Abstract
Cas9-mediated, high-throughput, saturating in situ mutagenesis permits fine-mapping of function across genomic segments. Disease- and trait-associated variants from genome-wide association studies largely cluster in regulatory DNA. Here we demonstrate the use of multiple designer nucleases and variant-aware library design to interrogate trait-associated regulatory DNA at high resolution. We developed a computational tool for the creation of saturating mutagenesis libraries with single or combinatorial nucleases with incorporation of variants. We applied this methodology to the HBS1L-MYB intergenic region, a locus associated with red blood cell traits, including fetal hemoglobin levels. This approach identified putative regulatory elements that control MYB expression. Analysis of genomic copy number highlighted potential false positive regions, which emphasizes the importance of off-target analysis in design of saturating mutagenesis experiments. Taken together, these data establish a widely applicable high-throughput and high-resolution methodology to reliably identify minimal functional sequences within large regions of disease- and trait-associated DNA.
INTRODUCTION
Genome-wide association studies (GWAS) are a powerful approach for the identification of disease- and trait-associated variants. Greater than 90% of GWAS variants lie within non-coding DNA1. However, linkage disequilibrium often obscures the causal variant and thus the biological mechanisms producing the trait association. Reliable methods to identify the underlying functional sequences remain elusive. The clustered regularly interspaced short palindromic repeats (CRISPR)-based genome editing systems have emerged as highly efficient tools to study regulatory DNA. Targeted deletion provides a valuable tool for loss of function2,3. However, targeted deletion has limited throughput, efficiency, and resolution4. Alternatively, the homology-directed repair (HDR) pathway can be exploited following cleavage by a designer nuclease to insert putative causal variants into endogenous DNA sequence using a customized extrachromosomal template. However, HDR to insert variants is low-throughput and limited by efficiency. Furthermore, individual trait-associated variants may underestimate the effect of the underlying haplotype, which in turn may underestimate the biological importance of the harboring genetic element2,3,5.
Saturating a region with insertions/deletions (indels) by using every available PAM-restricted single guide RNA (sgRNA) constitutes a powerful strategy to identify minimal functional sequences within regulatory DNA3. Saturating mutagenesis relies on pooled screening to take advantage of the typical indel spectrum following non-homologous end joining (NHEJ) repair of 1–10 bp deletions3,4,6–9. The ability to saturate a region with indels is a function of protospacer adjacent motif (PAM) availability. Moreover, genomic variants that attenuate sgRNA activity may reduce resolution through false negatives. We hypothesized that combining multiple nucleases with unique PAM sequences would enhance mutagenesis resolution and that incorporating variants into sgRNA library design would minimize false negatives associated with libraries based on the reference genome. To test the hypothesis, we applied this methodology to the genetically implicated HBS1L-MYB intergenic region.
RESULTS
The HBS1L-MYB intergenic region is associated with erythroid traits
GWAS, quantitative trait loci (QTL), and other human genetic studies of fetal hemoglobin (HbF) level (or the related trait F-cell number) have implicated the HBS1L-MYB interval10–17. The HBS1L-MYB interval has also been associated with erythroid traits including hemoglobin (Hb), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), mean corpuscular volume (MCV), packed cell volume (PCV), and red blood cell count (RBC)18–23. These associations have been suggested to reflect changes in the expression of MYB due to distant variants localizing kilobases away and approximately equidistant to the HBS1L gene15. Genotyping in multiple cohorts of sickle cell disease (SCD) individuals (n=2,222) was conducted to refine the genetic association with HbF-levels (Fig. 1a; Supplementary Table 1).
Figure 1. Trait-associations of the HBS1L-MYB intergenic region.
a, Meta-analysis of HbF-associated SNPs from SCD cohorts (n = 2,222). rs66650371 (green triangle) and rs9494142/rs11154792 (red diamond) have been previously implicated as possible functional SNPs affecting MYB expression15. The larger dot (gray) corresponds to the top HbF-associated SNP, rs9389269. Super-enhancer region is indicated by a black horizontal bar. Genome-wide significance is indicated by a horizontal dotted line (P<5×10−8). Schematic of the HBS1L-MYB interval region (hg19) with erythroid DNase I hypersensitivity and H3K27ac is shown above meta-analysis. b, Previously published meta-analyses of SNPs associated with erythroid traits including hemoglobin (Hb, red), mean corpuscular hemoglobin (MCH, green), mean corpuscular hemoglobin concentration (MCHC, gray), mean corpuscular volume (MCV, blue), packed cell volume (PCV, purple), and red blood cell count (RBC, orange)22. Only SNPs P<10−6 are displayed. Super-enhancer region is indicated by a black horizontal bar. Genome-wide significance is indicated by a horizontal dotted line (P<5×10−8). The larger dots correspond to the top HbF-associated SNP, rs9389269. The diamonds correspond to rs9494142/rs11154792. Schematic of the HBS1L-MYB interval region (hg19) with erythroid DNase I hypersensitivity and H3K27ac is shown above meta-analysis. Abbreviations: hemoglobin (Hb), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), mean corpuscular volume (MCV), packed cell volume (PCV), and red blood cell count (RBC).
This HbF meta-analysis identified SNPs with a similar clustering to a previously published meta-analysis of variants associated with erythroid traits (Fig. 1b; Supplementary Table 2)22. Due to extensive linkage disequilibrium (LD) and limited sample size, conditional analysis of HbF-associated SNPs could not confidently pinpoint a specific set of causal variants (Supplementary Table 3). Recent studies have utilized lineage-restricted expression patterns, clustering of erythroid transcription factor binding sites affecting MYB expression, and chromatin conformation capture to suggest that HbF-associated variants modulate MYB expression by altering GATA1 or GATA1/TAL1 motifs within regulatory elements −71 and −84 kb upstream of the MYB transcriptional start site (TSS)15. However, our meta-analysis, the largest to date for HbF levels in SCD patients, was unable to discriminate between the previously reported causal variant (rs66650371) and other markers in strong LD.
The HBS1L-MYB region is comprised of 98 DNase hypersensitive sites (DHSs) as identified from erythroid precursors (Fig. 1)2. The trait-associated single nucleotide polymorphisms (SNPs) from both meta-analyses are concentrated in an 83 kilobase intergenic super-enhancer (Fig. 1a, b; Supplementary Fig. 1). In order to interrogate the HBS1L-MYB locus in comprehensive fashion, each of the 98 DHSs was subject to saturating mutagenesis.
Distribution of PAM sequences in the genome and outline of the DNA Striker algorithm
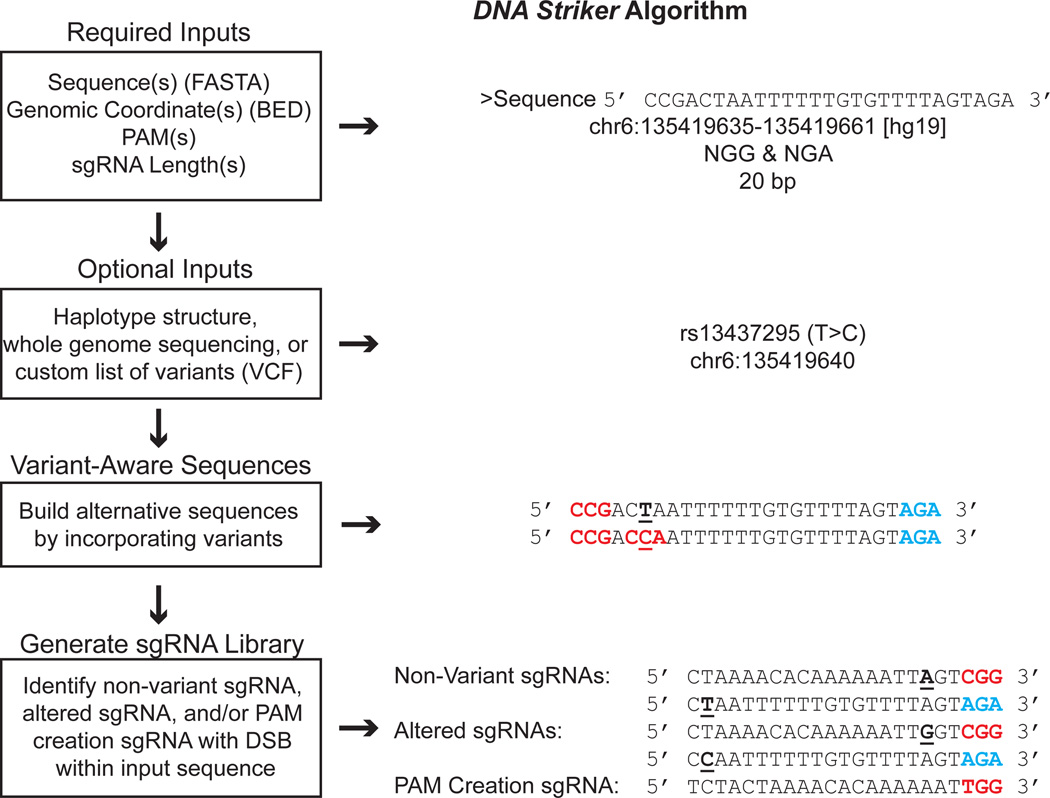
Maximizing the degree of saturating mutagenesis depends on minimizing genomic distance between potential adjacent cleavages. In order to functionally fine-map the HBS1L-MYB intergenic region, we reasoned that usage of multiple highly saturating nucleases in combination could increase resolution. We further hypothesized that variant-aware saturating mutagenesis library design could limit false negatives resulting from diminished sgRNA activity due to variants present in the cells used for study, which is a consideration highlighted by the region’s trait association with common genetic variants. To perform variant-aware saturating mutagenesis library design using multiple nucleases, we created the DNA Striker computational tool (Fig. 2; Supplementary Fig. 2). It facilitates design of saturating mutagenesis libraries employing single or multiple designer nucleases, and alternative sgRNAs based on haplotype structure, whole-genome sequencing (WGS), or a custom list of variants. The algorithm is summarized in Fig. 2 (see Methods for additional details).
Figure 2. DNA Striker algorithm.
Description of the DNA Striker algorithm for sgRNA design, which allows to create variant-aware saturating mutagenesis libraries from haplotype structure, WGS, or custom lists of variants. DNA Striker can output libraries using any combination of PAM sequences. NGG and NGA library design is shown as a representative example. In this example, NGG PAMs are shown in red, NGA PAMs are shown in blue, and the position of variants are underlined.
Saturating mutagenesis library design
CRISPR-associated nucleases with unique PAM recognition sequences have been reported for genome editing6,7,24–29. The frequency of each PAM varies in the genome (Fig. 3a; Supplementary Fig. 3; Supplementary Table 4). Given the sequence-dependence of PAM availability, feature-specific variation in cleavage density for each nuclease is observed in DNase I hypersensitivity sites (DHS), enhancers, and repressed regions as well as genes (Supplementary Figs. 4–7; Supplementary Table 5).
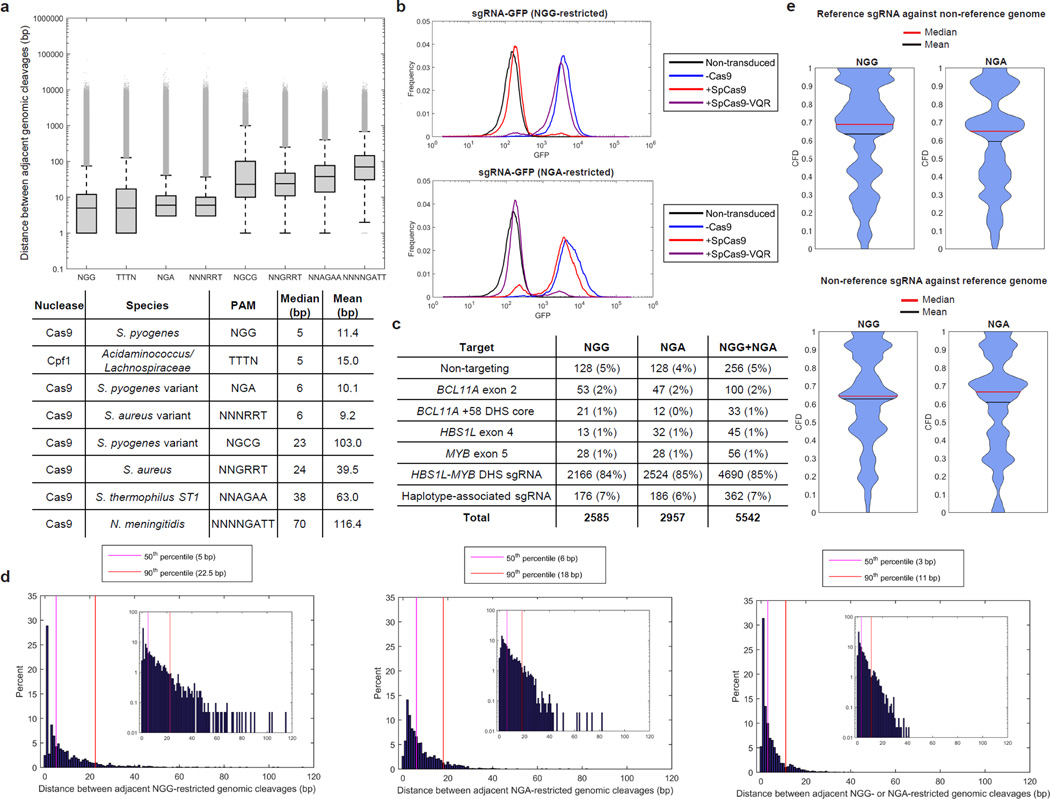
Figure 3. Pooled saturating mutagenesis screening of the HBS1L-MYB region using NGG- and NGA Cas9s and variants from 1,000 Genomes haplotypes.
a, Distances between adjacent genomic cleavages to assess genome-wide PAM availability and distribution. For each box-and-whisker plot, the three lines of box represent the 25th, 50th and 75th percentile. The upper and lower whiskers represent the 99th and 1st percentile, respectively. Outliers, defined as above the 99th percentile or below the 1st percentile are plotted as individual points. Lower whiskers are omitted if the 1st percentile is 0. b, Cells stably expressing SpCas9 (red), SpCas9-VQR (purple), or without Cas9 (blue) were transduced with a Cas9 activity reporter, which contained GFP and either an NGG- (top) or NGA-restricted (bottom) GFP-targeting sgRNA. A non-transduced sample (black) was included as a negative control. c, Library composition for NGG-restricted sgRNA library only, NGA-restricted sgRNA library only, as well as NGG- and NGA-restricted sgRNA libraries together. d, For the HBS1L-MYB intergenic region DHSs, the genomic cleavage density of using NGG-only (left panel), NGA-only (middle panel), and NGG and NGA combined (right panel) libraries. e, Violin plots of CFD analysis for haplotype-associated sgRNA with reference genomic sequence and for non-variant sgRNA with haplotype-variants present.
We reasoned that combining multiple species of Cas9 with unique PAM sequences would enhance the resolution of saturating mutagenesis. To evaluate this approach, we used the regions of each DHS summit (peak of DNase I sensitivity) ± 200 bp within the HBS1L-MYB intergenic region for saturating mutagenesis. NGG- and NGA-PAM restricted sgRNA were chosen because these PAM sequences result in the lowest mean and median gap distance between adjacent genomic cleavages in DHS (Supplementary Fig. 4; Supplementary Table 5).
To demonstrate feasibility of using these nucleases, as well as evaluate the specificity and efficiency of S. pyogenes Cas9 (SpCas9; NGG PAM) and S. pyogenes VQR variant Cas9 (SpCas9-VQR; NGA PAM)28, we used Cas9 reporter constructs that delivered GFP as well as either an NGG-restricted or NGA-restricted sgRNA targeting GFP. Cells stably expressing SpCas9, SpCas9-VQR, or no Cas9 were transduced with the reporter construct at low multiplicity and selected for 14 days. The analysis demonstrated that the SpCas9 and SpCas9-VQR Cas9 proteins were both specific and efficient nucleases as SpCas9 only led to GFP reduction with the NGG-restricted sgRNA, and SpCas9-VQR only led to GFP reduction with the NGA-restricted sgRNA (Fig. 3b).
Therefore, we used DNA Striker to design a high resolution saturating mutagenesis library consisting of all 20-mer sequences upstream of an NGG or NGA PAM sequence on the top or bottom strand within the HBS1L-MYB region DHS as well as controls including BCL11A exon 2, the core of the +58 DHS within the BCL11A enhancer3, HBS1L exon 4, and MYB exon 5 (Fig. 3c; Supplementary Tables 6–7). The median and 90th percentile gap distance between adjacent genomic cleavages using SpCas9 was 5 bp and 22.5 bp, respectively while it was 6 bp and 18 bp for SpCas9-VQR (Fig. 3d). The combination of both SpCas9 and SpCas9-VQR nucleases led to a reduction in the median and 90th percentile gap between adjacent genomic cleavages to 3 bp and 11 bp, respectively (Fig. 3d). Furthermore, use of both nucleases reduced the maximum gap size from 115 bp for SpCas9 and 82 bp for SpCas9-VQR to a maximum of 41 bp for the combination (Supplementary Fig. 8). Therefore, the inclusion of sgRNAs restricted by two distinct nucleases resulted in higher resolution by reducing the 50th and 90th percentile of distances between adjacent genomic cleavages as well as reducing the maximum gap between adjacent cleavages. Multiple nucleases allows for minimization of distance of double strand breaks (DSBs) to SNPs and motifs of interest, which allows for enhanced functional interrogation of regions of interest (Supplementary Fig. 9).
To construct a variant-informed library, phased variants within these regions were taken from the 1,000 Genomes Project database from all populations and incorporated into sgRNA design by DNA Striker to identify potential altered sgRNAs and novel sgRNAs resulting from variant-induced PAM creation (Fig. 2, 3c). Haplotype-associated sgRNA were included in the library if they were present at a frequency ≥1% (NGG: 176/1,350 haplotype-associated sgRNA; NGA: 186/1,551 haplotype-associated sgRNA) (Fig. 2, 3c; Supplementary Fig. 10a, b). Both NGG- and NGA-restricted sgRNA libraries were synthesized and successfully batch cloned into lentiviral constructs (Supplementary Fig. 10c, d).
Cutting frequency determination (CFD) has been previously used for imperfect-match sgRNA activity assessment30. We used CFD analysis to assess the predicted activity of the haplotype-associated (non-reference) sgRNA in the presence of the reference genome and the predicted activity of the non-variant (reference) sgRNA in the presence of the haplotype-derived variants (non-reference genome). This analysis demonstrated a reduction in CFD for reference sgRNA in the presence of a non-reference genome, which suggests utility of variant-aware library design (Fig. 3e, top panel). Furthermore, CFD analysis suggested that the majority of non-reference sgRNA had diminished activity against the reference genome (Fig. 3e, bottom panel).
Functional saturating mutagenesis screens using SpCas9 and SpCas9-VQR
For the HBS1L-MYB saturating mutagenesis experiments, we employed the immortalized human erythroid cell line HUDEP-2, which was previously used to examine erythroid maturation and HbF regulation3,31. Briefly, HUDEP-2 cells stably expressing SpCas9 or SpCas9-VQR were transduced at low multiplicity with the NGG-restricted or NGA-restricted sgRNA library, respectively. Cells were expanded, differentiated, sorted for high and low HbF-expression, and deep sequenced to enumerate sgRNA present within the HbF-high and HbF-low pools3. Three independent experiments were performed for both libraries. Surprisingly, sgRNAs targeting HBS1L exon 4 and MYB exon 5 failed to show significant HbF enrichment, although the positive control sgRNAs targeting BCL11A exon 2 and BCL11A DHS +58 enriched in the HbF-high pool as expected (Fig. 4a). Interestingly, sgRNA targeting MYB showed a preponderance to dropout (reduce in abundance) of the screen consistent with MYB’s known essential role in erythropoiesis32 (Fig. 4b). BCL11A +58 DHS targeted sgRNA were not underrepresented, whereas BCL11A exon 2 sgRNA showed modest dropout, consistent with previous findings3 (Fig. 4b). In addition, sgRNAs targeting HBS1L coding sequences did not dropout, suggesting this gene does not contribute to the fitness of the HUDEP-2 cells. Mann-Whitney testing showed no difference in the dropout between SpCas9 and SpCas9-VQR species (p > 0.05).
Figure 4. Mapping NGG- and NGA-restricted sgRNA dropout scores to genomic cleavage position identifies putative functional elements.
a, Mapping HbF enrichment scores to associated genomic loci. Non-targeting sgRNA are pseudo-mapped with 5-bp spacing. b, Mapping and dropout scores to associated genomic loci. Non-targeting sgRNA are pseudo-mapped with 5-bp spacing. c,d, Quantile-quantile plots of NGG and NGA sgRNA library dropout scores. e, Mapping NGG-restricted and NGA-restricted dropout scores to associated genomic loci identifies functional elements. The elements with the most potent dropout scores, −126, −83, −71, −36, and −7, are indicated by green highlight. Erythroid DNase I hypersensitivity, H3K27ac, GATA1 binding, and TAL1 binding are shown. HMM designation as active (green), repressive (red), and neutral (gray) are shown for each DHS. The coordinates for each DHS in the HBS1L-MYB interval on chromosome 6 in hg19 coordinates are −126 DHS: 135376369–135376770; −83 DHS:135419396–135419797; −71 DHS: 135431355–135431756; −7 DHS:135495667–135496068; −84 DHS: 135418448–135418849; −36 DHS is composed of two DHS: 135466090–135466491 and 135466671–135467072.
To orthogonally validate these findings, we evaluated HUDEP-2 cells for MYB dependence. Three shRNAs efficiently depleted MYB and led to a cellular proliferation defect in HUDEP-2 cells, consistent with the results of the CRISPR screen and indicative of MYB dependence (Supplementary Fig. 11a, b). We also examined the effect of MYB depletion in primary human CD34+ hematopoietic stem and progenitor cells (HSPCs) from G-CSF mobilized healthy adult donors subject to erythroid differentiation conditions. The same shRNAs targeting MYB demonstrated a profound cellular proliferation defect in CD34+ HSPC derived human erythroblasts (Supplementary Fig. 11c). Erythroid differentiation was assessed at days 10, 14, and 18 of culture by surface expression of CD71 (transferrin receptor) and CD235a (glycophorin A) erythroid markers. A severe differentiation block was observed upon MYB knockdown, consistent with previous reports33 (Supplementary Fig. 11d).
Introduction of an sgRNA targeting MYB coding sequence into HUDEP-2 cells stably expressing Cas9 resulted in an impairment of cellular proliferation, further indicating that HUDEP-2 cells rely on MYB for cell growth (Supplementary Fig. 11e). The same sgRNA targeting MYB also demonstrated a cellular proliferation defect in CD34+ HSPC derived human erythroblasts (Supplementary Fig. 11f). Notably, targeting MYB coding sequence in CD34+ derived human erythroblasts resulted in a significantly greater percentage of in-frame mutations as compared to BCL11A and HBS1L coding sequence targeting, suggesting strong selective pressure against loss-of-function MYB alleles (Supplementary Fig. 11g). Furthermore, targeting MYB led to a reduction in MYB expression (Supplementary Fig. 11h). Taken together, shRNA-mediated knockdown and CRISPR-mediated knockout of MYB resulted in proliferation defects in both HUDEP-2 cells and CD34-derived erythroblasts, indicative of MYB dependence of these cells. These data suggested that the HBS1L-MYB DHS CRISPR screen data could be analyzed with cellular dropout as opposed to HbF enrichment as the phenotype. Analysis of the library for dropout demonstrated that the majority of sgRNAs in both the NGG- and NGA-restricted libraries did not dropout, suggesting a neutral effect on cell growth (Fig. 4c, d). Notably, specific sgRNAs with significant dropout were identified in both libraries (Fig. 4c, d).
Variant aware, high resolution saturating mutagenesis of the HBS1L-MYB interval
The presence of multiple colocalizing top-scoring sgRNAs within in situ saturating mutagenesis screens suggests the position of minimal functional sequences3. Upon mapping the library sgRNAs to their associated genomic loci, the most potent dropout sgRNAs colocalized to discrete loci for both the NGG- and NGA-restricted libraries. A Hidden Markov Model (HMM) segmentation with three states (Neutral, Repressive and Active) was applied to the merged NGG and NGA dropout scores to identify functional sequence (Fig. 4e). The HMM analysis led to the identification of multiple regions of regulatory potential. These DHS were termed −126, −83, −71, −36 (composed of 2 adjacent DHS), and −7 based on their distance in kilobases from the MYB TSS (Fig. 4e; Supplementary Figs. 12–15).
Notably, the utilization of SpCas9 and SpCas9-VQR species together led to enhanced resolution at these identified DHS through reduction of gaps between adjacent genomic cleavages (Supplementary Fig. 16). In addition, higher density of sgRNA enhanced the reliability of functional sequence detection by HMM analysis. Of note, the −83 and −71 DHS fall within an annotated super-enhancer region and each of these five DHS co-localize with GATA1 and/or GATA1/TAL1 binding (Fig. 4e; Supplementary Figs. 12–13). These identified DHS suggest regulatory potential for MYB expression.
Previous reports have nominated possible causal variants within the −84 and −71 DHSs that influence MYB expression15. While saturating mutagenesis identified the −71 DHS as containing functional sequence, it suggested functional sequence localized to the −83 DHS as opposed to the −84 DHS (Fig. 4e; Supplementary Figs. 13–14). rs9389268, which is highly associated with erythroid traits, is located within the −83 DHS (Fig. 5; Supplementary Fig. 13). Interestingly, the 545 bp interval between −83 and −84 (chr6: 135418850–135419395, hg19) has several HbF- and erythroid-associated SNPs (Fig. 5a, c; Supplementary Fig. 17). This region is DNase insensitive in erythroid cells so it was not included in the library design, although recently functional elements have been identified by CRISPR mutagenesis that lack typical epigenetic or chromatin characteristics34. The top-scoring sgRNAs at the −71 element specified a cleavage about 200 bp from the peak of DNase I sensitivity and GATA1/TAL1 binding (Fig. 4e; Supplementary Fig. 12)34. The highly trait-associated SNP within the −71 DHS that disrupts a GATA1 motif, rs9494142/rs1115479215, localizes closer to the peak of DNase I sensitivity as compared to the putative functional sequence by approximately 100 bp (Supplementary Fig. 12). rs66650371 is a 3 bp indel that disrupts a TAL1 motif within the −84 DHS and localizes to the peak of DNase I sensitivity. However, application of the HMM designated the entire DHS as neutral (Fig. 4e; Supplementary Fig. 14).
Figure 5. Trait-associated SNPs mark essential enhancer elements.
a, b Genome-wide HbF-associated SNPs localize to identified regions. Genome-wide significant SNPs (P<5×10−8) are indicated: rs66650371 (−84 DHS, green triangle), rs61028892 (−84 DHS, gray circle), and rs9494142/rs11154792 (−71 DHS, red diamond). rs9494142/rs11154792 and rs66650371 have been previously associated with altering MYB expression15. −84 DHS (chr6:135,418,307–135,418,807, hg19) is highlighted in blue. −71 DHS (chr6:135431355–135431756, hg19) and −83 DHS (chr6:135419396–135419797, hg19) are highlighted in green. c, d Genome-wide RBC-associated SNPs localize to identified regions. Genome-wide significant SNPs (P<5×10−8) are indicated: rs7775698 (−84 DHS), rs9389268 (−83 DHS), and rs9494142/rs11154792 (−71 DHS, red diamond). rs7775698, rs9389268, and rs9494142/rs11154792 are associated with all 6 RBC-traits at genome-wide significance (P<5×10−8). rs9494142/rs11154792 has been previously associated with altering MYB expression15. −84 DHS (chr6:135,418,307–135,418,807, hg19) is highlighted in blue. −71 DHS (chr6:135431355–135431756, hg19) and −83 DHS (chr6:135419396–135419797, hg19) are highlighted in green. Abbreviations: hemoglobin (Hb), mean corpuscular hemoglobin (MCH), mean corpuscular hemoglobin concentration (MCHC), mean corpuscular volume (MCV), packed cell volume (PCV), and red blood cell count (RBC).
Stratification by off-target scores alters identification of functional sequences and implicates −36 and −84 DHS
Recent studies suggest a correlation between genomic copy number and dropout upon CRISPR targeting protein coding sequences35,36. Genomic copy number was evaluated for all sgRNA in the SpCas9 and SpCas9-VQR associated libraries. This analysis identified highly repetitive sequence within the HBS1L-MYB interval DHS that produced a wide distribution of the number of genomic matches for each sgRNA (Fig. 6a, b; Supplementary Fig. 18). shRNA-mediated knockdown of MYB expression demonstrated that loss of MYB reduces cellular fitness in HUDEP-2 and CD34+ derived erythroblasts (Supplementary Fig. 11b, c). This is further supported by sgRNAs targeting MYB exon 5, which all have a single genomic match, induced dropout, and a reduction in MYB expression (Fig. 4b; Supplementary Fig. 11e–h; Fig. 6a, b; Supplementary Fig. 18). However, increased genomic matches for a given sgRNA is also predicted to reduce cellular fitness35,36. Our data suggest a correlation between number of genomic matches and dropout. However, this trend is incompletely predictive, as numerous sgRNA with ≥10 genomic matches do not result in dropout (sgRNA with ≥10 genomic matches and dropout score; R2 = 0.076) (Fig. 6b). This might reflect sgRNA-specific variation in editing or cellular response.
Figure 6. The HBS1L-MYB intergenic region contains highly repetitive genomic sequences.
a, Histogram of the number of genomic matches for each sgRNA in the full library. b, Correlation between number of genomic matches and dropout score. HMM sgRNA (red) indicate sgRNA located in regions designated as Active by HMM analysis. c, MYB expression in HUDEP-2 cells after 14 days of culture (normalized to GAPDH). d, Proliferation rates of HUDEP-2 cells with sgRNAs targeting MYB enhancer elements. e, GATA1 binding in HUDEP-2 cells by ChIP-qPCR after six days of culture. Error bars represent standard deviation and each sample is n = 3 independent experiments. Samples were compared using unpaired two-sided t-tests. One asterisks signifies P<0.01, two asterisks signifies P<0.001, and three asterisks signify P<0.0001.
Off-target scores were calculated, as previously described, except using all possible 20mers upstream of an NG motif, which led to a reduction in the overall scores as compared to published values9,37 (Supplementary Fig. 19; see Methods for additional details). Off-target scores using this methodology ranged from 0–100 with a higher score signifying fewer predicted off-targets. Stratification of the library sgRNAs by off-target score >10 abolished the dropout signal from the −71 and −7 DHS; however, the signal was retained at the −126, −83, and −36 DHS (Supplementary Fig. 20). To validate the filtered screen data, we chose to focus on the −36 DHS site as it had lower off-target potential as compared to −83 and −126 (Supplementary Figs. 12–15). We used sgRNA 1910, which had the maximal off-target score (indicative of lower off-target potential) in the −36 region (Supplementary Fig. 15a; Supplementary Table 6). Editing with sgRNA 1910 resulted in lower MYB expression and reduced proliferation in HUDEP-2 cells (Fig. 6c, d), consistent with MYB regulatory potential within −36 DHS. sgRNA-1910 did not overlie a predicted GATA1 motif (Supplementary Fig. 15) and its target sequence was demonstrated to lack GATA1 binding by ChIP-qPCR (data not shown).
In addition, we aimed to evaluate the sequences flanking the previously implicated SNPs in the −71 and −84 DHS15,38. We used an NGG-restricted guide targeting the −71 DHS (sgRNA 1582) that produced a DSB directly adjacent to rs9494142/rs11154792/GATA1 motif (Supplementary Fig. 21). Targeting this motif in CD34+ derived erythroblasts resulted in successful mutagenesis (Supplementary Fig. 21a–d), but did not alter cellular proliferation or MYB expression (Supplementary Fig. 21e, f).
An NGA-restricted guide targeting the −84 DHS (sgRNA 1500) was used with a DSB position 1 bp from the implicated TAL1 motif15. Targeting this motif in CD34+ derived erythroblasts resulted in moderate levels of editing (Supplementary Fig. 22a–d), but did not alter cellular proliferation (Supplementary Fig. 22e). MYB expression trended toward reduction; however, this effect did not reach statistical significance (Supplementary Fig. 22f). Notably, sgRNA 1500 resulted in a predominance of indels sparing the adjacent GATA1 motif (Supplementary Fig. 22c–d). It is possible that selection against alleles disrupting key binding sites may have limited overall functional impact.
Lastly, we used an NGG-restricted sgRNA (sgRNA 1321) with a DSB position within the −84 DHS directly adjacent to a GATA1 motif. In addition, the DSB position was 3 bp upstream of rs61028892 (seventh highest association with HbF levels from the HbF meta-analysis) (Supplementary Fig. 9); this sgRNA demonstrated significant dropout in the saturating mutagenesis screen (Supplementary Figs. 14, 22; Supplementary Table 6). Of note, this GATA1 motif corresponds to the peak of GATA1 binding at this DHS and is 14 bp downstream from the previously implicated GATA1::TAL1 motif (Supplementary Figs. 14, 22). Targeting this motif resulted in downregulation of MYB expression and reduced proliferation in HUDEP-2 cells (Fig. 6c, d). Furthermore, mutagenesis resulted in a reduction of GATA1-binding in HUDEP-2 cells by ChIP-qPCR (Fig. 6e). Taken together, these data suggest MYB regulatory potential in the −84 DHS mediated by GATA1. These data also demonstrate the utility of multiple species of Cas9, which allows for more precise mutagenesis of motifs and putative casual variants. The lack of identification of −84 DHS in the screen may suggest this element has a modest effect on MYB expression or a narrow region of regulatory DNA, which would require an even higher density of co-localizing dropout sgRNAs for detection by HMM analysis.
Putative MYB enhancer activity of −126, −83, −71, and −7 DHS confounded by off-target effects
The saturating mutagenesis screen implicated −126, −83, −71, and −7 to potentially harbor functional sequence. HMM segmentation further identified sub-regions within these four DHS with dropout scores significantly diverging from the baseline, which revealed potential discrete minimal active sequences (Fig. 4e; Supplementary Figs. 12–13). All four of these DHS contain repetitive sequence (Supplementary Figs. 12–13, 20). We chose individual sgRNAs targeting the −126, −83, −71, and −7 that exhibited the most significant dropout, but also had poor off-target scores (sgRNA 0841 in −126, sgRNA 1449 in −83, sgRNA 5093 in −71, and sgRNA 2281 in −7). A set of negative control sgRNAs (sgRNA 5430 at DHS −49, HBS1L targeting, and BCL11A targeting sgRNAs) were also included.
HUDEP-2 and CD34+ HSPCs were transduced with the CRISPR/Cas9 components and subjected to erythroid differentiation conditions. Targeting the −126, −83, −71, and −7 DHS led to a severe proliferation defect in HUDEP-2 cells (Supplementary Fig. 23a). Similarly, a cellular proliferation defect was observed in the CD34+ derived erythroblasts (Supplementary Fig. 23b). Targeting MYB coding sequence had an intermediate phenotype by comparison in CD34+ derived erythroblasts. Targeting HBS1L and BCL11A coding sequence, −84 DHS (1329), −49 DHS (5430), and −71 DHS (1582) again showed no impact on cellular proliferation (Supplementary Fig. 23b). After 18 days of erythroid differentiation, MYB levels were significantly reduced upon targeting the four enhancer elements and MYB coding sequence, consistent with the observed cellular proliferation defects (Supplementary Fig. 23c). HBS1L expression levels were unchanged (Supplementary Fig. 23d). A moderate differentiation block was also observed when targeting the −126, −83, −71, and −7 DHS (Supplementary Fig. 23e).
Reduction in MYB expression upon targeting the sequences within −126, −83, −71, and −7 DHS implicated by the saturating mutagenesis screen suggested these regions may harbor MYB enhancer activity; however, these results are confounded by the repetitive sequence causing increased off-target cleavage potential. Therefore, the significance of these regions remains unclear. Current genome editing technology has limited ability to unambiguously target a single site when an sgRNA has multiple genomic matches.
DISCUSSION
The functional sequences responsible for most GWAS identified trait associations have remained unclear due to the paucity of methods to interrogate the function of trait-associated non-coding sequences in a high-throughput manner. Comprehensive mutagenesis by HDR to introduce every possible base within a segment may be the most stringent test of the functional impact of individual variants39; however, this approach is limited by throughput and efficiency. We propose that high-resolution, variant-informed CRISPR-based saturating mutagenesis can provide a powerful tool with which to investigate variant-decorated regulatory DNA. Notably, previous studies of the HBS1L-MYB intergenic region associated with HbF level and other erythroid traits had focused on two functional regions, −71 and −8415. Our approach allowed for high resolution functional mapping of all DHS in an approximately 300 kb locus, which identified multiple putative functional regions. This analysis suggests MYB enhancer function in the previously known −84 DHS and identified a novel MYB enhancer at −36. Furthermore, we identified potential function for the −7, −71, −83, and −126 elements. Our data confirms the genetic association of the −84 DHS region with MYB expression levels and nominates rs61028892 as a potential causal variant.
It is intriguing that the screen identified the −71 DHS as a site for potential MYB enhancer activity. Notably, mutagenesis of the GATA1 motif modified by the genetically implicated rs9494142/rs11154792 did not alter MYB expression. However, the significance of the identified repetitive region in close proximity to rs9494142/rs11154792 remains unclear, but may be essential for MYB regulation in this region. Our data identifying repetitive elements in close proximity to genetically implicated variants suggest the unique context of a repetitive sequence may influence its function.
This work highlights the challenge of repetitive sequences present in non-coding regions. Experimental methods to circumvent the issue of targeting a repetitive sequence are limited. One possibility is to engender deletion of an entire repetitive region; however, this approach suffers from low throughput and low resolution. Our work suggests that genomic match and off-target analysis need to be considered in execution of noncoding dropout screens to rule out off-target cleavages as a source of cellular toxicity. In addition, it may be important to consider that SNPs present in cell lines used for study can create novel off-target genomic matches40. Our data suggest that thorough off-target analysis can reduce ambiguity and allow for reliable assignment of regulatory potential even in the setting of repetitive regions.
We created DNA Striker to streamline design of variant-aware saturating mutagenesis libraries using multiple nucleases. We have included a computational algorithm to calculate off-target scores for these sgRNA libraries. Taken together, our data establish a methodology for high-resolution, variant-informed, off-target-aware, saturating mutagenesis as a powerful and high-throughput approach for identification of functional sequences at disease- and trait-associated regulatory DNA.
Methods
No statistical methods were used to predetermine sample size.
HUDEP-2 cell culture
HUDEP-2 cells were used as previously described3,31, which tested negative for mycoplasma contamination. HUDEP-2 cells were expanded in SFEM (Stem Cell Technologies) supplemented with 100 ng/mL stem cell factor (R&D), 3 UI/mL erythropoietin (Amgen), 10−6 M dexamethasone (Sigma), 1 µ/mL of doxycycline (Sigma), and 2% penicillin-streptomycin (Thermo Fisher). HUDEP-2 cells were differentiated in Iscove’s Modified Dulbecco’s Medium (IMDM) supplemented with 330 µg/mL holo-human transferrin (Sigma), 10 µg/mL recombinant human insulin (Sigma), 2 IU/mL heparin (Sigma), 5% human solvent detergent pooled plasma AB (Rhode Island Blood Center), 3 IU/mL erythropoietin (Amgen), 100 ng/mL human stem cell factor (SCF) (R&D), 1 µg/mL doxycycline (Sigma), 1% L-glutamine (Life Technologies), and 2% penicillin/streptomycin (Life Technologies).
HUDEP-2 SpCas9 and HUDEP-2 SpCas9-VQR cells
NGG Cas9 lentivirus was prepared as described below using LentiCas9-Blasticidin (Addgene plasmid ID 52962). Cells were transduced with LentiCas9-Blasticidin lentivirus and maintained with 10 µg/mL blasticidin (Sigma). The LentiCas9-Blasticidin (Addgene plasmid ID 52962) plasmid was modified to include the VQR mutations as described in Kleinstiver et al28 (Addgene plasmid ID 87155). SpCas9-VQR lentivirus was prepared as described below using VQR-modified LentiCas9-Blast lentivirus. Cells were transduced with VQR-modified LentiCas9-Blast and maintained with 10 µg/mL blasticidin (Sigma).
SpCas9 and SpCas9-VQR Cas9 activity reporters
To assess Cas9 activity, lentiviral reporters were used that included green florescent protein (GFP) and either an NGG-restricted or NGA-restricted sgRNA targeting GFP sequence. The NGG Cas9 activity reporter has been previously described41 (Supplementary Table 8). In order to construct an NGA Cas9 activity reporter, pLentiGuide-Puro (Addgene plasmid ID 52963) was modified to express GFP and an NGA-restricted sgRNA targeting GFP sequence (Addgene plasmid ID 87156; Supplementary Table 8).
Lentivirus production
HEK293T cells were cultured with Dulbecco’s Modified Eagle’s Medium (DMEM) (Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Omega Scientific) and 2% penicillin-streptomycin (Life Technologies). HEK29T were transfected at 80% confluence in 15 cm tissue culture treated petri dishes with 16.25 µg psPAX2, 8.75 µg VSV-G, and 25 µg of the lentiviral construct plasmid of interest using 150 µg of branched polyethylenimine (Sigma). Medium was refreshed 16–24 hours after transfection. Lentiviral supernatant was collected at 48 and 72 hours post-transfection. Viral supernatant was concentrated by ultracentrifugation (24,000 rpm for 2 hours at 4°C; Beckman Coulter SW 32 Ti rotor).
Non-targeting sgRNA design and calculation of off-target scores
In order to design sgRNAs that do not target the human (hg19) and mouse (mm9) genomes, we first extracted all possible 20 bp sequences immediately preceding NG PAM motifs in both genomes. We created 5,000 random 20 bases sgRNA sequences that we compared to all 20 bp reference sequences. We calculated a targeting score dependent on the number and position of mismatches between both sequences using the methodology of Sanjana et al37. The score ranges from 0 (non-targeting) to 1 (perfect match). We assigned a score of 0 to sequences with more than 4 mismatches. Reference sequences with score > 0 were considered potential off-targets. For each random guide, we derived an aggregated score from all possible off-targets, as per Sanjana et al37:
Where n is the number of potential off-target “hits”, and Shit(hi) is the targeting score of the possible off-target sequence hi. In this situation, an aggregated score of 100 corresponds to no possible targets in the genome. Multiple off-targets or the presence of hi-scoring off targets will lower the score towards 0. We defined guides with an aggregated score > 90 as non-targeting (n=128). This formula was also applied to all sgRNA in both NGG- and NGA-restricted libraries to calculate a predicted off-target score. This produces scores between 0 and 100 with a higher score indicating a decreased probability of off-target effects. This tool is publically available for download.
Pooled CRISPR/Cas9 library design for high resolution, variant-informed functional mapping of the HBS1L-MYB intergenic region
The summit of every DNase hypersensitive site (DHS) within the HBS1L-MYB region (n = 98) was identified from fetal- and adult-derived CD34+ subject to erythroid differentiation2. The regions of DHS summit +/− 200 bp were chosen for saturating mutagenesis based on previous work that suggested functional sequence tended to be located within 200 bp of the peak of DNase hypersensitivity3. Using the DNA Striker tool, every 20-mer sequence upstream of an NGG or NGA PAM sequence on the sense or anti-sense strand was identified for each HBS1L-MYB region DHS as well as BCL11A exon 2, the core of the +58 DHS within the BCL11A enhancer3, HBS1L exon 4, MYB exon 5 (Figure 2a–c; Supplementary Tables 6–7). Phased variants within these region were taken from the 1,000 Genomes Project database in VCF file format using all individuals available by August 2015 (2,504 individuals; 5,008 alleles)42. Using the 1,000 Genomes variants, the variants feature within DNA Striker was used to identify sgRNAs altered by variants or new sgRNA resulting from PAM sequences created by variants. Variant-associated sgRNA were included in the library if they were present at a frequency (“guide frequency”) of ≥ 1% (Supplementary Fig. 14). Guide frequency was used as a surrogate for variant frequency. After filtering non-unique sgRNA, the NGG library was composed of 2,166 sgRNAs targeting HBS1L-MYB DHS, 176 variant-associated sgRNAs, 13 sgRNAs targeting HBS1L exon 4, 28 sgRNAs targeting MYB exon 5, 21 sgRNAs targeting the BCL11A enhancer +58 DHS core, 53 sgRNAs targeting BCL11A exon 2, and 128 non-targeting sgRNAs for a total of 2,585 sgRNAs. After filtering non-unique sgRNA, the NGA library was composed of 2,524 sgRNAs targeting HBS1L-MYB DHS, 186 variant-associated sgRNAs, 32 sgRNAs targeting HBS1L exon 4, 28 sgRNAs targeting MYB exon 5, 12 sgRNAs targeting the BCL11A enhancer +58 DHS core, 47 sgRNAs targeting BCL11A exon 2, and 128 non-targeting sgRNAs for a total of 3,018 sgRNAs. Each of these 20-mer oligos were synthesized as previously described3,37,43,44 and cloned using a Gibson Assembly master mix (New England Biolabs) into pLentiGuide-Puro (Addgene plasmid ID 52963). Plasmid libraries were deep sequenced to confirm representation.
DNA Striker computational tool
DNA Striker allows users to create high resolution, variant aware saturating mutagenesis libraries and provides quantification of the degree of saturation and visualization of the distribution of sgRNA across the region(s) of interest. DNA Striker is a computational DNA Striker includes support for any combination of 3’ PAM sequences such as utilized for Cas9 from various species (such as SpCas9, SaCas9, NmCas9, et cetera) or 5’ PAM sequences such as utilized for the Cpf1 nuclease6,7,29. Briefly, uploaded DNA sequence(s) are analyzed for all selected PAM(s) sequences using a sliding window approach. The sgRNA length can be customized for each PAM sequence in the library given that optimal sgRNA length varies for different CRISPR-associated nucleases6,7,24,26,29. Variant-aware sgRNA library design involves identifying sgRNAs altered by variants and novel sgRNAs resulting from PAM sequences created by the presence of variants (Fig. 2).
Variant analysis for whole genome sequencing or a custom list of variants occurs by creating multiple versions of the sliding window: the non-variant version, versions with each variant in the window inserted in isolation (and all combinations of up to three variants in each window for custom variant lists). Variant analysis for haplotype data occurs by creating each individual allele present in the haplotype data provided. The output includes a list of oligonucleotides for full library design and two figures demonstrating the distribution of cleavages within the uploaded sequence(s) (Supplementary Fig. 2).
Cutting frequency determination (CFD)
CFD scores were calculated to evaluate the effect of mismatches on sgRNA activity. Published CFD scores were obtained from Doench et al30, which provides tables of CFD for all possible combinations of sgRNA and DNA single mismatches. For the calculation of >1 mismatches, single mismatch CFD scores were multiplied together.
Pooled CRISPR/Cas9 screen for high resolution, variant-informed functional mapping of the HBS1L-MYB intergenic region
HUDEP-2 cells with stable SpCas9 or SpCas9-VQR Cas9 expression were transduced at low multiplicity with the corresponding NGG or NGA sgRNA library lentivirus pool while in expansion medium (NGG and NGA screens were performed independently). 10 µg/mL blasticidin (Sigma) and 1 µg/mL puromycin (Sigma) were added 24 hours after transduction to select for lentiviral library integrants in cells with Cas9. The screens for fetal hemoglobin expression in HUDEP-2 cells were performed as previously described3. Briefly, HUDEP-2 cells were differentiated and intracellularly stained for HbF (clone HbF-1 with APC conjugation; Life Technologies). 0.2 µg HbF antibody was used per 500,000–5 million cells. An HbF-stained non-targeting sgRNA sample was used as a negative control to set a sorting gate for the HbF-high population (approximately top 5% of HbF-expressing cells). A corresponding percentage of cells from the HbF-low population were also sorted. After sorting the HbF-high and HbF-low pools, library preparation and deep sequencing was performed as previously described3,45. 6.6 µg of DNA per sample were submitted for Illumina MiSeq paired end sequencing with Nextera sequencing primers. Guide sequences present in the HbF-high and HbF-low pools were enumerated. HbF enrichment was determined by the log2 transformation of the median number of occurrences of a particular sgRNA in the HbF-high pool divided by the median number of occurrences of the same sgRNA in the HbF-low pool across the 3 biological screen replicates for each PAM-restricted library. Dropout scores were calculated by the ratio of normalized reads in the cells at end of experiment (average of reads in the HbF-high and HbF-low pools) to reads in the plasmid pool for the median of the 3 biological screen replicates for each PAM-restricted library followed by log2 transformation. Enrichment and dropout scores were converted to Z-scores using the Z-score function in MATLAB software. sgRNA sequences were mapped to the human genome (hg19). The plasmid library was deep sequenced to confirm representation using the same methodology. A quantile-quantile (Q-Q) plot was made using the dropout scores prior to Z-score normalization with a line fitted through the first and third quantiles using MATLAB software.
Determination of PAM distributions
Repeat-masked regions of the human genome (hg19) were removed. Non-repeat-masked repeats were parsed out separately to avoid creating false genomic junctions. PAMs were identified and the associated double strand break site for each potential sgRNA was determined. sgRNA with double strand break positions outside of these regions were excluded from analysis. Double strand break positions were compiled from sgRNA on both the plus and minus strands. The difference between adjacent genomic double strand break sites was calculated. Promoters (transcriptional start site +/− 2 kilobases), exons, and introns were determined from RefSeq annotations. Enhancer and DNase hypersensitive regions for GM12878, H1 hESC, HepG2, HMEC, HSMM, HUVEC, K562, NHEK, and NHLF cell lines were taken from publically available databases46. Repressed regions were used from previously published data47.
Super-enhancer analysis
Human H3K27ac ChIP-seq was obtained from a previously published dataset48. The ROSE algorithm was used to perform super enhancer analysis49.
GATA1/TAL1 chromatin immunoprecipitation sequencing (ChIP-seq) and chromatin immunoprecipitation quantitative PCR (ChIP-qPCR)
ChIP-sequencing was obtained from primary human erythroblasts from CD34+ HSPCs subject to erythroid differentiation conditions using a GATA1 antibody (ab11852; Abcam), TAL1 antibody (clone C-21; Santa Cruz), and H3K27Ac antibody (Abcam, ab4729). ChIP-qPCR data was obtained from HUDEP-2 cells six days after lentiviral transduction with CRISPR/Cas9 reagents.
Erythroid DNase I hypersensitivity
Erythroid DNase I hypersensitivity was obtained from a previously published dataset2.
Transcription factor motif analysis
Motif analysis was performed using FIMO software to scan putative transcription factor binding sites within the identified elements within the HBS1L-MYB intergenic region (p-value cutoff of 10−4)50. The most recent version of the JASPAR database using hg19 sequences was used for the analysis51.
Hidden Markov Model (HMM) analysis
HMM analysis to identify repressive, active, and neutral sequences was performed as previously described3.
Red blood cell traits meta-analysis
Red cell associated SNPs were taken from a previously published meta-analysis22. Only SNPs with P<10−6 are publically available.
Genotyping of SCD patients
Briefly, genotyping of 1,139 African Americans from the Cooperative Study of Sickle Cell Disease (CSSCD) was performed on the Illumina Human610-Quad array as previously described52. We further genotyped 353 independent samples form the CSSCD, 57 samples from the Multicenter Study of Hydroxyurea in Sickle Cell Anemia (MSH) study, 398 samples from GENMOD, 186 from the Sickle Cell Center at Georgia Health Sciences University, and 89 from the Jamaica Sickle Cell Cohort Study (JSCCS) using the Illumina Infinium HumanOmni2.5Exome-8v1.1 array. We performed quality control using PLINK, removing SNPs with Hardy-Weinberg P<1×10−7 and genotyping rate < 90%. After quality control, a total of 1,083 samples with available HbF measures and genotyping success rate >99.8% remained. We conducted genotype imputation on 1,000 genomes phase 3 haplotypes (version 5, hg19) using Minimac3 (v1.0.11). After imputation, both datasets contained around 47 million markers. We restricted the analysis to markers with an imputation r2>0.3 and falling inside the HBS1L-MYB intergenic region (chr6:135,281,517–135,540,311, hg19). In total, 2,763 markers were included in the analysis. We transformed HbF measures to Z-scores corrected for age and sex. We derived HbF association P-values independently for both datasets using RVtests (v.20140416), further correcting for the top 10 principal components. We meta-analyzed P-values using Raremetals (v.6.0).
Conditional analysis
Stepwise conditional analysis was performed until the top SNP had a P-value < 3.15 × 10−5. This P-value represents the Bonferroni-corrected P-value for the number of independent SNPs in the MYB region. The number of independent SNPs in the African 1000 genomes data was calculated using the PLINK option --indep 200 5 2, which found 1,587 independent SNPs from a total of 2,743 SNPs.
Deep sequencing indel quantification and frameshift analysis
Locus-specific deep sequencing was performed using a two PCR strategy as previously described3,45. Briefly, genomic DNA was extracted using the Qiagen Blood and Tissue kit. For PCR #1, Herculase PCR reaction (Agilent) was performed using locus-specific primers that included Illumina Nextera handle sequences. The PCR conditions were as follows: Herculase II reaction buffer (1×), forward and reverse primers (0.5 µM each), dimethyl sulfoxide (DMSO) (8%), deoxynucleotide triphosphates (dNTPs) (0.25 mM each), Herculase II Fusion DNA Polymerase (0.5 reactions) using the following PCR cycling parameters: 95°C for 2 minutes; 20 cycles of 95°C for 15 seconds, 60°C for 20 seconds, 72°C for 30 seconds; 72°C for 5 minutes. For PCR #2, the PCR #1 reaction product was diluted (1:10) and subjected to PCR using handle-specific primers to add adaptors and indexes to each sample using the following conditions3,45: Herculase II reaction buffer (1×), forward and reverse primers (0.5 µM each), dNTPs (0.25 mM each), Herculase II Fusion DNA Polymerase (0.5 reactions) with the following cycling parameters: 95°C for 2 minutes; 25 cycles of 95°C for 15 seconds, 60°C for 20 seconds, 72°C for 30 seconds; 72°C for 5 minutes. Products of the expected size from PCR #2 were gel purified and subjected to Illumina MiSeq 150 bp paired end sequencing. Quantification of indels and analysis of frameshift and in-frame mutations from the deep sequencing data was performed using CRISPResso (www.crispresso.rocks)38.
Sequencing
Sanger sequencing of the −126, −84, −83, −71, −36 and −7 DHSs identified a single variant in HUDEP-2 cells, which was heterozygousity for rs144062313 in −126 DHS. rs144062313 has a minor allele frequency <1% in the 1,000 Genomes Database so was not included in library design.
shRNA-mediated knockdown of MYB
shRNA constructs cloned into the pLKO.1-puromycin lentiral vector were acquired from Sigma Mission shRNA library. Three shRNA targeted against MYB were obtained (Supplementary Table 9): MYB shRNA 1 (TRCN0000295917), MYB shRNA 2 (TRCN0000040058), and MYB shRNA 3 (TRCN0000040060). A scrambled sequence shRNA was used as a non-targeting control. Lentivirus for each shRNA was produced as described above. MYB knockdown was confirmed by western blot for three shRNA constructs in HEK293T cells as they do not require MYB for cellular fitness. HEK293T cells were transduced with shRNA lentivirus. Successful transductants were selected for 24 hours after lentivirus administration using 1 µg/mL puromycin. Western blots were performed using a MYB antibody (1:1000 dilution; EP769Y; Abcam) and GAPDH (1:2000 dilution; FL-335; Santa Cruz).
Erythroid differentiation of primary human CD34+ hematopoietic stem and progenitor cells (HSPCs)
Primary human CD34+ HSPCs from de-identified, healthy adult donors following G-CSF mobilization were acquired from the Excellence in Molecular Hematology at the Fred Hutchinson Cancer Research Center (Seattle, Washington). CD34+ HSPCs were subjected to erythroid differentiation conditions using a three phase culture system as previously described3,53. Erythroid differentiation medium (EDM) was created as follows: IMDM (CellGro) supplemented with 330 µg/mL holo-human transferrin (Sigma), 10 µg/mL recombinant human insulin (Sigma), 2 IU/mL heparin (Sigma), 5% human solvent detergent pooled plasma AB (Rhode Island Blood Center), 3 IU/mL erythropoietin (Amgen), 1% L-glutamine (Life Technologies), and 2% penicillin/streptomycin (Life Technologies). Phase I medium consisted of EDM supplemented with 10−6 M hydrocortisone (Sigma), 100 ng/mL human SCF (R&D), and human IL-3 (R&D). Phase II medium consisted of EDM supplemented with 100 ng/mL SCF. Phase III medium consisted of EDM without additional supplementation. CD34+ HSPCs were thawed into Phase I medium and were maintained in this medium for the first 7 days of culture. Cells were switched to Phase II medium for days 7–11 of culture. Cells were switched to Phase III medium for days 11–18 of culture.
Transduction of CD34+ HSPCs with CRISPR/Cas9
CD34+ HSPCs were thawed into Phase I medium on day 0. On day 1, 10 µM prostaglandin E2 (PGE2) (Cayman Chemical) was added to culture medium in conjunction with Cas9 lentivirus (LentiCas9-Blast; Addgene plasmid ID 52962). On day 2, medium was refreshed and 10 µM prostaglandin E2 (PGE2) (Cayman Chemical) was added to the fresh Phase I culture medium in conjunction with sgRNA lentivirus (LentiGuide-Puro; Addgene plasmid ID 52963). On day 3, medium was refreshed and fresh Phase I medium was supplemented with 10 µg/mL blasticidin (Invivogen) and 1 µg/mL puromycin (Sigma) to select for successful transductants. Blasticidin selection persisted for 5 days and puromycin selection persisted for 14 days.
Transduction of CD34+ HSPCs with shRNA
CD34+ HSPCs were thawed into Phase I medium on day 0. On day 2, 10 µM prostaglandin E2 (PGE2) (Cayman Chemical) was added to culture medium in conjunction with shRNA lentivirus. On day 3, medium was refreshed and fresh Phase I medium was supplemented with 1 µg/mL puromycin (Sigma) to select for successful transductants. Puromycin selection persisted for 14 days.
Assessment of erythroid differentiation
Success of erythroid differentiation of CD34+ HSPCs was assessed at three time points during the 18 day three phase culture (days 10, 14, and 18) using the transferrin receptor (CD71; Clone OKT9 with FITC conjugation; eBioscience) and glycophorin A (CD235; Clone HIR2 with PE conjugation; eBioscience). To assess hemoglobinization and hemoglobin composition, flow cytometry was performed following intracellular staining for HbF (clone HbF-1 with APC conjugation; Life Technologies) and β-hemoglobin antibody (clone 37-8 with PE conjugation; Santa Cruz).
Assessment of cellular proliferation
Cell proliferation was assessed using the Countess automated cell counter (Invitrogen) with trypan blue exclusion.
Statistical tests
Unpaired two-sided Mann-Whitney testing was used to compare dropout between SpCas9 and SpCas9-VQR species (α = 0.05). All other statistical testing was performed using unpaired two-sided t-tests (α = 0.05).
Supplementary Material
Acknowledgments
We thank Z. Herbert, M. Berkeley, and M. Vangala at the Dana-Farber Cancer Institute Molecular Biology Core Facility for sequencing, Falong Lu at the HHMI Sequencing facility, and members at the Hematologic Neoplasia Flow Cytometry and the Flow Cytometry Core facilities at the Dana-Farber Cancer Institute for cell sorting. We also thank John Doench, Maximilian Haeussler, Jean-Paul Concordet, Robert Barretto, Vijay Sankaran, and Jian Xu for helpful discussions. M.C.C. is supported by a National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Award (F30DK103359-01A1). L.P. is supported by a National Human Genome Research Institute (NHGRI) Career Development Award (K99HG008399). S.L. is funded by a Canadian Institutes of Health research Banting doctoral scholarship. E.N.S. is supported by a Hematology Opportunities for the Next Generation of Research Scientists (HONORS) award from the American Society of Hematology. G.C.Y. is supported by awards from the National Heart, Lung, and Blood Institute (NHLBI) (R01HL119099). G.L. is funded by the Canada Research Program, the Montreal Heart Institute Foundation, and the Canadian Institute of Health Research (MOP123382). A portion of the DNA genotyping was performed as part of the Biogen Sickle Cell Disease Consortium. D.E.B. is supported by NIDDK (K08DK093705, R03DK109232), NHLBI (DP2OD022716), Burroughs Wellcome Fund, Doris Duke Charitable Foundation Innovations in Clinical Research Award, ASH Scholar Award, Charles H. Hood Foundation Child Health Research Award, and Cooley’s Anemia Foundation fellowship. S.H.O. is supported by an award from the NHLBI award (P01HL032262) and an award from the NIDDK (P30DK049216, Center of Excellence in Molecular Hematology).
Footnotes
AUTHOR CONTRIBUTIONS
M.C.C., D.E.B., and S.H.O. conceived this study. M.C.C. developed the DNA Striker computational tool and performed computational analysis of degrees of PAM saturation. M.C.C., Y.W., E.S., A.N., D.D.C., P.P.D., M.C., and J.Z. performed the experiments. S.L, Y.I., F.G., C.B., A.K., C.M., M.R., and G.L. performed the genotyping and genetic analysis. R.K. and Y.N. provided the HUDEP-2 cell line. M.C.C., S.L., Y.I., L.P., G.C.Y., G.L. performed computational and statistical analysis. D.E.B. and S.H.O. supervised this work. M.C.C., D.E.B., and S.H.O. wrote the manuscript with input from all authors.
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
URLs. DNA Striker, https://github.com/mcanver/DNA-Striker; CRISPR Off-Target Tool, http://www.mhi-humangenetics.org/en/resources; 1,000 Genomes Project, http://www.internationalgenome.org/; R Statistical Computing and Graphics, https://cran.r-project.org/; CRISPResso, http://crispresso.rocks/; MATLAB, https://www.mathworks.com/; PLINK, http://pngu.mgh.harvard.edu/~purcell/plink/; Minimac3, http://genome.sph.umich.edu/wiki/Minimac3, Raremetals, http://genome.sph.umich.edu/wiki/RareMETALS; RVtests, http://genome.sph.umich.edu/wiki/RvTests; Off-target formula, http://crispr.mit.edu/about.
Data availability. GATA1, TAL1, and H3K27Ac ChIP-sequencing experiments are publicly available from NCBI under accession GSE93372.
Code availability. DNA Striker was developed in MATLAB software. The Matlab .m file and a stand-alone version (.exe) for DNA Striker are available for download along with user instructions and example input/output datasets.
References
- 1.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer DE, et al. An Erythroid Enhancer of BCL11A Subject to Genetic Variation Determines Fetal Hemoglobin Level. Science. 2013;342:253–257. doi: 10.1126/science.1242088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Canver MC, et al. Characterization of Genomic Deletion Efficiency Mediated by Clusted Regularly Interspaced Palindromic Repeats (CRISPR)/Cas9 Nuclease System in Mammalian Cells. J. Biol. Chem. 2014;289:21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Corradin O, et al. Combinatorial effects of multiple enhancer variants in linkage disequilibrium dictate levels of gene expression to confer susceptibility to common traits. Genome Res. 2014;24:1–13. doi: 10.1101/gr.164079.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mali P, et al. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ran FA, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hsu PD, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uda M, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc. Natl. Acad. Sci. U. S. A. 2008;105:1620–1625. doi: 10.1073/pnas.0711566105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lettre G, et al. DNA polymorphisms at the BCL11A, HBS1L-MYB, and Beta-globin loci associate with fetal hemoglobin levels and pain crises in sickle cell disease. Proc. Natl. Acad. Sci. U. S. A. 2008;105:11869–11874. doi: 10.1073/pnas.0804799105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thein SL, et al. Intergenic variants of HBS1L-MYB are responsible for a major quantitative trait locus on chromosome 6q23 influencing fetal hemoglobin levels in adults. Proc. Natl. Acad. Sci. U. S. A. 2007;104:11346–11351. doi: 10.1073/pnas.0611393104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galarneau G, et al. Fine-mapping at three loci known to affect fetal hemoglobin levels explains additional genetic variation. Nat. Genet. 2010;42:1049–1051. doi: 10.1038/ng.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farrell JJ, et al. A 3-bp deletion in the HBS1L-MYB intergenic region on chromosome 6q23 is associated with HbF expression. Blood. 2011;117:4935–4945. doi: 10.1182/blood-2010-11-317081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stadhouders R, et al. HBS1L-MYB intergenic variants modulate fetal hemoglobin via long-range MYB enhancers. J. Clin. Invest. 2014;124:1699–1710. doi: 10.1172/JCI71520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mtatiro SN, et al. Genome wide association study of fetal hemoglobin in sickle cell anemia in Tanzania. PLoS One. 2014;9:e111464. doi: 10.1371/journal.pone.0111464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bae HT, et al. Meta-analysis of 2040 sickle cell anemia patients: BCL11A and HBS1L-MYB are the major modifiers of HbF in African Americans. Blood. 2012;120:1961–1962. doi: 10.1182/blood-2012-06-432849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganesh SK, et al. Multiple loci influence erythrocyte phenotypes in the CHARGE Consortium. Nat. Genet. 2009;41:1191–1198. doi: 10.1038/ng.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Soranzo N, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat. Genet. 2009;41:1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamatani Y, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat. Genet. 2010;42:210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 21.Menzel S, Garner C, Rooks H, Spector TD, Thein SL. HbA2 levels in normal adults are influenced by two distinct genetic mechanisms. Br. J. Haematol. 2013;160:101–105. doi: 10.1111/bjh.12084. [DOI] [PubMed] [Google Scholar]
- 22.van der Harst P, et al. Seventy-five genetic loci influencing the human red blood cell. Nature. 2012;492:369–375. doi: 10.1038/nature11677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen Z, et al. Genome-wide association analysis of red blood cell traits in African Americans: the COGENT Network. Hum. Mol. Genet. 2013;22:2529–2538. doi: 10.1093/hmg/ddt087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Esvelt KM, et al. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat. Methods. 2013;10:1116–1121. doi: 10.1038/nmeth.2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mali P, Esvelt KM, Church GM. Cas9 as a versatile tool for engineering biology. Nat. Methods. 2013;10:957–963. doi: 10.1038/nmeth.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kleinstiver BP, et al. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat. Biotechnol. 2015;33:1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleinstiver BP, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523:481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zetsche B, et al. Cpf1 Is a Single RNA-Guided Endonuclease of a Class 2 CRISPR-Cas System. Cell. 2015;163:759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doench JG, et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016;34:184–191. doi: 10.1038/nbt.3437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kurita R, et al. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS One. 2013;8:e59890. doi: 10.1371/journal.pone.0059890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Canver MC, Orkin SH. Customizing the genome as therapy for the β-hemoglobinopathies. Blood. 2016;127:2536–2545. doi: 10.1182/blood-2016-01-678128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sankaran VG, et al. MicroRNA-15a and-16-1 act via MYB to elevate fetal hemoglobin expression in human trisomy 13. Proc. Natl. Acad. Sci. U. S. A. 2011;108:1519–1524. doi: 10.1073/pnas.1018384108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rajagopal N, et al. High-throughput mapping of regulatory DNA. Nat. Biotechnol. 2016;34:167–174. doi: 10.1038/nbt.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Munoz DM, et al. CRISPR screens provide a comprehensive assessment of cancer vulnerabilities but generate false-positive hits for highly amplified genomic regions. Cancer Discov. 2016;6:900–913. doi: 10.1158/2159-8290.CD-16-0178. [DOI] [PubMed] [Google Scholar]
- 36.Aguirre AJ, et al. Genomic copy number dictates a gene-independent cell response to CRISPR-Cas9 targeting. Cancer Discov. 2016;2641:617–632. doi: 10.1158/2159-8290.CD-16-0154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sanjana NE, Shalem O, Zhang F. Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods. 2014;11:783–784. doi: 10.1038/nmeth.3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pinello L, et al. Analyzing CRISPR genome-editing experiments with CRISPResso. Nat. Biotechnol. 2016;34:695–697. doi: 10.1038/nbt.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Findlay GM, Boyle EA, Hause RJ, Klein JC, Shendure J. Saturation editing of genomic regions by multiplex homology-directed repair. Nature. 2014;513:120–123. doi: 10.1038/nature13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yang L, et al. Targeted and genome-wide sequencing reveal single nucleotide variations impacting specificity of Cas9 in human stem cells. Nat. Commun. 2014;5:5507. doi: 10.1038/ncomms6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Doench JG, et al. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shalem O, Sanjana NE, Zhang F. High-throughput functional genomics using CRISPR–Cas9. Nat. Rev. Genet. 2015 doi: 10.1038/nrg3899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen S, et al. Genome-wide CRISPR Screen in a Mouse Model of Tumor Growth and Metastasis. Cell. 2015;160:1246–1260. doi: 10.1016/j.cell.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shalem O, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science. 2014;343:84–87. doi: 10.1126/science.1247005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunham I, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pinello L, Xu J, Orkin SH, Yuan G-C. Analysis of chromatin-state plasticity identifies cell-type-specific regulators of H3K27me3 patterns. Proc. Natl. Acad. Sci. U. S. A. 2014;111:E344–E353. doi: 10.1073/pnas.1322570111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xu J, et al. Combinatorial assembly of developmental stage-specific enhancers controls gene expression programs during human erythropoiesis. Dev. Cell. 2012;23:796–811. doi: 10.1016/j.devcel.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mathelier A, et al. JASPAR 2014: An extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2014;42:142–147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Solovieff N, et al. Fetal hemoglobin in sickle cell anemia: genome-wide association studies suggest a regulatory region in the 5’ olfactory receptor gene cluster. Blood. 2010;115:1815–1822. doi: 10.1182/blood-2009-08-239517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Giarratana M, et al. Proof of principle for transfusion of in vitro generated red blood cells. Blood. 2011;118:5071–5079. doi: 10.1182/blood-2011-06-362038. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.