Abstract
Extracellular matrix fibrils of fibronectin (FN) are highly elastic, and are typically stretched three to four times their relaxed length. The mechanism of stretching has been controversial, in particular whether it involves tension-induced unfolding of FNIII domains. Recent studies have found that ~5 pN is the threshold isometric force for unfolding various protein domains. FNIII domains should therefore not be unfolded until the tension approaches 5 pN. Integrins have been reported to generate forces ranging from 1 to > 50 pN, but I argue that studies reporting 1-2 pN are the most convincing, This is not enough to unfold FNIII domains. Even if domains were unfolded, 2 pN would only extend the worm-like-chain to about twice the length of the folded domain. Overall I conclude that stretching FN matrix fibrils involves primarily the compact to extended conformational change of FN dimers, with minimal contribution from unfolding FNIII domains.
Introduction
Fibronectin (FN) is a large molecule with a modular structure comprising multiple copies of three types of domains (Fig. 1). The smaller FNI and FNII domains contain internal disulfide bonds that stabilize their structure. The most abundant domain is the FNIII, which is a peptide of about 90 amino acids (aa's) folded into a beta sandwich structure, with three beta strands on one side and four on the other. FNIII domains have no disulfides. Two FN monomers are connected by disulfide bonds near their C termini to make the FN dimer. FN dimers are assembled into fibrils that constitute the primordial extracellular matrix in embryonic tissues and wound healing.
Fig. 1.
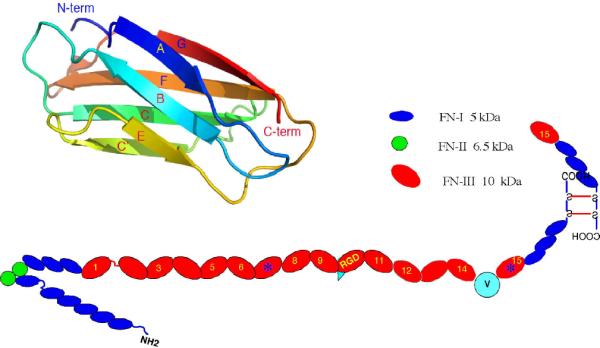
A model of the FN dimer showing the arrangement of FNI, FNII and FNIII domains. The upper left shows a cartoon diagram of the beta sheet structure of an FNIII domain.
FN matrix fibrils are generally quite straight, rather than wavy or bent. The FN molecules themselves have several hinge points [1], so one might expect matrix fibrils to be flexible. That they are always straight suggests that they are under tension. The source of the tension is the cells to which the FN fibrils are attached, whose actin cytoskeleton generates a contractile force that is transmitted to FN via transmembrane integrin receptors.
Tension does more than just straighten matrix fibrils – it also stretches them. The original observation was made with FN-GFP, in which a GFP (green fluorescent protein) module was inserted between FN-III domains 3 and 4 [2,3]. When matrix fibrils happened to sever, they were seen to contract about 3- to 4-fold from their stretched length. This elasticity of the FN matrix has been confirmed in studies incorporating fluorescently labeled FN into cell cultures [4] and into Xenopus embryos [5].
The mechanism of stretching has been controversial, in particular whether tension causes unfolding of FNIII domains. Here I review recent studies of FN itself, and then consider developments indirectly related to FN. How much tension is needed to unfold protein domains and prevent their refolding? How much tension do integrins generate on the extracellular matrix? If an FNIII domain is unfolded to give a worm-like-chain (wlc), how far would this tension extend it? Recent answers to these questions suggest that unfolding FNIII domains contributes minimally to stretching FN matrix fibrils.
Stretching fibronectin – two potential mechanisms
One model for FN fibril elasticity is based on the conformational change that the FN molecule undergoes in different solution conditions. In high ionic strength or high pH FN molecules assume an extended conformation, whereas at physiological ionic strength the molecule folds into a pretzel-like compact conformation [6,7]. Studies of proteolytic and recombinant protein fragments suggested that the compact conformation is stabilized by an electrostatic contact between FN-III domains 2–3 of one subunit of the dimer, and domains 12–14 of the other subunit [8]. The N-terminal FNI domains are also important. See [1] for a comprehensive review of the compact conformation and its dynamics.
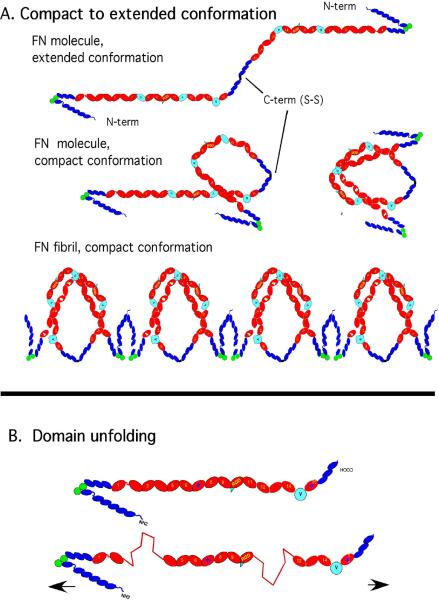
Fig. 2 shows a model of an FN molecule in the extended conformation, and the formation of the compact conformation by folding first one subunit and then the other to make the electrostatic contacts to the other subunit. The third panel illustrates how FN molecules in the compact conformation might be linked to make one strand of a fibril. Stretching fibrils would involve breaking the electrostatic bonds and pulling some or all molecules into the extended conformation. Importantly, the ends of the extended molecules are 3.5 times farther apart than they are in the compact conformation, in good agreement with the 3- to 4-fold contraction observed for broken FN fibrils in vivo.
Fig. 2.
Two models for the mechanism of stretching FN matrix fibrils. The top model is based on the compact-to-extended conformational change (modified from [17]). FN dimers in physiological solution fold into a compact pretzel-like shape. This may involve domains III-2-3 of one subunit forming electrostatic bonds to III-12-14 of the other subunit [8]; the N-terminal FNI domains probably also play a role [1]. If molecules are connected by their N-terminal FNI domains, initially in the compact conformation, extending them fully would generate a 3.5-fold extension. The bottom model shows the proposal for tension progressively unfolding FNIII domains and extending them to produce the stretch.
The second model assumes that the FN molecules are already extended as they are incorporated into the FN fibril, and that the stretching involves force-induced unraveling of FN-III domains. The concept that force could unravel FN-III and Ig domains was first proposed as a theoretical model [9,10], and has been demonstrated experimentally by stretching single molecules of titin [11], tenascin [12], and FN [13] with the atomic force microscope (AFM).
The model that FNIII domains in FN fibrils are unfolded by tension is supported primarily by experiments using an FN FRET reporter [14,15,16]. I have argued previously that this probe is poorly designed to detect domain unfolding [17]. It has two acceptor fluors per FN monomer, attached to the free Cys in III-7 and III-15; and it has 3.5 donor fluors randomly attached to amines. Smith et al [16] suggested that FRET changes could be detected for donors within 2Ro = 12 nm of the acceptors. However, at 2Ro FRET efficiency is only 1.5%. If an acceptor fluor at this distance moved farther out of range of the donor, the FRET of that acceptor-donor pair would drop from 1.5% to zero. I don't think this tiny change could be detected in the background of noise from multiple sources. I suggest that a more realistic model would draw the circles at 7 nm, 1.17Ro, where FRET efficiency should be 37%. This is twice the 3.5 nm length of an FNIII domain. 3/4 of the amines of FN are outside these smaller circles; these can only report the compact-to-extended conformational change, and otherwise contribute noise to the FRET signal. Overall, I suggest that this FN-FRET construct cannot reliably demonstrate domain unfolding, and studies using it should be interpreted as reporting primarily the compact-to-extended conformational change.
One recent study has made a seminal advance in addressing the conformation of FN dimers in matrix fibrils. Früh et al **[18] attached a fluorophore at the two ends of the FN dimer, and incorporated these sparsely into matrix fibrils. They then used super-resolution light microscopy to image single molecules and measure the separation of the ends. They found that the FN dimers were extended 133 nm on average, somewhat less than the 190 nm contour length. If domain unfolding were a major contribution to the stretch, one might expect the dimers to be stretched beyond their 190 nm contour length. The 133 nm length of the molecule measured by Früh et al. suggests that the FN dimers are extended, but there is no excess length from domain unfolding.
In the following sections I collect and evaluate recent studies of protein domain unfolding and integrin tension, and relate these to the question of tension-induced FNIII unfolding in FN fibrils.
Spontaneous unfolding and refolding of FNIII domains in the absence of force
PDB files of protein structures give a deceptive impression of permanence, with all atoms having a fixed structure. However, it is well known that globular protein domains spontaneously unfold and refold. Jackson [19] has tabulated the unfolding and refolding kinetics data for a wide range of proteins. Table 1 shows data for a selection of protein domains, ranked according to folding rate, and compares these to a selection of FNIII domains, mostly from FN **[20]. The globular proteins have a broad range of folding rates, from 8 to 10,000 s−1. The FNIII domains are remarkable for having the slowest folding rates of all these proteins, with kf mostly in the range of 0.1 – 1 s−1. Most of the FNIII domains are relatively stable, with lifetimes (1/ku) of the folded state from 1,000 – 10,000 s. Since they refold in 1–10 s (1/kf) they spend most of their life folded. However two domains are remarkably unstable. III-11C (the C refers to a Cys mutation used for the kinetics assay) spontaneously opens every 63 s and remains open for 16 s; III-3C opens every 520 s and remains open for 2 s. These domains should be most susceptible to tension, which would extend the spontaneously unfolded polypeptide and slow the refolding.
Table 1.
Rates of spontaneous unfolding and refolding of various protein domains. The first seven protein domains are taken from Table 1 of Jackson [19], which has original references to kinetics data. Data for the FNIII domains are from Shah et al **[20]. The C following the domain number indicates a Cys mutation introduced to measure kinetics by thiol exchange.
| Protein | Structure | PDB | ku s−1 | kf s−1 |
|---|---|---|---|---|
| Arc repressor | α/β | 1ARR | 1.5 | 10,600 |
| Monomeric λ repressor (λ6–85) | α | 1LMB | 30 | 4,900 |
| Villin 14T (headpiece) | α/β | 1VIK | 0.061 | 900 |
| CspA (B. subtilis) | ß-barrel | 1MEF | 4.2 | 200 |
| SH3 domain (PI3 kinase) | ß | 1PKS | 0.001 | 94 |
| Chymotrypsin inhib 2 | α/β | 1COA | 0.00018 | 48 |
| SH3 domain | ß | 1AEY | 0.045 | 8.1 |
| FN III-2C | ß | 2HA1 | 0.00027 | 4.8 |
| FN III-3C | ß | 2N1K | 0.0019 | 0.55 |
| FN III-6C | ß | 0.000013 | 0.14 | |
| FN III-7 | ß | 1FNF | 0.00007 | 1.4 |
| FN III-9C | ß | 1FNF | 0.00003 | - |
| FN III-11C | ß | 0.016 | 0.06 | |
| FN III-12C | ß | 1FNH | 0.00023 | 0.3 |
| Irisin | ß | 4LSD | 0.0021 | 1.9 |
| TNfn3C | ß | 1TEN | 0.006 | 0.72 |
What isometric force will unfold a globular protein?
If one can grab the N and C termini of a globular protein or domain and pull on it, the protein can be unraveled. This has been studied extensively by using the AFM to stretch proteins [21]. When a multi-domain protein is stretched, the force vs extension shows a classic sawtooth wave. The force at each peak is designated the unfolding force for the globular domain. This is a stochastic event, but if multiple ruptures are observed one can determine the most probable unfolding force. This unfolding force depends on the pulling speed – in most models it is directly proportional to the log of pulling speed. In AFM experiments pulling speeds are typically varied from 100 to 10,000 nm/s, and globular domains are seen to unravel at forces from 50 to 200 pN.
In many biological systems, including the FN matrix, the pulling speed is essentially zero. The term “isometric tension” has been used in the muscle field to describe a static force at zero extension speed. This raises an important question – what isometric tension can a globular protein sustain without unraveling? Alternatively, and just as important, what isometric tension can prevent an unfolded protein from refolding.
In recent years AFM and optical and magnetic tweezers have been developed on very stable platforms, These can achieve very low pulling speeds, down to 1 nm/s, or even isometric forces (zero pulling speed), suitable for observing the refolding pathway. In several studies a zone of force has been found where the protein domain oscillates, on a time scale of seconds, between folded and unfolded. I will call this the isometric unfolding force. Specific examples are given in Table 2.
Table 2.
The isometric force at which the indicated protein oscillates between folded and unfolded states.
| Protein domain | Isometric force pN | Method | ref |
|---|---|---|---|
| RNase H (molten glob -> unfold) | 5.5 | optical tw | Cecconi…Marqusee 2005 [22] |
| apomyoglobin (molten glob -> unfold) | 4.5 | optical tw | Elms…Marqusee 2012 [23] |
| titin I27 | 5.4 | magnetic tw | Chen…Yan 2015 *[24] |
| barnase | 4.0 | optical tw | Alemany…Ritort 2016 [25] |
| NuG2 | 8.5 | AFM | He…Li 2015 [26] |
| calmodulin | 6 (folding) 12 (unfolding) | AFM | Junker…Rief 2009 [27] |
| calmodulin | 10.5 | optical tw | Stigler…Rief 2011 [28] |
| villin HP35 | 7 for wt; 10 for stabilized mutant | optical tw | Zoldak…Rief 2013 [29] |
These proteins cover a wide range of structures from all beta sheet (titin I27), all alpha helix (apomyoglobin, calmodulin) and mixed (RNase H, barnase and NuG2), suggesting that they apply generally to small globular proteins and domains. The first four domains in the table all had an isometric unfolding force of ~5 pN. NuG2 and calmodulin were higher, 8.5 and 11 pN. From this limited survey I would conclude that most protein domains have an isometric unfolding force of ~5 pN. The titin I27 domain *[24] is especially important for the present discussion because its ß-sandwich structure is very similar to that of FNIII domains.
The transition zone where domains oscillate between folded and unfolded is generally quite narrow; at forces 20% above or below the threshold, domains are largely unfolded or folded respectively [22–24]. If the isometric unfolding force is 5 pN, most domains will be folded at 4 pN or less, and unfolded at 6 pN or more. Extrapolating from the generalization, it would seem that forces of 5 pN and above should rarely exist in a biological system. If they did, the myosin molecules generating force on actin, and the integrins transmitting the force to the FN, would likely themselves unfold.
What tension can a single integrin generate on the ECM ligand?
Cell adhesion is mediated by integrins binding to ECM ligands, and it has been well established that the cell can generate force on the substrate. Earlier studies used deformable substrates to measure the force generated by whole focal adhesions (FAs). With an estimate of ~500 integrins per FA these gave forces ~1 pN per integrin on RGD-peptide substrates (summarized in **[30]). More recently several force sensors have been designed to measure the force at the level of single integrin molecules. These have given values that range from 1-2 pN to >50 pN, summarized in Table 3. This is an amazing range – they can't all be right. Here I argue that the lower force estimate of Morimatsu, Dunn and colleagues **[30] is most convincing.
Table 3.
Forces reported to be generated by single integrins on extracellular ligands. RGD and cRGD are linear or cyclic peptides containing the integrin ligand RGD. FNIII9-10 is the two-domain protein segment that is the natural ligand for α5ß1 integrin.
| Ligand | Force per integrin, pN | comments | Force sensor | reference |
|---|---|---|---|---|
| cRGD | 1 | Range 1–15 pN | FRET unquench; PEG-82 wlc | Liu…Salaita 2013 [31] |
| cRGD, RGD | ~15 | Sensors crowded, 10 nm apart, maybe few are engaged | FRET unquench; PEG-24 wlc | Chang…Salaita 2016 [32] |
| RGD, cRGD | > 4.7, > 13 | Only ~10% of sensors over FA were engaged | FRET unquench; DNA zipper hairpin | Zhang…Salaita 2014 [33] |
| RGD FNIII9-10 | 36, 110 | 36 pN from I27 unfolding 110 from S-S reduction | FRET unquench; unfold I27 | Galior…Salaita 2015 **[34] |
| RGD | > 6, > 17 | Fraction of sensors engaged not deter-mined, may be small | FRET unquench; DNA zipper hairpin | Blakeley…Chen 2014 [35] |
| RGD | 40 | Before FA formation, 5–30 min | DNA zipper-release | Wang and Ha 2013 [36] |
| cRGD | > 54 | Within established FA, 2 hr | DNA zipper-release | Wang…Ha 2015 [37] |
| RGD | 2 | Sensors 25 nm apart, 90% of sensors in FA were engaged | FRET index/efficiency; peptide wlc | Morimatsu…Dunn 2013 **[30] |
| RGD FNIII9-10 | 1–3 (60–80%) 3–7 (20–30%) >7 (10%) | Single molecule FRET distinguishes sensors engaged | FRET efficiency; peptide wlc | Chang…Dunn 2016 **[38] |
If the true tension of integrins on the RGD ligand is 2 pN, how can we explain the much higher values reported in other studies? The short answer is we can't. Explanations can be sought in the calibration of the sensors, and perhaps in the fraction of sensors engaged. For example, the DNA zippers of Wang and Ha [36,37] were based on a calibration of a 2 second duration [39]. Wang and Ha measured attached cells after 5–30 min, or 2 hrs for fully formed FAs, so these detachments may have reflected rare excursions that exceeded the DNA threshold only briefly in the much longer time. In some studies only a small fraction of probes were engaged at the reported high forces. This would be consistent with high forces being transient excursions, and with 90% of sensors exerting a force below the sensor threshold.
High integrin forces were also reported in several studies by Salaita and colleagues. Several of these used DNA ziper hairpins that might be able to refold reversibly. However it is possible that once unfolded by a transient high force, a much weaker force would suffice to prevent unfolding. Gailor et al **[34] used a sensor based on I27, a beta sandwich domain very similar to FNIII. This domain was unfolded by integrin force, which would suggest a force >5 pN, based on the isometric unfolding force of Chen et al *[24]. However, Gailor et al also measured kinetics of fluorescence development. The half time of ~4 min for loss of FRET gave a force of 36 pN based on kinetics parameters from AFM stretching of I27 [40]. A separate experiment measured the kinetics of breaking disulfide bonds, which gave an even higher force estimate. This was calibrated by another AFM study that is somewhat less direct. This study is one of the most convincing indications of very high integrin forces.
Notwithstanding the several studies reporting very high forces, I think the most convincing measurements are those of Morimatsu, Dunn and colleagues **[30]. They used a FRET probe with donor and acceptor fluorophores at the ends of a 40-aa spider silk peptide that behaves as a worm-like-chain (wlc) entropic spring. In contrast to probes based on FRET unquenching, which only measure emission from the donor, Morimatsu et al measured fluorescence of both donor and acceptor and calculated a FRET index that was closely related to FRET efficiency. Their FRET index of ~0.4 for unengaged sensors dropped to ~0.25 over the FAs, consistent with a large fraction of probes engaged. This FRET drop from 0.4 to 0.25 corresponds exactly to a 2 pN force calibrated in vitro for a similar (F50) probe [41]. The global FRET from whole FAs was supported by elegant single-molecule FRET, which reported very similar forces for single integrins.
More recently Chang, Dunn and colleagues have extended this analysis using the same wlc-FRET force sensor, and focusing on single molecule measurements of dispersed sensors **[38]. They confirmed that the majority of sensors engaged to an integrin had forces of 1-3 pN. A smaller number had forces of 3-7 pN, but importantly, a small fraction had very low FRET, indicating forces >7 pN. This population might explain the release of DNA-based sensors of Wang and Ha [37]. The new study of Chang et al **[38] also extended the measurements to the full adhesion domain of FN, FNIII9-10. The forces to FNIII9-10 were almost identical to those on the simpler RGD peptide.
The 2 pN force exerted on the RGD adhesion ligand is similar to the 1-2 pN forces reported on the cytoplasmic side, for a similar wlc-FRET probe incorporated into vinculin [42]. Similar small forces were reported when this wlc probe was incorporated into talin [43]. It should be noted, however, that a separate study reported talin forces of 7-10 pN [44]. This group used a very different tension probe, the villin headpiece35. This is a 35 aa peptide is similar in length to the 40 aa wlc, but HP35 folds into a globular domain that unfolds at isometric tension of 7-10 pN [29]. This is a significant discrepancy that needs to be resolved.
One might also consider the possibility that multiple motors might operate in series on the actin connected to the integrin. This seems possible but would run into the problem of denaturing and/or detaching the talin and other molecules linking the actin to the integrin. On the extracellular side integrins attached to FN dimers in series might generate higher forces on more distal dimers, but it seems too crowded to have multiple integrins attached to a single FN.
The studies of Dunn and colleagues have no obvious flaws, and seem convincing to me. However, the several studies reporting much higher forces used sophisticated probes, and also have no obvious flaw. Resolving the discrepancy between 1-3 pN and >50 pN is a major challenge for the field of mechanobiology.
If FNIII domains are unraveled, how much would they stretch?
Let's assume that the most unstable FNIII domain is opened spontaneously and subjected to tension. How much does it stretch? When protein domains are unraveled by stretching in the AFM, they behave as a wlc, acting as an entropic spring to further stretch. The formula describing the stretch of a wlc was originally developed to fit the stretching of DNA [45,46], and it has been applied with excellent fits to the polypeptides released when a domain unravels in the AFM [21].
F(x) is the force needed to separate the ends of the peptide by a distance x. k is Boltzmann's constant, T is absolute temperature (kT = 4.1 pN nm). There are two parameters that determine the spring strength. L is the contour length, 30.6 nm for the 90 aa FNIII domains. P is the persistence length, which has been estimated from AFM stretching experiments to be 0.4 to 0.8 nm [24,40]. We will use a compromise value P = 0.55 nm, which was also the most frequent value deduced for intrinsically disordered peptides in relaxed solution conditions [47]. The extension at increasing force is given in Table 4.
Table 4.
Extension of a 90 aa wlc in response to force.
| x/L | x nm | F(x) pN |
|---|---|---|
| 0.1 | 3.1 | 1.2 |
| 0.2 | 6.1 nm | 2.5 |
| 0.33 | 10.1 | 4.7 |
| 0.4 | 12.2 nm | 6.3 |
| 0.6 | 18.4 nm | 14.3 |
| 0.8 | 24.5 nm | 50.7 |
The length of a folded FNIII domain is 3.5 nm. A force of 2.5 pN, the average integrin force indicated by the wlc tension sensors, would extend an unfolded domain less than twice the length of the folded domain. If a force of 4.7 pN could be maintained, it would only extend the 90 aa peptide to 10 nm, which is less than 3 times the length of the folded domain. Extension beyond this length requires forces that are unlikely to be biologically relevant, since they would unravel the domains of the motors and connectors.
The main conclusion of this analysis is that even if FNIII domains are unfolded, the 1-2 pN forces generated by integrins cannot stretch them more than twice the length of the folded domain. Even if all domains were unfolded, this could not account for the four-fold stretch observed for FN fibrils. If only a few domains are unfolded, their contribution to fibril stretching would be insignificant.
Conclusions
I conclude that tension-induced unfolding of FNIII domains is likely rare and does not contribute significantly to the stretch of FN matrix fibrils. Typical protein domains unfold at an isometric tension of ~5 pN, which is well above the ~2 pN force generated by integrins on the extracellular matrix. FNIII domains that do unfold, either spontaneously or assisted by tension, would be extended less than twice the length of the folded domain by a 2 pN force. A recent study **[18] reported that FN dimers were extended only 70% of their contour length in stretched matrix fibrils. This is consistent with FN fibril stretching involving the compact-to-extended conformational, with no indication of additional extension from domain unfolding.
Highlights.
The isometric tension needed to unfold various protein domains is ~5 pN.
Single integrins generate a force of 1-2 pN on the ECM ligand.
2 pN would only extend an unfolded FNIII (wlc) from 3.5 to 6 nm, not 3–4 fold.
I conclude that unfolding FNIII domains contributes minimally to stretching FN fibrils.
The compact to extended conformation can explain the stretching of FN fibrils.
Acknowledgments
Funding: This work was supported by the National Institutes of Health grant CA47056.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Maurer LM, Ma W, Mosher DF. Dynamic structure of plasma fibronectin. Crit Rev Biochem Mol Biol. 2016;51:213–227. doi: 10.1080/10409238.2016.1184224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ohashi T, Kiehart DP, Erickson HP. Dual labeling of the fibronectin matrix and actin cytoskeleton with green fluorescent protein variants. J Cell Sci. 2002;115:1221–1229. doi: 10.1242/jcs.115.6.1221. [DOI] [PubMed] [Google Scholar]
- 3.Ohashi T, Kiehart DP, Erickson HP. Dynamics and elasticity of the fibronectin matrix in living cell culture visualized by fibronectin-green fluorescent protein. Proc Natl Acad Sci USA. 1999;96:2153–2158. doi: 10.1073/pnas.96.5.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sivakumar P, Czirok A, Rongish BJ, Divakara VP, Wang YP, Dallas SL. New insights into extracellular matrix assembly and reorganization from dynamic imaging of extracellular matrix proteins in living osteoblasts. J Cell Sci. 2006;119:1350–1360. doi: 10.1242/jcs.02830. [DOI] [PubMed] [Google Scholar]
- 5.Davidson LA, Dzamba BD, Keller R, Desimone DW. Live imaging of cell protrusive activity, and extracellular matrix assembly and remodeling during morphogenesis in the frog, Xenopus laevis. Dev Dyn. 2008;237:2684–2692. doi: 10.1002/dvdy.21600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rocco M, Carson M, Hantgan R, McDonagh J, Hermans J. Dependence of the shape of the plasma fibronectin molecule on solvent composition. J Biol Chem. 1983;258:14545–14549. [PubMed] [Google Scholar]
- 7.Erickson HP, Carrell NA. Fibronectin in extended and compact conformations. Electron microscopy and sedimentation analysis. J Biol Chem. 1983;258:14539–14544. [PubMed] [Google Scholar]
- 8.Johnson KJ, Sage H, Briscoe G, Erickson HP. The compact conformation of fibronectin is determined by intramolecular ionic interactions. J Biol Chem. 1999;274:15473–15479. doi: 10.1074/jbc.274.22.15473. [DOI] [PubMed] [Google Scholar]
- 9.Soteriou A, Clarke A, Martin S, Trinick J. Titin folding energy and elasticity. Proc.R.Soc.Lond.[Biol.] 1993;254:83–86. doi: 10.1098/rspb.1993.0130. [DOI] [PubMed] [Google Scholar]
- 10.Erickson HP. Reversible unfolding of FN3 domains provides the structural basis for stretch and elasticity of titin and fibronectin. Proc Natl Acad Sci USA. 1994;91:10114–10118. doi: 10.1073/pnas.91.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rief M, Gautel M, Oesterhelt F, Fernandez JM, Gaub HE. Reversible unfolding of individual titin immunoglobulin domains by AFM. Science. 1997;276:1109–1112. doi: 10.1126/science.276.5315.1109. [DOI] [PubMed] [Google Scholar]
- 12.Oberhauser AF, Marszalek PE, Erickson HP, Fernandez JM. The molecular elasticity of the extracellular matrix protein tenascin. Nature. 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 13.Oberhauser AF, Badilla-Fernandez C, Carrion-Vazquez M, Fernandez JM. The mechanical hierarchies of fibronectin observed with single-molecule AFM. J Mol Biol. 2002;319:433–447. doi: 10.1016/S0022-2836(02)00306-6. [DOI] [PubMed] [Google Scholar]
- 14.Baneyx G, Baugh L, Vogel V. Coexisting conformations of fibronectin in cell culture imaged using fluorescence resonance energy transfer. Proc Natl Acad Sci U S A. 2001;20:20. doi: 10.1073/pnas.251422998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baneyx G, Baugh L, Vogel V. Fibronectin extension and unfolding within cell matrix fibrils controlled by cytoskeletal tension. Proc Natl Acad Sci U S A. 2002;99:5139–5143. doi: 10.1073/pnas.072650799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith ML, Gourdon D, Little WC, Kubow KE, Eguiluz RA, Luna-Morris S, Vogel V. Force-induced unfolding of fibronectin in the extracellular matrix of living cells. PLoS Biol. 2007;5:e268. doi: 10.1371/journal.pbio.0050268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Erickson HP. Stretching fibronectin. J Muscle Res Cell Motil. 2002;23:575–580. doi: 10.1023/a:1023427026818. [DOI] [PubMed] [Google Scholar]
- **18.Früh SM, Schoen I, Ries J, Vogel V. Molecular architecture of native fibronectin fibrils. Nat Commun. 2015;6:7275. doi: 10.1038/ncomms8275. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used elegant super-resolution light microscopy to measure the end-to-end distance of FN dimers in naturally stretched matrix fibrils. The average distance was 133 nm, about 70% of the 190 nm contour length of the dimer. This is consistent with the compact to extended conformational change, with no need to invoke additional extension from domain unfolding.
- 19.Jackson SE. How do small single-domain proteins fold? Fold Des. 1998;3:R81–91. doi: 10.1016/S1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- **20.Shah R, Ohashi T, Erickson HP, Oas TG. Spontaneous Unfolding-Refolding of Fibronectin Type III Domains Assayed by Thiol Exchange: Thermodynamic Stability Correlates with Rates of Unfolding Rather than Folding. J. Biol. Chem. 2017 doi: 10.1074/jbc.M116.760371. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study measured the kinetics of spontaneous unfolding and refolding of a dozen FNIII domains. Domains 3 and 11 of FN are especially weak and spend a significant fraction of time unfolded. These might be subject to limited extension (Table 4) by tension on the fibrils.
- 21.Hughes ML, Dougan L. The physics of pulling polyproteins: a review of single molecule force spectroscopy using the AFM to study protein unfolding. Rep Prog Phys. 2016;79:076601. doi: 10.1088/0034-4885/79/7/076601. [DOI] [PubMed] [Google Scholar]
- 22.Cecconi C, Shank EA, Bustamante C, Marqusee S. Direct observation of the three-state folding of a single protein molecule. Science. 2005;309:2057–2060. doi: 10.1126/science.1116702. [DOI] [PubMed] [Google Scholar]
- 23.Elms PJ, Chodera JD, Bustamante C, Marqusee S. The molten globule state is unusually deformable under mechanical force. Proc Natl Acad Sci U S A. 2012;109:3796–3801. doi: 10.1073/pnas.1115519109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen H, Yuan G, Winardhi RS, Yao M, Popa I, Fernandez JM, Yan J. Dynamics of equilibrium folding and unfolding transitions of titin immunoglobulin domain under constant forces. J Am Chem Soc. 2015;137:3540–3546. doi: 10.1021/ja5119368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alemany A, Rey-Serra B, Frutos S, Cecconi C, Ritort F. Mechanical Folding and Unfolding of Protein Barnase at the Single-Molecule Level. Biophys J. 2016;110:63–74. doi: 10.1016/j.bpj.2015.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.He C, Hu C, Hu X, Hu X, Xiao A, Perkins TT, Li H. Direct Observation of the Reversible Two-State Unfolding and Refolding of an alpha/beta Protein by Single-Molecule Atomic Force Microscopy. Angew Chem Int Ed Engl. 2015;54:9921–9925. doi: 10.1002/anie.201502938. [DOI] [PubMed] [Google Scholar]
- 27.Junker JP, Ziegler F, Rief M. Ligand-dependent equilibrium fluctuations of single calmodulin molecules. Science. 2009;323:633–637. doi: 10.1126/science.1166191. [DOI] [PubMed] [Google Scholar]
- 28.Stigler J, Ziegler F, Gieseke A, Gebhardt JC, Rief M. The complex folding network of single calmodulin molecules. Science. 2011;334:512–516. doi: 10.1126/science.1207598. [DOI] [PubMed] [Google Scholar]
- 29.Zoldak G, Stigler J, Pelz B, Li H, Rief M. Ultrafast folding kinetics and cooperativity of villin headpiece in single-molecule force spectroscopy. Proc Natl Acad Sci U S A. 2013;110:18156–18161. doi: 10.1073/pnas.1311495110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **30.Morimatsu M, Mekhdjian AH, Adhikari AS, Dunn AR. Molecular tension sensors report forces generated by single integrin molecules in living cells. Nano Lett. 2013;13:3985–3989. doi: 10.1021/nl4005145. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study uses a wlc FRET sensor and concludes that the average integrin force is about 2 pN. A separate measure of single molecule FRET gave a similar low force.
- 31.Liu Y, Yehl K, Narui Y, Salaita K. Tension sensing nanoparticles for mechanoimaging at the living/nonliving interface. J Am Chem Soc. 2013;135:5320–5323. doi: 10.1021/ja401494e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang Y, Liu Z, Zhang Y, Galior K, Yang J, Salaita K. A General Approach for Generating Fluorescent Probes to Visualize Piconewton Forces at the Cell Surface. J Am Chem Soc. 2016;138:2901–2904. doi: 10.1021/jacs.5b11602. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study follows up that of Morimatsu and Dunn, focusing on single molecule FRET of dilute and highly dispersed sensors. They find most sensors report 1-3 pN, some report 3-7 pN, and a small fraction report forces of >7 pN, beyond the limit of their sensor.
- 33.Zhang Y, Ge C, Zhu C, Salaita K. DNA-based digital tension probes reveal integrin forces during early cell adhesion. Nat Commun. 2014;5:5167. doi: 10.1038/ncomms6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **34.Galior K, Liu Y, Yehl K, Vivek S, Salaita K. Titin-Based Nanoparticle Tension Sensors Map High-Magnitude Integrin Forces within Focal Adhesions. Nano Lett. 2016;16:341–348. doi: 10.1021/acs.nanolett.5b03888. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study used a sensor based on unfolding the I27 beta sandwich domain. A kinetic analysis suggested forces ~36 pN. I think this force is much too high, but there is lots of clever technology here and no obvious flaw.
- 35.Blakely BL, Dumelin CE, Trappmann B, McGregor LM, Choi CK, Anthony PC, Duesterberg VK, Baker BM, Block SM, Liu DR, et al. A DNA-based molecular probe for optically reporting cellular traction forces. Nat Methods. 2014;11:1229–1232. doi: 10.1038/nmeth.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang X, Ha T. Defining single molecular forces required to activate integrin and notch signaling. Science. 2013;340:991–994. doi: 10.1126/science.1231041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X, Sun J, Xu Q, Chowdhury F, Roein-Peikar M, Wang Y, Ha T. Integrin Molecular Tension within Motile Focal Adhesions. Biophys J. 2015;109:2259–2267. doi: 10.1016/j.bpj.2015.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **38.Chang AC, Mekhdjian AH, Morimatsu M, Denisin AK, Pruitt BL, Dunn AR. Single Molecule Force Measurements in Living Cells Reveal a Minimally Tensioned Integrin State. ACS Nano. 2016 doi: 10.1021/acsnano.6b03314. 10.1021/acsnano.6b03314. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study follows up that of Morimatsu and Dunn, focusing on single molecule FRET of dilute and highly dispersed sensors. They find most sensors report 1-3 pN, some report 3-7 pN, and a small fraction report forces of >7 pN, beyond the limit of their sensor.
- 39.Hatch K, Danilowicz C, Coljee V, Prentiss M. Demonstration that the shear force required to separate short double-stranded DNA does not increase significantly with sequence length for sequences longer than 25 base pairs. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;78:011920. doi: 10.1103/PhysRevE.78.011920. [DOI] [PubMed] [Google Scholar]
- 40.Carrion-Vazquez M, Oberhauser AF, Fowler SB, Marszalek PE, Broedel SE, Clarke J, Fernandez JM. Mechanical and chemical unfolding of a single protein: a comparison. Proc Natl Acad Sci U S A. 1999;96:3694–3699. doi: 10.1073/pnas.96.7.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brenner MD, Zhou R, Conway DE, Lanzano L, Gratton E, Schwartz MA, Ha T. Spider Silk Peptide Is a Compact, Linear Nanospring Ideal for Intracellular Tension Sensing. Nano Lett. 2016;16:2096–2102. doi: 10.1021/acs.nanolett.6b00305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Grashoff C, Hoffman BD, Brenner MD, Zhou R, Parsons M, Yang MT, McLean MA, Sligar SG, Chen CS, Ha T, et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature. 2010;466:263–266. doi: 10.1038/nature09198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar A, Ouyang M, Van den Dries K, McGhee EJ, Tanaka K, Anderson MD, Groisman A, Goult BT, Anderson KI, Schwartz MA. Talin tension sensor reveals novel features of focal adhesion force transmission and mechanosensitivity. J Cell Biol. 2016;213:371–383. doi: 10.1083/jcb.201510012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Austen K, Ringer P, Mehlich A, Chrostek-Grashoff A, Kluger C, Klingner C, Sabass B, Zent R, Rief M, Grashoff C. Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat Cell Biol. 2015;17:1597–1606. doi: 10.1038/ncb3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bustamante C, Marko JF, Siggia ED, Smith S. Entropic elasticity of lambda-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 46.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 47.Ohashi T, Galiacy SD, Briscoe G, Erickson HP. An experimental study of GFP-based FRET, with application to intrinsically unstructured proteins. Protein Sci. 2007;16:1429–1438. doi: 10.1110/ps.072845607. [DOI] [PMC free article] [PubMed] [Google Scholar]