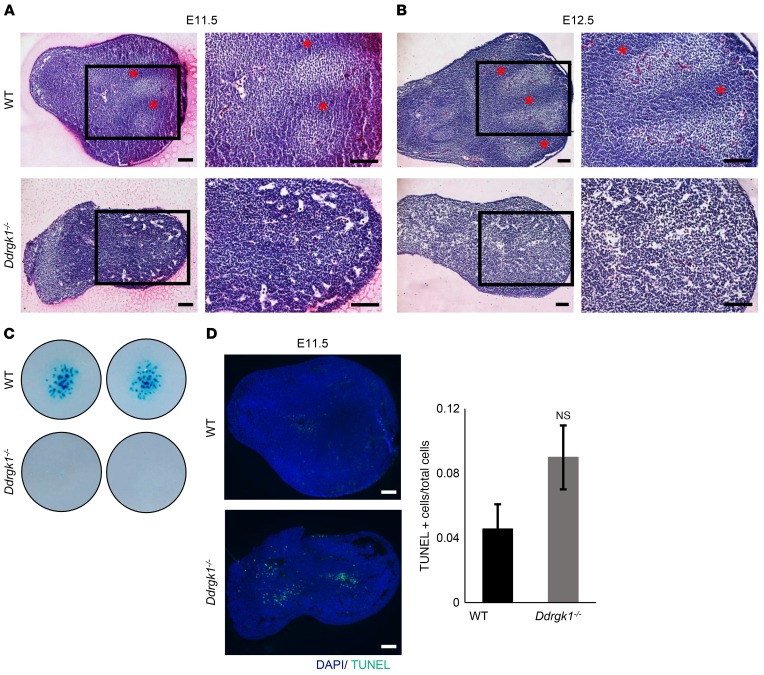
To evaluate the consequences of Ddrgk1 loss during chondrogenesis, we assessed chondrogenic mesenchymal condensation from Ddrgk1–/– limb buds at E11.5 and E12.5. We observed proper patterning of digit formation in WT limb buds at E11.5 and E12.5 by H&E staining (Figure 3, A and B). However, condensation of mesenchymal cells did not occur in Ddrgk1–/– limb buds up to E12.5, which suggests that Ddrgk1 deletion causes a delay or absence of the early stages of chondrogenesis (Figure 3, A and B). These results are consistent with those obtained from ex vivo chondrogenic differentiation assay. After 7 days, E11.5 Ddrgk1–/– mesenchymal cells generated fewer Alcian blue–positive cartilaginous nodules than E11.5 WT cells (Figure 3C). Additionally, deletion of Ddrgk1 increased apoptosis and cell death, trending toward significance, in E11.5 limb buds (Figure 3D). Taken together, the phenotypes of ddrgk1 knockdown zebrafish and Ddrgk1 null mouse indicate that Ddrgk1 is required for chondrocyte differentiation from mesenchymal cells, and furthermore, they suggest that loss of DDRGK1 reduces the chondrogenic potency of mesenchymal cells, causing the cartilage phenotype seen in Shohat-type SEMD.
Figure 3. Deletion of Ddrgk1 delays chondrogenic mesenchymal condensation and increases apoptosis in mouse limb buds.
(A and B) Mesenchymal condensation is detected in WT, but not in Ddrgk1–/–, limb buds stained with H&E at (A) E11.5 and (B) E12.5. Red asterisks indicate regions undergoing mesenchymal condensation. Images on the right are higher magnification depictions of the boxed regions of the images to the left. (C) After 7 days, micromass cultures of dissociated Ddrgk1–/– (n = 2) mesenchymal cells produced fewer Alcian blue–positive cartilaginous nodules than WT (n = 2) mesenchymal cells. Images are of 2 independent experiments. (D) TUNEL staining reveals Ddrgk1–/– limb buds (n = 3) have increased apoptosis compared with WT limb buds (n = 4) at E11.5, trending toward significance. Values are represented as mean ± SEM. Two-tailed t test. Scale bars: 100 μM.
DDRGK1 regulates SOX9 protein stability.
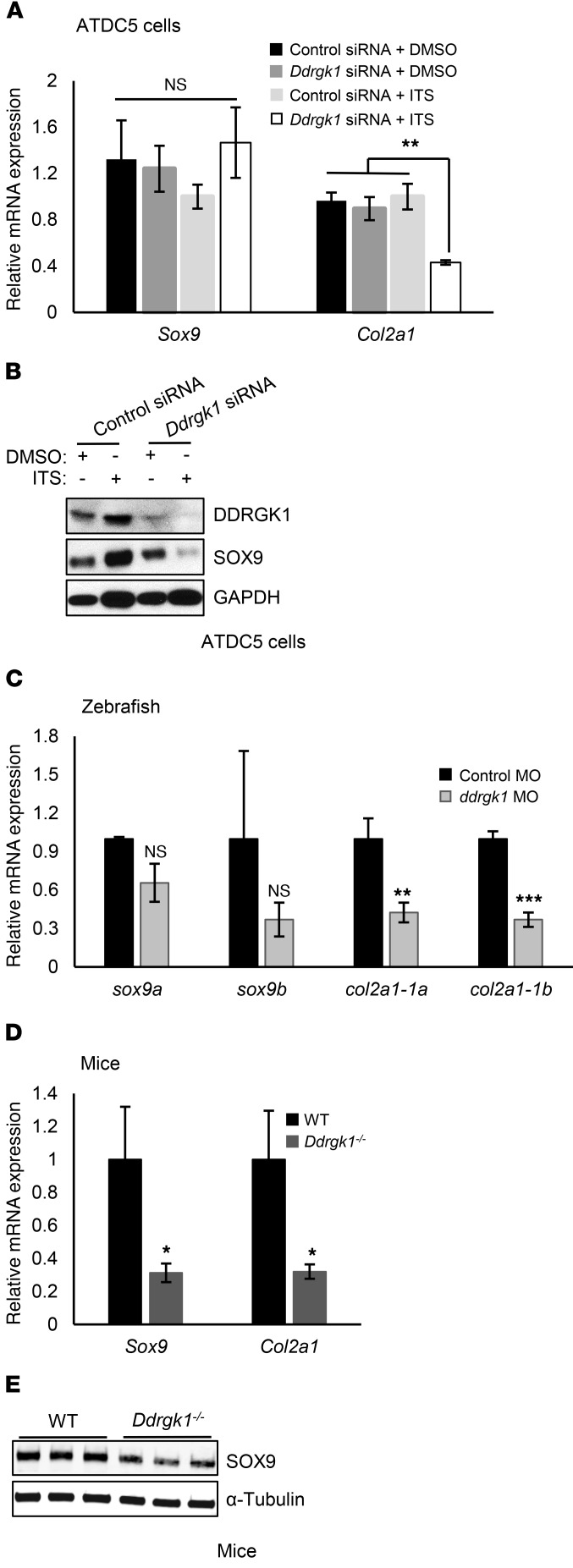
We observed that Ddrgk1 deficiency partially phenocopies the skeletal phenotype of Sox9 deficiency (21–23). Similar to ddrgk1 morphants, sox9a-deficient zebrafish embryos lack Meckel’s cartilage, ceratohyal cartilage, ceratobranchial cartilage, and neurocranial cartilage at 96 h.p.f. (21, 22). Also, there is an absence of mesenchymal condensation in the limb buds of conditional Sox9 mice deleted for Sox9 in their limb buds (Sox9fl/fl Prx1-Cre), which is comparable to the Ddrgk1–/– limb bud phenotype (23). These phenotypic similarities led us to test the epistasis of DDRGK1 and SOX9. Initially to address this question, we knocked down Ddrgk1 in undifferentiated and differentiated ATDC5 chondrogenic cells using siRNAs. Knockdown of Ddrgk1 did not alter the mRNA expression of Sox9, but did decrease SOX9 protein levels in ATDC5 cells after 7 days of differentiation induced by insulin, transferrin, and selenium (ITS) treatment (Figure 4, A and B). Ddrgk1 knockdown in differentiated ATDC5 cells also reduced mRNA expression of Col2a1 (Figure 4A). We found similar alterations in ddrgk1 morphants and Ddrgk1–/– mice. At 72 h.p.f., ddrgk1 morphants had decreased transcript levels of col2a1-1a and col2a1-1b, but not sox9a or sox9b, compared with control morphants (Figure 4C). Also, at E11.5, Ddrgk1–/– mice had decreased protein expression of Sox9 and reduced Col2a1 mRNA levels in their limb buds (Figure 4, D and E). Overall, these results demonstrate that SOX9 is a downstream target of DDRGK1.
Figure 4. Ddrgk1 deficiency decreases SOX9 protein and Col2a1 mRNA expression.
(A and B) ATDC5 cells were transiently transfected with control or Ddrgk1 siRNA and treated with either DMSO or ITS to induce differentiation 24 hours later. The cells were harvested 7 days after treatment. (A) RT-PCR of total RNA from ATDC5 cells treated with control siRNA + DMSO (n = 3), Ddrgk1 siRNA + DMSO (n = 3), control siRNA + ITS (n = 3), and Ddrgk1 siRNA + ITS (n = 3). Values are represented as mean ± SEM. **P < 0.01, 2-way ANOVA followed by Tukey’s post-hoc test. (B) Immunoblots of total cell lysates from ATDC5 cells. The immunoblots are representative of 3 independent experiments. (C) ddrgk1 morphants have less mRNA expression of col2a1, but not of sox9, than control morphants. Zebrafish embryos were injected with 5 pg control MO (n = 3) or 5 pg ddrgk1 MO (n = 3), and total RNA was collected for RT-PCR 72 h.p.f. later. Values are represented as mean ± SEM. **P < 0.01; ***P < 0.001, 2-tailed t test. (D and E) Deletion of Ddrgk1 decreases transcript levels of Col2a1 via SOX9 protein reduction in E11.5 limb buds. (D) RT-PCR of total RNA from E11.5 WT (n = 3) and Ddrgk1–/– (n = 3) limb buds. Values are represented as mean ± SEM. *P < 0.05, 2-tailed t test. (E) Immunoblots of total cell lysates from E11.5 WT (n = 3) and Ddrgk1–/– (n = 3) limb buds.
To further validate sox9 as a downstream target of ddrgk1 in zebrafish, we performed rescue experiments overexpressing sox9a in ddrgk1 zebrafish morphants. Overexpressing sox9a mRNA caused 60% lethality in zebrafish embryos by 120 d.p.f. (data not shown), which is consistent with the 80% postnatal lethality in Col2a1Sox9/+ mice overexpressing Sox9 in the chondrocytes (24). When we coinjected ddrgk1 MO with sox9a mRNA into zebrafish embryos, it rescued the ddrgk1 craniofacial defects in a dosage-dependent manner (Figure 5, A and B). Only 43% of ddrgk1 morphants had normal craniofacial features; however, as we increased sox9a mRNA in ddrgk1 morphants from 50 pg to 150 pg, the proportion of ddrgk1 morphants with normal craniofacial structures increased from 48% to 75% (Figure 5B). The rescue of the ddrgk1 cartilage phenotype by sox9a overexpression confirms sox9 as a downstream target of ddrgk1. These results suggest that in Shohat-type SEMD patients, the c.408+1G>A DDRGK1 loss-of-function mutation exerts its effects by decreasing SOX9 in the skeleton.
Figure 5. Expression of sox9a mRNA rescues the ddrgk1 knockdown cartilage phenotype in zebrafish.
(A) Ventral view at 120 h.p.f. of Alcian blue–stained embryos injected with either control MO or ddrgk1 MO with or without sox9a mRNA. Scale bars: 100 μM. (B) Quantification of the craniofacial phenotype in the embryos. The categories of craniofacial features are described in Figure 2. Control MO, n = 36; 5 pg ddrgk1 MO, n = 42; 5 pg ddrgk1 MO + 50 pg ddrgk1 mRNA, n = 31; 5 pg ddrgk1 MO + 150 pg ddrgk1 mRNA, n = 41. *P < 0.05; ***P < 0.001, Kruskal-Wallis rank-sum test followed by Wilcoxon’s rank-sum test with continuity correction.
DDRGK1 directly interacts with SOX9 to inhibit SOX9 ubiquitination.
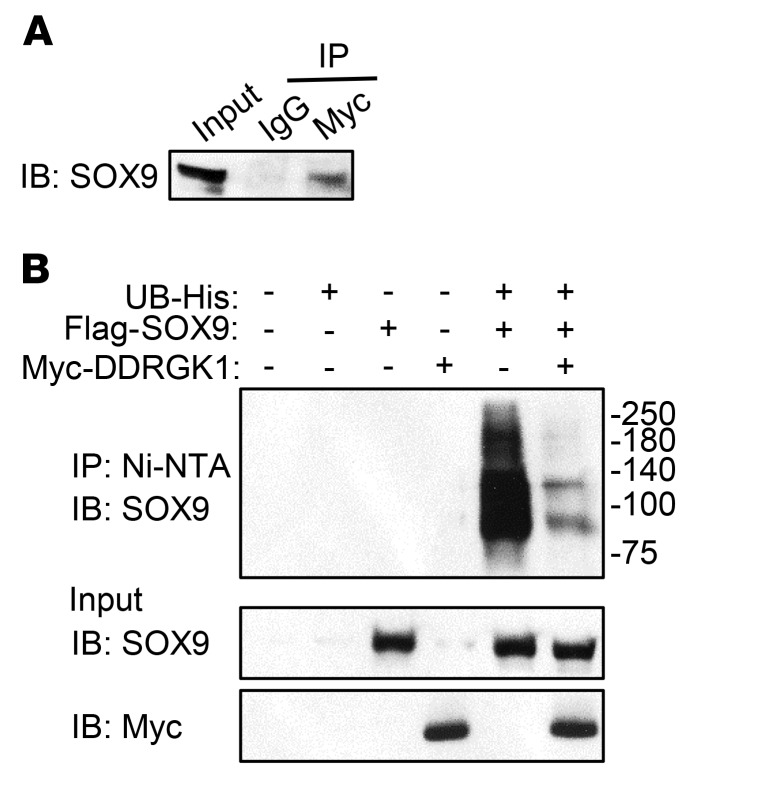
It has been reported that, mechanistically, DDRGK1 interacts with its downstream targets such as IκB and ASC1 to regulate posttranslational modification and protein stability (9, 10). With this in mind, we investigated whether DDRGK1 and SOX9 directly interacted in a coimmunoprecipitation assay by expressing Myc-tagged DDRGK1 and FLAG-tagged SOX9 in HEK293T cells. We found that SOX9 forms a complex with DDRGK1 (Figure 6A). After detecting a protein-protein interaction between SOX9 and DDRGK1, we assessed whether DDRGK1 regulates SOX9 stability by modulating its posttranslational modification, such as ubiquitination. As previously shown, SOX9 polyubiquitination can only be detected in vitro when proteasome inhibitors are added, thus demonstrating that SOX9 undergoes ubiquitin-dependent proteasomal degradation (25, 26). We confirmed SOX9 polyubiquitination by overexpressing Sox9 and Ub in HEK293T cells and treating the cells with the proteasome inhibitor MG132 for 6 hours (Figure 6B). Interestingly, the levels of ubiquitinated SOX9 decreased when Sox9 and Ub were coexpressed with Ddrgk1 in HEK293T cells (Figure 6B). This in vitro assay demonstrated that DDRGK1 forms a complex with SOX9 to regulate SOX9 ubiquitin-dependent proteasomal degradation during cartilage development. We then evaluated whether DDRGK1 can regulate the translation of SOX9 by assaying for DDRGK1 effects in the presence of the protein synthesis inhibitor cycloheximide. We coexpressed Sox9 and Ub with or without Ddrgk1 in HEK293T and then treated the cells 24 hours later with cycloheximide for 6 hours. Even when translation was halted by cycloheximide, overexpression of Ddrgk1 increased Sox9 protein expression, which indicates that inhibiting translation does not diminish the effects of DDRGK1 on SOX9 levels (Supplemental Figure 3). These results suggest that DDRGK1 regulates SOX9 primarily at the posttranslational level, but not at the translational level. Mechanistically, we have demonstrated that DDRGK1 is a critical upstream regulatory factor of SOX9 ubiquitination during the mesenchymal condensation stage of chondrocyte differentiation. Furthermore, the consequence of DDRGK1 absence illustrates the importance of its in vivo role in cartilage development seen in Shohat-type SEMD.
Figure 6. DDRGK1 forms a complex with SOX9 and inhibits SOX9 ubiquitination.
(A) SOX9 forms a complex with DDRGK1. 293T cells were transiently transfected with plasmids expressing Myc-tagged Ddrgk1 and FLAG-tagged Sox9. After 48 hours, SOX9 was detected by immunoblotting with a FLAG-specific antibody after immunoprecipitation of cell lysates with anti-Myc or IgG antibody. As a control, the input was immunoblotted with the FLAG-specific antibody. The immunoblot is representative of 3 independent experiments. (B) Ddrgk1 overexpression decreases SOX9 ubiquitination in 293T cells. 293T cells were transiently transfected with plasmids expressing His-tagged UB, FLAG-tagged SOX9, and Myc-tagged DDRGK1. Twenty-four hours later, the cells were treated with 20 μM of proteasome inhibitor MG132 for 6 hours. Afterwards, we pulled down His-tagged ubiquitinated proteins using Ni-NTA beads, and ubiquitinated SOX9 was probed using anti-SOX9 antibody. As controls, the input was immunoblotted with anti-SOX9 and anti-Myc antibodies. The immunoblots are representative of 3 independent experiments.
Discussion
In this study, we identified a homozygous c.408+1G>A donor splice site mutation in DDRGK1 as a genetic cause of Shohat-type SEMD. Specifically, this mutation causes aberrant splicing and a loss of DDRGK1 in patient LCLs. Similarly to what is observed in humans, we demonstrated that Ddrgk1 deficiency disrupts chondrogenesis and cartilage formation in mice and zebrafish; however, unlike in Shohat-type SEMD patients, loss of Ddrgk1 in mice is embryonic lethal. Though previous studies have shown that Ddrgk1 deficiency causes defective erythropoiesis in mice (13), we and others have not observed this phenotype in patients with Shohat-type SEMD. At the same time, a GWAS identified a link between DDRGK1 variants and thrombocytopenia in hepatitis C patients treated with pegylated interferon and ribavirin (27). Taken together, these data suggest that, unlike in mice, mutations or variants in DDRGK1 affect erythrocyte differentiation in humans only when secondary to other pathologies or therapeutic interventions, perhaps due to physiological functional redundancy in humans. In the future, it would be interesting to identify these redundant pathways and to assess whether they also have a role in chondrogenesis during cartilage development.
During development, there are numerous master regulators that orchestrate and control central tissue-specific developmental pathways. To list a few, RUNX2, MYOD, and TAL1 are master transcription factors that regulate the differentiation of osteoblast, skeletal muscle, and erythrocytes, respectively (28–30). On the other hand, SOX9 is a unique master transcription factor because of its essential role in regulating multiple developmental pathways, including chondrogenesis, gonadogenesis, gliogenesis, epithelial hair follicle stem cell specification, normal development of cardiac valve, septum, lung, pancreas, etc. (21, 23, 31–33). Dysregulation of SOX9 contributes to the development of different malignant cancers and causes developmental disorders (31, 34–36). For example, haploinsufficiency for Sox9 causes campomelic dysplasia (CMD) (31, 37–39), a rare neonatal chondrodysplasia characterized by bowing and angulation of long bones and many other skeletal and extraskeletal features (31). The few CMD survivors have disproportionate short stature, progressive kyphoscoliosis, and hip dislocation, among other findings (39), thus sharing phenotypic overlap with Shohat-type SEMD patients.
In both mice and zebrafish, SOX9 deficiency impairs chondrogenesis (21–23). sox9a knockout zebrafish show malformed Meckel’s cartilage and ceratohyal cartilage and lack neurocranial cartilage and ceratobranchial cartilage (21, 22), whereas loss of Sox9 in mouse limb buds prevents chondrogenic mesenchymal condensation (23). Interestingly, many of the phenotypic features of the SOX9-deficient animal models were also observed in our DDRGK1-deficient animal models. For this reason, we assessed changes in expression of SOX9 and its downstream target gene, Col2a1. Using CRISPR/CAS9-generated Ddrgk1–/– mice and differentiated ATDC5 cells, we found that Ddrgk1 deficiency decreases Sox9 protein levels and the expression of its target gene, Col2a1. We then rescued the craniofacial phenotype in ddrgk1 knockdown zebrafish by overexpressing sox9a to confirm that Sox9 is an important epistatic downstream target of Ddrgk1. Though the reduction in Sox9 protein levels is less than 50% in Ddrgk1–/– mice, these mice have a more severe skeletal phenotype than Sox9+/– mice haploinsufficient for Sox9 (38). One possible explanation for the more severe phenotype in Ddrgk1–/– mice may be that DDRGK1 regulates multiple targets involved in chondrogenesis in addition to SOX9. Another interesting finding that requires further investigation is that knockdown of Ddrgk1 decreases SOX9 in differentiated ATDC5 cells, but not in undifferentiated ATDC5 cells. These results suggest that the control of SOX9 protein stability by DDRGK1 is temporally regulated. The function of this potential temporal regulation, the factors that govern this regulation, and how they are regulated are interesting subjects for future studies.
The clinical overlap among CMD, the SED spectrum of disorders, and Shohat-type SEMD supports that their pathogenesis is caused by mutations in the same pathway. The SED spectrum phenotypes are caused by mutations altering the structure and trafficking of COL2A1, whereas CMD and Shohat-type SEMD are both caused by mutations affecting SOX9 in its regulation of COL2A1. Our study emphasizes the importance of studying phenocopies of disease because of the potential to reveal novel components of a developmental pathway. Investigating Shohat-type SEMD thus identified DDRGK1 as an indirect regulator of COL2A1 expression through its ability to inhibit SOX9 ubiquitin-dependent proteasomal degradation.
In the present study, we show that DDRGK1 interacts in a complex with SOX9 to regulate its ubiquitination; however, the underlying mechanism mediating this inhibition remains unknown. DDRGK1 is known to be required for ufmylation, a ubiquitin-like posttranslational modification that covalently binds ubiquitin-like fold modifier 1 (UFM1) to substrates (10, 12, 40, 41). DDRGK1 forms a complex with UFM1 substrates and UFM1-specific ligase 1 (UFL1), an UFM1 E3 ligase, and Ddrgk1 knockdown prevents ASC1 umfylation (10, 12). Therefore, based on these studies and our findings, it is possible that SOX9 may be ufmylated, a process for which DDRGK1 is required; SOX9 ufmylation in turn may prevent its ubiquitination and proteasomal degradation. Alternatively, another potential mechanism for DDRGK1 inhibition of SOX9 ubiquitination may be independent of its role in ufmylation. DDRGK1 has a proteasome, COP9, initiation factor 3 (PCI) domain (ref. 42 and Figure 1F), which can also be found in large multisubunit protein complexes, including the proteasome, COP9 signalosome, and eukaryotic translation initiation factor 3 (43–45). The PCI domain facilitates protein-protein interactions, and it is thought to facilitate interactions in complexes containing this domain (43–45). Therefore, DDRGK1 may interact with the proteasome complex to directly mediate SOX9 ubiquitin-dependent proteasomal degradation.
In conclusion, our results demonstrate that the homozygous c.408+1G>A loss-of-function mutation in DDRGK1 causes Shohat-type SEMD by loss of DDRGK1 inhibition of SOX9 ubiquitin-dependent proteasomal degradation, thus impairing chondrogenesis. By using a rapid multimodel system (a combination of morpholino-knockdown zebrafish, CRISPR/Cas9-generated mouse genetic models, and in vitro assays) to study the pathogenicity of a rare genetic variant, we identified the genetic cause of Shohat-type SEMD. Previous studies of DDRGK1 investigated its cellular and in vivo functions in mice, but our study addressed, for what we believe is the first time, its physiological role in the context of a human disease and skeletogenesis. Furthermore, this work reveals a mechanism regulating the developmental pathway of chondrogenesis.
Methods
WES.
Total genomic DNA was extracted from whole blood from 3 affected individuals. WES was performed on 3 affected individuals from families 1 and 2 and the data analyzed, as previously described (14). Afterwards, conventional Sanger sequencing was performed on all living patients to confirm the DDRGK1 variant identified in the WES analysis, as previously described (14).
Cell culture, transfection, and treatment.
LCLs were extracted from the proband and a control who did not have Sutcliffe type of spondylometaphyseal dysplasia. LCLs were grown at 37°C in αMEM (HyClone) plus 10% FBS and with 100 units/ml of penicillin and 1 μg/ml of streptomycin. HEK293T cells (ATCC) were grown at 37°C in DMEM (HyClone) supplemented with 100 units/ml of penicillin, 1 μg/ml of streptomycin, and 10% FBS. 293T cells were transiently transfected with expression plasmids for 48 hours, using Lipofectamine 2000 (Invitrogen) according to the manufacturer’s protocol. ATDC5 cells (ATCC) were grown at 37°C in DMEM/F12 (HyClone) supplemented with 100 units/ml of penicillin, 1 μg/ml of streptomycin, and 5% FBS. For the in vitro knockdown experiments, ATDC5 cells were transiently transfected with either 50 nM siGENOME mouse Ddrgk1 siRNA or 50 nM siGENOME Non-Targeting siRNA Pool #1 (Dharmacon) using Lipofectamine 2000 (Invitrogen). To differentiate ATDC5 cells, cells were treated 24 hours after siRNA treatment with 1× ITS Liquid Media Supplement (100×, Sigma-Aldrich) for 7 days. For the ubiquitination assays, 293T cells were treated with 20 μM of proteasome inhibitor MG132 6 hours before harvesting. To inhibit translation, 293T cells were treated with 10 μg/ml of protein synthesis inhibitor cycloheximide for 6 hours before harvesting.Antibodies. The rabbit anti-DDRGK1 antibody was a gift from Frans Shuit (12). Anti-SOX9 antibody (catalog AB5535), anti–c-Myc antibody (catalog sc-40), anti-FLAG antibody (catalog F1804), anti-BrdU antibody (catalog Ab1893), and normal rabbit IgG (catalog 2729) were purchased from Millipore, Santa Cruz Biotechnology Inc., Sigma-Aldrich, Abcam, and Cell Signaling Technology, respectively. The anti-SOX9 antibody (AB5535) and anti-BrdU antibody (AB1893) were purchased from Abcam. The secondary antibodies used were goat anti-mouse IgG-HRP (H+L) conjugate (catalog 1721011, Bio-Rad), goat anti-mouse antibody (catalog 926-68070, Licor), goat anti-rabbit antibody (catalog 926-32211, LiCor), donkey anti-sheep antibody Alexa Fluor 594 (catalog Ab150180, Abcam), and anti-rabbit IgG, HRP-linked whole antibody (catalog NA934, GE Healthcare Life Sciences).
Expression plasmids.
We previously generated the pcDNA3.1_FLAG-SOX9 plasmid (46); the pCMV_Myc-DDRGK1 and His6-UB plasmids were gifts from Honglin Li (11) and Dae-Sik Lim (47), respectively. The ddrgk1 and sox9a cDNAs were amplified from zebrafish cDNA using the primers listed in Supplemental Table 2 and then cloned into the pCS2+ vector by In-Fusion Cloning (Clontech). Afterwards, to prevent ddrgk1 MO targeting of exogenous ddrgk1 mRNA, 5 nucleotide sequences of ddrgk1 pCS2+ vector were mutated by QuikChange (Agilent Technologies) site-directed mutagenesis without altering the protein sequence (Supplemental Table 2).
Microinjection and Alcian blue staining of zebrafish embryos.
From 3 to 5 pg ddrgk1 translational MO or ddrgk1 control MO (Gene Tools) was injected into embryos at the 1-cell stage. For rescue experiments, mutated ddrgk1 mRNA and sox9a mRNA were transcribed from the pCS2+ vectors using the mMessage mMachine SP6 kit (Ambion). Amounts ranging from 25 to 150 pg were used for the injections (Supplemental Table 2). At 120 h.p.f., embryos were fixed in 2% PFA and then stained in 0.1% Alcian blue solution at 4°C for 24 hours. After 24 hours, the embryos were bleached in 3% H2O2 and 0.5% KOH.
CRISRP/Cas9 generation of Ddrgk1+/– mice.
Ddrgk1 sgRNA (Supplemental Table 2) was designed using the online CRISPR Design Tool and cloned into the hSpCas9 vector as previously described (48). After cloning, the sgRNA template was generated by PCR amplification, which simultaneously added a T7 promoter in front of the Ddrgk1 sgRNA sequence (Supplemental Table 2). RNA was transcribed from the template using the MEGAshortscript T7 Transcription Kit (Ambion) and cleaned up with the MEGAclear Transcription Clean-Up Kit (Ambion). 17 ng/μl Ddrgk1 sgRNA and 50 ng/μl Cas9 mRNA were coinjected into C57/B6J mouse embryos (Jackson Laboratory). The founder mice from these injections were screened for mutations in the Ddrgk1 gene (Supplemental Table 2), and the desired line was backcrossed 5 generations.
Histology of mouse limb buds.
E11.5 and E12.5 limb buds were collected and fixed in 4% PFA at 4°C for 24 hours. The limb buds were then embedded in paraffin and sectioned 7-μm thick. To examine morphology, sections were stained with H&E (Sigma-Aldrich). To assess apoptosis, sections were stained with the ApopTag Fluorescein In Situ Apoptosis Detection Kit (Milipore) according to the manufacturer’s protocol.
Coimmunoprecipitation.
293T cells were transiently transfected with 3 μg FLAG-Sox9 vector and 3 μg Myc-Ddrgk1. After 48 hours, the cells were lysed in lysis buffer (50 mM Tris HCl, pH 8, 150 mM NaCl, 0.1% NP-40, 1 mM EDTA, 2 mM N-ethylmaleimide and proteinase inhibitor cocktail; GenDEPOT), vortexed, and then incubated on ice for 20 minutes. After centrifugation at 13,000 g for 20 minutes at 4°C, the supernatant was precleared by adding protein G agarose (Roche) and rotating at 4°C for 20 minutes. The supernatant was then incubated with anti-MYC antibody (Sigma-Aldrich) or normal IgG (Santa Cruz Biotechnology Inc.) and bound to protein G beads (Roche). After washing the immunoprecipitated samples 3 times with lysis buffer, proteins were separated by SDS-PAGE, and SOX9 was detected with anti-FLAG antibody.
Ubiquitination assay.
293T cells were transiently transfected with 2 μg FLAG-Sox9 vector, His-Ub vector, and 4 μg Myc-Ddrgk1 plasmid. After 48 hours, the His-tagged ubiquitinated proteins were pulled down with Ni-NTA agarose beads (QIAGEN) as previously described (49). Following Ni-NTA pulldown, proteins were separated by SDS-PAGE and SOX9 was probed with anti-SOX9 antibody.
qRT-PCR.
Total RNA was extracted from tissues and cells using TRIzol reagent (Invitrogen) and cDNA was synthesized from total RNA using the SuperScript III First Strand RT-PCR kit (Invitrogen). The qRT-PCR was performed on a LightCycler instrument (Roche).
Statistics.
All the data are shown as mean ± SEM. Statistical analyses used were a type 2-tailed Student’s t test, 2-way ANOVA followed by a Tukey’s post-hoc test, and Kruskal-Wallis rank-sum test followed by Wilcoxon’s rank-sum test with continuity correction. P < 0.05 was considered significant.
Study approval.
Prior to their participation in this study, the families provided written, informed consent to participate in the study approved by the IRB at Rabin Medical Center, Petah Tikva, Israel. The families underwent physical examinations, and we obtained radiographs of the patients. WT adult zebrafish strain (AB) and Ddrgk1+/– mice were both maintained under standard laboratory conditions. Zebrafish embryos were obtained after natural crosses between males and females, and mouse embryos were obtained by timed mating. All animal studies were performed under protocols reviewed and approved by IACUCs at the University of Texas Medical School at Houston or Baylor College of Medicine.
Author contributions
NM, ARR, and MS were involved in patient care, patient diagnoses, and human data collection. ATE, YB, ECS, PMC, and BHL designed the research studies. JTL, RAG, and PMC conducted the bioinformatics analyses. SC performed the statistical analyses. ATE, YB, MMJ, DSL, YCE, TB, LN, and PMC conducted experiments and acquired data. ATE, YB, ECS, DHC, PMC, and BHL analyzed data. ECS, DHC, and BHL provided the reagents. ATE wrote the manuscript, and YB, TB, ECS, DHC, PMC, MS, and BHL edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the Baylor College of Medicine Intellectual and Developmental Disabilities Research Center (HD024064) from the Eunice Kennedy Shriver National Institute of Child Health and Human Development, the Baylor College of Medicine Advanced Technology Cores with funding from the NIH (AI036211, CA125123, and RR024574), the Rolanette and Berdon Lawrence Bone Disease Program of Texas, and the Baylor College of Medicne Center for Skeletal Medicine and Biology. It was also supported by NIH grant R01AR062651 and the Adler chair for Pediatric Cardiology, Tel Aviv University.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(4):1475–1484.https://doi.org/10.1172/JCI90193.
Contributor Information
Yangjin Bae, Email: bae@bcm.edu.
Ming-Ming Jiang, Email: mjiang@bcm.edu.
Terry Bertin, Email: tbertin@bcm.tmc.edu.
Shan Chen, Email: shanc@bcm.edu.
Lisette Nevarez, Email: lnevarez@ucla.edu.
Nurit Magal, Email: NMagal@clalit.org.il.
Annick Raas-Rothschild, Email: Annick.rothschild@sheba.health.gov.il.
Richard A. Gibbs, Email: agibbs@bcm.edu.
Mordechai Shohat, Email: mshohat1@gmail.com.
References
- 1.Spranger J, Winterpacht A, Zabel B. The type II collagenopathies: a spectrum of chondrodysplasias. Eur J Pediatr. 1994;153(2):56–65. doi: 10.1007/BF01959208. [DOI] [PubMed] [Google Scholar]
- 2.Kuivaniemi H, Tromp G, Prockop DJ. Mutations in fibrillar collagens (types I, II, III, and XI), fibril-associated collagen (type IX), and network-forming collagen (type X) cause a spectrum of diseases of bone, cartilage, and blood vessels. Hum Mutat. 1997;9(4):300–315. doi: 10.1002/(SICI)1098-1004(1997)9:4<300::AID-HUMU2>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 3.Wirth T. [Spondyloepiphyseal and metaphyseal dysplasia] Der Orthopäde. 2008;37(1):8–16. doi: 10.1007/s00132-007-1179-z. [DOI] [PubMed] [Google Scholar]
- 4.Cormier-Daire V. Spondylo-epi-metaphyseal dysplasia. Best Pract Res Clin Rheumatol. 2008;22(1):33–44. doi: 10.1016/j.berh.2007.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Figuera LE, Ramírez-Dueñas ML, Gallegos-Arreola MP, Cantú JM. Spondyloepimetaphyseal dysplasia (SEMD) Shohat type. Am J Med Genet. 1994;51(3):213–215. doi: 10.1002/ajmg.1320510307. [DOI] [PubMed] [Google Scholar]
- 6.Shohat M, Lachman R, Carmi R, Bar Ziv J, Rimoin D. New form of spondyloepimetaphyseal dysplasia (SEMD) in Jewish family of Iraqi origin. Am J Med Genet. 1993;46(4):358–362. doi: 10.1002/ajmg.1320460403. [DOI] [PubMed] [Google Scholar]
- 7.Venditti R, et al. Sedlin controls the ER export of procollagen by regulating the Sar1 cycle. Science. 2012;337(6102):1668–1672. doi: 10.1126/science.1224947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee B, Vissing H, Ramirez F, Rogers D, Rimoin D. Identification of the molecular defect in a family with spondyloepiphyseal dysplasia. Science. 1989;244(4907):978–80. doi: 10.1126/science.2543071. [DOI] [PubMed] [Google Scholar]
- 9.Xi P, Ding D, Zhou J, Wang M, Cong YS. DDRGK1 regulates NF-κB activity by modulating IκBα stability. PLoS One. 2013;8(5):e64231. doi: 10.1371/journal.pone.0064231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoo HM, et al. Modification of ASC1 by UFM1 is crucial for ERα transactivation and breast cancer development. Mol Cell. 2014;56(2):261–274. doi: 10.1016/j.molcel.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 11.Wu J, Lei G, Mei M, Tang Y, Li H. A novel C53/LZAP-interacting protein regulates stability of C53/LZAP and DDRGK domain-containing Protein 1 (DDRGK1) and modulates NF-κB signaling. J Biol Chem. 2010;285(20):15126–15136. doi: 10.1074/jbc.M110.110619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemaire K, et al. Ubiquitin fold modifier 1 (UFM1) and its target UFBP1 protect pancreatic β cells from ER stress-induced apoptosis. PLoS One. 2011;6(4):e18517. doi: 10.1371/journal.pone.0018517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cai Y, et al. UFBP1, a key component of the Ufm1 conjugation system, is essential for ufmylation-mediated regulation of erythroid development. PLoS Genet. 2015;11(11):e1005643. doi: 10.1371/journal.pgen.1005643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campeau PM, et al. Whole-exome sequencing identifies mutations in the nucleoside transporter gene SLC29A3 in dysosteosclerosis, a form of osteopetrosis. Hum Mol Genet. 2012;21(22):4904–4909. doi: 10.1093/hmg/dds326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lopez IA, et al. Slc4a11 gene disruption in mice: cellular targets of sensorineuronal abnormalities. J Biol Chem. 2009;284(39):26882–26896. doi: 10.1074/jbc.M109.008102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vithana EN, et al. Mutations in sodium-borate cotransporter SLC4A11 cause recessive congenital hereditary endothelial dystrophy (CHED2) Nat Genet. 2006;38(7):755–757. doi: 10.1038/ng1824. [DOI] [PubMed] [Google Scholar]
- 17. Thisse B, Thisse C. Fast release clones: a high throughput expression analysis. ZFIN direct data submiss. ZFIN web site. https://zfin.org/ZDB-PUB-040907-1 Accessed February 3, 2017.
- 18.Javidan Y, Schilling TF. Development of cartilage and bone. Methods Cell Biol. 2004;76:415–436. doi: 10.1016/S0091-679X(04)76018-5. [DOI] [PubMed] [Google Scholar]
- 19.Kimmel CB, et al. The shaping of pharyngeal cartilages during early development of the zebrafish. Dev Biol. 1998;203(2):245–263. doi: 10.1006/dbio.1998.9016. [DOI] [PubMed] [Google Scholar]
- 20.Schilling TF, Kimmel CB. Musculoskeletal patterning in the pharyngeal segments of the zebrafish embryo. Development. 1997;124(15):2945–2960. doi: 10.1242/dev.124.15.2945. [DOI] [PubMed] [Google Scholar]
- 21.Yan YL, et al. A zebrafish sox9 gene required for cartilage morphogenesis. Development. 2002;129(21):5065–5079. doi: 10.1242/dev.129.21.5065. [DOI] [PubMed] [Google Scholar]
- 22.Yan YL, et al. A pair of Sox: distinct and overlapping functions of zebrafish sox9 co-orthologs in craniofacial and pectoral fin development. Development. 2005;132(5):1069–1083. doi: 10.1242/dev.01674. [DOI] [PubMed] [Google Scholar]
- 23.Akiyama H, Chaboissier MC, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16(21):2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akiyama H, et al. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004;18(9):1072–1087. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Akiyama H, et al. The transcription factor Sox9 is degraded by the ubiquitin-proteasome system and stabilized by a mutation in a ubiquitin-target site. Matrix Biol. 2005;23(8):499–505. doi: 10.1016/j.matbio.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 26.Hattori T, et al. E6-AP/UBE3A protein acts as a ubiquitin ligase toward SOX9 protein. J Biol Chem. 2013;288(49):35138–35148. doi: 10.1074/jbc.M113.486795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tanaka Y, et al. Genome-wide association study identified ITPA/DDRGK1 variants reflecting thrombocytopenia in pegylated interferon and ribavirin therapy for chronic hepatitis C. Hum Mol Genet. 2011;20(17):3507–3516. doi: 10.1093/hmg/ddr249. [DOI] [PubMed] [Google Scholar]
- 28.Komori T. Runx2, a multifunctional transcription factor in skeletal development. J Cell Biochem. 2002;87(1):1–8. doi: 10.1002/jcb.10276. [DOI] [PubMed] [Google Scholar]
- 29.Weintraub H, et al. Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc Natl Acad Sci U S A. 1989;86(14):5434–5438. doi: 10.1073/pnas.86.14.5434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86(1):47–57. doi: 10.1016/S0092-8674(00)80076-8. [DOI] [PubMed] [Google Scholar]
- 31.Jo A, et al. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 2014;1(2):149–161. doi: 10.1016/j.gendis.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akiyama H, et al. Essential role of Sox9 in the pathway that controls formation of cardiac valves and septa. Proc Natl Acad Sci U S A. 2004;101(17):6502–6507. doi: 10.1073/pnas.0401711101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kang P, et al. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74(1):79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang H, McKnight NC, Zhang T, Lu ML, Balk SP, Yuan X. SOX9 is expressed in normal prostate basal cells and regulates androgen receptor expression in prostate cancer cells. Cancer Res. 2007;67(2):528–536. doi: 10.1158/0008-5472.CAN-06-1672. [DOI] [PubMed] [Google Scholar]
- 35.Kopp JL, et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22(6):737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwok C, et al. Mutations in SOX9, the gene responsible for Campomelic dysplasia and autosomal sex reversal. Am J Hum Genet. 1995;57(5):1028–1036. [PMC free article] [PubMed] [Google Scholar]
- 37.Bi W, Deng JM, Zhang Z, Behringer RR, de Crombrugghe B. Sox9 is required for cartilage formation. Nat Genet. 1999;22(1):85–89. doi: 10.1038/8792. [DOI] [PubMed] [Google Scholar]
- 38.Bi W, et al. Haploinsufficiency of Sox9 results in defective cartilage primordia and premature skeletal mineralization. Proc Natl Acad Sci U S A. 2001;98(12):6698–6703. doi: 10.1073/pnas.111092198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mansour S, Offiah AC, McDowall S, Sim P, Tolmie J, Hall C. The phenotype of survivors of campomelic dysplasia. J Med Genet. 2002;39(8):597–602. doi: 10.1136/jmg.39.8.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tatsumi K, et al. A novel type of E3 ligase for the Ufm1 conjugation system. J Biol Chem. 2010;285(8):5417–5427. doi: 10.1074/jbc.M109.036814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komatsu M, et al. A novel protein-conjugating system for Ufm1, a ubiquitin-fold modifier. EMBO J. 2004;23(9):1977–1986. doi: 10.1038/sj.emboj.7600205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neziri D, et al. Cloning and molecular characterization of Dashurin encoded by C20orf116, a PCI-domain containing protein. Biochim Biophys Acta. 2010;1800(4):430–438. doi: 10.1016/j.bbagen.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 43.Ellisdon AM, Stewart M. Structural biology of the PCI-protein fold. Bioarchitecture. 2012;2(4):118–123. doi: 10.4161/bioa.21131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dessau M, Halimi Y, Erez T, Chomsky-Hecht O, Chamovitz DA, Hirsch JA. The Arabidopsis COP9 signalosome subunit 7 is a model PCI domain protein with subdomains involved in COP9 signalosome assembly. Plant Cell. 2008;20(10):2815–2834. doi: 10.1105/tpc.107.053801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hofmann K, Bucher P. The PCI domain: a common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23(6):204–205. doi: 10.1016/S0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- 46.Zhou G, et al. Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc Natl Acad Sci U S A. 2006;103(50):19004–19009. doi: 10.1073/pnas.0605170103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song MS, Song SJ, Kim SY, Oh HJ, Lim DS. The tumour suppressor RASSF1A promotes MDM2 self-ubiquitination by disrupting the MDM2-DAXX-HAUSP complex. EMBO J. 2008;27(13):1863–1874. doi: 10.1038/emboj.2008.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mahanic CS, Budhavarapu V, Graves JD, Li G, Lin WC. Regulation of E2 promoter binding factor 1 (E2F1) transcriptional activity through a deubiquitinating enzyme, UCH37. J Biol Chem. 2015;290(44):26508–26522. doi: 10.1074/jbc.M115.659425. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.