Abstract
Objective
Body fat mass (BFM) is more homogeneous and accurate than body total mass in measuring obesity, but has rarely been studied. Aiming to uncover the genetic basis of fat-induced obesity, we performed a genome-wide association meta-analysis of BFM, after adjustment by body lean mass, in the European population.
Methods
Three samples of European ancestry were included into the meta-analysis: the Framingham heart study (FHS, N=6,004), the Kansas-city osteoporosis study (KCOS, N=2,207) and the Omaha osteoporosis study (OOS, N=968).
Results
At genome-wide significance level (α=5.0×10−8), we identified a cluster of 10 SNPs at chromosomal region 20p11 that were associated with BFM (lead SNP rs2069126 p=1.82×10−9, closest gene SLC24A3) in 9,179 subjects. One of the top SNPs rs6046308 (p=3.74×10−8) is nominally significant for body fat percentage in another independent study (p=0.03, N=75,888), and is reported to trans-regulate the expression of MPZ gene at 1q23.3 (unadjusted p=9.78×10−6, N=1,490). Differential gene expression analysis demonstrates that SLC24A3 and CFAP61 at the identified locus are differentially expressed in tissues of people with versus without obesity (p=3.40×10−5 and 8.72×10−4, N=126 and 70), implying their potential role in fat development.
Conclusions
Our results may provide new insights into the biological mechanism that underlies fat-induced obesity pathology.
Keywords: obesity, body fat mass, genome-wide association study, 20p11
Introduction
Obesity is a chronic metabolic disease characterized by an increase of body fat stores. It is a serious public health problem associated with an increased risk of type 2 diabetes, hypertension, cardiovascular disease and certain forms of cancer (1). It has become one of the leading causes of disability and death worldwide. In the United States, over two-thirds of the adults are overweight and over one third are obesity (2). The total annual economic cost associated with obesity in the US is in excess of $215 billion (3).
Body mass index (BMI, unit kg/m2), defined as the body weight divided by the square of height, is currently the standard measure of obesity. Twin, family and adoption studies have established the strong genetic determination of human obesity and the reported heritability for BMI is over 40% (4). To date, a variety of genome-wide association studies (GWASes) and meta-analyses have been conducted and hundreds of genetic variants have been identified (5, 6). Despite these fruitful discoveries, the cumulative explained heritability of BMI is less than 5% (5). On the other hand, it is estimated that over 20% of the obesity variation can be accounted for by common genomic variants (5). Therefore, the vast majority of genetic locus underlying obesity remains to be discovered.
Though BMI is popularly studied, it is not an ideal phenotype to measure obesity. This is because body mass is a composite combination of various types of body composition including fat mass, lean mass, bone mass and other soft tissues. Of them, fat mass is the only composition that gives rise to obesity that yields severe clinical outcomes (7). On the other hand, it is the lean mass instead of fat mass that takes up the vast majority of body mass. Therefore, BMI is an composite index of body composition, rather than of fat mass alone. Some more specific measures, such as body fat percentage and waist-to-hip ratio, have been extensively studied (8, 9, 10).
For the purpose of mapping fat-induced obesity genes, fat mass is the only accurate and ideal phenotype to model. Nonetheless, it has been studied very sparsely, partly because the phenotype is difficult to collect accurately. In the present study, aiming to uncover the genetic basis of fat-induced obesity, we conduct a genome-wide association meta-analysis of body fat mass in the European population. Specifically, we conduct a -meta-analysis in three independent samples. Our findings may be helpful in uncovering the genetic basis of fat mass development and of obesity.
Methods
This study was approved by local institutional review boards of all participating institutions. All participants provided informed consent before being enrolled into the study. The Framingham heart study (FHS) was accessed through the database of genotype and phenotype (dbGAP) repository (11). Neither phenotyping nor genotyping in the FHS was performed by the authors. Instead, they were acquired through the dbGAP.
FHS sample
Details of the FHS sample were described elsewhere (12). Briefly, the FHS is a longitudinal and prospective cohort comprising >16,000 related participants spanning three generations of European ancestry. The original generation was comprised of 5,209 participants living in the town of Framingham, Massachusetts. The offspring generation was comprised of 5,124 participants who were adult children of original cohort members or were spouses of these offspring, and the third generation was comprised of over 4,000 participants who were grandchildren of the original generation.
The original generation participants underwent bone densitometry scan by dual-energy X-ray absorptiometry (DXA) during their 22nd or 24th examination. The offspring participants underwent DXA scan during their 6th or 7th examination, and the third generation participants underwent DXA scan during their second examination. Body fat mass (BFM) and total soft tissue body mass were measured by the DXA scanners. Body lean mass (BLM) was calculated by subtracting BFM from total soft tissue mass.
OOS sample
The Omaha osteoporosis study (OOS) is a cross-sectional study of osteoporosis with 1,000 unrelated participants of European ancestry living in Omaha, NE, USA, and its surrounding areas. The participants were normal healthy subjects defined by a comprehensive suite of exclusion criteria, as described elsewhere (6). BFM and BLM were measured by DXA scanner (Hologic Inc., Bedford, MA, USA), following the manufacturer's protocol.
KCOS sample
The Kansas City osteoporosis study (KCOS) is a cross-sectional study of osteoporosis with 2,286 unrelated participants of European ancestry living in Kansas City, MO, USA, and its surrounding areas. The participants were normal healthy subjects defined by a comprehensive suite of exclusion criteria, as described elsewhere (13). BFM and BLM were measured again by DXA scanner (Hologic Inc., Bedford, MA, USA), following the manufacturer's protocol.
Phenotype measurements and modeling
In all samples, covariates, including gender, age, age squared, height, height squared and the first five principal components derived from genome-wide genotype data (14), were screened for significance with the step-wise linear regression model implemented in the stepAIC function in R package MASS. To correct for the effect of lean mass, BLM was also included as covariate. Raw BFM was adjusted by significant covariates, and the residuals were then normalized by inverse quantiles of standard normal distribution. Normalized residuals were used for subsequent association analysis.
Genotyping and quality control
All GWAS samples were genotyped by high-throughput SNP genotyping arrays (Affymetrix Inc., Santa Clara, CA) following the manufacturer’s protocols. Quality control (QC) within each sample was implemented at both individual level and SNP level. At the individual level, sex compatibility was checked by imputing sex from X-chromosome genotype data with Plink (15). Individuals of ambiguous imputed sex or with inconsistent imputed sex with the reported one were removed. At the SNP level, SNPs violating the Hardy-Weinberg equilibrium (HWE) rule (p-value<1.0×10−5) were removed. In the familial sample FHS, genotypes having the Mendel error were set to missing. To examine the population outliers, if present, genotype-derived principal components were monitored. Population outliers would be reported and removed.
Genotype imputation
All samples were imputed by the 1000 genomes project phase 3 sequence variants (as of May, 2013) (16). Haplotypes representing 240 individuals of European ancestry were downloaded from the project download site. Haplotypes of bi-allelic variants, including SNPs and deletion/insertion variants (DIVs), were extracted to form reference panel for imputation. As a QC procedure, variants with fewer than two copies (zero or one) of minor allele were removed.
Prior to imputation, a consistency test of allele frequency between GWAS and reference samples was examined with the chi-square test. To correct for potential mis-strandedness, GWAS SNPs that failed the consistency test (p<1.0×10−6) were transformed into the reverse strand. SNPs that again failed consistency test were removed from the GWAS sample. Imputation was performed with FISH (17), an algorithm that we developed to impute the diploid genome fast and accurately without the need to phasing genotype into haplotype. The software parameters were set by default.
Association testing in individual samples
In the FHS sample, the association between normalized phenotype residual and genotyped and imputed genotype was examined under an additive mode of inheritance. A mixed linear model was used in which the effect of genetic relatedness within each pedigree was taken into account (18). Briefly, within-family relatedness was modeled as random effects while genotype effect was modeled as fixed effects. Association test was examined within the variance-components framework.
In both OOS and KCOS samples, association was examined by a linear regression model with MACH2QTL (19), in which allele dosage was taken as the predictor for the phenotype.
Meta-analysis
Association signals from individual samples were combined for meta-analysis. The meta-analysis was performed with the sample size weighted fixed-effects model implemented in the software METAL (20). The genomic control inflation factor was estimated, and was used to correct for association signals when necessary.
As a QC step, only well-imputed, which was defined as the SNP whose imputation certainty measure r2 was larger than 0.3 in at least two of the three samples, and common (minor allele frequency, MAF>0.05) SNPs were included into analysis.
Replicability analysis of previously identified SNPs for BMI and body fat percentage
We checked if the previously identified SNPs for BMI and body fat percentage (BFP) could be replicated by the present study for BFM. For BMI, we searched the NHGRI-EBI GWAS catalog by the trait "body mass index" (as of June 26, 2016) (21), and identified a total of 109 SNPs locating into 95 genomic regions that were reported previously. For BFP, we combined the results of two GWAS meta-analysis studies and identified a total of 13 lead SNPs (8, 9).
Functional annotation
We annotated the functional relevance of the identified SNPs with HaploReg (22). HaploReg annotates SNPs into different functional categories according to the information from a variety of large experiment projects. These categories include conservation sites, DNase hypersensitivity region, transcription factor binding sites, promoter, enhancer, and others.
Differential gene expression analysis
To provide biological support for the relevant genes, we examined their differential expressions in expression data sets accessed through the Gene Expression Omnibus (GEO) repository (http://www.ncbi.nlm.nih.gov/geo/). We restricted the datasets to be fat or adipose tissue related human subjects/tissues. We searched the GEO datasets using each of the following terms "adipose tissue", "adiposity" and "adipose stem cell" and identified 30 datasets. After careful examination of the retrieved items, 7 datasets were left for analysis. In each dataset, we grouped subjects into case (obesity) group and control (non-obesity) group. Genome-wide gene expressions were logarithm-transformed and normalized. Normalized expression levels were compared between the two groups using the GEO online tool GEO2R (ncbi.nlm.nih.gov/geo/geo2r/). The expression difference was characterized by logarithm-transformed fold change (logFC) and p-value. P-values from individual datasets were further combined into fixed-effects meta-analysis with METAL, where the weight was proportional to sample size and the sign of logFC served as effect direction.
Results
Meta-analysis
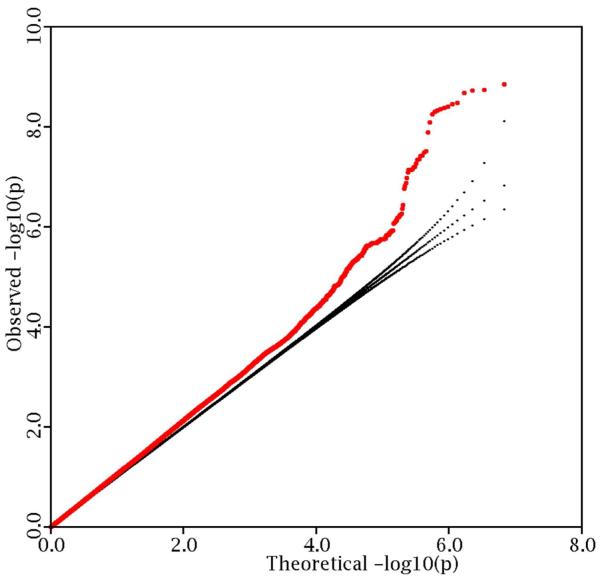
A total of 9,179 subjects are included into the meta-analysis. Basic characteristics of the samples are listed in Table 1. Sixty-two percent of the participants are women. Principal component analysis (PCA) was applied to each individual sample, and no population outliers were observed. Imputation by the 1000 genomes sequencing project generated 12,403,269 bi-allelic variants. After removing variants of low-frequency (MAF<0.05) or of poor imputation certainty (r2<0.3 in two or three samples), 6,817,908 variants are qualified for analysis; 88.3 percent (6,018,982) of them are SNPs, and the remaining 11.7 percent (798,926) are DIVs. After adjusting the phenotype by PCA in each individual study, the genomic control inflation factor of the meta-analysis is 1.08. A p-value logarithmic quantile-quantile (QQ) plot shows the concordance with the expected line (y=x) in p-value range [10−2, 1], indicating limited apparent population stratification (Figure 1).
Table 1.
Basic characteristics of the studied samples.Sample
| Female | Age | Weight | Height | BFM | BLM | ||
|---|---|---|---|---|---|---|---|
|
|
|||||||
| N | (%) | (kg) | (cm) | (kg) | (kg) | ||
| OOS | 968 | 49.3 | 50.5(18.3) | 81.0(17.8) | 170.9(9.7) | 24.9(9.8) | 56.1(13.2) |
| KCOS | 2207 | 76.7 | 51.5(13.7) | 75.0(17.3) | 166.2(8.4) | 24.2(10.6) | 51.4(11.2) |
| FHS | 6004 | 58.0 | 55.3(13.6) | 74.8(15.9) | 168.8(9.8) | 26.6(10.0) | 46.2(11.2) |
Notes: OOS, the Omaha osteoporosis study; KCOS, Kansas-city osteoporosis study; FHS, Framingham heart study; BFM, body fat mass; BLM, body lean mass. Presented was mean (s.d.). N was sample size after quality control.
Figure 1.
Logarithmic quantile–quantile (QQ) plot of the association p-values in the meta-analysis.
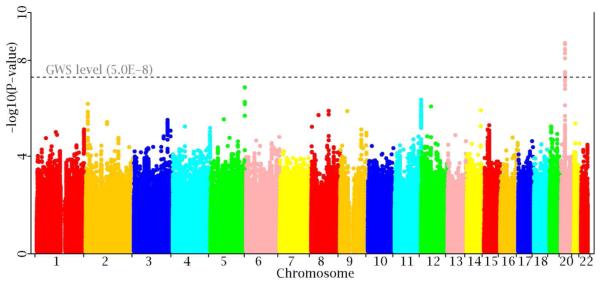
At the genome-wide significance (GWS, 5.0×10−8) level, a total of 20 SNPs are significant. These SNPs are located into the following 3 loci: 2q24.3 (rs10170665, p=3.22×10−8), 17q21 (rs4635383, p=4.40×10−8), and 20p11 (18 SNPs, lead SNP rs62203173 p=1.38×10−9). Here, an independent locus was defined as a genomic region of 1Mb at either side of the variant showing the strongest association.
Ten of the above associated SNPs, including the 3 lead SNPs, present strong heterogeneity effects (I2>50%). After applying the random-effects meta-analysis model to these SNPs, their signals become non-significant. The remaining 10 SNPs are located into the single locus 20p11 with the lead SNP being rs2069126 (p=1.82×10−9, I2=0.0%). They are generally clustered into one haplotype block, but with various levels of linkage disequilibrium (LD) pattern (r2=0.35-1.00, Supplementary Figure 1). Manhattan plot of the GWAS meta-analysis is displayed in Figure 2, and the main results are listed in Table 2.
Figure 2.
Manhattan plot of the meta-analysis.
Table 2.
Main results of significant SNPs at 20p11.
| FHS (N=6004) | KCOS (N=2207) | OOS (N=968) | Meta (N=9179) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|||||||||
| rs# | Position | Alleles | EAF | beta | p | beta | p | beta | p | beta | p | I2 (%) |
| rs910983 | 19738164 | C/T | 0.36 | −0.11 | 1.00×10−6 | −0.07 | 0.02 | −0.12 | 0.02 | −0.10 | 4.94×10−9 | 0.0 |
| rs111678496 | 19744988 | M/W | 0.64 | 0.11 | 4.25×10−7 | 0.07 | 0.02 | 0.12 | 0.02 | 0.10 | 2.08×10−9 | 0.0 |
| rs16981092 | 19745011 | A/G | 0.64 | 0.11 | 6.18×10−7 | 0.07 | 0.03 | 0.12 | 0.01 | 0.10 | 3.49×10−9 | 0.0 |
| rs6112563 | 19745561 | C/T | 0.36 | −0.11 | 5.94×10−7 | −0.07 | 0.03 | −0.12 | 0.02 | −0.10 | 3.35×10−9 | 0.0 |
| rs6112565 | 19748442 | A/G | 0.68 | 0.11 | 4.04×10−7 | 0.09 | 0.01 | 0.10 | 0.07 | 0.10 | 1.84×10−9 | 0.0 |
| rs2069126 | 19749957 | A/G | 0.36 | −0.11 | 3.26×10−7 | −0.07 | 0.03 | −0.12 | 0.02 | −0.10 | 1.82×10−9 | 0.0 |
| rs78194863 | 19751524 | C/G | 0.76 | 0.12 | 4.52×10−8 | 0.05 | 0.14 | 0.07 | 0.21 | 0.10 | 2.99×10−8 | 30.3 |
| rs6046308 | 19755673 | G/T | 0.26 | −0.12 | 1.95×10−8 | −0.05 | 0.17 | −0.04 | 0.38 | −0.09 | 3.74×10−8 | 49.1 |
| rs6046310 | 19757284 | A/G | 0.74 | 0.13 | 6.22×10−9 | 0.05 | 0.15 | 0.06 | 0.27 | 0.10 | 8.03×10−9 | 47.5 |
| rs12481515 | 19777184 | C/T | 0.84 | 0.14 | 5.87×10−8 | 0.08 | 0.05 | 0.03 | 0.66 | 0.11 | 4.48×10−8 | 40.7 |
Notes: The first and second alleles represent the effect and alternative alleles. EAF was the effect allele frequency. rs111678496 is an DIV, where "M" and "W" represent mutant (TGCTAGTCAGAT) and wild (-) alleles, respectively. Beta is the regression coefficient of normalized phenotypic residual on the effect allele.GWS significant p-values were marked in bold.
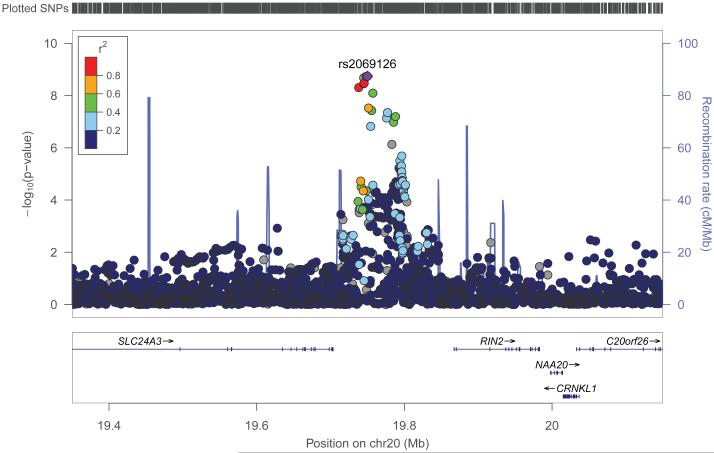
The lead SNP rs2069126 is an imputed SNP with high imputation certainty (r2=0.85-0.96 across samples). Allele A at this SNP is associated with decreased BFM consistently in all samples. Its p-value is extremely low in the FHS sample (3.26×10−7), and is nominally significant in both the OOS (p=0.02) and the KCOS (p=0.03) samples. Regional plot was drawn by LocusZoom (23), as displayed in Figure 3.
Figure 3.
Regional plot of 20p11.
Regional plot of the lead SNP rs2069126 and its flanking 400 kb region to either side was displayed.
Replication analysis with BMI and BFP
As different measures of obesity, BFM (after adjustment by BLM), BMI and BFP are correlated with each other. The correlation coefficients of adjusted BFM with BMI and BFP are 0.61 and 0.70, respectively, in the FHS sample. We cross-replicated the SNPs identified in the present study by the publicly accessible summary results for BMI and BFP. The BMI results were accessed from the GIANT consortium, and the BFP results were accessed from the reference (9). Of the 10 identified SNPs, the same 5 are present in both the BMI and BFP summary results. For BMI, none of the SNPs is significant, but all have consistent effect directions. For BFP, rs6046308 is nominally significant (p=0.03), and the other 4 SNPs have consistent effect directions (Supplementary Table 1).
We then checked if the present study could replicate the previously identified SNPs for BMI and BFP. For BMI, we searched the GWAS catalog and identified a total of 109 SNPs locating into 95 genomic loci. Two of the 109 SNPs are not present in the present study. The association signals of the remaining 107 SNPs are listed in Supplementary Table 2. Overall, we observed an excess of significant signals. For example, a total of 18 SNPs are significant at the nominal level 0.05, tripling the expected number when the null hypothesis of no association at all holds (pbinomial=6.08×10−6). Of the remaining 89 non-significant SNPs, an enrichment of effects in the same direction was also observed. Specifically, up to 61 SNPs have consistent effect directions with BFM (pbinomial=6.10×10−4), implying that small sample size may be the cause of non-replicability for BMI.
For BFP, a total of 13 lead SNPs were identified in two recent GWAS meta-analysis studies (8, 9). Of them, 6 are nominally significant in the present study (pbinomial=1.98×10−5), and 11 are consistent in effect direction (pbinomial=3.47×10−13), as listed in Supplementary Table 3. Overall, an excess of significant and consistent associations were observed too.
Association with other metabolic traits
To evaluate the association signals of our identified SNPs with other metabolic traits, we looked-up the large-scale GWAS meta-analysis results released by the following consortium: GIANT (waist-hip-ratio) (10), MAGIC (fasting glucose and two hour glucose adjusted by BMI) (24), LIPIDS (lipid) (25) and DIAGRAM (type 2 diabetes) (26). The results are listed in Supplementary Table 1. Only the level of lipid is weakly associated with rs16981092 and rs2069126 (both p=0.05), while none of the other traits is significant.
Functional annotation
We annotated the cluster of significant SNPs at the novel locus 20p11 through Haploreg. The lead SNP rs2069126 is located in the intergenic region between RIN2 (Ras and Rab interactor 2, 120.3kb to rs2069126) gene and SLC24A3 (Solute Carrier Family 24 Member 3, 46.4 kb to rs2069126) gene. Among its neighboring SNPs, rs6046308 (meta p=3.74×10−8) is annotated to trans-regulate the expression of MPZ (Myelin Protein Zero) gene located at 1q23.3 in peripheral blood monocyte cells extracted from 1,490 unrelated subjects (27). However, the p-value 9.78×10−6 is a little bit higher than the significance level when considering the Bonferroni correction of the 12,808 tested transcripts (α=3.90×10−6).
Several other genes, including NAA20, CRNKL1 and CFAP61, are also located at the novel locus 20p11. Their gene expression profiles in the GTEx project samples are displayed in Supplementary Figure 2. The three histone marks (H3K4me1, H3K4me3 and H3K27Ac) in seven ENCODE cell lines (GM12878, H1-hESC, HSMM, HUVEC, K562, NHEK and NHLF) at this locus were accessed from the UCSC genome browser and are displayed in Supplementary Figure 3.
Differential gene expression analysis
To provide biological insight in the role of relevant genes for fat development, we explored their differential expressions in data sets accessed through the GEO repository. Gene expression results are listed in Table 3. Of the 6 genes analyzed, SLC24A3 and CFAP61 are differentially expressed in tissues of obesity versus non-obesity human subjects (meta p=3.40×10−5 and 8.72×10−4). The effect directions are consistent across all the 6 GEO datasets: subjects with obesity tend to have decreased gene expressions than subjects without obesity have.
Table 3.
Differential gene expression analyses.
| GEO Series | N | RIN2 | MPZ | SLC24A3 | CRNKL1 | NAA20 | CFAP61 | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| logFC | p | logFC | p | logFC | p | logFC | p | logFC | p | logFC | p | ||
| GSE2508 | 9:10 | −0.34 | 0.05 | −0.01 | 0.95 | - | - | - | - | - | - | - | - |
| GSE48964 | 3:3 | 0.11 | 0.66 | 0.14 | 0.07 | −0.15 | 0.29 | 0.27 | 0.02 | 0.07 | 0.49 | −0.02 | 0.74 |
| GSE29718 | 10:10 | −0.12 | 0.23 | 0.58 | 0.10 | −0.33 | 0.06 | 0.12 | 0.12 | 0.01 | 0.93 | −0.15 | 0.05 |
| GSE27951 | 17:16 | 0.22 | 0.19 | −0.18 | 0.12 | −0.47 | 0.04 | 0.17 | 0.35 | 0.21 | 0.28 | −0.15 | 0.03 |
| GSE9624 | 5:6 | −1.05 | 0.08 | −0.01 | 0.98 | −0.99 | 2.74×10−3 | −0.24 | 0.28 | 0.18 | 0.62 | −1.13 | 0.08 |
| GSE15524 | 21:7 | 0.11 | 0.81 | −0.29 | 0.27 | −0.11 | 0.78 | −0.09 | 0.74 | 0.91 | 0.13 | - | - |
| GSE2510 | 14:14 | 0.05 | 0.76 | 0.11 | 0.71 | −0.45 | 0.02 | 0.02 | 0.80 | - | - | - | - |
|
| |||||||||||||
| Meta | 79:66 | 0.49 | 0.91 | 3.40×10−5 | 0.21 | 0.07 | 8.72×10−4 | ||||||
Notes: Seven datasets were accessed through the GEO repository. N was the sample size (case:control), where cases were obese human subjects and control were non-obese human subjects. logFC, the logarithm of fold change in cases to controls. "-": not available. Nominally significant
Discussion
In this study, aiming to identify additional genomic variants associated with human fat-induced obesity, we performed an imputation-based association meta-analysis in three independent samples of European ancestry. We identified a novel locus at 20p11 that was associated with BFM after adjustment by BLM.
The chromosome region 20p11 is important for human development. The inverted duplication deletion of this region results in a rare chromosomal disorder with clinical outcome of delayed developmental milestones and craniofacial dysmorphism (28). In another case report, a female patient suffering from several disorders including obesity was found to have a copy loss at 20p13 co-existing with a copy gain at 20p13-20p11.22 (29). Common genetic variants at this locus are also associated with complex disorders, such as type 2 diabetes susceptibility (30).
Several genes are located at the 20p11 locus, among which RIN2 was previously reported to associate with obesity risk. Using pig models, Kogelman et al. integrated transcriptomic and genomic data from subcutaneous adipose tissue and identified multiple causal genes including RIN2 for obesity (31). In another study of human childhood obesity, Comuzzie et al. found that RIN2 gene was significantly associated with free T4 (free thyroxine) (32), where the increment of free T4 was in turn associated with obesity risk. Other gene of interest in this region are SLC24A3 and CFAP61. These two genes are differentially expressed in human subjects with versus without obesity in our analysis of the GEO datasets. SLC24A3 was previously identified as having significantly decreased expression in diet-sensitive women with obesity (33). Its regulatory effect was proposed to achieve by mediating the level of glucose in the bloodstream (34). CFAP61 is highly expressed in skeletal muscle (Supplementary Figure 2). In a previous study, CFAP61 in skeletal muscle was differentially expressed between young and older human subjects (35). Its function may be related to calcium signaling and energy conversion (36).
Of the associated SNPs, rs6046308 is successfully replicated by the body fat percentage in another large-scale independent study. This SNP also trans-regulates the expression of MPZ gene at 1q23.3. Note that the p-value 9.78×10−6 is higher than the Bonferroni-corrected significance level (α=3.90×10−6), therefore this trans-eQTL activity may also be a false positive signal, among others. Literature review of the MPZ gene function implies its role in fat development. This gene provides instructions for making a protein called myelin protein zero. It is the most abundant protein in myelin, a protective substance that covers nerves and promotes the efficient transmission of nerve impulses (37). The protein is required for the proper formation and maintenance of myelin. One SNP rs4657015 near MPZ gene is reported to associate with fat deposition in visceral adipose tissue adjusted for BMI (38). Brain tissues from obese mice have significantly lower amount of myelin than the normal mice have (39). Moreover, diet-induced obesity decreases its expression (40). Therefore, one possible regulation mechanism of variants at 20p11 is the trans-regulatory effect to the MPZ gene expression.
It is notable that the vast majority (~85%) of the previously reported BMI-associated SNPs are not replicated in the present study. Considering the fact that these SNPs are convincingly associated with BMI, the non-replicability could be due to the small replication sample size. When analyzing effect direction, up to 79 of the 107 SNPs are indeed consistent.
Conclusion
In summary, by performing an imputation-based genome-wide association meta-analysis in the European population, we identified a novel locus at 20p11 for BFM. Functional annotation and gene expression analysis demonstrate that the associated variants may affect fat development by trans-regulating MPZ gene expression and/or cis-regulating nearby genes. Our results may provide new insights into the biology mechanism that underlies fat-induced obesity.
Supplementary Material
Study Importance Questions.
Question 1. What is already known about this subject?
1) Obesity is a major public health problem with strong genetic determination;
2) The vast majority of genetic loci underlying obesity remain to be discovered;
3) Body fat mass is more homogeneous and accurate than body total mass in measuring obesity. Nonetheless, it has been studied very sparsely.
Question 2. What does this study add?
1) By performing a genome-wide association meta-analysis of body fat mass in the European population, we identified a novel locus 20p11 (closest gene SLC24A3);
2) Functional annotations show that one of the top SNPs rs6046308 trans-regulates the expression of MPZ gene at 1q23.3. Differential gene expression analysis demonstrates that SLC24A3 and CFAP61 are differentially expressed in tissues of people with versus without obesity.
Acknowledgements
We are grateful to the two anonymous referees for their constructive comments that greatly improve this manuscript. We are also grateful to the GIANT consortium, the DIAGRAM consortium, the LIPID consortium, the MAGIC consortium, the GLGC consortium, and the body fat percentage study led by Dr. Ruth J.F. Loos for releasing the GWAS summary results. We thank study participants who participate into this study. Computing service was partially provided by the university of Shanghai for science and technology computing cluster.
The Framingham Heart Study is conducted and supported by the National Heart, Lung, and Blood Institute (NHLBI) in collaboration with Boston University (Contract No. N01-HC-25195). This manuscript was not prepared in collaboration with investigators of the Framingham Heart Study and does not necessarily reflect the opinions or views of the Framingham Heart Study, Boston University, or NHLBI. Funding for SHARe Affymetrix genotyping was provided by NHLBI Contract N02-HL-64278. SHARe Illumina genotyping was provided under an agreement between Illumina and Boston University. Funding support for the Framingham Whole Body and Regional Dual X-ray Absorptiometry (DXA) dataset was provided by NIH grants R01 AR/AG 41398. The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?db=gap through dbGaP accession phs000342.v14.p10.
Funding: This study was partially supported by the national natural science foundation of China (31501026 to Y.F.P., 31571291 and 31100902 to L.Z.), the natural science foundation of Jiangsu province of China (BK20150323 to Y.F.P.), the Scienti?c Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry (to Y.F.P.), the NIH (P50AR055081, R01AG026564, R01AR050496, RC2DE020756, R01AR057049, and R03TW008221 to H.W.D.), the Franklin D. Dickson/Missouri Endowment and the Edward G. Schlieder Endowment (to H.W.D.), the startup funding project of Soochow university (Q413900114 to Y.F.P. and Q413900214 to L.Z.), the undergraduate innovation program of Jiangsu province (201610285040Z to A.P.F.) and a project of the priority academic program development of Jiangsu higher education institutions (to Y.H.Z.).
Footnotes
DISCLOSURE: The authors declared no conflict of interest.
Author contributions: L.Z. designed the study. L.Z., Y.F.P, H.S., Q.T., and H.W.D. collected the data. Y.F.P., H.G.R. and L.Z. carried out the experiments. Y.F.P. and L.Z. analyzed the data. L.L., X.L., C.F., Y.H., W.W.K., A.P.F., W.Z.H., X.Y.Y. and W.Z. performed literature search. Y.F.P., H.G.R., C.F., Y.H., L.Z. and H.W.D. interpreted the data. Y.F.P. and L.Z. generated the figures. Y.F.P. drafted the early version of the manuscript. L.Z. and H.W.D. supervised the study. All authors were involved in writing the paper and had final approval of the submitted and published versions.
References
- 1.Kopelman PG. Obesity as a medical problem. Nature. 2000;404:635–643. doi: 10.1038/35007508. [DOI] [PubMed] [Google Scholar]
- 2.Yang L, Colditz GA. Prevalence of Overweight and Obesity in the United States, 2007-2012. JAMA Intern Med. 2015;175:1412–1413. doi: 10.1001/jamainternmed.2015.2405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammond RA, Levine R. The economic impact of obesity in the United States. Diabetes Metab Syndr Obes. 2010;3:285–295. doi: 10.2147/DMSOTT.S7384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bell CG, Walley AJ, Froguel P. The genetics of human obesity. Nat Rev Genet. 2005;6:221–234. doi: 10.1038/nrg1556. [DOI] [PubMed] [Google Scholar]
- 5.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pei YF, Zhang L, Liu Y, Li J, Shen H, Liu YZ, et al. Meta-analysis of genome-wide association data identifies novel susceptibility loci for obesity. Hum Mol Genet. 2014;23:820–830. doi: 10.1093/hmg/ddt464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Srikanthan Preethi, Horwich Tamara B., Tseng CH. Relation of Muscle Mass and Fat Mass to Cardiovascular Disease Mortality. The American Journal of Cardiology. 2016;117:1355–1360. doi: 10.1016/j.amjcard.2016.01.033. [DOI] [PubMed] [Google Scholar]
- 8.Kilpelainen TO, Zillikens MC, Stancakova A, Finucane FM, Ried JS, Langenberg C, et al. Genetic variation near IRS1 associates with reduced adiposity and an impaired metabolic profile. Nat Genet. 2011;43:753–760. doi: 10.1038/ng.866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lu Y, Day FR, Gustafsson S, Buchkovich ML, Na J, Bataille V, et al. New loci for body fat percentage reveal link between adiposity and cardiometabolic disease risk. Nature communications. 2016;7:10495. doi: 10.1038/ncomms10495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shungin D, Winkler TW, Croteau-Chonka DC, Ferreira T, Locke AE, Magi R, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawber TR, Meadors GF, Moore FE., Jr. Epidemiological approaches to heart disease: the Framingham Study. Am J Public Health Nations Health. 1951;41:279–281. doi: 10.2105/ajph.41.3.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei YF, Zhang L, Yang TL, Han Y, Hai R, Ran S, et al. Genome-wide association study of copy number variants suggests LTBP1 and FGD4 are important for alcohol drinking. PLoS One. 2012;7:e30860. doi: 10.1371/journal.pone.0030860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang L, Pei YF, Fu X, Lin Y, Wang YP, Deng HW. FISH: fast and accurate diploid genotype imputation via segmental hidden Markov model. Bioinformatics. 2014;30:1876–1883. doi: 10.1093/bioinformatics/btu143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang L, Li J, Pei YF, Liu Y, Deng HW. Tests of association for quantitative traits in nuclear families using principal components to correct for population stratification. Ann Hum Genet. 2009;73:601–613. doi: 10.1111/j.1469-1809.2009.00539.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genet Epidemiol. 2010;34:816–834. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2011;40:D930–934. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan J, et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44:991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willer CJ, Schmidt EM, Sengupta S, Peloso GM, Gustafsson S, Kanoni S, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–1283. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, et al. Genetics and beyond--the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leclercq S, Maincent K, Baverel F, Tessier DL, Letourneur F, Lebbar A, et al. Molecular cytogenetic characterization of the first reported case of inv dup del 20p compatible with a U-type exchange model. American journal of medical genetics Part A. 2009;149A:437–445. doi: 10.1002/ajmg.a.32640. [DOI] [PubMed] [Google Scholar]
- 29.Trachoo O, Assanatham M, Jinawath N, Nongnuch A. Chromosome 20p inverted duplication deletion identified in a Thai female adult with mental retardation, obesity, chronic kidney disease and characteristic facial features. European journal of medical genetics. 2013;56:319–324. doi: 10.1016/j.ejmg.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 30.Hayes MG, Pluzhnikov A, Miyake K, Sun Y, Ng MC, Roe CA, et al. Identification of type 2 diabetes genes in Mexican Americans through genome-wide association studies. Diabetes. 2007;56:3033–3044. doi: 10.2337/db07-0482. [DOI] [PubMed] [Google Scholar]
- 31.Kogelman LJ, Zhernakova DV, Westra HJ, Cirera S, Fredholm M, Franke L, et al. An integrative systems genetics approach reveals potential causal genes and pathways related to obesity. Genome Med. 2015;7:105. doi: 10.1186/s13073-015-0229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Comuzzie AG, Cole SA, Laston SL, Voruganti VS, Haack K, Gibbs RA, et al. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7:e51954. doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gerrits MF, Ghosh S, Kavaslar N, Hill B, Tour A, Seifert EL, et al. Distinct skeletal muscle fiber characteristics and gene expression in diet-sensitive versus diet-resistant obesity. Journal of lipid research. 2010;51:2394–2404. doi: 10.1194/jlr.P005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Logsdon BA, Hoffman GE, Mezey JG. Mouse obesity network reconstruction with a variational Bayes algorithm to employ aggressive false positive control. BMC bioinformatics. 2012;13:53. doi: 10.1186/1471-2105-13-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hangelbroek RW, Fazelzadeh P, Tieland M, Boekschoten MV, Hooiveld GJ, van Duynhoven JP, et al. Expression of protocadherin gamma in skeletal muscle tissue is associated with age and muscle weakness. Journal of cachexia, sarcopenia and muscle. 2016;7:604–614. doi: 10.1002/jcsm.12099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dymek EE, Smith EF. A conserved CaM- and radial spoke associated complex mediates regulation of flagellar dynein activity. The Journal of cell biology. 2007;179:515–526. doi: 10.1083/jcb.200703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shy ME. Peripheral neuropathies caused by mutations in the myelin protein zero. J Neurol Sci. 2006;242:55–66. doi: 10.1016/j.jns.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 38.Fox CS, Liu Y, White CC, Feitosa M, Smith AV, Heard-Costa N, et al. Genome-wide association for abdominal subcutaneous and visceral adipose reveals a novel locus for visceral fat in women. PLoS Genet. 2012;8:e1002695. doi: 10.1371/journal.pgen.1002695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sena A, Sarlieve LL, Rebel G. Brain myelin of genetically obese mice. J Neurol Sci. 1985;68:233–243. doi: 10.1016/0022-510x(85)90104-2. [DOI] [PubMed] [Google Scholar]
- 40.Jayaraman A, Lent-Schochet D, Pike CJ. Diet-induced obesity and low testosterone increase neuroinflammation and impair neural function. J Neuroinflammation. 2014;11:162. doi: 10.1186/s12974-014-0162-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.