Abstract
Systemic lupus erythematosus (SLE) is an inflammatory autoimmune disease characterized by immune complex formation with multi-organ manifestations. Lupus nephritis (LN) is one of the most severe types of organ damage in SLE, and it clearly contributes to increased morbidity and mortality due to SLE. LN occurs more frequently and is more severe in non-European ancestral backgrounds, although the cause of this disparity remains largely unknown. Genetic factors play an important role in the pathogenesis of SLE. Although many SLE susceptibility genes have been identified, the genetic basis of LN is not as well understood. While some of the established general SLE susceptibility genes are associated with LN, recent discoveries highlight a number of genes with renal functions that are specifically associated with LN. Some of these genes associated with LN help to explain the disparity in the prevalence of nephritis between individuals with SLE, and also partially explain differences in LN between ancestral backgrounds. Moreover, not only the gene mutations, but also post-translational modifications seem to play important roles in the pathogenesis of LN. Overall it seems likely that a combination of general SLE susceptibility genes cooperate with LN specific risk genes to result in the genetic propensity for LN. In this review, we will outline the genetic contribution to LN and describe possible roles of LN susceptibility genes.
Keywords: Systemic lupus erythematosus, Lupus nephritis, Genetics, APOL1, PDGFR, HAS2
1. Introduction
Systemic lupus erythematosus (SLE) is an inflammatory autoimmune disease characterized by multi-organ system involvement. It affects women 9 times more often than men, and often is diagnosed in early adulthood, frequently during the childbearing years.
Genetic and environmental factors play important roles in the pathogenesis of SLE [1]. The incidence and prevalence of SLE is higher in non-European ancestry, especially in African ancestry. The severity of SLE also varies among the ethnic groups, being more severe in non-European populations [2–4]. Supporting a genetic contribution to disease, monozygotic twins are much more likely to be concordant for SLE than dizygotic twins (concordance rate 24% and 2%, respectively) [5]. Familial aggregation of SLE has also been clearly documented, and most pedigrees support a non-Mendelian complex inheritance [6]. These facts strongly support notion of a polygenic genetic contribution to SLE pathogenesis.
Among the various organ manifestations of SLE, lupus nephritis (LN) is one of the most feared, potentially resulting in organ damage and renal insufficiency that results in poor clinical outcomes despite recent improvements in SLE treatment.
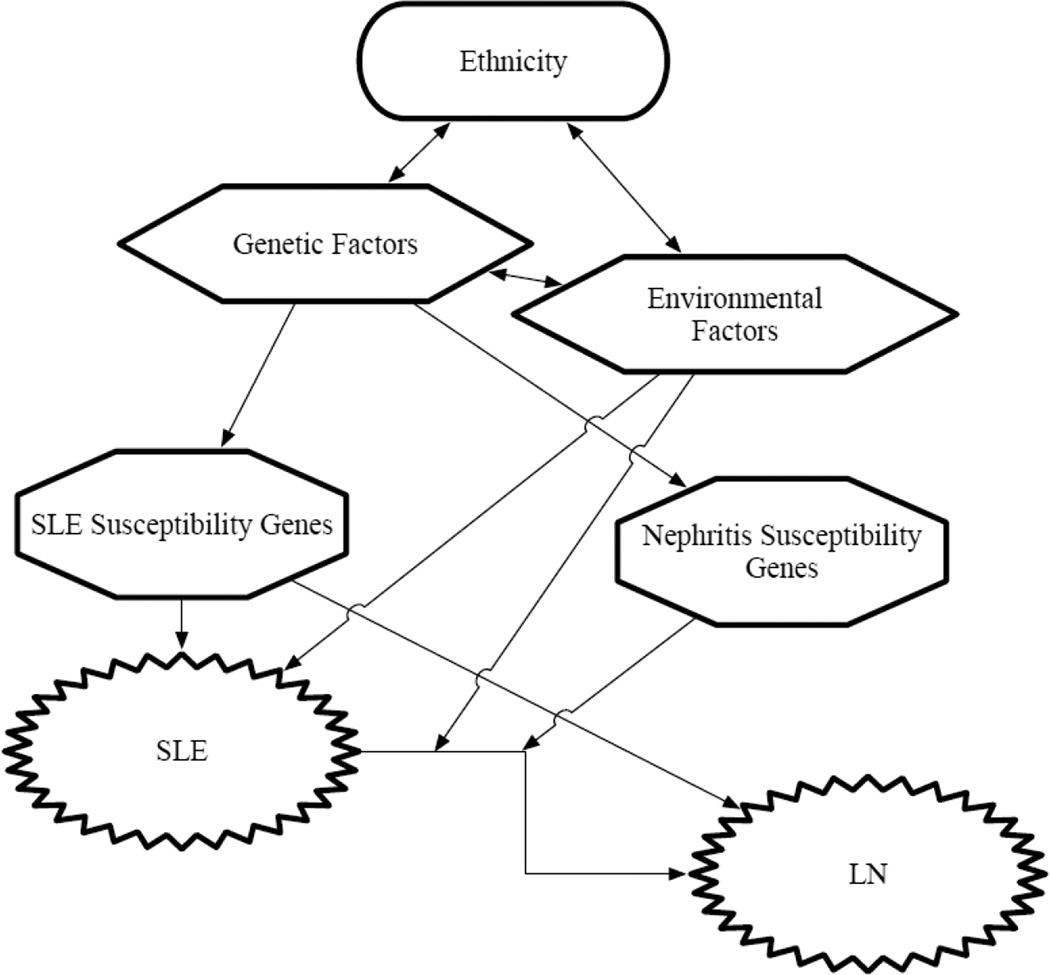
Genome-Wide-Association Studies (GWAS) and candidate gene association studies have revealed numerous risk genes for SLE, including loci which contain genes that function in the innate and adaptive immune system [7, 8]. Some of these genes are also closely associated with LN. However, most of these previous studies were not primarily focused on the nephritis phenotype, and less is known about which genes predispose to LN. Some recent studies which have focused on identifying the genes specifically responsible for LN have identified intrarenal genes that are associated with LN, but not associated with the susceptibility of SLE in general [9, 10]. While the exact functional mechanisms of these renal-related candidate genes remains unclear, it seems that the genetic basis of LN involves a combination of general SLE susceptibility genes which function in the immune system and genes which are more renal-specific that predispose specifically to LN (Figure 1). In this review, we outline the genetic contribution to LN and also discuss possible roles of each candidate genes in the pathogenesis of LN.
Figure 1. Interactions between multiple factors predisposing to LN.
Interactions between genetic factors and environmental factors predispose to SLE and LN.
LN, lupus nephritis; SLE, systemic lupus erythematosus
2. Difference in incidence rate and severity of LN between ethnicities
Among various organ manifestations of SLE, LN is one of the most severe, and can progress to end-stage renal disease (ESRD) leading to increased morbidity and mortality. LN affects about 40% of SLE patients throughout their lifetime [11]. Despite recent advances in treatment, patients with LN still have higher morbidity and mortality compared with those without LN [11–13]. It is well known that African, Asian, and Hispanic populations are more likely to develop LN as compared to European ancestry [3, 11, 14–16]. There are also differences in severity of LN among the ethnicities, with LN being more severe in non-European populations [3, 11]. This disparity between ancestral backgrounds could be related to genetic or environmental factors [17]. To investigate the importance of genetic factors, Sanchez et al. conducted a study evaluating the genetic impact of the proportion of Amerindian vs. European genetic ancestry in admixed populations living in South America. This is an informative way to study the contribution of genetics to LN with some control over environment, as different individuals living in the same population and same location will have different proportional genetic ancestry. This study revealed that an increased proportion of Amerindian genetic ancestry correlated with increased risk of developing LN [2]. Another study demonstrated familial clustering of ESRD African ancestry SLE patients with LN, suggesting shared genetic factors contributing to LN in these families [18]. These studies support the idea that genetic factors contribute to the pathogenesis of LN.
3. LN risk genes and possible functions
Although GWAS studies have identified numerous SLE risk genes, thus far fewer genes have been identified as predisposing to the LN phenotype. Most of the reported risk genes for LN have been studied in candidate gene studies, after being identified as SLE risk factors in GWAS studies [7, 8], with the hypothesis that SLE susceptibility genes may also be LN risk genes. More recently, studies have examined the LN phenotype directly, comparing patients that have developed LN to those patients who have not developed LN.
3.1. SLE susceptibility genes associated with LN
3.1.1. HLA-DR
The major histocompatibility complex (MHC) region which contains human leukocyte antigen (HLA) genes is located on human chromosome 6. This locus contains more than 200 genes, many of which function in the immune system. The HLA Class II region contains HLA-DR, -DQ, and –DP genes. They are highly expressed on antigen-presenting cells and are important for the activation of CD4+ T cells and other immune responses. Polymorphisms in the HLA region were among the first to be discovered as SLE risk factors, and this locus remains the strongest common genetic risk factor for SLE [19].
Among the HLA alleles, HLA-DR2 and HLA-DR3 are the most established SLE susceptibility loci proven in previous studies [7, 20]. Large phenotypic association studies also revealed HLA-DR3 [9, 21, 22] as a susceptibility gene for LN in European ancestry. Additionally, meta-analysis of multiethnic case-control studies identified 7 HLA Class II candidate alleles in the association of LN. HLA-DR15 (part of the older HLA-DR2 serotype group) and -DR3 were found as risk alleles for LN, and interestingly -DR4 and -DR11 were associated with protection from LN [23]. Despite the strong association of HLA alleles with both SLE and LN, the actual mechanism of how these HLA-DR haplotypes predispose to LN has not been elucidated. Some studies support the idea that particular MHC molecules may permit presentation of self peptides in an abnormal way. One study identified a dominant HLA-DR3 restricted T cell epitope on the SmD autoantigen, and those mimicry peptides were able to activate T cells that were reactive to this epitope [24]. This HLA restricted autoreactive T cell activation may then lead to stimulation of autoreactive B cells, resulting in the production of autoantibodies. Another group showed the association of HLA-DR3 and the presence of anti-Ro autoantibodies in SLE patients, which may also support the above mechanism [20].
3.1.2. ITGAM
The ITGAM gene is located on human chromosome 16. It encodes CD11b-integrin (alpha M) which is a subunit composing alpha M beta-2 integrin (also called complement receptor 3 or Mac-1). Mac-1 is highly expressed in granulocytes, macrophages and dendritic cells. Complement (e.g. iC3b) is one of the ligands of Mac-1, and iC3b-coated particles such as apoptotic cells will induce phagocytosis by phagocytes via engagement to Mac-1. Mac-1 also controls leukocyte migration to inflammation site and facilitate adherence to vascular endothelium[25].
A non-synonymous SNP (rs1143679) in the ITGAM gene results in a change of arginine (R) to histidine (H) at position 77 (R77H). Human primary monocytes carrying 77H allele showed reduced ability of phagocytosis to iC3b-opsonized particles [26]. Those monocytes with 77H allele also failed to repress the production of inflammatory cytokines such as interleukin (IL)-1b, IL-6 and tumor necrosis factor alpha, when stimulated with Toll-like receptor (TLR) 7/8 ligands. Association of the ITGAM gene with SLE susceptibility has been well established in several studies [7, 8, 27, 28]. Associations of ITGAM variants with renal involvement in SLE patients has also been shown in several studies [9, 21, 29–31]. Yang et al. reported higher prevalence of LN in SLE patients carrying the R77H variant in Hong Kong Chinese SLE patients, as compared to SLE patients without LN (OR = 3.35, p = 0.029). Kim-Howard et al also demonstrated association of the R77H variant with LN in large European ancestry SLE cohort (OR = 2.15, p = 4.69 × 10−22). Recently, two large studies, primarily designed to identify the association between genetic variants and LN, have also confirmed ITGAM as a susceptibility gene for LN [9, 31]. Collectively, defective function of the complement receptor due to these variants may result in incomplete clearance of glomerular deposits, and facilitate a pro-inflammatory environment in the kidney which leads to LN. Interestingly, while many of the other SLE susceptibility genes are shared with other autoimmune diseases, ITGAM seems to be an SLE-specific genetic risk factor (with the possible exception of systemic sclerosis)[32]. This could indicate a specific role for ITGAM in the pathogenesis of SLE and LN, rather than a more general pro-inflammatory role or global alteration in phagocytic function, but further investigation is needed.
3.1.3. FCGR
The FCGR gene locus is located on human chromosome 1 and encodes Fc gamma receptors (FcgRs). One key role of FcgRs is to remove immune complexes (ICs) [33], and FcgR2A and FcgR3A are expressed on antigen presenting cells (e.g. macrophage and dendritic cells). Defective clearance of apoptotic cells is identified in SLE patients, and is thought to contribute to disease pathogenesis [34, 35]. Failure of appropriate phagocytosis will allow apoptotic cells to proceed to secondary necrosis resulting in the release of nuclear self-antigens, which should lead to greater IC formation when combined with SLE-associated anti-nuclear antibodies. There is one high-affinity receptor (e.g. FcgR1 ) and two low-affinity receptors (e.g. FcgR2A and FcgR3) which are activating-type FcgRs, and there is one inhibitory-type FcgR known as FcgR2b. Each of these receptors has a different binding affinity to IgG subclasses and are present in most of myeloid cells [36]. Genetic associations between variants of low-affinity FcgR (i.e. FcgR2A and FcgR3A) and SLE have been reported [7, 37]. FcgR2A and FcgR3A are also associated with LN which have been proven in several individual studies among multiple ethnicities [38–43].
A non-synonymous SNP (rs1801274) of FCGR2A encoding a G-to-A allele change at position 131 results in amino-acid change from arginine (R) to histidine (H). These alleles, 131R and 131H, have different binding capacity for IgG2. Cells which are homozygous for the histidine allele (131H/H) strongly bind IgG2, and cells that are homozygous for the arginine allele (131R/R) show a reduction in binding capacity for IgG2 opsonized ICs (131H/R has intermediate binding capacity) [44]. In a study of anti-C1q antibody positive SLE patients, presence of FCGR2A 131R was associated in the increased risk for LN [45]. Zuniga et al. identified that the frequency of FCGR2A 131R allele was significantly higher in LN patients with intense IgG2 deposition in the kidney [46]. Also, the frequency of homozygous FCGR2A 131R/R was increased in the LN population as compared to non-proliferative idiopathic glomerulonephritis[39]. These data suggest that the FCGR2A 131R allele results in dysfunctional clearance of IgG2-opsonized ICs, which could allow greater IC deposition in the kidney and increased inflammation.
Also, a non-synonymous SNP (rs396991) of FCGR3A encoding T-to-G allele change at nucleotide 559 results in an amino-acid conversion from phenylalanine (F) to valine (V) [33]. FcgR3A-158V allele binds to IgG1, IgG3 and IgG4 in higher affinity compared to the 158F allele [33]. Several reports including meta-analysis article revealed that FCGR3A 158F allele is strongly associated with LN [40–43]. Although, in the recent large multiethnic lupus cohort, patients with SLE who were homozygous for FCGR3A 158V/V progressed to ESRD more frequently than compared to those with the 158F/V or 158F/F genotypes (21.9% versus 7.5%, p=0.0175) [47]. Low IgG binding capacity FCGR3A (i.e. 158F/F) may be needed to initiate LN, while high IgG binding FCGR3A (i.e. 158V/V) may play an important role in accelerating the inflammation of LN towards ESRD to explain the above discrepancy.
3.1.4. IRF5
The IRF5 gene is located on human chromosome 7 and encodes interferon regulatory factor 5 (IRF5), which is a transcription factor which induces the expression of inflammatory genes and type I interferon (IFN)-related genes downstream of TLR engagement. The genetic association between IRF5 and SLE was first reported by Sigurdsson et al. in 2005 [48]. Further studies demonstrated that a haplotype which contains multiple functional genetic elements was strongly associated with SLE [49, 50]. Type I interferon is known to play a critical role in the pathogenesis of SLE [51] and risk haplotype of IRF5 was associated with higher IFN alpha activity in the presence with auto-antibodies (i.e. anti-RNA binding protein or anti-double-stranded DNA autoantibodies) [52]. SLE patients carrying the risk haplotype of IRF5 also have elevated expression of IRF5 protein [53]. Moreover, the IRF5 gene has been associated with autoantibody formation in SLE patients and healthy individuals [54, 55]. This same risk haplotype was also identified as a LN susceptibility locus strongly associated with proliferative nephritis and severe renal insufficiency in patients with SLE [21]. While it is not known exactly how IRF5 genotype contributes to nephritis, this gene impacts on both autoantibody formation and type I IFN, and these factors may contribute.
3.1.5. TNIP1
The TNIP1 gene is located on human chromosome 5 and encodes the protein A20 binding and inhibitor of NF-kappaB 1 (ABIN1). The ubiquitin-binding protein ABIN1 is an adaptor protein of the ABIN family that cooperates with A20/TNFAIP3 to limit inflammatory responses. A20 is a ubiquitin-editing protein that inhibits transcription factor NF-kappaB [56, 57] which is a key player of inflammation in immune system.
In mice, Caster et al. reported that the knockin mouse expressing ABIN1[D485N], an inactive form of ABIN1, develops glomerulonephritis similar to class III and IV in human LN with enhanced activation of NF-kappaB and MAPK (mitogen-activated protein kinase) [58].
In humans, variants in TNIP1 gene were detected as susceptibility loci for SLE in multi-ethnic populations (rs7708392 and rs10036748) [59–61]. TNIP1 variants were also identified as LN susceptibility loci in Swedish Caucasian (rs7708392) [21], European American population (rs7708392) [58] and African American population (rs4958881) [58], supporting the important role of ABIN1 in the pathogenesis of LN observed in mice experiment. Another study from Japan also revealed the association with LN in Japanese population (rs7708392) [62]. Two partial loss-of-function risk haplotypes of TNIP1, accompanied with reduced ABIN1 protein expression was identified in SLE patients among European population [63]. Therefore, TNIP1 variants may partly be involved in the pathogenesis of LN by reducing ABIN1 protein expression and causing the dysregulation of NF-kappaB pathways.
3.1.6. STAT4
The STAT4 gene is located on human chromosome 2 and encodes signal transducer and activators of transcription-4 (STAT4). STAT4 is known as a susceptibility gene for SLE [64]. In 2008, large study conducted by Taylor et al. showed the associations between STAT4 (rs7574865) and the presence of LN and anti-dsDNA antibody in European ancestry SLE cohorts [65]. Additionally, a recent study in 2013 further confirmed the association of STAT4 with the development of proliferative nephritis, a severe subtype of LN [21]. A report from Japan confirmed the association of STAT4 (rs7574865) in both SLE patients with LN and in those without in this population [66]. The association was stronger in the SLE patients with LN and anti-dsDNA antibody, but small sample size precluded strong statistical significance for this comparison [66].
STAT4 is a transcriptional factor which is important for T cell signaling. STAT4 is activated by interleukin (IL)-12, leading naïve CD4+ T cells to differentiate to Th1 and Th17 cells which then play an important roles in inflammation and immune responses. IL-17 producing T cells are present in the kidney of LN patients, suggesting that these T cells play a role in the pathogenesis of LN [67]. STAT4 is also activated by type I IFN [68], and SLE patients carrying the STAT4 risk allele rs7574865 have an increased sensitivity to type I IFN supported by the increased expression of IFN inducible genes [69]. The functions of STAT4 in T cell differentiation, the type I IFN pathway, and the association of this risk allele with anti-dsDNA antibody formation could all play a role in considering how this risk allele predisposes to LN.
3.1.7. TNFSF4
The TNFSF4 gene is located on human chromosome 1 and encodes the TNF Super Family 4 (TNFSF4) protein. TNFSF4, also known as OX40L or CD252 is expressed on antigen presenting cells (APC) (e.g. monocytes, dendritic cells and B cells) and is a co-stimulatory factor for T cell activation. A genetic variant in TNFSF4 (rs2205960) was associated with LN in a European ancestry SLE cohort (OR = 1.14, p = 0.0013) [31], and this association was also observed in a Chinese population (rs2205960 and rs10489265) [70]. Serum levels of TNFSF4 are significantly higher among SLE patients with LN than those without LN [71]. Also, the expression level of TNFSF4 receptor on CD4 T cells has been associated with nephritis and disease activity in patients with SLE [72].
3.2. Newly discovered genes associated with LN, but not related to SLE susceptibility
3.2.1. APOL1
As shown in Figure 2, while some general SLE susceptibility genes predispose to LN, there are also some examples of genes which are specifically related to LN and not general SLE susceptibility. APOL1 gene is one example of an LN specific gene. The APOL genes located on human chromosome 22 and encodes apolipoprotein L-1 (APOL1). It is a component of circulating HDL (high density lipoprotein) and also exists abundantly in many organs including the kidney [73]. APOL1 gene variants were known to have a protective effect against African Trypanosoma brucei rhodesiense (T. b. rhodesiense) infection [74]. Serum apolipoprotein L-1 from individuals with the APOL1 variant lyses Tripanosomes more efficiently than that from individuals without the APOL1 variant [74]. Thus, the high prevalence of APOL1 variants in individuals of African descent seems likely to have been driven by evolutionary pressure from T. b. rhodesiense infection [75], and this variant is almost absent in European ancestry. Interestingly, this APOL1 gene variant in African ancestry populations has also been associated with multiple kidney diseases, such as focal segmental glomerulonephritis, ESRD in non-diabetic nephropathy and human immunodeficiency virus-associated nephropathy [74, 76–78].
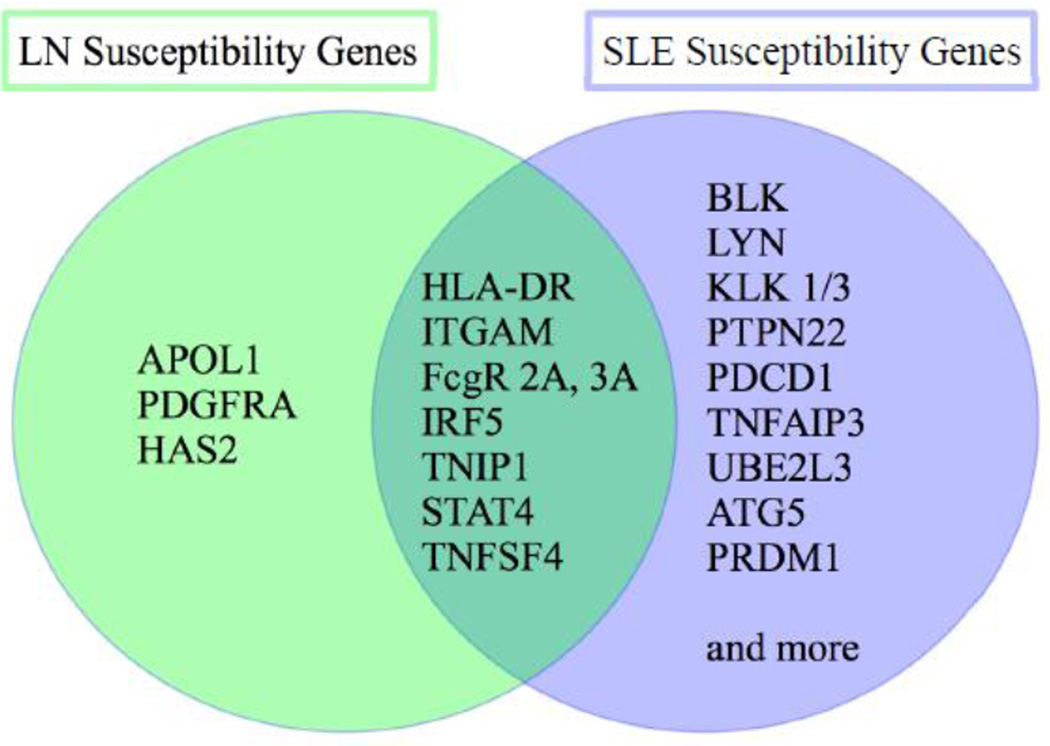
Figure 2. SLE and LN susceptibility genes.
Venn diagram of SLE and LN susceptibility genes.
LN, lupus nephritis; SLE, systemic lupus erythematosus
More recently, Freedman et al. reported the association of risk variants of APOL1 with ESRD in African Americans with LN [10]. SLE patients with LN having APOL1 risk alleles G1/G2 were much more likely to progress to ESRD (OR = 2.72, P = 6.23 × 10−6) as compared to those patients without risk alleles [10]. However, Lin et al. reported that APOL1 risk alleles were not associated with LN in European American population and also did not show significant relations with mild LN even in African American population [79]. Thus the association between APOL1 and LN could be limited to severe LN which is likely to progress to ESRD. From the immunological aspect, APOL1 may also have roles in innate immunity and anti-viral activities. Nichols et al. have shown that APOL1 is induced by TLR3 agonists and interferons (interferon-alpha, -beta and –gamma) [80]. Additionally, APOL1 is involved in autophagy pathway [81]. Of note, APOL1 has not been identified as a general SLE susceptibility gene, so it appears to be a modifier gene, important to nephritis risk and progression in African ancestry populations. Thus, this gene may partially explain the increased incidence and severity of LN in African ancestry SLE patients.
3.2.2. PDGFRA
In a recent study, a variant in the PDGFRA gene (rs1364989) was found to be strongly associated with LN in the European population [9]. The PDGFRA gene is located on human chromosome 4 and encodes platelet-derived growth factor receptor alpha (PDGFRA). PDGF and PDGFR (e.g. PDGFRA) are constitutively expressed in most renal cells (e.g. mesangial cells, fibroblasts and vascular smooth muscle cells), and are involved in normal kidney development. Alteration of the PDGF system is common in human kidney diseases [82]. PDGF-BB (one of the isoforms of PDGF), which is a ligand for PDGFRA, is an important factor that promotes mesangioproliferative disease and renal interstitial fibrosis [83]. In LN, increased PDGFRA mRNA expression was observed in the kidney (glomeruli and tubulointerstitial compartment) of LN patients [9].
Imatinib is a tyrosine-kinase inhibitor that has been used in the treatment of Philadelphia chromosome-positive chronic myelogenous leukemia, KIT-positive gastrointestinal stromal tumors, and other diseases. Imatinib is also known to inhibit PDGFR signaling [82]. Administration of Imatinib in several experimental studies in lupus mouse models showed a significant decrease in LN progression confirmed by histological findings[84, 85], and reduced PDGFR expression [84]. However, some animal experiments using trapidil, a broad PDGF antagonist, have failed to treat nephritis [86]. These data could support the idea of targeting PDGFR in LN, although it seems that the type of inhibitor may be important, as the murine studies are not consistent across all types of PDGFR blockade.
3.2.3. HAS2
The HAS2 gene is located on human chromosome 8 that encodes Hyaluronan synthase 2 (HAS2); an enzyme responsible for hyaluronan (HA) synthesis. HA plays a crucial role in fibrosis in various organs. Renal fibrosis is the principle pathologic process that moves LN toward chronic kidney disease and to ESRD. Several reports provide evidence that intra- and extraglomerular mesangial cells respond to various cytokines and growth factors (e.g. IL-1 beta, PDGF) and result in induction of HA, accompanied by up-regulation of HAS2 [87–90]. Yung et al. have shown that increased expression of HA in the mesangium and tubular portion of the kidney in renal biopsy specimens from LN patients [91]. Serum HA concentration was higher in the active phase of LN than in inactive LN. They also identified that anti-DNA antibody induced IL-1 beta, which then generates HA via overexpression of HAS2 in human mesangial cells [91]. Recent genetic association studies have demonstrated association of a variant in the HAS2 gene (rs7834765) with LN (OR = 3.15) [9]. Again, HAS2 has not been shown to be associated with SLE susceptibility in general, but seems to play a role in LN pathogenesis, possibly via HA production and a contribution to fibrosis.
4. Epigenetics in LN
Not only genetic changes, but epigenetic changes (i.e. post-translational modifications) also play an important role in the pathogenesis of SLE [92]. DNA methylation is one of the important post-translational regulatory modifications, typically occuring at CG dinucleotides. DNA methylation results in gene silencing by tightening the chromatin structure and limiting the access of transcriptional factors, while DNA hypomethylation induce transcripition of genes.
Impaired DNA methylation status in CD4+ T cells of SLE patients was reported more than 20 years ago [93]. As next-generation sequencing technology has advanced, genome-wide methylation studies have demonstrated the differences in methylation profiles of CD4 T cells in SLE patients compared to those of healthy controls. Some studies have shown a difference in methylation profiles between different groups of SLE patients [94, 95]. Of note, hypomethylation of type I IFN-regulated genes known to play important roles in the pathogenesis of SLE are reported in SLE patients [96, 97]. More recently, Coit et al. identified that there are more robust differences in methylation status of type I IFN-regulated genes when compared between SLE patients with LN and SLE patients without LN [98].
These studies shed light to another aspect of genetic involvement in the pathogenesis of SLE and LN, although there is still much work to be done to clarify their specific role to LN, and take advantage of this knowledge to design treatments.
5. Conclusions
Genetic factors play an important role in predisposing SLE patients to LN. The genetic factors discussed above are summarized in Table 1. The prevalence and severity of LN differs among individuals and ethnicities, and there are some examples of differences in genetic factors predisposing to LN in different populations. Despite the identification of numerous general SLE susceptibility genes, only a small number of these general SLE genes are associated with LN. Of note, the magnitude of the OR for general SLE susceptibility genes as risk factors for LN was lower than their impact upon SLE in general. This supports the idea that these general SLE susceptibility genes impact SLE more broadly than their impact upon LN alone. The more recent discovery of genes with renal function that are associated specifically with LN is fascinating and provides additional insight into the pathogenesis of LN. It seems that the predisposition to LN involves a combination of general SLE susceptibility genes and genes with renal function that are associated specifically with LN. Figure 3 shows some potential functional relationships between the loci discussed in this review. Moreover, new generation sequencing technique has revealed not only the genome mutation, but the post-translational modification may contribute in the pathogenesis of SLE and LN. Further epidemiologic and functional studies will better define the relationships of these genes with LN pathogenesis, and likely will identify additional gene loci involved in this serious disease manifestation.
Table 1.
Confirmed Susceptibility Loci Associated with lupus Nephritis
| Genes associated with SLE and LN | ||||
|---|---|---|---|---|
| Gene name | Chromosome | SNP associated with LN | Known fuction | Reference |
| HLA-DR | 6 | - | Antigen presentation | [9, 21–23] |
| ITGAM | 16 | rs1143679 rs9888739 |
Clearance of immune complex | [9, 21, 29–31] |
| FcgR 2A FcgR 3A |
1 | rs1801274 (H131/R131 polymorphism) rs396991 (F158/V158 polymorphism) |
Clearance of immune complex | [38, 39, 46] [40–43, 47] |
| IRF5 | 7 | rsl0488631 rs2070197 |
Innate immunity, type I IFN pathway | [21] |
| TNIP1 | 5 | rs7708392 rs4958881 rs6889239 |
Inhibition of NF-kappaB pathway | [21, 58, 62] |
| STAT4 | 2 | rs7574865 rs7568275 rs7582694 rs11889341 |
T cell signaling | [21, 65] |
| TNFSF4 | 1 | rs 22D5960 rs10489265 |
Costimulatory factor for T cell activation | [9, 31, 70] |
| Genes not associated with SLE, but associated with LN | ||||
| Gene name | Chromosome | SNP associated with LN | Known function | Reference |
| APOL1 | 22 | Gl/G2 haplotype | Innate immunity autophagy | [10] |
| PDGFRA | 4 | rs1364989 | Kidney development | [9] |
| HAS2 | 8 | rs7834765 | Organ fibrosis | [9] |
APOL1, apolipoprotein L-1; FCGR, Fc gamma receptor; F158, phenylalanine residue at position 158; H131, histidine residue at position 131; HAS2, hyaluronan synthase 2; ITGAM, integrin alpha M; LN, lupus nephritis; PDGFRA, platelet-derived growth factor receptor alpha; R131, arginine residue at position 131; SLE, systemic lupus erythematosus; SNP, single nucleotide polymorphism; STAT4, signal transducers and activators of transcription 4; TNFSF4, tumor necrosis factor super family 4; TNIP1, TNFAIP3-interacting protein 1; V158, valine residue at position 158
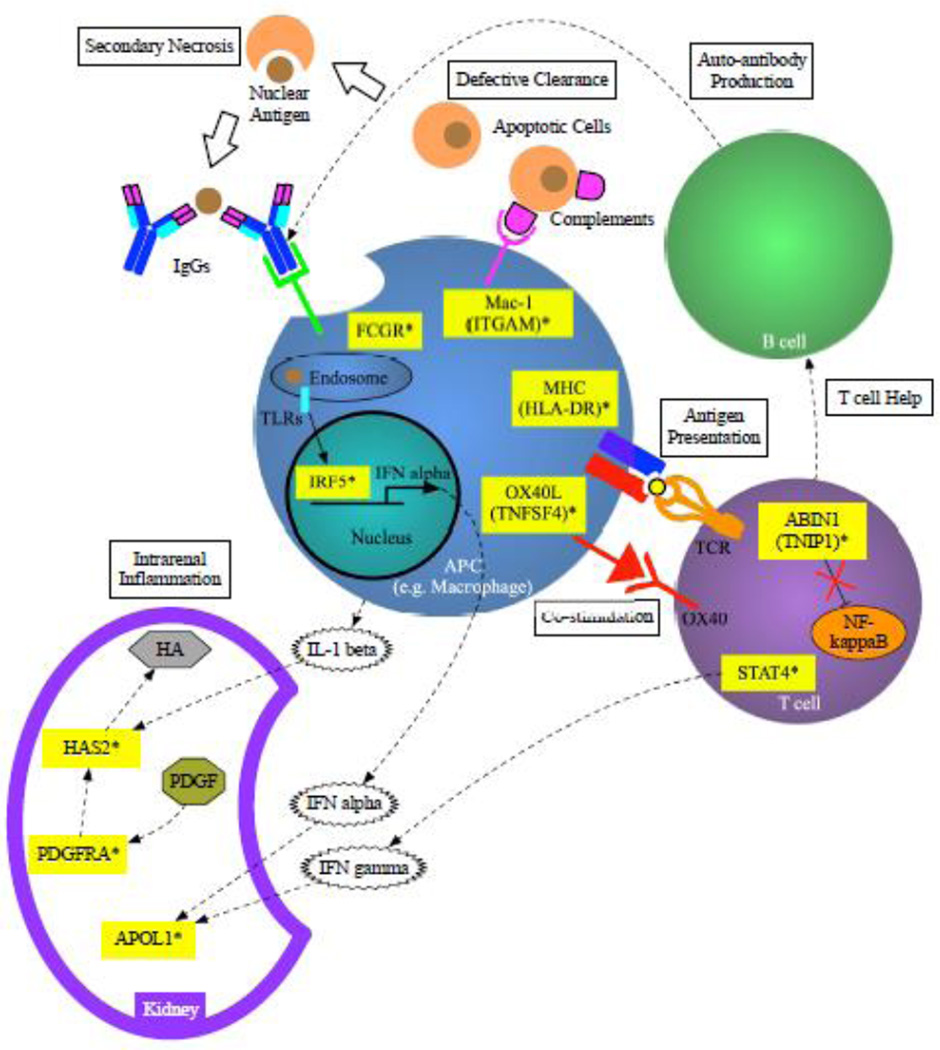
Figure 3. Genetic contribution to immune system in the pathogenesis of LN.
Altered functions of complement receptors (Mac-1) and FcgR will result in defective clearance of apoptotic cells and move towards secondary necrosis. Phagocyted nuclear antigens will activate TLRs and altered IRF5 leads to increased production of IFN alpha. Co-stimulatory factors (i.e. HLA and OX40L) activate T cells and result in IFN gamma production via STAT4 activation. Altered ABIN1 function, caused by TNIP1 variant leads to dysregulation of NF-kappaB activation. Auto-antibodies will be produced by B cells with the help of activated T cells. IFNs will induce APOL1 expression in the kidney. IL-1 beta, secreted by activated APC (e.g. macrophages) and the signal of PDGF via PDGFR will eventually lead to increased expression of HA via HAS2.
ABIN1, A20 binding and inhibitor of NF-kappaB 1; APC, antigen presenting cell; APOL1, apolipoprotein L-1; FCGR, Fc gamma receptor; HA, hyaluronan; HAS2, hyaluronan synthase 2; HLA, human leukocyte antigen; IFN, interferon; IL, interleukin; IRF5, interferon regulatory factor 5; ITGAM, integrin alpha M; OX40L, OX40 ligand; PDGFRA, platelet-derived growth factor receptor alpha; STAT4, signal transducers and activators of transcription 4; TLRs, Toll-like receptors; TNFSF4, tumor necrosis factor super family 4;
* Susceptible genes.
Highlights.
Some general SLE susceptibility genes predispose to LN
APOL1, PDGFR and HAS2 are novel LN susceptibility genes
Combination of general SLE and LN specific genes contribute to LN
Acknowledgments
Funding Sources: NIH grants (AR060861, AR057781, AR065964, AI071651), Rheumatology Research Foundation, CureJM Foundation, the Mayo Clinic Foundation, the Lupus Research Institute, and the Lupus Foundation of Minnesota to TBN.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Tsokos GC. Systemic lupus erythematosus. The New England journal of medicine. 2011;365:2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez E, Rasmussen A, Riba L, Acevedo-Vasquez E, Kelly JA, Langefeld CD, Williams AH, Ziegler JT, Comeau ME, Marion MC, Garcia-De La Torre I, Maradiaga-Cecena MA, Cardiel MH, Esquivel-Valerio JA, Rodriguez-Amado J, Moctezuma JF, Miranda P, Perandones CE, Castel C, Laborde HA, Alba P, Musuruana JL, Goecke IA, Anaya JM, Kaufman KM, Adler A, Glenn SB, Brown EE, Alarcon GS, Kimberly RP, Edberg JC, Vila LM, Criswell LA, Gilkeson GS, Niewold TB, Martin J, Vyse TJ, Boackle SA, Ramsey-Goldman R, Scofield RH, Petri M, Merrill JT, Reveille JD, Tsao BP, Orozco L, Baca V, Moser KL, Gaffney PM, James JA, Harley JB, Tusie-Luna T, Pons-Estel BA, Jacob CO, Alarcon-Riquelme ME. Impact of genetic ancestry and sociodemographic status on the clinical expression of systemic lupus erythematosus in American Indian-European populations. Arthritis and rheumatism. 2012;64:3687–3694. doi: 10.1002/art.34650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borchers AT, Leibushor N, Naguwa SM, Cheema GS, Shoenfeld Y, Gershwin ME. Lupus nephritis: a critical review. Autoimmunity reviews. 2012;12:174–194. doi: 10.1016/j.autrev.2012.08.018. [DOI] [PubMed] [Google Scholar]
- 4.Borchers AT, Naguwa SM, Shoenfeld Y, Gershwin ME. The geoepidemiology of systemic lupus erythematosus. Autoimmunity reviews. 2010;9:A277–A287. doi: 10.1016/j.autrev.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 5.Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, Walker A, Mack TM. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis and rheumatism. 1992;35:311–318. doi: 10.1002/art.1780350310. [DOI] [PubMed] [Google Scholar]
- 6.Alarcon-Segovia D, Alarcon-Riquelme ME, Cardiel MH, Caeiro F, Massardo L, Villa AR, Pons-Estel BA. Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis and rheumatism. 2005;52:1138–1147. doi: 10.1002/art.20999. [DOI] [PubMed] [Google Scholar]
- 7.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO, James JA, Merrill JT, Gilkeson GS, Seldin MF, Yin H, Baechler EC, Li QZ, Wakeland EK, Bruner GR, Kaufman KM, Kelly JA. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nature genetics. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, Lee AT, Chung SA, Ferreira RC, Pant PV, Ballinger DG, Kosoy R, Demirci FY, Kamboh MI, Kao AH, Tian C, Gunnarsson I, Bengtsson AA, Rantapaa-Dahlqvist S, Petri M, Manzi S, Seldin MF, Ronnblom L, Syvanen AC, Criswell LA, Gregersen PK, Behrens TW. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. The New England journal of medicine. 2008;358:900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 9.Chung SA, Brown EE, Williams AH, Ramos PS, Berthier CC, Bhangale T, Alarcon-Riquelme ME, Behrens TW, Criswell LA, Graham DC, Demirci FY, Edberg JC, Gaffney PM, Harley JB, Jacob CO, Kamboh MI, Kelly JA, Manzi S, Moser-Sivils KL, Russell LP, Petri M, Tsao BP, Vyse TJ, Zidovetzki R, Kretzler M, Kimberly RP, Freedman BI, Graham RR, Langefeld CD. Lupus nephritis susceptibility loci in women with systemic lupus erythematosus. Journal of the American Society of Nephrology : JASN. 2014;25:2859–2870. doi: 10.1681/ASN.2013050446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freedman BI, Langefeld CD, Andringa KK, Croker JA, Williams AH, Garner NE, Birmingham DJ, Hebert LA, Hicks PJ, Segal MS, Edberg JC, Brown EE, Alarcon GS, Costenbader KH, Comeau ME, Criswell LA, Harley JB, James JA, Kamen DL, Lim SS, Merrill JT, Sivils KL, Niewold TB, Patel NM, Petri M, Ramsey-Goldman R, Reveille JD, Salmon JE, Tsao BP, Gibson KL, Byers JR, Vinnikova AK, Lea JP, Julian BA, Kimberly RP. End-stage renal disease in African Americans with lupus nephritis is associated with APOL1. Arthritis & rheumatology (Hoboken, N.J.) 2014;66:390–396. doi: 10.1002/art.38220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanly JG, O’Keeffe AG, Su L, Urowitz MB, Romero-Diaz J, Gordon C, Bae SC, Bernatsky S, Clarke AE, Wallace DJ, Merrill JT, Isenberg DA, Rahman A, Ginzler EM, Fortin P, Gladman DD, Sanchez-Guerrero J, Petri M, Bruce IN, Dooley MA, Ramsey-Goldman R, Aranow C, Alarcon GS, Fessler BJ, Steinsson K, Nived O, Sturfelt GK, Manzi S, Khamashta MA, van Vollenhoven RF, Zoma AA, Ramos-Casals M, Ruiz-Irastorza G, Lim SS, Stoll T, Inanc M, Kalunian KC, Kamen DL, Maddison P, Peschken CA, Jacobsen S, Askanase A, Theriault C, Thompson K, Farewell V. The frequency and outcome of lupus nephritis: results from an international inception cohort study. Rheumatology (Oxford, England) 2016;55:252–262. doi: 10.1093/rheumatology/kev311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costenbader KH, Desai A, Alarcon GS, Hiraki LT, Shaykevich T, Brookhart MA, Massarotti E, Lu B, Solomon DH, Winkelmayer WC. Trends in the incidence, demographics, and outcomes of end-stage renal disease due to lupus nephritis in the US from 1995 to 2006. Arthritis and rheumatism. 2011;63:1681–1688. doi: 10.1002/art.30293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croca SC, Rodrigues T, Isenberg DA, Assessment of a lupus nephritis cohort over a 30-year period. Rheumatology (Oxford. England) 2011;50:1424–1430. doi: 10.1093/rheumatology/ker101. [DOI] [PubMed] [Google Scholar]
- 14.Calvo-Alen J, Reveille JD, Rodriguez-Valverde V, McGwin G, Jr, Baethge BA, Friedman AW, Alarcon GS. Clinical, immunogenetic and outcome features of Hispanic systemic lupus erythematosus patients of different ethnic ancestry. Lupus. 2003;12:377–385. doi: 10.1191/0961203303lu372oa. [DOI] [PubMed] [Google Scholar]
- 15.Seligman VA, Lum RF, Olson JL, Li H, Criswell LA. Demographic differences in the development of lupus nephritis: a retrospective analysis. The American journal of medicine. 2002;112:726–729. doi: 10.1016/s0002-9343(02)01118-x. [DOI] [PubMed] [Google Scholar]
- 16.Alarcon GS, McGwin G, Jr, Petri M, Reveille JD, Ramsey-Goldman R, Kimberly RP. Baseline characteristics of a multiethnic lupus cohort: PROFILE. Lupus. 2002;11:95–101. doi: 10.1191/0961203302lu155oa. [DOI] [PubMed] [Google Scholar]
- 17.Barr RG, Seliger S, Appel GB, Zuniga R, D’Agati V, Salmon J, Radhakrishnan J. Prognosis in proliferative lupus nephritis: the role of socio-economic status and race/ethnicity. Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association. 2003;18:2039–2046. doi: 10.1093/ndt/gfg345. [DOI] [PubMed] [Google Scholar]
- 18.Freedman BI, Wilson CH, Spray BJ, Tuttle AB, Olorenshaw IM, Kammer GM. Familial clustering of end-stage renal disease in blacks with lupus nephritis. American journal of kidney diseases : the official journal of the National Kidney Foundation. 1997;29:729–732. doi: 10.1016/s0272-6386(97)90126-8. [DOI] [PubMed] [Google Scholar]
- 19.Goldberg MA, Arnett FC, Bias WB, Shulman LE. Histocompatibility antigens in systemic lupus erythematosus. Arthritis and rheumatism. 1976;19:129–132. doi: 10.1002/art.1780190201. [DOI] [PubMed] [Google Scholar]
- 20.Graham RR, Ortmann W, Rodine P, Espe K, Langefeld C, Lange E, Williams A, Beck S, Kyogoku C, Moser K, Gaffney P, Gregersen PK, Criswell LA, Harley JB, Behrens TW. Specific combinations of HLA-DR2 and DR3 class II haplotypes contribute graded risk for disease susceptibility and autoantibodies in human SLE. European journal of human genetics : EJHG. 2007;15:823–830. doi: 10.1038/sj.ejhg.5201827. [DOI] [PubMed] [Google Scholar]
- 21.Bolin K, Sandling JK, Zickert A, Jonsen A, Sjowall C, Svenungsson E, Bengtsson AA, Eloranta ML, Ronnblom L, Syvanen AC, Gunnarsson I, Nordmark G. Association of STAT4 polymorphism with severe renal insufficiency in lupus nephritis. PloS one. 2013;8:e84450. doi: 10.1371/journal.pone.0084450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Taylor KE, Chung SA, Graham RR, Ortmann WA, Lee AT, Langefeld CD, Jacob CO, Kamboh MI, Alarcon-Riquelme ME, Tsao BP, Moser KL, Gaffney PM, Harley JB, Petri M, Manzi S, Gregersen PK, Behrens TW, Criswell LA. Risk alleles for systemic lupus erythematosus in a large case-control collection and associations with clinical subphenotypes. PLoS genetics. 2011;7:e1001311. doi: 10.1371/journal.pgen.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niu Z, Zhang P, Tong Y. Value of HLA-DR genotype in systemic lupus erythematosus and lupus nephritis: a meta-analysis. International journal of rheumatic diseases. 2015;18:17–28. doi: 10.1111/1756-185X.12528. [DOI] [PubMed] [Google Scholar]
- 24.Deshmukh US, Sim DL, Dai C, Kannapell CJ, Gaskin F, Rajagopalan G, David CS, Fu SM. HLA-DR3 restricted T cell epitope mimicry in induction of autoimmune response to lupus-associated antigen SmD. Journal of autoimmunity. 2011;37:254–262. doi: 10.1016/j.jaut.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fagerholm SC, MacPherson M, James MJ, Sevier-Guy C, Lau CS. The CD11b-integrin (ITGAM) and systemic lupus erythematosus. Lupus. 2013;22:657–663. doi: 10.1177/0961203313491851. [DOI] [PubMed] [Google Scholar]
- 26.Rhodes B, Furnrohr BG, Roberts AL, Tzircotis G, Schett G, Spector TD, Vyse TJ. The rs1143679 (R77H) lupus associated variant of ITGAM (CD11b) impairs complement receptor 3 mediated functions in human monocytes. Annals of the rheumatic diseases. 2012;71:2028–2034. doi: 10.1136/annrheumdis-2012-201390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, Chen W, Zhu C, McEver RP, Kimberly RP, Alarcon-Riquelme ME, Vyse TJ, Li QZ, Wakeland EK, Merrill JT, James JA, Kaufman KM, Guthridge JM, Harley JB. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nature genetics. 2008;40:152–154. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 28.Han S, Kim-Howard X, Deshmukh H, Kamatani Y, Viswanathan P, Guthridge JM, Thomas K, Kaufman KM, Ojwang J, Rojas-Villarraga A, Baca V, Orozco L, Rhodes B, Choi CB, Gregersen PK, Merrill JT, James JA, Gaffney PM, Moser KL, Jacob CO, Kimberly RP, Harley JB, Bae SC, Anaya JM, Alarcon-Riquelme ME, Matsuda K, Vyse TJ, Nath SK. Evaluation of imputation-based association in and around the integrin-alpha-M (ITGAM) gene and replication of robust association between a non-synonymous functional variant within ITGAM and systemic lupus erythematosus (SLE) Human molecular genetics. 2009;18:1171–1180. doi: 10.1093/hmg/ddp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang W, Zhao M, Hirankarn N, Lau CS, Mok CC, Chan TM, Wong RW, Lee KW, Mok MY, Wong SN, Avihingsanon Y, Lin IO, Lee TL, Ho MH, Lee PP, Wong WH, Sham PC, Lau YL. ITGAM is associated with disease susceptibility and renal nephritis of systemic lupus erythematosus in Hong Kong Chinese and Thai. Human molecular genetics. 2009;18:2063–2070. doi: 10.1093/hmg/ddp118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim-Howard X, Maiti AK, Anaya JM, Bruner GR, Brown E, Merrill JT, Edberg JC, Petri MA, Reveille JD, Ramsey-Goldman R, Alarcon GS, Vyse TJ, Gilkeson G, Kimberly RP, James JA, Guthridge JM, Harley JB, Nath SK. ITGAM coding variant (rs1143679) influences the risk of renal disease, discoid rash and immunological manifestations in patients with systemic lupus erythematosus with European ancestry. Annals of the rheumatic diseases. 2010;69:1329–1332. doi: 10.1136/ard.2009.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sanchez E, Nadig A, Richardson BC, Freedman BI, Kaufman KM, Kelly JA, Niewold TB, Kamen DL, Gilkeson GS, Ziegler JT, Langefeld CD, Alarcon GS, Edberg JC, Ramsey-Goldman R, Petri M, Brown EE, Kimberly RP, Reveille JD, Vila LM, Merrill JT, Anaya JM, James JA, Pons-Estel BA, Martin J, Park SY, Bang SY, Bae SC, Moser KL, Vyse TJ, Criswell LA, Gaffney PM, Tsao BP, Jacob CO, Harley JB, Alarcon-Riquelme ME, Sawalha AH. Phenotypic associations of genetic susceptibility loci in systemic lupus erythematosus. Annals of the rheumatic diseases. 2011;70:1752–1757. doi: 10.1136/ard.2011.154104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anaya JM, Kim-Howard X, Prahalad S, Chernavsky A, Canas C, Rojas-Villarraga A, Bohnsack J, Jonsson R, Bolstad AI, Brun JG, Cobb B, Moser KL, James JA, Harley JB, Nath SK. Evaluation of genetic association between an ITGAM non-synonymous SNP (rs1143679) and multiple autoimmune diseases. Autoimmunity reviews. 2012;11:276–280. doi: 10.1016/j.autrev.2011.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li X, Ptacek TS, Brown EE, Edberg JC. Fcgamma receptors: structure, function and role as genetic risk factors in SLE. Genes and immunity. 2009;10:380–389. doi: 10.1038/gene.2009.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ren Y, Tang J, Mok MY, Chan AW, Wu A, Lau CS. Increased apoptotic neutrophils and macrophages and impaired macrophage phagocytic clearance of apoptotic neutrophils in systemic lupus erythematosus. Arthritis and rheumatism. 2003;48:2888–2897. doi: 10.1002/art.11237. [DOI] [PubMed] [Google Scholar]
- 35.Gaipl US, Voll RE, Sheriff A, Franz S, Kalden JR, Herrmann M. Impaired clearance of dying cells in systemic lupus erythematosus. Autoimmunity reviews. 2005;4:189–194. doi: 10.1016/j.autrev.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 36.Takai T. Roles of Fc receptors in autoimmunity. Nature reviews. Immunology. 2002;2:580–592. doi: 10.1038/nri856. [DOI] [PubMed] [Google Scholar]
- 37.Edberg JC, Langefeld CD, Wu J, Moser KL, Kaufman KM, Kelly J, Bansal V, Brown WM, Salmon JE, Rich SS, Harley JB, Kimberly RP. Genetic linkage and association of Fcgamma receptor IIIA (CD16A) on chromosome 1q23 with human systemic lupus erythematosus. Arthritis and rheumatism. 2002;46:2132–2140. doi: 10.1002/art.10438. [DOI] [PubMed] [Google Scholar]
- 38.Salmon JE, Millard S, Schachter LA, Arnett FC, Ginzler EM, Gourley MF, Ramsey-Goldman R, Peterson MG, Kimberly RP. Fc gamma RIIA alleles are heritable risk factors for lupus nephritis in African Americans. The Journal of clinical investigation. 1996;97:1348–1354. doi: 10.1172/JCI118552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gelmetti AP, Freitas AC, Woronik V, Barros RT, Bonfa E, Monteiro RC. Polymorphism of the FcgammaRIIalpha IgG receptor in patients with lupus nephritis and glomerulopathy. The Journal of rheumatology. 2006;33:523–530. [PubMed] [Google Scholar]
- 40.Wu J, Edberg JC, Redecha PB, Bansal V, Guyre PM, Coleman K, Salmon JE, Kimberly RP. A novel polymorphism of FcgammaRIIIa (CD16) alters receptor function and predisposes to autoimmune disease. The Journal of clinical investigation. 1997;100:1059–1070. doi: 10.1172/JCI119616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Seligman VA, Suarez C, Lum R, Inda SE, Lin D, Li H, Olson JL, Seldin MF, Criswell LA. The Fcgamma receptor IIIA-158F allele is a major risk factor for the development of lupus nephritis among Caucasians but not non-Caucasians. Arthritis and rheumatism. 2001;44:618–625. doi: 10.1002/1529-0131(200103)44:3<618::AID-ANR110>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 42.Karassa FB, Trikalinos TA, Ioannidis JP. The Fc gamma RIIIA-F158 allele is a risk factor for the development of lupus nephritis: a meta-analysis. Kidney international. 2003;63:1475–1482. doi: 10.1046/j.1523-1755.2003.00873.x. [DOI] [PubMed] [Google Scholar]
- 43.Jonsen A, Gunnarsson I, Gullstrand B, Svenungsson E, Bengtsson AA, Nived O, Lundberg IE, Truedsson L, Sturfelt G. Association between SLE nephritis and polymorphic variants of the CRP and FcgammaRIIIa genes. Rheumatology (Oxford, England) 2007;46:1417–1421. doi: 10.1093/rheumatology/kem167. [DOI] [PubMed] [Google Scholar]
- 44.Bredius RG, de Vries CE, Troelstra A, van Alphen L, Weening RS, van de Winkel JG, Out TA. Phagocytosis of Staphylococcus aureus and Haemophilus influenzae type B opsonized with polyclonal human IgG1 and IgG2 antibodies. Functional hFc gamma RIIa polymorphism to IgG2. Journal of immunology (Baltimore, Md. : 1950) 1993;151:1463–1472. [PubMed] [Google Scholar]
- 45.Haseley LA, Wisnieski JJ, Denburg MR, Michael-Grossman AR, Ginzler EM, Gourley MF, Hoffman JH, Kimberly RP, Salmon JE. Antibodies to C1q in systemic lupus erythematosus: characteristics and relation to Fc gamma RIIA alleles. Kidney international. 1997;52:1375–1380. doi: 10.1038/ki.1997.464. [DOI] [PubMed] [Google Scholar]
- 46.Zuniga R, Markowitz GS, Arkachaisri T, Imperatore EA, D’Agati VD, Salmon JE. Identification of IgG subclasses and C-reactive protein in lupus nephritis: the relationship between the composition of immune deposits and FCgamma receptor type IIA alleles. Arthritis and rheumatism. 2003;48:460–470. doi: 10.1002/art.10930. [DOI] [PubMed] [Google Scholar]
- 47.Alarcon GS, McGwin G, Jr, Petri M, Ramsey-Goldman R, Fessler BJ, Vila LM, Edberg JC, Reveille JD, Kimberly RP. Time to renal disease and end-stage renal disease in PROFILE: a multiethnic lupus cohort. PLoS medicine. 2006;3:e396. doi: 10.1371/journal.pmed.0030396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sigurdsson S, Nordmark G, Goring HH, Lindroos K, Wiman AC, Sturfelt G, Jonsen A, Rantapaa-Dahlqvist S, Moller B, Kere J, Koskenmies S, Widen E, Eloranta ML, Julkunen H, Kristjansdottir H, Steinsson K, Alm G, Ronnblom L, Syvanen AC. Polymorphisms in the tyrosine kinase 2 and interferon regulatory factor 5 genes are associated with systemic lupus erythematosus. American journal of human genetics. 2005;76:528–537. doi: 10.1086/428480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, Ortmann WA, Koeuth T, Gonzalez Escribano MF, Pons-Estel B, Petri M, Daly M, Gregersen PK, Martin J, Altshuler D, Behrens TW, Alarcon-Riquelme ME. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- 50.Sigurdsson S, Goring HH, Kristjansdottir G, Milani L, Nordmark G, Sandling JK, Eloranta ML, Feng D, Sangster-Guity N, Gunnarsson I, Svenungsson E, Sturfelt G, Jonsen A, Truedsson L, Barnes BJ, Alm G, Ronnblom L, Syvanen AC. Comprehensive evaluation of the genetic variants of interferon regulatory factor 5 (IRF5) reveals a novel 5 bp length polymorphism as strong risk factor for systemic lupus erythematosus. Human molecular genetics. 2008;17:872–881. doi: 10.1093/hmg/ddm359. [DOI] [PubMed] [Google Scholar]
- 51.Lopez de Padilla CM, Niewold TB. The type I interferons: Basic concepts and clinical relevance in immune-mediated inflammatory diseases. Gene. 2016;576:14–21. doi: 10.1016/j.gene.2015.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Niewold TB, Kelly JA, Flesch MH, Espinoza LR, Harley JB, Crow MK. Association of the IRF5 risk haplotype with high serum interferon-alpha activity in systemic lupus erythematosus patients. Arthritis and rheumatism. 2008;58:2481–2487. doi: 10.1002/art.23613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng D, Stone RC, Eloranta ML, Sangster-Guity N, Nordmark G, Sigurdsson S, Wang C, Alm G, Syvanen AC, Ronnblom L, Barnes BJ. Genetic variants and disease-associated factors contribute to enhanced interferon regulatory factor 5 expression in blood cells of patients with systemic lupus erythematosus. Arthritis and rheumatism. 2010;62:562–573. doi: 10.1002/art.27223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cherian TS, Kariuki SN, Franek BS, Buyon JP, Clancy RM, Niewold TB. Brief Report: IRF5 systemic lupus erythematosus risk haplotype is associated with asymptomatic serologic autoimmunity and progression to clinical autoimmunity in mothers of children with neonatal lupus. Arthritis and rheumatism. 2012;64:3383–3387. doi: 10.1002/art.34571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Niewold TB, Kelly JA, Kariuki SN, Franek BS, Kumar AA, Kaufman KM, Thomas K, Walker D, Kamp S, Frost JM, Wong AK, Merrill JT, Alarcon-Riquelme ME, Tikly M, Ramsey-Goldman R, Reveille JD, Petri MA, Edberg JC, Kimberly RP, Alarcon GS, Kamen DL, Gilkeson GS, Vyse TJ, James JA, Gaffney PM, Moser KL, Crow MK, Harley JB. IRF5 haplotypes demonstrate diverse serological associations which predict serum interferon alpha activity and explain the majority of the genetic association with systemic lupus erythematosus. Annals of the rheumatic diseases. 2012;71:463–468. doi: 10.1136/annrheumdis-2011-200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heyninck K, De Valck D, Vanden Berghe W, Van Criekinge W, Contreras R, Fiers W, Haegeman G, Beyaert R. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. The Journal of cell biology. 1999;145:1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Verstrepen L, Carpentier I, Verhelst K, Beyaert R. ABINs: A20 binding inhibitors of NF-kappa B and apoptosis signaling. Biochemical pharmacology. 2009;78:105–114. doi: 10.1016/j.bcp.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 58.Caster DJ, Korte EA, Nanda SK, McLeish KR, Oliver RK, G’Sell RT, Sheehan RM, Freeman DW, Coventry SC, Kelly JA, Guthridge JM, James JA, Sivils KL, Alarcon-Riquelme ME, Scofield RH, Adrianto I, Gaffney PM, Stevens AM, Freedman BI, Langefeld CD, Tsao BP, Pons-Estel BA, Jacob CO, Kamen DL, Gilkeson GS, Brown EE, Alarcon GS, Edberg JC, Kimberly RP, Martin J, Merrill JT, Harley JB, Kaufman KM, Reveille JD, Anaya JM, Criswell LA, Vila LM, Petri M, Ramsey-Goldman R, Bae SC, Boackle SA, Vyse TJ, Niewold TB, Cohen P, Powell DW. ABIN1 dysfunction as a genetic basis for lupus nephritis. Journal of the American Society of Nephrology : JASN. 2013;24:1743–1754. doi: 10.1681/ASN.2013020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han JW, Zheng HF, Cui Y, Sun LD, Ye DQ, Hu Z, Xu JH, Cai ZM, Huang W, Zhao GP, Xie HF, Fang H, Lu QJ, Xu JH, Li XP, Pan YF, Deng DQ, Zeng FQ, Ye ZZ, Zhang XY, Wang QW, Hao F, Ma L, Zuo XB, Zhou FS, Du WH, Cheng YL, Yang JQ, Shen SK, Li J, Sheng YJ, Zuo XX, Zhu WF, Gao F, Zhang PL, Guo Q, Li B, Gao M, Xiao FL, Quan C, Zhang C, Zhang Z, Zhu KJ, Li Y, Hu DY, Lu WS, Huang JL, Liu SX, Li H, Ren YQ, Wang ZX, Yang CJ, Wang PG, Zhou WM, Lv YM, Zhang AP, Zhang SQ, Lin D, Li Y, Low HQ, Shen M, Zhai ZF, Wang Y, Zhang FY, Yang S, Liu JJ, Zhang XJ. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nature genetics. 2009;41:1234–1237. doi: 10.1038/ng.472. [DOI] [PubMed] [Google Scholar]
- 60.Gateva V, Sandling JK, Hom G, Taylor KE, Chung SA, Sun X, Ortmann W, Kosoy R, Ferreira RC, Nordmark G, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jonsen A, Bengtsson AA, Rantapaa-Dahlqvist S, Baechler EC, Brown EE, Alarcon GS, Edberg JC, Ramsey-Goldman R, McGwin G, Jr, Reveille JD, Vila LM, Kimberly RP, Manzi S, Petri MA, Lee A, Gregersen PK, Seldin MF, Ronnblom L, Criswell LA, Syvanen AC, Behrens TW, Graham RR. A large-scale replication study identifies TNIP1, PRDM1, JAZF1, UHRF1BP1 and IL10 as risk loci for systemic lupus erythematosus. Nature genetics. 2009;41:1228–1233. doi: 10.1038/ng.468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang C, Ahlford A, Jarvinen TM, Nordmark G, Eloranta ML, Gunnarsson I, Svenungsson E, Padyukov L, Sturfelt G, Jonsen A, Bengtsson AA, Truedsson L, Eriksson C, Rantapaa-Dahlqvist S, Sjowall C, Julkunen H, Criswell LA, Graham RR, Behrens TW, Kere J, Ronnblom L, Syvanen AC, Sandling JK. Genes identified in Asian SLE GWASs are also associated with SLE in Caucasian populations. European journal of human genetics : EJHG. 2013;21:994–999. doi: 10.1038/ejhg.2012.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kawasaki A, Ito S, Furukawa H, Hayashi T, Goto D, Matsumoto I, Kusaoi M, Ohashi J, Graham RR, Matsuta K, Behrens TW, Tohma S, Takasaki Y, Hashimoto H, Sumida T, Tsuchiya N Association of TNFAIP3 interacting protein 1. TNIP1 with systemic lupus erythematosus in a Japanese population: a case-control association study. Arthritis research & therapy. 2010;12:R174. doi: 10.1186/ar3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Adrianto I, Wang S, Wiley GB, Lessard CJ, Kelly JA, Adler AJ, Glenn SB, Williams AH, Ziegler JT, Comeau ME, Marion MC, Wakeland BE, Liang C, Kaufman KM, Guthridge JM, Alarcon-Riquelme ME, Alarcon GS, Anaya JM, Bae SC, Kim JH, Joo YB, Boackle SA, Brown EE, Petri MA, Ramsey-Goldman R, Reveille JD, Vila LM, Criswell LA, Edberg JC, Freedman BI, Gilkeson GS, Jacob CO, James JA, Kamen DL, Kimberly RP, Martin J, Merrill JT, Niewold TB, Pons-Estel BA, Scofield RH, Stevens AM, Tsao BP, Vyse TJ, Langefeld CD, Harley JB, Wakeland EK, Moser KL, Montgomery CG, Gaffney PM. Association of two independent functional risk haplotypes in TNIP1 with systemic lupus erythematosus. Arthritis and rheumatism. 2012;64:3695–3705. doi: 10.1002/art.34642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, de Bakker PI, Le JM, Lee HS, Batliwalla F, Li W, Masters SL, Booty MG, Carulli JP, Padyukov L, Alfredsson L, Klareskog L, Chen WV, Amos CI, Criswell LA, Seldin MF, Kastner DL, Gregersen PK. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. The New England journal of medicine. 2007;357:977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Taylor KE, Remmers EF, Lee AT, Ortmann WA, Plenge RM, Tian C, Chung SA, Nititham J, Hom G, Kao AH, Demirci FY, Kamboh MI, Petri M, Manzi S, Kastner DL, Seldin MF, Gregersen PK, Behrens TW, Criswell LA. Specificity of the STAT4 genetic association for severe disease manifestations of systemic lupus erythematosus. PLoS genetics. 2008;4:e1000084. doi: 10.1371/journal.pgen.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawasaki A, Ito I, Hikami K, Ohashi J, Hayashi T, Goto D, Matsumoto I, Ito S, Tsutsumi A, Koga M, Arinami T, Graham RR, Hom G, Takasaki Y, Hashimoto H, Behrens TW, Sumida T, Tsuchiya N. Role of STAT4 polymorphisms in systemic lupus erythematosus in a Japanese population: a case-control association study of the STAT1-STAT4 region. Arthritis research & therapy. 2008;10:R113. doi: 10.1186/ar2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Crispin JC, Oukka M, Bayliss G, Cohen RA, Van Beek CA, Stillman IE, Kyttaris VC, Juang YT, Tsokos GC. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. Journal of immunology (Baltimore, Md. : 1950) 2008;181:8761–8766. doi: 10.4049/jimmunol.181.12.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nguyen KB, Watford WT, Salomon R, Hofmann SR, Pien GC, Morinobu A, Gadina M, O’Shea JJ, Biron CA. Critical role for STAT4 activation by type 1 interferons in the interferon-gamma response to viral infection. Science (New York, N.Y.) 2002;297:2063–2066. doi: 10.1126/science.1074900. [DOI] [PubMed] [Google Scholar]
- 69.Kariuki SN, Kirou KA, MacDermott EJ, Barillas-Arias L, Crow MK, Niewold TB. Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo. Journal of immunology (Baltimore, Md. : 1950) 2009;182:34–38. doi: 10.4049/jimmunol.182.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhou XJ, Cheng FJ, Qi YY, Zhao MH, Zhang H. A replication study from Chinese supports association between lupus-risk allele in TNFSF4 and renal disorder. BioMed research international. 2013;2013:597921. doi: 10.1155/2013/597921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Farres MN, Al-Zifzaf DS, Aly AA, Abd Raboh NM. OX40/OX40L in systemic lupus erythematosus: association with disease activity and lupus nephritis. Annals of Saudi medicine. 2011;31:29–34. doi: 10.4103/0256-4947.75775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Patschan S, Dolff S, Kribben A, Durig J, Patschan D, Wilde B, Specker C, Philipp T, Witzke O. CD134 expression on CD4+ T cells is associated with nephritis and disease activity in patients with systemic lupus erythematosus. Clinical and experimental immunology. 2006;145:235–242. doi: 10.1111/j.1365-2249.2006.03141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Monajemi H, Fontijn RD, Pannekoek H, Horrevoets AJ. The apolipoprotein L gene cluster has emerged recently in evolution and is expressed in human vascular tissue. Genomics. 2002;79:539–546. doi: 10.1006/geno.2002.6729. [DOI] [PubMed] [Google Scholar]
- 74.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science (New York N.Y.) 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Thomson R, Genovese G, Canon C, Kovacsics D, Higgins MK, Carrington M, Winkler CA, Kopp J, Rotimi C, Adeyemo A, Doumatey A, Ayodo G, Alper SL, Pollak MR, Friedman DJ, Raper J. Evolution of the primate trypanolytic factor APOL1. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E2130–E2139. doi: 10.1073/pnas.1400699111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tzur S, Rosset S, Shemer R, Yudkovsky G, Selig S, Tarekegn A, Bekele E, Bradman N, Wasser WG, Behar DM, Skorecki K. Missense mutations in the APOL1 gene are highly associated with end stage kidney disease risk previously attributed to the MYH9 gene. Human genetics. 2010;128:345–350. doi: 10.1007/s00439-010-0861-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kopp JB, Nelson GW, Sampath K, Johnson RC, Genovese G, An P, Friedman D, Briggs W, Dart R, Korbet S, Mokrzycki MH, Kimmel PL, Limou S, Ahuja TS, Berns JS, Fryc J, Simon EE, Smith MC, Trachtman H, Michel DM, Schelling JR, Vlahov D, Pollak M, Winkler CA. APOL1 genetic variants in focal segmental glomerulosclerosis and HIV-associated nephropathy. Journal of the American Society of Nephrology : JASN. 2011;22:2129–2137. doi: 10.1681/ASN.2011040388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kruzel-Davila E, Wasser WG, Aviram S, Skorecki K. APOL1 nephropathy: from gene to mechanisms of kidney injury. Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association. 2016;31:349–358. doi: 10.1093/ndt/gfu391. [DOI] [PubMed] [Google Scholar]
- 79.Lin CP, Adrianto I, Lessard CJ, Kelly JA, Kaufman KM, Guthridge JM, Freedman BI, Anaya JM, Alarcon-Riquelme ME, Pons-Estel BA, Martin J, Glenn S, Adler A, Bae SC, Park SY, Bang SY, Song YW, Boackle SA, Brown EE, Edberg JC, Alarcon GS, Petri MA, Criswell LA, Ramsey-Goldman R, Reveille JD, Vila LM, Gilkeson GS, Kamen DL, Ziegler J, Jacob CO, Rasmussen A, James JA, Kimberly RP, Merrill JT, Niewold TB, Scofield RH, Stevens AM, Tsao BP, Vyse TJ, Langefeld CD, Moser KL, Harley JB, Gaffney PM, Montgomery CG. Role of MYH9 and APOL1 in African and non-African populations with lupus nephritis. Genes and immunity. 2012;13:232–238. doi: 10.1038/gene.2011.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Nichols B, Jog P, Lee JH, Blackler D, Wilmot M, D’Agati V, Markowitz G, Kopp JB, Alper SL, Pollak MR, Friedman DJ. Innate immunity pathways regulate the nephropathy gene Apolipoprotein L1. Kidney international. 2015;87:332–342. doi: 10.1038/ki.2014.270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Wan G, Zhaorigetu S, Liu Z, Kaini R, Jiang Z, Hu CA. Apolipoprotein L1, a novel Bcl-2 homology domain 3-only lipid-binding protein, induces autophagic cell death. The Journal of biological chemistry. 2008;283:21540–21549. doi: 10.1074/jbc.M800214200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Floege J, Eitner F, Alpers CE. A new look at platelet-derived growth factor in renal disease. Journal of the American Society of Nephrology : JASN. 2008;19:12–23. doi: 10.1681/ASN.2007050532. [DOI] [PubMed] [Google Scholar]
- 83.Boor P, Ostendorf T, Floege J. PDGF and the progression of renal disease, Nephrology, dialysis, transplantation : official publication of the European Dialysis and Transplant Association - European Renal Association. 2014;(29 Suppl 1):i45–i54. doi: 10.1093/ndt/gft273. [DOI] [PubMed] [Google Scholar]
- 84.Sadanaga A, Nakashima H, Masutani K, Miyake K, Shimizu S, Igawa T, Sugiyama N, Niiro H, Hirakata H, Harada M. Amelioration of autoimmune nephritis by imatinib in MRL/lpr mice. Arthritis and rheumatism. 2005;52:3987–3996. doi: 10.1002/art.21424. [DOI] [PubMed] [Google Scholar]
- 85.Zoja C, Corna D, Rottoli D, Zanchi C, Abbate M, Remuzzi G. Imatinib ameliorates renal disease and survival in murine lupus autoimmune disease. Kidney international. 2006;70:97–103. doi: 10.1038/sj.ki.5001528. [DOI] [PubMed] [Google Scholar]
- 86.Shinkai Y, Cameron JS. Trial of platelet-derived growth factor antagonist, trapidil, in accelerated nephrotoxic nephritis in the rabbit. British journal of experimental pathology. 1987;68:847–852. [PMC free article] [PubMed] [Google Scholar]
- 87.Jones S, Jones S, Phillips AO. Regulation of renal proximal tubular epithelial cell hyaluronan generation: implications for diabetic nephropathy. Kidney international. 2001;59:1739–1749. doi: 10.1046/j.1523-1755.2001.0590051739.x. [DOI] [PubMed] [Google Scholar]
- 88.Asselman M, Verhulst A, Van Ballegooijen ES, Bangma CH, Verkoelen CF, De Broe ME. Hyaluronan is apically secreted and expressed by proliferating or regenerating renal tubular cells. Kidney international. 2005;68:71–83. doi: 10.1111/j.1523-1755.2005.00382.x. [DOI] [PubMed] [Google Scholar]
- 89.Kastner S, Thomas GJ, Jenkins RH, Davies M, Steadman R. Hyaluronan induces the selective accumulation of matrix- and cell-associated proteoglycans by mesangial cells. The American journal of pathology. 2007;171:1811–1821. doi: 10.2353/ajpath.2007.070085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Michael DR, Phillips AO, Krupa A, Martin J, Redman JE, Altaher A, Neville RD, Webber J, Kim MY, Bowen T. The human hyaluronan synthase 2 (HAS2) gene and its natural antisense RNA exhibit coordinated expression in the renal proximal tubular epithelial cell. The Journal of biological chemistry. 2011;286:19523–19532. doi: 10.1074/jbc.M111.233916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yung S, Tsang RC, Leung JK, Chan TM. Increased mesangial cell hyaluronan expression in lupus nephritis is mediated by anti-DNA antibody-induced IL-1beta. Kidney international. 2006;69:272–280. doi: 10.1038/sj.ki.5000042. [DOI] [PubMed] [Google Scholar]
- 92.Zhang Y, Zhao M, Sawalha AH, Richardson B, Lu Q. Impaired DNA methylation and its mechanisms in CD4(+)T cells of systemic lupus erythematosus. Journal of autoimmunity. 2013;41:92–99. doi: 10.1016/j.jaut.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 93.Richardson B, Scheinbart L, Strahler J, Gross L, Hanash S, Johnson M. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis and rheumatism. 1990;33:1665–1673. doi: 10.1002/art.1780331109. [DOI] [PubMed] [Google Scholar]
- 94.Zhao M, Liu S, Luo S, Wu H, Tang M, Cheng W, Zhang Q, Zhang P, Yu X, Xia Y, Yi N, Gao F, Wang L, Yung S, Chan TM, Sawalha AH, Richardson B, Gershwin ME, Li N, Lu Q. DNA methylation and mRNA and microRNA expression of SLE CD4+ T cells correlate with disease phenotype. Journal of autoimmunity. 2014;54:127–136. doi: 10.1016/j.jaut.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 95.Renauer P, Coit P, Jeffries MA, Merrill JT, McCune WJ, Maksimowicz-McKinnon K, Sawalha AH. DNA methylation patterns in naive CD4+ T cells identify epigenetic susceptibility loci for malar rash and discoid rash in systemic lupus erythematosus. Lupus science & medicine. 2015;2:e000101. doi: 10.1136/lupus-2015-000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Absher DM, Li X, Waite LL, Gibson A, Roberts K, Edberg J, Chatham WW, Kimberly RP. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS genetics. 2013;9:e1003678. doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Coit P, Jeffries M, Altorok N, Dozmorov MG, Koelsch KA, Wren JD, Merrill JT, McCune WJ, Sawalha AH. Genome-wide DNA methylation study suggests epigenetic accessibility and transcriptional poising of interferon-regulated genes in naive CD4+ T cells from lupus patients. Journal of autoimmunity. 2013;43:78–84. doi: 10.1016/j.jaut.2013.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Coit P, Renauer P, Jeffries MA, Merrill JT, McCune WJ, Maksimowicz-McKinnon K, Sawalha AH. Renal involvement in lupus is characterized by unique DNA methylation changes in naive CD4+ T cells. Journal of autoimmunity. 2015;61:29–35. doi: 10.1016/j.jaut.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]