Abstract
Genetic variations in the ITGAM gene (encoding CD11b) strongly associate with risk for systemic lupus erythematosus (SLE). Here we have shown that 3 nonsynonymous ITGAM variants that produce defective CD11b associate with elevated levels of type I interferon (IFN-I) in lupus, suggesting a direct link between reduced CD11b activity and the chronically increased inflammatory status in patients. Treatment with the small-molecule CD11b agonist LA1 led to partial integrin activation, reduced IFN-I responses in WT but not CD11b-deficient mice, and protected lupus-prone MRL/Lpr mice from end-organ injury. CD11b activation reduced TLR-dependent proinflammatory signaling in leukocytes and suppressed IFN-I signaling via an AKT/FOXO3/IFN regulatory factor 3/7 pathway. TLR-stimulated macrophages from CD11B SNP carriers showed increased basal expression of IFN regulatory factor 7 (IRF7) and IFN-β, as well as increased nuclear exclusion of FOXO3, which was suppressed by LA1-dependent activation of CD11b. This suggests that pharmacologic activation of CD11b could be a potential mechanism for developing SLE therapeutics.
Introduction
Systemic lupus erythematosus (SLE, lupus) is a debilitating autoimmune disease that is characterized by hyperactive immune cells, serum autoantibodies, immune complex deposition, multiorgan damage, and accelerated vascular disease (1). Pathways downstream of various pattern recognition receptors — in particular, Toll-like receptors (TLRs) — are central to the aberrant immune responses contributing to SLE and result in elevated levels of inflammatory cytokines, such as type I interferons (IFN-I), IL-1β, IL-6, TNF-α, and IL-17, in patients (2). Elevated levels of IFN-I in circulation are a heritable risk factor for SLE and play a pathogenic role (3). Although the underlying mechanisms influencing IFN-I levels in SLE subjects and conferring a strong predisposition to disease remain to be fully characterized, genetic variations, in combination with environmental stressors, play a key role. Genome-wide association studies have identified single-nucleotide polymorphisms (SNPs) in a number of immune response genes as key contributors to disease activity, and SNPs in ITGAM (coding for CD11b or αM, the α chain of the β2 leukocytic integrin heterodimer CD11b/CD18) show highly significant correlation with SLE (4–6). Three ITGAM coding region SNPs (7, 8) (rs1143678, rs1143679, and rs1143683), which result in missense mutations P1146S (a C>T substitution), R77H (a G>A substitution), and A858V (a C>T substitution), respectively, in the protein, have strong correlation with the incidence of SLE and with SLE subphenotypes including lupus nephritis, discoid rash, and immunologic manifestations (reviewed in ref. 9). How these nonsynonymous mutations confer SLE risk is not clear, but they appear to reduce CD11b function, including integrin activation, ligand binding and cell adhesion, phagocytosis, and catch-bond formation (7, 10–13). Recent studies also show that CD11b acts as a negative regulator of TLR signaling pathways and of B cell autoreactivity (14, 15), implying that the ITGAM SNPs found in SLE patients may affect disease activity via a reduction in CD11b’s normal, antiinflammatory signaling activities in leukocytes, although the exact mechanism behind how these variants contribute to autoimmunity is unclear. In murine systems, while CD11b deficiency was shown to reduce neutrophil accumulation and glomerular injury in an acute model (16), it has also been shown to increase susceptibility to chronic inflammatory and autoimmune diseases (refs. 14, 17–21, and reviewed in ref. 22), including increased tissue infiltration of leukocytes and immune-mediated injury in lupus-prone mice (19, 20). These data further suggest that CD11b plays a protective role in SLE. Here, we show that SLE patients carrying SNPs in the ITGAM gene present with elevated IFN-I activity, molecularly linking reduced CD11b function with increased disease risk. Ex vivo, cells homozygous for ITGAM SNPs showed a basal increase in IFNB and IRF7 expression and a decreased level of nuclear FOXO3, which regulates their expression, suggesting reduced suppression of TLR signaling at a basal level. Pharmacologic activation of CD11b, protein product of ITGAM, reduced IFN-I responses ex vivo and in animals via suppression of TLR-dependent FOXO3/IRF7 pathway in a CD11b-dependent fashion. It also similarly protected lupus-prone mice from end-organ injury. Suppressing overactive immune responses is a key strategy for treating SLE, and our data suggest that allosteric activation of CD11b with small molecules could be a novel therapeutic strategy.
Results
SLE subjects carrying ITGAM SNPs have significantly elevated serum IFN-I activity.
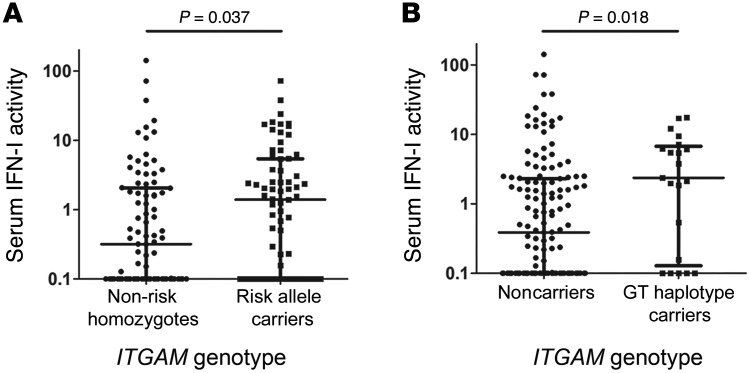
To test for a direct link between ITGAM SNPs and the IFN-I pathway in SLE, we measured the serum IFN-I activity (3) in 171 SLE subjects of European ancestry and determined their ITGAM genotype (Supplemental Table 1; supplemental material available online with this article; https://doi.org/10.1172/JCI88442DS1). This IFN assay was previously normalized using 100 healthy controls to establish normal values for serum IFN in healthy individuals (3). We found that the ITGAM risk alleles showed strong association with high IFN-I serum activity in patients (Figure 1A). Furthermore, analysis showed that the SNPs rs1143678 and rs1143683 were in complete linkage disequilibrium, forming a haplotype (not shown). Regression analysis demonstrated that alleles in rs1143679 and rs1143678/rs1143683 showed independent evidence for association with elevated IFN-I serum activity. Surprisingly, we found that a subset of these subjects (21/171) carried a haplotype containing the common SNP in rs1143679 (G) along with the rare SNP in rs1143683 (T). This GT haplotype was also associated with significantly elevated IFN-I activity (odds ratio = 3.57, P = 0.0068) (Figure 1B). To examine whether there was increased disease activity or flare in the ITGAM risk allele carriers at the time of blood draw that might account for this association, we examined their SLE Disease Activity Index (SLEDAI) scores, which were available for approximately 80% of the SLE patients at the time of blood draw. The average SLEDAI score in the overall cohort was 3.16, indicating mild to moderate average disease activity, typical of an outpatient SLE cohort. Next, we examined the SLEDAI scores in the same categories as those in Figure 1, A and B (Supplemental Figure 1). While there seems to be a trend toward higher disease activity, we did not find any significant increase in SLEDAI in the ITGAM risk allele carriers versus noncarriers. Furthermore, we examined it in a multivariate model, by performing a logistic regression analysis to assess the impact of SLEDAI score on the relationship between the ITGAM SNPs and IFN-I levels. SNPs were entered as predictor variables, with IFN as the outcome, and the model was assessed with and without inclusion of SLEDAI score. The β coefficients and odds ratios for the associations between SNP genotype and IFN did not change significantly between the 2 models (<10%), and the SLEDAI variable did not show significant association with the outcome in the overall model (odds ratio = 1.04), further supporting the idea that SLEDAI was not confounding the relationship observed between SNP genotype and IFN-I.
Figure 1. ITGAM SNPs correlate with elevated IFN-I activity in SLE patients.
(A) Functional IFN-I activity, as determined using reporter cell assay (3, 78), in serum samples from 171 SLE subjects genotyped for 3 ITGAM SNPs (rs1143678, rs1143679, and rs1143683) and its association with minor allele carriers compared with carriers of major alleles, stratified by high versus low IFN-I levels. Each dot represents a unique sample. (B) Functional association with serum IFN-I activity in patients carrying ITGAM haplotype rs1143679(G)-rs1143683(T) versus noncarriers.
An issue that commonly arises in the population cytokine data is that some of the data appear to fit a bimodal pattern, and the serum IFN levels in patients with the ITGAM GT genotype appear to show a similar behavior. To address this, we have previously reported an independent methodology for data analyses that uses binning of the data into high and low bins followed by categorical statistical analyses (3), in addition to the rank-order nonparametric t test of the quantitative data. Additionally, in such analyses, we have previously successfully used a value of 2.0 as a cutoff for high versus low, which corresponds to 2 SD above the mean of healthy donors. Applying this cutoff to the data in Figure 1, A and B, and using categorical analyses, we arrive at results similar to those of the nonparametric t test (Figure 1A, P = 0.037 by χ2, odds ratio = 2.01; Figure 1B, P = 0.0083 by Fisher’s exact test, odds ratio = 3.49). These analyses, for the first time to our knowledge, suggest a direct link between reduced CD11b function as a result of these SNPs and the increased IFN-I activity in SLE. Indeed, recent studies with macrophages from CD11b–/– mice show significantly higher IFN-β production upon stimulation with the TLR4 agonist LPS in comparison with WT macrophages (14), further supporting the idea that functional deficit in CD11b is directly linked to increased IFN-I activity in SLE.
CD11b agonist LA1 suppresses proinflammatory cytokine secretion by TLR-activated leukocytes.
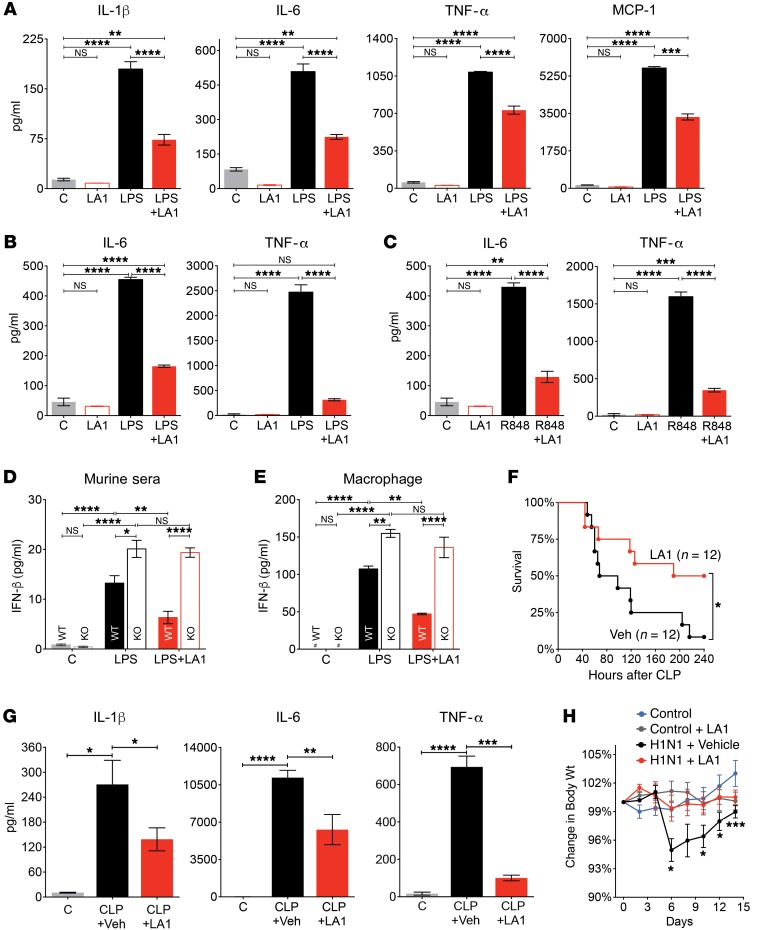
CD11b/CD18 ligation and clustering mediate proinflammatory signaling in leukocytes, including synergistic potentiation of signaling by other receptors (23–31). Conversely, they have also been shown to negatively regulate signaling by TLRs, TNF receptors, and cytokine receptors (14, 17, 23, 24, 32–34), which could suppress downstream IFN-I pathways. To test whether pharmacologic activation of CD11b could suppress TLR-dependent responses, and thus be a therapeutic strategy in SLE, we used our recently discovered small-molecule CD11b agonist LA1 as a tool (35, 36). LA1 binds to the CD11bA/I domain (CD11bA) in an allosteric pocket (Supplemental Figure 1), enhancing ligand binding. Molecular dynamics and isothermal calorimetry studies with CD11bA and fluid shear stress studies with cell surface–expressed full-length integrin CD11b/CD18 (Supplemental Methods; Supplemental Tables, 2 and 3; and Supplemental Figures 2–4) further established that LA1 binding promoted partial CD11bA activation, inducing an intermediate conformation in CD11bA (37) and no large conformational changes in the integrin heterodimer (38), defining this as its preferred binding mode. Given that the SLE-associated CD11b mutations are located outside of the ligand-binding αA domain (Supplemental Figure 2, A and B), it is unclear whether they would affect LA1 binding to the αA domain, and thus affect LA1’s activity. To assess, we generated cells stably expressing either full-length WT integrin CD11b (and its partner CD18) or the CD11b R77H mutant and used them in adhesion assay. The results show that both cell types displayed a similar level of LA1-mediated adhesion to immobilized Fg (Supplemental Figure 5), with similar EC50 values, suggesting that LA1 is able to activate the WT and the mutant integrin equally well and its binding is unaffected by the ITGAM SNP. The results also corroborate a recently published study, which also showed that the functional defects in CD11b R77H are rescued by integrin activation (13). Next, we treated primary murine macrophages and neutrophils with LPS in the absence or presence of LA1 and measured secretion of IL-1β, IL-6, TNF-α, and monocyte chemoattractant protein-1 (MCP-1) in cellular supernatants. LA1 significantly suppressed levels of all 4 in macrophages (Figure 2A) and neutrophils (Supplemental Figure 6A) in a dose-dependent manner (Supplemental Figure 6B). Similarly, TNF-α–stimulated IL-6 production was also significantly reduced (Supplemental Figure 6C). LA1 treatment also suppressed levels of IL-6 and TNF-α in cell culture supernatants of human macrophages stimulated with the TLR agonist LPS (Figure 2B) or R848 (Figure 2C), further validating the role of LA1-mediated integrin activation as suppressor of proinflammatory signaling by heterologous receptors in leukocytes. As TLR activation also induces IFN-I and downstream IFN-I signaling pathways (2), we tested the effects of TLR stimulation on IFN-I generation in vivo. LPS administration significantly upregulated IFN-β levels, with the level of increase significantly higher in the CD11b–/– animals as compared with the WT animals (Figure 2D), suggesting that CD11b activation–dependent modulation of TLR-signaling extends to IFN-I pathways. Importantly, we found that CD11b activation with LA1 significantly suppressed this upregulation of IFN-β in WT mice, but not in the mice lacking CD11b (CD11b–/–; Figure 2D), suggesting that the in vivo effects of LA1 here are mediated via CD11b. In vitro, LPS treatment also resulted in significantly increased production of IFN-β by CD11b–/– macrophages, as compared with the WT cells (Figure 2E), in accordance with the published studies (14). Furthermore, while LA1 significantly reduced the LPS-mediated upregulation in IFN-β levels in the cultured macrophages from WT animals, it failed to suppress IFN-β upregulation in the CD11b–/– cells (Figure 2E). These data, in combination with our previously published results showing that LA1 (and other analogs) significantly reduces peritoneal infiltration of CD11b+ leukocytes in response to thioglycollate-induced peritonitis in WT but not in CD11b–/– animals (35), strongly suggest that LA1 is primarily mediating its effects here via CD11b.
Figure 2. LA1 induces intermediate-affinity conformation in CD11bA and reduces TLR-stimulated proinflammatory cytokines.
(A) IL-1β, IL-6, TNF-α, and MCP-1 levels in primary mouse macrophage supernatants treated with vehicle DMSO (C), LA1 (20 μM), LPS (50 ng/ml), or LPS+LA1 for 8 hours (MCP-1) or 12 hours (IL-1β, IL-6, and TNF-α). Bars show mean ± SEM (n = 3) from 1 of at least 3 independent experiments. (B) IL-6 and TNF-α levels in supernatants of human macrophages treated with vehicle DMSO (C), LPS (50 ng/ml), or LPS+LA1 for 12 hours. Bars show mean ± SEM (n = 3) from 1 of at least 2 independent experiments. (C) IL-6 and TNF-α levels in supernatants of human macrophages treated with vehicle DMSO (C), R848, or R848+LA1 for 12 hours. Bars show mean ± SEM (n = 3) from 1 of 2 independent experiments. (D) IFN-β levels in the sera of WT or CD11b–/– mice treated with DMSO (C), LPS (100 ng/ml), or LPS+LA1 for 4 hours. Bars show mean ± SEM (n = 3). (E) IFN-β levels in supernatants of primary WT or CD11b–/– macrophages treated with DMSO (C), LA1 (20 μM), LPS (100 ng/ml), or LPS+LA1 for 12 hours. Bars show mean ± SEM (n = 3) from a representative experiment. #Not detectable. (F) Survival of C57BL/6 mice (n = 12) subjected to CLP and treated with vehicle (1% DMSO in saline) or LA1 (2 mg/kg/d). (G) IL-1β, IL-6, and TNF-α in plasma obtained 24 hours after C57BL/6 mice were subjected to either sham surgery (C) or CLP and treated with either vehicle (CLP+Veh) or LA1 (CLP+LA1). Bars show mean ± SEM. (H) Body weight loss (percentage of the initial weight) over 14 days in C57BL/6 mice (n = 20 per group) infected with WT H1N1 virus and treated daily with either vehicle (H1N1 + Vehicle) or LA1 (H1N1 + LA1). Weight loss in noninfected animals (control) and noninfected animals treated with LA1 (Control + LA1) is also shown. Each data point represents mean ± SEM (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001; A–E, 1-way ANOVA, Tukey’s test; F–H, Student’s t test).
LA1 treatment suppresses inflammatory cytokines in vivo and protects mice from severe sepsis.
To study the effects of CD11b activation with LA1 on TLR-stimulated cytokine generation in vivo, we subjected WT C57BL/6 mice to cecal ligation and puncture (CLP) (39), a model of polymicrobial sepsis, and treated them with LA1 or vehicle control. Among the control, vehicle-treated mice, 75% died within 120 hours (Figure 2F), and more than 90% (11/12) of them died within 240 hours. Surprisingly, more than 60% of LA1-treated mice were alive for 120 hours, and 50% of them survived more than 240 hours. Vehicle-treated mice showed high serum levels of the 3 hallmark cytokines (IL-1β, IL-6, and TNF-α) that play a significant role in cytokine storm–mediated septic shock (40), whereas LA1-treated animals showed a significant reduction in these cytokines (Figure 2G). Significantly, serum bacterial count 24 hours after CLP showed few remaining bacteria in LA1-treated mice as compared with high levels in the vehicle-treated group (Supplemental Figure 7), which suggests that LA1 treatment also suppresses bacterial load and correlates with previous data that LA1 enhances phagocytosis (35). Viral infections also lead to exacerbated leukocyte activation and cytokine storm, which prospectively predicts morbidity and mortality in patients (41). To determine whether LA1-mediated integrin activation in leukocytes would similarly suppress excessive immune response upon severe viral infection, we infected C57BL/6 mice via intranasal administration of influenza A virus (42) and monitored the transient loss in body weight, which is a primary manifestation of infection in these animals (41, 43). While the vehicle-treated animals exhibited significant weight loss within 3 days after viral infection (Figure 2H), reaching a maximum weight loss around day 6, before regaining it, LA1-treated animals did not show any loss of body weight after infection. These data suggest that LA1 blunts cytokine storm in vivo, improving survival.
LA1-mediated CD11b activation suppresses proinflammatory TLR and IFN-I pathways via MyD88/NF-κB and AKT/FOXO3/IRF3/7 axes.
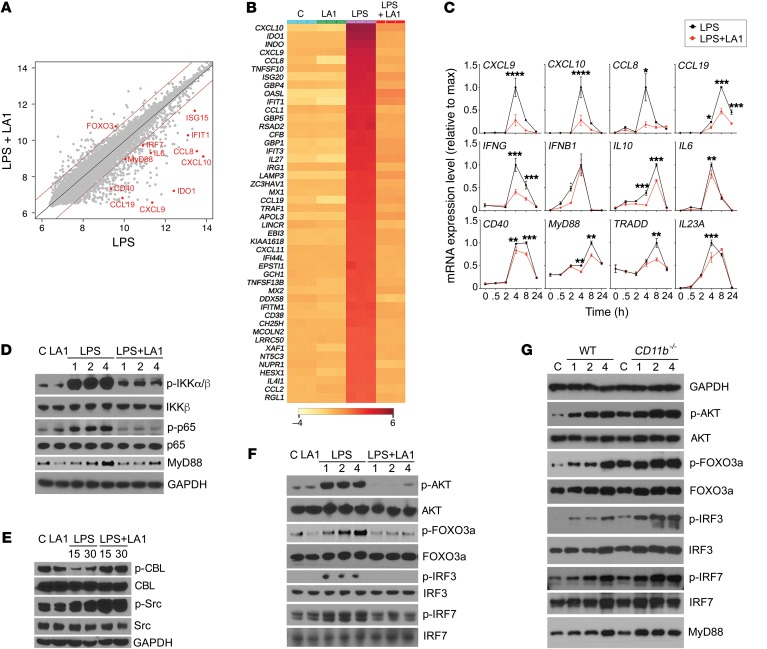
To identify TLR signaling components that are regulated by LA1-mediated CD11b activation, we performed genome-wide mRNA expression profiling of human macrophages stimulated with LPS in the absence or presence of LA1 (Figure 3A). Our analysis showed that 416 genes were significantly affected by LA1 treatment (Supplemental Table 4) and that LA1 normalized a majority of the LPS-regulated genes (Figure 3B). In addition to the known genes downstream of TLR engagement (MYD88, IL6, IL1A, CXCL1), pathway analysis showed that the genes in the TLR-associated canonical antiviral response (P < 1.3 × 10–12, overlap 13/52) and IFN-I pathways (P < 2.5 × 10–9, overlap 8/24) were highly enriched (Supplemental Figure 8). This includes CXCL9, CXCL10, IDO1, CCL5, IFIT1, IRF7, and ISG15. We used a multiplex mRNA counting method to measure the expression of a number of these genes in macrophages treated with LPS or LPS plus LA1 over time (Figure 3C), which validated the expression level changes induced by LA1 in the genes in the TLR and IFN-I pathways. Curiously, the microarray data also showed a concomitant increase in FOXO3 transcription, which was recently shown to suppress IFN-I pathway by limiting IFN regulatory factor 7 (IRF7) autoregulation (44). Additional known direct targets of FOXO3, including IFIH1, DDX58, RSAD2, MX2, and CMPK2, were also reduced by LA1. This suggests that LA1-mediated CD11b activation suppresses TLR signaling via MyD88-dependent NF-κB pathway and downstream IFN-I signaling via the FOXO3/IRF pathway.
Figure 3. LA1-mediated CD11b activation suppresses IFN signaling via AKT/FOXO3/IRF3/7 axis.
(A) Scatter plot comparing global mRNA expression profiles from human macrophages treated with LPS (50 ng/ml) (x axis) or LPS (50 ng/ml) plus LA1 (20 μM) (y axis) for 4 hours. The dark red lines indicate a 2-fold cutoff for the ratio of expression levels. Data represent the average of 3 experiments and are on the log2 scale. The numbers of genes that are significantly changed by LA1 are labeled on the graph (red dots). (B) Hierarchical clustering of the gene expression data for the top 50 genes downregulated by LA1. (C) mRNA levels of CXCL9, CXCL10, CCL8, CCL19, IFNG, IFNB1, IL10, IL6, CD40, MYD88, TRADD, and IL23a in LPS-stimulated macrophages in the absence or presence of LA1 at various time points. Data are mean ± SEM from a triplicate experiment (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, Student’s t test). (D–F) Immunoblot analysis of various phosphorylated (p-) and total proteins in lysates of RAW macrophages stimulated with LPS (50 ng/ml) for 0–4 hours (D and F) or 0–30 minutes (E) in the absence or presence of LA1. GAPDH (in D for D and F, in E for E) was used as loading control. (G) Immunoblot analysis of various phosphorylated (p-) and total proteins in lysates of primary mouse macrophages from WT and CD11b–/– animals. Macrophages in culture were stimulated with LPS (100 ng/ml) for 0–4 hours, lysed, and analyzed. GAPDH was used as loading control.
Western blot (WB) analysis confirmed that LA1 suppressed the LPS-stimulated NF-κB pathway (reduced levels of phosphorylated IKKα/β and p65) (Figure 3D), which is also in line with previous studies showing that CD11b negatively regulates TLR-dependent NF-κB pathway, by increasing degradation of MyD88, and that deficiency of CD11b exacerbates this TLR-dependent proinflammatory signaling by failing to reduce the MyD88 levels (14). Furthermore, active CD11b downregulates MyD88 protein levels by inducing Src and CBL phosphorylation (14), and the WB analyses show increased pSrc and pCBL by LA1 (Figure 3E), suggesting that this CD11b agonist may use a similar mechanism. Densitometric quantification of WBs for the level of MyD88, the most upstream TLR signaling event, confirmed that LPS treatment increased MyD88 levels that were significantly reduced by LA1-mediated CD11b activation (Supplemental Figure 9), suggesting it to be a key node. Transcriptional induction of IFN-I is mediated by transcription factors IRF3 and IRF7 that require phosphorylation for nuclear entry and activity (45). Under basal conditions, nuclear FOXO3 acts as a negative regulator of IFN-I pathway via suppression of IRF7 (44). Upon TLR stimulation, FOXO3 is excluded from the nucleus and degraded, via its AKT-mediated phosphorylation, thereby derepressing IRF7 and IRF7-dependent genes, including the IFN-I pathway. As TLR stimulation in macrophages induces MyD88-dependent AKT phosphorylation (46), we hypothesized that LA1-mediated CD11b activation mechanistically suppresses IFN-I pathway by reducing AKT phosphorylation, thereby reducing FOXO3 phosphorylation and, thus, enhancing nuclear FOXO3. Indeed, WB analysis showed that CD11b activation with LA1 suppressed the LPS-stimulated upregulation of pAKT and pFOXO3 in macrophages (Figure 3F). Reduced pFOXO3 levels also correlated with reduced phosphorylation of IRF3 and IRF7, suggesting that CD11b activation blunts IFN-I signaling via the AKT/FOXO3/IRF3/7 axis. To further confirm that CD11b regulates these pathways, we measured the levels of these proteins in LPS-stimulated primary macrophages from WT and CD11b–/– animals using WB (Figure 3G). Whereas the WT cells showed an expected time-dependent increase in the levels of MyD88, pAKT, pFOXO3, pIRF3, and pIRF7, the CD11b–/– cells showed a larger increase in the levels of these proteins at baseline and upon LPS stimulation, suggesting that the AKT/FOXO3/IRF3/7 signaling pathway is exacerbated in the CD11b-deficient macrophages in a manner similar to the previously reported exacerbation of the MyD88/IKK/NF-κB pathway in these cells (14).
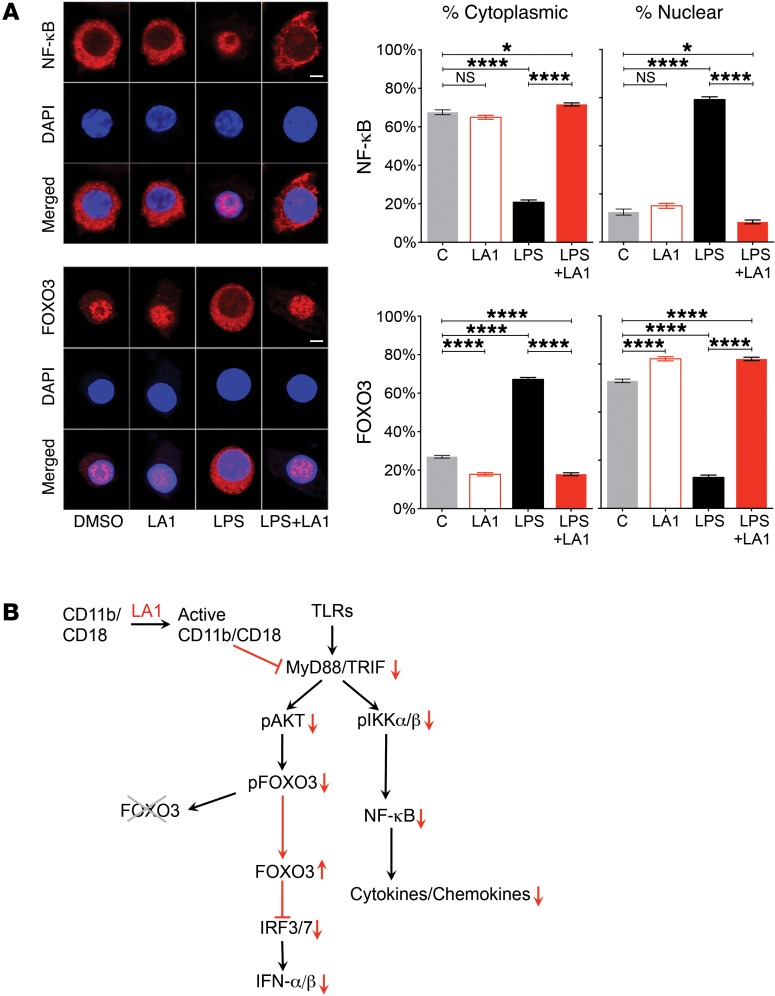
Cells stimulated with TLR3 agonist pI:C also showed that LA1 reduced upregulation of pAKT and pFOXO3 in macrophages (Supplemental Figure 10), and reduced pIRF3. Given that TLR3 is MyD88-independent and is known to regulate the IFN-I pathway via IRF3 (47, 48), this suggests that LA1 suppress TLR-dependent signaling via IRF3 in the MyD88-independent arm. Cells stimulated with other TLR agonists (R848 or CpG) also showed similar results (not shown), suggesting that this might be a general mechanism of downmodulating TLR signaling by CD11b. Since the downstream transcriptional activity of FOXO3 is mediated via its nuclear retention (49) and because LA1-mediated CD11b activation affects FOXO3 phosphorylation, we predicted that LA1 treatment would affect FOXO3’s subcellular localization. Confocal microscopy–based imaging showed that FOXO3 was mostly inside the nucleus under basal conditions in RAW macrophages (Figure 4A). As a control, we stained for NF-κB, which expectedly showed a mostly cytoplasmic localization. Upon stimulation with LPS, FOXO3 expectedly translocated out of the nucleus and was primarily in the cytoplasm (49), whereas NF-κB showed the opposite pattern. Strikingly, cotreatment with LA1 suppressed the translocation of FOXO3 from the nucleus into the cytoplasm, suggesting that LA1-mediated CD11b activation reduces IFN-I signaling pathway via protection of FOXO3 levels in the nucleus. In contrast, NF-κB showed the exact opposite results, suggesting that partial activation of CD11b is sufficient to limit the TLR-dependent IFN-I pathway in cells via an AKT/FOXO3/IRF3/7 circuit (Figure 4B).
Figure 4. CD11b activation preserves retention of FOXO3 in the nucleus.
(A) Representative immunofluorescence images of subcellular localization of NF-κB (red, top panels) and FOXO3 (red, bottom panels) in RAW macrophages treated with vehicle DMSO, LA1 (20 μM), LPS (50 ng/ml), or LPS+LA1 for 4 hours. Nuclei were stained with DAPI (blue). Scale bars: 10 μm. Bar graphs represent quantification of the cytoplasmic and nuclear fraction of NF-κB (top) and FOXO3 (bottom) in cells (n = 30). Data are mean ± SEM (*P < 0.05, ****P < 0.0001, 1-way ANOVA, Tukey’s test). (B) A schematic showing a working model of how CD11b activation with LA1 suppresses TLR and IFN-I signaling.
LA1 suppresses inflammatory injury and end-organ pathology in lupus-prone mice.
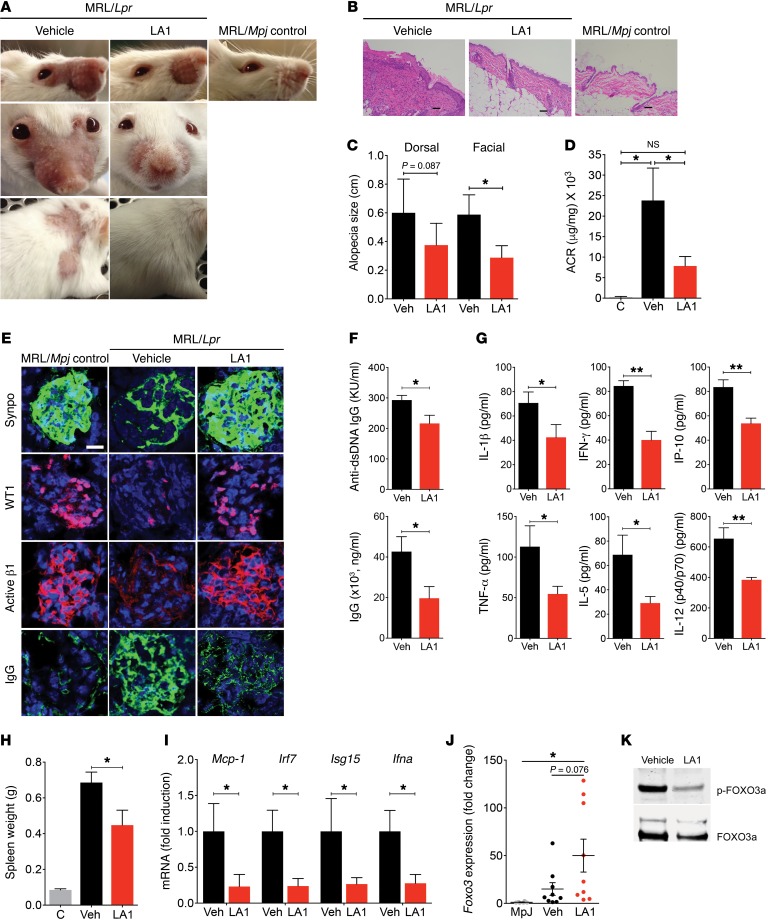
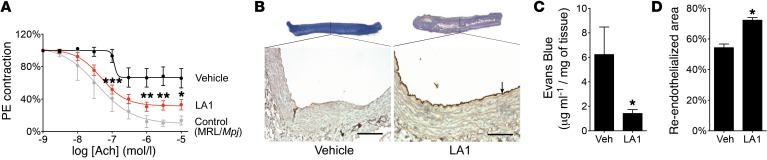
To test the efficacy of LA1 in a lupus model, we used the MRL/Lpr mice that develop IFN-I–dependent multiorgan lupus, which is similar to human lupus, including skin, vascular, and renal inflammation (50), and involves a leukocyte-mediated inflammatory process (51). There was no evidence of overt toxicity in these animals, with no differences in total body weight throughout treatment (not shown). Vehicle-treated MRL/Lpr mice (but not the haplotype-matched MRL/Mpj) lost whiskers and developed skin lesions and alopecia, identifiable within the intrascapular area and on the ears and face (muzzle) by 14 weeks of age (refs. 52, 53, and Figure 5, A–C). Increased leukocyte infiltration was identified in the dermis of these patches, along with enhanced epidermal thickness (Figure 5B). Treatment with LA1 significantly reduced alopecia, leukocyte infiltration, and epidermal thickness (Figure 5, A–C). In addition, vehicle-treated MRL/Lpr mice developed significant features consistent with lupus glomerulonephritis, including increased urine albumin/creatinine ratio and IgG renal immune complex deposition, which were significantly reduced with treatment with LA1 (Figure 5, D and E). LA1-treated animals also showed decreased glomerular damage (Figure 5E and Supplemental Figure 11). Consistent with this amelioration in lupus phenotype, LA1 treatment resulted in significant amelioration in levels of serum anti-dsDNA antibodies and total IgG, various inflammatory cytokines and chemokines, and spleen weight, in comparison with vehicle-treated mice (Figure 5, F–H), which correlates with reduced inflammation in LA1-treated mice. While the differences in numbers of splenic double-negative T cells (not shown), activated T and B cells, or plasma cells, DCs, macrophages, and neutrophils did not achieve statistical significance between LA1-treated and vehicle-treated mice (Supplemental Figures 12 and 13), the trends showed decreases in splenic macrophages, activated B cells, and plasma cells, which may explain the differences in autoantibody synthesis, as previously described (54, 55). To investigate whether the efficacy of LA1 in vivo was associated with modulation of the IFN-I pathway, we isolated mRNA from MRL/Lpr splenocytes and quantified relative expression of several candidate genes. Indeed, cells from LA1-treated animals showed significantly lower expression of Mcp-1, Irf7, Isg15, and Ifna (Figure 5I) and, while not statistically significant, a trend for increased Foxo3 mRNA levels (Figure 5J). Consistent with the hypothesis that increased phosphorylation of FOXO3 drives IFN-I signaling, bone marrow monocytes from LA1-treated animals showed significantly reduced levels of pFOXO3 (Figure 5K). These data suggest that LA1-mediated CD11b activation mitigates lupus phenotype and end-organ damage in association with significant suppression of the AKT/FOXO3/IRF7 pathway and modulation of IFN-I activity. Since aberrant leukocyte activation and enhanced IFN-I activity contribute to the development of endothelial dysfunction in SLE (56), we asked whether the endothelial dysfunction inherent to MRL/Lpr mice might also be reduced following LA1 treatment. Aortic rings were isolated from 19-week-old MRL/Lpr mice after 10-week treatment with LA1 or vehicle, and acetylcholine-dependent relaxation triggered after phenylephrine precontraction was determined as described in Methods. Indeed, treatment with LA1 significantly improved endothelium-dependent vasorelaxation, indicating a vasoprotective effect on the lupus vasculature (Figure 6A). Examination of vascular reendothelialization in balloon-injured Fisher male rats also showed significant reendothelialization at the site of balloon injury with LA1 treatment (Figure 6, B–D, and Supplemental Methods), suggesting that LA1 may accelerate the process of endothelial regeneration (57). Increased neutrophil adhesion via LA1 to HUVECs did not show any damage to the HUVEC monolayer (Supplemental Figure 14, A–C, and Supplemental Methods). Together, these results show that LA1 treatment does not harm the endothelium, similarly to previous studies using knock-in animals that express constitutively active mutants of integrins CD11a/CD18 (58, 59) or α4β7 (60) that also did not report any signs of vascular injury in the various experimental models, suggesting that integrin activation, per se, does not harm the vasculature or have any harmful consequences for animals under normal conditions.
Figure 5. LA1 protects mice from SLE.
(A) Erythema and alopecia in facial and dorsal areas in treated MRL/lpr mice. MRL/Mpj served as control. (B) Representative H&E images from skin of mice in A. Scale bar: 100µm. (C) Alopecia quantification from mice in A (n = 6/group). (D) Urinary albumin/creatinine ratio (ACR) from mice in A (n = 7 for C, n = 14 LA1-treated and n = 12 vehicle-treated). (E) Images of kidney glomeruli from treated, 19-week-old mice, stained with antibodies against synaptopodin (Synpo, green), WT1 (red), and integrin β1 activation epitope (Active β1, red), or with anti-IgG (IgG, green). Nuclei were stained with DAPI (blue). Scale bar: 20μm. (F) Quantification of serum anti-ds-DNA antibodies and total IgG in treated mice, at 19-weeks of age (n = 7 LA1-treated and n = 8 vehicle-treated). (G). Quantification of pro-inflammatory cytokines in the sera from 12-week-old MRL/lpr mice treated with either vehicle or LA1 for 4 weeks (n = 6/group). (H) Spleen weight from treated mice, at 19-weeks of age. MRL/Mpj served as controls (n = 7 LA1-treated and n = 8 vehicle-treated). (I) Relative levels of IFN-I-inducible genes in splenocytes from 19-week-old mice, expressed as fold-change in LA1-treated mice over vehicle-treated mice (normalized to 1) (n = 6 per group). (J) Foxo3 mRNA levels in splenic monocytes 19-week-old mice, expressed as fold-change in LA1 treated mice over MRL/Mpj (normalized to 1) (n = 9 per group in MRL/lpr and n = 6 in MRL/Mpj). (K) Immunoblot analysis of phosphorylated (p-) FOXO3a protein level in pooled lysates of bone marrow monocytes from 19-week-old treated mice (n = 6 per group). Total FOXO3a was used as loading control. (Data shown in all the graphs are mean ± SEM. *P <0.05, **P <0.01, D, 1-way ANOVA, Tukey’s test; C and F–J, Student’s t test or Mann-Whitney test).
Figure 6. LA1 improves vasorelaxation in mice.
(A) Quantification of acetylcholine-dependent (Ach-dependent) relaxation after phenylephrine (PE) precontraction in aortic rings from 19-week-old MRL/Lpr mice after 10-week treatment with LA1 or vehicle. Control data are from MRL/Mpj. Data are mean ± SEM (n = 7 LA1-treated, n = 8 vehicle-treated). (*P < 0.05, **P < 0.01, ***P < 0.001, comparing LA1 vs. controls.) Curves were first analyzed using an asymmetric (5 parameters) logistic equation; significance was determined by 2-way ANOVA. (B) Representative photomicrographs of balloon-injured arteries from rats treated with vehicle or LA1. Images of arteries stained with Evans blue vital staining in situ before sacrifice are shown at the top. Photomicrograph of cross section of arteries stained with anti-vWF antibody to label the endothelium (arrow) is presented below. Scale bars: 50 μm. (C) Quantification of the amount of Evans blue extracted from the balloon-injured arteries from rats treated with vehicle or LA1 (as in B). Data are mean ± SEM (n = 7 vehicle-treated, n = 8 LA1-treated). (D) Quantification of the extent of reendothelialization 7 days after balloon injury from the balloon-injured arteries from rats treated with vehicle or LA1 after staining with anti-vWF antibody (as in B). Data shown are mean ± SEM (n = 7 vehicle-treated, n = 8 LA1-treated) (C and D, *P < 0.05, Student’s t test).
Cells homozygous for CD11b SNPs show basal level increase in IFNB and IRF7 expression and increased FOXO3 nuclear exclusion that is suppressed by LA1 treatment.
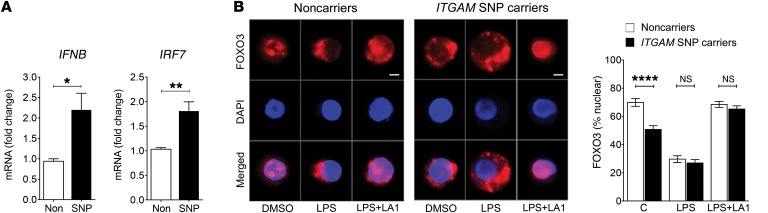
To examine the molecular link between ITGAM SNPs and IFNB in human cells, we used quantitative reverse transcriptase PCR to measure the expression level of IFNB and IRF7 in peripheral blood mononuclear cells (PBMCs) from donors carrying either major or minor ITGAM alleles at the rs1143679 or rs1143678/rs1143683 loci and found that the cells carrying a minor allele consistently showed increased expression of both transcripts (Figure 7A). Given that their expression is modulated by nuclear levels of FOXO3, we also quantified the levels of FOXO3 in the nucleus versus cytoplasm and found that, under basal conditions, the cells carrying a minor allele showed significantly reduced levels of FOXO3 in the nucleus, as compared with the cells carrying the common allele (Figure 7B). Recent reports show that TLR pathway is basally activated in SLE (61), and the data presented here suggest that ITGAM SNPs likely reduce a CD11b-dependent “tonic” suppression of TLR signaling in SLE, due to a failure of the mutant CD11b in maintaining levels of nuclear FOXO3, resulting in reduced suppression of the IRF7 (44) and increased IFN-I. Future research will further clarify this connection. We also found that LPS treatment of cells resulted in rapid movement of FOXO3 outside of the nucleus in both cell types (Figure 7B). Both cell types showed a similar level of LPS-dependent nuclear exclusion of FOXO3. Since the assays were performed at a single dose level of LPS, future LPS-dose-dependent studies will reveal whether the 2 types of cells have a finer regulation of FOXO3 levels in the nucleus in response to LPS stimulation. Furthermore, to determine whether CD11b activation via LA1 could suppress the nuclear exclusion of FOXO3, we stimulated the donor cells from either the major or the minor allele carriers with LPS and LA1 and quantified the nuclear levels of FOXO3 in those cells (Figure 7B). The data show that LA1 suppressed LPS-dependent relocation of FOXO3 under both conditions. Together, these results suggest that, while the ITGAM SNP–carrying cells show a slightly elevated TLR pathway at baseline, the IFN-I pathway is significantly upregulated upon LPS-mediated TLR activation in both types of cells, and that LA1-mediated activation of CD11b is able to reduce this in both types of cells, suggesting that LA1 can be equally effective on both WT and mutant CD11b protein. This implies that partial CD11b activation via pharmacologic agents could abrogate abnormal IFN-I signaling and be a potentially novel therapeutic strategy for SLE.
Figure 7. LA1 reduces IFN-I levels and the nuclear exclusion of FOXO3 in LPS-stimulated patient cells.
(A) Real-time qRT-PCR–based measurement of IFNB and IRF7 in PBMCs from normal donors carrying nonvariant or variant alleles of ITGAM. The data shown are expressed as fold change of ITGAM variant SNP carriers over noncarriers (normalized to 1). Data shown are mean ± SEM (major allele carriers, n = 5, and minor allele carriers, n = 6). (B) Representative immunofluorescence images of subcellular localization of FOXO3 (red, bottom panels) in human macrophages from donors carrying nonvariant (Noncarriers) or variant alleles (SNP carriers) of ITGAM treated with vehicle DMSO, LPS (50 ng/ml), or LPS (50 ng/ml) plus LA1 (20 μM) for 4 hours. Nuclei were stained with DAPI (blue). Scale bars: 10 μm. Bar graphs represent quantitation of the nuclear fraction of FOXO3 in cells (n = 30) from 3 unique donors in each group. Data are mean ± SEM (*P < 0.05, **P < 0.01, ****P < 0.0001, Student’s t test).
Discussion
The association between specific ITGAM SNPs and SLE risk is one of the strongest and most specific lupus genetic risk factors across various ethnic groups. However, how these polymorphisms in ITGAM that result in decreased function of CD11b functionally predispose to lupus and its associated organ damage remained to be fully characterized. Integrin CD11b combines with the β2 chain (ITGB2, CD18) to form Mac-1, an integrin heterodimer with crucial effects on myeloid cell adherence to stimulated endothelium. In addition to its role in inflammation-induced leukocyte tissue recruitment, prior studies have also highlighted a protective role of CD11b (reviewed in ref. 22), including maintenance of autoreactive B cell tolerance (15), suppression of DC maturation and function, and restriction of Th17 differentiation (62). CD11b (along with ITAM receptors) has also been shown to negatively regulate TLR4-triggered inflammatory responses (14, 23, 63). However, it has remained unclear whether aberrant function of CD11b in individuals with ITGAM polymorphisms can promote IFN-I responses and whether modulation of CD11b in SLE, by restoring integrin activation, can modify IFN-I signaling and modulate autoimmunity. Here, we have uncovered a direct link between an integrin mutation and a pathogenic circulating factor that may predispose to SLE and promote immune dysregulation characteristic of this disease. We found that LA1 enhanced CD11b-dependent ligand binding and cell adhesion by inducing an intermediate affinity conformation in the ligand-binding CD11bA domain. Together with our previously published studies that showed that LA1 induces increased binding of mAb24 (38, 64, 65) (which detects an activation-dependent neoepitope in the βA domain), but not KIM127 (66) (which detects an integrin extension–dependent neoepitope), this further suggests that LA1 binding does not induce an extended integrin conformation (38). Such LA1-mediated integrin activation significantly blunted generation of TLR-stimulated proinflammatory cytokines and chemokines in leukocytes, suppressing TLR-dependent intracellular signaling pathways in vitro and in vivo, and further explaining the observed strong antiinflammatory effects of LA1 in vivo. Importantly, in addition to suppressing the MyD88-dependent NF-κB pathways in leukocytes, LA1 also dampened the IFN-I signaling, indicating an important mechanism by which it may modulate immune dysregulation in SLE. Detailed molecular analysis showed that LA1 reduced phosphorylation of AKT and its substrate FOXO3a, thereby increasing the levels of FOXO3a in the nucleus that suppresses IRF3/7–dependent gene expression (44). IRF3 and IRF7 are members of the IFN regulatory transcription factor family, a master regulator of the IFN-I–inducible pathway and a key player in innate immunity (67). As such, our findings directly link integrin biology with crucial pathways in IFN-I responses that may predispose to SLE and promote immune dysregulation characteristic of this disease.
Given that the TLR and IFN-I pathways play a significant role in the pathogenesis of SLE, the significance of these findings was tested in murine lupus. LA1 reduced IFN-I signaling and protected animals from end-organ injury and vascular dysfunction, by similarly modulating the CD11b/FOXO3 axis in leukocytes. Importantly, the relevance of these findings is also translated into humans in that the subjects carrying missense ITGAM SNPs were found to have dysregulation of FOXO3 in leukocytes and higher IFN-I activity. This, for the first time to our knowledge, suggests a direct link between an integrin mutation and a pathogenic circulating factor. As the binding site for LA1 on CD11b does not overlap with the SLE-associated mutations in CD11b, cell-based studies also showed that LA1 is able to induce cell adhesion via the WT and the mutant R77H integrin equally well. Indeed, LA1-mediated CD11b activation in ex vivo donor leukocytes suppressed TLR-stimulated cytokines and IFN-I levels, by correcting the signaling change in the AKT/FOXO3/IRF3/7 axis. Published findings and these observations suggest that CD11b plays an important role in SLE and that CD11b activation should be explored as a potential novel therapeutic target in this autoimmune disease, particularly as a personalized approach in patients identified as carriers of specific genetic polymorphisms.
Methods
Full experimental details can be found in the Supplemental Methods.
Human samples and measurement of serum IFN-I activity.
We analyzed 171 SLE cases from multiple study centers consisting of patients of self-reported European ancestry that had serum available for IFN-I analysis (Supplemental Table 1). Informed consent was obtained from all patients in both cohorts included in this study, and the study was approved by the institutional review boards at the respective institutions. Patients were genotyped at coding change SNPs in ITGAM (rs1143678, rs1143679, and rs1143683) using custom-designed Applied Biosystems TaqMan primers and probes on an Applied Biosystems 7900HT PCR machine with greater than 98% genotyping success. Additional genotyped human PBMCs from donors carrying ITGAM nonrisk or risk variant for cell-based immunofluorescence and other assays were provided by the Genotype and Phenotype Registry at the Tissue Donation Program at the Feinstein Institute for Medical Research. Measurement of IFN-I in human sera was performed using our previously published WISH cell assay (68, 69). Logistic regression analysis was carried out using PLINK version 1.07 software (70). Enrichment P values were calculated using a Fisher’s exact test. Graphs were also generated showing the IFN values in patients segregated by genotype, with Mann-Whitney U test used to determine statistical significance.
SLE mouse model.
All mice were obtained from the Jackson Laboratory. Female MRL/Lpr were treated with LA1 (2 mg/kg/d) or vehicle (1% Tween-20 in sterile saline) for 11 weeks by daily i.p. injection, beginning at 8 weeks of age until euthanasia at 19 weeks of age (56). Haplotype-matched female MRL/Mpj mice were used as phenotypic controls. Urinary albumin and creatinine concentrations were measured using a mouse albumin ELISA (Bethyl Laboratories) and a creatinine assay (R&D Systems), respectively. Serum creatinine was determined by HPLC as previously described (71). Kidneys and affected skin samples were harvested after perfusion with PBS. Regions of alopecia were measured on the face and dorsum, with greatest diameter determined to the nearest millimeter with calipers. Paraffin-embedded sections (4 μm) were stained with H&E, periodic acid–Schiff, or Masson’s trichrome, and frozen sections were used for immunofluorescence studies.
Flow cytometry of mouse splenocytes for immunophenotyping.
A single-cell suspension of mouse splenocytes was prepared and resuspended in FACS buffer (2% FBS in PBS) at 1.0 × 106 cells/ml for analysis by flow cytometry. After blocking for 15 minutes, cells were resuspended in 100 μl FACS buffer and incubated with 2 μl each of respective antibodies or isotype control for 30 minutes, fixed with 2% paraformaldehyde, and quantified by flow cytometry using a BD FACSCanto research use only (RUO), and data were analyzed using FlowJo software (FlowJo LLC). Results are reported as number of cell subsets per million splenocytes.
Cecal ligation and puncture model.
Cecal ligation and puncture (CLP) was performed as described previously (72) using B6 WT mice (8–12 weeks old). Vehicle control (1% DMSO in PBS) or LA1 (2 mg/kg body weight, in PBS) was administered i.p. 2 hours before CLP and then daily until the end of the experiment. The serology and cytokine measurements were performed as previously described (72) using plasma from blood samples obtained 24 hours after CLP. Lactate dehydrogenase (LDH), creatinine kinase (CK), alanine aminotransferase (ALT), and urea levels were measured using BioAssay Systems kits according to the company’s protocol. Levels of IL-1, IL-6, and TNF-α were measured using the murine ELISA kits (R&D Systems) according to the company’s protocol.
Primary macrophage assays.
Human PBMCs were isolated from fresh blood collected from healthy volunteers under an IRB-approved protocol and using RosetteSep (human monocyte Enrichment Cocktail, catalog 15068, Stemcell Technologies) followed by Ficoll-Hypaque density gradient centrifugation according to published protocols (73). Cells adherent to the tissue culture flask were cultured for 5–7 days at 37°C in 5% CO2 to promote their full differentiation into monocyte-derived macrophages (74, 75).
Microarray analysis.
For microarray analyses, human macrophages (2 × 106) were cultured in 6-well tissue culture plates and were treated with LPS (50 ng/ml) in the absence or presence of LA1 (20 μM) for 4 hours as previously described (35). Cells were washed with ice-cold PBS and then lysed in TRIzol (Thermo Fisher Scientific) for total RNA isolation. RNA sample quality was analyzed using a 2100 Bioanalyzer (Agilent). Whole-genome transcriptional profiling was performed using Illumina HT-12 human gene expression arrays and analyzed using GeneSpring software version 13. Only significant genes (P ≤ 0.05 and fold change ≥ 2) were considered for pathway analysis. We deposited all microarray data in the Gene Expression Omnibus database in the series GSE76802. Expression levels of selected genes in human macrophages stimulated with LPS (50 ng/ml) for various amounts of time (0, 0.5, 2, 4, 8, and 24 hours) in the absence or presence of LA1 (20 μM) were measured using an inflammation panel set and the nCounter Digital Analyzer (Nanostring Technologies) as previously described (76).
Isolation of murine primary neutrophils and macrophages.
Thioglycollate-elicited neutrophils and macrophages were isolated from B6 WT mice or B6 CD11b–/– mice (Jackson Laboratory), as described previously (35). Neutrophils were collected 4 hours after thioglycollate injection followed by 1-hour adherence purification before collection of nonadherent cells. The macrophages were collected 4 days after thioglycollate injection and adherence-purified for 1 hour, followed by a wash with PBS to remove nonadherent cells. For the bone marrow–derived macrophages, 2 × 106 cells per well of harvested cells were plated in 6-well plates in RPMI containing 10% FBS and were cultured for 5–7 days. Macrophages were generated from bone marrow–derived monocytes as described in the literature (77).
Statistics.
To calculate statistical significance, a 2-tailed student’s t test was used, unless otherwise specified. The differences in the cytokine concentrations were statistically analyzed using the Mann-Whitney test, and plotted with the GraphPad Prism software package. For multiple comparisons, 1-way ANOVA with Tukey’s test was used. A value of P less than 0.05 was considered statistically significant. For endothelium-dependent vasorelaxation, curves were first analyzed using an asymmetric (5 parameters) logistic equation, and significance of each individual data point was determined by 2-way ANOVA. Unless otherwise specified, results are represented as mean ± SEM.
Study approval.
The studies on human samples were reviewed and approved by the Mayo Clinic IRB in Rochester, Minnesota, USA. All subjects provided appropriate prior informed consent to participate in the study. The respective studies on C57BL/6 WT and CD11b–/– animals were reviewed and approved by the IACUC at Rush University Medical Center or the IACUC at University of Miami School of Medicine, or in accordance with Portuguese guidelines and regulations after approval by the local ethics committee at Instituto Gulbenkian de Ciência, Oeiras, and the national regulating agency (Direcção Geral de Alimentação e Veterinária). All experiments using MRL/Lpr and MRL/Mpj mice were approved by the Animal Ethics Committee (ACUC) at the National Institute of Arthritis and Musculoskeletal and Skin Diseases/NIH, Bethesda, Maryland, USA.
Author contributions
MHF, SQK, and WZ designed and performed in vitro and in vivo experiments. EM and CCR performed animal experiments. KZ, VK, ARA, RD, DS, VH, SJK, PB, A.M. Santander, MTK, MMA, XL, SD, KHK, HWL, JSR, JR, and JFC contributed to in vitro experiments. JMD, A.M. Schnaith, YGP, MJ, EMB, JSR, BS, SKN, and TBN contributed to human sample analyses. TZ and RP contributed to molecular dynamics simulations and data analysis. BI analyzed the microarray data and helped build pathways. LFM and AZ contributed to CLP and virus infection experiments, respectively. MP, JIC, and CCR performed immunophenotyping of lupus mice and protein FOXO3 assessments in lupus. VH performed scoring of lupus murine skin. DJC performed scoring of lupus murine kidneys. PSY performed HPLC analyses for mouse creatinine. WLT performed cytokine work in lupus mice. VG, MJK, and RIVP codesigned and cosupervised the study, and cowrote the paper.
Supplementary Material
Acknowledgments
We thank Dony Maiguel, Nicholas J. Tardy, Hatem Elshabrawy, Yuntao Wei, and Dania Mateu for generous technical help and helpful discussions. We also thank Tanya Mayadas for valuable input and discussions. We thank S. Fagerholm (University of Helsinki, Helsinki, Finland) for providing the CD11b R77H plasmid. This work was supported in part by grants from the NIH (R01DK084195, R01DK106512, and R21CA176055 to VG; R01HL109582 to VG and RIVP; R01AR060366 to SN), a postdoctoral fellowship from the American Heart Association to SQK, and resources from the Rush University Medical Center. This work was also supported in part by the Intramural Research Program at the National Institute of Arthritis and Musculoskeletal and Skin Diseases/NIH (1ZIAAR041199 to MJK and by the Office of Science and Technology), the European Research Council (ERC-2014-CoG 647888-iPROTECTION to LFM), and the National Basic Research Program of China (2014CB541905).
Footnotes
Conflict of interest: V. Gupta is an inventor of pending patents related to this study (WO2013159082) and has the potential for financial benefit from their future commercialization. V. Gupta is also a cofounder and a member of the scientific advisory board of Adhaere Pharmaceuticals Inc., a company developing novel therapeutics for inflammation, cancer, and autoimmune diseases.
Reference information:J Clin Invest. 2017;127(4):1271–1283.https://doi.org/10.1172/JCI88442.
Contributor Information
Samia Q. Khan, Email: samia_khan@rush.edu.
Wenpu Zhao, Email: wenpu@med.umich.edu.
Ha Won Lee, Email: hawon_lee@rush.edu.
Kun Zhang, Email: zimu1993@163.com.
Vinay Kumar, Email: kvinay0000@gmail.com.
Carmelo Carmona-Rivera, Email: carmelo.carmona-rivera@nih.gov.
Xiaobo Li, Email: Xiaobo_Li@rush.edu.
Yogita Ghodke-Puranik, Email: ghodke.yogita@mayo.edu.
Erica Moore, Email: emoore1010@gmail.com.
Monica Purmalek, Email: monicapurmalek@gmail.com.
Tingting Zhang, Email: t.zhang7@umiami.edu.
Rachael Day, Email: rchldy@dordt.edu.
Darren Stoub, Email: darren.stoub@dordt.edu.
Victoria Hoffmann, Email: hoffmanv@ors.od.nih.gov.
Shehryar Jehangir Khaliqdina, Email: shehryar_khaliqdina@rush.edu.
Prachal Bhargava, Email: bhargavprachal@gmail.com.
Marta Torroella-Kouri, Email: mtorroella@med.miami.edu.
Biju Issac, Email: bissac@med.miami.edu.
Andrew Zloza, Email: az255@cinj.rutgers.edu.
Rajeev Prabhakar, Email: rpr@miami.edu.
Shashank Deep, Email: sdeep@iitd.ac.in.
Meenakshi Jolly, Email: meenakshi_jolly@rush.edu.
Kwi Hye Koh, Email: kwi_koh@rush.edu.
JianFeng Chen, Email: jfchen@sibcb.ac.cn.
Wanxia Li Tsai, Email: liwan@mail.nih.gov.
Bhupinder Singh, Email: bhupinder-singh@omrf.org.
Jochen Reiser, Email: jochen_reiser@rush.edu.
Mariana J. Kaplan, Email: mariana.kaplan@nih.gov.
Vineet Gupta, Email: vineet_gupta@rush.edu.
References
- 1.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- 3.Niewold TB, Hua J, Lehman TJ, Harley JB, Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8(6):492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.International Consortium for Systemic Lupus Erythematosus Genetics (SLEGEN) et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40(2):204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hom G, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358(9):900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 6.Nath SK, et al. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet. 2008;40(2):152–154. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 7.Zhou Y, et al. Multiple lupus-associated ITGAM variants alter Mac-1 functions on neutrophils. Arthritis Rheum. 2013;65(11):2907–2916. doi: 10.1002/art.38117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim-Howard X, et al. ITGAM coding variant (rs1143679) influences the risk of renal disease, discoid rash and immunological manifestations in patients with systemic lupus erythematosus with European ancestry. Ann Rheum Dis. 2010;69(7):1329–1332. doi: 10.1136/ard.2009.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fagerholm SC, MacPherson M, James MJ, Sevier-Guy C, Lau CS. The CD11b-integrin (ITGAM) and systemic lupus erythematosus. Lupus. 2013;22(7):657–663. doi: 10.1177/0961203313491851. [DOI] [PubMed] [Google Scholar]
- 10.MacPherson M, Lek HS, Prescott A, Fagerholm SC. A systemic lupus erythematosus-associated R77H substitution in the CD11b chain of the Mac-1 integrin compromises leukocyte adhesion and phagocytosis. J Biol Chem. 2011;286(19):17303–17310. doi: 10.1074/jbc.M110.182998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhodes B, et al. The rs1143679 (R77H) lupus associated variant of ITGAM (CD11b) impairs complement receptor 3 mediated functions in human monocytes. Ann Rheum Dis. 2012;71(12):2028–2034. doi: 10.1136/annrheumdis-2012-201390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maiti AK, et al. Combined protein- and nucleic acid-level effects of rs1143679 (R77H), a lupus-predisposing variant within ITGAM. Hum Mol Genet. 2014;23(15):4161–4176. doi: 10.1093/hmg/ddu106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rosetti F, et al. A lupus-associated Mac-1 variant has defects in integrin allostery and interaction with ligands under force. Cell Rep. doi: 10.1016/j.celrep.2015.02. [published online ahead of print March 11, 2015]. https://doi.org/10.1016/j.celrep.2015.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Han C, Jin J, Xu S, Liu H, Li N, Cao X. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat Immunol. 2010;11(8):734–742. doi: 10.1038/ni.1908. [DOI] [PubMed] [Google Scholar]
- 15.Ding C, et al. Integrin CD11b negatively regulates BCR signalling to maintain autoreactive B cell tolerance. Nat Commun. 2013;4:2813. doi: 10.1038/ncomms3813. [DOI] [PubMed] [Google Scholar]
- 16.Tang T, et al. A role for Mac-1 (CDIIb/CD18) in immune complex-stimulated neutrophil function in vivo: Mac-1 deficiency abrogates sustained Fcgamma receptor-dependent neutrophil adhesion and complement-dependent proteinuria in acute glomerulonephritis. J Exp Med. 1997;186(11):1853–1863. doi: 10.1084/jem.186.11.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao C, Gao Y, Li Y, Antalis TM, Castellino FJ, Zhang L. The efficacy of activated protein C in murine endotoxemia is dependent on integrin CD11b. J Clin Invest. 2010;120(6):1971–1980. doi: 10.1172/JCI40380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu JR, Han X, Soriano SG, Yuki K. The role of macrophage 1 antigen in polymicrobial sepsis. Shock. 2014;42(6):532–539. doi: 10.1097/SHK.0000000000000250. [DOI] [PubMed] [Google Scholar]
- 19.Kevil CG, et al. Loss of LFA-1, but not Mac-1, protects MRL/MpJ-Fas(lpr) mice from autoimmune disease. Am J Pathol. 2004;165(2):609–616. doi: 10.1016/S0002-9440(10)63325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosetti F, et al. Human lupus serum induces neutrophil-mediated organ damage in mice that is enabled by Mac-1 deficiency. J Immunol. 2012;189(7):3714–3723. doi: 10.4049/jimmunol.1201594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kopf M, Abel B, Gallimore A, Carroll M, Bachmann MF. Complement component C3 promotes T-cell priming and lung migration to control acute influenza virus infection. Nat Med. 2002;8(4):373–378. doi: 10.1038/nm0402-373. [DOI] [PubMed] [Google Scholar]
- 22.Rosetti F, Mayadas TN. The many faces of Mac-1 in autoimmune disease. Immunol Rev. 2016;269(1):175–193. doi: 10.1111/imr.12373. [DOI] [PubMed] [Google Scholar]
- 23.Wang L, et al. Indirect inhibition of Toll-like receptor and type I interferon responses by ITAM-coupled receptors and integrins. Immunity. 2010;32(4):518–530. doi: 10.1016/j.immuni.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huynh L, Wang L, Shi C, Park-Min KH, Ivashkiv LB. ITAM-coupled receptors inhibit IFNAR signaling and alter macrophage responses to TLR4 and Listeria monocytogenes. J Immunol. 2012;188(7):3447–3457. doi: 10.4049/jimmunol.1102211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan ST, Edgington TS. Coupling of the adhesive receptor CD11b/CD18 to functional enhancement of effector macrophage tissue factor response. J Clin Invest. 1991;87(1):50–57. doi: 10.1172/JCI115000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whitlock BB, Gardai S, Fadok V, Bratton D, Henson PM. Differential roles for α(M)β(2) integrin clustering or activation in the control of apoptosis via regulation of akt and ERK survival mechanisms. J Cell Biol. 2000;151(6):1305–1320. doi: 10.1083/jcb.151.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rezzonico R, Imbert V, Chicheportiche R, Dayer JM. Ligation of CD11b and CD11c β(2) integrins by antibodies or soluble CD23 induces macrophage inflammatory protein 1alpha (MIP-1α) and MIP-1β production in primary human monocytes through a pathway dependent on nuclear factor-κB. Blood. 2001;97(10):2932–2940. doi: 10.1182/blood.V97.10.2932. [DOI] [PubMed] [Google Scholar]
- 28.Guha M, Mackman N. The phosphatidylinositol 3-kinase-Akt pathway limits lipopolysaccharide activation of signaling pathways and expression of inflammatory mediators in human monocytic cells. J Biol Chem. 2002;277(35):32124–32132. doi: 10.1074/jbc.M203298200. [DOI] [PubMed] [Google Scholar]
- 29.Rubel C, Gómez S, Fernández GC, Isturiz MA, Caamaño J, Palermo MS. Fibrinogen-CD11b/CD18 interaction activates the NF-κB pathway and delays apoptosis in human neutrophils. Eur J Immunol. 2003;33(5):1429–1438. doi: 10.1002/eji.200323512. [DOI] [PubMed] [Google Scholar]
- 30.Kettritz R, Choi M, Rolle S, Wellner M, Luft FC. Integrins and cytokines activate nuclear transcription factor-κB in human neutrophils. J Biol Chem. 2004;279(4):2657–2665. doi: 10.1074/jbc.M309778200. [DOI] [PubMed] [Google Scholar]
- 31.Giancotti FG, Ruoslahti E. Integrin signaling. Science. 1999;285(5430):1028–1032. doi: 10.1126/science.285.5430.1028. [DOI] [PubMed] [Google Scholar]
- 32.Yee NK, Hamerman JA. β(2) integrins inhibit TLR responses by regulating NF-κB pathway and p38 MAPK activation. Eur J Immunol. 2013;43(3):779–792. doi: 10.1002/eji.201242550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Means TK, Luster AD. Integrins limit the Toll. Nat Immunol. 2010;11(8):691–693. doi: 10.1038/ni0810-691. [DOI] [PubMed] [Google Scholar]
- 34.Whitlock BB, Gardai S, Fadok V, Bratton D, Henson PM. Differential roles for α(M)β(2) integrin clustering or activation in the control of apoptosis via regulation of akt and ERK survival mechanisms. J Cell Biol. 2000;151(6):1305–1320. doi: 10.1083/jcb.151.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maiguel D, et al. Small molecule-mediated activation of the integrin CD11b/CD18 reduces inflammatory disease. Sci Signal. 2011;4(189):ra57. doi: 10.1126/scisignal.2001811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faridi MH, et al. Identification of novel agonists of the integrin CD11b/CD18. Bioorg Med Chem Lett. 2009;19(24):6902–6906. doi: 10.1016/j.bmcl.2009.10.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Salas A, Shimaoka M, Phan U, Kim M, Springer TA. Transition from rolling to firm adhesion can be mimicked by extension of integrin αLβ2 in an intermediate affinity state. J Biol Chem. 2006;281(16):10876–10882. doi: 10.1074/jbc.M512472200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Faridi MH, Altintas MM, Gomez C, Duque JC, Vazquez-Padron RI, Gupta V. Small molecule agonists of integrin CD11b/CD18 do not induce global conformational changes and are significantly better than activating antibodies in reducing vascular injury. Biochim Biophys Acta. 2013;1830(6):3696–3710. doi: 10.1016/j.bbagen.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Doi K, Leelahavanichkul A, Yuen PS, Star RA. Animal models of sepsis and sepsis-induced kidney injury. J Clin Invest. 2009;119(10):2868–2878. doi: 10.1172/JCI39421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Angus DC, van der Poll T. Severe sepsis and septic shock. N Engl J Med. 2013;369(9):840–851. doi: 10.1056/NEJMra1208623. [DOI] [PubMed] [Google Scholar]
- 41.Teijaro JR, et al. Endothelial cells are central orchestrators of cytokine amplification during influenza virus infection. Cell. 2011;146(6):980–991. doi: 10.1016/j.cell.2011.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rutigliano JA, et al. Highly pathological influenza A virus infection is associated with augmented expression of PD-1 by functionally compromised virus-specific CD8+ T cells. J Virol. 2014;88(3):1636–1651. doi: 10.1128/JVI.02851-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bouvier NM, Lowen AC. Animal models for influenza virus pathogenesis and transmission. Viruses. 2010;2(8):1530–1563. doi: 10.3390/v20801530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Litvak V, et al. A FOXO3-IRF7 gene regulatory circuit limits inflammatory sequelae of antiviral responses. Nature. 2012;490(7420):421–425. doi: 10.1038/nature11428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18(5):2986–2996. doi: 10.1128/MCB.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laird MH, et al. TLR4/MyD88/PI3K interactions regulate TLR4 signaling. J Leukoc Biol. 2009;85(6):966–977. doi: 10.1189/jlb.1208763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Doyle S, et al. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17(3):251–263. doi: 10.1016/S1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 48.Kawai T, Akira S. TLR signaling. Cell Death Differ. 2006;13(5):816–825. doi: 10.1038/sj.cdd.4401850. [DOI] [PubMed] [Google Scholar]
- 49.Lee JC, et al. Human SNP links differential outcomes in inflammatory and infectious disease to a FOXO3-regulated pathway. Cell. 2013;155(1):57–69. doi: 10.1016/j.cell.2013.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Andrews BS, et al. Spontaneous murine lupus-like syndromes. Clinical and immunopathological manifestations in several strains. J Exp Med. 1978;148(5):1198–1215. doi: 10.1084/jem.148.5.1198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pérez de Lema G, et al. Chemokine expression precedes inflammatory cell infiltration and chemokine receptor and cytokine expression during the initiation of murine lupus nephritis. J Am Soc Nephrol. 2001;12(7):1369–1382. doi: 10.1681/ASN.V1271369. [DOI] [PubMed] [Google Scholar]
- 52.Kanauchi H, Furukawa F, Imamura S. Characterization of cutaneous infiltrates in MRL/lpr mice monitored from onset to the full development of lupus erythematosus-like skin lesions. J Invest Dermatol. 1991;96(4):478–483. doi: 10.1111/1523-1747.ep12470176. [DOI] [PubMed] [Google Scholar]
- 53.Knight JS, et al. Peptidylarginine deiminase inhibition disrupts NET formation and protects against kidney, skin and vascular disease in lupus-prone MRL/lpr mice. Ann Rheum Dis. 2015;74(12):2199–2206. doi: 10.1136/annrheumdis-2014-205365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Furumoto Y, et al. Tofacitinib ameliorates murine lupus and its associated vascular dysfunction. Arthritis Rheum. 2017;69(1):148–160. doi: 10.1002/art.39818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ozgor L, Brandl C, Shock A, Nitschke L. Epratuzumab modulates B-cell signaling without affecting B-cell numbers or B-cell functions in a mouse model with humanized CD22. Eur J Immunol. 2016;46(9):2260–2272. doi: 10.1002/eji.201646383. [DOI] [PubMed] [Google Scholar]
- 56.Thacker SG, et al. Type I interferons modulate vascular function, repair, thrombosis, and plaque progression in murine models of lupus and atherosclerosis. Arthritis Rheum. 2012;64(9):2975–2985. doi: 10.1002/art.34504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen D, et al. Antibody blockade of thrombospondin accelerates reendothelialization and reduces neointima formation in balloon-injured rat carotid artery. Circulation. 1999;100(8):849–854. doi: 10.1161/01.CIR.100.8.849. [DOI] [PubMed] [Google Scholar]
- 58.Semmrich M, et al. Importance of integrin LFA-1 deactivation for the generation of immune responses. J Exp Med. 2005;201(12):1987–1998. doi: 10.1084/jem.20041850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park EJ, et al. Distinct roles for LFA-1 affinity regulation during T-cell adhesion, diapedesis, and interstitial migration in lymph nodes. Blood. 2010;115(8):1572–1581. doi: 10.1182/blood-2009-08-237917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park EJ, et al. Aberrant activation of integrin alpha4beta7 suppresses lymphocyte migration to the gut. J Clin Invest. 2007;117(9):2526–2538. doi: 10.1172/JCI31570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.O’Gorman WE, et al. Single-cell systems-level analysis of human Toll-like receptor activation defines a chemokine signature in patients with systemic lupus erythematosus. J Allergy Clin Immunol. 2015;136(5):1326–1336. doi: 10.1016/j.jaci.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ehirchiou D, Xiong Y, Xu G, Chen W, Shi Y, Zhang L. CD11b facilitates the development of peripheral tolerance by suppressing Th17 differentiation. J Exp Med. 2007;204(7):1519–1524. doi: 10.1084/jem.20062292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ivashkiv LB. How ITAMs inhibit signaling. Sci Signal. 2011;4(169):pe20. doi: 10.1126/scisignal.2001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dransfield I, Hogg N. Regulated expression of Mg2+ binding epitope on leukocyte integrin alpha subunits. EMBO J. 1989;8(12):3759–3765. doi: 10.1002/j.1460-2075.1989.tb08552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gupta V, et al. The beta-tail domain (βTD) regulates physiologic ligand binding to integrin CD11b/CD18. Blood. 2007;109(8):3513–3520. doi: 10.1182/blood-2005-11-056689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ortlepp S, Stephens PE, Hogg N, Figdor CG, Robinson MK. Antibodies that activate β2 integrins can generate different ligand binding states. Eur J Immunol. 1995;25(3):637–643. doi: 10.1002/eji.1830250302. [DOI] [PubMed] [Google Scholar]
- 67.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434(7034):772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 68.Hua J, Kirou K, Lee C, Crow MK. Functional assay of type I interferon in systemic lupus erythematosus plasma and association with anti-RNA binding protein autoantibodies. Arthritis Rheum. 2006;54(6):1906–1916. doi: 10.1002/art.21890. [DOI] [PubMed] [Google Scholar]
- 69.Niewold TB, Hua J, Lehman TJ, Harley JB, Crow MK. High serum IFN-alpha activity is a heritable risk factor for systemic lupus erythematosus. Genes Immun. 2007;8(6):492–502. doi: 10.1038/sj.gene.6364408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yuen PS, Dunn SR, Miyaji T, Yasuda H, Sharma K, Star RA. A simplified method for HPLC determination of creatinine in mouse serum. Am J Physiol Renal Physiol. 2004;286(6):F1116–F1119. doi: 10.1152/ajprenal.00366.2003. [DOI] [PubMed] [Google Scholar]
- 72.Figueiredo N, et al. Anthracyclines induce DNA damage response-mediated protection against severe sepsis. Immunity. 2013;39(5):874–884. doi: 10.1016/j.immuni.2013.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Fuss IJ, Kanof ME, Smith PD, Zola H. Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr Protoc Immunol. 2009;Chapter 7:Unit 7.1. doi: 10.1002/0471142735.im0701s85. [DOI] [PubMed] [Google Scholar]
- 74.Lacey DC, et al. Defining GM-CSF- and macrophage-CSF-dependent macrophage responses by in vitro models. J Immunol. 2012;188(11):5752–5765. doi: 10.4049/jimmunol.1103426. [DOI] [PubMed] [Google Scholar]
- 75.Mia S, Warnecke A, Zhang XM, Malmström V, Harris RA. An optimized protocol for human M2 macrophages using M-CSF and IL-4/IL-10/TGF-β yields a dominant immunosuppressive phenotype. Scand J Immunol. 2014;79(5):305–314. doi: 10.1111/sji.12162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chevrier N, et al. Systematic discovery of TLR signaling components delineates viral-sensing circuits. Cell. 2011;147(4):853–867. doi: 10.1016/j.cell.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang X, Goncalves R, Mosser DM. The isolation and characterization of murine macrophages. Curr Protoc Immunol. 2008;Chapter 14:Unit 14.1. doi: 10.1002/0471142735.im1401s83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kariuki SN, et al. Genetic analysis of the pathogenic molecular sub-phenotype interferon-α identifies multiple novel loci involved in systemic lupus erythematosus. Genes Immun. 2015;16(1):15–23. doi: 10.1038/gene.2014.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.