Abstract
Immobilized antibody systems are the key to develop efficient diagnostics and separations tools. In the last decade, developments in the field of biomolecular engineering and crosslinker chemistry have greatly influenced the development of this field. With all these new approaches at our disposal, several new immobilization methods have been created to address the main challenges associated with immobilized antibodies. Few of these challenges that we have discussed in this review are mainly associated to the site-specific immobilization, appropriate orientation, and activity retention. We have discussed the effect of antibody immobilization approaches on the parameters on the performance of an immunoassay.
Keywords: Site-selective, Site-specific, Orientation, Immobilization, Antibody, Antibody Fragments, Click Chemistry, Affinity Tags, Protein A, Nucleotide Binding Protein, Metal Binding Protein, Staudinger Ligation, Cycloaddition, Antibody-binding protein, Physical adsorption, Diagnostics, Therapeutics, Separation
1. Introduction
Immobilized biomolecular systems have revolutionized how we analyze biological/biochemical matrices. However, in such complex matrices, capturing an analyte of interest with high specificity and sensitivity is crucial. Therefore, biorecognition elements, such as receptors and antibodies, are required for capturing a specific analyte out of a complex biological sample. Antibody is one such category of biorecognition molecules that specifically binds to their corresponding antigen. This leads to the core of our long-standing interest in the development of immunodiagnostics. In addition to specificity, lower limits of detection and high sensitivity are the key features of an ideal immunoassay and can be achieved by employing antibodies as capture agents. For this, antibodies are immobilized on the surface of a solid support.[1] Thus, a suitable immobilization approach is always sought that preserves maximum antibody functionality by site-directed and oriented molecular presentation. In this review, we are discussing antibody immobilization strategies that provide (i) site-specific capture guided through various tags and functional groups on antibodies and (ii) orientation achieved through pre-capture biomolecules.
1.1. Antibody
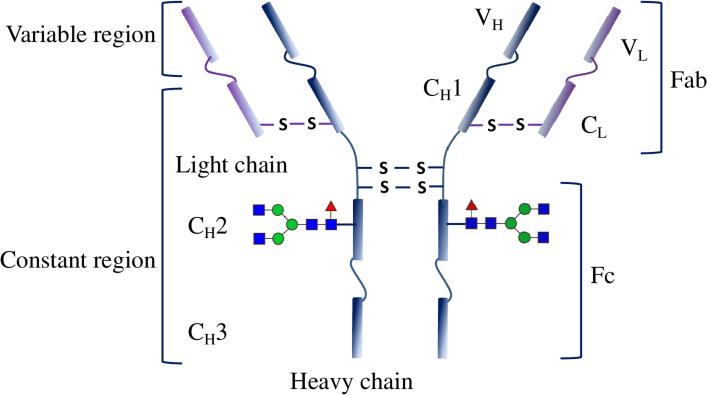
An antibody that is the key constituent of an assay is an immunoglobulin (Ig), typically type G (IgG). It has a molecular weight of ~150 kDa with molecular dimensions of approximately 142 × 85 × 45 Å3 [2], as shown in Figure 1.
Figure 1.
Illustration showing detailed structure of an antibody depicting various regions. Adapted from ref. [3] with permission from Macmillan Publishers Ltd: [Nature Chemistry], copyright (2016).
Structurally, an IgG is a homodimeric protein with two identical pairs of heavy and light chains linked by disulfide bonds.[4] It is worth noting that the structural and molecular composition can vary according to class/isotype (e.g., IgA, IgD, IgE, IgG, and IgM) and even subclass (e.g., IgG1, IgG2a and 2b, IgG3 and IgG4) of antibodies.[5] Like any other protein, chemically an antibody possesses carboxyl, amine, hydroxyl, sulfhydryl, alkyl, and aryl functional groups.[6] Researchers, however, have customized antibodies by adding different functional groups via molecular engineering. In addition, several engineered short variants of these full-length antibodies have been developed. Those antibody derivatives include, but are not limited , Fab (antigen binding fragment), single-chain variable fragment (scFv), and single domain antibody (sdAb). A Fab is a part of an IgG, which contains a whole light chain, and the variable region and the first constant region of heavy chain (Figure 1). ScFv is composed of variable regions of the heavy and light chains of IgGs, with a linker peptide. An sdAb only contains a monomeric variable region without any linker peptide. It is derived from either camelid antibodies (called VHH fragment) or novel antigen receptors IgNAR (VNAR fragment), both of which lack the light chains in their structure. The antigen binding capacity is completely gained by the VHH and VNAR fragment.
Antibodies possess highest binding affinity for their corresponding antigen, even if there are additional receptors that may recognize the given antigen. Therefore, antibodies make excellent probes for immunoanalysis. In addition to these intrinsic properties, antibodies must be immobilized on a solid support with intact structure and functional activity. It is well known that immobilization of antibodies results in activity loss. Therefore, it is crucial to achieve comparative functionality of the immobilized antibodies with respect to those in solution. This allows us to make an educated choice of a suitable immobilization strategy. Such strategies must encourage the use of mild chemistries that don't affect the antigen-binding activity and specificity of antibodies along with their compatibility with different surfaces. In addition, these strategies should provide effective orientation to the immobilized antibodies in order to have their antigen-binding sites freely presented to interact with the analyte in the biological matrix.
2. Applications of Antibodies
Antibodies in the development of biosensors and other systems have revolutionized the areas of medical diagnosis, therapeutics, and separation and purification sciences. Immobilized antibody systems are regularly being used in food & drug industry, clinical diagnosis, and environmental monitoring in the form of different analytical technologies. A continuous effort to improve the form and factor of immobilized antibody systems has been made since the development of first plate-based immunoassay in 1980 for increasing the application coverage. Microplate-based conventional immunoassays, such as enzyme-linked immunosorbent assay (ELISA) and immuno/histochemistry (IHC which is performed in the plates with either glass slide bottom or glass slides are kept at the well bottom for reaction), constitute the biggest fraction of in-vitro analysis. A few of the most important applications of antibodies are summarized in this section.
2.1. Diagnostics
Core component in clinical disease diagnosis is antibody-based immunoassays. Traditional immunoassays are mainly ELISA and IHC. Capture antibodies immobilized on solid supports facilitate efficient analyte capture in these assays. A second antibody conjugated to a type of enzyme (peroxidase and phosphatase) is used for catalyzing a colorimetric reaction. Such colorimetry-based immunoassays are multistep procedures and thus are usually time consuming. However, recently, several important developments have been registered in the field of plate-based ELISA.[7,8] Dixit and colleagues have designed and developed a new approach for generating fast sandwich assays with human fetuin A (HFA) as the model analyte.[7,8] Their reported assays were completed within 3 hours from the scratch against routine ELISA methods that usually take 6 hours or more to completion. They have achieved at least 20-fold better sensitivity (~23 pg/mL) compared to the routine approach (625 pg/mL).
Although work from Dixit and colleagues has led to new frontiers in the field of plate-based immunoassay development, the need of faster point-of-care systems is the current trend.[7,8] In modern immunoassays, such as lateral flow assays, the antibodies for capture and detection are stored within the diagnostics system.[9] Both the events are allowed to take place simultaneously on the support within the moving liquid front, which makes these assays very fast. Signal generation has also changed in form factor where a preferable method would be direct colorimetry using colored particles over enzyme-mediated. A pregnancy strip test for human chorionic gonadotropin (hCG) is an excellent example of modern immunoassay featuring immobilized antibodies.
Researchers are trying to develop highly integrated and multiplexed immunoassays in several different biosensor formats for detecting important diseases and disorders. Vashist et al. have reported the development of integrated fast immunoassays for human fetuin A in real-time label free mode by employing BIAcore 3000-based surface plasmon resonance (SPR) measurement.[10] Several optical biosensor-based assays have been reported for fast and sensitive detection of important proteins and analytes in biological fluids. Modani et al. (2016) reported a microarray-based detection of shiga toxin producing E. coli using an SPR imager for fast real time analysis.[11] Such hybrid cross-platform immunoassays are now getting more common as they produce fast results allowing for real time monitoring at the same time.
Rusling and colleagues have reported highly multiplexed ultra-sensitive sandwich immunoassays based on electrochemical sensing.[12] Eight biomarker proteins captured by 1 μm magnetic beads immobilized with 400, 000 enzyme labels and 120, 000 antibodies were assayed on multi-electrode system with biomarker-specific antibodies crosslinked to the gold nanoparticles electrostatically grafted on the electrode surface. They have achieved detection of these biomarkers with subpicogram sensitivity.
In-vivo imaging is another very important diagnostics domain. Antibodies conjugated to contrast agents, such as nanoparticles [13–15] and gadolinium liposomes [16], permit selective and sensitive imaging via specific antigen-antibody interaction, providing a targeted molecular imaging. Quantum dots (QDs) have become a good in-vivo imaging choice due to their high quantum yields, good biostability and photostability, and availability of approaches for conjugating them with antibodies.[17] Wang et al. fabricated antibody-functionalized Ag2S QDs featuring high near infrared (NIR) fluorescence intensity, small size and low in vivo toxicity.[18] The Ag2S QDs were covalently immobilized with antibodies against vascular endothelial growth factor (VEGF). These conjugates were successfully demonstrated for targeted imaging of VEGF positive human glioblastoma tumors within mice.
2.2. Therapeutics
There are approximately 30 FDA approved/under review antibody-based drugs for therapeutic use in various diseases including cancer.[19] These therapeutic antibodies may be employed as either standalone drug or as payloads with pro/drugs conjugated to them. Direct conjugation of antibodies may affect the potential biological activity of these drugs; thus, these drugs are usually encapsulated in a payload vehicle, such as polymeric nanoparticles. Antibodies can be grafted on drug-encapsulating polymeric nanoparticles for targeted delivery. Also, these constructs reduce the overall toxicity of the encapsulated drugs by minimizing systemic exposure to cells, while improving the stability of drugs.
Several reviews have described many effective conjugation strategies for generating various antibody-drug/nanoparticle conjugates.[3,20] Conjugation strategies are similar to those employed for antibody immobilization on solid support (Figure 2). Polymeric nanoparticles offer a broad range of covalent attachment chemistries for antibodies. Qian and group made a maleimide-functionalized amphiphilic diblock copolymer encapsulating arsenite ions.[21] They further decorated such arsenite-containing micelles with anti-CD44v6 scFvs for targeted delivery of arsenite to tumor cells. Unlike post-labelling the antibody to a well formed nanoparticle, Goodall & colleagues have recently reported a pre-labelling method by covalently incorporating scFv antibodies to poly(N-isopropylacrylamide) chains before their assembly onto a thermoresponsive polymeric nanoparticle.[22] The scFv maintained its immunoreactivity before and after assembly, even after glutaraldehyde cross-linking for stabilizing the immunoparticles.
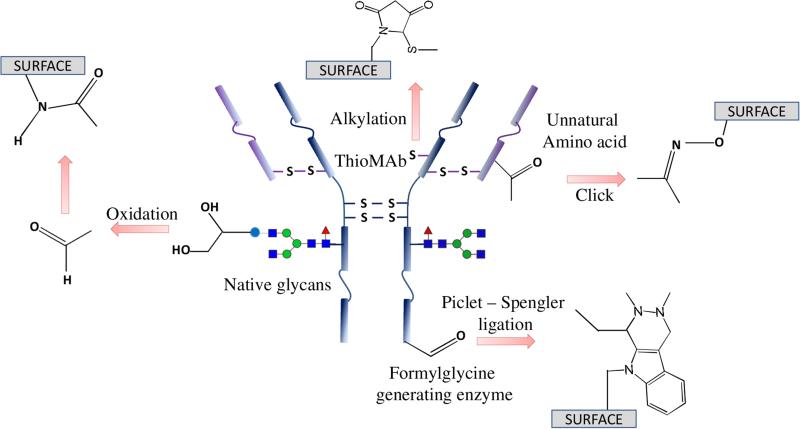
Figure 2.
Illustration showing various functional groups on an antibody that can potentially be employed for immobilization. Modified from ref. [3] with permission from Macmillan Publishers Ltd: [Nature Chemistry], copyright (2016).
2.3. Separation and purification sciences
Immunoaffinity separation for purification of a variety of analytes is a standard procedure in industrial and academic settings. An analyte-specific antibody immobilized on a solid chromatographic support, such as superparamagnetic microparticles,[23,24] agarose microbeads,[25] and monoliths [26–28], can extract the analyte of interest from the complex sample matrix, which is mostly cell lysates. Target analytes (antigen) can be captured via specific antigen-antibody interaction by highly specific monoclonal antibodies.
Immunoaffinity chromatography (IAC) is another way for the identification, quantification, or purification of antigens using solid support matrix immobilized with corresponding antibodies (Figure 3).[29,30] Monoclonal antibodies are preferred and their immobilization onto supports can be through either covalent linkage or non-covalent interaction such as biotin-(strept)avidin systems.[30] Due to the specificity of antigen-antibody interaction, high degree of protein purification can often be achieved in one step by IAC.[31] More recently, IAC was demonstrated for ultrafast immunoextractions that are capable of operating on the millisecond time scale. Operation in such short time manner can guarantee that all the extracted molecules (drugs or hormones) are in the unbound forms from proteins of choice.
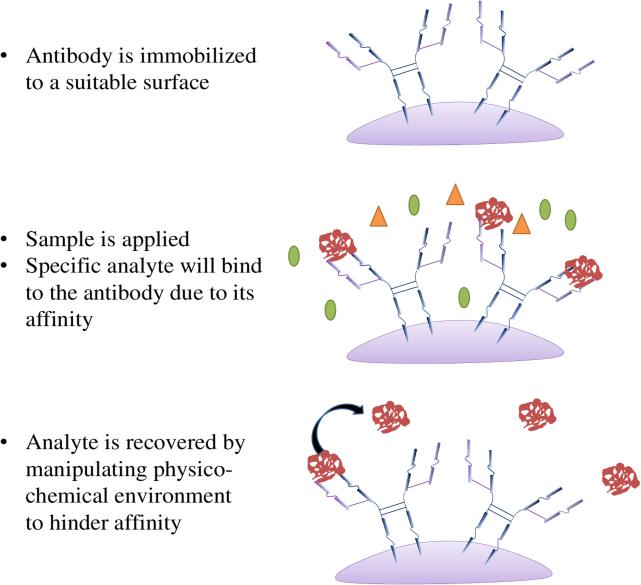
Figure 3.
Affinity chromatography at work. The illustration demonstrates the work flow of affinity capture.
For most of the antibody applications, receptor antibody must be immobilized to a solid support. There are several approaches to graft antibodies on such supports and we will discuss those in next section.
3. Antibody Immobilization Chemistries
3.1. Passive adsorption/Passivation
Passive adsorption of antibodies onto a support is the simplest approach for immobilization. There is no prerequisite of modification of either antibodies or the surface. This makes passivation of antibodies on the surface the most common approach employed to develop clinical immunoassays. However, a precise control over the orientation of antibodies is one of the major concerns (Figure 4).
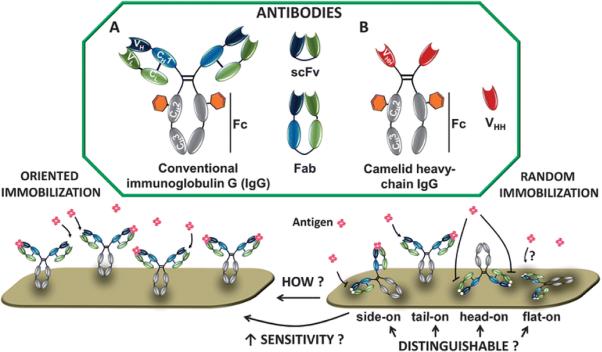
Figure 4.
Antibody and its variants in various orientations on the surface when physisorbed. Adapted from ref [32] published by The Royal Society of Chemistry.
Chemically antibodies are proteins. During passivation various non-covalent interactions govern the adsorption chemistry of. Hydrophobic, van der Waals, and pi-pi interactions are the key forces in immobilizing antibodies onto the surface of a solid support, which is usually a hydrophobic plastic. It involves various non-covalent interactions (e.g. hydrophobic forces and electrostatic interactions - a case of chemisorption), thus pH and ionic strength are critical for the antibody adsorption and the following antigen binding.[33]
Passive adsorption results in randomly orientated antibodies, which may depend on the dipole moments of antibodies and surface charges of the supports.[34–37] Improved oriented antibody adsorption has been reported using different pretreatment to either antibodies or to the supports, including UV light,[38] electric field, [39] electrochemistry,[40] and plasma-immersion ion implantation [18]. Jian and group has described that certain degree of orientation can be achieved for passively adsorbed IgG1 and IgG2a type antibodies on charged surfaces. They have observed that at low surface charge density and high ionic strength of the solution, van der Waals interaction dominates resulting in multiple orientations. At high surface charge density and low ionic strength they demonstrated that immobilization takes place via preferred positively or negatively charged domains on an antibody resulting in certain degree of orientation.[35,41]
In addition, adsorption is a dynamic process between solution and bound phases of antibodies. Apart from the random orientation, immobilized antibodies can easily leach out in presence of another protein possessing higher charge or more number of hydrophobic pockets. This is attributed to relatively weak and reversible interactions, thus inefficient antigen binding can happen even if higher amount of antibodies are adsorbed on the supports.[42] In addition, randomly adsorbed antibodies have poor affinity toward their antigens with relatively low antigen holding capacity. Ishihara group demonstrated a significant 100-fold reduction in antigen – antibody reaction equilibrium dissociation constant (Kd) with an approximate antigen holding capacity of 1.8 per antibody.[43] O'Kennedy and colleagues have demonstrated using surface plasmon resonance biosensor that mass density of orientated anti-human fetuin A (HFA) antibodies on protein A was 1.3 – fold higher than the randomly immobilized antibodies. Ordered presentation of antibodies has also allowed for a ~2 – fold higher HFA mass capture with respect to that captured by randomly immobilized antibodies.[10] To address those issues, antibody immobilization strategies via covalent binding or alternative specific, strong, and directional non-covalent interactions are preferred. These immobilization strategies have been already employed in the fields of ELISA and lateral flow assay, which were exclusively based on passive adsorption of antibody.[7,44]
3.2. Crosslinker mediated
Crosslinker is a class of chemicals that possess a reactive center at each terminal and are able to bind two corresponding functionalities. Based on the terminal reactive center, these could be either homo or heterobifunctional. Such linkers are widely used for developing convenient immobilization strategies, which is attributed to the commercial availability of the crosslinkers in various compositions, lengths, and physico-chemical properties.[51] The crosslinkers mainly aim at the primary amine (−NH2) and carboxyl (−COOH) groups since they are abundant and well distributed over the antibody surface. However, due to the same reason, the reactions between crosslinkers and these functional groups are not selective, resulting in a random orientation and in some instances loss of functional activity of the immobilized antibodies. Antibodies also possess sulfhydryl (−SH) groups in the form of disulfide bonds, which are responsible for their structural stability.[52] Researchers, via a selective partial reduction of hinge-region disulfide bonds, generated free pendent sulfhydryl groups, which can be attached to a variety of solid supports using sulfhydryl-specific heterobifunctional linkers in a very selective site-specific manner.[53] Antibodies also possess an unusual functionality in form of carbohydrate/sugar moieties at each of the two CH2 regions. Specific oxidation of such moieties yields aldehyde (−CHO) groups, which are highly reactive toward amine groups resulting in Schiffs base. Apart from that specific amine-terminated crosslinkers, such as α,Ω-bis aminoPEG, can also be used for immobilization.
All of the popular crosslinkers are listed in Table 1; such as, glutaraldehyde for amine-to-amine conjugation,[54] carbodiimide for carboxyl-to-amine linking, maleimide for sulfhydryl groups, and hydrazides for aldehyde groups. The site-specificity of the amine- and carboxyl-specific crosslinkers decreases reciprocally with the increasing number of respective functional groups, leading to the loss of orientation.[55,56] However, linkage via sulfhydryl or aldehyde groups on the antibodies is more selective, and provides better orientation. Since, generating sulfhydryl and aldehyde groups on the antibodies requires relatively aggressive conditions, thus it may lead to some unwanted reactions and may affect the binding of antibodies to their target antigens.[57–59]
Table 1.
The list of some popular protein crosslinkers used in antibody immobilization
| Cross-linking targets | Crosslinker reactive groups | Example products | Ref |
|---|---|---|---|
| Carboxyl reactive | Carbodiimide | EDC | [45] |
| DCC | |||
| Amine reactive | NHS ester | BS3 | [46] |
| Imidoester | DMP | ||
| Aldehyde | glutaraldehyde | ||
| Sulfhydryl reactive | Maleimide | [47] | |
| Aldehyde reactive | Hydrazide | [48] | |
| Alkoxyamine | |||
| Photo-reactive | Diazirine | [49] | |
| Aryl azide | |||
| to amine or sulfhydryl | Epoxide | [50] |
3.3. Site-directed chemistries
3.3.1. Antibody thiols/sulfhydryls
As mentioned above, site-specific sulfhydryl groups can be generated via selective reduction of disulfide bonds in hinge region. The free sulfhydryl groups can be reactive towards supports bearing gold,[60–64] maleimides,[65] pyridyl disulfides,[66,67] and others[68]. However, undesired reduction of other disulfide bonds may take place, although the resulting antibody fragments still might maintain their biological functions on the sensor surface.[69] Dixit et al. have thiolated antibodies with the reaction of 2-iminothiolane.HCl at primary amines without reducing their disulfide bonds without noticing any substantial activity loss.[70] Protein engineering strategies to introduce free sulfhydryl group(s) onto antibodies offer extreme control over the number and location of sulfhydryl group(s) in the construct.[71–73] Hortigüela et al. fused cysteine-containing peptide linkers to scFv antibody fragments at the position distal to the antigen binding sites for immobilization on maleimide-activated supports in covalent and orientated manner.[72] There are several approaches to fabricate antibody drug conjugates (ADCs) based upon site-specific crosslinking of small drug molecules through their sulfhydryls. These strategies can be potentially employed with certain thiol-reactive reagents for site-specific antibody immobilization. These approaches include genetic substitution of available serine or other residues to cysteine [74,75]; reduction of the number of interchain cysteines to make the leftover cysteines clearly defined [76]; covalent re-bridging of the disulfide bond using bis(sulfone) reagents, special maleimides, and others [5,77,78]; and genetic incorporation of selenocysteine to introduce conjugation selectivity to maleimides [79].
3.3.2. Antibody sugar chain
Periodate-mediated oxidation of polysaccharide moieties of antibodies yields aldehyde groups that can be employed for immobilizing antibodies either via hydrazide-derived crosslinkers or amine surfaces.[80–84] Alongside polysacchrides, some critical amino acids such as methionine, tryptophan, and histidine may also get oxidized during periodate reaction, thus affecting the site-selectivity of this approach. This slight non-specificity in oxidation may also affect the binding of antibodies to their target antigens.[57–59] In addition, the aldehyde groups might be reactive towards amine and sulfhydryl groups, resulting in inter-antibody cross-linking and thus aggregation.
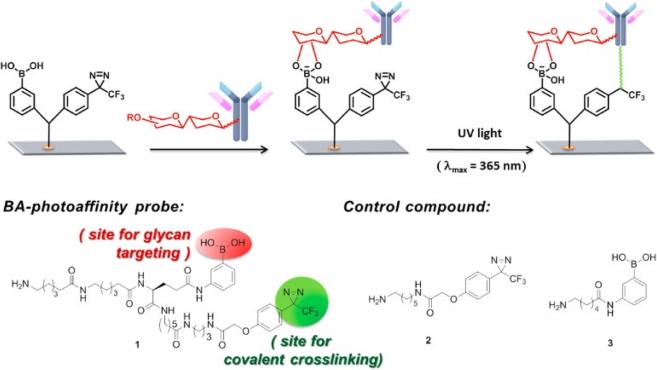
Boronic acids, unlike oxidizing the sugar moieties, interact with vicinal diol in polysaccharide molecules and form boronate esters.[85,86] Therefore, boronic acid-coated surfaces can equally be used to capture antibodies via boron-polysaccharide interactions and are good alternative. Many reports demonstrated the use of aminophenylboronic acid-coated support to immobilize antibodies in a site-directed manner.[63,87,88] However, boronate ester chemistry is reversible at physiological pH, and thus any glycoproteins in the sample may compete for the antibody immobilization. Adak et al. employed a crosslinker to overcome these drawbacks.[49] This crosslinker was a branched structure and possesses boronic acid and diazirine-containing photoreactive moiety (Figure 5). Antibodies were captured on the surface functionalized with this bifunctional crosslinker. At the first step boronic acid part reacts with the di-ols of the polysaccharide moieties resulting in weakly captured antibodies; while at the second step, the diazirine part forms a covalent linkage with the carboxy end of the antibodies when photoactivated with light at 360 nm wavelength. This results in site-specific covalent immobilization of antibodies onto the surface. In some conjugation methods immobilization was facilitated by site-specific modification of antibodies via polysaccharide moieties. These approaches mainly included generation of aldehyde groups via enzymatic methods and/or glycoengineering, [89–91] and incorporation of unnatural fucose-derivatives (e.g. thiol-fucose) to polysaccharide moieties [92].
Figure 5.
Fabrication of antibody microarrays by light-induced covalent and orientated immobilization. Reprinted from ref [49]. Copyright (2014) American Chemical Society.
3.3.3. Nucleotide binding site (NBS)
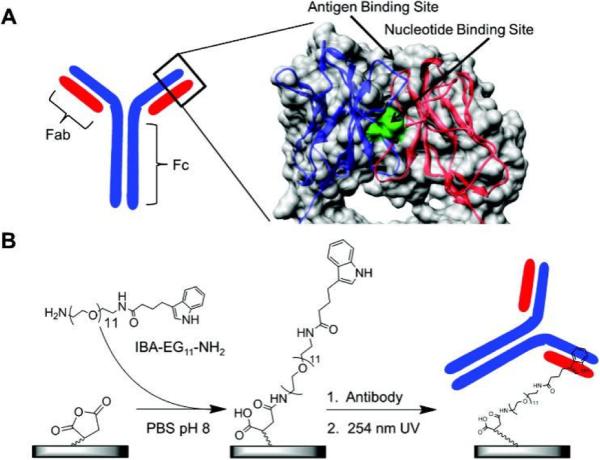
Nucleotide binding site (NBS) is a conserved region in the variable domain of all immunoglobulins. As its name suggests, NBS exhibits affinity for nucleotides and some aromatic amino acids, since there are several amino acids with aromatic side chains in the conserved regions of NBS and π-π stacking interactions might contribute to the affinity.[93,94] Handlogten et al. performed a computational screening for better NBS binding candidates and found several top hits.[94] The follow-up experimental investigation showed indole-3-butryic acid (IBA) had the best binding affinity for NBS with Kd of 1~8 μM. Alves et al. [95], in subsequent studies, demonstrated how high site-specificity allowed for ordered antibody capture on IBA-terminated surfaces (Figure 6). Following the initial capture of antibodies, photo-crosslinking was used to create a covalent bond between antibody and surface, similar to that presented by Adak et al. [49]. They also used the same photo-cross-linking strategy to prepare biotin labeled antibodies and coated such conjugates onto a streptavidin plate for enhanced antigen detection.[96,97] Although NBS is present in the variable region of an antibody but it is still distant from the antigen binding site (Figure 6),[93,94] and doesn't affect antigen binding capacity of the antibodies. In addition to NBS, other unconventional binding sites on antibodies, which are specific for small molecules, have been identified and may offer alternative conjugation/immobilization strategies.[98,99]
Figure 6.
Orientated surface immobilization of antibodies at the conserved nucleotide binding site for enhanced antigen detection. Reproduced from [96] with permission from Elsevier.
3.4. Bi-orthogonal covalent chemistries
3.4.1. Diels-Alder reaction
Diels-Alder (D-A) reaction is a classical organic reaction widely used in synthetic organic chemistry. It is a [4+2] cycloaddition between a conjugated diene and a double bond, forming a 6-membered unsaturated ring. The half-life of D-A reaction at ambient temperature is ~2 hours [100] and aqueous surroundings can largely accelerate the reaction rate [101], thus making it promising for the immobilization of biomolecules onto the solid supports. Shi et al. developed self-assembled micellar nanoparticles presenting furan groups at their surface, which reacted with the maleimide-modified antibodies via D-A reactions. They employed these immunopolymeric nanoparticles for targeted drug delivery.[102] There are other potential strategies that were developed to immobilize proteins and can be further extended to covalently capture antibodies. Stamos et al. site-specifically immobilized five individual proteins onto microarray supports via the D-A reaction to study the protein-protein interactions.[103] In their design, a 3-amino-L-tyrosine was incorporated to each protein, which was further oxidized to an o-iminoquinone for linking to the acryloyl-derivatized supports via D-A reactions.[103,104]
3.4.2. Staudinger ligation
Staudinger ligation is developed from classical Staudinger reaction by Saxon and Bertozzi in 2000,[105] and has been utilized for labeling, synthesis and for immobilization [106–108]. Staudinger ligation is a reaction between azide-containing proteins and phosphine-containing esters or thioesters of the surface, resulting in covalent amide bonds. Although, Staudinger ligation is mostly used for small molecules, peptides, and protein immobilization,[109–113] it can be potentially used for antibody immobilization. For instance, Soellner et. al. first reported a site-specific immobilization of a truncated ribonuclease S (RNAse S’) onto a glass slide.[109] They installed an azido group into a 15-mer peptide derived from RNAse S’ and linked the resulting peptide onto the surface presenting diphenylphosphine moieties by Staudinger ligation.
They further incubated the peptide coated surface with the rest of RNAses S’ to generate a surface of active RNAses S’. A major drawback of this method is that it requires protein engineering to incorporate active azide analogs, such as methionine analog azidohomoalanine [114] or unnatural amino acid p-azido-L-phenylalanine [115] at specific sites in the proteins, which may affect their final activity.
3.4.3. Click chemistry
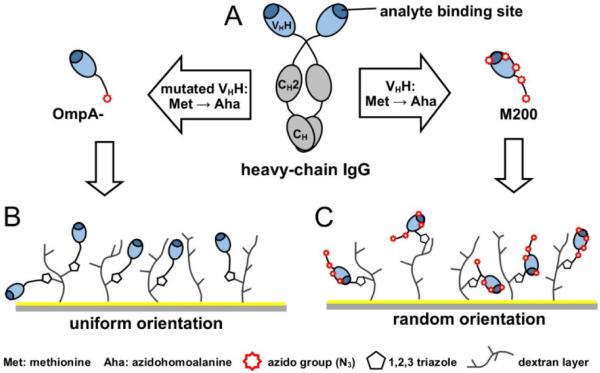
“Click chemistry” is a term introduced by Sharpless et al. to describe a series of reactions that meets a set of stringent criteria such as high yields, simple reaction conditions, simple product isolation, etc.[116] The most popular click chemistry reaction is the copper(I)-catalyzed [3+2] azide-alkyne cycloaddition (CuAAC) where an azide reacts with an alkyne to form a five-membered heteroatom ring in the presence of copper(I). For example, Finetti et al. successfully immobilized an azido modified anti-mouse IgG antibody on alkyne-functionalized gold nanoparticles via CuAAC click chemistry.[117] Due to the cytotoxicity of copper(I) catalyst, some copper-free click reactions have been also developed, such as strain-promoted azide-alkyne cycloaddition (SPAAC) where the alkyne part is a highly strained cyclic alkyne (e.g. cyclooctyne).[118,119] Trilling et al. introduced the azido group(s) to a VHH antibody that recognizes foot-and-mouth disease virus (FMDV) and attached the VHH antibodies to a cyclooctyne functionalized SPR chip via both the CuAAC and SPAAC reactions (Figure 7).[120] Comparing VHH antibodies bearing a single azido group (site-directed immobilization) and five azido groups (relatively random immobilization), the authors found that the site-directed strategy significantly improved the analytical performance of SPR. They were able to increase detection sensitivity by a factor of 800.
Figure 7.
Orientation of llama antibodies strongly increases sensitivity of biosensors. Reproduced from [120] with permission from Elsevier.
3.5. Site-directed Capture
3.5.1. Small affinity tags
Small affinity tags are receptor-ligand type biomolecule pairs with highly specific binding partners. These mainly include biotin-strep/avifin, polyhistidine/(Metal)+2-nitrolotriacetic acid, and other peptide affinity tags. Due to their small size they can be easily crosslinked or genetically fused at any noncritical locations of antibodies without affecting conformation or immunoreactivity. The immobilization of antibodies via biotin-(strep)avidin interaction is one of the most common immobilization approaches featured with good stability, high efficiency, high specificity and extremely high binding affinity (kd = ~fM). Biotins are usually attached to the antibodies in a random or site-specific fashion using above mentioned chemical conjugation methods or enzymatic approaches,[121–123] while (strep)avidins are often coated on the supports via passive adsorption or other chemistries [124,125].
Polyhistidine tag (His-tag), typically hexahistidine, is a well-known genetically encoded affinity tag having an affinity for certain transition metal ions such as Ni2+, Co2+ and Cu2+. His-tagged recombinant antibodies are immobilized on Ni2+ chelated nitrilotriacetic acid (NTA) supports.[71] However, the binding affinity between His-tag and Ni2+ (kd = ~μM) is relatively low and continuous efforts to improve the affinity are being made. The low affinity can be enhanced by using multivalent NTAs,[126,127] increasing histidine numbers per tag to 10~12 residues, introducing tandem His-tags,[128,129] or introducing thioalkane chelators [130]. Although, His tags are constantly being employed for antibody purification and immobilization,[131] the overall immobilization efficiency may significantly decrease due to the competition with other metal-binding endogenous proteins [132]. Ericsson and colleagues proposed that a stable binding on Ni-NTA surface can be achieved by introducing photo-reactive crosslinkers. In the presence of Ni2+, human IgG with C-terminus His-tag was first captured on such surface, followed by covalent bonding via photo-reactive crosslinkers.[133]
Besides His-tag, other peptide affinity tags such as FLAG tag are also used for immobilization. FLAG-tag, consisting of the sequence motif DYKDDDDK, can be genetically incorporated into proteins and such FLAG-tagged proteins can be further captured by a support bearing anti-FLAG antibodies. Such protein immobilization via FLAG-tag is based on antigen-antibody interaction, instead of chelation, so stronger affinity and better specificity can be expected.[134]
3.5.2. Enzyme-substrate
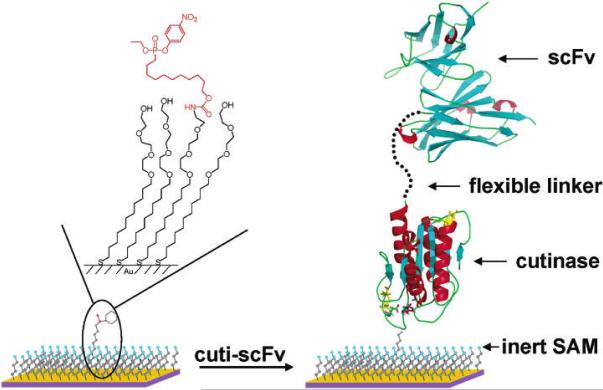
Certain enzyme inhibitors can form covalent bonding with the active site of corresponding enzymes and thus irreversibly inhibit the enzyme activity. Inspired by this irreversible inhibition mechanism, such enzymes or their active domains can be genetically fused with desired proteins for highly site-specificity immobilization. Such constructs can be selectively and covalently linked to a surface presenting enzyme substrate/inhibitor. Cutinase and its phosphonate inhibitors are one of the widely used enzyme-inhibitor pairs that fall into this catalog. Cutinase is a serine esterase which can covalently link its Serine residue in the active site with phosphonate ligands via esterification reaction. It is a relatively small enzyme (22 kD) with its active site far away from both N- and C-termini, offering some flexibility for designing the whole fusion protein. Kwon et al. fused antibody fragments (e.g. scFv and VHH) to cutinase and used phosphonate ligands presented supports to capture the fusion proteins (Figure 8).[135] A flexible 15-mer (GGGGS)3 polypeptide linker was inserted between antibody fragments and cutinase, which facilitated individual protein folding. The immobilized antibodies exhibited high affinity and specificity for their target antigens, as elucidated via a series of SPR and fluorescence studies.
Figure 8.
Antibody arrays prepared by cutinase-mediated immobilization on self-assembled monolayers. Reproduced from [135]. Copyright (2004) American Chemical Society.
Beside enzyme-inhibitor pairs, it is also a good idea to create covalent bonds between two reactants in an enzyme-mediated fashion. In this method, one reactant is introduced in the antibody while other is grafted on the surface, and in presence of a specific enzyme a covalent bond is created between them. Antibodies, unlike the previous enzyme-based methods, antibodies will most likely retain their functional activity due to minimal hindrance in their protien folding event. In addition, the amounts of enzymes used in this method are relatively smaller than the previous approach. Biotin ligase and numerous other enzymes participating in posttranslational covalent modification of proteins in vivo, including transferases (e.g. transglutaminases and peptidases) and oxidoreductases (e.g. tyrosinases and peroxidases), are few systems employed for such immobilizations.[136–139] Recently, Sortase A (Srt A)-mediated ligation strategy has been established.[140,141] SrtA is a transpeptidase which recognizes a penta-peptide motif LPXTG (where X is any amino acid). Upon recognition, the S-atom of Cys-184 of SrtA is attached to the threonine of the motif via nucleophilic substitution, resulting in a stable LPXT-thioacyl-SrtA intermediate. This intermediate would undergo another nucleophilic substitution where the α-amino group of an oligoglycine attacks and makes the enzyme SrtA as a leaving group. Using such SrtA-mediated reaction, radioactive metal complexes, highly potent anti-tumor drugs, and certain enzymes could be site-specifically labelled to antibodies for different purposes such as in vivo imaging, drug delivery and antibody-antigen detection.[142–145]
3.5.3. Immunoglobulin (Ig)-binding proteins
Protein A and protein G are derived from bacteria Staphylococcus aureus and Streptococcus species, respectively. These proteins possess multiple binding domains specific for the Fc portion of the mammalian Igs. Their binding properties are different and dependent on the subclasses of Igs and their species of origin. Protein G has binding affinity for a broader range of Igs than protein A; however, native protein G also has binding regions for other molecules (e.g. albumin), for which researchers have developed genetically truncated protein G with only Fc-binding capabilities. Protein A/G, a recombinant fusion protein consisting of Fc-binding domains from protein A and protein G, was also developed and binds to the broadest range of Igs. There is an additional Staphylococcal protein that binds to Igs via the kappa light chains and is called protein L. All of these Ig binding proteins have been successfully used in biosensors and immunoseparations to achieve orientated immobilization of intact antibodies, which significantly improved the experiment performance compared with random antibody immobilization.[10,62,146] Although, a very homogeneous and functional layer of antibodies have been achieved on several occasions with the use of Ig-binding proteins, there are few concerns that may be potentially associated with this strategy.
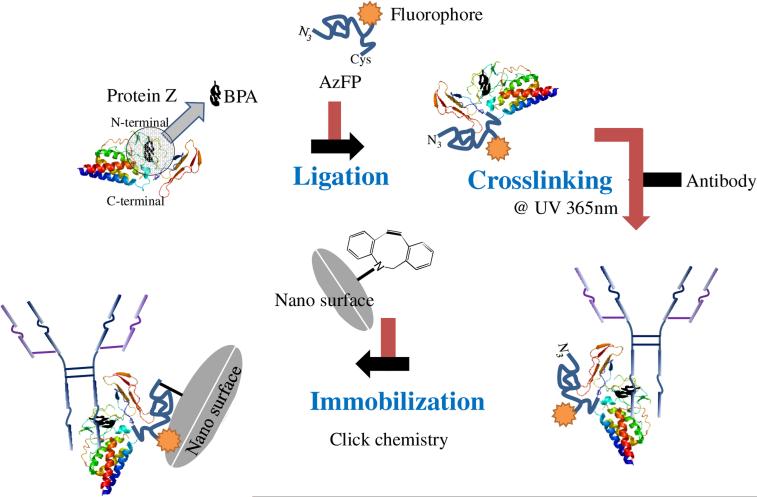
Orientating Ig-binding proteins on surface due to their 3D structure and orientation [147] is a primary challenge as the problem of protein orientation now simply shifted from one protein (antibody) to the other (protein A or G). The orientation of Ig-binding proteins can also be accomplished via either site-specific fusion of cysteine residues,[148,149] gold binding peptide,[150,151] His-tag [152–154] to protein A or G, or enzymatic conjugation,[43] which can provide a well-ordered protein A or G scaffold. The unclear stoichiometry of Ig binding to protein A or G having multiple Ig binding domains with different binding affinities presents another crucial challenge for capturing antibodies with these proteins. Given this, only two antibodies can stoichiometrically bind at the maximum binding capacity of protein A, G, or A/G.[10] Nilsson and group has developed engineered analog of protein A, the Z domain [155] that was demonstrated for Igs binding [156–159]. Miyao et al. constructed solid surfaces bearing either one or two Z domains for antibody immobilization and they found that the tandem Z domains (i.e. ZZ domains) captured the antibody more efficiently than single Z domain, suggesting multiple Ig binding domains may adopt a cooperative manner for Ig binding.[160] Ig binding via protein A or G is a non-covalent and reversible binding, which is good for protein A or G regeneration, thus improving their reusability.[161] However, the binding reversibility poses a problem for the stability of protein-antibody adduct that may not be suitable for certain applications. In this scenario, for applications like immunoprecipitation, antibodies of protein A-Ig adduct may co-elute with the other antibodies or proteins. Therefore, scientists usually crosslink Igs captured on protein A or G via chemical crosslinkers such as dimethyl pimelimidate (DMP) and bis(sulfosuccinimidyl) suberate (BS3).[162–165] However, crosslinkers like DMP may also modify the amine groups on the antigen biding sites, leading to a decrease in antigen binding efficiency.[166] Konrad et. al. [167] and Yu et. al. [168] have separately addressed this challenge by incorporating photo-reactive crosslinkers into Z domain to create UV light-induced covalent bond between Z domain and Igs at the Fc regions. Following such principle, Hui et al. fabricated a bifunctional Z domain protein bearing photo-reactive crosslinker and azido group.[169] They first captured IgGs onto Z domain followed by UV-induced crosslinking. This adduct has free azido group facilitating direct immobilization on cyclooctyne functionalized nanoparticles via SPAAC click chemistry (Figure 9). Another example is covalent immobilization of antibodies through directly loading antibodies onto the surface bearing Ig binding protein with photo-reactive crosslinker, followed by washing and UV irradiation.[170]
Figure 9.
Facile method for the site-specific, covalent attachment of full-length IgG onto nanoparticles through a combination of recombinant protein Z domain crosslinked to IgG via BPA binding immobilized on nanoparticle surface using click chemistry.
3.6. Miscellaneous
3.6.1. Calixarene derivatives
Calixarene is a widely studied organic host system which is a cup-shaped cyclic oligomer. Chemical architecture of the calixarene molecules can be adjusted for the polarity and other physico-chemical properties of the cuplike structure, making calixarenes interact with a wide range of guest species.[171] Lee et al. synthesized two novel calixarene derivatives, ProLinker A and ProLinker B, for protein immobilization on the microarrays.[172] Further studies showed the ProLinkers efficiently orientated the antibodies in a site-directed manner,[173,174] and such strategy got even more orientated antibodies than crosslinker or protein G mediated immobilization strategies [175]. The mechanism of antibody orientation is not well understood; host-guest interactions, hydrophobic forces, and dipole-dipole interactions are suggested for the highly stable immobilization and proper orientation of antibodies.[173–175] Calixarene-mediated antibody immobilization has been adopted by many research groups for developing immunosensors based on all kinds of techniques, including SPR and LSPR,[175] ECL,[176] and electrochemical impedance spectroscopy [177].
3.6.2. Material binding peptide
Researchers have identified affinity peptides for certain materials, such as polystyrene (PS), polymethyl methacrylate (PMMA), gold, silver, and others using phage display and computer simulations.[178–180] These peptide ligands can be site-specifically fused to recombinant antibodies and thus orient the fusion proteins on the solid support.[181] For example, a polystyrene-binding peptide (PS-tag) fused VHH was constructed and used for ELISA,[42] where PS-Tag facilitated not only adsorption but also orientated adsorption on PS support via hydrophobic forces.[42,182–184] Besides PS-tag, PMMA- [185] and spider silk protein [186] tags have been reported as fusion proteins to recombinant antibodies for orientated immobilization on corresponding solid supports.
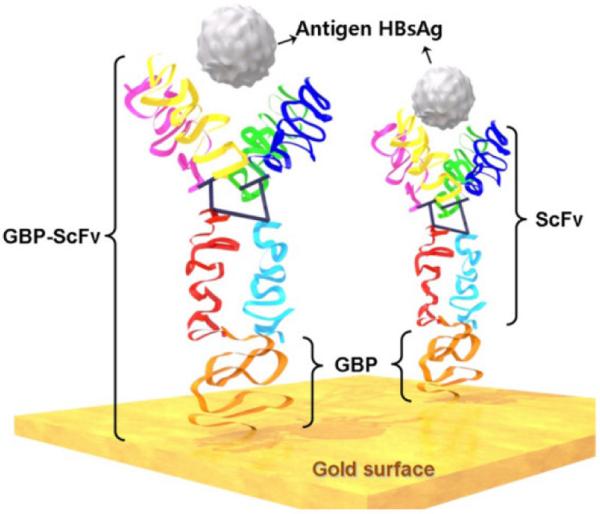
Researchers have reported polypeptides generated through combinatorial peptide selection that can bind to the metal surface, especially gold, with high affinity.[179,180] Interestingly, gold binding polypeptide (GBP) has its inherent binding chemistry as it carries no cysteine residue. Park and group have developed GBP- fused with single-chain antibody (ScFv) for detecting viral antigens (Figure 10).[187] Several other groups have also reported GBP-Ig fusion proteins for achieving highly orientated immobilization of antibodies.[150,151,188] Hattori and colleagues reported the development of GBP fused antibodies to achieve one-step antibody immobilization.[189] Moreover, antibodies that can specifically bind to gold surface have been developed.[190,191] These antibodies were further constructed as bispecific fragments consisting of fragments of two different monoclonal antibodies that can recognize gold and the analyte proteins.[192] Such bispecific antibodies can be immobilized on gold surface in one-step manner and exhibited an increased immunoreactivity to target analyte.
Figure 10.
Immobilization of scFv via fused gold-binding protein has a high affinity for the gold surface. GBPs have no sulfhydryls that may facilitate gold binding; however, it is mainly the affinity of the protein towards metal as described in ref [187]. Copyright (2012) MDPI.
3.6.3. Oligonucleotide-antibody conjugates
Oligonucleotide labelled antibodies can be immobilized onto a solid support pre-loaded with complimentary oligomers via highly specific Watson-Crick base pairing. The oligonucleotides are very stable and can be customized with different sequence and reactive groups. The oligonucleotide labelling strategies are usually non-selective.[193,194] However, the site-specificity during immobilization can be achieved via unnatural amino acid [195,196] or oligonucleotide labelled protein G [197]. The immobilization via oligonucleotide hybridization is simple but efficient approach and suitable for surface regeneration in a timely manner.[194] It also has possibility to simply convert an oligonucleotide array to an antibody array for high-throughput analysis.[198]
3.6.4. Unnatural amino acid
Incorporation of unnatural amino acids to proteins provides potential site-specific modification. Therefore, orientated protein immobilization could be expected via unique chemistries brought by unnatural amino acid.[199,200] As mentioned before, a 3-amino-L-tyrosine was incorporated to proteins for D-A reactions.[103,104] Introduction of a reactive azido group into the protein can be achieved by several aminoacid derivatives, such as azidohomoalanine,[114,120] p-azido-L-phenylalanine,[201] p-propargyloxyphenylalanine,[115] and others. In addition, antibody with site-specifically labelled oligonucleotides can be produced using p-acetylphenylalanine incorporated antibody and aminoxy-functionalized single-stranded oligonucleotide, which resulted in a stable oxime linkage.[195,196] It has been shown that few such amino acids, such as p-benzoyl-L-phenylalanine, can work as photo-reactive crosslinker for covalently coupling antibody and its affinity partner, where the photo-reactive crosslinker could be incorporated into either antibody or its affinity partner.[169,202]
3.6.5. Molecular imprinting
Molecular imprinting technique can offer artificial receptor sites for various molecules with antibody-like specificity.[203–207] Bereli and colleagues have developed MIPs with Fc fragment templated with poly(hydroxyethyl methacrylate) cryogel and used it for immobilizing anti-IgG in a site-directed fashion.[208] However, while immobilizing the template protein on the particle surface, it can still be randomly bound. This may result in an incorrect or poor affinity MIP. Corman et al. synthesized L-lysine imprinted nanoparticles and claimed for its ability to bind to the L-lysine molecules of the C-terminus on the antibodies.[209] L-lysine however doesn't represent a specific epitope and may introduce great variability. Therefore, Zhou group used borate-assisted molecular imprinting method targeting polysaccharide-containing Fc portion of the antibodies via boron-polysaccharide interactions.[210] They were able to capture anti HIV1 antibodies specifically via their carbohydrate regions, which were later detected via ECL.
3.6.7. Ig binding peptide
Combinatorial peptide selection can generate not only material-binding peptides, but also peptides having affinity for certain proteins, such as Fc region of IgG [211–213] and enzymes (namely horseradish peroxidase, alkaline phosphatase and β-Gal β-galactosidase) [214–216]. Jung et al. demonstrated that highly oriented layer of antibodies was obtained when immobilized on gold SPR chips coated with Fc-specific peptides.[211] They have shown that such peptides bound with human IgG1 and rabbit IgG with near nanomolar affinity. Orientation obtained with these peptides has significantly improved the antigen binding capacity with respect to the antibodies immobilized via EDC/NHS-mediated crosslinking.[211]
3.6.9. Carbohydrate-binding module
Carbohydrate-binding modules (CBMs) are protein domains derived from carbohydrate related enzymes, which exhibit their function for specific recognition of certain carbohydrate.[217,218] The most used CBMs are those that have affinity for cellulose, which can be used as affinity tags for purification and immobilization of peptides and proteins.[219–221] Ofir et al. prepared a series of scFv antibody fragments coupled with cellulose-binding domains and used them to construct a CBM-based microarray for HIV serodiagnosis.[222,223]
3.7. Conclusions and Perspective
On concluding remarks, in the past decade there has been tremendous progress on the front of orientated and site-specific antibody immobilization. However, there is a limited understanding of several of the methods and approaches employed for achieving orientation and site-specificity. Staphylococcal Ig-binding proteins, including protein A, G, L, and recombinant A/G, offer a significant orientation and are very simple to use, such that an untrained person can perform immobilization with these proteins. It is attributed to the five affinity domains present on each protein, which simply omits any special considerations regarding orientating these precapture proteins ahead of immobilizing the incoming antibodies. Nonetheless, the biggest challenge with staphylococcal precapture proteins is variable affinity of the binding domains. If a high affinity domain is involved in the interaction with the surface then immobilization will be sub-optimal.
On the contrary, highly site-specific strategies were developed that involve covalent chemical bonding. Biorthogonal chemistries, which are mainly cycloadditions at [4+2] and [3+2], are very straight forward like the Ig-binding proteins. However, these chemistries are superior to antibody-binding proteins due to their high site-specificity and covalent linkage that they offer. In spite, the degree of orientation obtained with these cycloadditions may not be as comparable as for Ig-binding proteins. Similarly, sugar chain-specific capture, affinity peptides, NBS-based immobilization, and other strategies offer unique advantages with respect to each other. There is not a single strategy which offers all the desired advantages, such as orientation, site-specificity, high package density on the surface, and preserved antibody activity.
In summary, obtaining an ideal antibody immobilization regimen is nearly impossible. However, a combination of approaches may be introduced, such as antibody orientation on covalently immobilized protein A or polysaccharide-based antibody capture stabilized with photoreactive crosslinking, and others. Multifunctional polymer coatings in tandem with synthetic or genetically engineered ligands for obtaining robust, site selective, orientated, high density, and homogeneous antibody immobilization should be the futuristic path, without affecting their functional activity.
Highlights.
Methods for site-selective orientated antibody immobilization are discussed, in particular to unconventional approaches
Focus on methods employing site-specific chemical tools for creating covalent interactions
Potential applications of the immobilized antibody systems are described
Acknowledgments
This work was supported financially by grant no. EB016707 and EB014586 from the National Institute of Biomedical Imaging and Bioengineering (NIBIB), NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu Y, Yu J. Oriented immobilization of proteins on solid supports for use in biosensors and biochips: a review. Microchim. Acta. 2015;183:1–19. doi:10.1007/s00604-015-1623-4. [Google Scholar]
- 2.Dong Y, Shannon C. Heterogeneous immunosensing using antigen and antibody monolayers on gold surfaces with electrochemical and scanning probe detection. Anal. Chem. 2000;72:2371–2376. doi: 10.1021/ac991450g. doi:10.1021/ac991450g. [DOI] [PubMed] [Google Scholar]
- 3.Chudasama V, Maruani A, Caddick S. Recent advances in the construction of antibody-drug conjugates. Nat. Chem. 2016;8:114–119. doi: 10.1038/nchem.2415. doi:10.1038/nchem.2415. [DOI] [PubMed] [Google Scholar]
- 4.Liu JKH. The history of monoclonal antibody development – Progress, remaining challenges and future innovations. Ann. Med. Surg. 2014;3:113–116. doi: 10.1016/j.amsu.2014.09.001. doi:10.1016/j.amsu.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salfeld JG. Isotype selection in antibody engineering. Nat. Biotechnol. 2007;25:1369–1372. doi: 10.1038/nbt1207-1369. doi:10.1038/nbt1207-1369. [DOI] [PubMed] [Google Scholar]
- 6.Maruani A, Smith MEB, Miranda E, Chester KA, Chudasama V, Caddick S. A plug-and-play approach to antibody-based therapeutics via a chemoselective dual click strategy. Nat. Commun. 2015;6:6645. doi: 10.1038/ncomms7645. doi:10.1038/ncomms7645 http://www.nature.com/articles/ncomms7645#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dixit CK, Vashist SK, O'Neill FT, O'Reilly B, MacCraith BD, O'Kennedy R. Development of a high sensitivity rapid sandwich ELISA procedure and its comparison with the conventional approach. Anal. Chem. 2010;82:7049–7052. doi: 10.1021/ac101339q. [DOI] [PubMed] [Google Scholar]
- 8.Dixit CK, Vashist SK, MacCraith BD, O'Kennedy R. Multisubstrate-compatible ELISA procedures for rapid and high-sensitivity immunoassays. Nat. Protoc. 2011;6:439–445. doi: 10.1038/nprot.2011.304. doi:10.1038/nprot.2011.304. [DOI] [PubMed] [Google Scholar]
- 9.Engels JF, Roose J, Zhai DS, Yip KM, Lee MS, Tang BZ, Renneberg R. Aggregation-induced emissive nanoparticles for fluorescence signaling in a low cost paper-based immunoassay. Colloids Surf. B Biointerfaces. 2016;143:440–446. doi: 10.1016/j.colsurfb.2016.03.051. doi:10.1016/j.colsurfb.2016.03.051. [DOI] [PubMed] [Google Scholar]
- 10.Vashist SK, Dixit CK, MacCraith BD, O'Kennedy R. Effect of antibody immobilization strategies on the analytical performance of a surface plasmon resonance-based immunoassay. The Analyst. 2011;136:4431–4436. doi: 10.1039/c1an15325k. doi:10.1039/c1an15325k. [DOI] [PubMed] [Google Scholar]
- 11.Mondani L, Delannoy S, Mathey R, Piat F, Mercey T, Slimani S, Fach P, Livache T, Roupioz Y. Fast detection of both O157 and non-O157 shiga-toxin producing Escherichia coli by real-time optical immunoassay. Lett. Appl. Microbiol. 2016;62:39–46. doi: 10.1111/lam.12503. doi:10.1111/lam.12503. [DOI] [PubMed] [Google Scholar]
- 12.Rusling JF. Multiplexed electrochemical protein detection and translation to personalized cancer diagnostics. Anal. Chem. 2013;85:5304–5310. doi: 10.1021/ac401058v. doi:10.1021/ac401058v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karmani L, Labar D, Valembois V, Bouchat V, Nagaswaran PG, Bol A, Gillart J, Levêque P, Bouzin C, Bonifazi D, Michiels C, Feron O, Grégoire V, Lucas S, Borght TV, Gallez B. Antibody-functionalized nanoparticles for imaging cancer: influence of conjugation to gold nanoparticles on the biodistribution of 89Zr-labeled cetuximab in mice. Contrast Media Mol. Imaging. 2013;8:402–408. doi: 10.1002/cmmi.1539. doi:10.1002/cmmi.1539. [DOI] [PubMed] [Google Scholar]
- 14.Abakumov MA, Nukolova NV, Sokolsky-Papkov M, Shein SA, Sandalova TO, Vishwasrao HM, Grinenko NF, Gubsky IL, Abakumov AM, Kabanov AV, Chekhonin VP. VEGF-targeted magnetic nanoparticles for MRI visualization of brain tumor. Nanomedicine Nanotechnol. Biol. Med. 2015;11:825–833. doi: 10.1016/j.nano.2014.12.011. doi: http://dx.doi.org/10.1016/j.nano.2014.12.011. [DOI] [PubMed] [Google Scholar]
- 15.Bates D, Abraham S, Campbell M, Zehbe I, Curiel L. Development and characterization of an antibody-labeled super-paramagnetic iron oxide contrast agent targeting prostate cancer cells for magnetic resonance imaging. PLOS ONE. 2014;9:e97220. doi: 10.1371/journal.pone.0097220. doi:10.1371/journal.pone.0097220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozlowska D, Biswas S, Fox EK, Wu B, Bolster F, Edupuganti OP, Torchilin V, Eustace S, Botta M, O'Kennedy R, Brougham DF. Gadolinium-loaded polychelating amphiphilic polymer as an enhanced MRI contrast agent for human multiple myeloma and non Hodgkin's lymphoma (human Burkitt's lymphoma) RSC Adv. 2014;4:18007–18016. doi:10.1039/C3RA45400B. [Google Scholar]
- 17.Michalet X, Pinaud FF, Bentolila LA, Tsay JM, Doose S, Li JJ, Sundaresan G, Wu AM, Gambhir SS, Weiss S. Quantum dots for live cells, in vivo imaging, and diagnostics. Science. 2005;307:538–544. doi: 10.1126/science.1104274. doi:10.1126/science.1104274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Yan X-P. Fabrication of vascular endothelial growth factor antibody bioconjugated ultrasmall near-infrared fluorescent Ag2S quantum dots for targeted cancer imaging in vivo. Chem. Commun. 2013;49:3324–3326. doi: 10.1039/c3cc41141a. doi:10.1039/C3CC41141A. [DOI] [PubMed] [Google Scholar]
- 19.Reichert JM. Marketed therapeutic antibodies compendium. mAbs. 2012;4:413–415. doi: 10.4161/mabs.19931. doi:10.4161/mabs.19931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Agarwal P, Bertozzi CR. Site-specific antibody–drug conjugates: The nexus of bioorthogonal chemistry, protein engineering, and drug development. Bioconjug. Chem. 2015;26:176–192. doi: 10.1021/bc5004982. doi:10.1021/bc5004982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qian C, Wang Y, Chen Y, Zeng L, Zhang Q, Shuai X, Huang K. Suppression of pancreatic tumor growth by targeted arsenic delivery with anti-CD44v6 single chain antibody conjugated nanoparticles. Biomaterials. 2013;34:6175–6184. doi: 10.1016/j.biomaterials.2013.04.056. doi: http://dx.doi.org/10.1016/j.biomaterials.2013.04.056. [DOI] [PubMed] [Google Scholar]
- 22.Goodall S, Howard CB, Jones ML, Munro T, Jia Z, Monteiro MJ, Mahler S. An EGFR targeting nanoparticle self assembled from a thermoresponsive polymer. J. Chem. Technol. Biotechnol. 2015;90:1222–1229. doi:10.1002/jctb.4509. [Google Scholar]
- 23.Matsunaga T, Takahashi M, Yoshino T, Kuhara M, Takeyama H. Magnetic separation of CD14+ cells using antibody binding with protein A expressed on bacterial magnetic particles for generating dendritic cells. Biochem. Biophys. Res. Commun. 2006;350:1019–1025. doi: 10.1016/j.bbrc.2006.09.145. doi:10.1016/j.bbrc.2006.09.145. [DOI] [PubMed] [Google Scholar]
- 24.Plouffe BD, Murthy SK, Lewis LH. Fundamentals and application of magnetic particles in cell isolation and enrichment. Rep. Prog. Phys. Phys. Soc. G. B. 2015;78:16601. doi: 10.1088/0034-4885/78/1/016601. doi:10.1088/0034-4885/78/1/016601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mejía-Manzano LA, González-Valdez J, Mayolo-Deloisa K, Escalante-Vázquez EJ, Rito-Palomares M. Covalent immobilization of antibodies for the preparation of immunoaffinity chromatographic supports. Sep. Sci. Technol. 2016;51:1736–1743. doi:10.1080/01496395.2016.1174264. [Google Scholar]
- 26.Liu J, Chen C-F, Chang C-W, DeVoe DL. Flow-through immunosensors using antibody-immobilized polymer monoliths. Biosens. Bioelectron. 2010;26:182–188. doi: 10.1016/j.bios.2010.06.007. doi:10.1016/j.bios.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mallik R, Hage DS, Affinity monolith chromatography J. Sep. Sci. 2006;29:1686–1704. doi: 10.1002/jssc.200600152. doi:10.1002/jssc.200600152. [DOI] [PubMed] [Google Scholar]
- 28.Jiang T, Mallik R, Hage DS. Affinity monoliths for ultrafast immunoextraction. Anal. Chem. 2005;77:2362–2372. doi: 10.1021/ac0483668. doi:10.1021/ac0483668. [DOI] [PubMed] [Google Scholar]
- 29.Moser AC, Hage DS. Immunoaffinity chromatography: an introduction to applications and recent developments. Bioanalysis. 2010;2:769–790. doi: 10.4155/bio.10.31. doi:10.4155/bio.10.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hennion M-C, Pichon V. Immuno-based sample preparation for trace analysis. J. Chromatogr. A. 2003;1000:29–52. doi: 10.1016/s0021-9673(03)00529-6. doi: http://dx.doi.org/10.1016/S0021-9673(03)00529-6. [DOI] [PubMed] [Google Scholar]
- 31.Springer TA. Curr. Protoc. Mol. Biol. John Wiley & Sons, Inc.; 2001. Immunoaffinity Chromatography. [DOI] [PubMed] [Google Scholar]
- 32.Trilling AK, Beekwilder J, Zuilhof H. Antibody orientation on biosensor surfaces: a minireview. The Analyst. 2013;138:1619–1627. doi: 10.1039/c2an36787d. doi:10.1039/c2an36787d. [DOI] [PubMed] [Google Scholar]
- 33.Zhao X, Pan F, Garcia-Gancedo L, Flewitt AJ, Ashley GM, Luo J, Lu JR. Interfacial recognition of human prostate-specific antigen by immobilized monoclonal antibody: effects of solution conditions and surface chemistry. J R Soc Interface. 2012;9:2457–67. doi: 10.1098/rsif.2012.0148. doi:10.1098/rsif.2012.0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen S, Liu L, Zhou J, Jiang S. Controlling antibody orientation on charged self-assembled monolayers. Langmuir. 2003;19:2859–2864. doi:10.1021/la026498v. [Google Scholar]
- 35.Zhou J, Tsao H-K, Sheng Y-J, Jiang S. Monte Carlo simulations of antibody adsorption and orientation on charged surfaces. J. Chem. Phys. 2004;121:1050–1057. doi: 10.1063/1.1757434. doi:doi: http://dx.doi.org/10.1063/1.1757434. [DOI] [PubMed] [Google Scholar]
- 36.Cooper CD, Clementi NC, Barba LA. Probing protein orientation near charged nanosurfaces for simulation-assisted biosensor design. J. Chem. Phys. 2015;143:124709. doi: 10.1063/1.4931113. doi:doi: http://dx.doi.org/10.1063/1.4931113. [DOI] [PubMed] [Google Scholar]
- 37.Wiseman ME, Frank CW. Antibody adsorption and orientation on hydrophobic surfaces. Langmuir. 2012;28:1765–1774. doi: 10.1021/la203095p. doi:10.1021/la203095p. [DOI] [PubMed] [Google Scholar]
- 38.Della Ventura B, Schiavo L, Altucci C, Esposito R, Velotta R. Light assisted antibody immobilization for bio-sensing. Biomed. Opt. Express. 2011;2:3223–3231. doi: 10.1364/BOE.2.003223. doi:10.1364/BOE.2.003223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Emaminejad S, Javanmard M, Gupta C, Chang S, Davis RW, Howe RT. Tunable control of antibody immobilization using electric field. Proc. Natl. Acad. Sci. U. S. A. 2015;112:1995–1999. doi: 10.1073/pnas.1424592112. doi:10.1073/pnas.1424592112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Um H-J, Kim M, Lee S-H, Min J, Kim H, Choi Y-W, Kim Y-H. Electrochemically oriented immobilization of antibody on poly-(2-cyano-ethylpyrrole)-coated gold electrode using a cyclic voltammetry. Talanta. 2011;84:330–334. doi: 10.1016/j.talanta.2011.01.013. doi:10.1016/j.talanta.2011.01.013. [DOI] [PubMed] [Google Scholar]
- 41.Zhou J, Chen S, Jiang S. Orientation of adsorbed antibodies on charged surfaces by computer simulation based on a united-residue model. Langmuir. 2003;19:3472–3478. doi:10.1021/la026871z. [Google Scholar]
- 42.Harmsen MM, Fijten HPD. Improved functional immobilization of llama single-domain antibody fragments to polystyrene surfaces using small peptides. J. Immunoassay Immunochem. 2012;33:234–251. doi: 10.1080/15321819.2011.634473. doi:10.1080/15321819.2011.634473. [DOI] [PubMed] [Google Scholar]
- 43.Tajima N, Takai M, Ishihara K. Significance of antibody orientation unraveled: well-oriented antibodies recorded high binding affinity. Anal. Chem. 2011;83:1969–1976. doi: 10.1021/ac1026786. doi:10.1021/ac1026786. [DOI] [PubMed] [Google Scholar]
- 44.Holstein CA, Chevalier A, Bennett S, Anderson CE, Keniston K, Olsen C, Li B, Bales B, Moore DR, Fu E, Baker D, Yager P. Immobilizing affinity proteins to nitrocellulose: a toolbox for paper-based assay developers. Anal. Bioanal. Chem. 2016;408:1335–1346. doi: 10.1007/s00216-015-9052-0. doi:10.1007/s00216-015-9052-0. [DOI] [PubMed] [Google Scholar]
- 45.Roy S, Dixit CK, Woolley R, O'Kennedy R, McDonagh C. Synthesis and characterization of a noble metal enhanced optical nanohybrid (neon): A high brightness detection platform based on a dye-doped silica nanoparticle. Langmuir. 2012;28:8244–8250. doi: 10.1021/la3016854. doi:10.1021/la3016854. [DOI] [PubMed] [Google Scholar]
- 46.Sung D, Yang S, Park JW, Jon S. High-density immobilization of antibodies onto nanobead-coated cyclic olefin copolymer plastic surfaces for application as a sensitive immunoassay chip. Biomed. Microdevices. 2012;15:691–698. doi: 10.1007/s10544-012-9732-x. doi:10.1007/s10544-012-9732-x. [DOI] [PubMed] [Google Scholar]
- 47.Zimmermann JL, Nicolaus T, Neuert G, Blank K. Thiol-based, site-specific and covalent immobilization of biomolecules for single-molecule experiments. Nat. Protoc. 2010;5:975–985. doi: 10.1038/nprot.2010.49. doi:10.1038/nprot.2010.49. [DOI] [PubMed] [Google Scholar]
- 48.Meier SM, Bomgarden R, Etienne C, Opperman K, Kaboord B. Improved antibody immobilization using aniline-catalyzed aldehyde-hydrazide chemistry for the study of protein-protein interactions. FASEB J. 2012;26:776.9–776.9. [Google Scholar]
- 49.Adak AK, Li B-Y, Huang L-D, Lin T-W, Chang T-C, Hwang KC, Lin C-C. Fabrication of antibody microarrays by light-induced covalent and oriented immobilization. ACS Appl. Mater. Interfaces. 2014;6:10452–10460. doi: 10.1021/am502011r. doi:10.1021/am502011r. [DOI] [PubMed] [Google Scholar]
- 50.Hu W, Liu Y, Chen T, Liu Y, Li CM. Hybrid ZnO Nanorod-Polymer Brush Hierarchically Nanostructured Substrate for Sensitive Antibody Microarrays. Adv. Mater. 2015;27:181–185. doi: 10.1002/adma.201403712. doi:10.1002/adma.201403712. [DOI] [PubMed] [Google Scholar]
- 51.E Hemaprabha. Chemical crosslinking of proteins: a review. J. Pharm. Sci. Innov. 2012 [Google Scholar]
- 52.Liu H, May K. Disulfide bond structures of IgG molecules. mAbs. 2012;4:17–23. doi: 10.4161/mabs.4.1.18347. doi:10.4161/mabs.4.1.18347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Makaraviciute A, Ramanaviciene A. Site-directed antibody immobilization techniques for immunosensors. Biosens. Bioelectron. 2013;50:460–471. doi: 10.1016/j.bios.2013.06.060. doi:10.1016/j.bios.2013.06.060. [DOI] [PubMed] [Google Scholar]
- 54.Huy TQ, Hanh NTH, Van Chung P, Anh DD, Nga PT, Tuan MA. Characterization of immobilization methods of antiviral antibodies in serum for electrochemical biosensors. Appl. Surf. Sci. 2011;257:7090–7095. doi: http://dx.doi.org/10.1016/j.apsusc.2011.03.051. [Google Scholar]
- 55.Pei Z, Anderson H, Myrskog A, Dunér G, Ingemarsson B, Aastrup T. Optimizing immobilization on two-dimensional carboxyl surface: pH dependence of antibody orientation and antigen binding capacity. Anal. Biochem. 2010;398:161–168. doi: 10.1016/j.ab.2009.11.038. doi: http://dx.doi.org/10.1016/j.ab.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 56.Yuan X, Fabregat D, Yoshimoto K, Nagasaki Y. Development of a high-performance immunolatex based on “soft landing” antibody immobilization mechanism. Colloids Surf. B Biointerfaces. 2012;99:45–52. doi: 10.1016/j.colsurfb.2011.09.040. doi: http://dx.doi.org/10.1016/j.colsurfb.2011.09.040. [DOI] [PubMed] [Google Scholar]
- 57.Wei Z, Feng J, Lin H-Y, Mullapudi S, Bishop E, Tous GI, Casas-Finet J, Hakki F, Strouse R, Schenerman MA. Identification of a single tryptophan residue as critical for binding activity in a humanized monoclonal antibody against respiratory syncytial virus. Anal. Chem. 2007;79:2797–2805. doi: 10.1021/ac062311j. doi:10.1021/ac062311j. [DOI] [PubMed] [Google Scholar]
- 58.Ji JA, Zhang B, Cheng W, Wang YJ. Methionine, tryptophan, and histidine oxidation in a model protein, PTH: Mechanisms and stabilization. J. Pharm. Sci. 2009;98:4485–4500. doi: 10.1002/jps.21746. doi:10.1002/jps.21746. [DOI] [PubMed] [Google Scholar]
- 59.Bertolotti-Ciarlet A, Wang W, Lownes R, Pristatsky P, Fang Y, McKelvey T, Li Y, Li Y, Drummond J, Prueksaritanont T, Vlasak J. Impact of methionine oxidation on the binding of human IgG1 to FcRn and Fc receptors. Mol. Immunol. 2009;46:1878–1882. doi: 10.1016/j.molimm.2009.02.002. doi: http://dx.doi.org/10.1016/j.molimm.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 60.Dixit CK, Kumar A, Kaushik A. Nanosphere lithography-based platform for developing rapid and high sensitivity microarray systems. Biochem. Biophys. Res. Commun. 2012;423:473–477. doi: 10.1016/j.bbrc.2012.05.144. doi:10.1016/j.bbrc.2012.05.144. [DOI] [PubMed] [Google Scholar]
- 61.Yoshimoto K, Nishio M, Sugasawa H, Nagasaki Y. Direct observation of adsorption-induced inactivation of antibody fragments surrounded by mixed-peg layer on a gold surface. J. Am. Chem. Soc. 2010;132:7982–7989. doi: 10.1021/ja910372e. doi:10.1021/ja910372e. [DOI] [PubMed] [Google Scholar]
- 62.Kausaite-Minkstimiene A, Ramanaviciene A, Kirlyte J, Ramanavicius A. Comparative study of random and oriented antibody immobilization techniques on the binding capacity of immunosensor. Anal. Chem. 2010;82:6401–6408. doi: 10.1021/ac100468k. doi:10.1021/ac100468k. [DOI] [PubMed] [Google Scholar]
- 63.Ho JA, Hsu W-L, Liao W-C, Chiu J-K, Chen M-L, Chang H-C, Li C-C. Ultrasensitive electrochemical detection of biotin using electrically addressable site-oriented antibody immobilization approach via aminophenyl boronic acid. Biosens. Bioelectron. 2010;26:1021–1027. doi: 10.1016/j.bios.2010.08.048. doi: http://dx.doi.org/10.1016/j.bios.2010.08.048. [DOI] [PubMed] [Google Scholar]
- 64.Balevicius Z, Ramanaviciene A, Baleviciute I, Makaraviciute A, Mikoliunaite L, Ramanavicius A. Evaluation of intact- and fragmented-antibody based immunosensors by total internal reflection ellipsometry. Sens. Actuators B Chem. 2011;160:555–562. doi: http://dx.doi.org/10.1016/j.snb.2011.08.029. [Google Scholar]
- 65.Billah MM, Hodges CS, Hays HCW, Millner PA. Directed immobilization of reduced antibody fragments onto a novel SAM on gold for myoglobin impedance immunosensing. Bioelectrochemistry. 2010;80:49–54. doi: 10.1016/j.bioelechem.2010.08.005. doi: http://dx.doi.org/10.1016/j.bioelechem.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 66.Iwasaki Y, Omichi Y, Iwata R. Site-specific dense immobilization of antibody fragments on polymer brushes supported by silicone nanofilaments. Langmuir. 2008;24:8427–8430. doi: 10.1021/la801327a. doi:10.1021/la801327a. [DOI] [PubMed] [Google Scholar]
- 67.Bonroy K, Frederix F, Reekmans G, Dewolf E, De Palma R, Borghs G, Declerck P, Goddeeris B. Comparison of random and oriented immobilisation of antibody fragments on mixed self-assembled monolayers. J. Immunol. Methods. 2006;312:167–181. doi: 10.1016/j.jim.2006.03.007. doi: http://dx.doi.org/10.1016/j.jim.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 68.Vikholm-Lundin I, Albers WM. Site-directed immobilisation of antibody fragments for detection of C-reactive protein. Biosens. Bioelectron. 2006;21:1141–1148. doi: 10.1016/j.bios.2005.04.011. doi: http://dx.doi.org/10.1016/j.bios.2005.04.011. [DOI] [PubMed] [Google Scholar]
- 69.Sharma H, Mutharasan R. Half antibody fragments improve biosensor sensitivity without loss of selectivity. Anal. Chem. 2013;85:2472–2477. doi: 10.1021/ac3035426. doi:10.1021/ac3035426. [DOI] [PubMed] [Google Scholar]
- 70.Dixit CK, Kaushik A. Nano-structured arrays for multiplex analyses and Lab-on-a-Chip applications. Biochem. Biophys. Res. Commun. 2012;419:316–320. doi: 10.1016/j.bbrc.2012.02.018. [DOI] [PubMed] [Google Scholar]
- 71.Baio JE, Cheng F, Ratner DM, Stayton PS, Castner DG. Probing orientation of immobilized humanized anti-lysozyme variable fragment by time-of-flight secondary-ion mass spectrometry. J. Biomed. Mater. Res. A. 2011;97A:1–7. doi: 10.1002/jbm.a.33025. doi:10.1002/jbm.a.33025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hortiguela MJ, Wall JG. Improved detection of domoic acid using covalently immobilised antibody fragments. Mar Drugs. 2013;11:881–95. doi: 10.3390/md11030881. doi:10.3390/md11030881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hortigüela MJ, Aumailley L, Srivastava A, Cunningham C, Anandakumar S, Robin S, Pandit A, Hu X, Wall JG. Engineering recombinant antibodies for polymer biofunctionalization. Polym. Adv. Technol. 2015;26:1394–1401. doi:10.1002/pat.3619. [Google Scholar]
- 74.Bhakta S, Raab H, Junutula JR. Engineering THIOMABs for site-specific conjugation of thiol-reactive linkers. In: Ducry L, editor. Antib.-Drug Conjug. Humana Press; Totowa, NJ: 2013. pp. 189–203. [DOI] [PubMed] [Google Scholar]
- 75.Junutula JR, Raab H, Clark S, Bhakta S, Leipold DD, Weir S, Chen Y, Simpson M, Tsai SP, Dennis MS, Lu Y, Meng YG, Ng C, Yang J, Lee CC, Duenas E, Gorrell J, Katta V, Kim A, McDorman K, Flagella K, Venook R, Ross S, Spencer SD, Lee Wong W, Lowman HB, Vandlen R, Sliwkowski MX, Scheller RH, Polakis P, Mallet W. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat Biotech. 2008;26:925–932. doi: 10.1038/nbt.1480. doi: http://www.nature.com/nbt/journal/v26/n8/suppinfo/nbt.1480_S1.html. [DOI] [PubMed] [Google Scholar]
- 76.McDonagh CF, Turcott E, Westendorf L, Webster JB, Alley SC, Kim K, Andreyka J, Stone I, Hamblett KJ, Francisco JA, Carter P. Engineered antibody-drug conjugates with defined sites and stoichiometries of drug attachment. Protein Eng Sel. 2006;19:299–307. doi: 10.1093/protein/gzl013. doi:10.1093/protein/gzl013. [DOI] [PubMed] [Google Scholar]
- 77.Badescu G, Bryant P, Bird M, Henseleit K, Swierkosz J, Parekh V, Tommasi R, Pawlisz E, Jurlewicz K, Farys M, Camper N, Sheng X, Fisher M, Grygorash R, Kyle A, Abhilash A, Frigerio M, Edwards J, Godwin A. Bridging disulfides for stable and defined antibody drug conjugates. Bioconjug. Chem. 2014;25:1124–1136. doi: 10.1021/bc500148x. doi:10.1021/bc500148x. [DOI] [PubMed] [Google Scholar]
- 78.Schumacher FF, Nunes JPM, Maruani A, Chudasama V, Smith MEB, Chester KA, Baker JR, Caddick S. Next generation maleimides enable the controlled assembly of antibody-drug conjugates via native disulfide bond bridging. Org. Biomol. Chem. 2014;12:7261–7269. doi: 10.1039/c4ob01550a. doi:10.1039/C4OB01550A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hofer T, Thomas JD, Burke TR, Rader C. An engineered selenocysteine defines a unique class of antibody derivatives. Proc. Natl. Acad. Sci. 2008;105:12451–12456. doi: 10.1073/pnas.0800800105. doi:10.1073/pnas.0800800105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fischer-Durand N, Salmain M, Rudolf B, Dai L, Jugé L, Guérineau V, Laprévote O, Vessières A, Jaouen G. Site-specific conjugation of metal carbonyl dendrimer to antibody and its use as detection reagent in immunoassay. Anal. Biochem. 2010;407:211–219. doi: 10.1016/j.ab.2010.08.027. doi: http://dx.doi.org/10.1016/j.ab.2010.08.027. [DOI] [PubMed] [Google Scholar]
- 81.Yeritsyan HE, Gasparyan VK. Homogeneous immunoassay for human IgG using oriented hen egg IgY immobilized on gold sol nanoparticles. Microchim. Acta. 2012;176:117–122. doi:10.1007/s00604-011-0703-3. [Google Scholar]
- 82.Hu X, Hortigüela MJ, Robin S, Lin H, Li Y, Moran AP, Wang W, Wall JG. Covalent and oriented immobilization of scFv antibody fragments via an engineered glycan moiety. Biomacromolecules. 2013;14:153–159. doi: 10.1021/bm301518p. doi:10.1021/bm301518p. [DOI] [PubMed] [Google Scholar]
- 83.Prieto-Simón B, Saint C, Voelcker NH. Electrochemical biosensors featuring oriented antibody immobilization via electrografted and self-assembled hydrazide chemistry. Anal. Chem. 2014;86:1422–1429. doi: 10.1021/ac401747j. doi:10.1021/ac401747j. [DOI] [PubMed] [Google Scholar]
- 84.Han HJ, Kannan RM, Wang S, Mao G, Kusanovic JP, Romero R. Multifunctional dendrimer-templated antibody presentation on biosensor surfaces for improved biomarker detection. Adv. Funct. Mater. 2010;20:409–421. doi: 10.1002/adfm.200901293. doi:10.1002/adfm.200901293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang X, Xia N, Liu L. Boronic Acid-based approach for separation and immobilization of glycoproteins and its application in sensing. Int J Mol Sci. 2013;14:20890–912. doi: 10.3390/ijms141020890. doi:10.3390/ijms141020890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lacina K, Skládal P, James TD. Boronic acids for sensing and other applications - a mini-review of papers published in 2013. Chem. Cent. J. 2014;8:1–17. doi: 10.1186/s13065-014-0060-5. doi:10.1186/s13065-014-0060-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Moreno-Guzmán M, Ojeda I, Villalonga R, González-Cortés A, Yáñez-Sedeño P, Pingarrón JM. Ultrasensitive detection of adrenocorticotropin hormone (ACTH) using disposable phenylboronic-modified electrochemical immunosensors. Biosens. Bioelectron. 2012;35:82–86. doi: 10.1016/j.bios.2012.02.015. doi: http://dx.doi.org/10.1016/j.bios.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 88.Zhang X, Wu Y, Tu Y, Liu S. A reusable electrochemical immunosensor for carcinoembryonic antigenvia molecular recognition of glycoproteinantibody by phenylboronic acid self-assembly layer on gold. Analyst. 2008;133:485–492. doi: 10.1039/b714896h. doi:10.1039/B714896H. [DOI] [PubMed] [Google Scholar]
- 89.Henderson GE, Isett KD, Gerngross TU. Site-specific modification of recombinant proteins: a novel platform for modifying glycoproteins expressed in E. coli. Bioconjug. Chem. 2011;22:903–912. doi: 10.1021/bc100510g. doi:10.1021/bc100510g. [DOI] [PubMed] [Google Scholar]
- 90.Solomon B, Koppel R, Schwartz F, Fleminger G. Enzymic oxidation of monoclonal antibodies by soluble and immobilized bifunctional enzyme complexes. J. Chromatogr. A. 1990;510:321–329. doi: 10.1016/s0021-9673(01)93766-5. doi: http://dx.doi.org/10.1016/S0021-9673(01)93766-5. [DOI] [PubMed] [Google Scholar]
- 91.Zhou Q, Stefano JE, Manning C, Kyazike J, Chen B, Gianolio DA, Park A, Busch M, Bird J, Zheng X, Simonds-Mannes H, Kim J, Gregory RC, Miller RJ, Brondyk WH, Dhal PK, Pan CQ. Site-specific antibody–drug conjugation through glycoengineering. Bioconjug. Chem. 2014;25:510–520. doi: 10.1021/bc400505q. doi:10.1021/bc400505q. [DOI] [PubMed] [Google Scholar]
- 92.Okeley NM, Toki BE, Zhang X, Jeffrey SC, Burke PJ, Alley SC, Senter PD. Metabolic engineering of monoclonal antibody carbohydrates for antibody–drug conjugation. Bioconjug. Chem. 2013;24:1650–1655. doi: 10.1021/bc4002695. doi:10.1021/bc4002695. [DOI] [PubMed] [Google Scholar]
- 93.Rajagopalan K, Pavlinkova G, Levy S, Pokkuluri PR, Schiffer M, Haley BE, Kohler H. Novel unconventional binding site in the variable region of immunoglobulins. Proc Natl Acad Sci U A. 1996;93:6019–24. doi: 10.1073/pnas.93.12.6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Handlogten MW, Kiziltepe T, Moustakas DT, Bilgiçer B. Design of a Heterobivalent Ligand to Inhibit IgE Clustering on Mast Cells. Chem. Biol. 2011;18:1179–1188. doi: 10.1016/j.chembiol.2011.06.012. doi: http://dx.doi.org/10.1016/j.chembiol.2011.06.012. [DOI] [PubMed] [Google Scholar]
- 95.Alves NJ, Kiziltepe T, Bilgicer B. Oriented surface immobilization of antibodies at the conserved nucleotide binding site for enhanced antigen detection. Langmuir ACS J. Surf. Colloids. 2012;28:9640–9648. doi: 10.1021/la301887s. doi:10.1021/la301887s. [DOI] [PubMed] [Google Scholar]
- 96.Alves NJ, Mustafaoglu N, Bilgicer B. Oriented antibody immobilization by site-specific UV photocrosslinking of biotin at the conserved nucleotide binding site for enhanced antigen detection. Biosens. Bioelectron. 2013;49:387–393. doi: 10.1016/j.bios.2013.05.052. doi:10.1016/j.bios.2013.05.052. [DOI] [PubMed] [Google Scholar]
- 97.Mustafaoglu N, Alves NJ, Bilgicer B. Site-specific fab fragment biotinylation at the conserved nucleotide binding site for enhanced ebola detection. Biotechnol. Bioeng. 2015;112:1327–1334. doi: 10.1002/bit.25558. doi:10.1002/bit.25558. [DOI] [PubMed] [Google Scholar]
- 98.Lin C-P, Saito K, Boysen RI, Campi EM, Hearn MTW. Static and dynamic binding behavior of an IgG2 monoclonal antibody with several new mixed mode affinity adsorbents. Sep. Purif. Technol. 2016;163:199–205. doi: http://dx.doi.org/10.1016/j.seppur.2016.02.048. [Google Scholar]
- 99.Mustafaoglu N, Alves NJ, Bilgicer B. Oriented immobilization of Fab fragments by site-specific biotinylation at the conserved nucleotide binding site for enhanced antigen detection. Langmuir. 2015;31:9728–9736. doi: 10.1021/acs.langmuir.5b01734. doi:10.1021/acs.langmuir.5b01734. [DOI] [PubMed] [Google Scholar]
- 100.Pozsgay V, Vieira NE, Yergey A. A method for bioconjugation of carbohydrates using diels−alder cycloaddition. Org. Lett. 2002;4:3191–3194. doi: 10.1021/ol026179v. doi:10.1021/ol026179v. [DOI] [PubMed] [Google Scholar]
- 101.Rideout DC, Breslow R. Hydrophobic acceleration of Diels-Alder reactions. J. Am. Chem. Soc. 1980;102:7816–7817. doi:10.1021/ja00546a048. [Google Scholar]
- 102.Shi M, Wosnick JH, Ho K, Keating A, Shoichet MS. Immuno-polymeric nanoparticles by Diels–Alder chemistry. Angew. Chem. Int. Ed. 2007;46:6126–6131. doi: 10.1002/anie.200701032. doi:10.1002/anie.200701032. [DOI] [PubMed] [Google Scholar]
- 103.Stamos B, Loredo L, Chand S, Phan TV, Zhang Y, Mohapatra S, Rajeshwar K, Perera R. Biosynthetic approach for functional protein microarrays. Anal. Biochem. 2012;424:114–123. doi: 10.1016/j.ab.2012.02.019. doi: http://dx.doi.org/10.1016/j.ab.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 104.Ray S, Chand S, Zhang Y, Nussbaum S, Rajeshwar K, Perera R. Implications of active site orientation in myoglobin for direct electron transfer and electrocatalysis based on monolayer and multilayer covalent immobilization on gold electrodes. Electrochimica Acta. 2013;99:85–93. doi: http://dx.doi.org/10.1016/j.electacta.2013.03.080. [Google Scholar]
- 105.Saxon E, Bertozzi CR. Cell surface engineering by a modified staudinger reaction. Science. 2000;287:2007–2010. doi: 10.1126/science.287.5460.2007. doi:10.1126/science.287.5460.2007. [DOI] [PubMed] [Google Scholar]
- 106.Köhn M, Breinbauer R. the Staudinger ligation—A gift to chemical biology. Angew. Chem. Int. Ed. 2004;43:3106–3116. doi: 10.1002/anie.200401744. doi:10.1002/anie.200401744. [DOI] [PubMed] [Google Scholar]
- 107.van Berkel SS, van Eldijk MB, van Hest JCM. Staudinger ligation as a method for bioconjugation. Angew. Chem. Int. Ed. 2011;50:8806–8827. doi: 10.1002/anie.201008102. doi:10.1002/anie.201008102. [DOI] [PubMed] [Google Scholar]
- 108.Schilling CI, Jung N, Biskup M, Schepers U, Brase S. Bioconjugation viaazide-Staudinger ligation: an overview. Chem. Soc. Rev. 2011;40:4840–4871. doi: 10.1039/c0cs00123f. doi:10.1039/C0CS00123F. [DOI] [PubMed] [Google Scholar]
- 109.Soellner MB, Dickson KA, Nilsson BL, Raines RT. Site-specific protein immobilization by staudinger ligation. J. Am. Chem. Soc. 2003;125:11790–11791. doi: 10.1021/ja036712h. doi:10.1021/ja036712h. [DOI] [PubMed] [Google Scholar]
- 110.Köhn M, Wacker R, Peters C, Schröder H, Soulère L, Breinbauer R, Niemeyer CM, Waldmann H. Staudinger ligation: A new immobilization strategy for the preparation of small-molecule arrays. Angew. Chem. Int. Ed. 2003;42:5830–5834. doi: 10.1002/anie.200352877. doi:10.1002/anie.200352877. [DOI] [PubMed] [Google Scholar]
- 111.Köhn M, Gutierrez-Rodriguez M, Jonkheijm P, Wetzel S, Wacker R, Schroeder H, Prinz H, Niemeyer CM, Breinbauer R, Szedlacsek SE, Waldmann H. A microarray strategy for mapping the substrate specificity of protein tyrosine phosphatase. Angew. Chem. Int. Ed. 2007;46:7700–7703. doi: 10.1002/anie.200701601. doi:10.1002/anie.200701601. [DOI] [PubMed] [Google Scholar]
- 112.Kalia J, Abbott NL, Raines RT. General method for site-specific protein immobilization by Staudinger ligation. Bioconjug. Chem. 2007;18:1064–1069. doi: 10.1021/bc0603034. doi:10.1021/bc0603034. [DOI] [PubMed] [Google Scholar]
- 113.Köhn M. Immobilization strategies for small molecule, peptide and protein microarrays. J. Pept. Sci. 2009;15:393–397. doi: 10.1002/psc.1130. doi:10.1002/psc.1130. [DOI] [PubMed] [Google Scholar]
- 114.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proc. Natl. Acad. Sci. 2002;99:19–24. doi: 10.1073/pnas.012583299. doi:10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Bundy BC, Swartz JR. Site-specific incorporation of p-propargyloxyphenylalanine in a cell-free environment for direct protein—protein click conjugation. Bioconjug. Chem. 2010;21:255–263. doi: 10.1021/bc9002844. doi:10.1021/bc9002844. [DOI] [PubMed] [Google Scholar]
- 116.Kolb HC, Finn MG, Sharpless KB. Click chemistry: Diverse chemical function from a few good reactions. Angew Chem Int Ed Engl. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 117.Finetti C, Sola L, Pezzullo M, Prosperi D, Colombo M, Riva B, Avvakumova S, Morasso C, Picciolini S, Chiari M. Click chemistry immobilization of antibodies on polymer coated gold nanoparticles. Langmuir. 2016;32:7435–7441. doi: 10.1021/acs.langmuir.6b01142. doi:10.1021/acs.langmuir.6b01142. [DOI] [PubMed] [Google Scholar]
- 118.Jewett JC, Bertozzi CR. Cu-free click cycloaddition reactions in chemical biology. Chem. Soc. Rev. 2010;39:1272–1279. doi: 10.1039/b901970g. doi:10.1039/B901970G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Debets MF, van Berkel SS, Dommerholt J, Dirks AJ, Rutjes FPJT, van Delft FL. Bioconjugation with strained alkenes and alkynes. Acc. Chem. Res. 2011;44:805–815. doi: 10.1021/ar200059z. doi:10.1021/ar200059z. [DOI] [PubMed] [Google Scholar]
- 120.Trilling AK, Hesselink T, van Houwelingen A, Cordewener JHG, Jongsma MA, Schoffelen S, van Hest JCM, Zuilhof H, Beekwilder J. Orientation of llama antibodies strongly increases sensitivity of biosensors. Biosens. Bioelectron. 2014;60:130–136. doi: 10.1016/j.bios.2014.04.017. doi: http://dx.doi.org/10.1016/j.bios.2014.04.017. [DOI] [PubMed] [Google Scholar]
- 121.Cho I-H, Paek E-H, Lee H, Kang JY, Kim TS, Paek S-H. Site-directed biotinylation of antibodies for controlled immobilization on solid surfaces. Anal. Biochem. 2007;365:14–23. doi: 10.1016/j.ab.2007.02.028. doi: http://dx.doi.org/10.1016/j.ab.2007.02.028. [DOI] [PubMed] [Google Scholar]
- 122.Franco EJ, Hofstetter H, Hofstetter O. A comparative evaluation of random and site-specific immobilization techniques for the preparation of antibody-based chiral stationary phases. J. Sep. Sci. 2006;29:1458–1469. doi: 10.1002/jssc.200600062. doi:10.1002/jssc.200600062. [DOI] [PubMed] [Google Scholar]
- 123.Trilling AK, Harmsen MM, Ruigrok VJB, Zuilhof H, Beekwilder J. The effect of uniform capture molecule orientation on biosensor sensitivity: Dependence on analyte properties. Biosens. Bioelectron. 2013;40:219–226. doi: 10.1016/j.bios.2012.07.027. doi: http://dx.doi.org/10.1016/j.bios.2012.07.027. [DOI] [PubMed] [Google Scholar]
- 124.Park J-W, Cho I-H, Moon DW, Paek S-H, Lee TG. ToF-SIMS and PCA of surface-immobilized antibodies with different orientations. Surf. Interface Anal. 2011;43:285–289. doi:10.1002/sia.3440. [Google Scholar]
- 125.Cho I-H, Park J-W, Lee TG, Lee H, Paek S-H. Biophysical characterization of the molecular orientation of an antibody-immobilized layer using secondary ion mass spectrometry. Analyst. 2011;136:1412–1419. doi: 10.1039/c0an00672f. doi:10.1039/C0AN00672F. [DOI] [PubMed] [Google Scholar]
- 126.Lata S, Piehler J. Stable and functional immobilization of histidine-tagged proteins via multivalent chelator headgroups on a molecular poly(ethylene glycol) brush. Anal. Chem. 2005;77:1096–1105. doi: 10.1021/ac048813j. doi:10.1021/ac048813j. [DOI] [PubMed] [Google Scholar]
- 127.Bhagawati M, Lata S, Tampé R, Piehler J. Native laser lithography of his-tagged proteins by uncaging of multivalent chelators. J. Am. Chem. Soc. 2010;132:5932–5933. doi: 10.1021/ja1000714. doi:10.1021/ja1000714. [DOI] [PubMed] [Google Scholar]
- 128.Khan F, He M, Taussig MJ. Double-hexahistidine tag with high-affinity binding for protein immobilization, purification, and detection on ni–nitrilotriacetic acid surfaces. Anal. Chem. 2006;78:3072–3079. doi: 10.1021/ac060184l. doi:10.1021/ac060184l. [DOI] [PubMed] [Google Scholar]
- 129.Steinhauer C, Wingren C, Khan F, He M, Taussig MJ, Borrebaeck CAK. Improved affinity coupling for antibody microarrays: Engineering of double-(His)6-tagged single framework recombinant antibody fragments. Proteomics. 2006;6:4227–4234. doi: 10.1002/pmic.200600036. doi:10.1002/pmic.200600036. [DOI] [PubMed] [Google Scholar]
- 130.Kröger D, Liley M, Schiweck W, Skerra A, Vogel H. Immobilization of histidine-tagged proteins on gold surfaces using chelator thioalkanes. Biosens. Bioelectron. 1999;14:155–161. doi: 10.1016/s0956-5663(98)00116-x. doi:10.1016/S0956-5663(98)00116-X. [DOI] [PubMed] [Google Scholar]
- 131.Kumar A, Wahlund P-O, Kepka C, Galaev IY, Mattiasson B. Purification of histidine-tagged single-chain Fv-antibody fragments by metal chelate affinity precipitation using thermoresponsive copolymers. Biotechnol. Bioeng. 2003;84:494–503. doi: 10.1002/bit.10810. doi:10.1002/bit.10810. [DOI] [PubMed] [Google Scholar]
- 132.Sun X, Yu G, Xu Q, Li N, Xiao C, Yin X, Cao K, Han J, He Q-Y. Putative cobalt-and nickel-binding proteins and motifs in Streptococcus pneumoniae. Metallomics. 2013;5:928–935. doi: 10.1039/c3mt00126a. doi:10.1039/C3MT00126A. [DOI] [PubMed] [Google Scholar]
- 133.Ericsson EM, Enander K, Bui L, Lundstrom I, Konradsson P, Liedberg B. Site-specific and covalent attachment of his-tagged proteins by chelation assisted photoimmobilization: a strategy for microarraying of protein ligands. Langmuir. 2013;29:11687–94. doi: 10.1021/la4011778. doi:10.1021/la4011778. [DOI] [PubMed] [Google Scholar]
- 134.Dixit CK, Ó'Fágáin C, O'Kennedy R. Nanobiotechnology Sens. Appl. Lab Field, I. Apple Academic Press Inc.; Waretown, NJ, USA: Antibody immobilization chemistries for nanosurfaces; pp. 65–100. n.d. [Google Scholar]
- 135.Kwon Y, Han Z, Karatan E, Mrksich M, Kay BK. Antibody arrays prepared by cutinase-mediated immobilization on self-assembled monolayers. Anal. Chem. 2004;76:5713–5720. doi: 10.1021/ac049731y. doi:10.1021/ac049731y. [DOI] [PubMed] [Google Scholar]
- 136.La Clair JJ, Foley TL, Schegg TR, Regan CM, Burkart MD. Manipulation of carrier proteins in antibiotic biosynthesis. Chem. Biol. 2004;11:195–201. doi: 10.1016/j.chembiol.2004.02.010. doi: http://dx.doi.org/10.1016/j.chembiol.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 137.Heck T, Faccio G, Richter M, Thöny-Meyer L. Enzyme-catalyzed protein crosslinking. Appl. Microbiol. Biotechnol. 2013;97:461–475. doi: 10.1007/s00253-012-4569-z. doi:10.1007/s00253-012-4569-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Walper SA, Turner KB, Medintz IL. Enzymatic bioconjugation of nanoparticles: developing specificity and control. Curr. Opin. Biotechnol. 2015;34:232–241. doi: 10.1016/j.copbio.2015.04.003. doi: http://dx.doi.org/10.1016/j.copbio.2015.04.003. [DOI] [PubMed] [Google Scholar]
- 139.Gray CJ, Weissenborn MJ, Eyers CE, Flitsch SL. Enzymatic reactions on immobilised substrates. Chem. Soc. Rev. 2013;42:6378–6405. doi: 10.1039/c3cs60018a. doi:10.1039/C3CS60018A. [DOI] [PubMed] [Google Scholar]
- 140.Ritzefeld M. Sortagging: A robust and efficient chemoenzymatic ligation strategy. Chem. – Eur. J. 2014;20:8516–8529. doi: 10.1002/chem.201402072. doi:10.1002/chem.201402072. [DOI] [PubMed] [Google Scholar]
- 141.Tsukiji S, Nagamune T. Sortase-mediated ligation: a gift from gram-positive bacteria to protein engineering. ChemBioChem. 2009;10:787–798. doi: 10.1002/cbic.200800724. doi:10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- 142.Paterson BM, Alt K, Jeffery CM, Price RI, Jagdale S, Rigby S, Williams CC, Peter K, Hagemeyer CE, Donnelly PS. Enzyme-mediated site-specific bioconjugation of metal complexes to proteins: sortase-mediated coupling of copper-64 to a single-chain antibody. Angew. Chem. Int. Ed. 2014;53:6115–6119. doi: 10.1002/anie.201402613. doi:10.1002/anie.201402613. [DOI] [PubMed] [Google Scholar]
- 143.Massa S, Vikani N, Betti C, Ballet S, Vanderhaegen S, Steyaert J, Descamps B, Vanhove C, Bunschoten A, van Leeuwen FWB, Hernot S, Caveliers V, Lahoutte T, Muyldermans S, Xavier C, Devoogdt N. Sortase A-mediated site-specific labeling of camelid single-domain antibody-fragments: a versatile strategy for multiple molecular imaging modalities. Contrast Media Mol. Imaging. 2016 doi: 10.1002/cmmi.1696. n/a-n/a. doi:10.1002/cmmi.1696. [DOI] [PubMed] [Google Scholar]
- 144.Beerli RR, Hell T, Merkel AS, Grawunder U. Sortase enzyme-mediated generation of site-specifically conjugated antibody drug conjugates with high in vitro and in vivo potency. PLoS ONE. 2015;10:e0131177. doi: 10.1371/journal.pone.0131177. doi:10.1371/journal.pone.0131177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Ismail NF, Lim TS. Site-specific scFv labelling with invertase via Sortase A mechanism as a platform for antibody-antigen detection using the personal glucose meter. Sci. Rep. 2016;6:19338. doi: 10.1038/srep19338. doi:10.1038/srep19338 http://www.nature.com/articles/srep19338#supplementary-information. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Seo J, Lee S, Poulter CD. Regioselective covalent immobilization of recombinant antibody-binding proteins A, G, and L for construction of antibody arrays. J. Am. Chem. Soc. 2013;135:8973–8980. doi: 10.1021/ja402447g. doi:10.1021/ja402447g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Johnson BN, Mutharasan R. pH effect on protein g orientation on gold surfaces and characterization of adsorption thermodynamics. Langmuir. 2012;28:6928–6934. doi: 10.1021/la3009128. doi:10.1021/la3009128. [DOI] [PubMed] [Google Scholar]
- 148.Lee JM, Park HK, Jung Y, Kim JK, Jung SO, Chung BH. Direct immobilization of protein g variants with various numbers of cysteine residues on a gold surface. Anal. Chem. 2007;79:2680–2687. doi: 10.1021/ac0619231. doi:10.1021/ac0619231. [DOI] [PubMed] [Google Scholar]
- 149.Lee JH, Choi HK, Lee SY, Lim M-W, Chang JH. Enhancing immunoassay detection of antigens with multimeric protein Gs. Biosens. Bioelectron. 2011;28:146–151. doi: 10.1016/j.bios.2011.07.011. doi: http://dx.doi.org/10.1016/j.bios.2011.07.011. [DOI] [PubMed] [Google Scholar]
- 150.de Juan-Franco E, Caruz A, Pedrajas JR, Lechuga LM. Site-directed antibody immobilization using a protein A-gold binding domain fusion protein for enhanced SPR immunosensing. Analyst. 2013;138:2023–2031. doi: 10.1039/c3an36498d. doi:10.1039/C3AN36498D. [DOI] [PubMed] [Google Scholar]
- 151.Ko S, Park TJ, Kim H-S, Kim J-H, Cho Y-J. Directed self-assembly of gold binding polypeptide-protein A fusion proteins for development of gold nanoparticle-based SPR immunosensors. Biosens. Bioelectron. 2009;24:2592–2597. doi: 10.1016/j.bios.2009.01.030. doi: http://dx.doi.org/10.1016/j.bios.2009.01.030. [DOI] [PubMed] [Google Scholar]
- 152.Lee J, Chang JH. Facile and high-efficient immobilization of histidine-tagged multimeric protein G on magnetic nanoparticles. Nanoscale Res. Lett. 2014;9:1–6. doi: 10.1186/1556-276X-9-664. doi:10.1186/1556-276x-9-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Choi HW, Sakata Y, Ooya T, Takeuchi T. Reflectometric interference spectroscopy-based immunosensing using immobilized antibody via His-tagged recombinant protein A. J. Biosci. Bioeng. 2015;119:195–199. doi: 10.1016/j.jbiosc.2014.06.017. doi:10.1016/j.jbiosc.2014.06.017. [DOI] [PubMed] [Google Scholar]
- 154.Feng B, Huang S, Ge F, Luo Y, Jia D, Dai Y. 3D antibody immobilization on a planar matrix surface. Biosens. Bioelectron. 2011;28:91–96. doi: 10.1016/j.bios.2011.07.003. doi:10.1016/j.bios.2011.07.003. [DOI] [PubMed] [Google Scholar]
- 155.Nilsson B, Moks T, Jansson B, Abrahmsen L, Elmblad A, Holmgren E, Henrichson C, Jones TA, Uhlen M. A synthetic IgG-binding domain based on staphylococcal protein A. Protein Eng. 1987;1:107–13. doi: 10.1093/protein/1.2.107. [DOI] [PubMed] [Google Scholar]
- 156.Le Brun AP, Holt SA, Shah DSH, Majkrzak CF, Lakey JH. The structural orientation of antibody layers bound to engineered biosensor surfaces. Biomaterials. 2011;32:3303–3311. doi: 10.1016/j.biomaterials.2011.01.026. doi: http://dx.doi.org/10.1016/j.biomaterials.2011.01.026. [DOI] [PubMed] [Google Scholar]
- 157.Kanno S, Yanagida Y, Haruyama T, Kobatake E, Aizawa M. Assembling of engineered IgG-binding protein on gold surface for highly oriented antibody immobilization. J. Biotechnol. 2000;76:207–214. doi: 10.1016/s0168-1656(99)00186-8. doi: http://dx.doi.org/10.1016/S0168-1656(99)00186-8. [DOI] [PubMed] [Google Scholar]
- 158.Yang H-M, Bao R-M, Cheng Y-Z, Tang J-B. Site-specific covalent attachment of an engineered Z-domain onto a solid matrix: an efficient platform for 3D IgG immobilization. Anal. Chim. Acta. 2015;872:1–6. doi: 10.1016/j.aca.2015.03.005. doi:10.1016/j.aca.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 159.Iijima M, Kadoya H, Hatahira S, Hiramatsu S, Jung G, Martin A, Quinn J, Jung J, Jeong S-Y, Choi EK, Arakawa T, Hinako F, Kusunoki M, Yoshimoto N, Niimi T, Tanizawa, Kuroda S. Nanocapsules incorporating K. IgG Fc-binding domain derived from Staphylococcus aureus protein A for displaying IgGs on immunosensor chips. Biomaterials. 2011;32:1455–1464. doi: 10.1016/j.biomaterials.2010.10.057. doi: http://dx.doi.org/10.1016/j.biomaterials.2010.10.057. [DOI] [PubMed] [Google Scholar]
- 160.Miyao H, Ikeda Y, Shiraishi A, Kawakami Y, Sueda S. Immobilization of immunoglobulin-G-binding domain of Protein A on a gold surface modified with biotin ligase. Anal. Biochem. 2015;484:113–121. doi: 10.1016/j.ab.2015.05.010. doi:10.1016/j.ab.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 161.Dutta P, Sawoo S, Ray N, Bouloussa O, Sarkar A. Engineering bioactive surfaces with fischer carbene complex: protein a on self-assembled monolayer for antibody sensing. Bioconjug. Chem. 2011;22:1202–1209. doi: 10.1021/bc200073r. doi:10.1021/bc200073r. [DOI] [PubMed] [Google Scholar]
- 162.Sousa MM, Steen KW, Hagen L, Slupphaug G. Antibody cross-linking and target elution protocols used for immunoprecipitation significantly modulate signal-to noise ratio in downstream 2D-PAGE analysis. Proteome Sci. 2011;9:1–8. doi: 10.1186/1477-5956-9-45. doi:10.1186/1477-5956-9-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Castillo MJ, McShane AJ, Cai M, Shen Y, Wang L, Yao X. Nonisotopic reagents for a cost-effective increase in sample throughput of targeted quantitative proteomics. Anal. Chem. 2015;87:9209–9216. doi: 10.1021/acs.analchem.5b01727. doi:10.1021/acs.analchem.5b01727. [DOI] [PubMed] [Google Scholar]
- 164.Bereli N, ener G, Yavuz H, Denizli A. Oriented immobilized anti-LDL antibody carrying poly(hydroxyethyl methacrylate) cryogel for cholesterol removal from human plasma. Mater. Sci. Eng. C. 2011;31:1078–1083. doi: http://dx.doi.org/10.1016/j.msec.2011.03.008. [Google Scholar]
- 165.Brogan KL, Shin JH, Schoenfisch MH. Influence of surfactants and antibody immobilization strategy on reducing nonspecific protein interactions for molecular recognition force microscopy. Langmuir. 2004;20:9729–9735. doi: 10.1021/la048437y. doi:10.1021/la048437y. [DOI] [PubMed] [Google Scholar]
- 166.Bergström G, Mandenius C-F. Orientation and capturing of antibody affinity ligands: Applications to surface plasmon resonance biochips. Sens. Actuators B Chem. 2011;158:265–270. doi: http://dx.doi.org/10.1016/j.snb.2011.06.017. [Google Scholar]
- 167.Konrad A, Eriksson Karlström A, Hober S. Covalent immunoglobulin labeling through a photoactivable synthetic z domain. Bioconjug. Chem. 2011;22:2395–2403. doi: 10.1021/bc200052h. doi:10.1021/bc200052h. [DOI] [PubMed] [Google Scholar]
- 168.Yu F, Järver P, Nygren P-Å. Tailor-making a protein a-derived domain for efficient site-specific photocoupling to fc of mouse IgG1. PLoS ONE. 2013;8:e56597. doi: 10.1371/journal.pone.0056597. doi:10.1371/journal.pone.0056597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Hui JZ, Zaki AA, Cheng Z, Popik V, Zhang H, Luning Prak ET, Tsourkas A. Facile method for the site-specific, covalent attachment of full-length igg onto nanoparticles. Small. 2014;10:3354–3363. doi: 10.1002/smll.201303629. doi:10.1002/smll.201303629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Jung Y, Lee JM, Kim J, Yoon J, Cho H, Chung BH. Photoactivable antibody binding protein: Site-selective and covalent coupling of antibody. Anal. Chem. 2009;81:936–942. doi: 10.1021/ac8014565. doi:10.1021/ac8014565. [DOI] [PubMed] [Google Scholar]
- 171.Nimse SB, Kim T. Biological applications of functionalized calixarenes. Chem. Soc. Rev. 2013;42:366–386. doi: 10.1039/c2cs35233h. doi:10.1039/C2CS35233H. [DOI] [PubMed] [Google Scholar]
- 172.Lee Y, Lee EK, Cho YW, Matsui T, Kang IC, Kim TS, Han MH. ProteoChip: a highly sensitive protein microarray prepared by a novel method of protein immobilization for application of protein-protein interaction studies. Proteomics. 2003;3:2289–304. doi: 10.1002/pmic.200300541. doi:10.1002/pmic.200300541. [DOI] [PubMed] [Google Scholar]
- 173.Chen H, Liu F, Qi F, Koh K, Wang K. Fabrication of calix[4]arene derivative monolayers to control orientation of antibody immobilization. Int. J. Mol. Sci. 2014;15:5496–5507. doi: 10.3390/ijms15045496. doi:10.3390/ijms15045496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Chen H, Huang J, Lee J, Hwang S, Koh K. Surface plasmon resonance spectroscopic characterization of antibody orientation and activity on the calixarene monolayer. Sens. Actuators B Chem. 2010;147:548–553. doi: http://dx.doi.org/10.1016/j.snb.2010.03.033. [Google Scholar]
- 175.Soler M, Estevez M-C, Alvarez M, Otte MA, Sepulveda B, Lechuga LM. Direct detection of protein biomarkers in human fluids using site-specific antibody immobilization strategies. Sensors. 2014;14:2239–2258. doi: 10.3390/s140202239. doi:10.3390/s140202239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Kim Y-R, Seo H-J, Oh J-W, Lim H, Kim TH, Kim H. Immunosensor based on electrogenerated chemiluminescence using ru(bpy)32+-doped silica nanoparticles and calix[4]crown-5 self-assembled monolayers. Electroanalysis. 2013;25:1056–1063. doi:10.1002/elan.201200548. [Google Scholar]
- 177.Chen D, Shen M, Cao Y, Bo B, Chen Z, Shu Y, Li G. Electrochemical identification of hepatocellular carcinoma based on the assay of human cervical cancer oncoprotein-1 in serum. Electrochem. Commun. 2013;27:38–41. doi: http://dx.doi.org/10.1016/j.elecom.2012.10.042. [Google Scholar]
- 178.Seker UO, Demir HV. Material binding peptides for nanotechnology. Molecules. 2011;16:1426–51. doi: 10.3390/molecules16021426. doi:10.3390/molecules16021426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Braun R, Sarikaya M, Schulten K. Genetically engineered gold-binding polypeptides: structure prediction and molecular dynamics. J. Biomater. Sci. Polym. Ed. 2002;13:747–757. doi: 10.1163/156856202760197384. doi:10.1163/156856202760197384. [DOI] [PubMed] [Google Scholar]
- 180.Palafox-Hernandez JP, Tang Z, Hughes ZE, Li Y, Swihart MT, Prasad PN, Walsh TR, Knecht MR. Comparative Study of materials-binding peptide interactions with gold and silver surfaces and nanostructures: A thermodynamic basis for biological selectivity of inorganic materials. Chem. Mater. 2014;26:4960–4969. doi:10.1021/cm501529u. [Google Scholar]
- 181.Kumada Y. Site-specific immobilization of recombinant antibody fragments through material-binding peptides for the sensitive detection of antigens in enzyme immunoassays. Biochim. Biophys. Acta. 2014;1844:1960–1969. doi: 10.1016/j.bbapap.2014.07.007. doi:10.1016/j.bbapap.2014.07.007. [DOI] [PubMed] [Google Scholar]
- 182.Feng B, Dai Y, Wang L, Tao N, Huang S, Zeng H. A novel affinity ligand for polystyrene surface from a phage display random library and its application in anti-HIV-1 ELISA system. Biologicals. 2009;37:48–54. doi: 10.1016/j.biologicals.2008.10.002. doi: http://dx.doi.org/10.1016/j.biologicals.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 183.Kumada Y, Katoh S, Imanaka H, Imamura K, Nakanishi K. Development of a one-step ELISA method using an affinity peptide tag specific to a hydrophilic polystyrene surface. J. Biotechnol. 2007;127:288–299. doi: 10.1016/j.jbiotec.2006.07.011. doi: http://dx.doi.org/10.1016/j.jbiotec.2006.07.011. [DOI] [PubMed] [Google Scholar]
- 184.Tang J-B, Sun X-F, Yang H-M, Zhang B-G, Li Z-J, Lin Z-J, Gao Z-Q. Well-oriented ZZ-PS-tag with high Fc-binding onto polystyrene surface for controlled immobilization of capture antibodies. Anal. Chim. Acta. 2013;776:74–78. doi: 10.1016/j.aca.2013.03.017. doi:10.1016/j.aca.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 185.Kumada Y, Ishikawa Y, Fujiwara Y, Takeda R, Miyamoto R, Niwa D, Momose S, Kang B, Kishimoto M. Efficient refolding and immobilization of PMMA-tag-fused single-chain Fv antibodies for sensitive immunological detection on a PMMA plate. J. Immunol. Methods. 2014;411:1–10. doi: 10.1016/j.jim.2014.05.015. doi:10.1016/j.jim.2014.05.015. [DOI] [PubMed] [Google Scholar]
- 186.Thatikonda N, Delfani P, Jansson R, Petersson L, Lindberg D, Wingren C, Hedhammar M. Genetic fusion of single-chain variable fragments to partial spider silk improves target detection in micro- and nanoarrays. Biotechnol. J. 2016;11:437–448. doi: 10.1002/biot.201500297. doi:10.1002/biot.201500297. [DOI] [PubMed] [Google Scholar]
- 187.Heo NS, Zheng S, Yang M, Lee SJ, Lee SY, Kim H-J, Park JY, Lee C-S, Park TJ. Label-free electrochemical diagnosis of viral antigens with genetically engineered fusion protein. Sensors. 2012;12:10097–10108. doi: 10.3390/s120810097. doi:10.3390/s120810097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Park JH, Kim Y-P, Kim I-H, Ko S. Rapid detection of aflatoxin B1 by a bifunctional protein crosslinker-based surface plasmon resonance biosensor. Food Control. 2014;36:183–190. doi: http://dx.doi.org/10.1016/j.foodcont.2013.08.038. [Google Scholar]
- 189.Hattori T, Umetsu M, Nakanishi T, Sawai S, Kikuchi S, Asano R, Kumagai I. A High-affinity gold-binding camel antibody: antibody engineering for one-pot functionalization of gold nanoparticles as biointerface molecules. Bioconjug. Chem. 2012;23:1934–1944. doi: 10.1021/bc300316p. doi:10.1021/bc300316p. [DOI] [PubMed] [Google Scholar]
- 190.Watanabe R, Yokoi T, Kobayashi E, Otsuka Y, Shimojima A, Okubo T, Tatsumi T. Extension of size of monodisperse silica nanospheres and their well-ordered assembly. J. Colloid Interface Sci. 2011;360:1–7. doi: 10.1016/j.jcis.2010.09.001. doi:10.1016/j.jcis.2010.09.001. [DOI] [PubMed] [Google Scholar]
- 191.Jain P, Soshee A, Narayanan SS, Sharma J, Girard C, Dujardin E, Nizak C. Selection of arginine-rich anti-gold antibodies engineered for plasmonic colloid self-assembly. J. Phys. Chem. C. 2014;118:14502–14510. doi:10.1021/jp502118n. [Google Scholar]
- 192.Ibii T, Kaieda M, Hatakeyama S, Shiotsuka H, Watanabe H, Umetsu M, Kumagai I, Imamura T. Direct immobilization of gold-binding antibody fragments for immunosensor applications. Anal. Chem. 2010;82:4229–4235. doi: 10.1021/ac100557k. doi:10.1021/ac100557k. [DOI] [PubMed] [Google Scholar]
- 193.Boozer C, Ladd J, Chen S, Jiang S. DNA-directed protein immobilization for simultaneous detection of multiple analytes by surface plasmon resonance biosensor. Anal. Chem. 2006;78:1515–1519. doi: 10.1021/ac051923l. doi:10.1021/ac051923l. [DOI] [PubMed] [Google Scholar]
- 194.Hu J, Yu Y, Brooks JC, Godwin LA, Somasundaram S, Torabinejad F, Kim J, Shannon C, Easley CJ. A reusable electrochemical proximity assay for highly selective, real-time protein quantitation in biological matrices. J. Am. Chem. Soc. 2014;136:8467–8474. doi: 10.1021/ja503679q. doi:10.1021/ja503679q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Wold ED, McBride R, Axup JY, Kazane SA, Smider VV. Antibody microarrays utilizing site-specific antibody-oligonucleotide conjugates. Bioconjug. Chem. 2015;26:807–811. doi: 10.1021/acs.bioconjchem.5b00111. doi:10.1021/acs.bioconjchem.5b00111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Kazane SA, Sok D, Cho EH, Uson ML, Kuhn P, Schultz PG, Smider VV. Site-specific DNA-antibody conjugates for specific and sensitive immuno-PCR. Proc. Natl. Acad. Sci. 2012;109:3731–3736. doi: 10.1073/pnas.1120682109. doi:10.1073/pnas.1120682109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Jung Y, Lee JM, Jung H, Chung BH. Self-directed and self-oriented immobilization of antibody by protein G−DNA conjugate. Anal. Chem. 2007;79:6534–6541. doi: 10.1021/ac070484i. doi:10.1021/ac070484i. [DOI] [PubMed] [Google Scholar]
- 198.Meyer R, Giselbrecht S, Rapp BE, Hirtz M, Niemeyer CM. Advances in DNA-directed immobilization. Curr. Opin. Chem. Biol. 2014;18:8–15. doi: 10.1016/j.cbpa.2013.10.023. doi: http://dx.doi.org/10.1016/j.cbpa.2013.10.023. [DOI] [PubMed] [Google Scholar]
- 199.Hallam TJ, Smider VV. Unnatural amino acids in novel antibody conjugates. Future Med. Chem. 2014;6:1309–1324. doi: 10.4155/fmc.14.79. doi:10.4155/fmc.14.79. [DOI] [PubMed] [Google Scholar]
- 200.Hallam TJ, Wold E, Wahl A, Smider VV. Antibody conjugates with unnatural amino acids. Mol. Pharm. 2015;12:1848–1862. doi: 10.1021/acs.molpharmaceut.5b00082. doi:10.1021/acs.molpharmaceut.5b00082. [DOI] [PubMed] [Google Scholar]
- 201.Chin JW, Santoro SW, Martin AB, King DS, Wang L, Schultz PG. Addition of pazido-l-phenylalanine to the genetic code of Escherichia coli. J. Am. Chem. Soc. 2002;124:9026–9027. doi: 10.1021/ja027007w. doi:10.1021/ja027007w. [DOI] [PubMed] [Google Scholar]
- 202.Petersson L, Städe LW, Brofelth M, Gärtner S, Fors E, Sandgren M, Vallkil J, Olsson N, Larsen KL, Borrebaeck CAK, Duroux L, Wingren C. Molecular design of recombinant scFv antibodies for site-specific photocoupling to -cyclodextrin in solution and onto solid support. Biochim. Biophys. Acta. 2014;1844:2164–2173. doi: 10.1016/j.bbapap.2014.08.010. doi:10.1016/j.bbapap.2014.08.010. [DOI] [PubMed] [Google Scholar]
- 203.Bhakta S, Seraji MSI, Suib SL, Rusling JF. Antibody-like biorecognition sites for proteins from surface imprinting on nanoparticles. ACS Appl. Mater. Interfaces. 2015;7:28197–28206. doi: 10.1021/acsami.5b11650. doi:10.1021/acsami.5b11650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lv Y, Tan T, Svec F. Molecular imprinting of proteins in polymers attached to the surface of nanomaterials for selective recognition of biomacromolecules. Biotechnol. Adv. 2013;31:1172–1186. doi: 10.1016/j.biotechadv.2013.02.005. doi: http://dx.doi.org/10.1016/j.biotechadv.2013.02.005. [DOI] [PubMed] [Google Scholar]
- 205.Vasapollo G, Sole RD, Mergola L, Lazzoi MR, Scardino A, Scorrano S, Mele G. Molecularly imprinted polymers: present and future prospective. Int J Mol Sci. 2011;12:5908–45. doi: 10.3390/ijms12095908. doi:10.3390/ijms12095908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Chen L, Wang X, Lu W, Wu X, Li J. Molecular imprinting: perspectives and applications. Chem. Soc. Rev. 2016;45:2137–2211. doi: 10.1039/c6cs00061d. doi:10.1039/C6CS00061D. [DOI] [PubMed] [Google Scholar]
- 207.Wulff G. Fourty years of molecular imprinting in synthetic polymers: origin, features and perspectives. Microchim. Acta. 2013;180:1359–1370. doi:10.1007/s00604-013-0992-9. [Google Scholar]
- 208.Bereli N, Ertürk G, Tümer MA, Say R, Denizli A. Oriented immobilized anti-hIgG via F(c) fragment-imprinted PHEMA cryogel for IgG purification. Biomed. Chromatogr. BMC. 2013;27:599–607. doi: 10.1002/bmc.2833. doi:10.1002/bmc.2833. [DOI] [PubMed] [Google Scholar]
- 209.Çorman ME, Armutcu C, Uzun L, Say R, Denizli A. Self-oriented nanoparticles for site-selective immunoglobulin G recognition via epitope imprinting approach. Colloids Surf. B Biointerfaces. 2014;123:831–837. doi: 10.1016/j.colsurfb.2014.10.020. doi: http://dx.doi.org/10.1016/j.colsurfb.2014.10.020. [DOI] [PubMed] [Google Scholar]
- 210.Zhou J, Gan N, Li T, Hu F, Li X, Wang L, Zheng L. A cost-effective sandwich electrochemiluminescence immunosensor for ultrasensitive detection of HIV-1 antibody using magnetic molecularly imprinted polymers as capture probes. Biosens. Bioelectron. 2014;54:199–206. doi: 10.1016/j.bios.2013.10.044. doi: http://dx.doi.org/10.1016/j.bios.2013.10.044. [DOI] [PubMed] [Google Scholar]
- 211.Jung Y, Kang HJ, Lee JM, Jung SO, Yun WS, Chung SJ, Chung BH. Controlled antibody immobilization onto immunoanalytical platforms by synthetic peptide. Anal. Biochem. 2008;374:99–105. doi: 10.1016/j.ab.2007.10.022. doi: http://dx.doi.org/10.1016/j.ab.2007.10.022. [DOI] [PubMed] [Google Scholar]
- 212.Tsai C-W, Jheng S-L, Chen W-Y, Ruaan R-C. Strategy of Fc-recognizable Peptide ligand design for oriented immobilization of antibody. Anal. Chem. 2014;86:2931–2938. doi: 10.1021/ac4029467. doi:10.1021/ac4029467. [DOI] [PubMed] [Google Scholar]
- 213.Zhang Y, Islam N, Carbonell RG, Rojas OJ. Specificity and regenerability of short peptide ligands supported on polymer layers for immunoglobulin G binding and detection. ACS Appl. Mater. Interfaces. 2013;5:8030–8037. doi: 10.1021/am4021186. doi:10.1021/am4021186. [DOI] [PubMed] [Google Scholar]
- 214.Fu J, Cai K, Johnston SA, Woodbury NW. Exploring peptide space for enzyme modulators. J. Am. Chem. Soc. 2010;132:6419–6424. doi: 10.1021/ja100403a. doi:10.1021/ja100403a. [DOI] [PubMed] [Google Scholar]
- 215.Fu J, Reinhold J, Woodbury NW. Peptide-modified surfaces for enzyme immobilization. PLoS ONE. 2011;6:e18692. doi: 10.1371/journal.pone.0018692. doi:10.1371/journal.pone.0018692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Zhao J, Yan Y, Zhu L, Li X, Li G. An amperometric biosensor for the detection of hydrogen peroxide released from human breast cancer cells. Biosens. Bioelectron. 2013;41:815–819. doi: 10.1016/j.bios.2012.10.019. doi: http://dx.doi.org/10.1016/j.bios.2012.10.019. [DOI] [PubMed] [Google Scholar]
- 217.Oliveira C, Carvalho V, Domingues L, Gama FM. Recombinant CBM-fusion technology — Applications overview. Biotechnol. Adv. 2015;33:358–369. doi: 10.1016/j.biotechadv.2015.02.006. doi: http://dx.doi.org/10.1016/j.biotechadv.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 218.Shoseyov O, Shani Z, Levy I. Carbohydrate Binding Modules: Biochemical properties and novel applications. Microbiol. Mol. Biol. Rev. 2006;70:283–295. doi: 10.1128/MMBR.00028-05. doi:10.1128/mmbr.00028-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Wan W, Wang D, Gao X, Hong J. Expression of family 3 cellulose-binding module (CBM3) as an affinity tag for recombinant proteins in yeast. Appl. Microbiol. Biotechnol. 2011;91:789–798. doi: 10.1007/s00253-011-3373-5. doi:10.1007/s00253-011-3373-5. [DOI] [PubMed] [Google Scholar]
- 220.Shpigel E, Goldlust A, Eshel A, Ber IK, Efroni G, Singer Y, Levy I, Dekel M, Shoseyov O. Expression, purification and applications of staphylococcal Protein A fused to cellulose-binding domain. Biotechnol. Appl. Biochem. 2000;31:197–203. doi: 10.1042/ba20000002. doi:10.1042/BA20000002. [DOI] [PubMed] [Google Scholar]
- 221.Ramos R, Moreira S, Rodrigues A, Gama M, Domingues L. Recombinant expression and purification of the antimicrobial peptide magainin-2. Biotechnol. Prog. 2013;29:17–22. doi: 10.1002/btpr.1650. doi:10.1002/btpr.1650. [DOI] [PubMed] [Google Scholar]
- 222.Azriel-Rosenfeld R, Valensi M, Benhar I. A human synthetic combinatorial library of arrayable single-chain antibodies based on shuffling in vivo formed CDRs into general framework regions. J. Mol. Biol. 2004;335:177–192. doi: 10.1016/j.jmb.2003.10.053. doi:10.1016/j.jmb.2003.10.053. [DOI] [PubMed] [Google Scholar]
- 223.Ofir K, Berdichevsky Y, Benhar I, Azriel-Rosenfeld R, Lamed R, Barak Y, Bayer EA, Morag E. Versatile protein microarray based on carbohydrate-binding modules. Proteomics. 2005;5:1806–1814. doi: 10.1002/pmic.200401078. doi:10.1002/pmic.200401078. [DOI] [PubMed] [Google Scholar]