Abstract
Adjacent CpG sites in mammalian genomes can be co-methylated due to the processivity of methyltransferases or demethylases. Yet discordant methylation patterns have also been observed, and found related to stochastic or uncoordinated molecular processes. We focused on a systematic search and investigation of regions in the full human genome that exhibit highly coordinated methylation. We defined 147,888 blocks of tightly coupled CpG sites, called methylation haplotype blocks (MHBs) with 61 sets of whole genome bisulfite sequencing (WGBS) data, and further validated with 101 sets of reduced representation bisulfite sequencing (RRBS) data and 637 sets of methylation array data. Using a metric called methylation haplotype load (MHL), we performed tissue-specific methylation analysis at the block level. Subsets of informative blocks were further identified for deconvolution of heterogeneous samples. Finally, we demonstrated quantitative estimation of tumor load and tissue-of-origin mapping in the circulating cell-free DNA of 59 cancer patients using methylation haplotypes.
Introduction
Mammalian CpG methylation is a relatively stable epigenetic modification, which can be transmitted across cell division1 through DNMT1, and dynamically established, or removed by DNMT3 A/B and TET proteins. Due to the locally coordinated activities of these enzymes, adjacent CpG sites on the same DNA molecules can share similar methylation status, although discordant CpG methylation has been observed, especially in cancer2. The theoretical framework of linkage disequilibrium3, which was developed to model the co-segregation of adjacent genetic variants on human chromosomes in human populations, can be applied to the analysis of CpG co-methylation in cell populations. A number of studies related to the concepts of methylation haplotypes4, epi-alleles5, or epi-haplotypes6 have been reported, albeit at small numbers of genomic regions or limited numbers of cell/tissue types. Recent data production efforts, especially by large consortia7, have produced a large number of whole-genome, base-resolution bisulfite sequencing data sets for many tissue and cell types. These public data sets, in combination with additional WGBS data generated in this study, allowed us to perform full-genome characterization of locally coupled CpG methylation across the largest set of human tissue types available to date, and annotate these blocks of co-methylated CpGs as a distinct set of genomic features.
DNA methylation is cell-type specific, and the pattern can be harnessed for analyzing the relative cell composition of heterogeneous samples, such as different white blood cells in whole blood8, fetal components in maternal circulating cell-free DNA(cfDNA)9, or circulating tumor DNA (ctDNA) in plasma9. Most of these recent efforts relies on the methylation level of individual CpG sites, and are fundamentally limited by the technical noise and sensitivity in measuring single CpG methylation. Recently, Lehmann-Werman demonstrated a superior sensitivity with multi-CpG haplotypes in detecting tissue-specific signatures in cfDNA10, although based on the sparse genome coverage of Illumina 450k methylation arrays (HM450K). Here we performed an exhaustive search of tissue-specific methylation haplotype blocks across the full genome, and proposed a block-level metric, termed methylated haplotype load (MHL), for a systematic discovery of informative markers. Applying our analytic framework and identified markers, we demonstrated accurate determination of tissue origin and prediction of cancer status in clinical plasma samples from patients of lung cancer (LC) and colorectal cancer (CRC) (Fig. 1a).
Figure 1.
Identification and characterization of human methylation haplotype blocks (MHBs). (a) Schematic overview of data generation and analysis. (b) An example of MHB at the promoter of the gene APC. (c) Smooth scatterplots of methylation linkage disequilibrium within MHBs. Red indicate relative higher density and blue indicates relative low density. The yellow dotted lines and percentages highlight the reduction of high linkage disequilibrium (r2>0.9). (d) Co-localization of MHBs with known genomic features. (e) Enrichment of MHBs in known genomic features.
Results
Identification and characterization of methylation haplotype blocks
To investigate the co-methylation status of adjacent CpG sites along single DNA molecules, we extended the concept of genetic linkage disequilibrium3,4 and the r2 metric to quantify the degree of coupled CpG methylation among different DNA molecules. CpG methylation status of multiple CpG sites in single- or paired-end Illumina sequencing reads were extracted to form methylation haplotypes, and pairwise “linkage disequilibrium” of CpG methylation r2 was calculated from the fractions of different methylation haplotypes (see Methods).
We started with 51 sets of published WGBS data from human primary tissues11,12, as well as the H1 human embryonic stem cells, in vitro derived progenitors13 and human cancer cell line14,15. We also included an in-house generated WGBS dataset from 10 adult tissues of one human donor. Across these 61 samples (>2000x combined genome coverage) we identified a total of ~ 55 billion methylation haplotype informative reads that cover 58.2% of autosomal CpGs. The uncovered CpG sites were either in regions with low mappability, or CpG sparse regions where there are too few CpG sites within Illumina read pairs for deriving informative haplotypes. We partitioned the human genome into blocks of tightly coupled CpG methylation sites, called methylation haplotype blocks (MHBs, Fig. 1b), using a r2 cutoff of 0.5. We identified 147,888 MHBs at the average size of 95bp and minimum 3 CpGs per block, which represents ~0.5% of the human genome that tends to be tightly co-regulated on the epigenetic status at the level of single DNA molecules (Supplementary Table 1, Supplementary Fig. 1a, b). The majority of CpG sites within the same MHBs are near perfectly coupled (r2 ~1.0) regardless of the sample type. We found that the fraction of tightly coupled CpG pairs (r2 > 0.9, Fig. 1c) slightly decreased over CpG spacing from stem and progenitor cells (94.8%, mostly cultured cells) to somatic cells (91.2%, mixture of primary adult tissues) to cancer cells (87.8%, mixture of CRC tissues and LC cell lines). The loss of LD in cancer cells was validated by another independent WGBS data from primary kidney cancer tissues16 (Supplementary Fig. 2). Although the WGBS data came from different laboratories that might have batch technical differences, we found that that methylation LD extends further over CpG distance in stem and progenitor cells, which is consistent with our previous observations on 2,020 CpG islands4 for culture cell lines and with another report17. Interestingly, in cancer samples, we observed a reduction of perfectly coupled CpG pairs, which could be related to the pattern of discordant methylation recently reported in variable methylation regions (VMR)2,18. The cancer-specific decayed MHBs were enriched for cancer related pathways and functions (Supplementary Table 2). Nonetheless, the majority of MHBs in cancers still contains tightly coupled CpGs (87.8%), allowing us to harness the pattern for detecting tumor in plasma. We further validated the co-methylation of these MHBs in 101 ENCODE RRBS datasets and 637 TCGA HM450K datasets (Supplementary Note, Supplementary Fig. 3).
Co-localization of methylation haplotype blocks with known regulatory elements
The MHBs established by 61 sets of WGBS data represent a distinct type of genomic feature that partially overlaps with multiple known genomic elements (Fig. 1d). Among all MHBs, 60,828 (41.1%) located in intergenic regions while 87,060 (58.9%) regions in transcribed regions. These MHBs were significantly enriched (P <1.0×10−6) in enhancers, super enhancers, promoters, CpG islands and imprinted genes. In addition, we observed modest depletion in the lamina-associated domains (LAD)19 and the large organized chromatin K9 modifications (LOCK) regions20 modest enrichment in TAD21. Importantly, we observed a strong (26-fold) enrichment in VMR (Fig. 1e), suggesting that increased epigenetic variability in a cell population or tissue can be coordinated locally among hundreds of thousands of genomic regions22. We further examined a subset of MHBs that do not overlap with CpG islands, and observed a consistent enrichment pattern (Fig. 1e, Supplementary Fig. 1c), suggesting that local CpG density alone does not account for the enrichment. Previous studies on mouse and human23,24 demonstrated that dynamically methylated regions were associated with regulatory regions such as enhancer-like regions marked by H3K27ac and transcription factor binding sites. Using publicly histone mapping data for human adult tissues, we found co-localization of methylation haplotype blocks with marks for active promoters (H3K4me3 with H3K27ac), but not for active enhancers25 (no peak for H3K4me1) (Supplementary Fig. 4). We found that enhancers tend to overlap with CpG sparse MHBs, whereas the co-localization with super enhancers were independent of CpG density (Supplementary Fig. 1c). Therefore, MHBs likely capture the local coherent epigenetic signatures that are directly or indirectly coupled to transcriptional regulation.
Block-level analysis of human normal tissues and stem cell lines with methylation haplotype load
To enable quantitative analysis of the methylation patterns within individual MHBs across many samples, we need a single metric to define the methylated pattern of multiple CpG sites within each block. Ideally this metric is not only a function of average methylation level for all the CpG sites in the block, but also can capture the pattern of co-methylation on single DNA molecules. Therefore, we defined methylation haplotype load (MHL), a weighted mean of the fraction of fully methylated haplotypes and substrings at different lengths (i.e. all possible substrings, see Methods). Compared with other metrics used in the literature (methylation level, methylation entropy, epi-polymorphism and haplotypes counts), MHL is capable of distinguishing blocks that have the same average methylation but various degrees of coordinated methylation (Fig. 2). In addition, MHL is bounded between 0 and 1, which allows for direct comparison of different regions across many data sets.
Figure 2.
Comparison of methylation haplotype load with four other metrics used in the literatures. Five patterns of methylation haplotype combinations are used to illustrate the difference between methylation frequency, methylation entropy, epi-polymorphism and methylation haplotype load. MHL is the only metric that can discriminate all the five patterns.
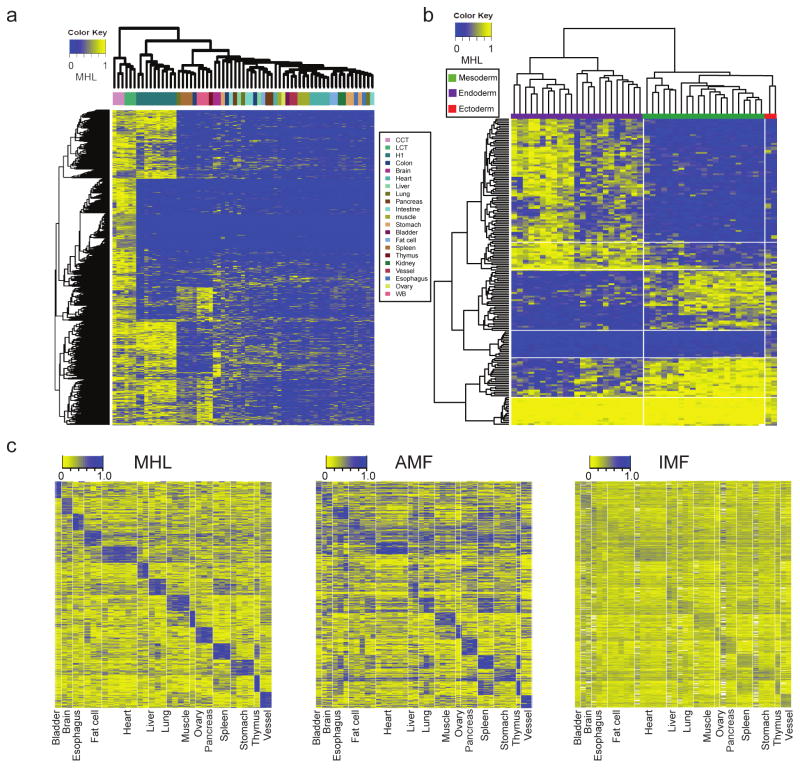
We next asked whether treating MHBs as individual genomic features and performing quantitative analysis based on MHL would provide an advantage over previous approaches using individual CpG sites or weighted (or unweighted) averaging of multiple CpG sites in certain genomic windows. Therefore, we clustered 65 WGBS data sets (including 4 additional colon and lung cancer WGBS sets26) from human solid tissues based on MHL. Unsupervised clustering with the 15% most variable MHBs showed that, regardless of the data sources, samples of the same tissue origin clustered together (Fig. 3a), while cancer samples and stem cell samples exhibit distinct patterns from human adult tissues. PCA analysis on all MHBs yielded a similar pattern (Supplementary Fig. 5). To identify a subset of MHBs for effective clustering of human somatic tissues, we calculated a tissue specific index (TSI) for each MHB. Feature selection using random forest identified a set of 1,365 tissue-specific MHBs (Supplementary Table 3) that can predict tissue type at an accuracy of 0.89 (95%CI: 0.84–0.93), although several tissue types share rather similar cell compositions (i.e. muscle vs. heart). Using these MHBs, we compared the performance between MHL, average methylation fraction(AMF) in the MHBs and all individual CpG methylation fraction(IMF). MHL and AMF provided similar tissue specificity, while MHL has a lower noise (background: 0.29, 95%CI: 0.23–0.35) compared with AMF (background: 0.4, 95%CI: 0.32–0.48). Clustering based on individual CpGs in the blocks has the worst performance, which might be due to higher biological or technical viability of individual CpG sites (Fig. 3c). Thus, block-level analysis based on MHL is advantageous over single CpG or local averaging of multiple CpG sites in distinguishing tissue types.
Figure 3.
Tissue clustering based on methylation haplotype load. (a) MHL based unsupervised clustering of human tissues using the 15% most variable regions. (b) Supervised clustering of germ-layer specific MHBs. (c) MHL exhibits better signal-to-noise ratio than AMF and IMF for sample clustering.
The human adult tissues that we used have various degrees of similarity amongst each other. We hypothesize that this is primarily defined by their developmental lineage, and that the related MHBs might reveal epigenetic insights relevant to germ layer speciation. We searched for MHBs that have differential MHL among data sets from the three germ layers. In total we identified 114 ectoderm-specific MHBs (99 hyper- and 15 hypo-methylated), 75 endoderm specific MHBs (58 hyper and 17 hypo-methylated) and 31 mesoderm specific MHBs (9 hyper and 22 hypo-methylated) (Supplementary Table 4). Cluster analysis based on layer specific MHBs shows expected aggregation among tissues of same the lineage (Fig. 3b). We speculated that some of these MHBs might capture binding events of transcription factors (TF) specific to developmental germ-layers. Overlapped with TF binding events identified from ENCODE TF binding data27, we observed patterns of TFs binding to layer specific MHBs. (Supplementary Fig. 6). For layer specific MHBs with low MHL, we identified 53 TFs in mesoderm specific MHBs, 71 in endoderm specific MHBs and 2 in ectoderm specific MHBs. Gene ontology analysis showed TFs binding to mesoderm exhibit negative regulator activity, while TFs binding to endoderm exhibited positive regulator activity (Supplementary Table 5). For layer specific MHBs with high MHL, we identified 38 TFs in mesoderm specific MHBs, 102 in endoderm specific MHB and 145 in ectoderm specific MHBs. Interestingly, ectoderm and endoderm shared few bounded TFs, while mesoderm tissues share multiple groups of TFs with ectoderm and endoderm. We identified two endoderm specific high-MHL regions, which are associated with ERR1 and NANOG. This is consistent with a previous finding that mouse ES cells differentiated spontaneously into visceral/parietal endoderm upon NANOG knock-out28. Mesoderm and endoderm shared low-MHL regions might have regulatory functions in the fate commitment towards multiple tissues, whereas ectoderm specific high-MHL regions might induce the ectoderm development by suppressing the path towards the immune lineage (Supplementary Fig. 6). These observations are indicative of two distinctive “push” and “pull” mechanisms in the transition of cell states that have been harnessed for the induction of pluripotency by over-expressing lineage specifiers29.
Methylation-haplotype based analysis of circulating cell-free DNA in cancer patients and healthy donors
A unique aspect of methylation haplotype analysis is that the pattern of co-methylation, especially within MHBs, is robust in capturing low-frequency alleles among a heterogeneous population of molecules or cells, in the presence of biological noise or technical variability (ie. incomplete bisulfite conversion or sequencing errors). To explore potential clinical applications, we next focused on the methylation haplotype analysis of cfDNA from healthy donors and cancer patients, of which various low fractions of DNA molecules were released from tumor cells and potentially carry epigenetic signatures different from blood. We isolated cfDNA from human plasma of 75 normal individuals (NCP), 29 lung cancer patients (LCP), and 30 colorectal cancer patients (CCP). Due to the limited DNA availability, we performed scRRBS30 and obtained an average of 13 million paired-end 150bp reads per sample. On average, 57.7% WGBS-defined MHBs were covered in our RRBS data set from clinical samples.
We queried the presence of tumor specific signatures in the plasma samples, using methylation haplotypes identified from tumor tissues as the reference and normal samples as the negative controls. For five LCP and five CCP samples, we obtained matched primary tumor tissues, and generated RRBS data from 100 ng of tumor genomic DNA. Focusing on the MHBs with low MHL in the blood, we identified cancer-associated highly methylated haplotypes (caHMHs). Such haplotypes were present only in the tumor tissues and the matched plasma from the same patient, but not in whole blood or any other non-cancer samples. We found caHMHs in all cancer patient plasma samples (Average=36, IQR=17, Supplementary Table 6). These caHMHs were associated with 183 genes, some of which are known to be aberrantly methylated in human cancers such as WDR37, VAX1, SMPD1 (Supplementary Table 6). Next, we extended the analysis to 49 additional cancer plasma samples that have no matched tumor samples, using 75 NCPs as the background. On average 60 (IQR=31) caHMHs were identified for each cancer plasma sample (Supplementary Table 6). Interestingly, a significant fraction (35%) of caHMHs called on matched tumor-plasma pairs were also detected the expanded set of cancer patient plasma samples. Most caHMHs were individual specific, while several caHMHs were present in at least 53% CCPs and 62% LCPs (Supplementary Fig. 7). Improving the sampling depth, by using more input cfDNA or reducing sample loss during the experiments, will likely increase the number of caHMHs commonly observed in multiple patients.
Next we sought to quantify the cancer DNA fraction in cancer plasma samples using deconvolution. We used the reference data from primary cancer biopsies (LCT and CCT) and from 10 normal tissues, and estimated that a predominant fraction, 72.0% (IQR=40%) in the cancer and normal plasma were contributed by white blood cells, which is consistent with the levels reported recently based on shallow WGBS (69.4%)9. Primary tumor and normal tissue-of-origin contributed at the similar levels of 2.3% (IQR=3.7%) and 3.0% (IQR=4.4%). In contrast, when we applied the same deconvolution analysis to normal plasma, we found only residual plasma fragments with a tumor signature (0.17%, IQR=2.9% for CCT and 1.0%, IQR=3.1% for LCT), which were significantly lower (P=3.4x10−5 and 5.2x10−10 for CCT and LCT, two-sample t-test) than cancer plasma. We also found that 23/30 CCPs and only 10/75 NCPs have detectable contribution from CCT while 26/29 LCPs and 20/75 NCPs have detectable contribution from LCT (Supplementary Fig. 8). Therefore, cfDNA contains a relatively stable fraction of molecules released from various normal tissues, whereas in cancer patients tumor cells released DNA molecules at higher levels than normal tissues (Supplementary Table 7). The fractions of white blood cells observed are lower than what was reported previously9, and is likely due to the inclusion of 10 normal tissue types in deconvolution.
Next, we searched for a small subset of MHBs among all the RRBS targets that have significantly higher levels of MHL in cancer plasma than in normal plasma. We found 81 and 94 MHBs with significantly higher MHL for CCPs and LCPs (Supplementary Table 8). The majority (71/81 for CCP and 83/94 for LCP) were also present in at least one of the matched primary tumor-plasma pairs. Some of these regions (such as HOXA3) have been shown aberrantly methylated in lung cancer and colorectal cancer. Using these MHBs as markers, the diagnostic sensitivity is 96.7% and 93.1% for colorectal cancer and lung cancer respectively at the specificity 94.6% and 90.6% respectively. As a comparison, we also performed a prediction based on average 5mC methylation level within these MHB regions, or based on single CpG sites. MHL was found to be superior to average 5mC methylation level (sensitivity of 90.0% and 86.2%; specificity of 89.3% and 90.6% for CCP and LCP respectively) and methylation of individual CpG site (sensitivity of 89.6% and 80.6%; specificity of 89.3% and 92.0%).
We then sought to use the information from normal human tissues, primary tumor biopsies and cancer cell lines to improve the detection of ctDNA. We started by selecting a subset of MHBs that show high MHL (>0.5) in primary cancer biopsies and low MHL (<0.1) in whole blood, then clustered these MHBs into three groups based on the MHL in all normal and cancer plasma, as well as cancer and normal tissues (Fig. 4a,b). We identified a subset (Group II) of MHBs that have high MHL in cancer tissues and low MHLs in normal tissues (Supplementary Table 9). Cancer plasma showed significantly higher MHL in these regions than normal plasma (P=1.4×10−12 and 6.2×10−8 for CCP and LCP). By computationally mixing the sequencing reads from cancer tissues and whole blood samples (WB), we created synthetic admixtures at various levels of tumor fraction. We found that MHL is 2–5 folder higher than the methylation level of individual CpG sites across the full range of tumor fractions (Supplementary Table 9). Remarkably, MHL provides additional gain of signal-to-noise ratio (mean divided by standard deviation) compared with AMF as the fraction of tumor DNA decreases below 10%, which is typical for clinical samples (Fig. 4c). We then took the individual plasma data sets, and predicted the tumor fraction based on the MHL distribution established by computational mixing (Fig. 4a,b). Except for a small number (N<5) of outliers, we observed significantly higher average MHL in cancer plasma than in normal plasma (Fig. 4d). Note that all Group II MHBs were selected without using any information from the plasma samples, and hence they should be generally applicable to other plasma samples. Interestingly, we also found that the estimated tumor DNA fractions were positively correlated with normalized cfDNA yield from the cancer patients (P=0.000023, Supplementary Fig. 9; Supplementary Table 10).
Figure 4.
Quantitative estimation of cancer DNA proportion in cell-free DNA based on MHL of informative MHBs. (a) Colorectal cancer (b) Lung cancer. Informative MHBs were selected based on the presence of high-MHL in cancer solid tissues (CT) and the absence of MHL in whole blood (WB). Group II regions have high MHL in cancer tissues (MHL>0.5) and cancer plasma while low MHL in WB and normal tissues (MHL<0.1), and hence were selected for further analysis. Bar-plots show average MHL in different groups of samples. MHL in cancer plasma (CCP and LCP) and normal plasma (NP) were compared with a two-tail t-test. NCT denotes normal colon tissues, NLT denotes normal lung tissues, and ONT denotes other normal tissues. (c) MHL has higher signal-to-noise ratio (Mean/SD) than individual 5mC levels as tumor fraction decreases. X-axis is the tumor fraction in synthetic mixtures. (d) Estimation of the cancer DNA proportions in plasma samples. CCP denotes colorectal cancer plasma, LCP denotes lung cancer plasma, and NP denotes normal plasma.
Recent studies9,10,31 have demonstrated that epigenetic information imbedded in cfDNA has the potential for predicting tumor’s tissue-of-origin. Consistently, we found that tissue-of-origin derived methylation haplotypes were the most abundant fraction in cancer plasma (Supplementary Table 6, 7). To predict tissue-of-origin with quantifiable sensitivity and specificity with MHBs, we compiled 43 WGBS and RRBS data sets for 10 human normal tissues that have high cancer incident rate, and identified a set of 2,880 tissue-specific MHBs (Supplementary Table 11). We then used these tissue-specific MHBs or subsets to predict the tissue-of-origin for the cancer plasma samples. Although we found many tissue-of-origin specific MHBs that have low MHL in normal plasma (Fig. 5a), the multiclass prediction based on random forest yielded limited power. This is likely due to the large number of the tissue classes (N=10). We then adopted an alternative approach by counting the number of methylated (or high MHL) tissue-specific MHBs in the plasma samples and comparing with all other tissues, to infer the most probable tissue-of-origin. At the cutoff of minimal 10 tissue-specific MHL signals per tissue type, we observed an average of 90% accuracy for mapping a data set from the primary tissue to its tissue type (Fig. 5b). We then applied this method to the plasma data, and achieved an average prediction accuracy of 82.8%, 88.5%, 91.2% for the CCP, LCP, and NCP samples respectively with 5-fold cross-validation (Fig. 5c, Supplementary Fig. 10, Supplementary Table 12). The classified samples were mainly due to the inclusion of samples with heterogeneous clinical status: 4 out of 5 CCP were from metastatic colorectal cancer patients while the fifth was in fact tubular adenoma; one misclassified LCP sample came from a patient with cryptococcal pulmonary infection.
Figure 5.
MHL-based prediction of cancer tissue-of-origin from plasma DNA. (a) Detection of tissue-specific MHL in the plasma of cancer patients, but not normal plasma or whole blood. Tissue specific MHL were visible in corresponding tissue and cancer plasma, indicating the feasibility for tissue-of-origin mapping. (b) Identification of informative MHBs for tissue prediction, using training data included WGBS and RRBS datasets from 10 human normal tissues. (c) Application of the prediction model to plasma samples from cancer patients and normal individuals.
Discussions
In this study, we extended a well-established concept in population genetics, linkage disequilibrium, to the analysis of co-methylated CpG patterns. While the mathematical representations are identical, there are two key differences. First, traditional linkage disequilibrium was defined on human individuals in a population, whereas in this study the analysis was performed on the diploid genome of individual cells in a heterogeneous cell population. Second, linkage disequilibrium in human populations depends on the mutation rate, frequency of meiotic recombination, effective population size and demographic history. The LD level decays typically over the range of hundreds of kilobases to megabases. In contrast, CpG co-methylation depends on DNA methytransferases and demethylases, which tend to have much lower processivity (if any), and, in the case of hemi-methyltransferases, much lower fidelity compared with DNA polymerases32. Therefore, methylation LD decays over much shorter distance in tens to hundreds of bases, with the exception of imprinting regions. Even if longer-read sequencing methods were used, we do not expect a radical change of the block-like pattern presented in this work, which is supported by another recent study33. Nonetheless, these short and punctuated blocks capture discrete entities of epigenetic regulation in individual cells widespread in the human genome. This phenomenon can be harnessed to improve the robustness and sensitivity of DNA methylation analysis, such as the deconvolution of data from heterogeneous samples including cfDNA.
While we demonstrated a superior power of MHL over single-CpG methylation level or average methylation level in classification and deconvolution using MHBs as features, the accuracy is slightly less than what has been reported on the deconvolution of blood cell types. One major difference is that each reference tissue type itself is a mixture of multiple cell types that might share various degrees of similarity with another reference tissue type. Furthermore, most solid tissues also contain blood vessels and blood cells. Given such background signals, the accuracy that we achieved is promising, and will be further improved once reference methylomes of pure adult cell types are available.
Practically, the amount of cfDNA per patient is rather limited, typically in the range of tens of nanogram. We therefore used 1–10 ng per patient for the scRRBS experiments. Considering the material losses during bisulfite conversion, library preparation, and the sequencing depth, there were most likely no more than 30 genome equivalents in each data set. Our data set is rather sparse, especially when the fraction of tumor DNA is low. Hence, the chance of finding cancer-specific methylation haplotypes in a specific region consistently across many samples is low. This is likely the reason that marker sets selected using random forest has limited sensitivity and specificity. However, epigenetic abnormalities tend to be more widespread across the genome (compared with somatic mutations), and hence, we managed to integrate the sparse coverage across many loci to achieve accurate prediction by direct counting of methylated haplotypes within the informative genomics regions. Importantly, we showed that, in cancer patients, plasma contains circulating DNA fragments from both normal and malignant cell types detectible with methylation haplotyping. This allowed us to detect the presence of cancer and map the tissue or organ of tumor growth. Interestingly, when we combined all the data from primary tumors and cancer cell lines as a “pan-cancer” tissue, and included it as the 11th reference for tissue-of-origin mapping, the detection sensitivity and specificity was further improved (Supplementary Fig. 11,12), suggesting that a joint analysis of cancer signature and tissue-of-origin signature is more sensitive than focusing on cancer signature alone. In summary, methylation haplotyping in plasma is a promising strategy for early detection of tumor and its primary growth site, as well as continuous monitoring of tumor progression and metastasis to multiple organs. With more plasma samples from patients at multiple clearly defined cancer stages and from healthy controls, it is possible to further improve the prediction sensitivity and specificity to a level adequate for clinical testing.
Online Methods
Normal and cancer samples
Ten human primary tissues were purchased from BioChain Institute Inc. Cancer patient tissues and plasma samples were purchased from UCSD Moores Cancer Center and normal plasma samples were obtained from UCSD Shirley Eye center under IRB protocols approved by UCSD Human Research Protections Program (HRPP). All data sets generated in this study or obtained from public databases were listed in Supplementary Table 13.
Generation of DNA libraries for sequencing
Extracted genomic DNA were prepared for bisulfite sequencing using published protocols. For whole genome bisulfite (WGBS) and reduced representation bisulfite sequencing (RRBS), the DNA fragments were adapted to barcoded methylated adaptors (Illumina). For WGBS, the adapted DNA were converted using the EZ DNA Methylation Lightning kit (Zymo Research) and then amplified for 10 cycles using iQ SYBR Green Supermix (BioRad). For RRBS, the adapted DNA were converted using the MethylCode™ Bisulfite Conversion kit (Thermo Fisher Scientific) and amplified using the PfuTurboCx polymerase (Agilent) for 12–14 cycles. Libraries were pooled and size selected using 6% TBE polyacrylamide gels. Libraries were sequenced using the Illumina HiSeq platform for paired-end 100–111 cycles, the Illumina MiSeq platform for paired-end 75 cycles, and the GAIIx (WGBS only) for single-end 36 cycles.
Read mapping
WGBS and RRBS data were processed in similar fashions. We first trimmed all PE or SE fastq files using trim-galore version 0.3.3 to remove low quality bases and biased read positions. Next, the reads were encoded to map to a three-letter genome via conversion of all C to T or G to A if the read appears to be from the reverse complement strand. Then the reads were mapped using BWA mem version 0.7.5a, with the options “-B2 -c1000” to both the Watson and Crick converted genomes. The alignments with mapping quality scores of less than 5 were discarded and only reads with a higher best mapping quality score in either Watson or Crick were kept. Finally, the encoded read sequences were replaced by the original read sequences in the final BAM files. Overlapping pair end reads were also clipped with bamUtils clipOverlap function.
Methylation haplotype blocks (MHBs)
Human genome was split into non-overlapping “sequenceable and mappable” segments using a set of in-house generated WGBS data from 10 tissues of a 25-year adult male donor. Mapped reads from WGBS data sets were converted into methylation haplotypes within each segment. Methylation linkage disequilibrium was calculated on the combined methylation haplotypes. We then partitioned each segment into methylation haplotype blocks (MHBs). MHBs were defined as the genomic region in which the r2 value of two adjacent CpG sites is no less than 0.5.
High methylation linkage regions defined based on ENCODE and TCGA data
We collected RRBS data from the ENCODE project (downloaded from UCSC Browser) and HM450K data from the TCGA project. Pearson correlation coefficient were calculated between adjacent CpG sites across all samples. The Takai and Jones’s sliding-window algorithm34 was used to identify blocks of highly correlated methylation. We set a 100-base window in the beginning of genomic position and move the window to the downstream when there are least 2 probes in the window. Calculate the total probes in extended regions until the last window does not meet the criteria. The regions covering at least 4 probes were defined as CpG dense regions, and the average Pearson correlation coefficients among all the probes in cancer and normal samples were calculated respectively. Simulation analysis to investigate the relationship between LD at the single-read level and correlation coefficients of average 5mC between two CpG sites were performed based on random sampling of 10 different methylation haplotypes from each of the 1000 individuals.
Enrichment analysis of methylation haplotype blocks for known functional elements
Enrichment analysis was performed by random sampling as previously described35. Genomic regions with same number (147,888), fragment length distribution and CpG ratios were randomly sampled within the mappable regions (genomic regions beyond CRG mappability blacklisted regions and non-cover regions in our WGBS dataset), and repeated 1,000,000 times. Statistical significance was estimated based on empirical p-value (P). Fold changes (enrichment factors) were calculated as the ratios of observation over expectation. Exon, intron, 5-UTR, 3-UTR were collected UCSC database. Enhancer definition was based on Andersson et al36, super enhancer was derived from Hnisz et al37 and promoter regions were based on the definition by Thurman et al38. All the genomic coordinates were based on GRCh37/hg19.
Methylation haplotype load (MHL)
We defined a methylated haplotype load (MHL) for each candidate region, which is the normalized fraction of methylated haplotypes at different length:
Where l is the length of haplotypes, P(MHi) is the fraction of fully successive methylated CpGs with i loci. For a haplotype of length L, we considered all the sub-strings with length from 1 to L in this calculation. wi is the weight for i-locus haplotype. Options for weights are wi = i or wi = i2 to favor the contribution of longer haplotyes. In the present study, wi = i was applied.
Following the concept of Shannon entropy H(x), methylation entropy (ME) for haplotype variable in specific genome region were calculated with the following formula:
For a genome region with b CpG loci and n methylation haplotype, P(Hi) represents the probability of observing methylation haplotype Hi, which can be calculated by dividing the number of reads carrying this haplotype by the total reads in this genomic region. ME is bounded between 0 and 1, and can be directly compared across different regions genome-wide and across multiple samples. Methylation entropy were widely used in the measurement of variability of DNA methylation in specific genome regions39.
Epi-polymorphism40 was calculated as
where Pi is the frequency of epi-allele i the population (with 16 potential epialleles representing all possible methylation states of the set of four CpGs).
Developmental germ layers and tissue specific MHBs
To investigate the germ layer and tissue specific MHBs, group specific index (GSI, see below) was defined. An empirical threshold GSI>0.6 was used define layer and tissue specific MHBs. Layer specific MHBs were selected again to show the ability to distinguish different development layers. Tissue specific MHBs were further used for tissue mapping and cancer diagnosis.
n indicates the number of the groups. MHL(j) denotes the average of MHL of jth group. MHL(max) denotes the average of MHL of highest methylated group.
Genome-wide methylation haplotype load matrix (MHL) analysis
Methylation haplotype load was calculated for all MHBs on each sample. The MHBs with top 15% MHL were included in the heatmap to investigate the tissue relationship. The Euclidean distance and Ward.D aggregation were used in the heatmap plot (R, gplots package41). PCA (R package prcomp42) was conducted with default setting of the corresponding R packages42. Before the PCA analysis, raw data were quantile normalized within same tissue/cell groups. Standardization (scale) and batch effect elimination (the Combat algorithm43) were also applied to decrease the random noise. MAF and IMF were extracted from BAM files with customized PileOMeth (https://github.com/dpryan79/PileOMeth). Differential MHL analysis between cancer plasma and normal plasma were based on two-tailed Student’s t-test or Wilcoxon rank sum test. Correction for multiple testing was based on false discovery rate (FDR). Statistic variations were estimated among different groups and therefore one-way ANOVA analysis could be conducted.
Simulation and real-data deconvolution analysis
Deconvolution analysis was performed on simulated and experimental datasets. The deconvolution references were constructed on data from human normal primary tissues, whole blood (WB), colorectal cancer tissues (CCT) and lung cancer tissues (LCT). For the simulation analysis, methylation haplotypes from CCT and WB were randomly mixed to generate a series of synthetic data sets with CCT factions ranging from 0.1% to 50%. We then plotted the expected and observed CCT factions. Although MHL is a non-linear metrics, when mixing CCT and WB, we found the deconvolution result is accurate with log-transform (median root-mean-square-error < 5%), which is within the acceptable region of the deconvolution method44 when the contribution of colorectal fraction is less than 20%. Tissue specific MHBs were selected features for deconvolution based on non-negative decomposition with quadratic programming9,44,45. MHL values were log-transformed before deconvolution.
Highly methylated haplotype in cancer plasma and normal tissues
Highly methylated haplotype (HMH) was defined as the methylation haplotype that have at least 2 methylated CpGs in the haplotype. Cancer-associated highly methylated haplotypes (caHMH) were the ones only found in cancer plasma samples but absence in any of the normal plasma samples and normal tissues. For the analysis of matched tumor-plasma data from the same individuals, caHMHs were the HMHs present in both the cancer plasma and the matched primary cancer tissues, but absence in all normal samples. In the analysis of plasma samples with no matched primary tumor tissue, we identified caHMHs by subtracting HMHs found in cancer plasma with those present in all normal tissues and all normal plasma samples.
Simulation of MHL in plasma mixture and comparison between MHL and 5mC in the plasma mixture
In evaluating caHMHs as potential markers for non-invasive diagnosis, we hypothesized that cfDNA in plasma is a mixture of DNA fragments from cancer cells and WB cells at different ratios (cancer DNA fragment from 0.1% to 50%). We created synthetic mixtures by random sampling of haplotypes in the Group II regions from cancer and WB data sets at different ratios, and repeated 1,000 times to empirically determined the mean and variance of MHL and 5mC levels at different fractions of cancer DNA. Once an empirical “standard curve” was constructed, we then used it to estimate the fraction cancer DNA in the plasma samples. In addition, we assessed the relationship between estimated cfDNA fraction and log-transformed normalized plasma cfDNA yield by linear regression. Signal-to-noise ratio to MHL and 5mC was conducted with the 1,000-time sampling procedure and then the average estimated tumor fraction as well as the variation (standard deviation) were recorded and the ratio was calculated to measure the performance of the metric.
Mapping cancer tissue-of-origin with plasma DNA
The workflow for data analysis is illustrated in Supplementary Fig. 13. Tissue specific methylation haplotype blocks (tsMHBs) were identified by a 2-tailed t-test with FDR correction. Additional statistical analyses with MHL were also conducted by 2-tailed t-test unless stated explicitly. CRC plasma and LC plasma distinguish prediction evaluation were applied random forecast therefore the test and validation sample were independent. Tissue-of-origin prediction was performed using a tsMHBs counting strategy, in which the tissue-of-origin of the plasma were assigned to the reference group with the maximum number of tsMHB fragments (assignment by maximum likelihood). Specifically, in the first stage, the tissue-specific MHBs were identified with WGBS and RRBS datasets from solid tissues in the training samples. tsMHBs (each tissue have ~ 300 MHBs) were identified with the cutoff GSI> 0.1. In the second stage, the predictions were validated with our own RRBS dataset that included 30 colorectal cancer plasma, 29 lung cancer plasma and 75 normal plasma samples. In the test dataset, we separated the samples into 5 parts so that 5-fold cross-validation could be applied to estimate the stability of the prediction, and the number of tissue-specific MHB features were iterating from 50 to 300. The minimum number of features was selected when the accuracy for cancer plasma is higher than 0.8 and the accuracy for normal plasma is higher than 0.9 since we require high specificity in clinical applications. The selected number of features were used in the remaining samples to measure the accuracy of tissue-mapping. The variations of sensitivity, specificity, and accuracy in different subsets of 5-fold cross-variation were low (training dataset standard deviation<0.04 while testing dataset standard deviation<0.14)
Joint analysis of tumor and normal tissue for non-invasive cancer detection in plasma
Cancer-specific markers (GSI scores derived from 8 CRC, 8 LC and 2 KC) and tissue-specific markers were integrated and considered as a “pan-cancer tissue”, and then together with the data sets from 10 normal tissues were applied for the tissue/reference-specific MHB identification. The top 200 MHBs specific to each of the 11 reference tissues were selected as the prediction features. The distribution for the reference specific MHBs in 75 normal plasma samples, 30 CRC plasma and 29 LC plasma samples were constructed for 11 references. The p-value of each reference in the plasma could be inferred by comparison with background distribution of the reference in normal plasma. Meanwhile, tissue-of-origin was assigned by maximum Z-scores among different references. With leave-one out cross-validation on normal plasma, the Type-1 error (FDR) for the corresponding Z-score threshold and sensitivity were estimated. Finally, setting a predefined Z-score threshold could be also used for tissue-of-origin assignment, meanwhile, ROC curve was built to show the performance of the predictors.
Data Availability
WGBS and RRBS data are available at the Gene Expression Omnibus (GEO) under accession GSE79279.
Code Availability
All codes and scripts developed for this study are available for non-commercial use at http://genome-tech.ucsd.edu/public/MONOD_NG_TR44413/.
Supplementary Material
Supplementary Table 1: A complete list of MHBs
Supplementary Table 2: Gene ontology of MHBs that loss methylation linkage in cancers.
Supplementary Table 3: Tissue specific MHBs for classification of normal tissues.
Supplementary Table 4: Layer specific MHBs with group specificity index.
Supplementary Table 5: Gene ontology analysis of TFs bound MHBs.
Supplementary Table 6: Cancer-associated High-Methylation-Haplotype (caHMH) regions.
Supplementary Table 7: Deconvolution of plasma samples by 10 normal tissues, LCT, and CCT.
Supplementary Table 8: Differential MHLs between cancer plasma and normal plasma.
Supplementary Table 9: Group II MHB regions for estimation of cancer DNA proportion.
Supplementary Table 10: Relationship between average cancer fraction and cell-free DNA extraction yield.
Supplementary Table 11: Predictors for colon cancer, lung cancer, and normal plasma.
Supplementary Table 12: Prediction accuracy based on tsMHB counting with 5-fold cross-validation.
Supplementary Table 13: Information of all samples used in this study.
Acknowledgments
This study was supported by NIH grants R01GM097253 (to K.Z.) and P30CA23100. We thank S. Kaushal for managing and handling patient samples in UCSD Moores Cancer Center BTTSR, S. Lippman, R. Liu and B. Ren for insightful discussions.
Abbreviation
- MHB
methylation haplotype load
- MHL
Methylation Haplotype Load
- cf-DNA
cell-free DNA
- RRBS
Reduced representation bisulfite sequencing
- scRRBS
single-cell reduced-representation bisulfite sequencing
- WGBS
genome-wide bisulfite sequencing
- TCGA
The Cancer Genome Atlas project
- ENCODE
the Encyclopedia of DNA Elements
- GEO
Gene Expression Omnibus
- LC
Lung Cancer
- CRC
Colorectal cancer
- ACC
Accuracy
- caHMH
cancer associated Highly Methylated Haplotype
- tsMHB
tissue specific methylation haplotype blocks
- CCT
Colorectal cancer tissue
- CCP
colorectal cancer plasma
- LCT
lung cancer tissue
- LCP
lung cancer plasma
- NP
normal plasma
Footnotes
Author’s Contributions
Ku.Z. conceived the initial concept and oversaw the study. S.G., D.D. and Ku.Z. performed bioinformatics analyses. N.P., D.D., and H.F. performed experiments. Ka. Z. contributed normal plasma samples. Ku. Z., S.G. and D.D. wrote the manuscript with inputs from all co-authors.
Competing Financial interests
S. Guo, D. Diep and Ku. Zhang were listed as inventors in patent applications related to the methods disclosed in this manuscript. Ku. Z. is a co-founder and scientific advisor of Singlera Genomics Inc.
References
- 1.Wigler M, Levy D, Perucho M. The somatic replication of DNA methylation. Cell. 1981;24:33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- 2.Landau DA, et al. Locally disordered methylation forms the basis of intratumor methylome variation in chronic lymphocytic leukemia. Cancer Cell. 2014;26:813–25. doi: 10.1016/j.ccell.2014.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slatkin M. Linkage disequilibrium--understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 2008;9:477–85. doi: 10.1038/nrg2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shoemaker R, Deng J, Wang W, Zhang K. Allele-specific methylation is prevalent and is contributed by CpG-SNPs in the human genome. Genome Res. 2010;20:883–9. doi: 10.1101/gr.104695.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones B. DNA methylation: Switching phenotypes with epialleles. Nat Rev Genet. 2014;15:572. doi: 10.1038/nrg3797. [DOI] [PubMed] [Google Scholar]
- 6.Schwartzman O, Tanay A. Single-cell epigenomics: techniques and emerging applications. Nat Rev Genet. 2015;16:716–26. doi: 10.1038/nrg3980. [DOI] [PubMed] [Google Scholar]
- 7.Stunnenberg HG, Hirst M International Human Epigenome, C. The International Human Epigenome Consortium: A Blueprint for Scientific Collaboration and Discovery. Cell. 2016;167:1897. doi: 10.1016/j.cell.2016.12.002. [DOI] [PubMed] [Google Scholar]
- 8.Houseman EA, et al. Reference-free deconvolution of DNA methylation data and mediation by cell composition effects. BMC Bioinformatics. 2016;17:259. doi: 10.1186/s12859-016-1140-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun K, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A. 2015;112:E5503–12. doi: 10.1073/pnas.1508736112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lehmann-Werman R, et al. Identification of tissue-specific cell death using methylation patterns of circulating DNA. Proc Natl Acad Sci U S A. 2016;113:E1826–34. doi: 10.1073/pnas.1519286113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schultz MD, et al. Human body epigenome maps reveal noncanonical DNA methylation variation. Nature. 2015;523:212–6. doi: 10.1038/nature14465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heyn H, et al. Distinct DNA methylomes of newborns and centenarians. Proc Natl Acad Sci U S A. 2012;109:10522–7. doi: 10.1073/pnas.1120658109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie W, et al. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell. 2013;153:1134–48. doi: 10.1016/j.cell.2013.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blattler A, et al. Global loss of DNA methylation uncovers intronic enhancers in genes showing expression changes. Genome Biol. 2014;15:469. doi: 10.1186/s13059-014-0469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heyn H, et al. Epigenomic analysis detects aberrant super-enhancer DNA methylation in human cancer. Genome Biol. 2016;17:11. doi: 10.1186/s13059-016-0879-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen K, et al. Loss of 5-hydroxymethylcytosine is linked to gene body hypermethylation in kidney cancer. Cell Res. 2016;26:103–18. doi: 10.1038/cr.2015.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao X, Zhang C, Sun MA, Lu X, Xie H. Deciphering the heterogeneity in DNA methylation patterns during stem cell differentiation and reprogramming. BMC Genomics. 2014;15:978. doi: 10.1186/1471-2164-15-978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen KD, et al. Increased methylation variation in epigenetic domains across cancer types. Nat Genet. 2011;43:768–75. doi: 10.1038/ng.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guelen L, et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature. 2008;453:948–51. doi: 10.1038/nature06947. [DOI] [PubMed] [Google Scholar]
- 20.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–50. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dixon JR, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–80. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pujadas E, Feinberg AP. Regulated noise in the epigenetic landscape of development and disease. Cell. 2012;148:1123–31. doi: 10.1016/j.cell.2012.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Irizarry RA, et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ziller MJ, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013;500:477–81. doi: 10.1038/nature12433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leung D, et al. Integrative analysis of haplotype-resolved epigenomes across human tissues. Nature. 2015;518:350–4. doi: 10.1038/nature14217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Heyn H, et al. Epigenomic analysis detects aberrant super-enhancer DNA methylation in human cancer. Genome Biol. 2016;17:11. doi: 10.1186/s13059-016-0879-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mitsui K, et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell. 2003;113:631–42. doi: 10.1016/s0092-8674(03)00393-3. [DOI] [PubMed] [Google Scholar]
- 29.Shu J, et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell. 2013;153:963–75. doi: 10.1016/j.cell.2013.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo H, et al. Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res. 2013;23:2126–35. doi: 10.1101/gr.161679.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Snyder MW, Kircher M, Hill AJ, Daza RM, Shendure J. Cell-free DNA Comprises an In Vivo Nucleosome Footprint that Informs Its Tissues-Of-Origin. Cell. 2016;164:57–68. doi: 10.1016/j.cell.2015.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Williams K, et al. TET1 and hydroxymethylcytosine in transcription and DNA methylation fidelity. Nature. 2011;473:343–8. doi: 10.1038/nature10066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saito D, Suyama M. Linkage disequilibrium analysis of allelic heterogeneity in DNA methylation. Epigenetics. 2015;10:1093–8. doi: 10.1080/15592294.2015.1115176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Takai D, Jones PA. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci U S A. 2002;99:3740–5. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timmons JA, Szkop KJ, Gallagher IJ. Multiple sources of bias confound functional enrichment analysis of global -omics data. Genome Biol. 2015;16:186. doi: 10.1186/s13059-015-0761-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–61. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–47. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thurman RE, et al. The accessible chromatin landscape of the human genome. Nature. 2012;489:75–82. doi: 10.1038/nature11232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xie H, et al. Genome-wide quantitative assessment of variation in DNA methylation patterns. Nucleic Acids Res. 2011;39:4099–108. doi: 10.1093/nar/gkr017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landan G, et al. Epigenetic polymorphism and the stochastic formation of differentially methylated regions in normal and cancerous tissues. Nat Genet. 2012;44:1207–14. doi: 10.1038/ng.2442. [DOI] [PubMed] [Google Scholar]
- 41.Warnes Gregory R, BB, Bonebakker Lodewijk, Gentleman Robert, Liaw Wolfgang Huber Andy, Lumley Thomas, Maechler Martin, Magnusson Arni, Moeller Steffen, Schwartz Marc, Venables Bill. gplots: Various R Programming Tools for Plotting Data. R package version 3.0.1. 2016 https://cran.r-project.org/package=gplots.
- 42.Team, R.C. R: A language and environment for statistical computing. R Foundation for Statistical Computing; Vienna, Austria: 2016. URL https://www.r-project.org/ [Google Scholar]
- 43.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–27. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 44.Houseman EA, et al. DNA methylation arrays as surrogate measures of cell mixture distribution. BMC Bioinformatics. 2012;13:86. doi: 10.1186/1471-2105-13-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gong T, Szustakowski JD. DeconRNASeq: a statistical framework for deconvolution of heterogeneous tissue samples based on mRNA-Seq data. Bioinformatics. 2013;29:1083–5. doi: 10.1093/bioinformatics/btt090. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1: A complete list of MHBs
Supplementary Table 2: Gene ontology of MHBs that loss methylation linkage in cancers.
Supplementary Table 3: Tissue specific MHBs for classification of normal tissues.
Supplementary Table 4: Layer specific MHBs with group specificity index.
Supplementary Table 5: Gene ontology analysis of TFs bound MHBs.
Supplementary Table 6: Cancer-associated High-Methylation-Haplotype (caHMH) regions.
Supplementary Table 7: Deconvolution of plasma samples by 10 normal tissues, LCT, and CCT.
Supplementary Table 8: Differential MHLs between cancer plasma and normal plasma.
Supplementary Table 9: Group II MHB regions for estimation of cancer DNA proportion.
Supplementary Table 10: Relationship between average cancer fraction and cell-free DNA extraction yield.
Supplementary Table 11: Predictors for colon cancer, lung cancer, and normal plasma.
Supplementary Table 12: Prediction accuracy based on tsMHB counting with 5-fold cross-validation.
Supplementary Table 13: Information of all samples used in this study.
Data Availability Statement
WGBS and RRBS data are available at the Gene Expression Omnibus (GEO) under accession GSE79279.