Abstract
SIRT2 is a cytoplasmic sirtuin that plays a role in various cellular processes, including tumorigenesis, metabolism, and inflammation. Since these processes require iron, we hypothesized that SIRT2 directly regulates cellular iron homeostasis. Here, we have demonstrated that SIRT2 depletion results in a decrease in cellular iron levels both in vitro and in vivo. Mechanistically, we determined that SIRT2 maintains cellular iron levels by binding to and deacetylating nuclear factor erythroid-derived 2–related factor 2 (NRF2) on lysines 506 and 508, leading to a reduction in total and nuclear NRF2 levels. The reduction in nuclear NRF2 leads to reduced ferroportin 1 (FPN1) expression, which in turn results in decreased cellular iron export. Finally, we observed that Sirt2 deletion reduced cell viability in response to iron deficiency. Moreover, livers from Sirt2–/– mice had decreased iron levels, while this effect was reversed in Sirt2–/– Nrf2–/– double-KO mice. Taken together, our results uncover a link between sirtuin proteins and direct control over cellular iron homeostasis via regulation of NRF2 deacetylation and stability.
Introduction
Iron is an integral component of numerous proteins, however, free iron is a major source of ROS and cellular injury (1). Thus, the levels of iron in cellular compartments are tightly regulated. Cellular iron is regulated through both import and export mechanisms. Under iron-deficient conditions, the iron-regulatory proteins IRP1 and IRP2 are activated to modulate the mRNA stability or translation rates of key iron transporters by binding to iron response elements (IREs) in the 3′- or 5′-UTRs of target mRNAs (2, 3). IRP1/2 binding to the 5′-UTR of ferritin (Ftl) and ferroportin (Fpn1) mRNAs results in reduced translation rates, which leads to the mobilization of iron stores and reduced iron export, respectively. Interaction of IRP1/2 with the 3′-UTR stabilizes target mRNAs, such as the iron importer transferrin receptor 1 (Tfrc), and enhances iron uptake (2). Overall, activation of the IRP1/2 system by iron deficiency restores iron balance through increased import and decreased export, while iron overload suppresses IRP1/2, favoring iron storage and removal from the cell. Iron homeostasis is controlled by other signaling pathways such as HIF, which regulates cellular iron import through transcriptional upregulation of Tfrc by binding to HIF-responsive elements in the promoter of Tfrc (4). In addition, our group recently discovered a pathway of iron conservation involving the RNA-binding protein tristetraprolin (TTP) (5).
Lysine acetylation/deacetylation has emerged as an important and physiologically significant posttranslational protein modification. Acetylation of proteins is carried out by histone acetyltransferases (HATs), while histone deacetylases (HDACs) remove acetyl groups from lysine residues (6). There are 4 classes of HDACs in mammals (classes I–IV), with the sirtuin family comprising the class III HDACs. Unlike other HDACs, sirtuins require NAD+ to carry out their enzymatic reaction (7). In mammals, 7 sirtuins (SIRT1-7) have been identified, each of which shares a conserved 275-amino-acid catalytic core domain. These sirtuins are generally categorized according to their subcellular localization: nuclear (SIRT1, SIRT6, and SIRT7); cytoplasmic (SIRT2); and mitochondrial (SIRT3, SIRT4, and SIRT5). SIRTs 1–3 have robust deacetylation activity, while SIRT4 is reported to display ADP-ribosyltransferase activity. SIRT5 may function as a protein desuccinylase and demalonylase, and SIRT6 and SIRT7 show weak deacetylase activity (8–11). Sirtuins have been implicated in a wide range of cellular processes including aging, apoptosis, response to stress and inflammation, control of energy efficiency, circadian clocks, and mitochondrial biogenesis (12, 13).
SIRT2, which is among the least-characterized SIRTs (14), contributes to the regulation of metabolism (15), inflammation (16, 17), cell-cycle progression (18), neurodegeneration (19), and tumorigenesis (20). Since iron is an essential component of each of these cellular processes (21–24), we hypothesized that an overarching function of SIRT2 activity may be to maintain cellular iron at levels sufficient to support the processes described above. Here, we demonstrate that SIRT2 increases cellular iron by binding to and deacetylating nuclear factor erythroid-derived 2–related factor 2 (NRF2), also known as NFE2L2, with a subsequent decrease in its nuclear levels. This, in turn, reduces the expression of Fpn1, which is directly regulated by NRF2 at the transcriptional level. We also show that the regulation of cellular iron by SIRT2 has physiological significance, as its deletion decreases cell survival in response to iron deficiency. We confirm our findings and the role of NRF2 in SIRT2 regulation of cellular iron using both in vitro and in vivo models.
Results
SIRT2 regulates cellular iron content.
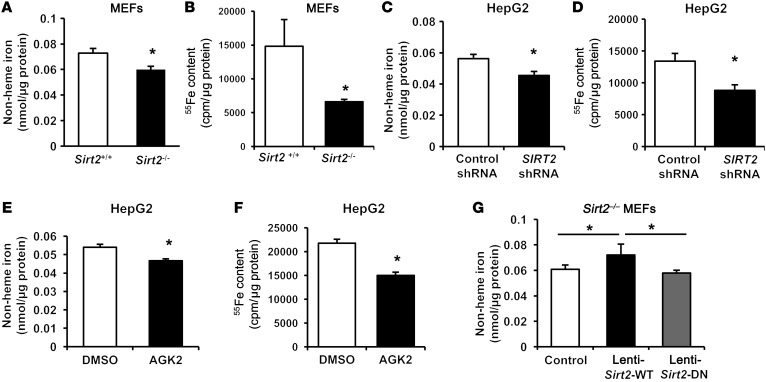
To test the hypothesis that SIRT2 directly regulates cellular iron homeostasis, we first measured iron content in Sirt2+/+ and Sirt2–/– mouse embryonic fibroblasts (MEFs). We found that cellular iron levels were significantly lower in Sirt2–/– MEFs than in Sirt2+/+ MEFs, as assessed by a non-heme iron assay, radioactive iron content analysis, and measurement of heme levels (Figure 1, A and B, and Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI88574DS1). Similarly, silencing SIRT2 in human HepG2 hepatoma cells using an shRNA lentivirus decreased cellular iron content (Figure 1, C and D), and pharmacological inhibition of the protein by the specific SIRT2 inhibitor AGK2 also reduced iron levels in HepG2 cells (Figure 1, E and F). To further confirm the observed effects of SIRT2 on iron regulation, we constructed 2 lentiviruses that overexpressed either the WT SIRT2 (lenti-Sirt2-WT) or a deacetylation-null SIRT2 mutant that lacked deacetylase activity (lenti-Sirt2-DN) (25). These viruses were infected into Sirt2–/– MEFs. Infection with lenti-Sirt2-WT, but not lenti-Sirt2-DN, reversed iron deficiency in Sirt2–/– MEFs (Figure 1G). These results indicate that SIRT2 regulates cellular iron homeostasis at least in part through its deacetylation activity.
Figure 1. SIRT2 regulates cellular iron content.
(A) Non-heme iron content in Sirt2+/+ and Sirt2–/– MEFs normalized to the protein concentration (n = 5 for each genotype). (B) Cellular content of 55Fe in Sirt2+/+ and Sirt2–/– MEFs after 48 hours of incubation with radioactive iron (n = 10–12 for each genotype). (C) Non-heme iron content in HepG2 cells infected with control lentivirus (lenti-control shRNA) or SIRT2 shRNA (lenti-SIRT2 shRNA) (n = 10–12 per group). (D) Cellular content of 55Fe in HepG2 cells infected with lenti-control shRNA or lenti-SIRT2 shRNA after 48 hours of incubation with radioactive iron (n = 5–6 per group). (E) Non-heme iron content in HepG2 cells treated with DMSO or the SIRT2 inhibitor AGK2 (n = 6 per group). (F) Cellular content of 55Fe in HepG2 cells treated with DMSO or the SIRT2 inhibitor AGK2 overnight after 48 hours of incubation with radioactive iron (n = 6 per group). (G) Non-heme iron content in Sirt2–/– MEFs infected with lenti-control, lenti-Sirt2-WT, or lenti-Sirt2-DN (n = 10–11 per group). Data are presented as the mean ± SEM. *P < 0.05, by Student’s t test (A–F) or 1-way ANOVA with Bonferroni’s correction for multiple comparisons (G).
SIRT2 mediates iron export through FPN1.
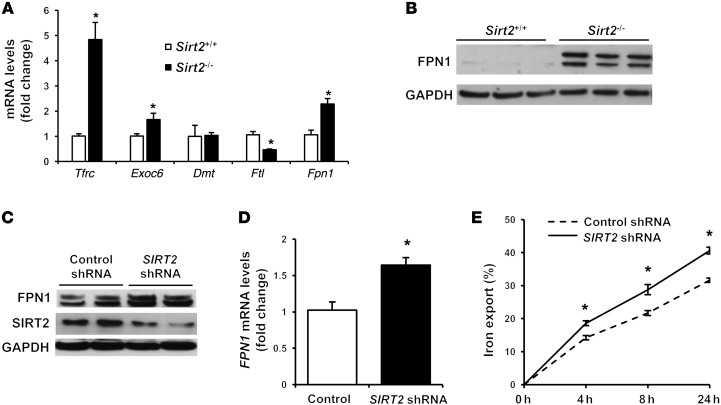
We next studied the mechanism by which SIRT2 alters cellular iron by measuring the expression of genes involved in iron regulation. Gene expression of the cellular iron import protein Tfrc was significantly higher and the light chain of the iron storage molecule ferritin (Ftl) was lower in Sirt2–/– MEFs than in Sirt2+/+ MEFs (Figure 2A). These changes are consistent with an iron deficiency signature; however, they were not the primary driver of the decrease in cellular iron in Sirt2–/– MEFs. Among other genes, the cellular iron export protein Fpn1 was noted to be significantly higher at both the mRNA and protein levels in Sirt2–/– MEFs (Figure 2, A and B). Likewise, FPN1 was upregulated in human HepG2 cells infected with lenti-SIRT2 shRNA as compared with those infected with lenti-control shRNA (Figure 2, C and D). HepG2 cells with SIRT2 knockdown also showed increased iron export, as measured by a radioactive iron–based export assay (Figure 2E). These results suggest that enhanced cellular iron export via increased Fpn1 expression in Sirt2–/– or Sirt2-knockdown conditions is likely the primary cause of cellular iron deficiency and that SIRT2 mediates iron export though the regulation of Fpn1.
Figure 2. SIRT2 mediates iron export through FPN1.
(A) mRNA levels of genes involved in iron homeostasis in Sirt2+/+ and Sirt2–/– MEFs (n = 5–9 for each genotype). Exoc6, exocyst complex component 6; Dmt1, divalent metal transporter 1; Ftl, ferritin light chain. (B) FPN1 protein levels in Sirt2+/+ and Sirt2–/– MEFs (n = 3 per group). (C) Representative blot from 3 independent experiments showing FPN1 levels in HepG2 cells treated with lenti-control shRNA or lenti-SIRT2 shRNA. (D) mRNA levels of FPN1 in HepG2 cells infected with lenti-control shRNA or lenti-SIRT2 shRNA (n = 5 per group). (E) Time-dependent 55Fe export in HepG2 cells treated with lenti-control shRNA or lenti-SIRT2 shRNA after 16 hours of incubation with radioactive iron (n = 6 per group). Data are presented as the mean ± SEM. *P < 0.05, by Student’s t test.
SIRT2 regulates FPN1 through NRF2.
The regulation of cellular iron has previously been shown to mostly occur at the posttranscriptional level via RNA-binding proteins such as IRP1/2 and TTP. Our data suggest that FPN1 mRNA and protein levels are regulated by SIRT2. To determine whether the increase in Fpn1 mRNA with SIRT2 deficiency is at the posttranscriptional level, we measured Fpn1 mRNA stability in Sirt2+/+ and Sirt2–/– MEFs and in HepG2 cells with or without SIRT2 knockdown after treatment with the transcriptional inhibitor actinomycin D. Our results demonstrated that SIRT2 deficiency does not alter Fpn1 mRNA stability (Supplemental Figure 2, A and B), suggesting that SIRT2 regulation of Fpn1 probably occurs at the transcriptional level.
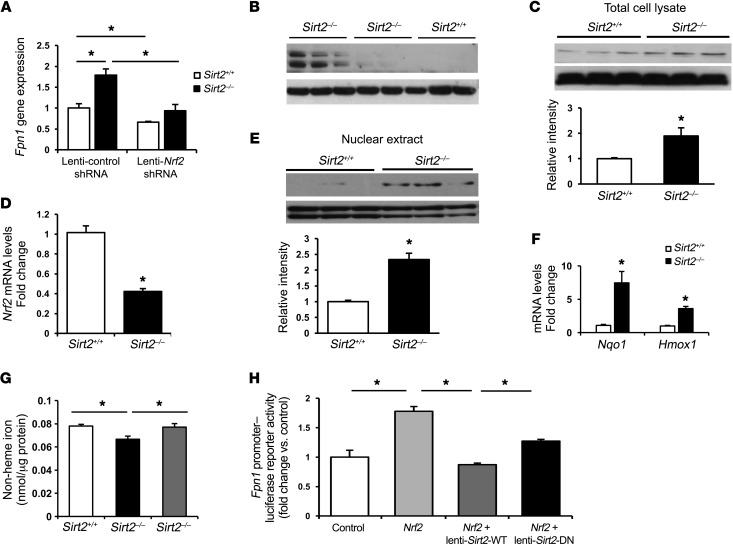
We next assessed the mechanism by which SIRT2 regulates Fpn1 transcription. Fpn1 has been shown to be transcriptionally regulated by the transcription factors HIF, metal-regulatory transcription factor 1 (MTF1), and NRF2 (26, 27). We asked whether these transcription factors are involved in SIRT2-mediated regulation of Fpn1. To determine the role of the HIF pathway in the regulation of Fpn1 by SIRT2, we downregulated SIRT2 in aryl hydrocarbon receptor nuclear translocator (Arnt, encoding the obligatory dimer for HIF1-α and HIF2-α) KO MEFs (Arnt–/– MEFs) and assessed Fpn1 mRNA levels. Since the change in Fpn1 expression with SIRT2 modulation was not altered in Arnt–/– MEFs (Supplemental Figure 2C), we concluded that SIRT2 regulation of Fpn1 is independent of the HIF pathway. Furthermore, Mtf1 knockdown in Sirt2–/– MEFs did not alter Fpn1 levels (Supplemental Figure 2D), indicating that MTF1 is also not involved in this process. However, the increase in mRNA and protein levels of FPN1 in Sirt2–/– MEFs was reversed with Nrf2 knockdown using an Nrf2 shRNA lentivirus (Figure 3, A and B, and Supplemental Figure 3A), indicating that NRF2 mediates SIRT2 regulation of Fpn1. Thus, we focused our studies on the mechanism of SIRT2 regulation of NRF2.
Figure 3. Regulation of Fpn1 by SIRT2 occurs through NRF2.
(A) Fpn1 mRNA levels in MEFs infected with lenti-control shRNA or lenti-Nrf2 shRNA (n = 5–6 per group). (B) Representative blot from 2 independent experiments showing FPN1 expression in Sirt2+/+ and Sirt2–/– MEFs infected with lenti-control shRNA or lenti-Nrf2 shRNA. (C) NRF2 protein levels in Sirt2+/+ and Sirt2–/– MEFs (n = 3 per genotype). (D) mRNA levels of Nrf2 in Sirt2+/+ and Sirt2–/– MEFs (n = 8 per genotype). (E) NRF2 protein levels in nuclei of Sirt2+/+ and Sirt2–/– MEFs (n = 3 per genotype). (F) NRF2 antioxidant target genes Nqo1 and Hmox1 mRNA levels in Sirt2+/+ and Sirt2–/– MEFs (n = 8–9 per genotype). (G) Non-heme iron levels in Sirt2+/+ and Sirt2–/– MEFs infected with lenti-control shRNA or lenti-Nrf2 shRNA (n = 6 per genotype). (H) Luciferase activity in HEK293T cells transfected with a murine Fpn1 promoter–luciferase reporter construct along with Nrf2 plasmid and WT Sirt2 or deacetylation-null Sirt2 plasmids (lenti-Sirt2-WT and lenti-Sirt2-DN). An empty vector was used as a control (n = 4–6 per group). Data are presented as the mean ± SEM. *P < 0.05, by ANOVA with Bonferroni’s correction for multiple comparisons (A, G, and H) or by Student’s t test (C, D, and F).
We found that NRF2 protein levels were higher in Sirt2–/– MEFs (Figure 3C), while its mRNA levels were lower (Figure 3D), which suggests that SIRT2 destabilizes NRF2 protein at the posttranslational level. The reduced Nrf2 mRNA levels in Sirt2–/– cells is probably a compensatory mechanism when NRF2 protein levels are elevated. Similarly, NRF2 protein levels were also increased in HepG2 cells with SIRT2 knockdown (Supplemental Figure 3B). Using a luciferase construct containing NRF2-binding sites (also known as antioxidant response elements [AREs]) of the murine Fpn1 promoter, we showed that NRF2 activates the Fpn1 promoter (Supplemental Figure 3C). Since NRF2 is a transcription factor and translocates into the nucleus upon activation (28), we measured nuclear levels of NRF2 and found them to be significantly higher in Sirt2–/– MEFs than in Sirt2+/+ MEFs (Figure 3E). Cytosolic levels of NRF2 were also increased in Sirt2–/– MEFs (Supplemental Figure 3D), reflecting the increased total levels of the proteins. The increase in nuclear levels of NRF2 suggests a higher transcriptional activity of the protein with SIRT2 knockdown. Thus, we next examined the effects of SIRT2 modulation on NRF2 transcriptional activity. In Sirt2–/– MEFs, we observed an increase in mRNA levels of the antioxidant genes regulated by NRF2, NAD(P)H quinone oxidoreductase 1 (Nqo1) and heme oxygenase 1 (Hmox1) (Figure 3F), suggesting that NRF2 transcriptional activity is increased when SIRT2 levels are reduced.
To confirm these results, we also conducted studies with overexpression of WT SIRT2 and deacetylation-null SIRT2 (lenti-Sirt2-DN). Overexpression of lenti-Sirt2-WT, but not lenti-Sirt2-DN, resulted in a significant decrease in both cellular and nuclear NRF2 (Supplemental Figure 4, A and B). Additionally, mRNA expression of Fpn1 and Tfrc was decreased with lenti-Sirt2-WT, but not lenti-Sirt2-DN (Supplemental Figure 4C).
We next asked whether NRF2 is involved in SIRT2-mediated iron homeostasis. Knockdown of Nrf2 significantly increased non-heme iron in Sirt2+/+ MEFs (Supplemental Figure 5A) and reversed the reduced iron levels in Sirt2–/– MEFs (Figure 3G). Furthermore, transcriptional activation of the Fpn1 promoter by NRF2 was reversed by lenti-Sirt2-WT, but not by lenti-Sirt2-DN (Figure 3H and Supplemental Figure 5B). These results indicate that the regulation of iron homeostasis and Fpn1 expression by SIRT2 is NRF2 dependent.
SIRT2 deacetylates NRF2 and decreases its stability.
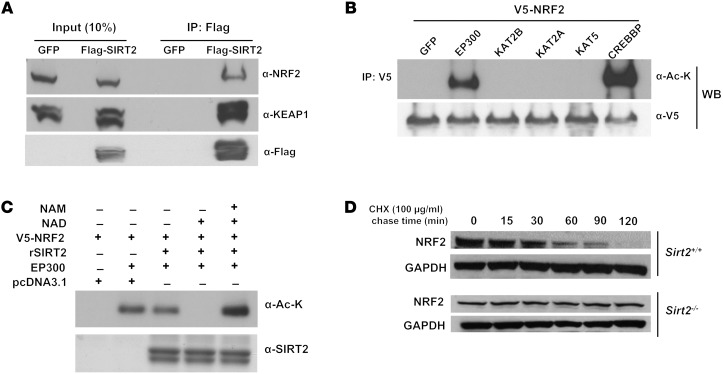
We next investigated how SIRT2 regulates NRF2 levels. Under normal conditions, Kelch-like ECH-associated protein 1 (KEAP1) sequesters NRF2 in the cytoplasm and promotes its degradation, while stress conditions lead to NRF2 release from KEAP1 and its translocation to the nucleus, with subsequent upregulation of antioxidant gene expression (29). Since NRF2 activity was regulated by SIRT2 and SIRT2 is localized to the cytoplasm, we proposed that SIRT2 interacts with either NRF2 or KEAP1 to alter their activity. To test the hypothesis, we performed co-IP studies of SIRT2 with NRF2 or KEAP1. Flag-tagged SIRT2 was transfected into cells and immunoprecipitated with an anti-Flag antibody, followed by Western blotting with NRF2 or KEAP1 antibody. The co-IP experiments showed interaction between SIRT2 and the NRF2-KEAP1 complex (Figure 4A).
Figure 4. SIRT2 deacetylates NRF2 and promotes its ubiquitination.
(A) Co-IP of SIRT2 and either NRF2 or KEAP1 in HeLa cells transfected with Flag-SIRT2. The label “GFP” refers to transfection with a control GFP plasmid, while Flag-SIRT2 was transfected with a Flag-SIRT2 construct. Endogenous NRF2 was detected in the input lane transfected with GFP. (B) Co-IP experiments of HEK293T cells transfected with V5-NRF2 and either GFP (control) or different HATs including EP300, KAT2B, KAT2A, KAT5, and CREBBP, using an anti-V5 antibody for IP and anti–acetyl-lysine (α-Ac-K) or anti-V5 (α-V5) antibodies for immunoblotting. WB, Western blot. (C) HEK293T cells cotransfected with p300 and V5-NRF2 were harvested, and NRF2 was purified by IP. Immunoprecipitated acetylated NRF2 was mixed with NAD+ and WT purified SIRT2 in the presence and absence of NAM, which was used as a deacetylase inhibitor. rSIRT2, recombinant SIRT2. (D) Time course of NRF2 protein stability in Sirt2+/+ and Sirt2–/– MEFs after treatment with 100 μg/ml CHX. All panels are representative blots from 2 independent experiments.
Since there was an interaction between the NRF2-KEAP1 complex and SIRT2, we then assessed whether NRF2 or KEAP1 could be deacetylated by SIRT2. Cells were cotransfected with V5-tagged NRF2, together with vectors containing either GFP (control) or different HATs including E1A-binding protein P300 (EP300), K(lysine) acetyltransferase 2B (KAT2B), KAT2A, KAT5, and CREB-binding protein (CREBBP), followed by IP using V5 antibody and blotting with anti–acetylated lysine antibody. These studies showed that EP300 and CREBBP acetylate NRF2 (Figure 4B), indicating that NRF2 is posttranslationally modified by acetylation. However, KEAP1 could not be acetylated by any of these HATs (Supplemental Figure 6A). Furthermore, SIRT2 removed NRF2 acetylation in the presence of NAD+ (Figure 4C), and nicotinamide (NAM), which inhibits sirtuins, reversed the deacetylation of NRF2 by SIRT2 (Figure 4C). Since NRF2 levels are increased with SIRT2 depletion, we assessed whether SIRT2 alters the stability of NRF2 protein. We found that NRF2 levels were significantly decreased 120 minutes after treatment with the protein synthesis inhibitor cycloheximide (CHX), while we did not observe this effect in Sirt2–/– MEFs (Figure 4D). Additionally, overexpression of lenti-Sirt2-WT, but not lenti-Sirt2-DN, resulted in the destabilization of NRF2 protein (Supplemental Figure 6C). There also was no change in SIRT2 levels with CHX treatment (Supplemental Figure 6B). These data indicate that SIRT2 deacetylates NRF2, resulting in its instability and shortened half-life.
Lysines 506 and 508 of NRF2 are the deacetylation targets of SIRT2.
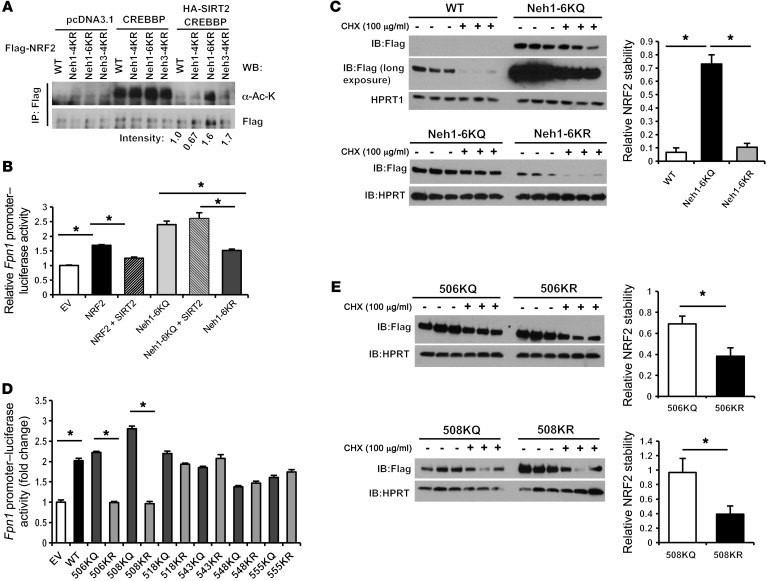
We next asked which lysine residue(s) of NRF2 are targets of SIRT2 and are required for the regulation of NRF2 by SIRT2. We found that Flag-tagged human NRF2 was overexpressed in Sirt2+/+ and Sirt2–/– MEFs and then performed IP with Flag antibody to isolate Flag-NRF2 protein and assessed the immunoprecipitated protein by mass spectrometry. Peptide identification and analysis of the precise localization of potential reversible lysine acetylation sites showed that multiple lysine residues in the highly conserved NRF2-ECH homology domains Neh1 and Neh3 were acetylated in Sirt2–/– MEFs, but not in deacetylated in Sirt2+/+ MEFs (Table 1 and Supplemental Figure 6D). To confirm these mass spectrometric findings and to narrow down the potential targets, we generated 3 different deacetylation mimetic mutants by mutating lysine (K) to arginine (R) in the following 3 regions: (a) the first 4 lysine residues in Neh1 (Neh1-4KR); (b) the remaining 6 lysine residues in Neh1 (Neh1-6KR); and (c) the 4 lysine residues in Neh3 (Neh3-4KR) (Supplemental Figure 6D). Deacetylation assays with these mutants demonstrated that CREBBP acetylated all of the mutant constructs. However, in the presence of SIRT2, Neh1-6KR and Neh3-4KR maintained their acetylation levels, suggesting that SIRT2 may target lysines within both regions (Figure 5A and Supplemental Figure 7). However, since Neh3 lysines have been shown to be potential targets of SIRT1 (30) and our goal was to study the impact of acetylation of the NRF2 DNA-binding domain on the function of NRF2, we focused our studies on Neh1-6.
Table 1. Acetylated/deacetylated lysine residues in FLAG-NRF2 overexpressed in MEFs detected by mass spectrometry.
Figure 5. Lysines 506 and 508 of NRF2 are targeted and deacetylated by SIRT2.
(A) Co-IP experiments in HEK293T cells overexpressing Flag-NRF2 or mutant plasmids and CREBBP together, with or without SIRT2. Lysates were immunoprecipitated with anti-Flag antibody and blotted with anti–acetyl-lysine (α-Ac-K) antibody. The numbers under the gel reflect the degree of protein acetylation in the presence of SIRT2 and CREBBP normalized to CREBBP only. pcDNA3.1 empty vector was used as a control. (B) Luciferase activity in HepG2 cells transfected with a murine Fpn1 promoter–luciferase reporter construct along with NRF2 plasmid or the indicated NRF2 deacetylation mutants with mutations in the indicated NRF2-ECH homology (Neh) domain. SIRT2 was coexpressed as indicated. Fpn1 promoter reporter activity was normalized to Renilla luciferase activity and empty vector (EV). Empty vector was used as a control (n = 6 per group). (C) Protein stability of overexpressed WT NRF2, 6KQ, and 6KR mutants after treatment with CHX in HepG2 cells. Samples were taken 0 and 90 minutes after CHX treatment. Since all of the samples could not be run on the same gel, they were processed on 2 separate gels, with Neh1-6KQ shared between them (n = 3 per group). (D) Luciferase activity in HepG2 cells transfected with a murine Fpn1 promoter–luciferase reporter construct along with NRF2 or NRF2 mutants. Luciferase activity was measured and analyzed as in B. An empty vector was used as a control, and pairwise comparisons were made between the glutamate and arginine mutants for each amino acid site (n = 6 per group). (E) Protein stability of overexpressed 506KQ, 506KR, 508KQ, and 508KR mutants after treatment with 100 μg/ml CHX in HepG2 cells. Samples were taken 0 and 90 minutes after CHX treatment (n = 3 per group). Data are presented as the mean ± SEM. *P < 0.05, by ANOVA with Bonferroni’s correction for multiple comparisons (B and C) or by Student’s t test (D and E).
To confirm that SIRT2-mediated deacetylation of the lysine residues in Neh1 has functional consequences, we assessed the effects of the Neh1-6KR mutant on Fpn1 promoter activity using the Fpn1 promoter–luciferase construct. For these studies, we also made group mutations of Neh1-6K to glutamine (Q) to mimic the acetylated form. A luciferase reporter assay showed that overexpression of NRF2 increased Fpn1 promoter activity, while concurrent overexpression of Sirt2 inhibited NRF2-induced transcriptional activity of the reporter, consistent with our earlier observation. Overexpression of the acetylated form of Neh-6K (i.e., Neh1-6KQ) showed an increase in Fpn1 promoter activity similar to that seen with WT NRF2, which was not decreased with concurrent SIRT2 overexpression, whereas the deacetylated form (i.e., Neh1-6KR) displayed loss of transcriptional activation analogous to Sirt2 overexpression (Figure 5B and Supplemental Figure 8). We also assessed the protein stability of the mutant constructs after treatment with CHX. Neh1-6KR displayed a shorter half-life compared with that of Neh1-6KQ (Figure 5C), suggesting that protein acetylation in at least 1 of these 6 lysine residues is important for NRF2 protein stability. These results further support the notion that acetylation of NRF2 increases its stability and transcriptional activity.
To determine which specific lysine residue(s) in the Neh1 region are targeted by SIRT2, we made individual mutations of the 6 lysine residues to either glutamine or arginine, followed by measurement of Fpn1 promoter activity and NRF2 protein stability. We found that only mutations of 506K and 508K had effects on Fpn1 promoter activity that were similar to the effects seen with the Neh1-6k group mutants; mutation of 506K to Q (506KQ) or 508K to Q (508KQ) maintained transcriptional activity at the same level as that detected in WT NRF2, while mutation of 506K to R (506KR) or 508K to R (508KR) resulted in a loss of transcriptional activation (Figure 5D). Mutation of other lysine residues in this region to either Q or R did not alter transcriptional activation of the Fpn1 promoter (Figure 5D). We confirmed these results by assessing the stability of the mutant proteins. Consistent with the results of our luciferase studies, only 506K and 508K mutants showed protein stability similar to that of the Neh1-6k group mutants (Figure 5E), while no difference was observed in protein stability between Q and R mutations of other lysine residues in this region in response to CHX (Supplemental Figure 9). Taken together, our data indicate that lysines 506 and 508 of NRF2, both of which are evolutionarily conserved across multiple species (Supplemental Figure 6D), are the functional deacetylation targets of SIRT2.
SIRT2 modulates cellular iron homeostasis in vivo, and its absence promotes cell death in iron deficiency.
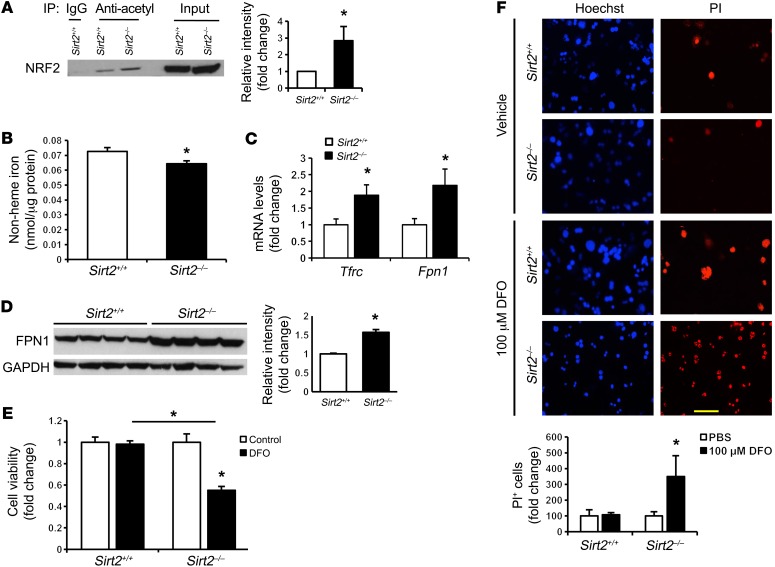
Our in vitro studies thus far suggest a role for SIRT2 in the regulation of cellular iron homeostasis through deacetylation of NRF2, which diminishes NRF2 stability and transcriptional activity. We then asked whether SIRT2 deficiency could regulate cellular iron homeostasis in vivo. We first demonstrated that NRF2 acetylation was increased in the livers of Sirt2–/– mice, confirming that NRF2 is a target of SIRT2 in mouse livers (Figure 6A). Consistent with the in vitro data, non-heme iron levels were significantly decreased in the livers of Sirt2–/– mice compared with levels in Sirt2+/+ littermate controls (Figure 6B), while FPN1 mRNA and protein levels increased significantly (Figure 6, C and D). The higher Tfrc mRNA levels may reflect activation of the IRP system under iron deficiency (Figure 6C), similar to our observation in tissue culture (Figure 2A).
Figure 6. SIRT2 regulates cellular iron homeostasis in vivo and protects against primary hepatocyte death in iron deficiency.
(A) NRF2 acetylation levels in the livers of Sirt2+/+ and Sirt2–/– mice (n = 3 per genotype). (B) Non-heme iron content in the livers of Sirt2+/+ and Sirt2–/– mice (n = 5 per genotype). (C) Tfrc and Fpn1 mRNA levels in the livers of Sirt2+/+ and Sirt2–/– mice (n = 8 per genotype). (D) FPN1 protein levels in the livers of Sirt2+/+ and Sirt2–/– mice (n = 4 per genotype). (E) Steady-state cell numbers in primary hepatocytes from Sirt2+/+ and Sirt2–/– mice, as measured by MTS assay after treatment with DFO (n = 1 per genotype; cells cultured in 5 to 6 individual wells per genotype). (F) Cell death in primary hepatocytes from Sirt2+/+ and Sirt2–/– mice measured by PI staining (n = 1 per genotype; cells were cultured in 5 or 6 individual wells per genotype). Original magnification, ×200; scale bar: 50 μm. Data are presented as the mean ± SEM. *P < 0.05, by Student’s t test (A–D) or ANOVA with Bonferroni’s correction for multiple comparisons (E and F).
Iron is essential for cell survival and is needed for many basic cellular processes such as energy production, DNA synthesis, and substrate conversion. Therefore, we assessed the physiological implications of our observations by assessing cell viability with iron deficiency and Sirt2 deletion. We observed decreased steady-state cell numbers (using the MTS assay) and increased cell death (as determined by positive staining of propidium iodide [PI]) in Sirt2–/– primary hepatocytes compared with littermate control primary hepatocytes in response to iron deficiency (Figure 6, E and F). These results suggest that Sirt2–/– primary hepatocytes are more susceptible to cell death in iron deficiency and that the regulation of cellular iron by SIRT2 may function to protect against cell death during iron deficiency.
Nrf2 deletion reverses iron deficiency in the livers of Sirt2–/– mice.
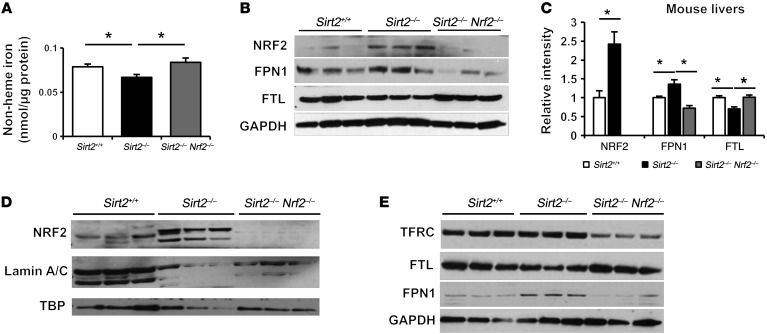
To determine whether iron deficiency in the livers of Sirt2–/– mice is NRF2 dependent, we generated Sirt2–/– Nrf2 double-KO (DKO) mice by crossing Sirt2–/– mice with Nrf2–/– mice. As expected, non-heme iron and FTL levels were lower (Figure 7, A and B) and NRF2 and FPN1 protein levels were higher (Figure 7B) in the livers of Sirt2–/– mice compared with levels in Sirt2+/+ mice. However, deletion of Nrf2 in Sirt2–/– mice reversed the decrease in liver iron and FTL levels and the increase in FPN1 protein levels (Figure 7, A and B). Sirt2–/– Nrf2–/– DKO mice also exhibited a reversal of the expected increase in the expression of Tfrc and Fpn1 genes as well as antioxidant gene targets of NRF2 (Supplemental Figure 10, A and B). We also found increased nuclear levels of NRF2 in the primary hepatocytes of Sirt2–/– mice compared with levels in Sirt2+/+ mice (Figure 7C). The effects of SIRT2 and NRF2 on FTL and FPN1 protein expression levels was further confirmed in isolated primary mouse hepatocytes (Figure 7D). Furthermore, we investigated the effects of Nrf2 deletion on hepatocyte cell survival in the absence of SIRT2 and found that deletion of Nrf2 rescued the increased cell death in primary hepatocytes from Sirt2–/– mice under iron deficiency (Supplemental Figure 11). Collectively, these results provide in vivo data demonstrating that NRF2 mediates the iron deficiency response in the livers of Sirt2–/– mice.
Figure 7. Nrf2 deletion reverses iron deficiency in the livers of Sirt2–/– mice.
(A) Non-heme iron content in the livers of Sirt2+/+, Sirt2–/–, and Sirt2–/– Nrf2–/– mice (n = 5 per group). (B) NRF2, FPN1, and FTL protein levels in the livers of Sirt2+/+, Sirt2–/–, and Sirt2–/– Nrf2–/– mice (n = 3 per genotype). Bar graph representing densitometry result were shown in (C). (D) Nuclear NRF2 levels in primary hepatocytes isolated from Sirt2+/+, Sirt2–/–, and Sirt2–/– Nrf2–/– mice (n = 1 per genotype; cells were cultured in 5 or 6 individual wells). (E) TFRC1, FTL, and FPN1 protein levels in primary hepatocytes isolated from Sirt2+/+, Sirt2–/–, and Sirt2–/– Nrf2–/– mice (n = 1 per genotype; cells were cultured in 3 individual wells). Data are presented as the mean ± SEM. *P < 0.05, by ANOVA with Bonferroni’s correction for multiple comparisons.
Finally, we investigated the physiological significance of our findings by measuring SIRT2 levels in mouse and human liver tissue with iron overload. We first showed that a reduction in cellular iron leads to an increase in SIRT2, while iron overload has the opposite effect (Supplemental Figure 12). We then assessed the levels of SIRT2 in the livers of 3 different animal models of iron overload: mice with deletion of the hemochromatosis (Hfe) gene, mice with deletion of the hemojuvelin (Hjv) gene, and mice with dietary iron overload (31). In all of these models, we noted a significant decrease in hepatic SIRT2 protein levels in the iron-overloaded livers (Supplemental Figure 13).
We also studied the effects of cellular iron on SIRT2 levels in 2 sets of human liver samples: (a) patients with various degrees of transfusion-associated iron overload due to thalassemia, and (b) infants with fatal neonatal hemochromatosis (NH). In the first group, liver iron content (LIC) was assessed by MRI, and the samples were divided into low, intermediate, and high iron groups on the basis of their LIC scores. Immunohistochemical studies showed that intermediate and high LIC scores were associated with a significant decrease in SIRT2 levels (Figure 8A). Additionally, liver samples from patients with NH showed significantly higher iron (as assessed by Prussian blue staining) and lower SIRT2 levels (Figure 8B), indicating that iron overload is associated with lower SIRT2 levels in human liver. The decrease in SIRT2 likely serves as a compensatory mechanism for the cell to reduce its intracellular iron levels by increasing FPN1 expression.
Figure 8. SIRT2 is decreased with iron overload in human liver.
(A) SIRT2 levels in liver biopsies from human patients with various degrees of secondary iron overload. LIC was determined by MRI. A summary of the measurements is shown underneath the images. Scale bar: 100 μm. Bars in the summary graph represent the group mean. *P < 0.05, by ANOVA with Bonferroni’s correction for multiple comparisons. (B) Representative immunostaining showing SIRT2 expression in human liver tissue from infants with NH or from nondiseased control infants. Quantification of iron content measured by Prussian blue staining (top right) and SIRT2 levels by immunofluorescence intensity (bottom right) from NH patients (n = 3–5). Black scale bar: 100 μm; white scale bar: 50 μm. *P < 0.05, by Student’s t test. (C) Model of iron regulation by SIRT2 and NRF2. Data are presented as the mean ± SEM.
Discussion
In this study, we provide the first evidence to our knowledge that cellular iron is regulated by a member of the sirtuin family, SIRT2. We show that SIRT2 deacetylates NRF2 and that deacetylation of NRF2 by SIRT2 leads to a decrease in both total cellular and nuclear levels of NRF2 (through its degradation). The decrease in nuclear NRF2 is associated with a reduction in its transcription of the target gene Fpn1. The reduction in FPN1 results in less iron export and increased cellular iron levels (Figure 8C). We confirmed this pathway both in vitro and in vivo using Sirt2–/– and Sirt2–/– Nrf2–/– DKO mouse models. Finally, we show that this pathway has physiological significance, as disruption of SIRT2-mediated cellular iron regulation is associated with decreased cell viability in response to iron deficiency. Thus, SIRT2 regulates cellular iron, and this regulation is relevant to the cellular response to iron deficiency.
Iron is an essential molecule for many cellular processes including metabolism, inflammation, and cell-cycle progression. Excessive cellular iron, as seen in conditions such as hemochromatosis, is associated with increased oxidative stress and cellular injury. Conversely, iron deficiency is the most common nutrient deficiency in the world, and a number of homeostatic cellular systems have been developed to regulate total body iron distribution and cellular iron levels. In response to iron deficiency, the IRP1/2 system is activated and increases cellular iron availability through 2 mechanisms: increased cellular iron uptake via transferrin receptor upregulation and decreased iron storage and export via ferritin and ferroportin downregulation. Additionally, we recently showed that iron deficiency is associated with activation of an “iron conservation” pathway through TTP, which prioritizes iron availability for essential proteins. These 2 parallel pathways (i.e., the “iron acquisition” pathway through IRPs and the “iron conservation” pathway through TTP) work together to ensure cell survival in the setting of iron deficiency. In this study, we show that SIRT2 directly regulates cellular iron by altering its cellular export and that its expression is regulated by cellular iron content, thus providing evidence for yet another pathway to ensure normal cellular physiology and survival in response to iron deficiency. Cellular responses to inflammation, injury, and proliferation require ample iron levels; iron deficiency (which is needed to activate the IRP or TTP pathway) under these conditions may hamper the response to injury or inflammation, which could be detrimental to the cell. Our data suggest that the SIRT2 pathway inhibits iron export to ensure adequate cellular iron retention, independently of the IRP or TTP pathway.
While other sirtuins are localized to various cellular locations, SIRT2 is the only sirtuin that resides predominantly in the cytoplasm. This cellular localization of SIRT2 is consistent with previously identified cytosolic iron–sensing systems (32). However, it is possible that other sirtuins also regulate cellular iron levels. Previously, SIRT3, a mitochondrial sirtuin, was also shown to be involved in cellular iron regulation; however, this regulation occurred through an indirect mechanism involving ROS production (33). While we have demonstrated that SIRT2 is the first direct modulator of cellular iron levels in the SIRT family, it remains to be determined whether other sirtuins also contribute to iron homeostasis.
Deacetylation of NRF2 by SIRT2 leads to a decrease in both the total cellular and nuclear levels of NRF2 (through its degradation). This dual effect of SIRT2 on NRF2 ensures that when NRF2 is deacetylated, its transcriptional activity is reduced, and FPN1 levels and iron export are decreased.
Another interesting finding is that SIRT2 does not raise the levels of cellular iron import proteins, such as TFRC (Figure 7D). This could be due to a number of factors. First, TFRC regulation occurs at the mRNA stability level rather than at the transcriptional level; SIRT2 is not known to alter mRNA degradation mechanisms. Additionally, when a cell becomes metabolically active, increases its rate of proliferation, or is stimulated by inflammation, the cell prioritizes the retention of existing iron prior to competing for systemic iron, which is potentially in short supply (34). Thus, the preferred mechanism for increasing cellular iron levels is to withhold the iron that is already inside the cell rather than to compete for the scarce systemic iron.
NRF2 has been shown to play a role in cellular antioxidant activity (35). Thus, it is not surprising that it also plays a role in cellular iron regulation. Iron is a source of ROS, and increasing iron export, which in turn leads to a decrease in cellular iron, would result in a reduction in the oxidant effects of iron. Thus, the role of NRF2 in iron regulation is in line with its antioxidant effects.
In summary, we have shown that cells have developed a system by which SIRT2 regulates iron, a substrate that is needed to carry out its various functions. This regulation of iron by SIRT2 has an effect on cellular viability in response to iron deficiency. Thus, in addition to the IRP pathway, other essential mechanisms exist to ensure that iron is available for important cellular processes.
Methods
Cell culture and reagents.
MEFs and HEK293T cells were grown in complete DMEM (cellgro; Corning) supplemented with 10% FBS (Invitrogen, Thermo Fisher Scientific) and 1% penicillin-streptomycin (P/S). Each of the genetically modified MEF lines was compared with the matched WT MEFs obtained from a littermate control mouse. HEK293T cells were purchased from ATCC. Sirt2+/+ and Sirt2–/– MEFs were obtained from David Gius (25). ARNT-KO and corresponding WT MEFs were a gift of Celeste Simon (University of Pennsylvania, Philadelphia, Pennsylvania, USA). HepG2 cells were purchased from ATCC and kept in MEM (cellgro; Corning) with 10% FBS and 1% P/S. HepG2 cells were treated with 10 μM AGK2 (Sigma-Aldrich) for 24 hours or with DMSO as a control. All cells were maintained in a 37°C incubator with 5% CO2 and were 70% to 90% confluent when collected for analyses unless otherwise noted. Isolation and culturing of primary hepatocytes are described in the Supplemental Experimental Procedures. Sex-matched littermates of the mice were used for the isolation of hepatocytes. The following antibodies were used in the studies: SIRT2 (Sigma-Aldrich; S8847); NRF2 (Abcam; ab31163); FPN1 (Abcam; 85370); GAPDH (Abcam ab9485); tubulin (Abcam; ab1086); lamin A/C (Cell Signaling Technology; 2032); KEAP1 (Cell Signaling Technology; 7705); V5 (Sigma-Aldrich; V8137); acetylated lysine (ImmuneChem; 100514); Flag (Sigma-Aldrich; F1804); hypoxanthine phosphoribosyltransferase (HPRT) (Proteintech; 15059-1-AP); TATA-binding protein (TBP) (Abcam; 63766); and TFRC (Abcam; ab1086).
Mouse studies.
All animals were kept in accordance with standard animal care requirements and maintained in a 22°C room on a 12-hour light/12-hour dark cycle and received food and drinking water ad libitum. Nrf2–/– mice were purchased from The Jackson Laboratory. For organ harvesting, mice were anesthetized with a 250-mg/kg dose of freshly prepared tribromoethanol (Sigma-Aldrich), and the harvested organs were flash-frozen in liquid nitrogen.
Plasmids, transfection, and lentiviral transduction.
The clones of SIRT2 shRNA against human SIRT2 and Nrf2 shRNA against mouse Nrf2 were obtained from the Northwestern University High Throughput Analysis Laboratory (Evanston, Illinois, USA). An 8.4-kb Fpn promoter luciferase construct was provided by Martina Muckenthaler (University of Heidelberg, Heidelberg, Germany). Cells were cotransfected with the Fpn promoter constructs and the Renilla luciferase vector (pRL-SV40). Forty-eight hours after transfection, luciferase activity was determined using the Dual-Luciferase Reporter Assay System (Promega) according to the manufacturer’s instructions, and firefly luciferase activity was normalized to Renilla luciferase activity to correct for differences in transfection efficiency.
For transient transfection, cells were transfected using Lipofectamine 2000 Transfection Reagent (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s instructions. WT SIRT2 and the SIRT2 deacetylation–null mutant (lenti-Sirt2-DN), which has a mutation in the domain of deacetylase activity, were obtained from David Gius. For stable overexpression or knockdown, HEK 293T cells were transfected with lentiviral vectors along with the packaging vectors pSPAX and pMD2.G. Culture media were collected 48 hours later, filtered through a 0.2-μM filter, and used to infect cells in the presence of polybrene (Santa Cruz Biotechnology Inc.). The infected cells were selected for using puromycin (4 μg/ml).
mRNA stability assay.
MEFs or HepG2 cells were grown to approximately 80% confluence and incubated in the complete medium supplemented with 5 μM actinomycin D (Sigma-Aldrich) for 0 to 8 hours. No drop in cell viability was observed at the end of the treatment. RNA was collected and mRNA levels were analyzed by quantitative reverse transcription PCR (qRT-PCR). Data were normalized to the 0-hour time point.
Protein stability assay.
Endogenous or overexpressed NRF2 protein degradation was analyzed by a protein stability assay. Briefly, HepG2 cells transfected with WT NRF2 or mutants for 48 hours or MEFs were incubated with 100 μg/ml CHX (Sigma-Aldrich) for indicated duration (Figures 4 and 5). NRF2 protein levels were analyzed by Western blotting. The results from Western blot analysis were quantified by densitometry and normalized against the 0-minute time point. See complete unedited blots in the supplemental material.
Non-heme and heme iron assays.
Non-heme iron was quantified using a commercial Iron Assay Kit (BioVision). Briefly, cell or tissue lysate was mixed with an acidic solution to release protein-bound iron, followed by a reduction of iron to its ferrous form and incubation with a ferene S compound to produce a colored complex. The absorbance was then measured on a SpectraMax Plus (Molecular Devices) microplate reader at 593 nm and normalized to the protein concentration of each sample. Heme was quantified as described previously (36). Briefly, equal amounts of protein were mixed with 2 M oxalic acid and heated to 95°C for 30 minutes to release iron from heme and generate protoporphyrin IX. Samples were then centrifuged for 10 minutes at 1,000 ×g at 4°C to remove debris, and the fluorescence of the supernatant was assessed at 405 nm/600 nm on a SpectraMax Gemini fluorescence microplate reader (Molecular Devices).
55Fe content studies.
55Fe content studies were performed as previously described (5). Briefly, 55Fe (PerkinElmer) was conjugated with nitriloacetic acid (NTA) (Sigma-Aldrich), dissolved to a final concentration of 100 nM in complete serum-containing medium, and added into cells for 48 hours. Cells were then washed 3 times with ice-cold 500 mM BPS in PBS to remove the membrane-associated 55Fe, lysed with 1% Triton X-100 in TBS, centrifuged at 5,000 × g for 10 minutes at 4°C to remove debris, and protein content analyzed by bicinchoninic acid (BCA) assay. The radioactivity of each sample was determined on a Beckman scintillation counter and normalized to the protein content of each sample. For iron export experiments, HepG2 cells were first incubated with 100 nM 55Fe-NTA for 16 hours to allow for iron loading and then washed 4 times with PBS and replaced with complete serum-containing medium. Iron export was calculated as follows: (iron medium)/(iron medium + iron in cells) × 100, as previously described (37).
Cell death and cell viability studies.
Permeability to PI (Sigma-Aldrich) was used as a fluorescent signal for cell death (38). Cells were treated with deferoxamine (DFO) for 16 hours, washed once with HBSS, and costained with PI and Hoechst 33342 (Invitrogen, Thermo Fisher Scientific). Excess dye was washed away with HBSS, and images were taken using a Zeiss Axio Observer.Z1 fluorescence microscope. Data were analyzed using ImageJ (NIH). Cell viability studies were performed using the CellTiter 96 Non-Radioactive Cell Proliferation Assay (MTS) (Promega) according to the manufacturer’s instructions.
Mass spectrometry–based proteomics and analysis.
Flag-NRF2 expression plasmids were transfected into SIRT2-WT and SIRT2-KO MEFs, and transfected cells were treated with 1 μM trichostatin A (TSA) (Sigma-Aldrich) for 12 hours. Flag-NRF2 protein was immunoprecipitated with Flag-conjugated beads and eluted. Samples were separated by SDS-PAGE (4%–20% acrylamide) and stained with colloidal Coomassie Brilliant Blue G-250 (Invitrogen, Thermo Fisher Scientific). A prominent band present in each gel lane was excised, destained with 50% acetonitrile in 50 mM ammonium bicarbonate, finely diced, reduced with 45 mM DTT for 20 minutes at 55°C, alkylated with 100 mM iodoacetamide for 30 minutes at room temperature, and digested overnight at 37°C with trypsin. Peptides were extracted with 2 rounds of 60% acetonitrile and 0.1% trifluoroacetic acid and were then lyophilized to dryness. Digests were solubilized in 15 μl of 0.1% formic acid and analyzed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) (39).
In vitro deacetylation assay.
For the in vitro deacetylation assay of NRF2, HEK-293T cells were transiently cotransfected with V5-NRF2 and p300/CBP and then treated with 1 μM TSA and 20 mM NAM for 12 hours. Cells were harvested and lysed with IP lysis buffer (25 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 5% glycerol) in the presence of 1 μM TSA. The acetylated NRF2 proteins were immunoprecipitated with anti-V5 agarose beads (Biotool, http://www.bimake.com/), and immunocomplexes were washed 5 times with IP buffer and eluted by competition with V5 peptide (Biotool). The eluted acetylated NRF2 proteins were concentrated by spin columns (10,000 molecular weight cutoff [MWCO] polyethersulfone [PES]; Sartorius Stedim Biotech). For purification of SIRT2, pCMV-Flag-SIRT2 was transiently transfected into HEK-293T cells, and Flag-SIRT2 was purified from cell lysate using Flag antibody–conjugated beads. The acetylated NRF2 proteins were resuspended in 20 μl deacetylation reaction buffer (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 1 mM NAD), with or without the addition of purified SIRT2, for 3 hours at 30°C. Reactions were stopped by the addition of 2× SDS buffer (Bio-Rad), and samples were analyzed by Western blotting with anti–acetyl lysine antibodies (ImmuneChem).
Human tissue samples.
Paraffin-embedded human liver tissue sections were obtained from the Pathology Department at the Ann and Robert H. Lurie Children’s Hospital of Chicago. Tissues from 5 full-term newborn infants with clinical or/and pathological confirmation of gestational autoimmune liver disease leading to NH were compared against tissues from age-matched, nondiseased control infants who expired from perinatal asphyxia. Tissue sections with different intracellular iron concentrations were selected from a cohort study investigating iron chelation in patients with β-thalassemia (40). Intracellular iron concentration in these samples was determined by MRI imaging.
Statistics.
Continuous variables were tested for normal distribution using the Kolmogorov–Smirnov test. The Mann-Whitney U test was used to analyze variables with skewed distribution. The means between 2 categories were compared with an unpaired, 2-tailed Student’s t test. The means among 3 or more categories in the same experiment were compared using 1-way ANOVA with Bonferroni’s correction for multiple comparisons. Statistical significance was assumed when a null hypothesis could be rejected at a P value of less than 0.05. Statistical analysis was performed using GraphPad Prism 5 (GraphPad Software).
Study approval.
All animal studies were approved by the Institutional Animal Care and Use Committee at Northwestern University (Chicago, Illinois) and were performed in accordance with guidelines from the National Institutes of Health. For human tissue samples, the deidentified release of the decedents’ material was reviewed and approved by the IRB of the Ann and Robert H. Lurie Children’s Hospital of Chicago.
Author contributions
XY, AV, DG, and HA designed the research study. XY, SHP, JS, KTS, CC, MS, and HCC performed the experiments and data analysis. XY, LW, PB, and HA wrote the manuscript, and all authors commented on it. CLE provided the human tissue slides and commented on the manuscript. MK and SJ provided liver samples from mice with iron overload and commented on the manuscript. HA supervised the project.
Supplementary Material
Acknowledgments
XY is supported by American Heart Association grant 14POST20490097. MK is supported by NIH grant DK080706. HA is supported by NIH grants K02 HL107448, R01 HL127646, and 1PO1 HL108795. DG is supported by NIH grants 2R01CA152601-A1, 1R01CA152799-01A1, 1R01CA168292-01A1; the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust; the Zell Family Foundation, and the Avon Foundation for Breast Cancer Research.
Footnotes
Conflict of interest: H. Ardehali receives consulting honoraria from the Gerson Lehrman Group.
Reference information:J Clin Invest. 2017;127(4):1505–1516.https://doi.org/10.1172/JCI88574.
Contributor Information
Xiaoyan Yang, Email: xyxiaoyanyang@gmail.com.
Seong-Hoon Park, Email: romad96@gmail.com.
Hsiang-Chun Chang, Email: hsiang-chun-chang@northwestern.edu.
Athanassios Vassilopoulos, Email: athanasios.vasilopoulos@northwestern.edu.
Chunlei Chen, Email: cchen95@hawk.iit.edu.
Meng Shang, Email: meng.shang@northwestern.edu.
Supak Jenkitkasemwong, Email: supakj@ufl.edu.
Mitchell Knutson, Email: mknutson@ufl.edu.
David Gius, Email: david.gius@northwestern.edu.
Hossein Ardehali, Email: h-ardehali@northwestern.edu.
References
- 1.De Domenico I, McVey Ward D, Kaplan J. Regulation of iron acquisition and storage: consequences for iron-linked disorders. Nat Rev Mol Cell Biol. 2008;9(1):72–81. doi: 10.1038/nrm2295. [DOI] [PubMed] [Google Scholar]
- 2.Hentze MW, Muckenthaler MU, Galy B, Camaschella C. Two to tango: regulation of Mammalian iron metabolism. Cell. 2010;142(1):24–38. doi: 10.1016/j.cell.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 3.Wang J, Pantopoulos K. Regulation of cellular iron metabolism. Biochem J. 2011;434(3):365–381. doi: 10.1042/BJ20101825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peyssonnaux C, et al. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs) J Clin Invest. 2007;117(7):1926–1932. doi: 10.1172/JCI31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bayeva M, et al. mTOR regulates cellular iron homeostasis through tristetraprolin. Cell Metab. 2012;16(5):645–657. doi: 10.1016/j.cmet.2012.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370(Pt 3):737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sauve AA, Wolberger C, Schramm VL, Boeke JD. The biochemistry of sirtuins. Annu Rev Biochem. 2006;75:435–465. doi: 10.1146/annurev.biochem.74.082803.133500. [DOI] [PubMed] [Google Scholar]
- 8.Du J, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334(6057):806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haigis MC, et al. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell. 2006;126(5):941–954. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- 10.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16(10):4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pan PW, Feldman JL, Devries MK, Dong A, Edwards AM, Denu JM. Structure and biochemical functions of SIRT6. J Biol Chem. 2011;286(16):14575–14587. doi: 10.1074/jbc.M111.218990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baur JA, Ungvari Z, Minor RK, Le Couteur DG, de Cabo R. Are sirtuins viable targets for improving healthspan and lifespan? Nat Rev Drug Discov. 2012;11(6):443–461. doi: 10.1038/nrd3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Preyat N, Leo O. Sirtuin deacylases: a molecular link between metabolism and immunity. J Leukoc Biol. 2013;93(5):669–680. doi: 10.1189/jlb.1112557. [DOI] [PubMed] [Google Scholar]
- 14.Gomes P, Outeiro TF, Cavadas C. Emerging role of sirtuin 2 in the regulation of mammalian metabolism. Trends Pharmacol Sci. 2015;36(11):756–768. doi: 10.1016/j.tips.2015.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Jiang W, et al. Acetylation regulates gluconeogenesis by promoting PEPCK1 degradation via recruiting the UBR5 ubiquitin ligase. Mol Cell. 2011;43(1):33–44. doi: 10.1016/j.molcel.2011.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang F, Tong Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARgamma. Mol Biol Cell. 2009;20(3):801–808. doi: 10.1091/mbc.E08-06-0647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rothgiesser KM, Erener S, Waibel S, Lüscher B, Hottiger MO. SIRT2 regulates NF-κB dependent gene expression through deacetylation of p65 Lys310. J Cell Sci. 2010;123(pt 24):4251–4258. doi: 10.1242/jcs.073783. [DOI] [PubMed] [Google Scholar]
- 18.Serrano L, et al. The tumor suppressor SirT2 regulates cell cycle progression and genome stability by modulating the mitotic deposition of H4K20 methylation. Genes Dev. 2013;27(6):639–653. doi: 10.1101/gad.211342.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Donmez G, Outeiro TF. SIRT1 and SIRT2: emerging targets in neurodegeneration. EMBO Mol Med. 2013;5(3):344–352. doi: 10.1002/emmm.201302451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park SH, et al. SIRT2 is a tumor suppressor that connects aging, acetylome, cell cycle signaling, and carcinogenesis. Transl Cancer Res. 2012;1(1):15–21. [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang C. Essential functions of iron-requiring proteins in DNA replication, repair and cell cycle control. Protein Cell. 2014;5(10):750–760. doi: 10.1007/s13238-014-0083-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wessling-Resnick M. Iron homeostasis and the inflammatory response. Annu Rev Nutr. 2010;30:105–122. doi: 10.1146/annurev.nutr.012809.104804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fischer-Fodor E, Miklasova N, Berindan-Neagoe I, Saha B. Iron, inflammation and invasion of cancer cells. Clujul Med. 2015;88(3):272–277. doi: 10.15386/cjmed-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ward RJ, Zucca FA, Duyn JH, Crichton RR, Zecca L. The role of iron in brain ageing and neurodegenerative disorders. Lancet Neurol. 2014;13(10):1045–1060. doi: 10.1016/S1474-4422(14)70117-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim HS, et al. SIRT2 maintains genome integrity and suppresses tumorigenesis through regulating APC/C activity. Cancer Cell. 2011;20(4):487–499. doi: 10.1016/j.ccr.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marro S, et al. Heme controls ferroportin1 (FPN1) transcription involving Bach1, Nrf2 and a MARE/ARE sequence motif at position –7007 of the FPN1 promoter. Haematologica. 2010;95(8):1261–1268. doi: 10.3324/haematol.2009.020123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nairz M, et al. Nitric oxide-mediated regulation of ferroportin-1 controls macrophage iron homeostasis and immune function in Salmonella infection. J Exp Med. 2013;210(5):855–873. doi: 10.1084/jem.20121946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hybertson BM, Gao B, Bose SK, McCord JM. Oxidative stress in health and disease: the therapeutic potential of Nrf2 activation. Mol Aspects Med. 2011;32(4–6):234–246. doi: 10.1016/j.mam.2011.10.006. [DOI] [PubMed] [Google Scholar]
- 29.Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1–Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16(2):123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- 30.Kawai Y, Garduño L, Theodore M, Yang J, Arinze IJ. Acetylation-deacetylation of the transcription factor Nrf2 (nuclear factor erythroid 2-related factor 2) regulates its transcriptional activity and nucleocytoplasmic localization. J Biol Chem. 2011;286(9):7629–7640. doi: 10.1074/jbc.M110.208173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jenkitkasemwong S, et al. SLC39A14 is required for the development of hepatocellular iron overload in murine models of hereditary hemochromatosis. Cell Metab. 2015;22(1):138–150. doi: 10.1016/j.cmet.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muhlenhoff U, et al. Cytosolic monothiol glutaredoxins function in intracellular iron sensing and trafficking via their bound iron-sulfur cluster (vol 12, pg 373, 2010) Cell Metab. 2014;20(4):696. doi: 10.1016/j.cmet.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeong SM, Lee J, Finley LW, Schmidt PJ, Fleming MD, Haigis MC. SIRT3 regulates cellular iron metabolism and cancer growth by repressing iron regulatory protein 1. Oncogene. 2015;34(16):2115–2124. doi: 10.1038/onc.2014.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao R, Georgieff MK. Iron in fetal and neonatal nutrition. Semin Fetal Neonatal Med. 2007;12(1):54–63. doi: 10.1016/j.siny.2006.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ma Q. Role of nrf2 in oxidative stress and toxicity. Annu Rev Pharmacol Toxicol. 2013;53:401–426. doi: 10.1146/annurev-pharmtox-011112-140320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward JH, Jordan I, Kushner JP, Kaplan J. Heme regulation of HeLa cell transferrin receptor number. J Biol Chem. 1984;259(21):13235–13240. [PubMed] [Google Scholar]
- 37.Bories G, et al. Liver X receptor activation stimulates iron export in human alternative macrophages. Circ Res. 2013;113(11):1196–1205. doi: 10.1161/CIRCRESAHA.113.301656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brana C, Benham C, Sundstrom L. A method for characterising cell death in vitro by combining propidium iodide staining with immunohistochemistry. Brain Res Brain Res Protoc. 2002;10(2):109–114. doi: 10.1016/S1385-299X(02)00201-5. [DOI] [PubMed] [Google Scholar]
- 39.Vassilopoulos A, et al. SIRT3 deacetylates ATP synthase F1 complex proteins in response to nutrient- and exercise-induced stress. Antioxid Redox Signal. 2014;21(4):551–564. doi: 10.1089/ars.2013.5420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vichinsky E, et al. A randomised comparison of deferasirox versus deferoxamine for the treatment of transfusional iron overload in sickle cell disease. Br J Haematol. 2007;136(3):501–508. doi: 10.1111/j.1365-2141.2006.06455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.