Abstract
Alloimmune T cell responses induce graft-versus-host disease (GVHD), a serious complication of allogeneic bone marrow transplantation (allo-BMT). Although Notch signaling mediated by Delta-like 1/4 (DLL1/4) Notch ligands has emerged as a major regulator of GVHD pathogenesis, little is known about the timing of essential Notch signals and the cellular source of Notch ligands after allo-BMT. Here, we have shown that critical DLL1/4-mediated Notch signals are delivered to donor T cells during a short 48-hour window after transplantation in a mouse allo-BMT model. Stromal, but not hematopoietic, cells were the essential source of Notch ligands during in vivo priming of alloreactive T cells. GVHD could be prevented by selective inactivation of Dll1 and Dll4 in subsets of fibroblastic stromal cells that were derived from chemokine Ccl19-expressing host cells, including fibroblastic reticular cells and follicular dendritic cells. However, neither T cell recruitment into secondary lymphoid organs nor initial T cell activation was affected by Dll1/4 loss. Thus, we have uncovered a pathogenic function for fibroblastic stromal cells in alloimmune reactivity that can be dissociated from their homeostatic functions. Our results reveal what we believe to be a previously unrecognized Notch-mediated immunopathogenic role for stromal cell niches in secondary lymphoid organs after allo-BMT and define a framework of early cellular and molecular interactions that regulate T cell alloimmunity.
Introduction
Allogeneic bone marrow transplantation (allo-BMT) can cure hematological malignancies and other blood disorders. However, alloimmune T cell responses arising against foreign tissue antigens can trigger major complications such as graft-versus-host disease (GVHD) after allo-BMT (1–3). At the onset of GVHD, donor T cells are exposed to host tissue alloantigens in a highly inflammatory environment, inducing potent T cell immunoreactivity and subsequent pathogenicity. Current GVHD prophylactic and therapeutic strategies act through global immunosuppression and thus diminish both beneficial and detrimental aspects of T cell alloreactivity. Efforts to develop new selective therapies to dampen GVHD have focused on early microenvironmental signals to donor alloreactive T cells (4). Many of these signals, which include alloantigens, costimulatory ligands, and local inflammatory mediators, have been assumed to derive from hematopoietic antigen-presenting cells (APCs) (5–7). However, recent work demonstrated that CD4+ T cell–mediated alloresponses can occur in the absence of hematopoietic APCs as a source of alloantigens (8–10), suggesting that our current understanding of key early cellular and molecular events that drive donor T cell–mediated GVHD is incomplete.
The Notch pathway has emerged as a new, attractive therapeutic target to control the deleterious effects of T cell alloimmunity (11–17). Notch signaling is a conserved cell-to-cell communication pathway mediated by interactions between NOTCH1-4 receptors and their ligands Delta-like 1/3/4 (DLL1/3/4) or JAGGED1/2 (JAG1/2) (18, 19). During GVHD, DLL1/4 ligands in the host engage NOTCH1/2 receptors in T cells, and transient systemic blockade of DLL1/4 Notch ligands with neutralizing antibodies results in long-term protection from GVHD (14). Despite the central role of Notch signaling in alloreactivity, the timing of critical Notch signals, the cellular source of Notch ligands, and the microanatomical context in which alloreactive T cells are exposed to Notch signaling in vivo remain unknown.
Early studies showed that hematopoietic APCs such as DCs can express DLL1 and DLL4 ligands in a TLR-inducible manner (20, 21). These observations led to the widely accepted concept that hematopoietic APCs can simultaneously deliver antigen and Notch ligands to modulate T cell function. In vitro studies supported this model, as TLR agonist–stimulated antigen-pulsed DCs induced naive T cells to differentiate in a Notch-regulated manner (21, 22). Similarly, a subpopulation of CD11c+DLL4hi DCs was capable of delivering Notch signals to alloreactive T cells in mixed lymphocyte reactions when purified from GVHD animal models (23). However, the in vivo relevance of APC-derived Notch signals has not been rigorously tested, and their importance has been inferred indirectly on the basis of their capacity to modulate T cell responses in vitro. Nonhematopoietic cells also express Notch ligands in multiple contexts, including in primary and secondary lymphoid organs (SLOs). In the thymus, FOXN1+ thymic epithelial cells act as nonredundant transducers of DLL4-mediated signals during T cell development (24–26). Blood and lymphatic endothelial cells (BECs and LECs) express high levels of DLL1 and DLL4 (27–31). Finally, genetic studies identified fibroblastic stromal cells in SLOs as nonredundant sources of DLL1-mediated Notch signals to marginal-zone B cells and DCs that express high levels of endothelial cell–specific adhesion molecule high (ESAMhi DCs), as well as of DLL4-mediated signals to follicular helper T cells (32). Thus, multiple cellular sources have the potential to deliver Notch signals to T cells in vivo after allo-BMT, making it unclear whether critical signals are delivered in a defined microanatomical niche, and by hematopoietic or stromal cells.
To address these questions, we used a combination of systemic neutralizing antibodies and loss-of-function genetics to interrogate the in vivo spatial and temporal requirements for DLL1/4-mediated Notch signaling during GVHD. Surprisingly, we found that essential Notch signals were delivered to incoming T cells within 2 days of allo-BMT and that both donor and host hematopoietic cells were dispensable as a source of Notch ligands driving acute GVHD. In contrast, we found that subsets of nonhematopoietic fibroblastic stromal cells lineage traced with a Ccl19-Cre transgene functioned as the essential cellular source of Delta-like Notch ligands after allo-BMT. Interference with Notch ligands in fibroblastic stromal cells had selective effects on T cell alloreactivity and did not impair other functions of these cells in immune homeostasis. These findings change our understanding of the key early cellular and molecular events that condition the outcome of T cell alloimmunity. In addition, they pave the way toward the development of targeted therapeutic approaches to block Notch signaling and other stromal niche–derived pathogenic signals in GVHD and other T cell–mediated immune disorders.
Results
Early DLL1/4-mediated Notch signals drive T cell alloreactivity during acute GVHD.
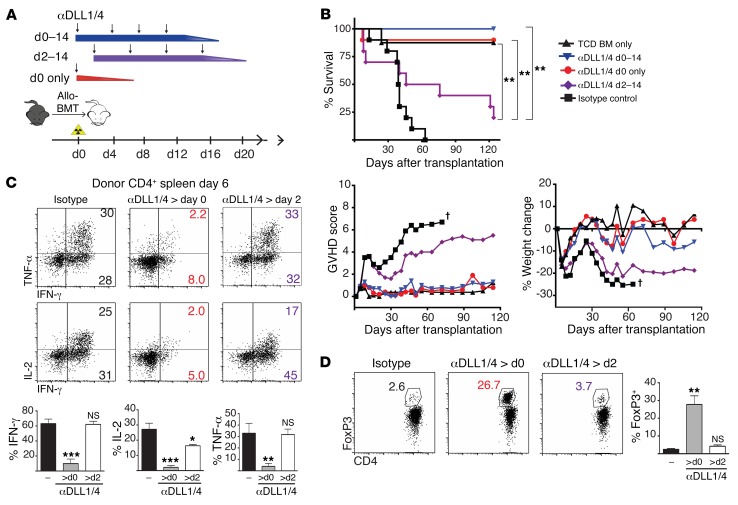
To understand the temporal requirement for Notch signaling after allo-BMT, we used neutralizing antibodies against DLL1 and DLL4 Notch ligands in an irradiation-dependent MHC-mismatched mouse allo-BMT model (Figure 1A). A single injection of DLL1- and DLL4-neutralizing antibodies before allo-BMT was sufficient for long-term protection from GVHD lethality and morbidity, decreased production of T cell proinflammatory cytokines by CD4+ and CD8+ T cells, and expanded FoxP3+ Tregs (Figure 1, B–D, Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/JCI89535DS1). In contrast, delayed initiation of antibody treatment by only 2 days resulted in loss of clinical protection, persistent IFN-γ and TNF-α production, and no increase in Treg numbers. DLL1/4 blockade inhibited cytokine production and Notch target gene expression in T cells recovered from both spleens and lymph nodes (LNs) (Supplemental Figure 1, B and C). Notch target gene expression was decreased in proliferating donor-derived alloreactive T cells isolated during the temporal window of Notch sensitivity (Supplemental Figure 1, D and E). These data identify an early pulse of Notch signaling delivered within 2 days of allo-BMT that programs T cells to a pathogenic state of reactivity.
Figure 1. An early pulse of Notch signaling is critical to drive pathogenic T cell alloreactivity after BMT.
(A) Dosing schedule of systemic neutralizing antibodies against DLL1 and DLL4 Notch ligands. (B) Survival, GVHD score, and weight of lethally irradiated (8.5 Gy) BALB/c mice transplanted with 5 × 106 T cell–depleted (TCD) B6 BM or 5 × 106 TCD B6 BM plus 5 × 106 allogeneic B6 splenocytes. Isotype control versus anti-DLL1/4 antibodies were injected i.p., as shown in A (n = 10 mice/group). (C) Intracellular cytokine production by donor CD4+ T cells after anti-CD3/CD28 restimulation on day 6 after transplantation (n = 5 mice/group). (D) Intracellular FoxP3 in donor CD4+ T cells on day 6 (n = 5 mice/group). *P < 0.05, **P < 0.01, ***P < 0.001, and NS = P > 0.05, by unpaired, 2-tailed Student’s t test with Sidak’s correction for multiple comparisons. Data are representative of at least 4 experiments; error bars indicate SD.
Donor and host hematopoietic cells are dispensable sources of Notch ligands during GVHD.
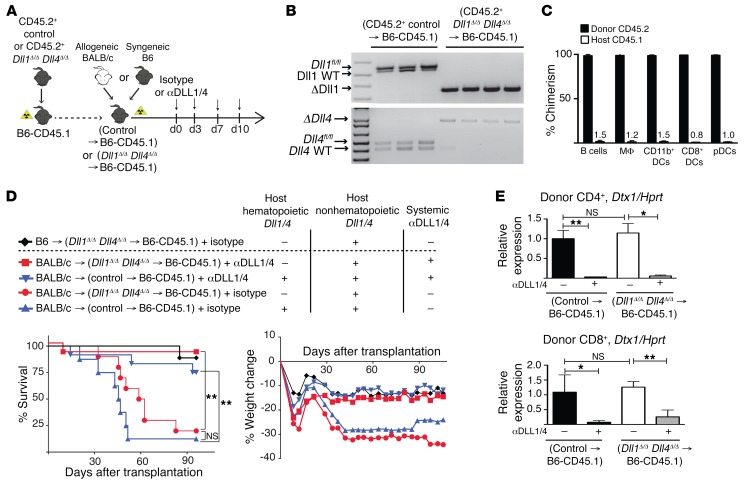
The early window of sensitivity to Notch inhibition suggested that T cells might receive Notch signals from residual host hematopoietic APCs, which can express both alloantigens and Notch ligands (21–23, 33–35). To test whether hematopoietic APCs were responsible for delivering Notch signals to donor alloreactive T cells, we generated BM chimeras lacking DLL1/4 only in hematopoietic cells with poly(I:C)-induced TgMx1-Cre+ Dll1Δ/Δ Dll4Δ/Δ BM (designated as Dll1Δ/Δ Dll4Δ/Δ→B6-CD45.1) (Figure 2A). This strategy led to efficient Cre-mediated recombination and high donor chimerism in all hematopoietic APCs (Figure 2, B and C). Allo-BMT into Dll1Δ/Δ Dll4Δ/Δ→B6-CD45.1 chimeras as compared with control BM chimeras did not protect mice from GVHD mortality or morbidity (Figure 2D). In contrast, systemic antibody-mediated DLL1/DLL4 blockade protected both Dll1Δ/Δ Dll4Δ/Δ→B6-CD45.1 and control BM chimeras from GVHD lethality. Consistent with persistent exposure to DLL1/4 ligands in these mice, donor-derived T cells expressed equivalent amounts of Notch target gene transcripts in Dll1Δ/Δ Dll4Δ/Δ→B6-CD45.1 and control chimeras, but transcript abundance remained sensitive to systemic anti-DLL1/4 antibodies (Figure 2E). Thus, host hematopoietic cells were dispensable as a source of Notch ligands after allo-BMT.
Figure 2. Host hematopoietic cells are dispensable as cellular sources of DLL1/4 Notch ligands in acute GVHD.
(A) Experimental strategy. BM chimeras were generated via transplantation of syngeneic B6-CD45.2+ poly(I:C)-induced TgMx1-Cre– littermate control or TgMx1-Cre+ Dll1Δ/Δ Dll4Δ/Δ BM into irradiated B6-CD45.1 recipients. After reestablishment of steady-state hematopoiesis 12 weeks later, BM chimeras were subjected to a second syngeneic or allogeneic transplant, with or without systemic anti-DLL1/4 blockade. (B) Quantification of Dll1 and Dll4 inactivation in sort-purified Gr1+CD11b+ blood myeloid cells from BM chimeras 12 weeks after transplantation (PCR). In this particular experiment, control BM chimeras were generated from poly(I:C)-induced TgMx1-Cre– Dll1fl/+ Dll4fl/+ donor mice. Each lane represents an individual mouse. (C) Donor chimerism (frequency of CD45.2+ donor cells) in the indicated splenic cell populations 12 weeks after transplantation. MΦ, macrophages; pDCs, plasmacytoid DCs. (D) Survival and weight loss of lethally irradiated (11 Gy) BM chimeras transplanted with 8 × 106 TCD BM plus 30 × 106 B6 splenocytes (syngeneic control) or 30 × 106 allogeneic BALB/c splenocytes (allo-BMT). Isotype control or anti-DLL1/4 antibodies were injected i.p. on days 0, 3, 7, and 10 (n = 10 mice/group). (E) Abundance of Dtx1 Notch target gene transcripts (qRT-PCR) in sort-purified donor CD4+ T cells and CD8+ cells on day 6 (n = 5 mice/group). *P < 0.05, **P < 0.01, and NS = P > 0.05, by unpaired, 2-tailed Student’s t test with Sidak’s correction for multiple comparisons. Data are representative of at least 2 experiments; error bars indicate SD.
To rule out redundant DLL1/4 expression in donor and host hematopoietic cells, we backcrossed TgMx1–Cre+ Dll1fl/fl Dll4fl/fl mice onto the BALB/c background and generated BM chimeras with poly(I:C)-induced TgMx1–Cre+ Dll1Δ/Δ Dll4Δ/Δ donors (Dll1Δ/Δ Dll4Δ/Δ→BALB/c) (Supplemental Figure 2A). Transplantation of Dll1Δ/Δ Dll4Δ/Δ→B6-CD45.1 donor cells into Dll1Δ/Δ Dll4Δ/Δ→BALB/c recipients (designated as Dll1Δ/Δ Dll4Δ/Δ→Dll1Δ/Δ Dll4Δ/Δ) had no impact on T cell cytokine production or Treg frequencies (Supplemental Figure 2, B–D), despite sensitivity to systemic DLL1/DLL4 blockade. Donor-derived T cells from Dll1Δ/Δ Dll4Δ/Δ→Dll1Δ/Δ Dll4Δ/Δ recipient mice retained abundant Notch target gene transcripts (Supplemental Figure 2E). Therefore, both donor and host hematopoietic cells were dispensable sources of DLL1/4 Notch ligands during GVHD.
Ccl19-Cre+ stromal cells are critical sources of Notch ligands during GVHD.
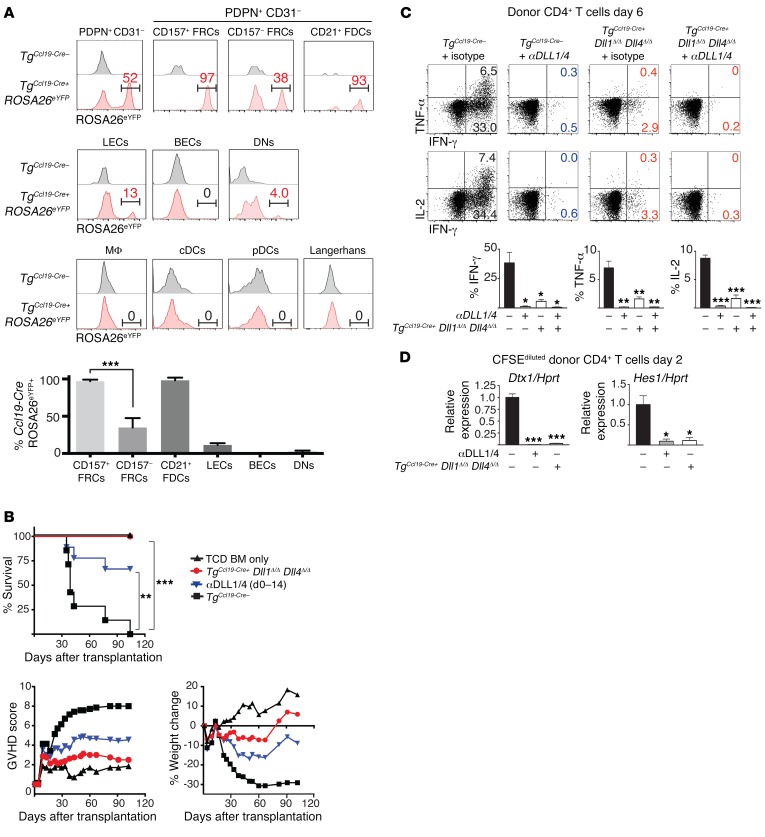
Among multiple radioresistant nonhematopoietic cells that express Notch ligands in SLOs, fibroblastic stromal cells lineage traced with Ccl19-Cre act as a non-motile source of Notch ligands to immune cells in steady-state conditions (32). To assess the importance of these cells during allo-BMT, we conditionally inactivated Dll1 and Dll4 with Ccl19-Cre (hereafter referred to as TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ) (36–38). After irradiation and allo-BMT, lineage tracing by Ccl19-Cre was detected in PDPN+CD31– fibroblastic stromal cells, as assessed by Cre-inducible enhanced yellow fluorescent protein (eYFP) expression from a ROSA26eYFP allele (Figure 3A and Supplemental Figure 3A). Further characterization of Ccl19-Cre activity within cellular subsets of PDPN+CD31– stromal cells showed eYFP in more than 95% of cells from the CD157hiCD21– subset (consistent with fibroblastic reticular cells [FRCs]) and from the CD157hiCD21+ subset (consistent with follicular dendritic cells [FDCs]). TgCcl19-Cre also lineage traced a smaller fraction of PDPN+CD31+ LECs and very few PDPN–CD31– stromal cells. In contrast, TgCcl19–Cre was inactive in PDPN–CD31+ BECs and in all hematopoietic cells, including professional APCs in SLOs and Langerhans cells isolated from the epidermis (Figure 3A and Supplemental Figure 3, A–D). Thus, TgCcl19-Cre had a pattern of activity suitable for inactivation of Dll1/4 selectively in subsets of fibroblastic stromal cells (including FRCs and FDCs) and in a small fraction of LECs.
Figure 3. Ccl19-Cre+ lineage–traced stromal cells are the critical cellular source of DLL1/4 Notch ligands during acute GVHD.
(A) LNs were collected on day 1.5 after transplantation from lethally irradiated TgCcl19-Cre+ ROSA26eYFP mice receiving allogeneic BALB/c splenocytes and enzymatically digested. (A, top) eYFP expression in LN-resident bulk fibroblastic stromal cells (PDPN+CD31–) as well as subfractionated CD157+ FRCs, CD157– FRCs, and CD21+ FDCs. (A, middle) eYFP in LECs, BECs, PDPN–CD31– stromal cells (DNs). (A, bottom) eYFP in macrophages, conventional DCs (cDCs), pDCs, and skin-derived Langerhans cells. Bars in histograms define gating for eYFP+ cells, and numbers indicate the percentage of gated eYFP+ cells within parental cell populations, as identified by flow cytometric analysis. Bar graph in A shows the mean percentage of eYFP expression in each indicated nonhematopoietic subset (n = 4 mice/group; error bars indicate SD). (B) Survival, GVHD score, and weight of lethally irradiated (12 Gy) littermate control TgCcl19-Cre– or TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ mice that were transplanted with 10 × 106 TCD BM only or 10 × 106 TCD BM plus 20 × 106 allogeneic BALB/c splenocytes. Isotype control or anti-DLL1/4–neutralizing antibodies were injected i.p. on days 0, 3, 7, and 10 (n = 10 mice/group). (C) Intracellular cytokines in donor CD4+ cells after anti-CD3/CD28 restimulation on day 6 (n = 5 mice/group). (D) Relative abundance of Dtx1 and Hes1 Notch target gene transcripts in donor CD4+ T cells sort purified from TgCcl19-Cre– plus isotype control, TgCcl19-Cre– plus anti-DLL1/4, or TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ recipient mice on day 2 after transplantation (n = 5 mice/group). *P < 0.05, **P < 0.01, and ***P < 0.001, by unpaired, 2-tailed Student’s t test with Sidak’s correction for multiple comparisons. Data are representative of at least 5 experiments; error bars indicate SD.
TgCcl19-Cre+Dll1Δ/ΔDll4Δ/Δ mice were highly protected from GVHD lethality and morbidity (Figure 3B). Markedly fewer donor T cells from TgCcl19-Cre+ Dll1Δ/ΔDll4Δ/Δ recipients produced proinflammatory cytokines, as observed with systemic DLL1/4 blockade (Figure 3C and Supplemental Figure 4). Notch target gene transcripts were profoundly decreased in donor-derived T cells isolated from TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ recipients (Figure 3D). As shown with anti-DLL1/4 antibodies (Supplemental Figure 1, B and C), the effects of Ccl19-Cre–mediated Dll1/4 loss were observed in the spleen (which lacks LECs) as well as in LNs (Figure 3, C and D, and data not shown). Thus, nonhematopoietic cells lineage traced by Ccl19-Cre were the dominant source of DLL1/DLL4 Notch ligands encountered in vivo by donor T cells after allo-BMT.
T cell cytokine production was preserved after allo-BMT into TgCcl19–Cre+ Dll1+/+ Dll4+/+ control recipients, ruling out Cre toxicity (Supplemental Figure 5). To assess whether DLL1/4 blockade might impact GVHD through effects that are independent of T cells, we established an experimental system that preserved stroma-derived Notch signals in T cells, but removed Notch-dependent non–T cells. Past work showed that NOTCH2 receptors engaged by stromal DLL1/4 ligands control the development and/or maintenance of ESAMhi DCs, CD8+ DCs, and marginal-zone B cells in the spleen (39–41). We generated syngeneic BM chimeras using poly(I:C)-induced TgMx1-Cre+ Notch2Δ/Δ or control TgMx1-Cre– Notch2fl/fl progenitors (Supplemental Figure 6A). These BM chimeras lacked marginal-zone B cells and had decreased numbers of NOTCH2-dependent CD8+ and ESAMhi DCs, as expected (Supplemental Figure 6B and refs. 39–41). We subjected these BM chimeras to allogeneic BMT with MHC-mismatched BALB/c BM and purified T cells. With this approach, recipient mice lacked NOTCH2-dependent host populations but had preserved stromal DLL1/4 expression (Supplemental Figure 6, A, C, and D). Systemic DLL1/4 inhibition blocked the production of IL-2, IFN-γ, and TNF-α by donor-derived T cells after allo-BMT. In contrast, TgMx1-Cre+ Notch2Δ/Δ chimeras showed preserved cytokine production in LNs and increased, but not decreased, production in donor-derived splenic T cells. These findings indicate that loss of NOTCH2-dependent host splenic cells cannot indirectly explain the impact of DLL1/4 inhibition during GVHD, which is consistent with the observed direct effects of DLL1/4 on donor T cells.
Genetic inactivation of DLL1/4 ligands within Ccl19-Cre+ stromal cells preserves immune homeostasis and classical functions of FRCs.
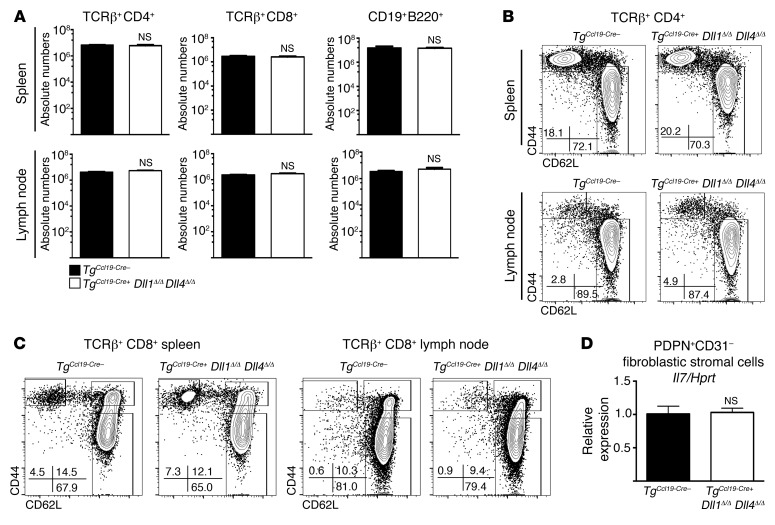
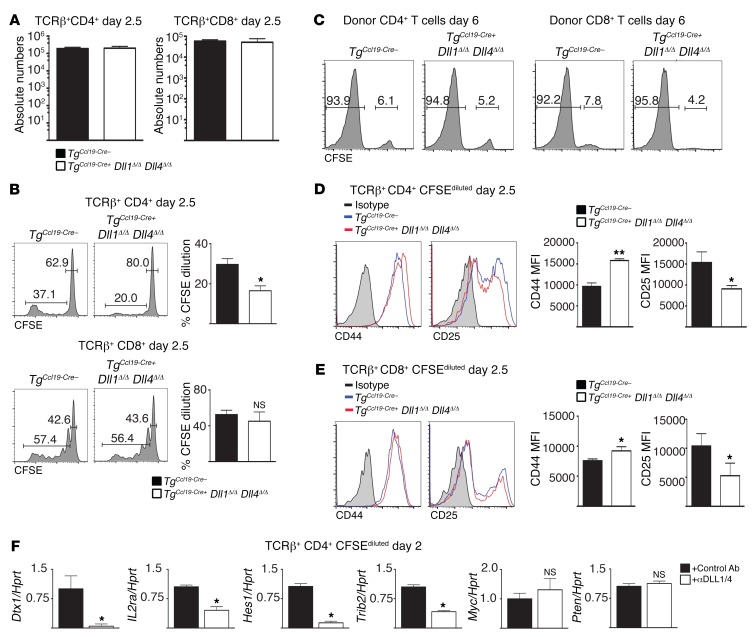
Among fibroblastic stromal cells, FRCs provide survival and migration cues to naive T cells (42), form conduits for antigen trafficking (43), and support the overall SLO infrastructure (37, 44–48). Physical loss of FRCs disrupts immune homeostasis, leading to profoundly impaired immune responses (37). To determine whether these FRC functions were preserved in TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ mice, we studied these mice during steady state and in the peri-transplantation period. SLOs from nonirradiated TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ mice had normal lymphocyte numbers, naive/memory T cell distribution, and Il7 expression in FRCs (Figure 4, A–D). After allo-BMT, normal numbers of donor T cells were recovered upon Dll1/4 inactivation in FRCs and showed no impact on T cell homing or survival (Figure 5A). Donor T cell proliferation was maintained in TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ recipients (Figure 5, B and C), or was only mildly decreased at early time points for CD4+ T cells (Figure 5B). Donor CD4+ and CD8+ T cells normally upregulated the early activation markers CD44 (Figure 5, D and E) and CD69 (not shown). CD25 was decreased in TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ hosts (Figure 5, D and E), consistent with reports that Notch can regulate IL-2 receptor subunit α (Il2ra) expression in T cells (49–51). We also observed a decreased abundance of the Notch target gene transcripts deltex 1, E3 ubiquitin ligase (Dtx1), tribbles pseudokinase 2 (Trib2), and hairy and enhancer of split 1 (Hes1) in donor CD4+ T cells (Figure 5F). However, phosphatase and tensin homolog (Pten) mRNA was unchanged, indicating that Hes1-mediated Pten repression did not operate in alloreactive T cells (52, 53). Other genes, including Myc, that were previously identified as Notch transcriptional targets in T cell acute lymphoblastic leukemia were expressed at normal levels (Figure 5F and refs. 54, 55). Altogether, DLL1/4 loss in FRCs maintained T cell recruitment and initial activation, which is consistent with our observation of preserved classical FRC functions but selective loss of DLL1/4-mediated instructive signals to incoming T cells.
Figure 4. Ccl19-Cre–mediated Dll1 and Dll4 inactivation preserves lymphocyte numbers and distribution in SLOs at steady state.
(A) Absolute numbers of CD4+ T cells, CD8+ T cells, and B cells in spleens and LNs of B6 littermate control TgCcl19-Cre– or TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ mice at steady state (n = 5 mice/group). (B and C) CD62L and CD44 expression in CD4+ (B) and CD8+ (C) T cells from spleens and LNs of TgCcl19-Cre– or TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ mice at steady state. (D) Abundance of Il7 transcripts (qRT-PCR) in sort-purified PDPN+CD31– fibroblastic stromal cells from TgCcl19-Cre– or TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ mice (n = 5 mice/group). NS = P > 0.05, by unpaired, 2-tailed Student’s t test. Data are representative of at least 3 experiments; error bars indicate SD.
Figure 5. Ccl19-Cre–mediated Dll1 and Dll4 inactivation does not impair T cell recruitment or proliferation in SLOs after irradiation, but selectively affects Notch target gene transcripts.
(A–E) Absolute numbers (A), proliferation (CFSE dilution) on day 2.5 (B) versus day 6 after transplantation (C), and expression of activation markers. Bars in histograms define gating for proliferated CFSElow and unproliferated CFSEhi cells, and numbers indicate the percentage of gated cells among parental cell populations, as identified by flow cytometric analysis. Bar graphs represent the mean percentage of proliferated (CFSElow) cells in each population (n=5 mice/group); error bars indicate SD. (D and E) by donor-derived CD4+ and CD8+ T cells after transplantation into lethally irradiated (12 Gy) littermate control TgCcl19-Cre– or TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ B6 recipient mice. Donor cells were isolated on day 2.5 or day 6 after transplantation (n = 5 mice/group). (F) Abundance of the indicated transcripts (qRT-PCR) in sort-purified donor-derived CFSEdiluted BALB/c CD4+ T cells on day 2 after transplantation into irradiated B6 hosts treated with or without anti-DLL1/4 antibodies. *P < 0.05, **P < 0.01, NS = P > 0.05, by unpaired, 2-tailed Student’s t test. Data are representative of at least 3 experiments; error bars indicate SD. MFI, mean fluorescence intensity.
Irradiation rapidly alters the microanatomy of SLOs.
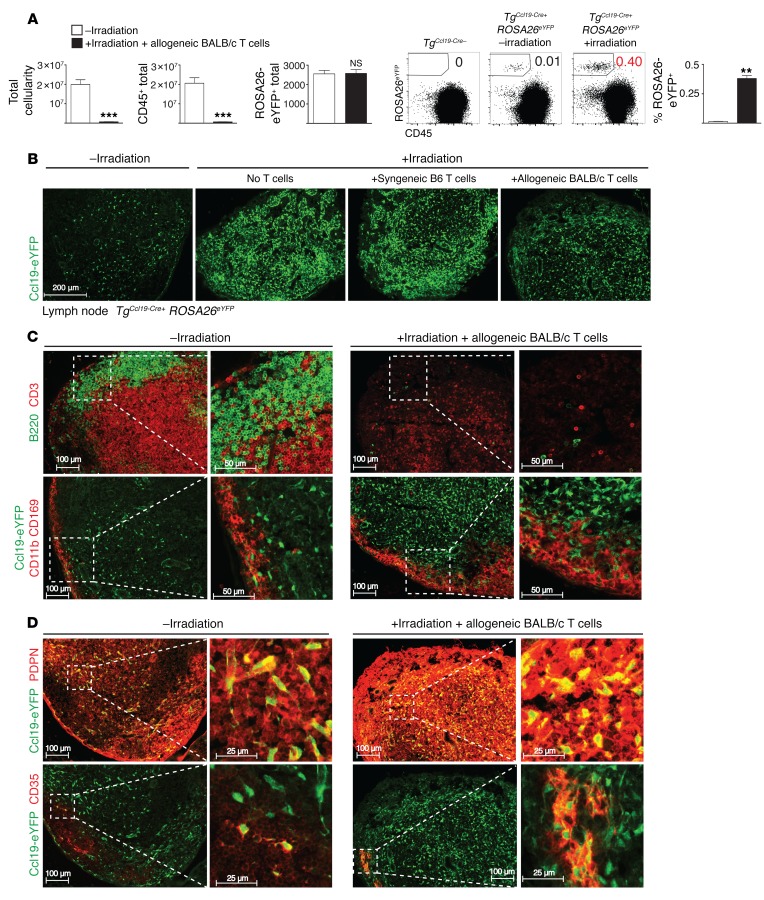
To assess the impact of allo-BMT on SLO organization with respect to Ccl19-Cre+ fibroblastic stromal cells, we studied TgCcl19-Cre+ ROSA26eYFP mice. We found that allo-BMT reduced LN cellularity due to loss of radiosensitive hematopoietic cells, while absolute numbers of eYFP+ cells were preserved (Figure 6A). Flow cytometric analysis and immunofluorescence microscopy revealed markedly increased relative frequencies of eYFP+ stromal cells in LN and spleen after allo-BMT (Figure 6, A and B, and Supplemental Figure 7A), concomitant with depletion of radiosensitive lymphocytes and thickening of the subcapsular macrophage layer (Figure 6C and Supplemental Figure 7B). eYFP+ fibroblastic cells from allo-BMT mice were located within both the T zone and B follicles, as delineated by the stromal markers podoplanin (PDPN) and CD35, respectively. T zone Ccl19-Cre+ fibroblastic cells showed increased staining intensity for PDPN, while CD35, CD157/BP3, and mucosal vascular addressin cell adhesion molecule 1 (MAdCAM-1) staining of stromal cells in B follicles was also highly apparent (Figure 6D and Supplemental Figure 7C). The increased staining intensity could be related at least in part to the rise in stromal cell density within a given surface area after irradiation (Figure 6B). Thus, allo-BMT rapidly altered the spatial organization of lymphoid organ–resident cells, exposing donor T cells to a dense network of stromal cells.
Figure 6. Irradiation changes the LN ultrastructure and increases the relative density of Ccl19-Cre+ stromal cells.
(A) Total cellularity, absolute numbers of CD45+ cells, and absolute numbers and frequencies of eYFP+ cells in LNs of unirradiated or lethally irradiated (12 Gy) TgCcl19-Cre+ ROSA26eYFP reporter mice receiving allogeneic BALB/c splenocytes (n = 6 mice/group). **P < 0.01, ***P < 0.001, NS = P > 0.05, by unpaired, 2-tailed Student’s t test. (B) Immunofluorescence microscopy images of LN cryosections from TgCcl19-Cre+ ROSA26eYFP mice stained for GFP. Cryosections were from unirradiated or lethally irradiated (12 Gy) mice receiving no T cells, syngeneic B6 CD4+ T cells, or allogeneic BALB/c CD4+ T cells. (C and D) Immunofluorescence microscopy images of LN cryosections from TgCcl19-Cre+ ROSA26eYFP mice stained for B220 and CD3 (C, top panel); eYFP, CD11b, and CD169 (C, bottom panel); eYFP and PDPN (D, top panel); and eYFP and CD35 (D, bottom panel). Cryosections were from unirradiated or lethally irradiated (12 Gy) mice receiving allogeneic BALB/c CD4+ T cells on day 1.5 after transplantation. Data are representative of 2 experiments.
Defined fibroblastic niches deliver Notch ligands to donor alloreactive T cells.
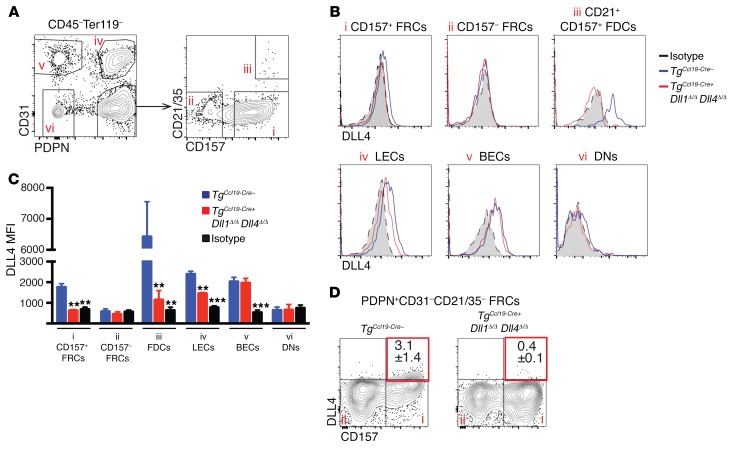
We next investigated the distribution of Notch ligands in SLOs after allo-BMT, focusing on DLL4, since it is the dominant Notch ligand driving GVHD (14). We examined cell-surface DLL4 within 6 distinct populations of Ter119–CD45– LN stromal cells, as defined by expression of CD31, PDPN, CD157/BP3, and CD21/35 (Figure 7). Comparison of control TgCcl19-Cre– and TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ mice identified cellular subsets expressing functionally relevant Notch ligands after allo-BMT. Interestingly, DLL4 expression was not uniformly distributed among all Ccl19-Cre+ stromal cells, but was observed in a fraction of PDPN+CD31– FRCs marked by high levels of CD157/BP3, and in PDPN+CD31–CD21/35hi cells that were immunophenotypically consistent with FDCs. In keeping with the capacity of Ccl19-Cre to lineage trace the majority of CD157hi FRCs and FDCs (Figure 3A), DLL4 expression was reduced to isotype control levels in these cell populations. LECs expressed slightly decreased levels of DLL4 in TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ mice, consistent with the Ccl19-Cre activity we observed in a small fraction of LECs (Figure 3A). We found that PDPN+CD31–CD157lo FRCs had no detectable DLL4, while BECs expressed DLL4 in both control and TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ mice (Figure 7).
Figure 7. DLL4 expression within different LN nonhematopoietic cell populations after allogeneic BMT.
(A) LN stromal cell subsets. TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ and TgCcl19-Cre– littermate control mice were lethally irradiated before infusion of allogeneic BALB/c splenocytes. LNs were collected on day 1.5 after transplantation and enzymatically digested. Dot plots show representative flow cytometric analysis of CD45–Ter119– nonhematopoietic stromal cells. (B) DLL4 expression in LN-resident nonhematopoietic cell subsets. Roman numerals refer to the cell populations shown in A. (C) MFI comparing DLL4 expression in stromal cell subsets from TgCcl19-Cre– versus TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ mice as well as isotype staining in controls. **P < 0.01 and ***P < 0.001, by unpaired 2-tailed Student’s t test with Sidak’s correction for multiple comparisons. (D) Representative flow cytometric plots of PDPN+CD31–CD21/35– FRCs showing DLL4 expression in the CD157hi subset. Data are representative of at least 3 experiments.
In the spleen, characterization of stromal cells by flow cytometry is more challenging than in LNs due to difficulties in isolation and the presence of many red pulp fibroblasts (39). TgCcl19-Cre expression induced ROSA26-eYFP expression in a high proportion of PDPN+CD31–CD157hi and PDPN+CD31–CD21/35hi stromal cells that were phenotypically similar to their LN counterparts (Supplemental Figure 8, A and B). Histological evaluation revealed that these cells were restricted to the white pulp cords and most likely represented T zone FRCs and B zone FDCs (Supplemental Figure 7, A–C). Both cell populations expressed surface DLL4 that was abrogated by Ccl19-Cre–mediated Dll4 inactivation (Supplemental Figure 8C). eYFP was also detected in fewer than 10% of the rare PDPN+CD31–CD157lo cells and in fewer than 2% of PDPN–CD31– cells (which are heterogeneous and most likely include red pulp–derived fibroblasts). However, flow cytometry did not detect surface DLL4 in these latter cell types, a finding consistent with our observation of restricted expression of DLL4 in fibroblast subsets of the white pulp.
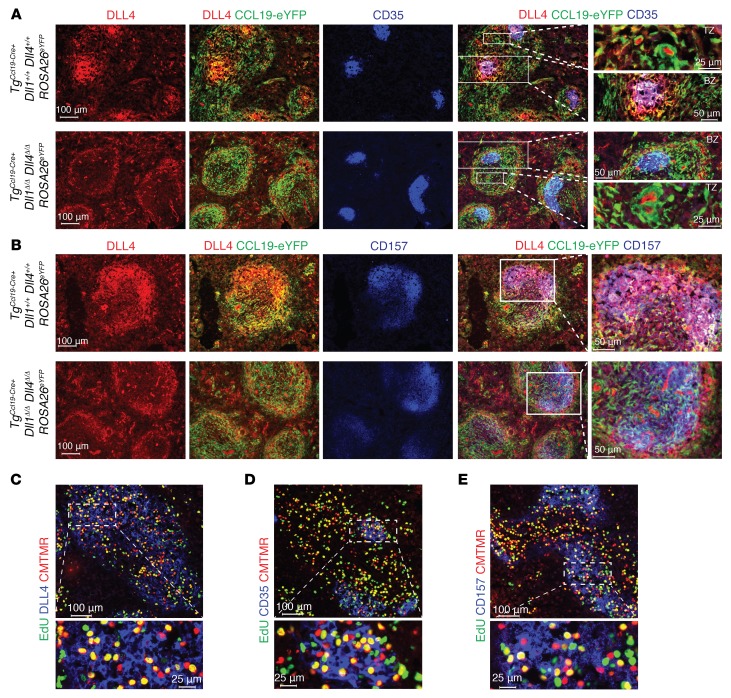
Immunofluorescence microscopy of spleens from TgCcl19-Cre+ Dll1+/+ Dll4+/+ ROSA26eYFP and TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ ROSA26eYFP allo-BMT mice provided anatomical landmarks and demonstrated that, within white pulp areas, only a subset of Ccl19-Cre lineage–traced eYFP+ cells expressed high levels of DLL4 protein. The coexpression of CD35 and CD157 and the organization in a follicular morphology point to FDC-like cells as being the highest expressors of DLL4. We observed loss of DLL4 expression in FDC-like cells, identified as eYFP+CD35+ (Figure 8A), and in eYFP+CD157+ cells (Figure 8B) from TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ ROSA26eYFP mice. Loss of DLL4 in FRCs could not be assessed histologically due to low signals but was supported by flow cytometric data (Supplemental Figure 8C). To assess whether alloantigen-specific T cells first localized next to DLL4-expressing stromal cells in SLOs after allo-BMT, we injected CD4+ 4C T cell receptor–transgenic (TCR-transgenic) T cells specific to recipient I-Ad MHC class II and tracked these alloantigen-specific T cells during their early window of Notch sensitivity. Using EdU incorporation, we detected proliferating donor 4C T cells only within white pulp cords, close to both DLL4-expressing CD157+ and CD35+ fibroblastic stromal cells (Figure 8, C–E). Altogether, our data suggest that fibroblastic subsets of the white pulp cords form specialized niches that provide DLL4 Notch ligands to incoming T cells and promote their pathogenic properties at the onset of GVHD.
Figure 8. Fibroblastic niches in spleen express DLL1/4 Notch ligands and localize next to alloreactive T cells.
(A and B) Immunofluorescence microscopy of splenic cryosections from TgCcl19-Cre+ Dll1+/+ Dll4+/+ ROSA26eYFP and TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ ROSA26eYFP mice stained for GFP, CD35, and DLL4 (A) or GFP, CD157, and DLL4 (B). (C–E) Immunofluorescence microscopy of splenic cryosections from lethally irradiated (8.5 Gy) BALB/c mice transplanted with CMTMR-labeled alloantigen-specific CD4+ 4C TCR–transgenic cells and pulsed with EdU 12 hours prior to organ collection to label proliferating cells. Cryosections were incubated with Alexa Fluor 488 picolyl azide to reveal EdU, along with anti-DLL4 (C), anti-CD35 (D), or anti-CD157 (E). Organs were collected on day 1.5 after transplantation. Data are representative of at least 2 experiments.
Discussion
Preclinical models identified donor and host hematopoietic APCs as critical cellular partners of donor T cells at the onset of GVHD (2, 5–7, 56). Early interactions between donor T cells and APCs, in concert with inflammatory signals from the microenvironment, are thought to provide essential instructive signals to induce T cell alloreactivity. Our findings revise this prevailing model of GVHD pathogenesis, as they identify LN- and spleen-resident Ccl19-Cre+ fate-mapped stromal cells as key initiators of alloimmune T cell pathogenicity through induction of Notch signals in donor-derived T cells. Contrary to past assumptions, donor and host hematopoietic APCs were not responsible for the delivery of Notch signals to donor T cells, as genetic inactivation of Delta-like Notch ligands within donor and host hematopoietic APCs failed to ameliorate GVHD. In contrast, selective inactivation of Notch ligands within stromal cells derived from Ccl19-Cre–expressing cells resulted in long-term protection from GVHD. Thus, specialized stromal niches are uniquely important in the context of alloimmunity.
In this study, several populations of stromal cells were lineage traced by Ccl19-Cre and should, given our findings, be considered potential essential sources of DLL1/4 Notch ligands available to alloreactive T cells at the onset of GVHD. Our findings identify spleen and LN fibroblastic stromal cells expressing high levels of PDPN and CD157 as prime candidates for sources of DLL1/4 Notch ligands. These DLL4-expressing fibroblasts are referred to as FRCs and are thought to reside predominantly in the T cell zone of SLOs. In addition, we detected high DLL4 expression in stromal cells with characteristics of FDCs. Although these cells are located within follicles that are not typically entered by naive T cells, post-irradiation changes may change trafficking rules in SLOs and render FDCs accessible to incoming alloreactive T cells, reminiscent of T follicular helper cells and memory CD8+ T cells (57). The small fraction of LECs lineage traced by Ccl19-Cre also represents a potential source of Notch ligands in LNs. However, we observed prominent DLL1/4-dependent effects on alloreactive T cells located in the spleen, even during the early peri-transplantation period, when T cell recirculation is unlikely. As the spleen contains hardly any LECs, these cells are unlikely to be the dominant source of DLL1/4 ligands in GVHD. We observed no DLL4 expression in red pulp cells, where very little Ccl19-Cre activity was detected. Aside from its activity in the spleen and LNs, Ccl19-Cre is also active in fibroblastic cell subsets from Peyer’s patches (58). Thus, Peyer’s patches and other gut-associated lymphoid structures could also be sites of DLL1/4-mediated Notch activation in T cells during GVHD.
Immunofluorescence microscopy and flow cytometric analysis of SLOs after allo-BMT demonstrated that Delta-like Notch ligand expression was not uniformly distributed throughout all fibroblastic stromal cells of the lymphoid compartment. Instead, we observed focal niches of Notch ligand expression within Ccl19-Cre+ cells marked by the stromal surface markers CD157 and CD21/35. Concomitant expression of chemokines and Delta-like Notch ligands in resident nonhematopoietic cells within defined SLO niches bears similarities to the coregulated expression of chemokines and DLL4 ligands in cortical thymic epithelial cells. In the thymus, Foxn1 controls expression of CCL21/25 and DLL4, which is critical to attract lymphoid progenitors and induce T lineage development (26). We speculate that SLOs rely on a similar organizational module to attract immune cells to defined niches with specialized immunological functions in which they are exposed to Notch ligands and other regulatory signals. This is consistent with the role of fibroblastic stromal cells as a source of Notch ligands for marginal-zone B cells, ESAMhi DCs, and T follicular helper cells (32). During alloimmune responses, direct interaction of T cells with these defined stromal niches may deliver important molecular cues in a temporally and spatially controlled fashion. An in-depth characterization of the cellular and humoral signals delivered within these defined niches should provide additional critical insight into the molecular pathogenesis of GVHD. Furthermore, upstream signals and transcription factors controlling the expression of Delta-like Notch ligands and chemokines in fibroblastic stromal cells remain to be discovered.
It is interesting to speculate as to why alloreactive T cells derive all their critical DLL1/4-mediated Notch signals from small subsets of fibroblastic cells, while Notch ligands are expressed by multiple other cell types in SLOs, including endothelial cells and hematopoietic cells. The precise pattern of alloreactive T cell trafficking in SLOs may be important in explaining this phenomenon. We speculate that CCL19 and other chemokines such as CCL21 and CXCL13 recruit incoming alloreactive T cells into specialized fibroblastic networks that resemble B and T zone niches, where T cells engage in durable interactions with fibroblastic cells that induce high-intensity Notch signals. Adhesion molecules and other concomitant signals may also play a role in T cell retention in this microenvironment. As only limited information is available to date about how alloantigen-specific T cells traffic in SLOs during their initial priming (59), additional investigations will be needed to address these questions.
Among fibroblastic stromal cells, FRCs support steady-state immune homeostasis through the secretion of IL-7, CCL19, and CCL21 (42, 60, 61). In vitro, FRCs enhance naive CD4+ or CD8+ T cell survival and viability in an IL-7/CCL19–dependent manner, while Ccl19 loss in vivo results in decreased total numbers of CD4+ and CD8+ T cells. Importantly, genetic Dll1/4 inactivation of Notch ligands with Ccl19-Cre did not negatively impact the ability of FRCs to maintain naive T cell homeostasis, as the total number of immune cells within LN and spleen and the distribution of naive/memory cells at steady state were unaffected. Consistent with this notion, Il7 transcripts were unaltered in sort-purified FRCs from TgCcl19-Cre+ Dll1Δ/Δ Dll4Δ/Δ mice, showing that DLL1/4 loss did not globally disrupt FRC function.
FRCs can regulate immunity through multiple mechanisms. FRCs form a scaffold to which DCs can adhere and present antigens to naive T cells, thus using the FRC network as a “road system” for their migration (45, 47, 62). This random migration is enhanced by the chemokines CCL19 and CCL21, which are constitutively expressed by FRCs. Physical removal of FRCs after SLO development prevents the efficient activation and proliferation of antigen-specific T cells (37, 63, 64). In contrast, in our studies, genetic Dll1/4 inactivation with Ccl19-Cre preserved T cell homing, as equivalent numbers of donor CD4+ and CD8+ T cells were isolated in SLOs. Furthermore, donor T cells upregulated the early activation markers CD69 and CD44 upon transfer into TgCcl19–Cre+ Dll1Δ/Δ Dll4Δ/Δ mice, suggesting their preserved ability to encounter and respond initially to alloantigens. Finally, donor T cells demonstrated no obvious impairment in their ability to proliferate in response to antigen. Thus, our observations differ from the broader dysfunction of SLOs observed upon physical elimination of Ccl19-Cre+ FRCs (37, 63, 64). Instead, our targeted genetic approach suggests that fibroblastic stromal cell subsets regulate specific aspects of immune cell biology through dedicated signaling pathways (e.g., Notch), consistent with the delivery of unique activation codes to immune cell subsets. Moreover, the post-transplantation environment may enhance the delivery of Notch signals to incoming T cells through activation of fibroblastic stromal cells after irradiation and loss of competing radiation-sensitive host lymphocytes.
The precise molecular mechanisms by which early Notch signals increase the pathogenicity of alloreactive T cells remain to be investigated further. We previously reported that Notch-deprived alloreactive T cells acquire a hyporesponsive phenotype with decreased TCR/CD28-mediated Ras/MAPK and NF-κB activation (65). As these changes were observed upon genetic interference with the Notch transcriptional activation complex (11, 14, 65), the effects of Notch signaling in alloreactive T cells must ultimately be mediated by canonical transcriptional mechanisms. Our observations now provide insights into the critical early window of time during which alloantigen-specific T cells are exposed to essential Notch signals. Gene expression analysis revealed that only small numbers of known Notch transcriptional targets, including Dtx1, Hes1, Il2ra, and Trib2, were downregulated in this context. More work is necessary to establish whether the profound effects of Notch signaling in alloreactive T cells are accounted for by novel effects of these known Notch target genes, or by other previously unknown transcriptional targets that are controlled by Notch in mature T cells.
Our in vivo results contrast with those of past work identifying a role for stromal cells as negative regulators of immune responses (66–68), as the dominant role of these cells after allo-BMT was to promote rather than restrain alloimmune reactivity. In previous studies, the addition of ex vivo–isolated FRCs to T cell/DC cocultures suppressed T cell proliferation in a dose-dependent fashion. Suppression was mediated by IFN-γ–dependent upregulation of NOS2 and COX1/2-dependent metabolites within FRCs. While Nos2 inactivation resulted in enhanced T cell proliferation in vivo, the lack of specificity of the genetic targeting strategy made it difficult to assess whether FRCs were in fact responsible for enforcing this suppressive mechanism. Furthermore, the expression of Notch ligands on FRCs was not examined in these in vivo or in vitro studies. In our in vivo studies, irradiation of allo-BMT recipients upregulated the expression of several surface markers on stromal cells, changed their morphology, and resulted in profound remodeling of SLO microanatomy within the first few days of transplantation. Thus, it is possible that the highly inflammatory environment that ensues after irradiation, in combination with the differences in the nature and availability of antigen, could account for the extreme proinflammatory functions of fibroblastic stromal cells after allo-BMT. It is also possible that alloreactive T cells gain unique access to fibroblastic niches from which T cells are typically excluded, such as FDCs or follicular FRCs residing in the outer B cell zone. Alternatively, our observations may reveal a previously unrecognized proinflammatory function of stromal cells mediated by Notch signaling that also operates in other contexts.
With regard to a temporal requirement for Notch signaling, systemic delivery of DLL1/4-blocking antibodies at the time of transplantation had profound effects on T cell cytokine production, Treg expansion, and overall T cell alloreactivity, but inhibition delayed by only 2 days failed to efficiently block GVHD. This narrow window of therapeutic sensitivity is significant in terms of translational applications. Indeed, short-term DLL1/4 inhibition during this critical time has emerged as an attractive therapeutic strategy for preventing GVHD without exposing recipients to the risks of long-term DLL1/4 blockade (69–71). Mechanistically, this narrow window might reflect the temporal delivery of a physical pulse of Notch signaling in defined microanatomical niches after allo-BMT. This physical pulse may result from transient exposure to cellular sources of Notch ligands that are typically inaccessible to T cells, or from prolonged initial contacts with fibroblastic stromal cell subsets that upregulate DLL1/4. Alternatively, our findings could reflect the existence of a unique window of T cell sensitivity to Notch signaling during early stages of priming and activation.
Altogether, our study reveals for the first time to our knowledge the existence of specialized subsets of host nonhematopoietic fibroblastic cells that deliver Notch signals to donor T cells at early stages after allo-BMT to program the pathogenicity of these T cells. Donor and host hematopoietic APCs were dispensable sources of Notch ligands, thus challenging the widely accepted hypothesis that motile APCs simultaneously provide both antigen and Notch ligands to prime T cells. These findings illustrate the importance of exploring Notch signaling in vivo using loss-of-function approaches, as relevant sources of Notch ligands may be missing from established in vitro experimental systems. Our work also highlights the utility of precisely targeting immunomodulatory pathways in stromal cells, as we uncovered what we believe to be a previously unrecognized pathogenic role for fibroblastic stromal cells independent of their functions in structural integrity and immune homeostasis. In GVHD, allograft rejection, and other immune disorders, we believe our findings pave the way toward selective inhibition of niche-derived signals that drive deleterious immune responses, without interfering with other essential immunological functions of the lymphoid environment.
Methods
Mice.
BALB/c (H-2d), C57BL/6 (H-2b, CD45.2) (B6), and B6-CD45.1 (H-2b, CD45.1) mice were bred at the University of Michigan. TgMx1-Cre, Dll1fl/fl, Dll4fl/fl, Notch2fl/fl, and 4C Rag1–/– TCR–transgenic mice were previously described (24, 72–75). TgMx1-Cre allows for IFN-inducible activation of Cre expression, e.g., via systemic administration of poly(I:C). 4C Rag1–/– mice have a monoclonal population of CD4+ T cells specific for I-Ad MHC class II alloantigens, which enables tracking of alloantigen-specific T cells in vivo (72). TgMx1-Cre Dll1fl/fl Dll4fl/fl mice were maintained on the B6 background and were backcrossed onto the BALB/c background for more than 5 generations. Ccl19-Cre BAC–transgenic mice (TgCcl19-Cre) express Cre recombinase in subsets of nonhematopoietic stromal cells under the control of a 90-kb regulatory sequence upstream of the Ccl19 transcriptional start site (32, 36–38). TgCcl19-Cre mice were crossed with B6 Dll4fl/fl Dll1fl/fl ROSA26eYFP mice (expressing Cre-inducible eYFP under control of the ROSA26 promoter).
Induction and assessment of GVHD.
Six- to ten-week-old BALB/c mice or eight- to twelve-week-old B6 recipients underwent allo-BMT as previously described (11, 14, 15). Both female and male mice were used as recipients and were distributed equally among experimental groups. BALB/c mice received 8.5–9 Gy irradiation (137Cs source) 4 hours prior to allo-BMT, while B6 mice received 2 doses of 6 Gy irradiation (137Cs) separated by 3 hours. T cell–depleted BM (TCD BM) was prepared with anti-Thy1.2 antibodies and complement (Cedar Lane Laboratories) (14), resulting in over 95% depletion of Thy1.2+ cells. BALB/c recipients received 5 × 106 to 10 × 106 TCD BM ± 5 × 106 donor splenocytes, while B6 recipients received 10 × 106 to 15 × 106 TCD BM ± 20 × 106 splenocytes. Clinical GVHD score and weight changes were monitored at least weekly, as described previously (11, 14, 15, 76).
Antibody-mediated inhibition of Delta-like Notch ligands.
Humanized IgG1-neutralizing antibodies against DLL1, DLL4, and isotype control were described previously (14, 70, 77). All antibodies were injected i.p. (5 mg/kg twice weekly). The potency and specificity of each antibody batch was verified by assessing their capacity to achieve in vivo depletion of DLL1-dependent marginal-zone B cells or DLL4-dependent thymocytes (14, 24, 73).
Generation of BM hematopoietic chimeras and GVHD induction.
B6-CD45.2 TgMx1-Cre+ Dll1fl/fl Dll4fl/fl donor mice or control littermates received 5 i.p. injections of 50 μg poly(I:C) (Amersham) every other day. Two weeks after the last poly(I:C) dose, six- to ten-week-old female B6-CD45.1 mice received 9 Gy of irradiation and were reconstituted with syngeneic BM from poly(I:C)-induced TgMx1-Cre+ Dll1Δ/Δ Dll4Δ/Δ mice or their control littermates. Donor chimerism was quantified 12 weeks later by determining the relative frequency of CD45.2+ cells within mature cell populations in blood and spleen. The efficiency of Cre-mediated Dll1 and Dll4 recombination was assessed by PCR in sort-purified blood myeloid cells using the following primers: Dll1 forward: CACACCTCCTACTTACCTGA; Dll1 reverse: GAGAGTACTGGATGGAGCAAG; Dll1 loxas: GGCGCTCAAAGGATATGGGA; Dll4 forward: GTGCTGGGACTGTAGCCACT; Dll4 reverse: TGTTAGGGATGTCGCTCTCC; and Dll4 revdel: CTCGTCTGTTCGCCAAATCTTAC. In other experiments, Notch2fl/fl mice were used to generate BM chimeras with poly(I:C)-induced TgMx1-Cre+ Notch2Δ/Δ or control TgMx1-Cre– Notch2fl/fl donor BM (75).
BM chimeras were rested at least 12 weeks to allow for establishment of steady-state hematopoiesis before GVHD induction. BM chimeras were irradiated with 2 doses of 5.5 Gy each (137Cs), separated by 3 hours, and received 8 × 106 TCD BM ± 30 × 106 splenocytes from BALB/c donors (or syngeneic B6 donors as a negative control). Clinical GVHD score and weight changes were monitored at least weekly (11, 14, 15). For Notch2Δ/Δ or Notch2+/+ BM chimeras, transplantation was performed with 107 BALB/c BM and 5 × 106 T cells (Thy1 selection; EasySep; STEMCELL Technologies).
LN and splenic stromal cell isolation.
Stromal cells were isolated from LNs using a modified version of a previously described protocol (32, 42). Briefly, peripheral LNs (cervical, axial, brachial, inguinal) from unirradiated or irradiated mice were coarsely chopped with a scalpel and incubated at 37°C for 15 minutes in digestion solution containing DMEM, 2% FBS, 1.2 mM CaCl2, penicillin/streptomycin, and 1.0 mg/ml collagenase IV (Invitrogen, Thermo Fisher Scientific) and 40 μg/ml DNAse I (Roche). Samples were pipetted gently and reincubated at 37°C until all solid material appeared to be dissolved. Suspensions were filtered through a 70-μM strainer and analyzed by flow cytometry. For spleen, chopped fragments were incubated at 37°C for 15 minutes with 1.0 mg/ml collagenase IV, 0.5 mg/ml collagenase D, 0.25 mg/ml dispase (Invitrogen, Thermo Fisher Scientific), and 40 μg/ml DNAse I (Roche). Samples were pipetted and macroscopic components allowed to settle while supernatant was filtered, washed, and resuspended in PBS and 2% FBS. The remaining solid components were subjected to sequential rounds of digestion, pipetting, and collection of supernatant until no macroscopic components remained.
Flow cytometric analysis.
The following antibodies were from BioLegend: CD4 (clone GK1.5); CD8α (clone 53-6.7); CD19 (clone 6D5); CD11c (clone N418); CD11b (clone M1/70); F4/80 (clone BM8); PDCA1 (clone 129C1); B220 (clone RA3-6B2); MHC II (clone M5/114.15.2); CD44 (clone IM7); CD62L (clone MEL-14); PDPN/gp38 (clone 8.1.1); CD31 (clone 390); Ter119 (clone TER-119); CD157 (clone BP-3); CD25 (clone PC61); CD21/35 (clone 7E9); CD45 (clone 30-F11); TCRβ (clone Η57-597); H-2Kb (clone AF6-88.5); H-2Kd (clone SF1-1.1); IFN-γ (clone XMG1.2); TNF-α (clone MP6-XT22); and IL-2 (clone JES6-5H4). Anti-FoxP3 (clone FJK-16s) and anti-CD207/anti-langerin (clone eBioL31) antibodies were from eBioscience. For flow cytometric analysis of DLL4 surface expression, samples were blocked in rat/mouse serum on ice for 15 minutes, stained on ice with biotinylated anti-DLL4 (clone 9A1.5; a gift of H.R. McDonald, Ludwig Institute for Cancer Research, Lausanne, Switzerland) (78) for 1 hour in NaN3 and EDTA-free FACS buffer, stained with surface markers for 30 minutes on ice, washed twice, and stained in streptavidin-allophycocyanin (streptavidin-APC) (1:100) for 30 minutes on ice. For assessment of T cell cytokine production, donor splenocytes were stimulated with plate-bound anti-CD3 (2.5 μg/ml; clone 145-2C11; BD Biosciences) and anti-CD28 (2.5 μg/ml; clone 37.51; BD Biosciences) for 2 to 3 hours prior to the addition of monensin (GolgiStop; BD Biosciences). Samples were fixed and stained for intracellular proteins according to the manufacturer’s instructions. Flow cytometric analysis was performed on a 3-laser Fortessa or 4-laser FACSAria II/III (both from BD). Dead cells were excluded with DAPI (Sigma-Aldrich) or Zombie Aqua Fixable Viability Dye (BioLegend).
Immunofluorescence microscopy.
LNs and spleens were fixed in 1% PFA for 2 hours, sunk in 30% sucrose overnight, and embedded and frozen in Tissue-Tek O.C.T. (Sakura Finetek) in an ethanol–dry ice bath. Staining of 6- to 8-μm-thin cryosections was performed for 60 minutes or overnight at room temperature using the following antibodies: monoclonal anti-CD3ε (145-2C11); B220 (RA3-6B2); CD35 (8C12); PDPN/gp38 (8.1.1); MAdCAM-1 (Meca-89); (all purified in house at the Institute of Biochemistry, University of Lausanne, Lausanne, Switzerland); and polyclonal goat anti-DLL4 (AF1389; R&D Systems). The following secondary antibodies were used: donkey anti–rat biotin; donkey anti–rat Cy3; donkey anti–rabbit Cy3; donkey anti–sheep APC (all from Jackson ImmunoResearch); and streptavidin Alexa Fluor 488 and donkey anti–rabbit Alexa Fluor 647 (both from Invitrogen, Thermo Fisher Scientific). For DLL4 labeling, primary antibodies were detected using a biotinylated donkey anti–goat IgG followed by HRP-coupled streptavidin (Jackson ImmunoResearch) and tyramide signal amplification (Invitrogen, Thermo Fisher Scientific) according to the manufacturer’s instructions, but using a borate buffer (0.1 M in PBS, pH 8.5) for tyramide dilution. eYFP was detected using a rabbit anti-GFP antibody followed by Alexa Fluor 488–conjugated donkey anti–rabbit IgG (both from Invitrogen, Thermo Fisher Scientific). Images were acquired on an AxioImager.Z1 microscope with an AxioCam and were processed in Adobe Photoshop. Specifically, image contrast was adjusted using the “levels” tool, while image sharpness was improved with the “unsharpen mask” command. Tissue sections were stained with 3 different fluorochromes, but processed images were often simplified into 2 color images with the ImageJ (NIH) “split channels” and “merge channels” functions.
EdU incorporation and detection.
Mice were injected i.p. with 2.5 mg/ml 5-ethynyl-2′-deoxyuridine (EdU) 12 hours prior to tissue collection. LNs and spleens were fixed in 1% PFA for 2 hours, sunk in 30% sucrose overnight, and embedded and frozen in Tissue-Tek (Sakura Finetek) in an ethanol–dry ice bath. EdU was detected with a Click-iT EdU Alexa Fluor 488 Imaging Kit (Molecular Probes, Thermo Fisher Scientific) according to the manufacturer’s instructions.
Quantitative real-time PCR.
Donor T cells or host stromal cells were sort purified directly into TRIzol Reagent (Invitrogen, Thermo Fisher Scientific). Total RNA was extracted with phenol/chloroform and purified with an RNeasy Micro Kit (QIAGEN). cDNA was generated with SuperScript II (Invitrogen, Thermo Fisher Scientific) and subjected to quantitative PCR with either TaqMan or SYBR Green PCR Master Mixes on an Eppendorf Mastercycler ep realplex (Applied Biosystems, Thermo Fisher Scientific). Gene expression analysis was performed using the following primers: Dtx1 (Mm00492297_m1, Applied Biosystems); Hes1 (Mm01342805_m1); Il2ra (Mm01340213_m1); Pten (Mm00477209_m1); Trib2 (Mm00454876_m1); Il7 forward: GTGCCACATTAAAGACAAAGAAG; Il7 reverse: GTTCATTATTCGGGCAATTACTATC; Myc forward: CAGCGACTCTGAAGAAGAGCAAG; Myc reverse: GGGTTTGCCTCTTCTCCACAG; Hprt forward: CTCCTCAGACCGCTTTTTGC; and Hprt reverse: TAACCTGGTTCATCATCGCTAATC. Relative gene expression was determined using the ΔΔCt method, with normalization to Hprt.
Statistics.
The sample size for in vivo mouse experiments was determined empirically on the basis of prior experience and used to calculate power with the “pwr” statistical package in R. Unless otherwise noted, all statistical tests were done using Prism software (GraphPad Prism7). Briefly, the means of 2 different groups were compared with an unpaired, 2-tailed Student’s t test. If the variances of 2 groups differed significantly by F test, a Welch’s correction for t tests was used. If multiple t tests were evaluated at once within a data set, the Holm-Sidak method was used to adjust for multiple comparisons, assuming α = 0.05. Survival curves were compared using a log-rank (Mantel-Cox) test. Bar graphs were generated with GraphPad Prism.
Study approval.
All experiments were performed according to NIH guidelines and approved by the Committee on the Use and Care of Animals of the University of Michigan.
Author contributions
JC designed and performed the majority of the experiments, analyzed data, and wrote the manuscript. CLE and EP designed, performed, and analyzed experiments. VR, AT, FA, SW, JF, AF, DG, and ITT performed and analyzed transplantation experiments. LS performed immunofluorescence microscopy. QC, LO, and BL generated and provided Ccl19-Cre–transgenic mice. MY and CWS provided DLL1/4-blocking antibodies. PR, BRB, AYH, and DKB participated in study design and data analysis. TVB generated and provided 4C-transgenic mice. UK and FR generated a floxed Dll4 allele, provided Dll1 and Dll4 conditional-KO mice, shared key unpublished information, and participated in the study design. SAL analyzed all immunofluorescence microscopy results, participated in study design, and provided technical assistance. IM supervised the research, designed experiments, analyzed data, and wrote the manuscript. All authors reviewed and edited the manuscript.
Supplementary Material
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases (NIAID) (R01-AI091627, to IM and R01-AI34495, to BRB); the Leukemia and Lymphoma Society (CDP 1227-14, to IM and TRP 6462-15, to IM and BRB); the Michigan Institute for Clinical and Health Research (to IM); and the Swiss National Science Foundation (to BL, FR, and SAL). Individual support included T32-GM007315 from the National Institute of General Medical Sciences (NIGMS) (to JC), T32-GM007863 from NIGMS (to JC and EP), and T32-HL007762 from the National Heart, Lung, and Blood Institute (to CLE); a research training award from the American Society of Hematology (to VR); and a Career Development Award from the American Society for Blood and Marrow Transplantation (to VR). We thank H.R. McDonald for providing anti-DLL4 antibodies.
Footnotes
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Conflict of interest: M. Yan and C.W. Siebel are employed by Genentech.
Reference information:J Clin Invest. 2017;127(4):1574–1588.https://doi.org/10.1172/JCI89535.
See the related Commentary at T cells take directions from supporting cast in graft-versus-host disease.
Contributor Information
Jooho Chung, Email: jooh@umich.edu.
Eric Perkey, Email: eperkey@umich.edu.
Vedran Radojcic, Email: vedran.radojcic@gmail.com.
Ute Koch, Email: ute.koch@epfl.ch.
Alexander Tong, Email: axt307@case.edu.
Frederick Allen, Email: ffa5@case.edu.
Sherri Wood, Email: sychan@med.umich.edu.
Jiane Feng, Email: jifeng@umich.edu.
Ann Friedman, Email: friedmna@umich.edu.
David Granadier, Email: dwgranad@umich.edu.
Qian Chai, Email: qianchai@moon.ibp.ac.cn.
Lucas Onder, Email: Lucas.Onder@kssg.ch.
Minhong Yan, Email: yan.minhong@gene.com.
Pavan Reddy, Email: reddypr@umich.edu.
Bruce R. Blazar, Email: blaza001@umn.edu.
Burkhard Ludewig, Email: burkhard.ludewig@kssg.ch.
Freddy Radtke, Email: freddy.radtke@epfl.ch.
Ivan Maillard, Email: imaillar@umich.edu.
References
- 1.Ferrara JL, Levine JE, Reddy P, Holler E. Graft-versus-host disease. Lancet. 2009;373(9674):1550–1561. doi: 10.1016/S0140-6736(09)60237-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blazar BR, Murphy WJ, Abedi M. Advances in graft-versus-host disease biology and therapy. Nat Rev Immunol. 2012;12(6):443–458. doi: 10.1038/nri3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shlomchik WD. Graft-versus-host disease. Nat Rev Immunol. 2007;7(5):340–352. doi: 10.1038/nri2000. [DOI] [PubMed] [Google Scholar]
- 4.McDonald-Hyman C, Turka LA, Blazar BR. Advances and challenges in immunotherapy for solid organ and hematopoietic stem cell transplantation. Sci Transl Med. 2015;7(280):280rv2. doi: 10.1126/scitranslmed.aaa6853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Koyama M, et al. Plasmacytoid dendritic cells prime alloreactive T cells to mediate graft-versus-host disease as antigen-presenting cells. Blood. 2009;113(9):2088–2095. doi: 10.1182/blood-2008-07-168609. [DOI] [PubMed] [Google Scholar]
- 6.Duffner UA, et al. Host dendritic cells alone are sufficient to initiate acute graft-versus-host disease. J Immunol. 2004;172(12):7393–7398. doi: 10.4049/jimmunol.172.12.7393. [DOI] [PubMed] [Google Scholar]
- 7.Shlomchik WD, et al. Prevention of graft versus host disease by inactivation of host antigen-presenting cells. Science. 1999;285(5426):412–415. doi: 10.1126/science.285.5426.412. [DOI] [PubMed] [Google Scholar]
- 8.Koyama M, et al. Recipient nonhematopoietic antigen-presenting cells are sufficient to induce lethal acute graft-versus-host disease. Nat Med. 2011;18(1):135–142. doi: 10.1038/nm.2597. [DOI] [PubMed] [Google Scholar]
- 9.Toubai T, et al. Induction of acute GVHD by sex-mismatched H-Y antigens in the absence of functional radiosensitive host hematopoietic-derived antigen-presenting cells. Blood. 2012;119(16):3844–3853. doi: 10.1182/blood-2011-10-384057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li H, et al. Profound depletion of host conventional dendritic cells, plasmacytoid dendritic cells, and B cells does not prevent graft-versus-host disease induction. J Immunol. 2012;188(8):3804–3811. doi: 10.4049/jimmunol.1102795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, et al. Notch signaling is a critical regulator of allogeneic CD4+ T-cell responses mediating graft-versus-host disease. Blood. 2011;117(1):299–308. doi: 10.1182/blood-2010-03-271940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Riella LV, et al. Blockade of Notch ligand δ1 promotes allograft survival by inhibiting alloreactive Th1 cells and cytotoxic T cell generation. J Immunol. 2011;187(9):4629–4638. doi: 10.4049/jimmunol.1004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roderick JE, et al. Therapeutic targeting of NOTCH signaling ameliorates immune-mediated bone marrow failure of aplastic anemia. J Exp Med. 2013;210(7):1311–1329. doi: 10.1084/jem.20112615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tran IT, et al. Blockade of individual Notch ligands and receptors controls graft-versus-host disease. J Clin Invest. 2013;123(4):1590–1604. doi: 10.1172/JCI65477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandy AR, et al. T cell-specific notch inhibition blocks graft-versus-host disease by inducing a hyporesponsive program in alloreactive CD4+ and CD8+ T cells. J Immunol. 2013;190(11):5818–5828. doi: 10.4049/jimmunol.1203452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Charbonnier LM, Wang S, Georgiev P, Sefik E, Chatila TA. Control of peripheral tolerance by regulatory T cell-intrinsic Notch signaling. Nat Immunol. 2015;16(11):1162–1173. doi: 10.1038/ni.3288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wood S, et al. Transient blockade of delta-like Notch ligands prevents allograft rejection mediated by cellular and humoral mechanisms in a mouse model of heart transplantation. J Immunol. 2015;194(6):2899–2908. doi: 10.4049/jimmunol.1402034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guruharsha KG, Kankel MW, Artavanis-Tsakonas S. The Notch signalling system: recent insights into the complexity of a conserved pathway. Nat Rev Genet. 2012;13(9):654–666. doi: 10.1038/nrg3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137(2):216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamaguchi E, et al. Expression of Notch ligands, Jagged1, 2 and Delta1 in antigen presenting cells in mice. Immunol Lett. 2002;81(1):59–64. doi: 10.1016/S0165-2478(01)00326-1. [DOI] [PubMed] [Google Scholar]
- 21.Amsen D, Blander JM, Lee GR, Tanigaki K, Honjo T, Flavell RA. Instruction of distinct CD4 T helper cell fates by different notch ligands on antigen-presenting cells. Cell. 2004;117(4):515–526. doi: 10.1016/S0092-8674(04)00451-9. [DOI] [PubMed] [Google Scholar]
- 22.Schaller MA, et al. Notch ligand Delta-like 4 regulates disease pathogenesis during respiratory viral infections by modulating Th2 cytokines. J Exp Med. 2007;204(12):2925–2934. doi: 10.1084/jem.20070661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mochizuki K, et al. Delta-like ligand 4 identifies a previously uncharacterized population of inflammatory dendritic cells that plays important roles in eliciting allogeneic T cell responses in mice. J Immunol. 2013;190(7):3772–3782. doi: 10.4049/jimmunol.1202820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koch U, et al. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J Exp Med. 2008;205(11):2515–2523. doi: 10.1084/jem.20080829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hozumi K, et al. Delta-like 4 is indispensable in thymic environment specific for T cell development. J Exp Med. 2008;205(11):2507–2513. doi: 10.1084/jem.20080134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Calderón L, Boehm T. Synergistic, context-dependent, and hierarchical functions of epithelial components in thymic microenvironments. Cell. 2012;149(1):159–172. doi: 10.1016/j.cell.2012.01.049. [DOI] [PubMed] [Google Scholar]
- 27.Tan JB, et al. Lunatic and manic fringe cooperatively enhance marginal zone B cell precursor competition for delta-like 1 in splenic endothelial niches. Immunity. 2009;30(2):254–263. doi: 10.1016/j.immuni.2008.12.016. [DOI] [PubMed] [Google Scholar]
- 28.Shutter JR, et al. Dll4, a novel Notch ligand expressed in arterial endothelium. Genes Dev. 2000;14(11):1313–1318. [PMC free article] [PubMed] [Google Scholar]
- 29.Hellström M, et al. Dll4 signalling through Notch1 regulates formation of tip cells during angiogenesis. Nature. 2007;445(7129):776–780. doi: 10.1038/nature05571. [DOI] [PubMed] [Google Scholar]
- 30.Zheng W, et al. Notch restricts lymphatic vessel sprouting induced by vascular endothelial growth factor. Blood. 2011;118(4):1154–1162. doi: 10.1182/blood-2010-11-317800. [DOI] [PubMed] [Google Scholar]
- 31.Niessen K, et al. The Notch1-Dll4 signaling pathway regulates mouse postnatal lymphatic development. Blood. 2011;118(7):1989–1997. doi: 10.1182/blood-2010-11-319129. [DOI] [PubMed] [Google Scholar]
- 32.Fasnacht N, et al. Specific fibroblastic niches in secondary lymphoid organs orchestrate distinct Notch-regulated immune responses. J Exp Med. 2014;211(11):2265–2279. doi: 10.1084/jem.20132528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laky K, Evans S, Perez-Diez A, Fowlkes BJ. Notch signaling regulates antigen sensitivity of naive CD4+ T cells by tuning co-stimulation. Immunity. 2015;42(1):80–94. doi: 10.1016/j.immuni.2014.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meng L, et al. The Notch ligand DLL4 defines a capability of human dendritic cells in regulating Th1 and Th17 differentiation. J Immunol. 2016;196(3):1070–1080. doi: 10.4049/jimmunol.1501310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maekawa Y, Ishifune C, Tsukumo S, Hozumi K, Yagita H, Yasutomo K. Notch controls the survival of memory CD4+ T cells by regulating glucose uptake. Nat Med. 2015;21(1):55–61. doi: 10.1038/nm.3758. [DOI] [PubMed] [Google Scholar]
- 36.Chai Q, et al. Maturation of lymph node fibroblastic reticular cells from myofibroblastic precursors is critical for antiviral immunity. Immunity. 2013;38(5):1013–1024. doi: 10.1016/j.immuni.2013.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cremasco V, et al. B cell homeostasis and follicle confines are governed by fibroblastic reticular cells. Nat Immunol. 2014;15(10):973–981. doi: 10.1038/ni.2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rodda LB, Bannard O, Ludewig B, Nagasawa T, Cyster JG. Phenotypic and morphological properties of germinal center dark zone Cxcl12-expressing reticular cells. J Immunol. 2015;195(10):4781–4791. doi: 10.4049/jimmunol.1501191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fasnacht N, et al. Specific fibroblastic niches in secondary lymphoid organs orchestrate distinct Notch-regulated immune responses. J Exp Med. 2014;211(11):2265–2279. doi: 10.1084/jem.20132528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lewis KL, et al. Notch2 receptor signaling controls functional differentiation of dendritic cells in the spleen and intestine. Immunity. 2011;35(5):780–791. doi: 10.1016/j.immuni.2011.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Saito T, et al. Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity. 2003;18(5):675–685. doi: 10.1016/S1074-7613(03)00111-0. [DOI] [PubMed] [Google Scholar]
- 42.Link A, et al. Fibroblastic reticular cells in lymph nodes regulate the homeostasis of naive T cells. Nat Immunol. 2007;8(11):1255–1265. doi: 10.1038/ni1513. [DOI] [PubMed] [Google Scholar]
- 43.Sixt M, et al. The conduit system transports soluble antigens from the afferent lymph to resident dendritic cells in the T cell area of the lymph node. Immunity. 2005;22(1):19–29. doi: 10.1016/j.immuni.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 44.Kaldjian EP, Gretz JE, Anderson AO, Shi Y, Shaw S. Spatial and molecular organization of lymph node T cell cortex: a labyrinthine cavity bounded by an epithelium-like monolayer of fibroblastic reticular cells anchored to basement membrane-like extracellular matrix. Int Immunol. 2001;13(10):1243–1253. doi: 10.1093/intimm/13.10.1243. [DOI] [PubMed] [Google Scholar]
- 45.Katakai T, Hara T, Sugai M, Gonda H, Shimizu A. Lymph node fibroblastic reticular cells construct the stromal reticulum via contact with lymphocytes. J Exp Med. 2004;200(6):783–795. doi: 10.1084/jem.20040254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clark SL., Jr The reticulum of lymph nodes in mice studied with the electron microscope. Am J Anat. 1962;110:217–257. doi: 10.1002/aja.1001100303. [DOI] [PubMed] [Google Scholar]
- 47.Bajénoff M, et al. Stromal cell networks regulate lymphocyte entry, migration, and territoriality in lymph nodes. Immunity. 2006;25(6):989–1001. doi: 10.1016/j.immuni.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chyou S, et al. Coordinated regulation of lymph node vascular-stromal growth first by CD11c+ cells and then by T and B cells. J Immunol. 2011;187(11):5558–5567. doi: 10.4049/jimmunol.1101724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mathieu M, Duval F, Daudelin JF, Labrecque N. The Notch signaling pathway controls short-lived effector CD8+ T cell differentiation but is dispensable for memory generation. J Immunol. 2015;194(12):5654–5662. doi: 10.4049/jimmunol.1402837. [DOI] [PubMed] [Google Scholar]
- 50.Adler SH, et al. Notch signaling augments T cell responsiveness by enhancing CD25 expression. J Immunol. 2003;171(6):2896–2903. doi: 10.4049/jimmunol.171.6.2896. [DOI] [PubMed] [Google Scholar]
- 51.Backer RA, et al. A central role for Notch in effector CD8(+) T cell differentiation. Nat Immunol. 2014;15(12):1143–1151. doi: 10.1038/ni.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wong GW, Knowles GC, Mak TW, Ferrando AA, Zúñiga-Pflücker JC. HES1 opposes a PTEN-dependent check on survival, differentiation, and proliferation of TCRβ-selected mouse thymocytes. Blood. 2012;120(7):1439–1448. doi: 10.1182/blood-2011-12-395319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Palomero T, et al. Mutational loss of PTEN induces resistance to NOTCH1 inhibition in T-cell leukemia. Nat Med. 2007;13(10):1203–1210. doi: 10.1038/nm1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Herranz D, et al. A NOTCH1-driven MYC enhancer promotes T cell development, transformation and acute lymphoblastic leukemia. Nat Med. 2014;20(10):1130–1137. doi: 10.1038/nm.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yashiro-Ohtani Y, et al. Long-range enhancer activity determines Myc sensitivity to Notch inhibitors in T cell leukemia. Proc Natl Acad Sci U S A. 2014;111(46):E4946–E4953. doi: 10.1073/pnas.1407079111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Magenau J, Runaas L, Reddy P. Advances in understanding the pathogenesis of graft-versus-host disease. Br J Haematol. 2016;173(2):190–205. doi: 10.1111/bjh.13959. [DOI] [PubMed] [Google Scholar]
- 57.Im SJ, et al. Defining CD8(+) T cells that provide the proliferative burst after PD-1 therapy. Nature. 2016;537(7620):417–421. doi: 10.1038/nature19330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gil-Cruz C, et al. Fibroblastic reticular cells regulate intestinal inflammation via IL-15-mediated control of group 1 ILCs. Nat Immunol. 2016;17(12):1388–1396. doi: 10.1038/ni.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin KL, et al. Intravital imaging of donor allogeneic effector and regulatory T cells with host dendritic cells during GVHD. Blood. 2014;123(10):1604–1614. doi: 10.1182/blood-2013-09-526020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Malhotra D, Fletcher AL, Turley SJ. Stromal and hematopoietic cells in secondary lymphoid organs: partners in immunity. Immunol Rev. 2013;251(1):160–176. doi: 10.1111/imr.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malhotra D, et al. Transcriptional profiling of stroma from inflamed and resting lymph nodes defines immunological hallmarks. Nat Immunol. 2012;13(5):499–510. doi: 10.1038/ni.2262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Katakai T, Hara T, Lee JH, Gonda H, Sugai M, Shimizu A. A novel reticular stromal structure in lymph node cortex: an immuno-platform for interactions among dendritic cells, T cells and B cells. Int Immunol. 2004;16(8):1133–1142. doi: 10.1093/intimm/dxh113. [DOI] [PubMed] [Google Scholar]
- 63.Scandella E, et al. Restoration of lymphoid organ integrity through the interaction of lymphoid tissue-inducer cells with stroma of the T cell zone. Nat Immunol. 2008;9(6):667–675. doi: 10.1038/ni.1605. [DOI] [PubMed] [Google Scholar]
- 64.Novkovic M, et al. Topological small-world organization of the fibroblastic reticular cell network determines lymph node functionality. PLoS Biol. 2016;14(7):e1002515. doi: 10.1371/journal.pbio.1002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sandy AR, et al. T cell-specific notch inhibition blocks graft-versus-host disease by inducing a hyporesponsive program in alloreactive CD4+ and CD8+ T cells. J Immunol. 2013;190(11):5818–5828. doi: 10.4049/jimmunol.1203452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Siegert S, et al. Fibroblastic reticular cells from lymph nodes attenuate T cell expansion by producing nitric oxide. PLoS One. 2011;6(11):e27618. doi: 10.1371/journal.pone.0027618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Khan O, Headley M, Gerard A, Wei W, Liu L, Krummel MF. Regulation of T cell priming by lymphoid stroma. PLoS One. 2011;6(11):e26138. doi: 10.1371/journal.pone.0026138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lukacs-Kornek V, et al. Regulated release of nitric oxide by nonhematopoietic stroma controls expansion of the activated T cell pool in lymph nodes. Nat Immunol. 2011;12(11):1096–1104. doi: 10.1038/ni.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yan M, et al. Chronic DLL4 blockade induces vascular neoplasms. Nature. 2010;463(7282):E6–E7. doi: 10.1038/nature08751. [DOI] [PubMed] [Google Scholar]
- 70.Ridgway J, et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444(7122):1083–1087. doi: 10.1038/nature05313. [DOI] [PubMed] [Google Scholar]
- 71.Yan M. Therapeutic promise and challenges of targeting DLL4/NOTCH1. Vasc Cell. 2011;3:17. doi: 10.1186/2045-824X-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brennan TV, et al. A new T-cell receptor transgenic model of the CD4+ direct pathway: level of priming determines acute versus chronic rejection. Transplantation. 2008;85(2):247–255. doi: 10.1097/TP.0b013e31815e883e. [DOI] [PubMed] [Google Scholar]
- 73.Hozumi K, et al. Delta-like 1 is necessary for the generation of marginal zone B cells but not T cells in vivo. Nat Immunol. 2004;5(6):638–644. doi: 10.1038/ni1075. [DOI] [PubMed] [Google Scholar]
- 74.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269(5229):1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 75.McCright B, Lozier J, Gridley T. Generation of new Notch2 mutant alleles. Genesis. 2006;44(1):29–33. doi: 10.1002/gene.20181. [DOI] [PubMed] [Google Scholar]
- 76.Cooke KR, et al. An experimental model of idiopathic pneumonia syndrome after bone marrow transplantation: I. The roles of minor H antigens and endotoxin. Blood. 1996;88(8):3230–3239. [PubMed] [Google Scholar]
- 77.Wu Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010;464(7291):1052–1057. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 78.Fiorini E, et al. Cutting edge: thymic crosstalk regulates delta-like 4 expression on cortical epithelial cells. J Immunol. 2008;181(12):8199–8203. doi: 10.4049/jimmunol.181.12.8199. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.