Abstract
The thalamus connects the cortex with other brain regions and supports sensory perception, movement, and cognitive functions via numerous distinct nuclei. However, the mechanisms underlying the development and organization of diverse thalamic nuclei remain largely unknown. Here we report an intricate ontogenetic logic of mouse thalamic structures. Individual radial glial progenitors in the developing thalamus actively divide and produce a cohort of neuronal progeny that exhibits striking spatial configuration and nuclear occupation related to functionality. While the anterior clonal cluster displays relatively more tangential dispersion and contributes predominantly to nuclei with cognitive functions, the medial ventral posterior clonal cluster forms prominent radial arrays and contributes mostly to nuclei with sensory/motor-related activities. Moreover, the first-order and higher-order sensory/motor nuclei across different modalities are largely segregated clonally. Notably, the Shh signaling activity influences clonal spatial distribution. Our study reveals lineage relationship to be a critical regulator of non-laminated thalamus development and organization.
Introduction
The thalamus, with its intricate cortical, subcortical, and cerebellar connections, is a pivotal network node in relaying and modulating sensory and motor signals to the cortex as well as supporting higher-order cognitive functions such as attention and consciousness 1-3. It receives inputs from diverse brain regions including the retina, medial lemniscus, inferior colliculus, basal ganglia, spinal cord, cerebellum, and cortex, and projects to multiple brain structures, especially the cortex 4. The extensive reciprocal connections between the cortex and thalamus allow the traveling of different sensory and motor information along separate pathways, as well as effective information integration.
The thalamus is composed of more than 30 cytoarchitectonically and functionally distinct nuclei, each of which has a different pattern of anatomical connectivity 1, 5-7. In particular, every sensory system (with the exception of olfaction) relies on a thalamic nucleus that receives sensory signals and sends them to the corresponding primary cortical area. Furthermore, each of the thalamic sensory relay nuclei also receives feedback connections from the cortex. The distinct sources and properties of inputs to the thalamus have led to the concept of first-order (FO) and higher-order (HO) thalamic nuclei linked to different sensory modalities 8. The FO nuclei relay peripheral or subcortical information of a particular type to a primary cortical area, whereas the HO nuclei relay information from one cortical area to another cortical area. For example, visual inputs from the retina are sent to the lateral geniculate nucleus (LGN) of the thalamus, which in turn projects to the primary visual cortex. In comparison, the pulvinar (i.e. lateral posterior, LP, in rodents) nucleus relays information between the primary and higher-order visual cortical areas, or between two higher-order cortical areas. Similarly, peripheral somatosensory inputs reach the primary and higher-order somatosensory cortical areas largely via the ventral posterior (VP) and posterior (PO) thalamic nuclei, respectively. Peripheral auditory inputs reach the primary and higher-order auditory cortical areas mostly via the ventral division (vMG) and dorsal division (dMG) of the medial geniculate nucleus in the thalamus, respectively. While the order-specific nuclear organization across different modalities provides an influential framework for understanding thalamic structure and function, very little is known about the mechanisms responsible for its establishment.
The thalamus emerges from the embryonic diencephalon 9, 10. It consists of glutamatergic excitatory neurons and gamma-aminobutyric acid (GABA)-ergic inhibitory interneurons. In rodents, the vast majority of thalamic nuclei contain exclusively excitatory neurons with the exception of the LGN that contains both excitatory and inhibitory neurons, whereas the thalamic reticular nucleus (TRN) harbors exclusively GABAergic interneurons that provide inhibition to all other thalamic nuclei 11. Previous genetic mapping studies in mice have demonstrated that the caudal progenitor domain of the developing thalamus (pTH-C) generates all thalamic excitatory neurons 12. On the other hand, the rostral progenitor domain (pTH-R) of the developing thalamus and prethalamus (PTh) produce GABAergic interneurons in the LGN and TRN 12, 13. While the progenitor domains of thalamic neurons have been delineated, the principles underlying complex nuclear formation and organization of the mammalian thalamus remain largely elusive.
Previous studies have demonstrated that lineage relationship plays an instructive role in guiding the structural and functional assembly of the cortex 14-18, a laminated structure intimately associated and reciprocally interconnected with the thalamus 8, 19, 20. It has previously been suggested that cell lineages in the chick diencephalon exhibit diverse migration routes in general 21, 22. However, it remains largely unclear whether lineage relationship influences the complex nuclear formation and organization of the generally non-laminated thalamus. To address this, we performed a systematic clonal analysis of the progenitor behavior and progeny organization in the developing mouse thalamus using mosaic analysis with double markers (MADM) 23, 24,as well as Cre recombinase-dependent retroviral labeling 25.
Results
MADM Labeling of Thalamic Clones
To label individual neural progenitors lining the third ventricle in the developing mouse thalamus, we introduced the Nestin-CreERT2 transgene 26, in which a tamoxifen (TM)-inducible Cre recombinase was selectively expressed in neural progenitors, including oligodendrocyte transcription factor 3 (OLIG3) 12 and brain lipid-binding protein (BLBP)-positive thalamic radial glial progenitors (RGPs) (Supplementary Fig. 1a), into the MADM11 system 27. In MADM, Cre recombinase-mediated inter-chromosomal recombination in dividing progenitors followed by X-segregation (G2-X) restores one fluorescent marker, enhanced green fluorescent protein (EGFP, green) or tandem dimer Tomato (tdTomato, red), in each of the two daughter cells 23 (Supplementary Fig. 1b). This results in permanent labeling of the two daughter cells and all their descendent lineages in two distinct colors. In addition, upon G2-Z recombination and segregation, or G1/0 recombination events, both EGFP and tdTomato are restored simultaneously in the same daughter cell, resulting in double-labeled (yellow) lineages.
We induced Cre recombinase activity to trigger recombination and labeling via a single dose of TM administered to timed pregnant female mice at one of the following embryonic (E) stages: E9, E10, E11, or E12, and analyzed the brains at the embryonic stages or postnatal (P) days 21-24 (Supplementary Fig. 1c). We found no labeling in the absence of TM treatment (n=3 mice). To ensure unequivocal clonal analysis, we titrated the TM dose to achieve very sparse labeling, i.e. on average 1-2 progenitors (i.e. clones) per thalamic hemisphere (n=120 from 60 embryonic brains) (Fig. 1). We sometimes observed labeled cells in the other brain regions such as the cortex (data not shown). Notably, OLIG3-expressing progenitors in the developing thalamus produced neurons exclusively located in the thalamus (Supplementary Fig. 2a), as previously shown 12. To recover all labeled cells in the thalamus, we performed serial sectioning, immunostaining, and three-dimensional (3D) reconstruction of individual thalami (METHODS).
Figure 1. Labeling of thalamic clones using MADM.
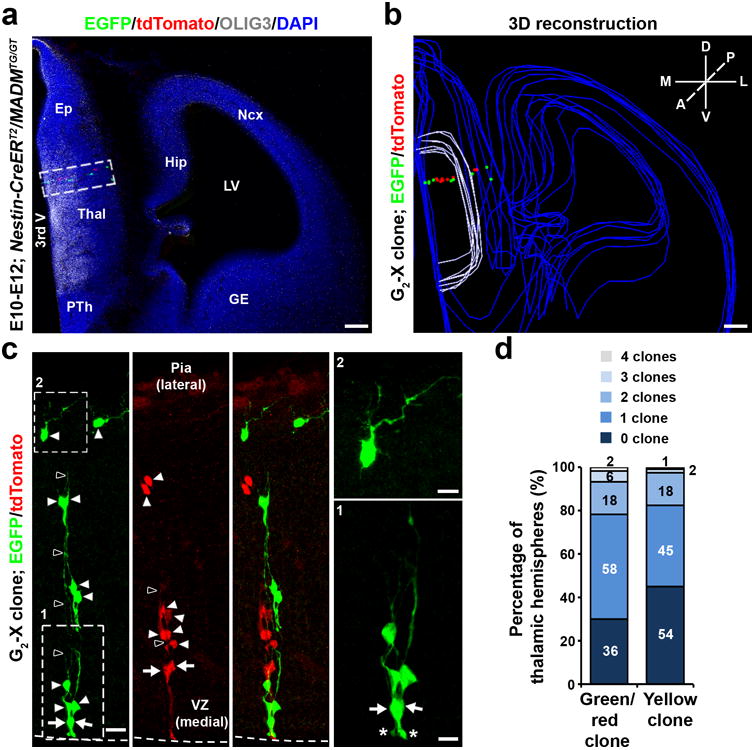
(a) Confocal image of a MADM-labeled E12 clone labeled by TM treatment at E10 and stained for EGFP (green), tdTomato (red), and OLIG3 (white), a transcription factor selectively expressed in developing thalamic progenitors, and with 4′,6-Diamidino-2-phenylindole (DAPI, blue). Scale bars: 100 μm. (b) 3D reconstructed image of the hemisphere in a. Blue lines indicate the contours of brain structures, white lines indicate the OLIG3-positive thalamic domain, and colored dots represent the cell bodies of labeled neurons. A, anterior; P, posterior; D, dorsal; V, ventral; M, medial; L, lateral. Scale bar: 100 μm. (c) High magnification confocal images of the clone in a. Arrows indicate the bipolar RGPs and arrowheads indicate the progeny that are arrayed along the radial glial fibers (open arrowheads). Broken lines indicate the surface of the VZ. Zoomed-in images of RGPs (area 1) and a progeny located near the pia with numerous branches (area 2) are shown to the right. The asterisks indicate the ventricular endfeet of RGPs. Ep, epithalamus; Thal, thalamus; PTh, prethalamus; Ncx, neocortex; Hip, hippocampus; GE, ganglionic eminence; LV, lateral ventricle; 3rd V, 3rd ventricle. Scale bars: 20 μm, 10 μm and 10 μm. (d) Percentage of the thalamic hemispheres with 0, 1, 2, 3, or 4 green/red clone or yellow clones.
We observed individual clusters of cells in green or red (Fig. 1a-c), or yellow (Supplementary Fig. 1d) fluorescence in the embryonic thalamus. These clusters were radially (i.e. mediolaterally) organized and contained bipolar RGPs with the defining morphological characteristics including a cell body in the ventricular zone (VZ) (arrows), a short process reaching the VZ surface with a large end-foot (asterisks), and a long fine radial glial fiber pointing towards the pial surface (open arrowheads), as well as a number of cells with short processes arrayed along the long radial glial fiber(s) (arrowheads) (Fig. 1c). There were no scattered fluorescent cells or mixed clusters of green or red fluorescent cells with yellow fluorescent cells (Fig. 1d). Notably, similar discrete radial clusters in the embryonic thalamus were also reliably labeled by in utero intraventricular injection of low titer Cre recombinase-dependent retroviruses expressing EGFP into the Olig3-Cre28 mouse embryos (Supplementary Fig. 2b), which selectively infected individual OLIG3-expressing thalamic progenitors dividing at the VZ surface and thereby labeled their progeny. The consistent observation of radial clusters with two distinct clonal labeling methods confirmed the clonal nature of individual radial clusters in the developing thalamus.
Cellular Composition of Embryonic Thalamic Clones
We next assessed the cell identity of individual radial clonal clusters and found that the bipolar RGPs were positive for neural progenitor marker NESTIN (area 1, Fig. 2a) and proliferation marker Ki-67 (area 1, Fig. 2b). We also observed short-processed cells outside of the VZ positive for Ki-67 (areas 2, Fig. 2b), suggesting the existence of intermediate progenitors (IPs, or basal progenitors), as previously suggested 29. Besides RGPs and IPs, individual radial clonal clusters also contained cells located further away from the VZ that grew relatively long processes with numerous branches (area 2, Fig. 1c) and expressed neuron-specific marker TUJ1 (area 3, Fig. 2c). Together, these results suggest that individual thalamic RGPs actively divide to produce IPs and post-mitotic neurons that are spatially organized along the mother radial glial fiber into radial clusters.
Figure 2. Cellular composition of MADM-labeled embryonic thalamic clones.
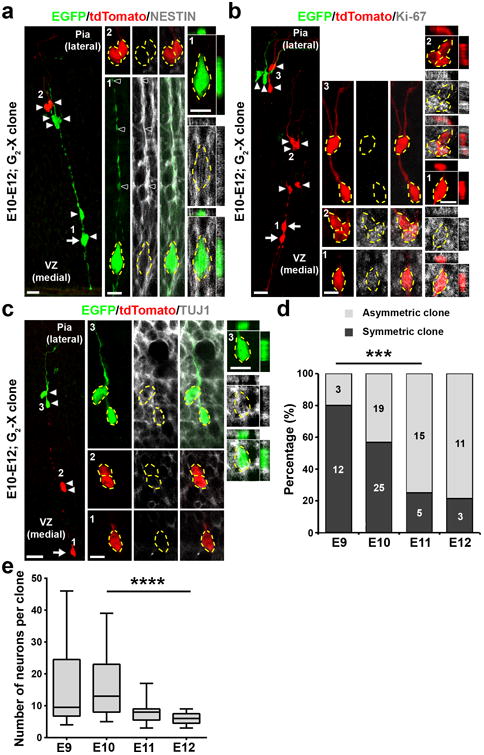
(a) Confocal image of an E12 G2-X clone labeled at E10 and stained for EGFP (green), tdTomato (red), and NESTIN (white), a neural progenitor marker. The arrow indicates the RGP and arrowheads indicate the progeny. High magnification images (areas 1 and 2) are shown in the middle and cross-section images are shown to the right. Broken lines indicate the cell bodies. Scale bars: 20 μm, 10 μm, and 10 μm. (b) Confocal image of an E12 G2-X clone labeled at E10 and stained for EGFP (green), tdTomato (red), and Ki-67 (white), a proliferation marker. Arrows indicate the bipolar RGPs and arrowheads indicate the progeny. High magnification images (areas 1-3) are shown in the middle and cross-section images are shown to the right. Broken lines indicate the cell bodies. Scale bars: 20 μm, 10 μm, and 10 μm. (c) Confocal image of an E12 G2-X clone labeled at E10 and stained for EGFP (green), tdTomato (red), and TUJ1 (white), a neuronal marker. The arrow indicates the RGP and arrowheads indicate the progeny. High magnification images (areas 1-3) are shown in the middle and cross-section images are shown to the right. Broken lines indicate the cell bodies. Scale bars: 20 μm, 10 μm, and 10 μm. (d) Percentage of symmetric (Sym.) vs. asymmetric (Asym.) thalamic clones labeled at different embryonic stages and examined two days later (E9, n=15 clones; E10, n=44 clones; E11, n=20 clones; E12, n=14 clones). ***, p=0.0006 (chi-square test for linear trend).(e) Quantification of the average number of neurons in individual green/red G2-X clones labeled at different embryonic stages and examined at P21-24 (E9, n=10 clones; E10, n=23 clones; E11, n=13 clones; E12, n=9 clones). Data are presented as median with interquartile range, and whiskers are the minimum and maximum. ****, p=2.5e-05 (linear regression analysis).
RGPs divide either symmetrically to generate two RGPs or asymmetrically to give rise to an RGP and a differentiating progeny such as an IP or a neuron. Consistent with this, we observed two types of G2-X green and red fluorescent clones. One type contained both green and red fluorescent RGPs as well as a cohort of green and red fluorescent progeny distributed radially along the radial glial fibers (Fig. 1b), representing the symmetrically dividing clone. In comparison, the other type contained a ‘majority’ population including RGP(s) in one color and a ‘minority’ population in the other color (Fig. 2a-c), representing the asymmetrically dividing clone. We found that as development proceeded, the frequency of observing symmetric clones decreased, while the frequency of observing asymmetric clones concurrently increased (Fig. 2d). Related to this, we observed a progressive decrease in the average neuronal number in clones labeled between E10-E12 (Fig. 2e). There was a small but significant decrease in the average neuronal number in clones labeled at E10, and examined at E15 and P21-24 (Supplementary Fig. 3a), indicating that some neurons are pruned away during development. We also occasionally observed small local clusters without RGPs (Supplementary Fig. 3b), likely representing dividing IP-derived clones.
Spatial Clustering of Thalamic Clones
The diverse thalamic functions are carried out by a large number of nuclei 1, 3. To test whether the progenitor origin and lineage relationship influence nuclear formation and organization, we systematically examined the spatial distribution of individual E9, E10, E11, and E12-labeled clones at P21-24 (Supplementary Table 1), when the nuclei are formed and functional connections are largely established. The vast majority of clones spanned several consecutive sections and consisted of NEUN-positive neurons with the characteristic morphological features of thalamic excitatory neurons (Fig. 3a and Supplementary Fig. 4a,b and 4g), as shown previously30. Moreover, within individual sections, neurons in individual clones were not broadly distributed, but restricted to a defined region (Fig. 3a,b). Notably, the overall spatial localization of progenitors labeled at different embryonic stages correlated well with the overall regional localization of clones observed at P21-24 (Supplementary Fig. 3c), suggesting that clones located in different regions of the mature thalamus are most probably derived from progenitors located in the corresponding regions of the embryonic thalamus.
Figure 3. Spatial clustering of clonally related neurons in the thalamus at the mature stage.
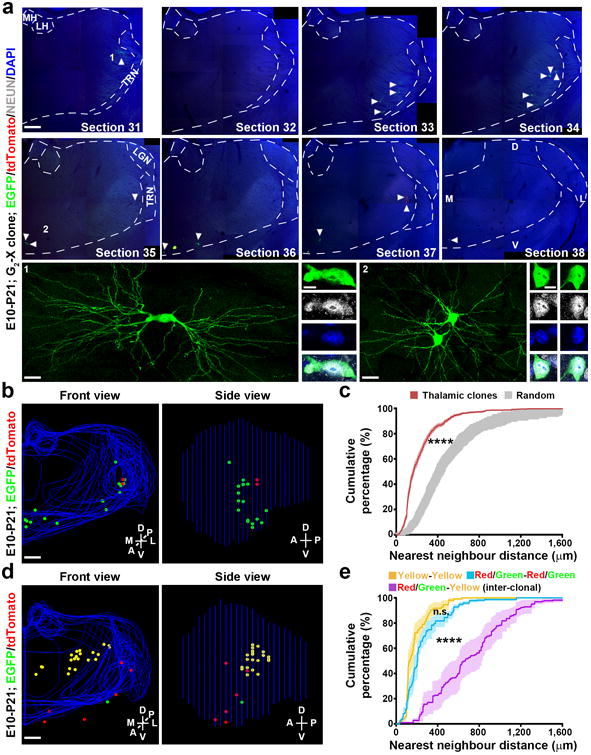
(a) Confocal images of a green/red G2-X clone in a P21 brain treated with TM at E10. Consecutive sections were stained for EGFP (green) and tdTomato (red), and with DAPI (blue). Broken lines indicate the contours of the thalamus and landmark nuclei including the medial and lateral habenula (MH, LH), lateral geniculate nucleus (LGN), and the thalamic reticular nucleus (TRN). Arrowheads indicate the labeled neurons. High magnification images of representative labeled neurons positive for neuronal marker NEUN (white) (areas 1 and 2) are shown at the bottom. Scale bars: 500 μm, 20 μm, 10μm, 20 μm, and 10μm. (b) 3D reconstructed images of the thalamic hemisphere containing the clone shown in a (Front view, left; Side view, right). Blue lines indicate the contours of the thalamus and landmark nuclei and colored dots represent the cell bodies of labeled neurons. M, medial; L, lateral; D, dorsal; V, ventral; A, anterior; P, posterior. Similar display is used in subsequent 3D reconstructed images. Scale bar: 500 μm. (c) Nearest neighbor distance (NND) analysis of MADM-labeled neuronal clones (n=77) in the P21-24 thalamus. Data are presented as mean±s.e.m. ****, p=1e-15 (unpaired t-test with Welch's correction). (d) 3D reconstructed images of a thalamic hemisphere containing both a green/red G2-X clone and a yellow clone. Scale bar: 500 μm. (e) NND analysis of MADM-labeled green/red and yellow neuronal clones in the same thalamic hemisphere. Note that the cumulative frequency of NND of green/red (blue, n=11) or yellow (yellow, n=11) fluorescent neurons (i.e. intra-clonal) is similar and significantly left-shifted than that of green/red and yellow (magenta, n=11) fluorescent neurons (i.e. inter-clonal). Data are presented as mean±s.e.m. ****, p=1e-15 (unpaired t-test with Welch's correction).
To quantitatively assess the spatial distribution and clonal relationship of labeled neurons, we applied nearest neighbor distance (NND) analysis 24, 31 to all the reconstructed P21-24 datasets labeled at E9-E12 (n=77 in total from over 45 mice). Compared with spatially random simulated datasets (gray), the experimental datasets of thalamic clones (red) exhibited similar and much shorter (i.e. left-shifted) NNDs (Fig. 3c and Supplementary Fig. 3d). Moreover, in the datasets containing both green/red and yellow fluorescent clones, the green/red fluorescent neurons (blue) or the yellow fluorescent neurons (yellow) (i.e. intra-clonal NNDs) exhibited similar and significantly shorter NNDs than the green/red and yellow fluorescent neurons (magenta, i.e. inter-clonal NNDs) (Fig. 3d,e). Together, these results demonstrate that clonally related neurons originating from sparsely labeled progenitors do not randomly mix, but form spatially segregated clusters in the thalamus. Thereby, each neuronal cluster represents a clone that arises from a single dividing thalamic progenitor.
We also observed NEUN-negative cells with a small soma/nucleus and short neurite processes in nearly half (34 out of 77) of the labeled clones (Fig. 4a-d). Additional immunohistochemsitry analyses revealed that cells with a small soma/nucleus and short bushy neurites typically expressed S100, an astrocyte marker (Supplementary Fig. 4c,d and 4g), and that cells with a small soma/nucleus and short parallel neurites characteristically expressed OLIG2, an oligodendrocyte marker (Supplementary Fig. 4e-g). These results suggest that NEUN-negative cells are glial cells. The labeled glia were typically located in close proximity to the labeled neurons (Fig. 4c); however, there was no obvious correlation between the numbers of glia and neurons at the clonal level (Fig. 4e). Clonally related glia also exhibited a clear clustering feature in spatial distribution (Fig. 4f). The fraction of glia-containing clones exhibited a progressive decrease between E9 and E11 (Supplementary Fig. 3e), likely reflecting the progressive decrease in the RGP number in individual clones as a result of the temporal switch of symmetric proliferative to asymmetric neurogenic division by RGPs (Fig. 2d).
Figure 4. Spatial clustering of clonally related glial cells in the thalamus at the mature stage.
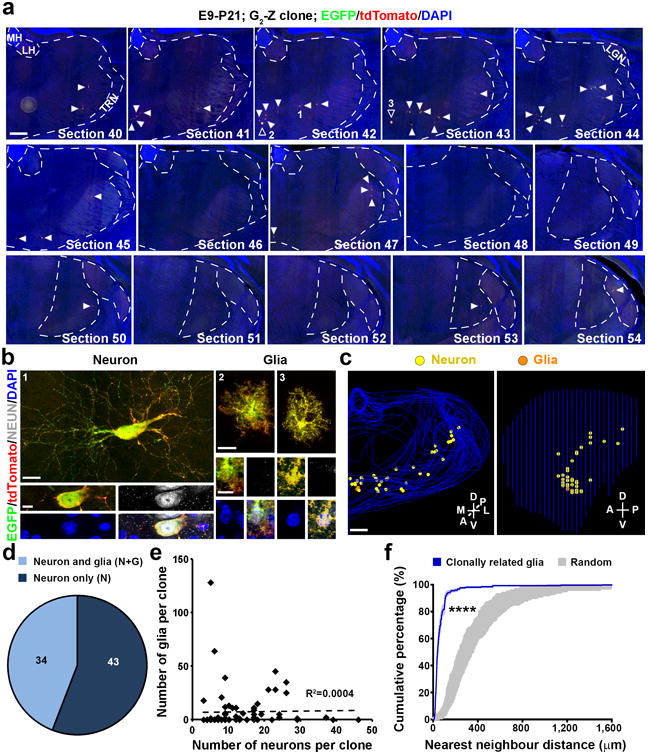
(a, b) Scanned images of a yellow G2-Z clone in a P21 brain treated with TM at E9. Consecutive sections were stained for EGFP (green) and tdTomato (red), and with DAPI (blue). Open arrowheads indicated the labeled NEUN-negative cells and arrowheads indicate the labeled neurons. High magnification images of the representative labeled neuron positive for NEUN (white) (area 1) and glial cells (areas 2 and 3) negative for NEUN are shown in b. Scale bars: 500 μm, 20 μm, 10μm, 20 μm, and 10 μm. (c) 3D reconstructed images of the thalamic hemisphere containing the clone shown in a (Front view, left; Side view, right). Scale bar: 500 μm. (d) Quantification of the fraction of P21-24 clones labeled at E9-12 containing neuron only (N) or neuron and glia (N+G). (e) No correlation between the number of neurons and the number of glia at the clonal level (n=77 clones). The broken line indicates the linear regression of the data. (f) NND analysis of MADM-labeled glial clones (n=25) in the P21-24 thalamus. Note that compared to random datasets simulated 100 times (gray), the cumulative frequency of NND of thalamic glial clones (blue) is significantly left-shifted towards shorter distances, indicating a spatial clustering. Data are presented as mean±s.e.m. ****, p=1e-15 (unpaired t-test with Welch's correction).
Progenitor Origin of Thalamic Nuclei
To reveal the specific nuclear occupation of neurons in individual clones, we systematically aligned individual experimental 3D datasets to the reference thalamus with a defined nuclear boundary based on the Allen Brain Atlas (ABA) by adopting a previously established method (Supplementary Fig. 5)6. Notably, the labeled clones as a whole covered a vast majority of thalamic nuclei representing over 98% of the total volume. Moreover, consistent with a regional restriction in spatial distribution, individual clones generally occupied a few nuclei (∼4.2±0.3 out of a total of 34 nuclei).
The regional occupation of individual clones raised an intriguing possibility that different nuclei may have distinct progenitor origins. To test this, we performed an unsupervised agglomerative hierarchical clustering analysis of all neurons in individual RGP clones labeled at E9-E12 (n=77 clones) on the basis of their distribution in different nuclei (Fig. 5a). Remarkably, we observed two major clonal clusters corresponding to excitatory neuron clones and inhibitory interneuron clones, respectively (left dendrogram, Fig. 5a). Clones occupying the TRN and zonaincerta (ZI) were exclusively GABAergic interneurons (Supplementary Fig. 6a,b) and segregated from the clones located in all other thalamic nuclei except the LGN (Fig. 5a), the only other rodent thalamic nucleus besides the TRN that contains inhibitory interneurons 13, suggesting a clear lineage separation of excitatory and inhibitory neurons in the thalamus at the clonal level. This lineage separation is consistent with the previous genetic mapping studies 12, 13 and strongly supports the reliability of our clonal labeling and clustering analysis. The remaining excitatory neuron clones fell into three main subclusters (Supplementary Fig. 7). They were located predominantly in the anterior (a), medial dorsal (md), or medial ventral posterior (mvp) regions, respectively. We thus termed them the ‘a’, ‘md’, and ‘mvp’ clonal clusters.
Figure 5. Nuclear localization of thalamic clones.
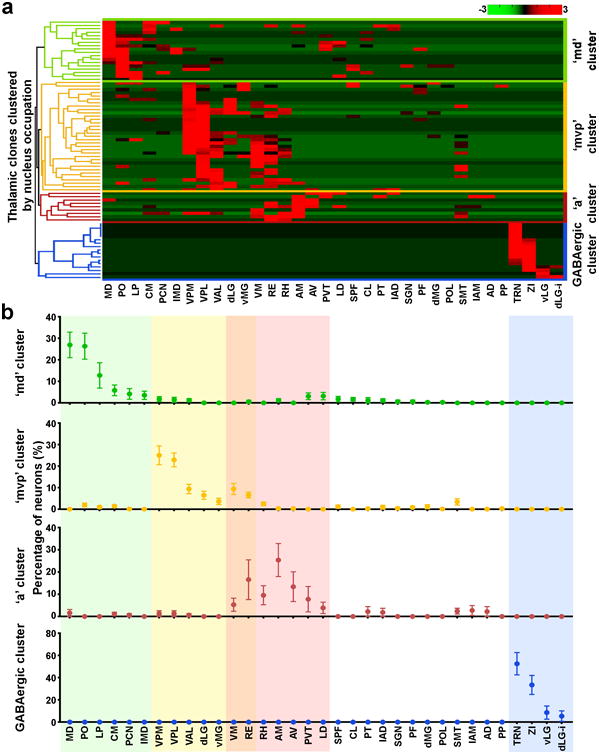
(a) The heat map of the distribution of individual clones labeled at E9-E12 in different thalamic nuclei based on an unsupervised agglomerative hierarchical clustering analysis. The dendrogram is shown to the left. Each row represents an RGP-derived thalamic clone (n=77 in total). dLG-i, interneurons in dLG. Note that there are two major clonal clusters corresponding to excitatory neuron clones and inhibitory interneuron clones (blue, n=17), respectively. Within the excitatory neuron clones, there are three main subclusters largely corresponding to the ‘md’ (green, n=33), ‘mvp’ (yellow, n=9), and ‘a’ (red, n=18), clones. (b) Quantification of the distribution of the four clonal clusters in different nuclei. Color shades highlight the top 6-7 dominant nuclei harboring over 80% of labeled neurons in the respective clonal clusters. Data are presented as mean±s.e.m. AD, anterodorsal; AM, anteromedial; AV, anteroventral; CL, central lateral; CM, central medial; IAD, interanterodorsal; IAM, interanteromedial; IMD, intermediodorsal; LD, lateral dorsal; dLG, dorsal lateral geniculate; vLG, ventral lateral geniculate; LP, lateral posterior; MD, mediodorsal; MG, medial genicualte; PCN, paracentral; PF, parafascicular; PO, posterior; POL, posterior limiting; PP, peripenduncular; PT, paratenial; PVT, paraventricular; RE, nucleus of reunions; RH, rhomboid; SGN, suprageniculate; SMT, submedial; SPF, subparafascicular; VAL, ventral anterior lateral; VM, ventral medial; VPL, ventral posterolateral; VPM, ventral posteromedial.
We then quantitatively analyzed the fraction of individual neuronal clones in these four clonal clusters located in different thalamic nuclei (Fig. 5b). As expected, the GABAergic neuron clonal cluster was exclusively located in the TRN and LGN, including both the ventral (vLG) and dorsal (dLG) regions, as well as the ZI (blue, Fig. 5b). The three excitatory neuron clonal clusters occupied largely non-overlapping nuclei, as reflected by the top 6-7 dominant nuclei that harbored over 80% of neurons in each clonal cluster (Fig. 5b). Specifically, the ‘md’ clonal cluster largely occupied the mediodorsal (MD), posterior (PO), lateral posterior (LP), central medial (CM), paracentral (PCN), and intermediodorsal (IMD) nuclei (green). The ‘mvp’ clonal cluster mostly occupied the ventral posteromedial (VPM), ventral posterolateral (VPL), ventral anterior lateral (VAL), dLG, ventral medial geniculate (vMG), ventral medial (VM), reunions (RE) nuclei (yellow). The ‘a’ clonal cluster predominantly occupied the anteromedial (AM), RE, anteroventral (AV), rhomboid (RH), paraventricular (PVT), VM, and lateral dorsal (LD) nuclei (red). These results suggest that individual progenitors produce neurons preferentially located to distinct sets of nuclei. Notably, the nuclei occupied by the ‘mvp’ clonal cluster largely overlap with the nuclei harboring the excitatory neuron progeny of OLIG2-positive progenitors in the pTH-C subdomain (Supplementary Fig. 8), indicating a lineal correlation between them. The four sets of a total of 20 dominant nuclei (not including ZI) account for >80% of the entire thalamic volume. Moreover, clonally labeled neurons occupied all 34 nuclei, including those non-dominant nuclei that are generally small (Fig. 5). These results suggest that our clonal labeling and clustering analysis effectively cover the vast majority, if not all, of the thalamus and that the small non-dominant nuclei are likely marginally formed by the three excitatory neuron clonal clusters.
Ontogenetic Segregation of Cognitive and Sensory/Motor-related Nuclei
Among the major nuclei occupied by excitatory neuron clones, VM and RE were partially occupied by both the ‘mvp’ and ‘a’ clonal clusters, raising the question as to whether the labeled neurons between these two clonal clusters in these two overlapping nuclei were the same or different populations. We found that neurons labeled in the ‘a’ clonal cluster were predominantly located in the anterior half of VM and RE, as well as RH, whereas neurons labeled in the ‘mvp’ clonal cluster were mostly located in the posterior half of these three nuclei (Supplementary Fig. 9a), suggesting a lineage-based segregation of the anterior and posterior parts of VM, RE, and RH.
Interestingly, our in-depth analysis of the previous thalamocortical axonal tracing data 6 showed that the anterior parts of VM, RE, and RH (aVM, aRE and aRH) predominantly projected to cognition-related HO areas of the cortex such as the anterior cingulated cortex (ACC) and prelimbic cortex (PrL) (asterisks, Supplementary Fig. 9b). In contrast, the posterior parts of VM, RE, and RH (pVM, pRE, pRH) mostly projected to the sensory and motor areas of the cortex, consistent with the notion that they are involved in multimodal sensory processing and motor activity 32, 33. Moreover, the remaining cells in the respective clones (n=18) were located in regions sharing a similar cognition-related HO versus sensory/motor-related cortical projection pattern (Supplementary Fig. 9b), indicating a clonal but not nuclear level of correlation in functionality. Together, these results suggest that lineage relationship influences the organization of thalamic structures associated with distinct functionality, even at a resolution beyond the conventional individual nucleus level.
To further test this, we systematically analyzed the properties of individual clones in the ‘mvp’ and ‘a’ clonal clusters with regard to their spatial configuration and nuclear occupation. We found that clones in the ‘mvp’ cluster were prominently arrayed radially along the mediolateral axis with a dorsoposterior bend (Fig. 6a left and 6b, and Supplementary Movie 1). In contrast, clones in the ‘a’ cluster were relatively more tangentially dispersed (Fig. 6a right and 6b, and Supplementary Movie 2). This distinction in clonal configuration appeared to be an intrinsic feature of the respective lineages, as we observed similar differences in clonal organization at the embryonic stage (Fig. 6c,d).
Figure 6. Distinct spatial configuration and functionality of the clonal clusters.
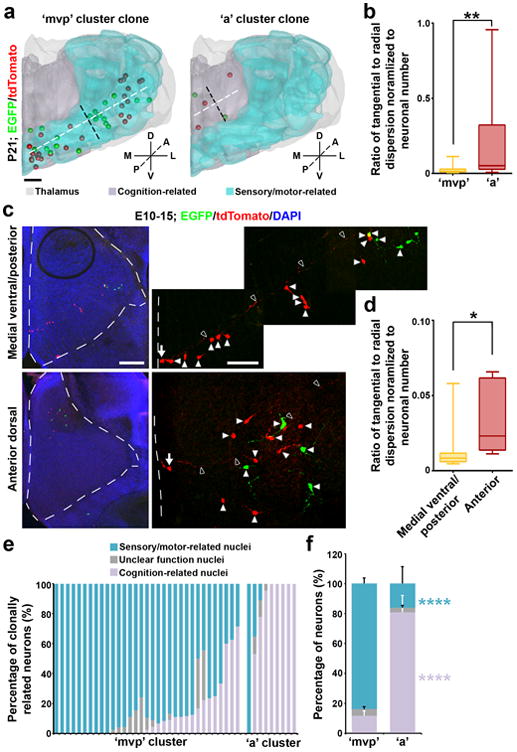
(a) 3D rendered images of a representative ‘mvp’ (left) or ‘a’ (right) cluster clone that predominantly occupies sensory/motor-related nuclei and cognition-related higher order nuclei, respectively. Broken lines indicate the radial (white) versus tangential (black) dispersion of the clone. Scale bar: 500 μm. (b) Quantification of the average ratio of the radial versus tangential dispersion normalized to the total number of neurons in individual clones at P21-24 (‘mvp’, n=33; ‘a’, n=9). Data are presented as median with interquartile range, and whiskers are the minimum and maximum. **, p=0.008 (Mann-Whitney test). (c) Confocal images of a representative E15 medial ventral/posterior (top) or anterior dorsal (bottom) clone labeled at E10. Broken lines indicate the contours of the thalamus. High magnification images of the clone are shown to the right. Arrows indicate the radial glial progenitors with a long radial glial fiber (open arrowheads) and arrowheads indicate the progeny. Scale bars: 200 μm and 50 μm. (d) Quantification of the average ratio of the radial versus tangential dispersion normalized to the total number of neurons in individual clones at E15-16 (‘mvp’, n=20; ‘a’, n=5). Data are presented as median with interquartile range, and whiskers are the minimum and maximum. *, p=0.012 (Mann-Whitney test). (e) Quantification of the percentage of individual ‘mvp’ and ‘a’ cluster clones located in sensory/motor-related, cognition-related, or unclear function nuclei. Each bar represents a clone. (f) Quantification of the percentage of all ‘mvp’ or ‘a’ cluster clones located in sensory/motor-related, cognition-related, or unclear function nuclei (‘mvp’: sensory/motor-related, 83.7±3.6%; cognition-related, 11.9±3.3%; unclear function, 4.4±1.6%; ‘a’: sensory/motor-related, 16.3±11.1.0%; cognition-related, 80.7±11.4%; unclear function, 3.0±1.7%). Data are presented as mean±s.e.m. ****, p=5e-05; ****, p=2.5e-05 (Mann-Whitney test).
There was also a tight association between the ‘mvp’ and ‘a’ clonal clusters and distinct functionalities. The ‘a’ clonal cluster mostly occupied the nuclei associated with cognition-related HO functions (e.g. AM, AV, aRE/aRH/aVM, IAM) (Fig. 6a, e, f right), whereas the ‘mvp’ clonal cluster predominantly occupied the nuclei associated with sensory/motor-related activities (e.g. VPL, VPM, VAL, dLG, vMG, pRE/pRH/pVM) (Fig. 6a,e,f left). Together, these results suggest that HO cognitive nuclei and sensory/motor-related nuclei are ontogenetically segregated with distinct progenitor origins. These results also further indicate that lineage relationship not only affects spatial configuration of thalamic neurons, but also contributes to a global nuclear organization of the thalamus related to functionality.
Lineage Distinction between FO and HO Sensory/Motor-related Nuclei
Clones in the ‘md’ clonal cluster appeared to be associated with either sensory/motor (e.g. PO and LP) or cognition-related (e.g. MD, CM, PCN, and IMD) activities (Fig. 7a,b). Interestingly, the sensory/motor activities mediated by the ‘md’ clonal cluster appeared to be fundamentally different from those mediated by the ‘mvp’ clonal cluster (Fig. 7c,d). The sensory/motor-related thalamic nuclei are well organized in a modality and hierarchy-based manner 1, 8. Notably, the clones occupying the sensory/motor-related FO and HO nuclei were largely segregated (Fig. 7c,d, and Supplementary Movie 3 and 4). The ‘mvp’ clonal cluster predominantly contributed to the FO sensory/motor-related nuclei such as VPL, VPM, VAL, dLG, vMG as well as a small fraction in pRE/pRH/SMT, whereas the ‘md’ clonal cluster mainly contributed to the HO sensory/motor-related nuclei such as PO and LP (Fig. 7d,f). These results suggest a strong lineage-related separation of the sensory/motor-related nuclei in hierarchy (i.e. order). On the other hand, individual clones were frequently found to occupy the FO or HO nuclei across different modalities (Fig. 7e,f). Together, these results support a progenitor origin of a cross-modal hierarchical framework of sensory/motor pathways in the thalamus.
Figure 7. Clonal segregation of the FO and HO thalamic nuclei.
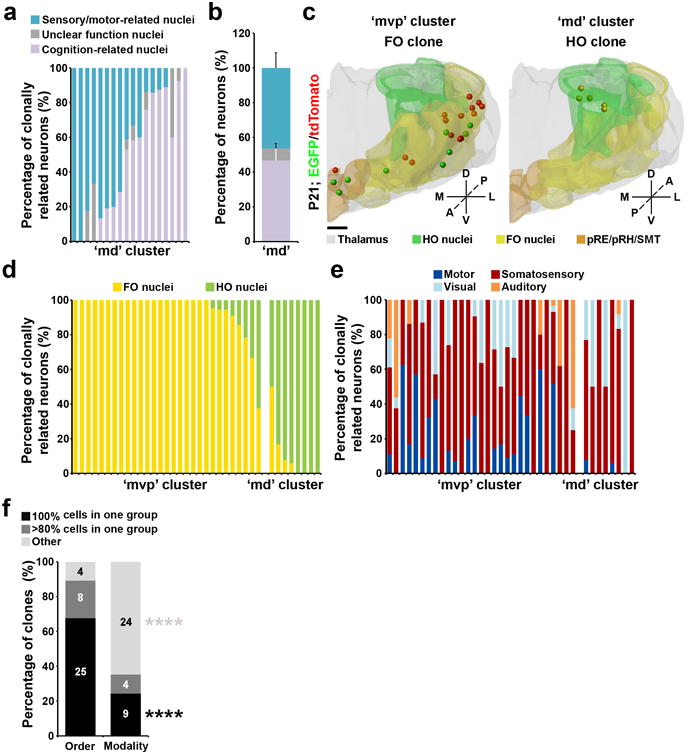
(a) Quantification of the percentage of individual ‘md’ cluster clones located in sensory/motor-related, cognition-related, or unclear function nuclei. Each bar represents a clone (n=18 in total). (b) Quantification of the percentage of all ‘md’ cluster clones located in sensory/motor-related, cognition-related, or unclear function nuclei (sensory/motor-related, 46.3±8.7%; cognition-related, 46.8±8.7%; unclear function, 6.8±2.8%). Data are presented as mean±s.e.m. (c) 3D rendered images of a representative ‘mvp’ (left) or ‘md’ (right) cluster clone that predominantly occupies the FO and HO nuclei, respectively. Scale bar: 500 μm.(d) Quantification of the percentage of clonally related neurons in sensory/motor-related nuclei that are in the FO or HO nuclei. Each bar represents a clone. (e) Quantification of the percentage of clonally related neurons in sensory/motor-related nuclei related to different modalities. (f) Quantification of the percentage of all clonally related neurons in sensory/motor-related nuclei according to the order (FO vs. HO) or modality (i.e. somatosensory, visual, auditory, or motor). ****, p=00019; ****, p=2e-06 (chi-square test).
Shh Signaling Influences Spatial Organization of Thalamic Clones
Previous studies suggested that Sonic hedgehog (Shh) signaling regulates thalamic progenitor identity and nuclear specification 28, 34. To explore the influence of Shh signaling on the development and organization of thalamic clones, we examined the size and spatial distribution of clones with a higher level of Shh signaling than normal. To achieve this, we performed MADM analysis in the R26SmoM2-EYFP/+(SmoM2) mice 35. As the R26SmoM2-EYFP and MADM-11 transgenes are located in different chromosomes, it prevents a genetic link between EGFP or tdTomato expression and SmoM2-EYFP expression for a strict mosaic analysis. Nonetheless, in the Nestin-CreERT2/MADM11/R26SmoM2-EYFP/+ animals, a single dose of TM treatment would trigger both MADM labeling upon inter-chromosomal recombination as well as the expression of SmoM2 upon intra-chromosomal recombination, as confirmed by the expression of SmoM2-EYFP in tdTomato-labeled RGPs (Supplementary Fig. 10, arrows).
Within the diencephalon, Shh is expressed in the basal plate and zonalimitans intrathalamica (ZLI) that are ventral to the developing thalamus 28, 36. As a result, the embryonic thalamus is exposed to a graded Shh activity. Coinciding with this Shh signaling gradient, clones in the anterior and medial dorsal regions were more tangentially distributed than clones in the medial ventral/posterior region (Fig. 6a-d), indicating that a high Shh signaling suppresses tangential distribution of clonally related neurons. Consistent with this notion, clones located in the anterior region of the SmoM2 thalamus became less tangentially and more radially distributed than similarly located wildtype control clones (Fig. 8a,b), whereas clones located in the medial ventral/posterior region did not exhibit any significant change in spatial distribution (Supplementary Fig. 11a,b). In addition, we observed an expansion of the expression of retinoic acid receptor-related orphan receptor α (RORα), a marker gene predominantly expressed in VP and dLG37, to the anterior region as well as dorsally (Supplementary Fig. 11d), indicating a change in nuclear boundary and functional organization of the thalamus, as previously shown 28. We did not observe any significant changes in the average neuronal number in individual clones between the wildtype and SmoM2 thalami (Supplementary Fig. 11c). However, while the average glial number in individual clones was not significantly different between the wildtype and SmoM2 thalami (Supplementary Fig. 12a), there was a substantial increase in the fraction of clones with glia in the SmoM2 thalamus (Supplementary Fig. 12b), indicating that Shh signaling promotes gliogenesis in the thalamus. The overall distribution of clonally related neurons and glia remained spatially clustered in the SmoM2 thalamus (Supplementary Fig. 11e and 12c). Together, these results suggest that Shh signaling influences the spatial organization of thalamic neurons at the clonal level, which likely contributes to distinct thalamic nuclear configuration and organization.
Figure 8. Shh signaling regulates the spatial distribution of thalamic clones depending on the localization.
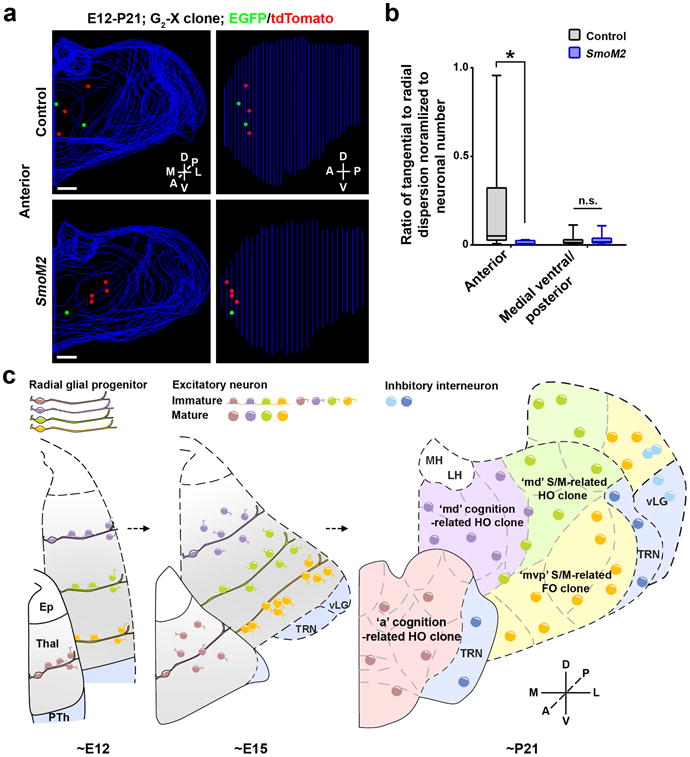
(a) Representative 3D reconstructed images of the thalamic hemispheres containing a MADM-labeled clone in the anterior region of the wildtype control (top) and SmoM2 (bottom) mice. Scale bars: 500 μmand 500 μm. (b) Quantification of the average ratio of the radial versus tangential dispersion normalized to the total number of neurons in individual P21-24 clones in the anterior and medial ventral/posterior region of the control and SmoM2 mouse (anterior: control, n=9 clones; SmoM2, n=4 clones; p=0.02; medial ventral/posterior: control, n=33 clones; SmoM2, n=20 clones; n.s., not significant, p=0.2; Mann-Whitney test). Data are presented as median with interquartile range, and whiskers are the minimum and maximum. (c) A diagram illustrating the ontogenetic origin and organization of thalamic nuclei and function. S/M, sensory/motor.
Discussion
Our findings reveal a previously unknown ontogenetic logic for structural development and functional organization of the mammalian thalamus (Fig. 8c and Supplementary Fig. 12d). RGPs in the VZ of the developing thalamus actively divide to produce a cohort of neuronal progeny. At the early embryonic stage, individual RGPs and their progeny are similarly organized radially along a mediolateral axis. As development proceeds, while clones located in the medial ventral posterior region remain tightly arrayed radially along the radial glial fiber with the early born cells progressively moving outward (i.e. laterally) and dorsoposteriorly, contributing mostly to the FO sensory/motor-related nuclei, clones located in the anterior and medial dorsal regions become more tangentially dispersed, occupying predominantly the HO cognitive and HO sensory/motor-related nuclei.
Diverse migratory patterns of lineages have been suggested in the embryonic chick diencephalon 21, 22. It has also been suggested that migrating cells in the developing rat thalamus follow both radial and non-radial glia pathways 38. However, it is unclear how different migratory patterns may be related to distinct nuclear formation and functionality in the thalamus. In this study, we carried out a systematic clonal analysis of mouse thalamic progenitor cells using MADM, which provides fine resolution in progenitor behavior and lineage progression in a temporal and spatial specific manner. MADM allows the progeny to be labeled throughout the development starting from the TM treatment. Our focus of E9-E12 coincides with the beginning phase of thalamic neurogenesis, thereby providing a comprehensive grasp of neuronal production and migration, and nuclear formation in the thalamus.
Interestingly, our unsupervised agglomerative hierarchical clustering analysis of the nuclear occupation of neurons in individual clones at the relatively mature stage (i.e. P21-24) not only reliably distinguished excitatory and inhibitory neuronal clones, but also identified three main excitatory neuron clonal subclusters that occupy largely non-overlapping sets of thalamic nuclei. These results suggest that the spatial configuration and nuclear occupation are robust and reliable features of thalamic progenitors and their neuronal progeny (i.e. lineages). Our finding of co-distribution of clonal clusters to certain thalamic nuclei at the single progenitor level is consistent with the previous population level observation 12, 28. Notably, additional subclusters may be revealed by further dissecting the hierarchical dendrogram. For example, in the ‘md’ clonal cluster, there appears to have two subclusters largely corresponding to the clones preferentially occupying the HO cognitive nuclei (e.g. MD, PVT, CM) and HO sensory/motor-related nuclei (e.g. PO, LP, dMG), respectively. On the other hand, the lineage-related organization is not absolute but preferential, as reflected in our quantitative analyses. While individual clonal clusters exhibit strong propensity in occupying distinct nuclei, clones in the same cluster show some variability.
The differences in clonal behavior and neuronal organization are likely related to morphogen gradients and transcriptional regulations that affect thalamic progenitor specification and neuronal differentiation. Consistent with this, distinct spatial configurations of clones can be observed at the embryonic stage. Previous studies have shown that Shh, Fgf, and Wnt act together to orchestrate the development and regionalization of the thalamus 9, 10, 28, 39-42. We found that neuronal clones in the medial ventral/posterior region with high levels of Shh activity are more radially organized, whereas neuronal clones in the anterior and mediodorsal regions with low levels of Shh activity are more tangentially organized. Moreover, an enhanced Shh activity by SmoM2 expression suppresses tangential and promotes radial organization of clones in the anterior and dorsal regions, suggesting that Shh signaling influences the spatial organization of thalamic clones. Therefore, the precise localization of progenitors and the corresponding signaling environment likely regulate lineage progression and clone organization.
Besides the spatial regulation by a combination of signaling pathways, there are also important temporal regulations. Individual RGPs are capable of producing a cohort of neuronal progeny in a sequential manner. The temporal dynamics of RGP and IP divisions of the corresponding clones likely dictate the size and configuration of thalamic nuclei in which the clones reside, in conjunction with neuronal migration. Notably, besides the relative radial versus tangential dispersion difference, the average neuronal number of the ‘mvp’ clonal cluster occupying the FO sensory/motor-related nuclei appears to be higher than that of the ‘a’ clonal cluster occupying the HO cognitive nuclei, which may contribute to the overall structural differences between these two general groups of nuclei. Therefore, while sharing close lineage relationship, neurons in the same clone likely also exhibit temporal differences in their maturation and properties, which may contribute to neuronal diversity and individual nuclear distinction in the thalamus.
The clonal relationship is not only coupled to structural formation, but also related to functional organization of the thalamus. The anterior and medial dorsal clones principally contribute to the nuclei of cognitive and sensory/motor-related HO functions, whereas the medial ventral posterior clones mostly contribute to the nuclei of sensory/motor-related FO functions. The thalamus is commonly demarcated into a large number of distinct nuclei with defined functions 1. However, little is known about the general principles underlying the complex structural and functional organization of thalamic nuclei. Moreover, accumulating evidence suggests that, although useful, these nuclear divisions may not always be the relevant functional unit 3. Our data showed that cognition-related HO nuclei in the anterior region share a similar progenitor origin, and sensory/motor-related FO nuclei in the medial ventral posterior region share a similar progenitor origin. In addition, sensory/motor-related HO nuclei share a close but largely distinguishable lineage relationship with cognition-related HO nuclei in the medial dorsal region. These results suggest that lineage relationship influences functional organization of the thalamus.
This lineage-related functional organization can manifest at the resolution beyond the conventional individual nuclear boundaries. For example, we found that neurons in the VM, RE, and RH exhibit two distinct progenitor/clonal origins. Neurons located in the anterior portion of these nuclei are largely derived from the ‘a’ clonal cluster, whereas neurons located in the posterior portion are predominantly originated from the ‘mvp’ clonal cluster. Interestingly, the anterior and posterior regions of VM, RE, and RH exhibit distinct thalamocortical axonal projection patterns. While the anterior regions predominantly innervate cognitive cortical areas such as the ACC and PLC, the posterior regions largely project to sensory/motor-related cortical areas. Taken together, these results demonstrate that clonal relationship predicts functionality of neurons both across different nuclei and within the same nucleus.
We also observed a clonal segregation between the FO and HO sensory/motor-related nuclei across different modalities. Notably, previous studies have shown that thalamic nuclei of the same order across different modalities are composed of neurons with extremely similar axonal arborization architectures 30. Consistent with the anatomical connectivity difference, the FO and HO nuclei also exhibit distinct properties in information processing 4, 43, 44. The distinct progenitor origin between neurons in the FO and HO nuclei likely contribute to their different morphologies, anatomical connections, and functional properties. In addition, we observed a lineal segregation of GABAergic interneurons in the TRN and LGN, which may have critical functional implications.
Limited intra-thalamic connectivity has been found to structurally or functionally organize the complex thalamic nuclei 45-47. Our findings suggest that lineage-related development and organization provides a fundamental blueprint for the assembly and function of the thalamus. Consistent with this, it has previously been shown that the overall topography of thalamocortical axonal projections appears to be normal in the absence of any evoked synaptic transmission 48. Moreover, our observation of a strong lineage segregation of the sensory/motor-related FO and HO nuclei across different modalities points to a model of the development of thalamic sensory/motor pathways in distinct phases. The developmental origin promotes the establishment of a generic hierarchical framework which is further specified into different modalities, likely dependent on distinctive inputs from the periphery and other brain regions. This is consistent with a recent study suggesting a cross-modal genetic framework for the development of sensory pathways in the thalamus 49. Additionally, changes in the periphery sensory input can also lead to changes in the functional subdivisions and the relevant corticofugal connectivity of different thalamic nuclei 50, indicating a degree of rewiring and plasticity of thalamic organization.
Methods
Animals and in utero intraventricular injection
MADM-11GT (JAX Stock No. 013749) and MADM-11TG (JAX Stock No. 013751) mice were produced as previously described 27. MADM11, Nestin-CreERT2 26, R26-SmoM2-EYFP35, and Olig3-Cre28 mice were kindly provided by Drs. Hippenmeyer, Kageyama, Joyner, and Nakagawa, respectively. Mice were bred and maintained according to guidelines established by the Institutional Animal Care and Use Committee of Memorial Sloan Kettering Cancer Center. For MADM labeling, Nestin-CreERT2+/-/MADM-11TG/TG mice were crossed with MADM-11GT/GT mice and the time of pregnancy was determined by the presence of the vaginal plug (E0). For clone induction, pregnant females were injected intraperitoneally with tamoxifen (T5648, Sigma) dissolved in corn oil (C8267, Sigma) at E9, E10, E11 and E12 at a dose of 5-25 μg/g body weight. In utero intraventricular injection of retrovirus was performed as previously described 15. In brief, uterine horns of pregnant mice were exposed in a clean environment. Cre-recombinase-dependent retrovirus expressing EGFP (∼1.0 μL) mixed with fast green (2.5 mg/mL, Sigma) was injected into the third ventricle through a beveled, calibrated glass micropipette (Drummond Scientific). After injection, the uterus was placed back in the abdominal cavity and the wound was surgically sutured. After surgery, the animal was placed in a recovery incubator under close monitoring until it fully recovered.
Serial Sectioning, immunohistochemistry and 3D Reconstruction
Both male and female mice were perfused intracardially with 4% paraformaldehyde (PFA) in phosphate-buffered saline (PBS, pH 7.4). Brains were removed and post-fixed overnight at 4°C. Serial coronal sections of individual brains were prepared using a vibratome or cryostat (Leica Microsystems) and subjected to immunohistochemistry. The following primary antibodies were used: chicken anti-GFP (1:1,000 dilution, GFP-1020, Aves Lab) 51, rabbit anti-RFP (1:1,000 dilution, 600-401-379, Rockland) 51, guinea pig anti-RFP (1:20,000 dilution, gift from J. Nicholas Betley, University of Pennsylvania), mouse anti-OLIG3 (1:200 dilution, MAB2456, R&D Systems) 12, rabbit anti-BLBP (1:200 dilution, ab32423, Abcam) 52, mouse anti-Ki-67 (1:500 dilution, 556003, BD Pharmingen) 52 mouse anti-TUJ1 (1:500 dilution, MMS-435P, Covance) 52, mouse anti-NESTIN (1:500 dilution, rat-401, Developmental Studies Hybridoma Bank) 52, mouse anti-NEUN (1:500 dilution, MAB377, Millipore) 53, rabbit anti-S100 (1:500 dilution, Z0311, Dako) 54, rabbit anti-OLIG2 (1:500 dilution, AB9610, Millipore)55, and mouse anti-PARVALBUMIN (1:500 dilution, MAB1572, Millipore) 52. Sections were mounted on glass slides, imaged using confocal microscopy (FV1000, Olympus or LSM700, Zeiss) and slide scanner (NanoZoomer 2.0-HT, Hamamatsu Photonics),and reconstructed using Neurolucida and StereoInvestigator (MBF Bioscience). Neurons, astrocytes, and oligodendrocytes were distinguished based on their morphology (e.g. nuclear size and neurite length) and marker expression.
For 3D reconstruction, each section was analyzed sequentially in anterior to posterior order. The boundaries of the entire thalamus as well as the landmark nuclei including MH, LH, LGN, TRN, and parts of MGN and VM were traced in each section and aligned. Individually labeled neurons were represented as colored dots (∼4 times the size of the cell body at P21-24 and ∼2 times at E12), respectively. The anterior region of the thalamus was defined as the part before the appearance of the major sensory/motor nuclei (i.e. ABA sections 56-61) and the posterior region as the part following the separation of the thalamic hemispheres (i.e. ABA sections 78-92). The remaining part was defined as the medial region (i.e. ABA sections 62-77). The distribution of the nearest neighbor distance (NND) reflects the spatial point pattern of the dataset, as previously described 31. Specifically, given N cells in a dataset, for each cell i the distance to its closest neighbor was measured and denoted as di, the NND for cell i. The indicator function f (y, d) was then calculated as:
Thus, the cumulative distribution function (CDF) of NND is:
Thalamic Alignment, nuclear registration, and clonal clustering
After 3D reconstruction, all serial sections of individual experimental thalami were aligned based on the midline. The cutting angle tiltation was corrected using a previously established method based on anatomical landmarks 6. All corrected sections were then aligned based on the dorsal boundary of the thalamus. To register known nuclei within our experimental thalami, the nuclei from the ABA were traced and linearly scaled to best fit the average experimental thalamus. The correspondence between individual experimental thalami and the ABA reference thalamus were further assessed by manual comparisons of the landmark structures (MH, LH, LGN, and TRN), and adjusted. The nuclear identities and corresponding numbers of labeled neurons of individual clones were then systematically inferred and manually confirmed. For clonal clustering, the fractions of individual clones located in different nuclei were calculated, standardized, and subjected to an unsupervised agglomerative hierarchical clustering analysis (Clustergram: Distance metric, Euclidean; Linkage, Average; MATLAB, MathWorks). The number of excitatory neuron clonal subclusters was validated by the Silhouette coefficient analysis (MATLAB, MathWorks). The criterion values (i.e. coefficients) in the Silhouette analysis peaked at three clusters within the range of number of clusters tested. For volume overlap estimation, the posterior domain of the thalamus (i.e. ABA sections 85-92) containing very few labeled neurons was not included. The volume of individual experimental thalami (Vexperimental) was compared to the volume of the scaled ABA reference thalamus (Vreference), and the volume overlap was estimated as
where ‘∩’ is the logical AND operator (i.e. overlap).
Based on the literature 1, the following nuclei were considered as sensory/motor-related: SPF, VAL, VPL, VPM, dLG, vMG, PO, LP, dMG, PF, pRE, pRH, and pVM; and the following nuclei as cognition-related higher order: AD, AM, AV, CM, IAD, IAM, IMD, MD, LD, PT, PVT, aRE, aRH, and aVM. For quantification of the order versus modality segregation, only clones with more than half of the neurons located in the well-characterized sensory/motor-related nuclei (i.e. VPM, VPL, PO, dLG, LP, dMG, vMG, VAL, and pVM) were included in the analysis. Specifically, VAL and pVM are motor-related nuclei; VPL, VPM, and PO are somatosensory-related nuclei; dLG and LP are visual related nuclei; vMG and dMG are auditory-related nuclei. Among them, VPL, VPM, dLG, vMG, VAL, and pVM are FO nuclei, and PO, LP, and dMG are HO nuclei.
Thalamocortical axonal projection labeling and analysis were done as previously described 6. In brief, P14-18 C57BL/6J mice were anesthetized and stabilized in a custom stereotaxic apparatus. Recombinant adeno-associated viruses expressing either EGFP or tdTomato were injected into the thalamus using a beveled sharp glass micropipette. Coordinates for injections ranged from: 0.5 to −1.6 anterior to posterior, 0–1.6 lateral, and 2.8–4.2 deep from the pia (in mm from bregma). Animals were perfused transcardially with PBS followed by 4% PFA 14 days after the surgery. The brains were collected, post-fixed and sectioned coronally on a cryostat at 50 μm thickness. All sections were imaged on the Nanozoomer slide scanner (Hamamatsu) and Zeiss Axio Imager. A suite of custom algorithms using MATLAB (MathWorks) was developed to analyze and compare thalamic injections across animals. Each thalamus was manually traced and each injection site was manually marked, to generate the binary thalamus masks and injection masks. All thalamic masks were normalized, corrected for variability in cutting angle, and aligned together to obtain the averaged model thalamus. Then, each injection site was mapped onto the model thalamus. Finally, injection and target information for the injections were combined to localize the precise thalamic origin of the cortical projections.
RNA in situ hybridization
Cryostat sections (18 μm) of P4 brains were prepared and used for RNA in situ hybridization, as previously described 56. In brief, the primer sequences for the RORα probe were obtained from Allen Brain Database. T7 or SP6 promoter sequences were added to the 5′ end of the reverse or forward primer sequences, respectively, which were then used for PCR amplification to generate the probe template. In vitro transcription of the resulted PCR product was performed to generate digoxigenin (DIG)-labeled antisense RNA probes. Hybridization was performed at 62 °C overnight, followed by post-hybridization washes. Sections were then incubated with alkaline phosphatase-coupled anti-DIG antibody (Roche, diluted 1:3000) at 4°C overnight. For the visualization of the reaction product, sections were incubated in the dark at room temperature with freshly prepared NTMT buffer (100 mM NaCl, 100 mM Tris-HCl pH 9.5, 50 mM MgCl2, 0.1% Tween20, 0.5 mg/mL Levamisol) containing NBT/BCIP (Roche). Once the color was fully developed, sections were washed, post-fixed with 4% PFA, dehydrated, and mounted with Permount (Fisher) for image acquisition using a slide scanner (NanoZoomer 2.0-HT, Hamamatsu Photonics).
Statistics
No statistical methods were used to predetermine sample sizes but our sample sizes are similar to those reported in previous publications 24, 51. Data collection and analysis were not randomized nor performed blind to the conditions of the experiments. No data points were excluded. Data are presented as median with interquartile range and whiskers as the minimum and maximum, or as mean±s.e.m., and statistical differences were determined using non-parametric Mann–Whitney test, unpaired t-test with Welch's correction (unequal variances), chi-square test, or linear regression analysis. Data distribution was not formally tested. Statistical significance was set at p<0.05.
Data and code availability
The data and scripts that support the findings of this study are available from the corresponding author upon request.
Supplementary Material
Acknowledgments
We thank Dr. Alexandra L. Joyner (Memorial Sloan Kettering Cancer Center, U.S.A), Dr. Ryoichiro Kageyama (Kyoto University, Japan), Dr. Simon Hippenmeyer (The Institute of Science and Technology, Austria), and Dr. Yasushi Nakagawa (University of Minnesota, U.S.A.) for kindly providing the R26-LSL-SmoM2-EYFP, Nestin-CreERT2, MADM11, and Olig3-Cre mouse lines, respectively, Dr. J. Nicholas Betley (University of Pennsylvania, U.S.A.) for guinea pig anti-RFP antibody, and Shi laboratory members for insightful discussion and input. This work was supported by the NIH grants (R01DA024681 and R01MH101382 to S.-H.S., and P30CA008748 to Memorial Sloan Kettering Cancer Center Core Facilities), the Human Frontier Science Program (RGP0053 to S.-H.S. and K.H.), and the National Natural Science Foundation of China (61572265 to Z.H. and 759881 to S.-H.S.).
Footnotes
Author Contributions: W.S. and S.-H.S. conceived the project; W.S. collected, reconstructed, and analyzed MADM and retroviral labeling data with help from A.X.; Z.H., X.T. and K.H. performed NND analysis; Z.L. generated retrovirus; A.X., Z.H. and K.H. performed alignment and nuclear identity inference and clustering analysis with input from H.Z. and T.M.; H.Z. and T.M. provided thalamocortical axonal projection data; W.S. and S.-H.S wrote the paper with inputs from all other authors.
References
- 1.Jones EG. The Thalamus. Cambridge University Press; New York: 2007. [Google Scholar]
- 2.Steriade M, Llinas RR. The functional states of the thalamus and the associated neuronal interplay. Physiological reviews. 1988;68:649–742. doi: 10.1152/physrev.1988.68.3.649. [DOI] [PubMed] [Google Scholar]
- 3.Sherman SM, Guillery RW. Exploring the Thalamus and its Role in Cortical Function. MIT Press; Cambridge, MA: 2006. [Google Scholar]
- 4.Sherman SM. Thalamus plays a central role in ongoing cortical functioning. Nat Neurosci. 2016;19:533–541. doi: 10.1038/nn.4269. [DOI] [PubMed] [Google Scholar]
- 5.O'Leary DD, Schlaggar BL, Tuttle R. Specification of neocortical areas and thalamocortical connections. Annu Rev Neurosci. 1994;17:419–439. doi: 10.1146/annurev.ne.17.030194.002223. [DOI] [PubMed] [Google Scholar]
- 6.Hunnicutt BJ, et al. A comprehensive thalamocortical projection map at the mesoscopic level. Nat Neurosci. 2014;17:1276–1285. doi: 10.1038/nn.3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oh SW, et al. A mesoscale connectome of the mouse brain. Nature. 2014;508:207–214. doi: 10.1038/nature13186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sherman SM, Guillery RW. Functional Connetions of Cortical Areas. MIT Press; Cambridge, MA: 2013. [Google Scholar]
- 9.Scholpp S, Lumsden A. Building a bridal chamber: development of the thalamus. Trends Neurosci. 2010;33:373–380. doi: 10.1016/j.tins.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakagawa Y, Shimogori T. Diversity of thalamic progenitor cells and postmitotic neurons. Eur J Neurosci. 2012;35:1554–1562. doi: 10.1111/j.1460-9568.2012.08089.x. [DOI] [PubMed] [Google Scholar]
- 11.Arcelli P, Frassoni C, Regondi MC, De Biasi S, Spreafico R. GABAergic neurons in mammalian thalamus: a marker of thalamic complexity? Brain research bulletin. 1997;42:27–37. doi: 10.1016/s0361-9230(96)00107-4. [DOI] [PubMed] [Google Scholar]
- 12.Vue TY, et al. Characterization of progenitor domains in the developing mouse thalamus. J Comp Neurol. 2007;505:73–91. doi: 10.1002/cne.21467. [DOI] [PubMed] [Google Scholar]
- 13.Inamura N, Ono K, Takebayashi H, Zalc B, Ikenaka K. Olig2 lineage cells generate GABAergic neurons in the prethalamic nuclei, including the zona incerta, ventral lateral geniculate nucleus and reticular thalamic nucleus. Dev Neurosci. 2011;33:118–129. doi: 10.1159/000328974. [DOI] [PubMed] [Google Scholar]
- 14.Rakic P. Specification of cerebral cortical areas. Science. 1988;241:170–176. doi: 10.1126/science.3291116. [DOI] [PubMed] [Google Scholar]
- 15.Yu YC, Bultje RS, Wang X, Shi SH. Specific synapses develop preferentially among sister excitatory neurons in the neocortex. Nature. 2009;458:501–504. doi: 10.1038/nature07722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu YC, et al. Preferential electrical coupling regulates neocortical lineage-dependent microcircuit assembly. Nature. 2012;486:113–117. doi: 10.1038/nature10958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Y, et al. Clonally related visual cortical neurons show similar stimulus feature selectivity. Nature. 2012;486:118–121. doi: 10.1038/nature11110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He S, Li Z, Ge S, Yu YC, Shi SH. Inside-Out Radial Migration Facilitates Lineage-Dependent Neocortical Microcircuit Assembly. Neuron. 2015;86:1159–1166. doi: 10.1016/j.neuron.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McCormick DA, McGinley MJ, Salkoff DB. Brain state dependent activity in the cortex and thalamus. Curr Opin Neurobiol. 2015;31:133–140. doi: 10.1016/j.conb.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lien AD, Scanziani M. Tuned thalamic excitation is amplified by visual cortical circuits. Nat Neurosci. 2013;16:1315–1323. doi: 10.1038/nn.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golden JA, Cepko CL. Clones in the chick diencephalon contain multiple cell types and siblings are widely dispersed. Development. 1996;122:65–78. doi: 10.1242/dev.122.1.65. [DOI] [PubMed] [Google Scholar]
- 22.Golden JA, Zitz JC, McFadden K, Cepko CL. Cell migration in the developing chick diencephalon. Development. 1997;124:3525–3533. doi: 10.1242/dev.124.18.3525. [DOI] [PubMed] [Google Scholar]
- 23.Zong H, Espinosa JS, Su HH, Muzumdar MD, Luo L. Mosaic analysis with double markers in mice. Cell. 2005;121:479–492. doi: 10.1016/j.cell.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 24.Gao P, et al. Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell. 2014;159:775–788. doi: 10.1016/j.cell.2014.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ciceri G, et al. Lineage-specific laminar organization of cortical GABAergic interneurons. Nat Neurosci. 2013;16:1199–1210. doi: 10.1038/nn.3485. [DOI] [PubMed] [Google Scholar]
- 26.Imayoshi I, Ohtsuka T, Metzger D, Chambon P, Kageyama R. Temporal regulation of Cre recombinase activity in neural stem cells. Genesis. 2006;44:233–238. doi: 10.1002/dvg.20212. [DOI] [PubMed] [Google Scholar]
- 27.Hippenmeyer S, et al. Genetic mosaic dissection of Lis1 and Ndel1 in neuronal migration. Neuron. 2010;68:695–709. doi: 10.1016/j.neuron.2010.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vue TY, et al. Sonic hedgehog signaling controls thalamic progenitor identity and nuclei specification in mice. J Neurosci. 2009;29:4484–4497. doi: 10.1523/JNEUROSCI.0656-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang L, Bluske KK, Dickel LK, Nakagawa Y. Basal progenitor cells in the embryonic mouse thalamus - their molecular characterization and the role of neurogenins and Pax6. Neural development. 2011;6:35. doi: 10.1186/1749-8104-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clasca F, Rubio-Garrido P, Jabaudon D. Unveiling the diversity of thalamocortical neuron subtypes. Eur J Neurosci. 2012;35:1524–1532. doi: 10.1111/j.1460-9568.2012.08033.x. [DOI] [PubMed] [Google Scholar]
- 31.Diggle PJ. Statistical Analysis of Spatial Point Patterns. Oxford University Press Inc.; New York: 2003. [Google Scholar]
- 32.Kuramoto E, et al. Ventral medial nucleus neurons send thalamocortical afferents more widely and more preferentially to layer 1 than neurons of the ventral anterior-ventral lateral nuclear complex in the rat. Cereb Cortex. 2015;25:221–235. doi: 10.1093/cercor/bht216. [DOI] [PubMed] [Google Scholar]
- 33.Van der Werf YD, Witter MP, Groenewegen HJ. The intralaminar and midline nuclei of the thalamus. Anatomical and functional evidence for participation in processes of arousal and awareness. Brain Res Brain Res Rev. 2002;39:107–140. doi: 10.1016/s0165-0173(02)00181-9. [DOI] [PubMed] [Google Scholar]
- 34.Szabo NE, Zhao T, Zhou X, Alvarez-Bolado G. The role of Sonic hedgehog of neural origin in thalamic differentiation in the mouse. J Neurosci. 2009;29:2453–2466. doi: 10.1523/JNEUROSCI.4524-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jeong J, Mao J, Tenzen T, Kottmann AH, McMahon AP. Hedgehog signaling in the neural crest cells regulates the patterning and growth of facial primordia. Genes Dev. 2004;18:937–951. doi: 10.1101/gad.1190304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puelles L, Rubenstein JL. Expression patterns of homeobox and other putative regulatory genes in the embryonic mouse forebrain suggest a neuromeric organization. Trends Neurosci. 1993;16:472–479. doi: 10.1016/0166-2236(93)90080-6. [DOI] [PubMed] [Google Scholar]
- 37.Nakagawa Y, O'Leary DD. Dynamic patterned expression of orphan nuclear receptor genes RORalpha and RORbeta in developing mouse forebrain. Dev Neurosci. 2003;25:234–244. doi: 10.1159/000072271. [DOI] [PubMed] [Google Scholar]
- 38.Frassoni C, Amadeo A, Ortino B, Jaranowska A, Spreafico R. Organization of radial and non-radial glia in the developing rat thalamus. J Comp Neurol. 2000;428:527–542. doi: 10.1002/1096-9861(20001218)428:3<527::aid-cne9>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 39.Nakagawa Y, O'Leary DD. Combinatorial expression patterns of LIM-homeodomain and other regulatory genes parcellate developing thalamus. J Neurosci. 2001;21:2711–2725. doi: 10.1523/JNEUROSCI.21-08-02711.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Suzuki-Hirano A, et al. Dynamic spatiotemporal gene expression in embryonic mouse thalamus. J Comp Neurol. 2011;519:528–543. doi: 10.1002/cne.22531. [DOI] [PubMed] [Google Scholar]
- 41.Kataoka A, Shimogori T. Fgf8 controls regional identity in the developing thalamus. Development. 2008;135:2873–2881. doi: 10.1242/dev.021618. [DOI] [PubMed] [Google Scholar]
- 42.Chatterjee M, Li JY. Patterning and compartment formation in the diencephalon. Front Neurosci. 2012;6:66. doi: 10.3389/fnins.2012.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saalmann YB, Kastner S. Cognitive and perceptual functions of the visual thalamus. Neuron. 2011;71:209–223. doi: 10.1016/j.neuron.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Roth MM, et al. Thalamic nuclei convey diverse contextual information to layer 1 of visual cortex. Nat Neurosci. 2015 doi: 10.1038/nn.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Crabtree JW, Collingridge GL, Isaac JT. A new intrathalamic pathway linking modality-related nuclei in the dorsal thalamus. Nat Neurosci. 1998;1:389–394. doi: 10.1038/1603. [DOI] [PubMed] [Google Scholar]
- 46.Shu Y, McCormick DA. Inhibitory interactions between ferret thalamic reticular neurons. J Neurophysiol. 2002;87:2571–2576. doi: 10.1152/jn.00850.2001. [DOI] [PubMed] [Google Scholar]
- 47.Lee SC, Patrick SL, Richardson KA, Connors BW. Two functionally distinct networks of gap junction-coupled inhibitory neurons in the thalamic reticular nucleus. J Neurosci. 2014;34:13170–13182. doi: 10.1523/JNEUROSCI.0562-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molnar Z, et al. Normal development of embryonic thalamocortical connectivity in the absence of evoked synaptic activity. J Neurosci. 2002;22:10313–10323. doi: 10.1523/JNEUROSCI.22-23-10313.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Frangeul L, et al. A cross-modal genetic framework for the development and plasticity of sensory pathways. Nature. 2016;538:96–98. doi: 10.1038/nature19770. [DOI] [PubMed] [Google Scholar]
- 50.Grant E, Hoerder-Suabedissen A, Molnar Z. The Regulation of Corticofugal Fiber Targeting by Retinal Inputs. Cereb Cortex. 2016;26:1336–1348. doi: 10.1093/cercor/bhv315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu HT, et al. Distinct lineage-dependent structural and functional organization of the hippocampus. Cell. 2014;157:1552–1564. doi: 10.1016/j.cell.2014.03.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown KN, et al. Clonal production and organization of inhibitory interneurons in the neocortex. Science. 2011;334:480–486. doi: 10.1126/science.1208884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ha GE, et al. The Ca2+-activated chloride channel anoctamin-2 mediates spike-frequency adaptation and regulates sensory transmission in thalamocortical neurons. Nature communications. 2016;7:13791. doi: 10.1038/ncomms13791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bombeiro AL, et al. MHC-I and PirB Upregulation in the Central and Peripheral Nervous System following Sciatic Nerve Injury. PLoS One. 2016;11:e0161463. doi: 10.1371/journal.pone.0161463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tan X, et al. Vascular Influence on Ventral Telencephalic Progenitors and Neocortical Interneuron Production. Dev Cell. 2016;36:624–638. doi: 10.1016/j.devcel.2016.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Blaess S, et al. Temporal-spatial changes in Sonic Hedgehog expression and signaling reveal different potentials of ventral mesencephalic progenitors to populate distinct ventral midbrain nuclei. Neural development. 2011;6:29. doi: 10.1186/1749-8104-6-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and scripts that support the findings of this study are available from the corresponding author upon request.


