Supplemental Digital Content is available in the text.
Keywords: cardiovascular diseases, genes, risk factors, RNAs, stroke
Abstract
Background and Purpose—
There is increasing interest in extracellular RNAs (ex-RNAs), with numerous reports of associations between selected microRNAs (miRNAs) and a variety of cardiovascular disease phenotypes. Previous studies of ex-RNAs in relation to risk for cardiovascular disease have investigated small numbers of patients and assayed only candidate miRNAs. No human studies have investigated links between novel ex-RNAs and stroke.
Methods—
We conducted unbiased next-generation sequencing using plasma from 40 participants of the FHS (Framingham Heart Study; Offspring Cohort Exam 8) followed by high-throughput polymerase chain reaction of 471 ex-RNAs. The reverse transcription quantitative polymerase chain reaction included 331 of the most abundant miRNAs, 43 small nucleolar RNAs, and 97 piwi-interacting RNAs in 2763 additional FHS participants and explored the relations of ex-RNAs and prevalent (n=63) and incident (n=51) stroke and coronary heart disease (prevalent=286, incident=69).
Results—
After adjustment for multiple cardiovascular disease risk factors, 7 ex-RNAs were associated with stroke prevalence or incidence; there were no ex-RNA associated with prevalent or incident coronary heart disease. Statistically significant ex-RNA associations with stroke were specific, with no overlap between prevalent and incident events.
Conclusions—
This is the largest study of ex-RNAs in relation to stroke using an unbiased approach in an observational cohort and the first large study to examine human small noncoding RNAs beyond miRNAs. These results demonstrate that when studied in a large observational cohort, extracellular miRNAs are associated with stroke risk.
Plasma extracellular noncoding RNAs (ex-RNAs) are a class of circulating RNA molecules that directly modulate networks of gene expression in target tissues. Given the stability and accessibility of ex-RNAs in plasma and their potential epigenetic role in pathogenesis of disease, there has been intense interest in identifying specific ex-RNAs as diagnostic, functional, and prognostic biomarkers of a variety of systemic disorders, including cardiovascular disease (CVD). Studies in animal models of stroke have provided mechanisms by which key ex-RNAs, specifically microRNAs (miRNAs), involved in inflammation and fibrosis may modulate organ-level phenotypes.1 In turn, the promise of these initial discoveries has engendered significant enthusiasm for ex-RNAs as novel biomarkers and therapeutic targets in patients with stroke.
Over the past 5 years, there has been burgeoning interest in ex-RNA translational investigation, with numerous reports of associations between selected miRNAs and a variety of CVD phenotypes, namely acute coronary syndromes, heart failure, coronary heart disease (CHD), and stroke. Although promising, most of these studies have relied on carefully selected populations with established disease and have largely assayed candidate miRNAs in relatively small study populations. Furthermore, to our knowledge, no human studies have investigated links between novel ex-RNAs known to modulate gene expression (eg, small nucleolar RNAs [snoRNAs] and piwi-interacting RNAs [piRNAs]) and stroke. Ultimately, the clinical translation of ex-RNAs as useful diagnostic biomarkers for stroke will rely on an unbiased evaluation in large populations.
To provide essential data for ex-RNA clinical translation, we investigated 2763 participants in FHS (Framingham Heart Study) and explored the relation of ex-RNAs to stroke and CHD. We used an unbiased 2-stage study design of next-generation RNA sequencing (RNASeq) for discovery followed by high-throughput reverse transcription quantitative polymerase chain reaction (RT-qPCR) to identify and quantify ex-RNAs and test their associations with stroke and CHD. To our knowledge, this report represents the largest population study identifying ex-RNAs associated with stroke or CHD.
Methods
Sample Population and Blood Collection
The FHS Offspring Cohort is a community-based, prospective study of CVD, with serial examinations every 4 to 8 years, with concomitant dense phenotyping of CVD and metabolic traits over multiple examinations. Subjects were diagnosed with stroke based on review of medical records, including relevant hospitalizations, and clinic-reported events by at least 2 neurologists agreeing on one of the following manifestations: definite cerebrovascular accident, atherothrombotic infarction of the brain, cerebral embolism, intracerebral hemorrhage, or subarachnoid hemorrhage. CHD was diagnosed if on review of the case, a panel of 3 investigators agreed on 1 of the following definite manifestations of CHD: myocardial infarction, coronary insufficiency, angina pectoris, sudden death from CHD, nonsudden death from CHD.
For the purposes of this analysis, prevalent cases of CHD or stroke were defined as any history recorded prior to the eighth visit for the Offspring Cohort (the second generation enrolled in the study), otherwise known as Exam 8, which is the date of the participants’ ex-RNA assessment. Incident cases were defined as any case occurring during 5 years after the Exam 8 visit date.
Blood samples previously collected in the FHS Offspring Cohort participants at Exam 8 (March 2005 to January 2008) were analyzed. Venipuncture was performed on study participants in a supine position after an overnight fast by using standard venipuncture techniques. Blood was collected into blood collection tubes with a liquid-buffered sodium citrate additive (0.105 mol/L), centrifuged, and plasma separated and immediately frozen at −80°C (all steps within 90 minutes of collection). An aliquot of 170 μL of plasma samples were transferred to our laboratory in March 2014 and stored at −80°C. To assess for cellular contamination, during the isolation phase, we carefully inspected all samples for overt hemolysis. In addition, RT-qPCR results were examined for hemolysis issue based on delta Cq values for miR-23a-miR-451. A delta Cq<5 suggests little or no hemolysis and >7 is significant hemolysis.2 For our samples, only 24 samples had a value between >7.0, representing <1% of all samples.
RNA Isolation, Library Preparation, and RNASeq
To obtain an unbiased screen of ex-RNAs from the Offspring Cohort, 40 total samples (21 with prevalent CVD and 19 age-/sex-matched FHS participants without CVD) were analyzed by RNASeq. Complete information for this data set and extensive and complete details of the specific methodology used, including quality controls for sample preparation and technologies, have been recently published.3 Briefly, RNA was isolated from plasma samples using a miRCURY RNA Isolation Kit–Biofluids (Exiqon) per the manufacturer’s protocol. A total of 130 μL plasma was used in each RNA isolation.
The Ion Total RNAseq Kit v2 (Life Technologies) was used for creating libraries for sequencing. Manufacturer’s instructions were followed, with a few exceptions as previously described.3 In collaboration with the Life Technologies R&D team, we used their underdevelopment Ion Adapter Mix to improve performance when using low template amounts. The Ion Chef System and Ion PI IC 200 kits were used for template preparation. The entire procedure was automated using the Ion Chef System. At the end of the template preparation, loaded PI Chips (Life Technologies) were placed on the sequencer. RNASeq was performed on the Ion Proton System with a sequencing depth of 10 million reads per sample. Sequencing reactions were performed on the Ion PI Chip Kit v2 BC and Ion Proton System (Life Technologies). Sequencing reads were at maximum 200 nucleotides.
Identification of Widely Expressed Ex-RNAs From RNASeq in FHS Participants
The Genboree Workbench (http://www.genboree.org/) developed by our collaborators at the National Institutes of Health as part of the Extracellular RNA Consortium was used for standard processing of small RNA reads from RNASeq. Small RNASeq reads were identified, aligned, and quantified for differential expression between CVD and non-CVD FHS participants using the exceRpt tool available on the Genboree Workbench, and full details of the package and processing were recently published.3
Each of the 40 small RNASeq samples from FHS participants was processed independently. The output of the pipeline is the expression values in terms of read counts for all annotated human miRNAs, piRNAs, snoRNAs, and transfer RNAs for each sample. The expression value for each sample was normalized to units of reads per million total mapped reads. Small ex-RNAs that had an average expression across 40 samples >1 reads per million total mapped reads were 669 human miRNAs, 144 human piRNAs, and 74 human snoRNAs.
Importantly, our RNASeq analysis of this limited sample size did not identify any plasma circulating ex-RNA significantly differentially expressed between prevalent CVD and non-CVD FHS participants after correction for multiple testing using the Benjamini and Hochberg false discovery rate. Given potential differences between RNASeq methodology and traditional PCR approaches used in most previous studies, we selected the most abundantly expressed ex-RNAs by RNASeq (331 miRNAs, 97 piRNAs, and 43 snoRNAs) for RT-qPCR analysis in the entire FHS Offspring Cohort as previously described.3
RT-qPCR of Human miRNAs, piRNAs, and snoRNAs in the Entire FHS Offspring 8 Cohort
Of the 2822 eligible subjects from the FHS Offspring Cohort at Exam 8 (baseline examination for this study), 59 (2%) subjects were excluded because of laboratory error (eg, inaccurate volume of plasma pipetted, N=31; poor protein precipitation performance, N=23; or potential contamination, N=5), resulting in 2763 subjects as a final study cohort. After removing poorly performing ex-RNA PCR assays (defined as consistent, nonvariable expression in samples, as well as no template reverse transcription control; N=15), we assayed expression of 331 human miRNAs, 97 human piRNAs, and 43 human snoRNAs. RNA was isolated using methods described earlier. RNAs were reverse transcribed as previously described3 using a Dynamic Array 96.96 GE (Fluidigm Corp). PCR reactions were stopped at 23 cycles (Cq≤23) based on manufacturer’s recommendation. It should be noted that because of the decreased volume using the Dynamic Array, this Cq is equivalent to a Cq of ≈30 by standard larger volume PCR methods with similar to superior reproducibility.
Statistical Analysis
As described earlier, we studied plasma samples from 2763 subjects. All statistical analyses were performed using STATA 13.0. Descriptive statistics are displayed as mean±SD for continuous variables and count (percentage) for categorical variables.
Model Fitting
Only those ex-RNA expressed in at least 100 FHS participants were included in RT-qPCR analyses (301 miRNAs, 59 piRNAs, and 38 snoRNAs; see Tables I and II in the online-only Data Supplement for lists of included and excluded ex-RNA). Ex-RNAs were considered expressed when Cq values were <23 (the upper limit for PCR reactions using the Biomark platform). Logistic regression models were used to identify ex-RNAs associated with prevalent CVD phenotypes, and Cox-proportional hazards models were used to identify ex-RNAs associated with incident cases occurring within 5 years of follow-up. The 5-year all-cause mortality rate in these data is 9.9 (8.6–11.5). Any impact of competing risk would likely be small because all cases of death and CVD onset are highly reviewed by the FHS, and biased results because of censoring would require very strong associations between mortality and also be strongly linked with CVD.
For both sets of analyses, multivariable regression models were fitted adjusting for known and suspected CVD risk factors (all assessed at the same examination and in fasting subjects). In addition to age and sex, these factors included smoking status (≥1 cigarette per day during the year prior to examination), systolic blood pressure, diastolic blood pressure, total cholesterol to high-density lipoprotein ratio, triglycerides, glucose level, diabetes mellitus (fasting plasma glucose ≥126 mg/dL or treatment with blood glucose–lowering medication4), hemoglobin A1C, C-reactive protein, lipid-lowering therapy, antihypertensive therapy, and regular aspirin use (at least 3× per week).
Hypothesis Testing
Bootstrapping was used to estimate unbiased test of statistical significance using resampling with replacement (N=50) for each of the multivariable regression models fitting CHD or stroke as a function of ex-RNA indicators of expression and confounding risk factors. To account for the number of statistical comparisons conducted for each of the 398 ex-RNAs assessed for the prevalence and incidence of stroke or CVD, we used false discovery rate correction within phenotype for logistic regression and Cox-proportional hazards models.
Results
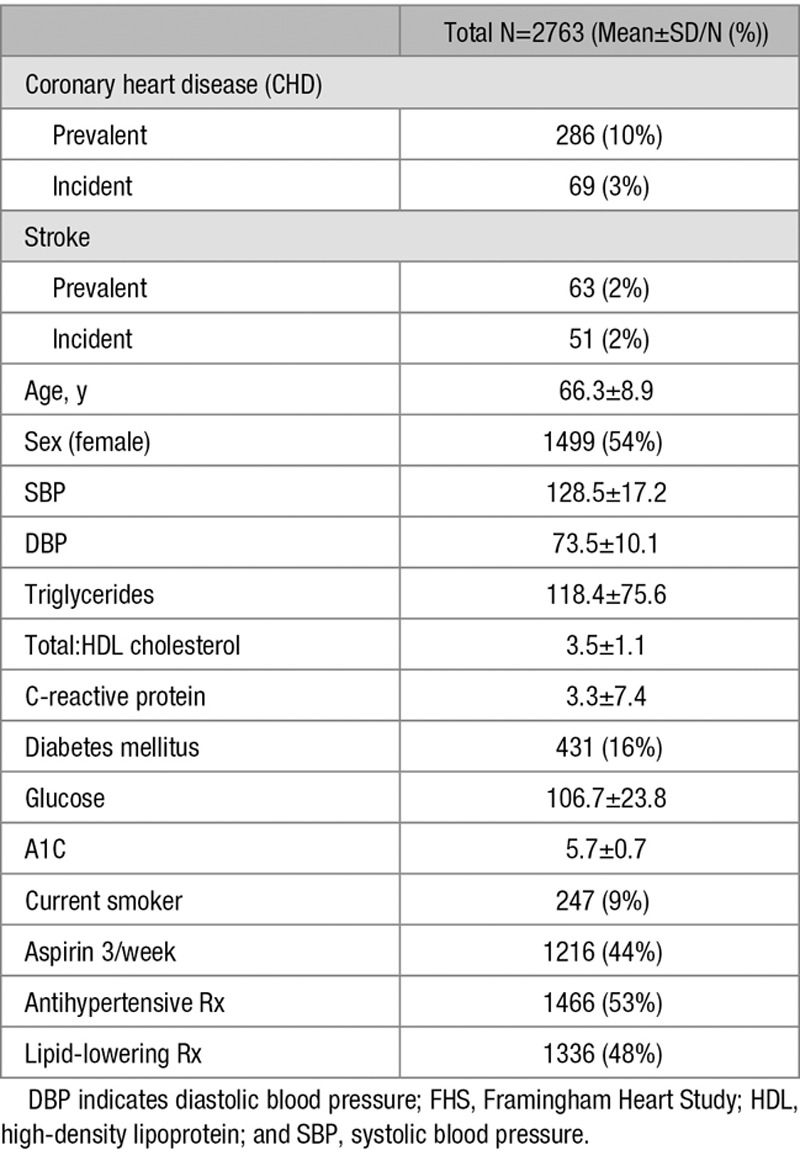
Characteristics of the 2763 FHS participants included in our validation analysis are shown in Table 1. Of the eligible individuals, 286 (10%) had prevalent CHD and 63 (2%) had prevalent stroke. The average duration from event diagnosis to the date of the baseline examination was 12.8±9.1 years for CHD events and 8.3±7.9 years for stroke events. A total of 69 (3%) at-risk participants developed an incident CHD event and 51 (2%) developed an incident stroke within 5 years of the baseline visit. The mean follow-up time to event was 2.2±1.3 years for CHD events and 2.5±1.6 years for stroke events.
Table 1.
Clinical and Demographic Characteristics of FHS Cohort (Offspring 8)
Ex-RNAs associated with prevalent and incident CHD or stroke are listed in Table 2, and the complete results for all ex-RNA modeled with multivariable regression is given in Table III in the online-only Data Supplement (full results for prevalent and incident CHD available in Table IV in the online-only Data Supplement). After adjustment for the number of ex-RNA assessed, there were no significant associations with prevalent CHD, but 3 miRNAs and 1 snoRNA were associated with prevalent stroke. We fit an additional adjusted multiple logistic regression model including all of the significant ex-RNA and found that each was independently associated with prevalent stroke: hsa-miR-877-5p (odds ratio, 0.18; 95% confidence interval [CI], 0.06–0.52; P=0.002), hsa-miR-124-3p (odds ratio, 0.29; 95% CI, 0.11–0.72; P=0.008), hsa-miR-320d (odds ratio, 0.33; 95% CI, 0.14–0.78; P=0.01), and SNO1402 (odds ratio, 0.20; 95% CI, 0.07–0.52; P=0.001). In each case, individuals with prevalent stroke at baseline were significantly less likely than those without stroke to express the ex-RNA.
Table 2.
Association With Ex-RNA Measured in Plasma at Exam 8 With the Incident and Prevalent Stroke or CHD in the FHS Cohort (Offspring 8)
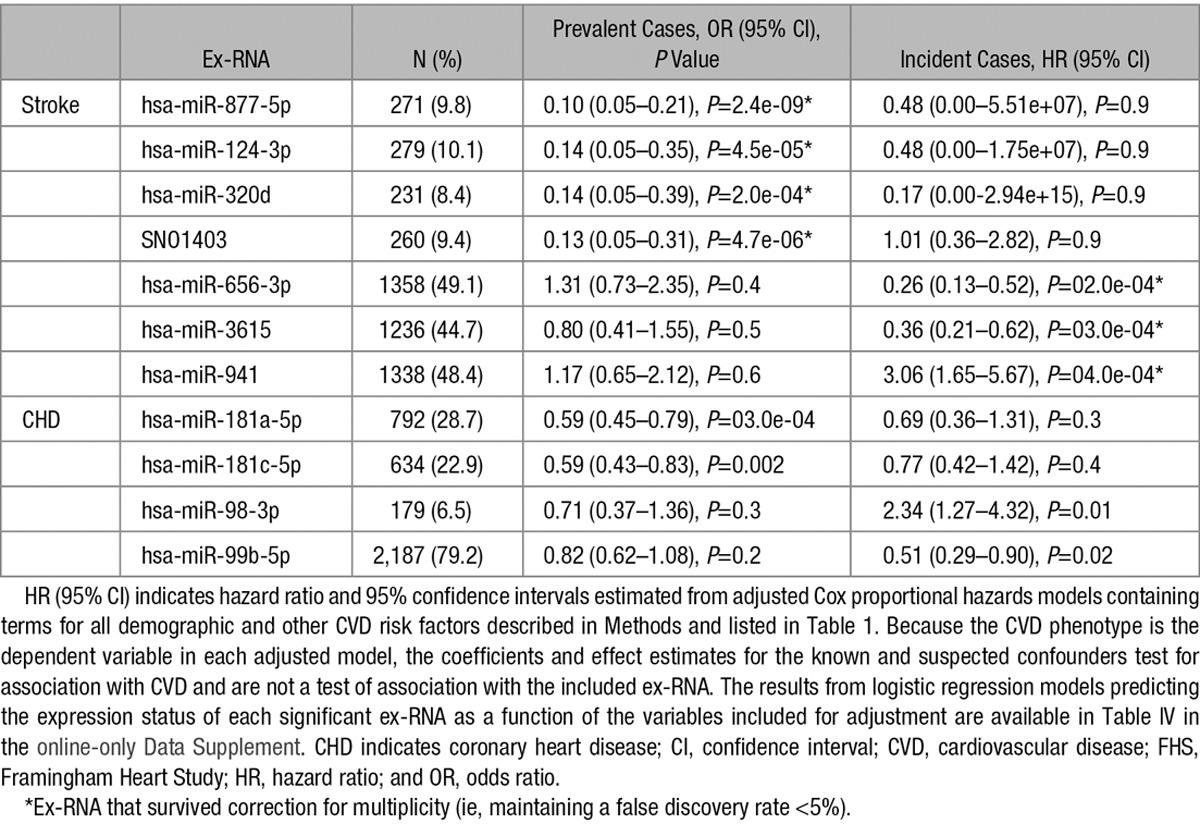
There were no ex-RNA associated with incident CHD, but 3 miRNAs were associated with incident stroke (Table 2). We fit additional Cox-proportional hazards models to assess the independence of hsa-miR-656-3p, hsa-miR-3615, and hsa-miR-941 with stroke risk. Hsa-miR-3615 was no longer statistically significant after adjusting for the other significant miRNAs, but both hsa-miR-656-3p (hazard ratio [HR], 0.33; 95% CI, 0.16–0.68; P=0.003) and hsa-miR-941 (HR, 2.2; 95% CI, 1.2–4.2; P=0.013) remained significantly associated with incident stroke risk adjusting for each other and potentially confounding clinical variables.
Both hsa-miR-941 and hsa-miR-656-3p were commonly expressed with 48% (N=1306) and 49% (N=1324) of the at-risk FHS samples with a Cq<23, but the distribution of expression values differed at baseline between stroke cases and controls (Figure [A]). To assess the impact of relative expression of hsa-miR-941 and hsa-miR-656-3p, we stratified the sample into those not expressing each miRNA and among those expressing the miRNA if there was greater than or less than average expression relative to the modified global mean normalized value 18.8 (Table 3). The distribution of hsa-miR-656-3p was not even, with only 2% (N=56) of participants expressing more hsa-miR-656-3p than the global mean value. The reduced risk for stroke associated with hsa-miR-656-3p was similar in those with low and high expression, but limited sample size of those with high expression strata led to wide confidence intervals and P>0.05 (Table 3). hsa-miR-941 was more normally distributed around the global mean expression values (Figure [A]); the increased risk for stroke was similar and statistically significant for those with high and low expression (Table 3). These results suggest that it is the absolute expression of these miRNAs rather than the relative expression that best predicts stroke risk.
Table 3.
Impact of Relative Expression Level of hsa-miR-656-3p and hsa-miR-941 on Association With Stroke Incidence
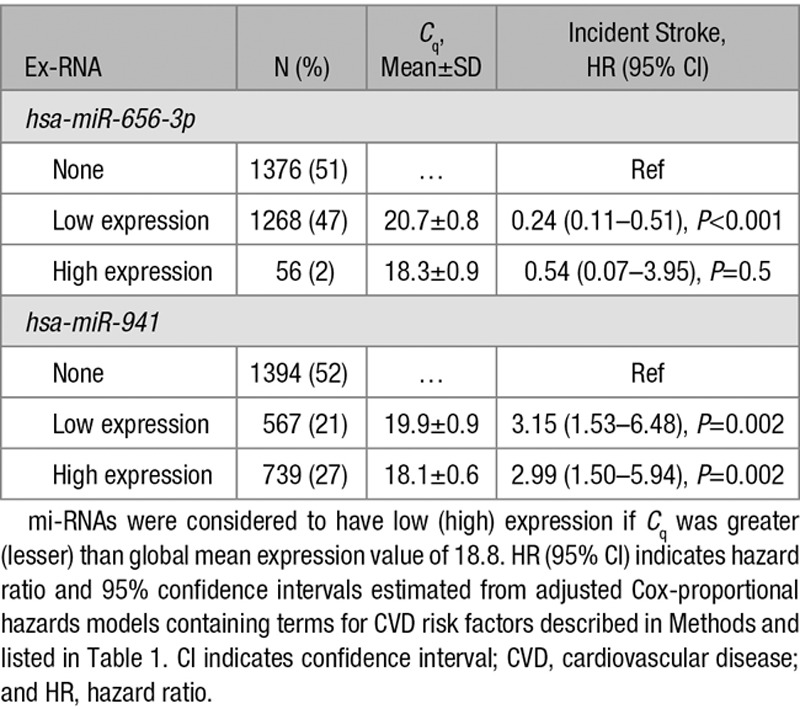
Figure.
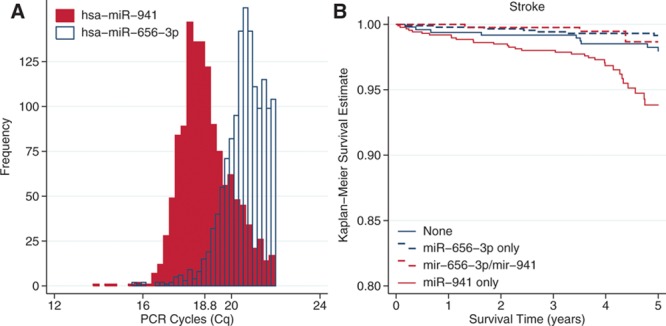
Novel ex-RNA associations with prevalent cardiovascular disease and cardiovascular risk factors in the Framingham Heart Study Offspring 8 Cohort. A, Represented as percent of total expressed. B, The survival rates observed in the 4 groups were statistically significantly different after adjustment for other risk factors (Wald’s X2df=3=22.7; P<0.001). PCR indicates polymerase chain reaction.
We stratified the sample into groups defined by the coexpression of hsa-miR-656-3p and hsa-miR-941 (Figure [B]). The greatest 5-year risk for stroke was observed in subjects expressing hsa-miR-941 but not hsa-miR-656-3p. There was no statistically significant differences in stroke risk among the remaining groups (P=0.3) and each—relative to those expressing only hsa-miR-941—were at reduced risk: those expressing only hsa-miR-656-3p (HR, 0.2; 95% CI, 0.07–0.4; P<0.001), those expressing neither (HR, 0.4; 95% CI, 0.2–0.8; P=0.01), and those expressing both (HR, 0.2; 95% CI, 0.07–0.7; P=0.01).
Discussion
The principal objective of our study was to identify and validate ex-RNA signatures of stroke from plasma in participants of the FHS. The results demonstrate that when studied in a large observational cohort, miRNAs are limited discriminators of prevalent and incident CHD but are significantly associated with stroke. The results suggest that varieties of ex-RNAs, in addition to miRNAs, may characterize stroke. Although there have been other studies using samples from known stroke biosamples, to our knowledge, this is the largest, unbiased, community-based report of association between plasma circulating ex-RNAs (miRNAs, piRNAs, and snoRNAs) and stroke.
The role of ex-RNAs in epigenetic control of gene expression networks has become an intense area of investigation. Studies in animal models of atherosclerosis have elucidated mechanistic roles for tissue expression of miRNAs in essential aspects of CVD pathogenesis, including endothelial dysfunction, inflammation, hypertrophy, fibrosis, and apoptosis. Given that ex-RNAs are released into circulation and may even function as molecular mediators at a distance, there has been a growing body of literature attempting to define a plasma ex-RNA signature of different CVD states. Most of the efforts in clinical translation in CVD have focused on microarray-based discovery of candidate ex-RNAs (nearly exclusively miRNAs) in well-defined disease subsets. However, most discovery efforts have been limited by small sample size and relied on careful cohort selection (eg, plasma profiles at a specific time after acute myocardial infarction) to minimize confounding.
In response, studies have been undertaken to clarify the diagnostic and prognostic ability of miRNA in CVD. In general, most measure associations between selected candidate miRNAs and CVD outcome or diagnosis, despite the well-established associations among miRNAs,5 promiscuous mRNA targets and complex regulation,6 and consequent biological functions within networks. Few have examined the association of ex-RNAs with stroke. Finally, whether the discriminatory ability of ex-RNAs (including those other than miRNAs) for stroke and CHD is generalizable to a large, undifferentiated, community-based cohort with and without chronic CVD remain undefined.
We sought to address these key limitations using a large, community-based cohort (the FHS) with an unbiased approach to discovery (RNASeq) and a broader profile of ex-RNAs, rather than single candidate miRNA associations. Although we did not identify miRNAs, piRNAs, or snoRNAs by RNASeq that were differentially expressed between patients with and without CVD, likely because of the size of the exploratory cohort, the significant expansion to the full cohort demonstrated significant associations with stroke.
When specifically considering CHD and miRNAs, our findings were not consistent with many previous studies that show significant associations. Reasons for this include lack of consistent miRNA detection in the literature, the use of limited candidate approaches for picking miRNAs, differences in methodology, and size of previous studies. Consistent with our modest miRNA–CHD associations, the use of miRNAs to predict short-term prognosis has been generally inferior to cardiac troponin7,8—perhaps owing to strong associations between miRNAs and troponin.9,10
Preclinical studies have demonstrated the differential expression of miRNAs in brain and blood after ischemic or hemorrhagic cerebral damage,11 and specific small RNAs, miRNAs-19b, -29b-2*, and -339-5p, show an early and sustained upregulation in ischemic models of stroke.1 Downregulation of miR-181b in mouse brain after ischemic stroke induces neuroprotection against ischemic injury through targeting heat shock protein A5 and ubiquitin carboxyl-terminal hydrolase isozyme L1.12 It has been shown that target mRNA expression is correlated with the regulation of miRNA.13 Bioinformatics show that DNA methyltransferase 3a is a major target of miR-29c. It has been suggested that miR-29c is a prosurvival miRNA, and its downregulation is a promoter of ischemic brain damage by acting through its target.14 Additionally, intracerebral hemorrhage alters both the abundance and the compartmentalization of several inflammation-related miRNAs in plasma.15 Importantly, treatment to reverse these effects has yet to be clearly established. Data have shown that there is no consistent change in any of the miRNAs tested between resveratrol and vehicle groups, indicating that miRNAs play a minimal role in resveratrol-mediated cerebral ischemic tolerance.16 One proposed mechanism for small RNA modulation of stroke is that miRNA-107 may contribute to poststroke angiogenesis by targeting Dicer-1.17 Finally, although we did not find an association for any of the piRNAs measured, it has been shown that many piRNAs are expressed in adult rodent brain, and several of them respond to focal ischemia.18
There have been smaller but highly informative clinical studies showing association between clinical stroke and miRNA expression. In 48 patients, several miRNA were differentially expressed in blood cells of patients with acute ischemic stroke.19 Another study of 197 patients showed that circulating miR-30a, miR-126, and let-7b were associated with ischemic stroke.20 In a study of 136 patients, elevated miR-106b-5P and miR-4306 and decreased miR-320e and, consistent with our findings, miR-320d in plasma were associated with acute stroke.21 Notably, no previous studies in stroke examined piRNAs or snoRNAs.
In our findings, incident stroke was significantly associated with miR-941. miR-941 was a commonly expressed ex-RNA (48%), and we found that both those with high and low expression were at increased risk of stroke over 5 years. This specific observation is of particular interest because we have found miR-941 to be highly heritable.22 Importantly, in a study that explored in human evolution,23 miR-941 emerged de novo in the human lineage and is highly expressed in brain, affecting genes involved in neurotransmitter signaling.
Our data show that 3 miRNAs (miR-877-5p, miR-124-3p, and miR-320d) were associated with prevalent stroke. miR-124-3p is highly expressed in the developing and adult vertebrate brain and is involved in a broad spectrum of biological functions in the central nervous system.24 Recently, miR-124-3p was associated with stroke and damage caused by ischemic injury in the acute setting.24 It has been shown to regulate cell proliferation in the setting of astrocytoma.25 miR-320d has been investigated in various cancers, and its expression has been shown to suppress cell growth, migration, and invasion and alter levels of MMP-2, MMP-9, N-cadherin, and Integrin-β1.26 Although abundant in the cerebellum, there has been no clear pathway target for miR-877-5p.
Our study also included novel ex-RNAs (piRNAs and snoRNAs) with potent modulatory capacity on gene expression that, to our knowledge, have not been reported in large cohorts with and without stroke and CHD, representing a completely open horizon for ex-RNA biomarker discovery. Although few of these novel ex-RNAs were found to be associated with stroke, these data highlight the importance of expanding transcriptomic studies as our knowledge of these other small RNA species expands. Although we found 1 snoRNA associated with prevalent stroke (SNO1402), the mechanistic implication of this finding is unclear at this time. Specifically, the role of snoRNAs in the circulation is currently not known. SnoRNAs are a class of small RNAs that primarily guide chemical modifications of other RNAs but may function, at times, like miRNAs.
Summary/Conclusions
The strength of our study lies in (1) the use of a large, well-phenotyped cohort of individuals with and without established CVD; (2) unbiased approaches to ex-RNA discovery (RNASeq) and validated methods for high-throughput PCR-based biomarker screening; and (3) inclusion of novel ex-RNAs (piRNAs and snoRNAs) heretofore uninvestigated in clinical at-risk populations. In selecting the ex-RNAs for high-throughput PCR screening in the full cohort, we selected only the most commonly expressed miRNAs and, therefore, did not measure rare miRNAs (including some identified in previous studies of CHD). Although we cannot comment on ex-RNA origin in the circulation with our approach (cellular, vesicular, protein-bound, or free), the use of plasma (a standard source for biomarker discovery) ensures generalizability of our results.
In conclusion, in 2763 participants of the FHS, we found specific ex-RNAs associated with selected CVD phenotypes, primarily stroke. These results suggest promise for the use of ex-RNAs in the setting of stroke and suggest a focusing on discovery for mechanism, other related phenotypes, minority populations, and the use of broader populations of ex-RNAs.
Sources of Funding
This work was supported by UH2TR000921 and U01HL126495 (to Dr Freedman), that are supported by the National Institutes of Health (NIH) Common Fund, through the Office of Strategic Coordination/Office of the NIH Director and from NHLBI, Framingham Heart Study (National Heart, Lung, and Blood Institute/NIH contract No HHSN268201500001I).
Disclosures
None.
Supplementary Material
Footnotes
Guest Editor for this article was Christopher L.H. Chen, FRCP.
Drs Mick and Shah contributed equally.
The online-only Data Supplement is available with this article at http://stroke.ahajournals.org/lookup/suppl/doi:10.1161/STROKEAHA.116.015140/-/DC1.
References
- 1.Dhiraj DK, Chrysanthou E, Mallucci GR, Bushell M. miRNAs-19b, -29b-2* and -339-5p show an early and sustained up-regulation in ischemic models of stroke. PLoS One. 2013;8:e83717. doi: 10.1371/journal.pone.0083717. doi: 10.1371/journal.pone.0083717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blondal T, Jensby Nielsen S, Baker A, Andreasen D, Mouritzen P, Wrang Teilum M, et al. Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods. 2013;59:S1–S6. doi: 10.1016/j.ymeth.2012.09.015. doi: 10.1016/j.ymeth.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 3.Freedman JE, Gerstein M, Mick E, Rozowsky J, Levy D, Kitchen R, et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat Commun. 2016;7:11106. doi: 10.1038/ncomms11106. doi: 10.1038/ncomms11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox CS, Pencina MJ, Meigs JB, Vasan RS, Levitzky YS, D’Agostino RB., Sr Trends in the incidence of type 2 diabetes mellitus from the 1970s to the 1990s: the Framingham Heart Study. Circulation. 2006;113:2914–2918. doi: 10.1161/CIRCULATIONAHA.106.613828. doi: 10.1161/CIRCULATIONAHA.106.613828. [DOI] [PubMed] [Google Scholar]
- 5.Baskerville S, Bartel DP. Microarray profiling of microRNAs reveals frequent coexpression with neighboring miRNAs and host genes. RNA. 2005;11:241–247. doi: 10.1261/rna.7240905. doi: 10.1261/rna.7240905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang Y, Li X, Hu H. Transcriptional regulation of co-expressed microRNA target genes. Genomics. 2011;98:445–452. doi: 10.1016/j.ygeno.2011.09.004. doi: 10.1016/j.ygeno.2011.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Devaux Y, Mueller M, Haaf P, Goretti E, Twerenbold R, Zangrando J, et al. Diagnostic and prognostic value of circulating microRNAs in patients with acute chest pain. J Intern Med. 2015;277:260–271. doi: 10.1111/joim.12183. doi: 10.1111/joim.12183. [DOI] [PubMed] [Google Scholar]
- 8.Olivieri F, Antonicelli R, Lorenzi M, D’Alessandra Y, Lazzarini R, Santini G, et al. Diagnostic potential of circulating miR-499-5p in elderly patients with acute non ST-elevation myocardial infarction. Int J Cardiol. 2013;167:531–536. doi: 10.1016/j.ijcard.2012.01.075. doi: 10.1016/j.ijcard.2012.01.075. [DOI] [PubMed] [Google Scholar]
- 9.Gidlöf O, Andersson P, van der Pals J, Götberg M, Erlinge D. Cardiospecific microRNA plasma levels correlate with troponin and cardiac function in patients with ST elevation myocardial infarction, are selectively dependent on renal elimination, and can be detected in urine samples. Cardiology. 2011;118:217–226. doi: 10.1159/000328869. doi: 10.1159/000328869. [DOI] [PubMed] [Google Scholar]
- 10.Gidlöf O, Smith JG, Miyazu K, Gilje P, Spencer A, Blomquist S, et al. Circulating cardio-enriched microRNAs are associated with long-term prognosis following myocardial infarction. BMC Cardiovasc Disord. 2013;13:12. doi: 10.1186/1471-2261-13-12. doi: 10.1186/1471-2261-13-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clancy L, Freedman JE. New paradigms in thrombosis: novel mediators and biomarkers platelet RNA transfer. J Thromb Thrombolysis. 2014;37:12–16. doi: 10.1007/s11239-013-1001-1. doi: 10.1007/s11239-013-1001-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng Z, Li J, Li Y, Yang X, Feng S, Han S, et al. Downregulation of miR-181b in mouse brain following ischemic stroke induces neuroprotection against ischemic injury through targeting heat shock protein A5 and ubiquitin carboxyl-terminal hydrolase isozyme L1. J Neurosci Res. 2013;91:1349–1362. doi: 10.1002/jnr.23255. doi: 10.1002/jnr.23255. [DOI] [PubMed] [Google Scholar]
- 13.Jeyaseelan K, Lim KY, Armugam A. MicroRNA expression in the blood and brain of rats subjected to transient focal ischemia by middle cerebral artery occlusion. Stroke. 2008;39:959–966. doi: 10.1161/STROKEAHA.107.500736. doi: 10.1161/STROKEAHA.107.500736. [DOI] [PubMed] [Google Scholar]
- 14.Pandi G, Nakka VP, Dharap A, Roopra A, Vemuganti R. MicroRNA miR-29c down-regulation leading to de-repression of its target DNA methyltransferase 3a promotes ischemic brain damage. PLoS One. 2013;8:e58039. doi: 10.1371/journal.pone.0058039. doi: 10.1371/journal.pone.0058039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo D, Liu J, Wang W, Hao F, Sun X, Wu X, et al. Alteration in abundance and compartmentalization of inflammation-related miRNAs in plasma after intracerebral hemorrhage. Stroke. 2013;44:1739–1742. doi: 10.1161/STROKEAHA.111.000835. doi: 10.1161/STROKEAHA.111.000835. [DOI] [PubMed] [Google Scholar]
- 16.Lopez MS, Dempsey RJ, Vemuganti R. Resveratrol preconditioning induces cerebral ischemic tolerance but has minimal effect on cerebral microRNA profiles. J Cereb Blood Flow Metab. 2016;36:1644–1650. doi: 10.1177/0271678X16656202. doi: 10.1177/0271678X16656202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, Mao L, Gao Y, Baral S, Zhou Y, Hu B. MicroRNA-107 contributes to post-stroke angiogenesis by targeting Dicer-1. Sci Rep. 2015;5:13316. doi: 10.1038/srep13316. doi: 10.1038/srep13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dharap A, Nakka VP, Vemuganti R. Altered expression of PIWI RNA in the rat brain after transient focal ischemia. Stroke. 2011;42:1105–1109. doi: 10.1161/STROKEAHA.110.598391. doi: 10.1161/STROKEAHA.110.598391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jickling GC, Ander BP, Zhan X, Noblett D, Stamova B, Liu D. microRNA expression in peripheral blood cells following acute ischemic stroke and their predicted gene targets. PLoS One. 2014;9:e99283. doi: 10.1371/journal.pone.0099283. doi: 10.1371/journal.pone.0099283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Long G, Wang F, Li H, Yin Z, Sandip C, Lou Y, et al. Circulating miR-30a, miR-126 and let-7b as biomarker for ischemic stroke in humans. BMC Neurol. 2013;13:178. doi: 10.1186/1471-2377-13-178. doi: 10.1186/1471-2377-13-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang W, Sun G, Zhang L, Shi L, Zeng Y. Circulating microRNAs as novel potential biomarkers for early diagnosis of acute stroke in humans. J Stroke Cerebrovasc Dis. 2014;23:2607–2613. doi: 10.1016/j.jstrokecerebrovasdis.2014.06.002. doi: 10.1016/j.jstrokecerebrovasdis.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 22.Huan T, Rong J, Liu C, Zhang X, Tanriverdi K, Joehanes R, et al. Genome-wide identification of microRNA expression quantitative trait loci. Nat Commun. 2015;6:6601. doi: 10.1038/ncomms7601. doi: 10.1038/ncomms7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu HY, He L, Fominykh K, Yan Z, Guo S, Zhang X, et al. Evolution of the human-specific microRNA miR-941. Nat Commun. 2012;3:1145. doi: 10.1038/ncomms2146. doi: 10.1038/ncomms2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji Q, Ji Y, Peng J, Zhou X, Chen X, Zhao H, et al. Increased brain-specific MiR-9 and MiR-124 in the serum exosomes of acute ischemic stroke patients. PLoS One. 2016;11:e0163645. doi: 10.1371/journal.pone.0163645. doi: 10.1371/journal.pone.0163645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deng D, Wang L, Chen Y, Li B, Xue L, Shao N, et al. MicroRNA-124-3p regulates cell proliferation, invasion, apoptosis, and bioenergetics by targeting PIM1 in astrocytoma. Cancer Sci. 2016;107:899–907. doi: 10.1111/cas.12946. doi: 10.1111/cas.12946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qin CZ, Lv QL, Yang YT, Zhang JM, Zhang XJ, Zhou HH. Down-regulation of microrna-320d predicts poor overall survival and promotes the growth and invasive abilities in glioma. Chem Biol Drug Des. 2016;12:12906. doi: 10.1111/cbdd.12906. doi: 10.1111/cbdd.12906. [DOI] [PubMed] [Google Scholar]


