Abstract
Detecting visual features in the environment such as motion direction is crucial for survival. The circuit mechanisms that give rise to direction selectivity in a major visual center, the superior colliculus (SC), are entirely unknown. Here, we optogenetically isolate the retinal inputs that individual direction-selective SC neurons receive and find that they are already selective as a result of precisely converging inputs from similarly-tuned retinal ganglion cells. The direction selective retinal input is linearly amplified by the intracollicular circuits without changing its preferred direction or level of selectivity. Finally, using 2-photon calcium imaging, we show that SC direction selectivity is dramatically reduced in transgenic mice that have decreased retinal selectivity. Together, our studies demonstrate a retinal origin of direction selectivity in the SC, and reveal a central visual deficit as a consequence of altered feature selectivity in the retina.
Introduction
Neurons in the brain are specialized in detecting unique features in the environment. In the visual system, many neurons at various stages of processing respond selectively to stimuli moving along specific directions or having particular orientations1–3. Such direction and orientation selectivity are critical for motion sensing and image processing, leading to visually-guided behaviors that are important for survival. Not surprisingly, the circuit mechanisms of direction and orientation selectivity have been extensively studied. However, these studies have mostly focused on the retina and primary visual cortex (V1)2,3, while neglecting the superior colliculus (SC), a major retinal target and vision center.
The SC, or optic tectum, is an evolutionarily conserved structure that receives direct retinal input in all vertebrates4–6. It was the most sophisticated visual center until the neocortex recently evolved in mammals. Even in mice, a mammalian species that has become a useful model in vision research7, 85–90% of retinal ganglion cells (RGCs) project to the SC8, making it the most prominent visual structure in this species. Although the SC is mostly known for its functions in initiating rapid gaze shift towards salient stimuli, neurons in its superficial layers (i.e., the visual layers), including the stratum griseum superficiale (SGS) and stratum opticum (SO), display diverse visual response properties. In particular, selectivity for motion direction has been observed in the superficial SC of all mammalian species that have been studied, including tree shrews9, cats10, hamsters11, squirrels12, rabbits13, rats14 and mice15. Direction selectivity has also been observed in the primate SC, although it is much less prominent16,17. However, despite that it has been almost half a century since direction selectivity was first described in the mammalian SC, its underlying circuit and synaptic mechanisms remain entirely unknown.
Several mechanisms could give rise to the direction selectivity seen in superficial SC neurons. First, they could inherit it directly from direction selective (DS) retinal inputs. Indeed, direction selective ganglion cells (DSGCs) have been discovered in the retina of a number of species, and most of the DSGCs project to the superficial SC18. In mice, several subtypes of DSGCs preferentially terminate their axons in the upper half of the SGS, while most of the non-DSGCs tend to project to the lower half19–22. Interestingly, DS collicular neurons are also organized in a depth-specific manner, where they are most concentrated in the topmost lamina of the SGS and become less prevalent with depth23. This correspondence between the anatomical and functional organization supports the idea that similarly tuned DSGCs could project to common targets, thereby providing SC neurons with a synaptic drive that is biased towards certain directions.
Second, direction selectivity in the SC could arise from a specific arrangement of retinal inputs that are not individually tuned. Reichardt proposed in his motion detector model that direction selectivity could be generated by non-selective inputs that have different temporal delays24. In such a model, these inputs are arranged in such a way that stimuli moving in the preferred direction would result in synchronous arrival of synaptic inputs and consequently a large depolarization in the postsynaptic cell onto which they converge. On the other hand, stimuli moving in the opposite direction would cause only a small depolarization because the inputs arrive asynchronously. This scenario was indeed shown to underlie direction selectivity in cat V1, as revealed by both extracellular and intracellular recordings25,26. Whether a similar mechanism exists for direction selectivity in the SC is not known.
Finally, direction selectivity in the SC could be computed de novo. Under such scenario, the combined retinal input that a DS SC neuron receives would be non-selective for motion direction. Direction selectivity could then arise via dynamic interactions within the local circuits, such as tuned or spatially-offset inhibition as originally proposed by Barlow and Levick for retinal direction selectivity27.
In this study, we set out to determine which of the above mechanisms could give rise to direction selectivity in the mouse SGS. In one set of experiments, we isolated the retinal inputs that individual SGS neurons receive, by combining in vivo whole-cell voltage clamp and optogenetic silencing. In another, we reduced the selectivity of DSGCs by genetically manipulating retinal circuits and then studied the impact on SGS direction selectivity using 2-photon calcium imaging. Together, these experiments demonstrate for the first time that direction selectivity in the SGS is inherited from DSGCs in the retina; and consequently, disrupting DSGCs’ tuning leads to altered feature selectivity in the SC.
Results
Vm-Spike transformation in SGS neurons
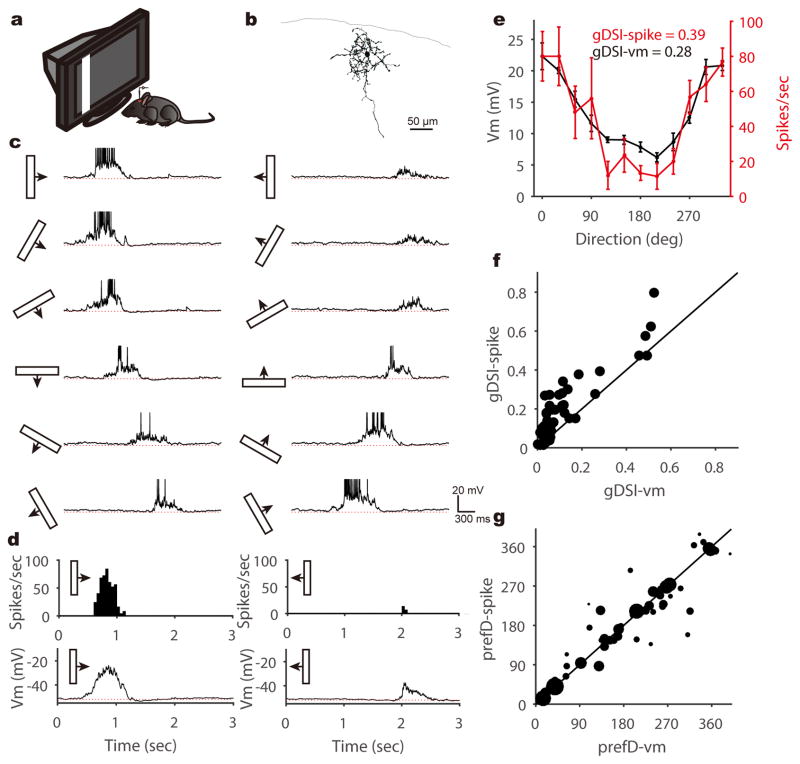
To study the synaptic mechanisms of direction selectivity in the superficial SC, we carried out in vivo whole-cell recording of SGS neurons in urethane-anesthetized mice with the entire V1 removed. First, we recorded SGS neurons under current clamp to reveal their spiking and the underlying membrane potential (Vm) changes in response to sweeping bars in different directions (Fig. 1a–c). Various degrees of direction selectivity were observed in the recorded cells, including a substantial population that was highly selective in its spiking responses (e.g., Fig. 1b–e). Using the normalized vector sum as an index (see Methods for details), which we refer to as global Direction Selectivity Index (gDSI), we found that 27% of the SGS neurons were highly DS in their spiking (gDSI-spike ≥ 0.25; n = 14/52 cells), consistent with our previous extracellular studies across the entire depth of the SGS28. The gDSI-spike and gDSI-Vm responses were overall correlated for individual cells, but gDSI-spike was greater in most cases (Fig. 1f), as expected from the non-linear effect of the spike threshold3. As the vast majority of selective SGS neurons (gDSI-spike ≥ 0.25) had gDSI-Vm ≥ 0.1 (n = 11/14 cells), we subsequently used 0.1 as a cutoff for classifying Vm and EPSC responses as highly DS. Importantly, the Vm-to-spike transformation in the SGS neurons was not as steep as seen in the DS cells in cat or mouse visual cortex. The ratio between DSI-spike and DSI-Vm was ~3 in cat V126 and ~6 mouse V129, but only ~1.2 in mouse SGS. In fact, a number of the recorded cells showed nearly identical tuning curves and gDSI values between their spiking and Vm responses (Fig. 1f and Supplementary Fig. 1). Consistently, some SGS cells showed very weak and barely detectable depolarization to bars moving along their non-preferred directions (e.g., Supplementary Fig. 1), a phenomenon rarely seen in cortical DS cells26,29. Furthermore, the preferred directions of SGS cells are similar for spike and Vm responses, especially for highly DS cells (Fig. 1g). These observations thus suggest that synaptic inputs likely play a more important role in determining direction selectivity in the mouse SGS.
Figure 1. SGS neurons’ membrane potential responses to sweeping bars.
a, A schematic of the experimental setup. b, Morphology of an example SGS neuron. c, This neuron’s membrane potential (Vm) traces in response to sweeping bars. The movement direction is diagramed by the bar and arrow to the left of each trace. Action potentials are truncated at −10mV to better reveal visually-evoked Vm responses. The red dotted lines indicate the resting membrane potential of −53 mV. d, This cell’s mean spike and Vm responses to bars moving along its preferred (left column) and opposite (right) directions. Peri-stimulus spike time histograms (top) and trial-averaged Vm (bottom) are shown. The red dotted lines indicate the resting Vm. e, This cell’s direction tuning curve for peak Vm (black, left axis) and spike rate (red, right axis). Note that the Vm tuning curve is slightly above the spike one around the non-preferred directions. Data are presented as mean ± SEM, and n = 5, 5, 5, 5, 5, 6, 6, 7, 6, 6, 5, 6 trials for the 12 directions, respectively. f, Scatter plot of gDSI for spikes (“gDSI-spike”) versus gDSI-Vm (n = 52 cells). Most cells’ gDSI-spike are greater than the gDSI-Vm, but some have nearly identical values (i.e., on the marked identity line). g, Scatter plot of the preferred direction (“prefD”) for spike versus that for Vm responses. The diameter of each dot is scaled to the gDSI-spike of that cell. The solid line is the line of identity. Note that the preferred directions are similar for spike and Vm responses, especially for highly selective cells.
SGS direction selectivity originates from individually-tuned retinal input
SGS neurons receive several sources of synaptic inputs, including both excitation and inhibition. To isolate the excitatory inputs, we next performed whole-cell voltage clamp recording. These experiments were done in transgenic mice that expressed Channelrhodopsin-2 (ChR2) in GABAergic inhibitory neurons. By illuminating the exposed SC with blue LED light, we were able to photoactivate local inhibitory neurons and suppress excitatory neurons in the SGS (Supplementary Fig. 2). This allowed us to achieve three goals. First, we could identify whether the recorded cells were excitatory or inhibitory by their responses to LED photoactivation (Supplementary Fig. 3). Second, we were able to determine the reversal potential of inhibitory currents in the recorded excitatory neurons individually (Supplementary Fig. 3). As expected from the fact that the same internal solution was used in all recordings, the inhibitory reversal potential was very consistent across cells (−64.92 ± 0.21 mV, n = 43 cells, Supplementary Fig. 3). We thus used the same holding potential (−65mV) for voltage clamp of inhibitory neurons even though the reversal potential could not be determined directly in these cells because of ChR2 activation. Finally, optogenetic activation of local inhibitory neurons in these mice could silence excitatory neurons in the SGS and remove local excitatory interactions, thereby exposing the retinal input to the recorded cells (Supplementary Fig. 2). This allowed us to analyze and compare the selectivity of retinal and total excitatory inputs to each SGS neuron individually.
We first analyzed the total excitatory postsynaptic currents (EPSCs) in the absence of LED photoactivation. Consistent with the above observation of highly-tuned Vm responses, the visually-evoked total EPSCs in many SGS neurons, including both excitatory and inhibitory neurons, were direction selective (e.g., Fig. 2a left, Fig. 2b top, and Supplementary Fig. 4). In fact, the gDSI distributions of visually-evoked peak excitation (0.13 ± 0.01, n = 87) and Vm responses (0.12 ± 0.02, n = 52) were very similar (Fig. 2d; see Methods for details of determining peak EPSCs and Vm responses). Furthermore, we recorded both Vm and EPSC responses in a small number of cells (n = 23), and found that their gDSI values were nearly identical (Fig. 2e). Specifically, for cells that were highly selective in their Vm (gDSI-Vm ≥ 0.1, above the horizontal dotted line in Fig. 2e), all of them (n = 7/7) received selective EPSCs (gDSI-EPSC ≥ 0.1). Conversely, all of the cells whose gDSI-Vm < 0.1 had gDSI-EPSC < 0.1 (n = 16/16). In other words, the excitatory input that individual SGS neurons receive is a deciding factor of their degree of direction selectivity. Similarly, the Vm’s preferred direction was also determined by the EPSC (Fig. 2f). These results thus suggest that local inhibition in the SGS does not generate direction selectivity de novo from non-selective excitatory input.
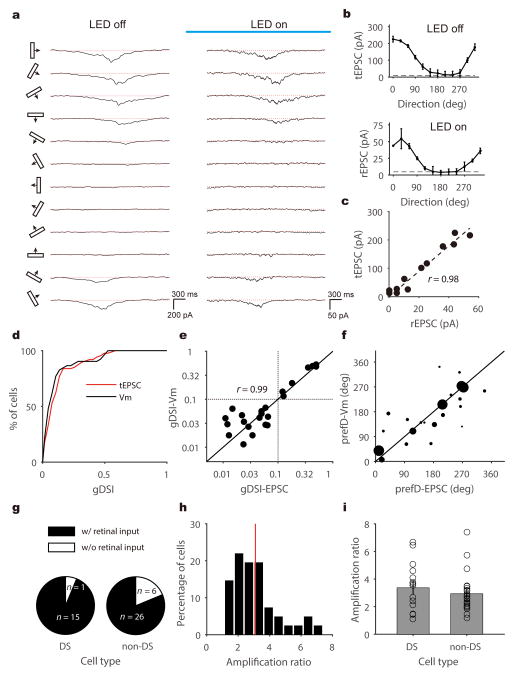
Figure 2. Voltage clamp recording and optogenetic silencing to isolate retinal excitation to SGS neurons.
a, Trial-averaged EPSC traces of an example SGS neuron to bars moving along 12 directions in the absence (left panel) or presence (right) of LED illumination (as indicated by the blue bar). The red dotted lines indicate the mean current level in the absence of visual stimulus. b, Direction tuning curves of this cell’s total EPSC (top panel, “tEPSC”) and retinal input (bottom, “rEPSC”). The dashed lines indicate the level that is 3 standard deviations above the mean current level as determined in the absence of visual stimulus. Data are presented as mean ± SEM, and n = 8, 7, 7, 7, 6, 7, 7, 7, 7, 7, 6, 6 trials in the top panel and n = 3 for all directions in the bottom panel. c, Scatter plot of this cell’s tEPSC versus its rEPSC. Peak rEPSC and tEPSC amplitudes are plotted for the responses to 12 directions of bars. The dotted line is the linear regression of the data points (R2 = 0.95, F(1,10) = 192.7, p < 0.001, linear regression; r = 0.98, p < 0.001, Pearson correlation). d, Cumulative distributions of gDSI for tEPSC (n = 87 cells from 58 mice, red) and for Vm (n = 52 cells from 41 mice, black), indicating that tEPSC and Vm have nearly identical gDSI distributions (p = 0.30 K-S stat = 0.17, K-S test). e, Scatter plot of gDSI values for tEPSC and Vm in the same cells (n = 23 cells from 21 mice). They are well correlated (r = 0.99, p < 0.001, Pearson correlation) and similar in values (p = 0.18, W = 90, Wilcoxon test). Note that the plot is shown in log-log axis to better illustrate the cells with low gDSI. The dotted lines indicate gDSI levels of 0.1. f, Scatter plot of the preferred direction (“prefD”) for Vm versus EPSC. The diameter of each dot is scaled to the gDSI- Vm of that cell. Note that the preferred directions are similar for EPSC and Vm responses, especially for highly selective cells. The solid lines in both panel e and f are the lines of identity. g, Percentage of cells that receive direct retinal input in DS (n = 15/16) and non-DS (n = 26/32) SGS neurons. h, Distribution of amplification ratio of all cells that receive direct retinal input (n = 41). The red line indicates the mean of the distribution. i, Similar amplification ratio between DS (3.37 ±0.46, n = 15 cells from 13 mice) and non-DS (2.94 ± 0.26, n = 26 cells from 20 mice) SGS neurons (p = 0.41, U = 164, Mann-Whitney U test). Data are presented as mean ± SEM.
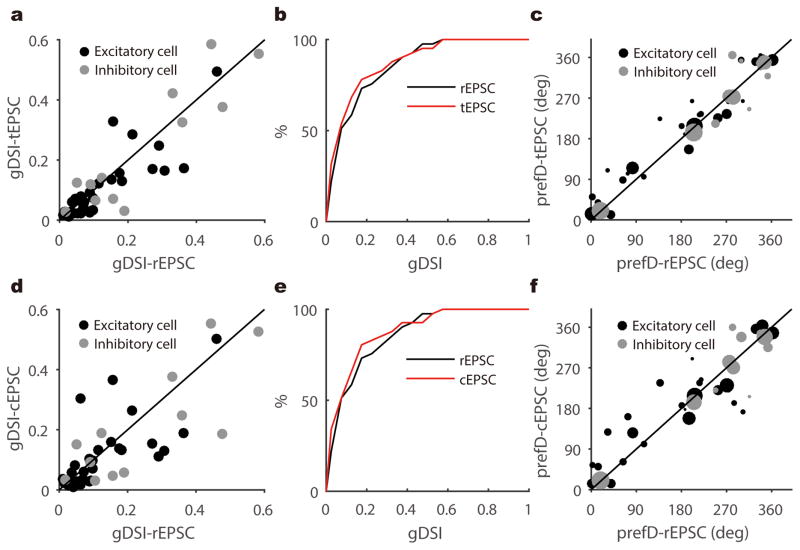
Most SGS neurons still showed visually-evoked excitation upon optogenetic silencing of local excitatory neurons (n = 41/48, 85.4%), including both excitatory and inhibitory neurons (Supplementary Fig. 5), indicating that they receive direct retinal inputs. In particular, a slightly larger portion of the DS cells (n = 15/16, 93.8%) were directly innervated by the retina than non-DS cells (n = 26/32, 81.3%, Fig. 2g). When comparing the peak amplitudes of total and retinal EPSCs in the same cells (referred to as “tEPSC” and “rEPSC” respectively), we found that the retinal inputs were amplified by the intracollicular circuits in a largely linear fashion (e.g., Fig. 2c), similar to the transformation from thalamus to visual cortex30,31. The amplification ratio ranged from 1.11 to 7.39, with a mean of 3.09 ± 0.24 (n = 41, Fig. 2h–i). Consistent with the largely linear amplification (Supplementary Fig. 6), rEPSC and tEPSC of the same cells were similarly tuned in their peak response amplitudes (e.g., Fig. 2a–b). Indeed, the gDSI values and preferred directions were well correlated between rEPSC and tEPSC, and this was the case for both excitatory and inhibitory neurons (Fig. 3a–c). In other words, the direction selective SGS cells, i.e., cells that have high gDSI values for their tEPSC and the resulting Vm, receive similarly-tuned retinal excitation. Furthermore, the intracollicular excitatory inputs, determined as the difference between tEPSC and rEPSC (Supplementary Fig. 7), displayed largely similar selectivity as the retinal inputs (Fig. 3d–f). These analyses thus indicate that the direction selectivity of SGS neurons is determined by their retinal inputs, which are further amplified by similarly-tuned intracollicular inputs to maintain the directional preference and level of selectivity.
Figure 3. Retinal excitation and total excitation are similarly tuned in SGS neurons.
a, Scatter plot of gDSI of the peak tEPSC versus that of peak rEPSC, indicating their gDSI values were well correlated (n = 41 cells from 28 mice, r = 0.89, p < 0.001, Pearson correlation). The gDSI values for tEPSCs and rEPSC were similar in cells that receive DS excitation (i.e., cells whose gDSI-tEPSC or gDSI-rEPSC ≥ 0.1, n = 22/41 cells, p = 0.34, W = 61, Wilcoxon test). b, Cumulative distribution of data shown in a. c, The preferred direction (“prefD”) of the peak tEPSC versus that of peak rEPSC. The diameter of each dot is scaled to the gDSI of that cell’s tEPSC. d–f, Same as a–c, but comparing collicular EPSC (“cEPSC”) versus rEPSC. In panel d, n = 41 cells from 28 mice, r = 0.77, p < 0.001, Pearson correlation. n = 23 cells whose gDSI-cEPSC or gDSI-rEPSC ≥ 0.1, p = 0.30, W = 70, Wilcoxon test. In panel f, the diameter of each dot is scaled to the gDSI of that cell’s peak rEPSC. The solid lines in panel a, c, d and f are the lines of identity.
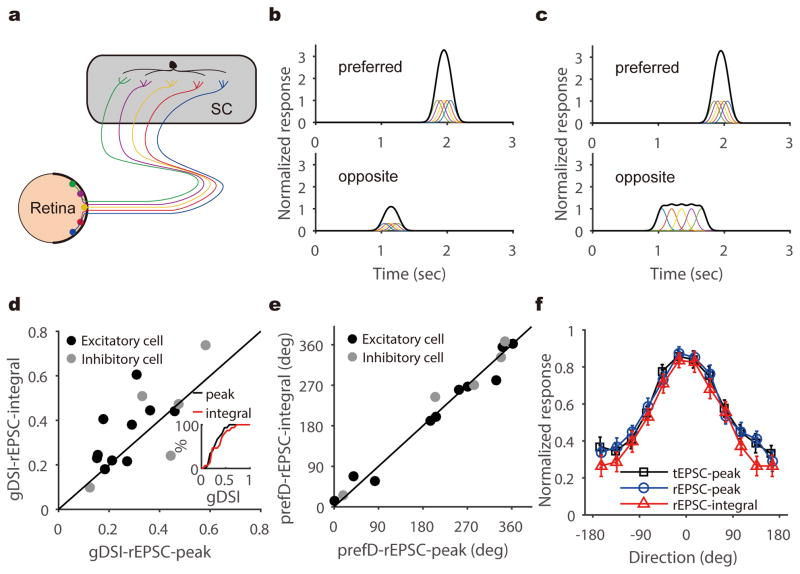
The selective retinal inputs could result from summing inputs that are individually tuned, such as from direction selective ganglion cells (DSGCs, Fig. 4a–b). Alternatively, individual retinal inputs may not be DS, but their integration in the postsynaptic neuron could take place in a precise spatiotemporal manner to generate larger EPSC peaks at the preferred direction than at the opposite direction. For example, a difference in response latency between the non-DS retinal inputs could cause them to arrive at the postsynaptic cell synchronously in response to the preferred direction but asynchronously to the opposite direction, thus resulting in different peak amplitudes (Fig. 4c), as proposed originally by Reichardt for motion detection24. In this latter scenario, the total charge of the retinal EPSCs would be much less selective than their peaks or even entirely un-tuned, which would be analogous to the emergence of orientation selectivity in the visual cortex30. To determine which of two scenarios is true for the retinocollicular transformation of direction selectivity, we calculated the integral of rEPSCs during responses evoked by the moving bars (see Methods for details) and compared its direction selectivity to that of the peak rEPSC. The integral of retinal input was in fact similarly selective or even more selective in a few cells, compared to the peak rEPSC (Fig. 4d–e and Supplementary Fig. 8), ruling out the aforementioned second scenario. The averaged tuning curves of peak rEPSC and integral rEPSC were very similar, and nearly identical to that of total EPSC (Fig. 4f), confirming that SC direction selectivity originates from individually-tuned retinal inputs. Finally, the same integral vs. peak analysis showed that the intracollicular excitatory inputs also followed the first scenario (Supplementary Fig. 7), indicating that DS SGS neurons are preferentially connected with other collicular neurons that prefer similar directions.
Figure 4. SGS direction selectivity originates from individually-tuned retinal inputs.
a–c, Two different scenarios where retinal inputs could give rise to direction selectivity in the SGS. In one (b), individual retinal inputs, as indicated by traces of different colors, are selective for similar directions, thus resulting in larger excitation to the preferred (top) than to the opposite direction (bottom). In the other scenario (c), individual retinal inputs respond with similar amplitudes to all directions. These non-DS retinal inputs arrive at the postsynaptic cell synchronously in response to the preferred direction (top) and asynchronously to the opposite direction (bottom), thus resulting in different peak amplitudes. In this scenario, the total charge of rEPSC would be much less selective than their peaks. d, gDSI of rEPSC integral and peak were correlated (r = 0.71, p = 0.003, Pearson correlation) and similar for individual DS neurons (p = 0.11, W = 58, Wilcoxon test, n = 15 cells from 13 mice), supporting the scenario shown in (b). Inset shows the cumulative distribution of the two. e, The integral and peak of retinal EPSCs prefer similar directions in DS SGS neurons. The solid lines in both panel d and e are the lines of identity. f, Averaged tuning curves for tEPSC-peak (black), rEPSC-peak (blue), and rEPSC-integral (red) in DS SGS neurons (n = 15 cells from 13 mice). Individual curves were normalized by their maximum responses and aligned to their preferred directions. They were then averaged for plotting. Error bars represent SEM.
Genetic disruption of retinal direction selectivity reduces selectivity in the SGS
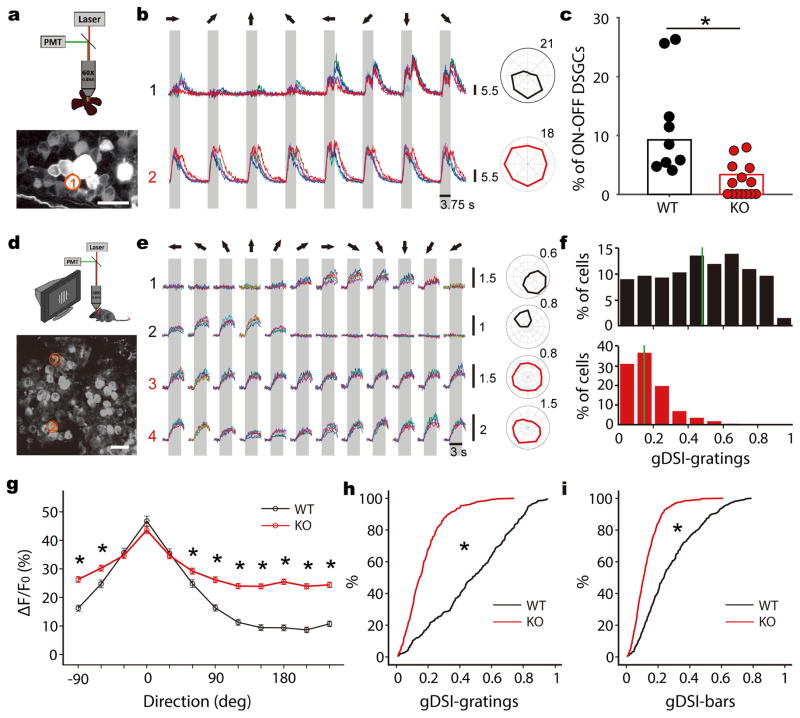
Our in vivo whole-cell experiments support the conclusion that the direction selectivity of SGS neurons originates from converging inputs of similarly-tuned DSGCs. If this is indeed the case, a reduction of retinal direction selectivity would compromise the selectivity in the SGS. We next tested this prediction using a genetic manipulation. GABAergic inhibition provided by starburst amacrine cells is a critical factor in generating direction selectivity in the retina1,2, and it can be eliminated by knocking out (KO) the vesicular GABA transporter (Vgat) gene Slc32a1 from these cells by crossing Slc32a1flox/flox (i.e. Vgatflox/flox) with choline acetyltransferase (ChAT)-IRES-Cre mice32. We then performed 2-photon calcium imaging in the ganglion cell layer of these KO mice using the genetically-encoded calcium indicator GCaMP6s33. In particular, we focused on the On-Off DSGCs because they are the ones that primarily project to the SGS18. In wildtype littermate controls, 9.3% of cells in the ganglion cell layer were On-Off DSGCs (Fig. 5a–c, n = 60/648 cells, from 9 mice), consistent with previous studies34,35. In contrast, in the KO mice, the percentage of cells that displayed On-Off DS responses was significantly reduced (Fig. 5c, n = 19/566 cells, 3.4%, from 14 mice. p <0.001, χ2 test). Because cholinergic inputs (i.e., ChAT+ cells) to the SC terminate in the intermediate and deep layers and do not co-release GABA, these “ChAT-Vgat” KOs mice provide us with a unique opportunity to study the effect of altered retinal direction selectivity on the visual response properties of superficial SC neurons.
Figure 5. Genetic disruption of retinal direction selectivity reduces selectivity in the SGS.
a, A schematic of 2-photon calcium imaging of retina (top). The bottom panel shows a max-intensity projection of GCaMP6 fluorescence in an example field of view. Scale bar is 25 μm. b, Top panel shows Ca2+ signals of the RGC circled in a to the presentation of moving bars in 8 directions (different colors represent separate trials). The gray shade corresponds to the time interval in which the bar stimulus sweeps across the field of view and the arrows represent the direction of movement in relation to the polar plot on the right. This cell showed DS responses to both leading and trailing edges of the moving bars, indicating that it was an On-Off DSGC. Bottom panel shows an On-Off cell from a Vgat conditional KO mouse. c, Summary plot showing the percentages of On-Off DSGCs in WT (black, n = 60/648 cells from 9 mice) and KO (red, n = 19/566 cells from 14 mice) retinas (p <0.001, χ2 = 17.3, χ2 test). Data points represent percentages of On-Off DSGCs in individual mice. d, A schematic of 2-photon imaging of the SGS (top) and an example field of view from a WT (bottom). Scale bar is 20 μm. e, Ca2+ signals of the two neurons (1 and 2) circled in d, and of two neurons from a Vgat KO (3 and 4), in response to drifting gratings. The gray boxes mark the duration of stimulation. The moving directions are represented by arrows on top. Corresponding polar plots are shown to the right. Scale bars for the Ca2+ signals and polar plots in b and e represent the change in fluorescence from baseline (ΔF/F0). f, gDSI distribution of WT (top, black) and KO (bottom, red) cells to drifting gratings. The solid green lines indicate the median of distributions. g, Average WT (black, n = 310 cells from 5 mice) and KO (red, n = 407 from 8 mice) tuning curves to gratings after aligning each cell’s preferred direction at 0. * indicate statistically significant difference between genotypes (all p-values < 0.001, Mann-Whitney U test). Data are presented as mean ± SEM. h, Cumulative distribution of the data shown in f (p < 0.001, K-S stat = 0.61, K-S test). i, Cumulative distribution of gDSI to sweeping bars (p < 0.001, K-S stat = 0.43, K-S test).
We first performed intrinsic imaging and found normal SC retinotopic maps in the KOs (Supplementary Fig. 9). Next, we performed 2-photon calcium imaging of the topmost SGS lamina, which we have previously shown to be enriched with DS neurons that have overlapping ON-OFF receptive fields23. The receptive field structures of the imaged cells were largely normal in the KOs, with subtle increase in subfield size and completely normal ON-OFF overlap (Supplementary Fig. 10). The small increase in subfield size was consistent with the reduced inhibition in their retina of the KOs. Also consistent with this was that slightly more cells were responsive in the KOs. When stimulated with drifting gratings or sweeping bars, 46.5% (310/667 cells to gratings) or 47.2% (315/667 to bars) were responsive in WT littermate controls, compared to 50.0% (407/821 cells, to gratings) or 61.5% (505/821, to bars) in the KOs. As expected, the vast majority of the responsive cells in this lamina were DS in WT (Fig. 5d–f, gDSI ≥ 0.25; n = 235/310 cells to gratings, 76%; and 146/315 to bars, 46%). Preferred motion directions were more widely represented than the 4 cardinal directions in the retina (Supplementary Fig. 11), presumably due to specifically combining inputs of DSGCs that prefer neighboring cardinal directions. Remarkably, in the KOs, much fewer cells were DS in this lamina (n = 84/407 to gratings, 21%; and 35/505 to bars, 7%. Fig. 5f). The mean and median gDSI values in the KOs (0.17 ± 0.01 and 0.15 to gratings, 0.13 ± 0.00 and 0.11 to bars) were significantly lower than in the controls (0.48 ± 0.01 and 0.48 to gratings, 0.28 ± 0.01 and 0.24 to bars, Fig. 5h and 5i, p < 0.001 for both gratings and bars, K-S test). Importantly, this reduction in direction selectivity was caused by increased responses to the non-preferred directions (Fig. 5g), consistent with decreased inhibitory inputs onto On-Off DSGCs in the retina. Together, these data demonstrate that disrupting DSGCs’ tuning leads to reduced selectivity in SGS neurons, thereby confirming the retinal origin of SGS direction selectivity.
Discussion
In this study, we have both isolated and manipulated the retinal input in order to study its role in generating SGS direction selectivity. The whole-cell recording and optogenetic silencing experiments demonstrate that DS SGS neurons receive retinal input that is already selective, generated by precisely converging inputs from similarly-tuned DSGCs. The selective retinal input is amplified by intracollicular circuits without changing its preferred direction or level of selectivity. The resulting membrane potential depolarization in the SGS neuron then leads to a slightly more selective spiking response because of the non-linearity of thresholding. Consistent with these results, we also found that SGS direction selectivity is reduced in mice that have altered retinal selectivity. Our studies thus demonstrate that SGS neurons inherit their direction selectivity from DSGCs in the retina, a finding that has important implications for understanding signal processing in the early visual system.
It is known that individual SGS neurons are innervated by several RGCs36. Consequently, in order to provide DS excitation to the postsynaptic neuron, the converging DSGCs must prefer similar directions. In addition, a new directional preference would emerge when the DSGCs that prefer neighboring cardinal directions precisely converge. Our findings therefore indicate that well-controlled developmental mechanisms must exist to ensure the precise and selective targeting of DSGCs in the SGS. Consistent with this idea, several subtypes of DSGCs have been found to project primarily to the upper SGS18, which contains more DS cells than the lower SGS23. How such depth-specific targeting is established during development, and how even more precise patterns of connectivity are generated at the level of individual cells, have not been studied.
Our data further show that the intracollicular excitation that DS SGS neurons receive is also tuned to similar directions, thereby amplifying the retinal inputs without changing their preferred direction or levels of selectivity. This result thus indicates that within the SGS, excitatory neurons that prefer similar directions are preferentially connected. A similar non-random connectivity has been revealed for orientation selective cells in the mouse visual cortex37–39, and its emergence requires visual experience40. The exact wiring diagram of the SGS circuits and its development have not yet been studied. Given that the mouse retinocollicular pathway is already a productive model for studying cell types and neural development4, future studies of this pathway will likely reveal the molecular and cellular mechanisms that establish the precise connections from the retina to the SGS and also within the SGS, which are necessary to generate and maintain feature selectivity in the superior colliculus.
Direction selectivity is an evolutionarily-conserved property seen in many visual structures and in various species. In zebrafish, for example, RGC subtypes that prefer different directions project to segregated layers in the optic tectum, and the tectal neurons with matching preferred directions arborize their dendrites in the corresponding layers41–44. This suggests that the DS retinal inputs could determine the direction preference of tectal neurons, just like what we discovered here in the mouse SC. In monkeys, a very small population of DS neurons was found in the SC16,17, largely consistent with the fact that DSGCs have so far remained elusive in primates. It is certainly possible that DSGCs may be discovered with new genetic and imaging techniques in the future and that they may give rise to observed SC direction selectivity. Alternatively, the weak DS responses in the primate SC could result from the excitatory input from visual cortex, which includes DS cells. This possibility has in fact been addressed in cats by lesioning or cooling the cortex, but unfortunately these studies yielded conflicting results45,46. On the other hand, cortical input does not appear to affect SC selectivity in rodents. For example, in ground squirrels, SC direction selectivity remain unchanged when visual cortex is removed, leading to the proposal that the DS cells receive their inputs from the retina47. We have recently shown that in mice cortical inputs do not affect the magnitude or looming speed tuning of SC responses under anesthesia and only increase the response magnitude in awake condition48. Our current study, where the animals were anesthetized and their V1 removed, demonstrates that retinal input is the origin of the direction selectivity in the mouse SC. Future studies will be needed to determine whether cortical input could modulate SC direction selective responses under certain behaviors in mice or even give rise to SC direction selectivity in primates.
In addition to direction selectivity, SGS neurons also display a number of other response properties, such as size preference, motion selectivity and speed tuning28,48,49. These properties could be generated by integrating inputs from DSGCs, other RGC subtypes, local intracollicular circuits, and afferent inputs from visual cortex. In terms of local circuits, the SGS contains a large population of inhibitory neurons. Inhibition could sharpen direction selectivity if it is tuned to the opposite direction or offset spatiotemporally, as shown for DS neurons in zebrafish tectum41 and mouse visual cortex29. Although we did not directly address the role of synaptic inhibition in this study, our data indicate that it is not required to generate direction selectivity in SGS neurons or sharpen its tuning. This is because the excitatory inputs that individual DS neurons receive are already selective and they are tuned to the same level as Vm. On the other hand, inhibitory neurons in the topmost SGS lamina are known to be DS23. It is thus conceivable that these inhibitory neurons may provide direction-specific interactions between stimulus center and surround in response to complicated visual scenes. Future studies will be needed to determine the spatial and direction tuning of inhibition in SGS neurons in order to reveal its functions in visual processing.
In conclusion, the diverse response properties in the SC are generated by selective and precise connections in the retinocollicular and intracollicular circuits. Using the genetic and optogenetic tools available in mice, we discovered the neuronal mechanism underlying one of the most important properties, direction selectivity. Given the fundamental importance of the SC in visually-guided behaviors, our discovery will motivate exciting future studies of visual system organization, function and development.
Online Methods
Animal Preparation
C57BL/6 wild type (WT) and transgenic mice of both sexes were used in this study. Gad2-IRES-cre (Stock no. 010802) and Ai32 (RCL-ChR2(H134R)/EYFP, Stock no. 012569) mice were acquired from Jackson Laboratory and crossed to generate heterozygous offspring that express ChR2 in glutamate decarboxylase 2 positive (GAD2+) cells for in vivo optogenetic experiments (n = 100). Additional WT mice were also used for in vivo whole-cell recording (n = 25). For 2-photon calcium imaging in the retina and SC, Slc32a1flox/flox (i.e. Vgatflox/flox) mice (Stock no. 012897) and ChAT-IRES-Cre mice (Stock no. 006410) were originally acquired from Jackson Laboratory and crossed to knock out (KO) vesicular GABA transporter (Vgat) gene (Slc32a1) from choline acetyltransferase (ChAT)+ cells. These strains were backcrossed to the C57BL/6 background32. Both KOs (n = 22) and littermate controls (n = 14) were used. All mice were kept on a 12hr light:12hr dark cycle, 1 to 5 mice per cage. All experimental procedures were approved by the Northwestern University and the University of Chicago Institutional Animal Care and Use Committees.
For in vivo experiments, mice between postnatal day 45 and 90 were anesthetized with urethane (1.2 g/kg in 10% saline solution, i.p.) and then sedated by chlorprothixene (10 mg/kg in water, i.m.) as described before23,48. Atropine (0.3 mg/kg in 10% saline) and dexamethasone (2 mg/kg in 10% saline) were administrated subcutaneously. The animal was then transferred onto a heating pad for recording or imaging. The animal’s body temperature was monitored through a rectal thermoprobe and maintained at 37 °C through a feedback heater control module (Frederick Haer Company, Bowdoinham, ME). Toe-pinch reflex was monitored during experiments to test for depth of anesthesia. Additional urethane (0.2–0.3 g/kg) was administered when necessary. After the mice were anesthetized, the scalp was shaved and skin removed to expose the skull. For whole-cell and single-unit recording, a metal plate was mounted on top of the skull with Metabond (Parkell, Edgewood, NY) mixed with black ink. The plate was then mounted to a steel stand on the vibration isolation table. A thin layer of silicon oil was applied on both eyes to prevent drying. A craniotomy (~4.0 × 2.0 mm2) was performed on the left hemisphere, and the tissues including the entire V1 overlaying the SC was removed by aspiration to expose the SC. The procedures for imaging are described below.
In vivo whole-cell recording
Blind whole-cell patch clamp was performed to record SGS neurons intracellularly. Glass pipettes had tip openings of 1.5–2μm (5–7MΩ). For current-clamp recordings, the K+-based internal solution contained 135 mM K-gluconate, 7 mM KCl, 0.5 mM EGTA, 10 mM HEPES, 10 mM Na-phosphocreatine, 4 mM Mg-ATP, 0.4 mM Na-GTP and 0.5% biocytin. The pH was adjusted to 7.25 with KOH. For voltage-clamp recordings, both K+-based (n = 66) and Cs+-based (n = 21) internal solutions were used. The Cs+-based internal solution contained 125mM Cs-gluconate, 2mM CsCl, 0.5mM EGTA, 10mM HEPES, 1mM QX-314, 5mM TEA-Cl, 10mM Na-phosphocreatine, 4mM Mg-ATP, 0.4mM Na-GTP and 0.5% biocytin. The pH was adjusted to 7.25 with CsOH. K+-based internal solution was used to try to record EPSCs and Vm from the same cells, and Cs+-based internal solution was used only for recording EPSCs. No difference was found between using K+-based and Cs+-based internal solutions for recording EPSCs. Glass pipettes were advanced perpendicularly to the horizontal plane of the mouse head until just touching the SC surface. 2% agarose in artificial cerebrospinal fluid solution (ACSF, containing 140mM NaCl, 2.5mM KCl, 11mM Glucose, 20mM HEPES, 2.5mM CaCl2, 3mM MgSO4, 1mM NaH2PO4) was then added onto the exposed SC to stabilize the brain. A small piece of wet gauze was placed onto the agarose to prevent drying. Pipettes were then inserted into the SC.
Electrical signals were amplified using MultiClamp 700B (Axon Instruments, CA), and acquired with System 3 workstation (Tucker Davis Technologies, FL) at 10 kHz. Pipette capacitance and the electrode resistance were compensated initially. For current-clamp recordings, only responsive cells with stable resting membrane potentials and series resistances lower than 80 MΩ across the duration of the recordings were included in our analysis. No current injection was applied during recordings except for measurement of series resistance50. For voltage-clamp recordings, neurons were clamped at the reversal potential of inhibition, which was determined by adjusting the holding potential to minimize the amplitude of the inhibitory postsynaptic current evoked by photostimulation of Gad2+ neurons (Supplementary Fig. 3). The mean reversal potential for inhibitory currents in excitatory cells was −64.92 ± 0.21 mV (n = 43). For recordings of inhibitory cells, −65 mV was used as the holding potential to isolate excitatory currents. Only responsive cells with stable current baselines and series resistance lower than 50 MΩ across the duration of the recordings were included in our analysis. Note that the reported values were not corrected for the junction potential. The depths of recorded cells were between 0 and 300 μm (reading from the micromanipulator) from the point where the pipette broke into the thin membrane on the SC surface as indicated by a large sudden and quickly recovered electrical signal change. The morphology of 9 biocytin-stained cells further confirmed the accuracy of the manipulator readings and that all cells were in the SGS.
Histology
After in vivo whole-cell recordings, mice were overdosed with euthanasia solution (150 mg/kg pentobarbital) and perfused with PBS and then 4% paraformaldehyde (PFA). The brain was immersed in 4% PFA overnight. Coronal slices of 150μm were cut from the fixed brain using a vibrating blade microtome (VT1000S, Leica Microsystems). The labeled cells were revealed by visualizing biocytin with streptavidin-Alex Fluor 488 conjugate (Invitrogen). Images were captured using a Zeiss LSM5 Pascal confocal microscope (Carl Zeiss, Jena, Germany) in z-series scanning and reconstructed in Filament Tracer of Imaris (Bitplane, Zurich, Switzerland).
In vivo extracellular recording
Tungsten electrodes (5–10 MΩ, FHC) were inserted perpendicularly into the SC. The SC surface was estimated visually under the microscope. 2% agarose in ACSF was then applied onto the exposed SC. SGS neurons were recorded between 0 and 300μm below the surface. The electrical signals were filtered between 0.3 and 5 kHz for spikes and between 10 and 300 Hz for local field potentials, and sampled at 25 kHz using a System 3 workstation (Tucker Davis Technologies, FL). The spike waveforms were further sorted offline in OpenSorter (Tucker Davis Technologies, FL) to isolate single-units as described before28,48.
Two-photon calcium imaging of SGS neurons
We followed our recently published procedures for 2-photon imaging of the superficial SGS neurons23. Briefly, a craniotomy was performed on the left hemisphere, starting at the lambda point and extending ~3 mm both laterally and rostrally. Tissues overlaying the SC were removed by aspiration. Once the SC was exposed, a glass pipette (inner diameter of 10–20 μm) filled with freshly-made solution containing the fluorogenic calcium-sensitive dye Cal-520AM51 (1.13 mM, ATT Bioquest) and SR101 (100 μM) was lowered into the tissue. Twenty pulses of 2.3 nl each (46 nl total volume), at 20 s interval, were delivered to inject the Cal-520 solution at a depth of 500 μm below the surface. The same procedure was repeated after retracting the pipette to a depth of 250 μm. The pipette was left in the tissue for 1–2 min before being slowly retracted. The SC was then covered by ACSF. Imaging was performed 1–2h after loading.
Once the injection procedure was complete, a small metal plate was mounted on the mouse’s head with Metabond (Parkell, Edgewood, NY), which, when clamped under the microscope, resulted in the imaged SC surface being largely flat and perpendicular to the objective. A shield was placed around the craniotomy to block light from the visual stimulus during imaging. The SC was covered by 3% agarose in ACSF for stability. Imaging was performed with a 2-photon microscope (2P-SGS, Bruker Nano Surface Division) and a Ti:sapphire laser (Coherent Chameleon Ultra II) at an excitation wavelength of 800 nm, and with a 40X/0.8NA objective (Leica). Data were acquired using PrairieView software with a spiral scan at 2X optical zoom, resulting in a circular field of view with a diameter of 135 μm. Image resolution was 256×256 pixels and the acquisition rate was 8.079 Hz.
Optogenetic silencing of SGS excitatory neurons
To photostimulate ChR2-expressing cells, we used an optic fiber (0.2 mm core diameter) driven by a blue LED (470 nm, Doric Lenses) placed ~0.5 mm above the exposed SC. The tip of the LED fiber was placed at a similar position in all mice. During recordings, it was buried in the agarose that was applied to reduce the pulsation of the brain and protect the tissue. To prevent direct photostimulation of the eyes by the LED light, the Metabond used for mounting the head plate was prepared with black ink. The agarose surface was painted with black ink, and a piece of thick black paper was carefully placed around the fiber to ensure that light could not be seen from the front and sides, as described before48. The LED was driven by a square wave starting from 500 ms before the onset of each visual stimulus and ending at 100 ms after the offset of each visual stimulus (3600 ms for sweeping bars, as described below). The intensity of LED light was ~160 mW/mm2 at the tip of the optic fiber in all recordings, which was confirmed to be reliably effective in silencing SGS excitatory neurons (Supplementary Fig. 2).
Visual stimulation
For in vivo experiments, visual stimuli were generated with Matlab Psychophysics toolbox52,53 on a LCD (37.5 cm × 30 cm, 60 Hz refresh rate, ~50 cd/m2 mean luminance) or a CRT monitor (40 cm × 30 cm, 60 Hz, ~35 cd/m2 luminance). The monitor was placed 25 cm away from the eye contralateral to the recording/imaging site (the right eye), and slightly adjusted for each cell so that its receptive field was completely covered. For 2-photon imaging, the screen was also tilted at an angle matching that of the mouse’s head, given that the mouse’s nose was slightly elevated to correct for the curvature of SC and allow imaging from a relatively flat surface. The screen was adjusted so that the imaged cells’ receptive fields were near the center of the screen. The ipsilateral eye was covered throughout the experiments.
Two types of visual stimuli were used to determine direction selectivity of SGS neurons. First, sweeping bars, 5° wide and drifting at a speed of 30°/s, were used in both physiology and imaging experiments. The drifting directions were varied between 0° and 330° (12 steps, 30° spacing), which were presented in a pseudorandom sequence together with a “blank stimulus” (gray screen at the mean luminance). In whole-cell recording, the inter-stimulus interval was 0.5 or 1 sec when there was no LED illumination, and 10 sec where LED was used in order to allow the stimulated inhibitory cells to recover. In 2-photon imaging, the inter-stimulus interval was 3 sec. Second, drifting sinusoidal gratings were also used in the imaging experiments, at 0.08 cpd, 2Hz, 100% contrast23. They were presented at 12 movement directions (0°–330°, with 30° increments) in a pseudorandom order within a circular window (32° in diameter and surrounded by a grey background) near the center of the imaged cells’ receptive fields (which was determined by flashing white or black squares as described in reference23. The stimulus duration was 3 sec and inter-stimulus interval 5 sec. Each stimulus was repeated 4–6 times for imaging and 3–8 times for recording.
To examine whether optogenetic activation of GABAergic neurons could affect retinal transmission by potentially acting presynaptically through GABAB receptors, we recorded visually-evoked local field potentials (LFP) in the SGS, before and 15–45 minutes after administration of the GABAB receptor antagonist CGP54626 (10 μM) to the SC surface. The visual stimulus was a 20° diameter circle flashing on and off, centered at the receptive field of the recorded site. 40–60 trials were recorded to calculate the average response for quantification.
Data analysis
Whole-cell recording data were first analyzed using a custom MATLAB program (originally written by a former lab member Dr. Xinyu Zhao). For whole-cell recording, data collection and analysis were not performed blind to the conditions of the experiments because it does not apply. For current-clamp data, spikes were detected by calculating the first derivative of raw voltage traces (dV/dt), and the start of a spike was the time point when dV/dt reached a manually set positive threshold. Individual traces were carefully inspected to ensure proper spike detection. Peri-stimulus spike time histograms (PSTHs) were calculated by trial-averaging the spike counts in each 50 ms time bin. Subthreshold Vm were extracted by removing spikes from the raw voltage traces by a 6 ms median filter. The subthreshold Vm traces were trial-averaged for each stimulus condition. The trial-averaged Vm trace for the blank stimulus (i.e., gray screen) was used to calculate the mean (Vm baseline) and standard deviation of spontaneous Vm fluctuations. The Vm baseline was then subtracted from the trial-averaged Vm trace for each visual stimulus condition, i.e., bars of certain direction.
For voltage-clamp data, the current traces (Im) were firstly smoothed by a 40 ms mean filter17, 19 and then trial-averaged for each stimulus condition. For visual stimulus conditions in the absence of LED photoactivation (i.e., LED-off), the EPSC baseline was calculated as the mean of the trial-averaged Im trace to the blank stimulus, and subtracted from the trial-averaged trace of each condition. For LED-on conditions, Im traces were similarly trial-averaged for each stimulus condition. For inhibitory cells, all traces showed LED-activated ChR2 currents in addition to visually-evoked EPSCs (Supplementary Fig. 5). For excitatory cells, the trace of the blank condition was flat in most cells (Supplementary Fig. 5); but in some cells, a slowly increasing inward current was seen during LED stimulation, possibly the “bystander currents” mediated by acid-sensing ion channels54. The dynamic of this slow current, when present, was always the same for all stimulus conditions of the same cell, including the blank stimulus. We therefore did a point-by-point subtraction of a further-smoothed trace of the blank condition from the trace of each condition. This was done for both excitatory and inhibitory cells to remove the LED-evoked currents, while keeping the fluctuations in each trace (including the blank condition, which was used to determine baseline fluctuation). Finally, the intracollicular EPSC traces were generated by a point-by-point subtraction of retinal EPSCs (LED-on) from the total EPSCs (LED-off) traces (Supplementary Fig. 7).
For analyzing Vm and EPSCs, we determined time windows of responses to the sweeping bars. This was necessary for finding “response” magnitude for directions that evoked weak or no responses. To do this, we first calculated a cutoff threshold, which was the Vm or EPSC level 2 standard deviations away from the baseline fluctuation (determined from the blank condition as described above). The widest segment of the traces that were above (for Vm) or below (for EPSCs) the threshold was determined as the response time window for each stimulus condition. This time window was expanded if there were any short above-threshold segments within 150ms of the 2 sides. Next, the conditions that evoked wider time windows were used to guide the analysis of other conditions, to ensure that the estimation of response window was not too conservative or inaccurate for non-preferred directions. Specifically, for conditions where the window was narrower than 1/3 of the widest window of this cell (or 333 ms if the widest window is bigger than 1s), the response time window determined from the opposite direction, reversed in timing, was used. All traces were checked visually to confirm that the time windows were determined properly. Peak Vm and spike rate were calculated for the response time window of each stimulus condition, subtracting the mean values of blank condition. For EPSCs, they could spontaneously fluctuate across the threshold, thus leading to an overestimation of the weak responses. Therefore, for data shown in the main text, if the peak EPSCs were within 3 standard deviations of the baseline, they were manually set to 0. We compared this with other methods of calculation (Supplementary Fig. 8) and found no difference in our conclusions. Finally, the EPSC charge integral for each stimulus condition was quantified as the time integral of the data points in the response time window as determined above.
For SC 2-photon imaging and data analysis, the experimenter was blind to the genotype of the mice. Animals that had visible tissue damage to their SC after dye loading, or where the dye failed to be incorporated into the cells were not subject to imaging. Data analysis was performed on all animals that were subject to imaging, and no data points were excluded from the resulting data sets. We followed our published procedures of analysis23. Briefly, regions of interest (ROIs) were drawn manually on the average images, and the intensity values for all pixels in each ROI were averaged for each frame to obtain the raw Ca2+ signal of each cell. From the raw signal, for each stimulus presentation, ΔF/F0 = (F−F0)/F0, was calculated, where F0 was the mean of the baseline signal over a fixed interval of 1.25 sec (for gratings) or 0.75sec (for bars) before stimulus onset; and F was the fluorescence signal from 250 ms after stimulus onset to 500 ms after stimulus offset. A cell was considered responsive if its mean F (for gratings) or peak F (for bars) was more than two standard deviations above F0 for at least one of the stimulus conditions. The mean (for gratings) or peak (for bars) value of ΔF/F0 for each of the stimulus conditions was then used to determine the direction tuning curves for every responsive cell.
To quantify the degree of direction selectivity, we calculated a global direction selectivity index (gDSI), which is the vector sum of responses normalized by the scalar sum of responses23,49: , where Rθ is the response magnitude of spikes, Vm, EPSC, or ΔF/F0, at θ direction of bars or gratings. The preferred direction is quantified as the angle of the vector sum of responses. Previous studies of direction selectivity mostly used , where Rpref is the cell’s maximal response, and Ropp is the cell’s response to the opposite direction. To facilitate comparing with such studies, we plotted the relationship between gDSI and DSI of individual neurons for both whole-cell and imaging data (Supplementary Fig. 12).
To classify whether a cell was direction selective in Figure 4, we used a criterion of gDSI-tEPSC ≥ 0.1. In the 19 cells that met this criterion and received direct retinal inputs, 4 cells showed in their tuning curves a small but obvious second peak at the direction opposite to the preferred direction, thus leading to a greater global orientation selectivity index ( ) than gDSI. For all of the other 15 cells, their gDSI was greater than gOSI. We therefore did not include these 4 cells in the DS group, but none of our conclusions would change if these 4 cells were included.
Retinal calcium imaging and data analysis
WT littermate controls and Vgat KO mice were injected intravitreally after eye opening at P18 with an AAV2 viral vector carrying GCaMP6s (University of Pennsylvania Vector Core). After 21 days, the injected mice were dark adapted for 1 hour and their retinas were dissected in the dark under infrared (IR) light. During dissection, the retina was cut into dorsal and ventral pieces following the procedure described by Wei et al. 201055 and the nasal direction for each piece was noted. The dissected retinas were kept in darkness at room temperature in Ames’ medium bubbled with 95% O2/5% CO2 until use (0–7 h). Cells were visualized with IR light (>900 nm) and an IR-sensitive video camera (Watec). Light responsive, GCaMP6s-positive cells were identified by a two-photon microscope (Bruker Nano Surface Division) and a Ti:sapphire laser (Coherent Chameleon Ultra II) tuned to 920 nm while presenting a flashing spot (660 um diameter) from a white organic light-emitting display (OLEDXL, eMagin; 800 × 600 pixel resolution, 60-Hz refresh rate). Imaging was performed at an excitation wavelength of 920 nm with a 60X objective while the field of view was presented with a positive contrast 990 μm × 220 μm moving bar stimulus in a dark background moving in 8 pseudorandomly chosen directions spaced at 45 degree intervals at a speed of 440 μm/sec on the retina (~15 °/sec in visual space). Data were acquired using PrairieView software in a 100 μm × 100 μm field of view. Image resolution was 256 × 256 pixels and the acquisition rate ~13 Hz.
Raw frames were uploaded onto ImageJ software in which regions of interest (ROIs) were manually drawn to enclose the soma of each GCaMP6s expressing cell and for a background region where there was no detectable GCaMP6s expression. Using the TimeSeries Analyzer plugin for ImageJ, we calculated the average intensity over time for all ROIs. In MATLAB, the background trace was subtracted from the light responsive somatic traces to remove noise. The background subtracted traces were then truncated and sorted by direction of the moving bar stimulus and the peak of each calcium transient was used to calculate ΔF/F0 as described above. For each responsive cell, we calculated gDSI and gOSI as described above and , where Rpref is the cell’s response at the preferred direction (i.e., maximal response), and Ropp is the cell’s response to the direction opposite to the preferred one. Cells showing responses to both the leading and trailing edge of the positive contrast moving bar and whose DSI > 0.2 and gDSI > gOSI were classified as ON-OFF DSGCs.
Statistics
All pooled data were presented as mean ± SEM. Statistical significance was calculated using non-parametric, two-sided, Mann-Whitney U test, Wilcoxon signed rank test, Kolmogorov-Smirnov (K-S) test, or χ2 test as mentioned in the text. All analyses and graph plotting were performed in MATLAB (MathWorks) or Prism (GraphPad Software Inc). No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those reported in the field. We did not randomly assign animals to groups for whole-cell recording because it does not apply to these experiments. For two-photon imaging experiments, KOs and littermate controls were assigned according to their genotypes.
Data and Code availability
The data that support the findings of this study and the custom Matlab code are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank Xinyu Zhao and Hui Chen for their help with data analysis. For the use of GCaMP6s, we gratefully acknowledge Vivek Jayaraman, Rex A. Kerr, Douglas S. Kim, Loren L. Looger, Karel Svoboda from the GENIE Project, Janelia Farm Research Campus, Howard Hughes Medical Institute. This research was supported by US National Institutes of Health (NIH) grants (EY026286 to J.C. and X.L., and EY024016 to W.W.), National Natural Science Foundation of China (NSFC) grant (81371049 to X.S.), China Scholarship Council (CSC) scholarship (201309120003 to X.S.) and Tianjin “131” Innovative Talent Project first-level talent scholarship (to X.S.).
Footnotes
Author Contributions: X.S., J.B., X.L., W.W. and J.C. designed the experiments. X.S. performed in vivo whole-cell recording experiments and analyzed the data. J.B. performed in vivo 2-photon imaging experiments and analyzed the data. H.A.L. and D.K. performed retinal imaging experiments and analyzed the data. J.C. performed intrinsic imaging. Y.J. performed histology. W.W. and J.C. guided data analysis and oversaw the project. All authors discussed the results and wrote the manuscript.
The authors declare no competing financial interests.
References
- 1.Wei W, Feller MB. Organization and development of direction-selective circuits in the retina. Trends Neurosci. 2011;34:638–645. doi: 10.1016/j.tins.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaney DI, Sivyer B, Taylor WR. Direction selectivity in the retina: symmetry and asymmetry in structure and function. Nat Rev Neurosci. 2012;13:194–208. doi: 10.1038/nrn3165. [DOI] [PubMed] [Google Scholar]
- 3.Priebe NJ, Ferster D. Mechanisms of neuronal computation in mammalian visual cortex. Neuron. 2012;75:194–208. doi: 10.1016/j.neuron.2012.06.011. S0896-6273(12)00571-5 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cang J, Feldheim DA. Developmental mechanisms of topographic map formation and alignment. Annu Rev Neurosci. 2013;36:51–77. doi: 10.1146/annurev-neuro-062012-170341. [DOI] [PubMed] [Google Scholar]
- 5.Gandhi NJ, Katnani HA. Motor functions of the superior colliculus. Annu Rev Neurosci. 2011;34:205–231. doi: 10.1146/annurev-neuro-061010-113728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.May PJ. The mammalian superior colliculus: laminar structure and connections. Prog Brain Res. 2006;151:321–378. doi: 10.1016/S0079-6123(05)51011-2. [DOI] [PubMed] [Google Scholar]
- 7.Huberman AD, Niell CM. What can mice tell us about how vision works? Trends Neurosci. 2011;34:464–473. doi: 10.1016/j.tins.2011.07.002. S0166-2236(11)00106-8 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellis EM, Gauvain G, Sivyer B, Murphy GJ. Shared and distinct retinal input to the mouse superior colliculus and dorsal lateral geniculate nucleus. J Neurophysiol. 2016;116:602–610. doi: 10.1152/jn.00227.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Albano JE, Humphrey AL, Norton TT. Laminar organization of receptive-field properties in tree shrew superior colliculus. J Neurophysiol. 1978;41:1140–1164. doi: 10.1152/jn.1978.41.5.1140. [DOI] [PubMed] [Google Scholar]
- 10.McIlwain JT, Buser P. Receptive fields of single cells in the cat’s superior colliculus. Exp Brain Res. 1968;5:314–325. doi: 10.1007/BF00235906. [DOI] [PubMed] [Google Scholar]
- 11.Rhoades RW, Chalupa LM. Directional selectivity in the superior colliculus of the golden hamster. Brain Res. 1976;118:334–338. doi: 10.1016/0006-8993(76)90721-6. [DOI] [PubMed] [Google Scholar]
- 12.Michael CR. Visual receptive fields of single neurons in superior colliculus of the ground squirrel. J Neurophysiol. 1972;35:815–832. doi: 10.1152/jn.1972.35.6.815. [DOI] [PubMed] [Google Scholar]
- 13.Masland RH, Chow KL, Stewart DL. Receptive-field characteristics of superior colliculus neurons in the rabbit. J Neurophysiol. 1971;34:148–156. doi: 10.1152/jn.1971.34.1.148. [DOI] [PubMed] [Google Scholar]
- 14.Fortin S, et al. Maturation of visual receptive field properties in the rat superior colliculus. Brain Res Dev Brain Res. 1999;112:55–64. doi: 10.1016/s0165-3806(98)00157-6. [DOI] [PubMed] [Google Scholar]
- 15.Drager UC, Hubel DH. Responses to visual stimulation and relationship between visual, auditory, and somatosensory inputs in mouse superior colliculus. J Neurophysiol. 1975;38:690–713. doi: 10.1152/jn.1975.38.3.690. [DOI] [PubMed] [Google Scholar]
- 16.Cynader M, Berman N. Receptive-field organization of monkey superior colliculus. J Neurophysiol. 1972;35:187–201. doi: 10.1152/jn.1972.35.2.187. [DOI] [PubMed] [Google Scholar]
- 17.Marrocco RT, Li RH. Monkey superior colliculus: properties of single cells and their afferent inputs. J Neurophysiol. 1977;40:844–860. doi: 10.1152/jn.1977.40.4.844. [DOI] [PubMed] [Google Scholar]
- 18.Dhande OS, Huberman AD. Retinal ganglion cell maps in the brain: implications for visual processing. Curr Opin Neurobiol. 2014;24:133–142. doi: 10.1016/j.conb.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huberman AD, et al. Architecture and activity-mediated refinement of axonal projections from a mosaic of genetically identified retinal ganglion cells. Neuron. 2008;59:425–438. doi: 10.1016/j.neuron.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huberman AD, et al. Genetic identification of an On-Off direction-selective retinal ganglion cell subtype reveals a layer-specific subcortical map of posterior motion. Neuron. 2009;62:327–334. doi: 10.1016/j.neuron.2009.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kay JN, et al. Retinal ganglion cells with distinct directional preferences differ in molecular identity, structure, and central projections. J Neurosci. 2011;31:7753–7762. doi: 10.1523/JNEUROSCI.0907-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim IJ, Zhang Y, Meister M, Sanes JR. Laminar restriction of retinal ganglion cell dendrites and axons: subtype-specific developmental patterns revealed with transgenic markers. J Neurosci. 2010;30:1452–1462. doi: 10.1523/JNEUROSCI.4779-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Inayat S, et al. Neurons in the most superficial lamina of the mouse superior colliculus are highly selective for stimulus direction. J Neurosci. 2015;35:7992–8003. doi: 10.1523/JNEUROSCI.0173-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reichardt W. In: Sensory Communication. Rosenblith W, editor. Wiley; 1961. pp. 303–317. [Google Scholar]
- 25.Saul AB, Humphrey AL. Temporal-frequency tuning of direction selectivity in cat visual cortex. Vis Neurosci. 1992;8:365–372. doi: 10.1017/s0952523800005101. [DOI] [PubMed] [Google Scholar]
- 26.Priebe NJ, Ferster D. Direction selectivity of excitation and inhibition in simple cells of the cat primary visual cortex. Neuron. 2005;45:133–145. doi: 10.1016/j.neuron.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 27.Barlow HB, Levick WR. The mechanism of directionally selective units in rabbit’s retina. J Physiol. 1965;178:477–504. doi: 10.1113/jphysiol.1965.sp007638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang L, Sarnaik R, Rangarajan K, Liu X, Cang J. Visual receptive field properties of neurons in the superficial superior colliculus of the mouse. J Neurosci. 2010;30:16573–16584. doi: 10.1523/JNEUROSCI.3305-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li YT, Liu BH, Chou XL, Zhang LI, Tao HW. Strengthening of Direction Selectivity by Broadly Tuned and Spatiotemporally Slightly Offset Inhibition in Mouse Visual Cortex. Cereb Cortex. 2015;25:2466–2477. doi: 10.1093/cercor/bhu049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lien AD, Scanziani M. Tuned thalamic excitation is amplified by visual cortical circuits. Nat Neurosci. 2013;16:1315–1323. doi: 10.1038/nn.3488. nn.3488 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li YT, Ibrahim LA, Liu BH, Zhang LI, Tao HW. Linear transformation of thalamocortical input by intracortical excitation. Nat Neurosci. 2013;16:1324–1330. doi: 10.1038/nn.3494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pei Z, et al. Conditional Knock-Out of Vesicular GABA Transporter Gene from Starburst Amacrine Cells Reveals the Contributions of Multiple Synaptic Mechanisms Underlying Direction Selectivity in the Retina. J Neurosci. 2015;35:13219–13232. doi: 10.1523/JNEUROSCI.0933-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen TW, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature. 2013;499:295–300. doi: 10.1038/nature12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sanes JR, Masland RH. The types of retinal ganglion cells: current status and implications for neuronal classification. Annu Rev Neurosci. 2015;38:221–246. doi: 10.1146/annurev-neuro-071714-034120. [DOI] [PubMed] [Google Scholar]
- 35.Baden T, et al. The functional diversity of retinal ganglion cells in the mouse. Nature. 2016;529:345–350. doi: 10.1038/nature16468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chandrasekaran AR, Shah RD, Crair MC. Developmental homeostasis of mouse retinocollicular synapses. J Neurosci. 2007;27:1746–1755. doi: 10.1523/JNEUROSCI.4383-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ko H, et al. Functional specificity of local synaptic connections in neocortical networks. Nature. 2011;473:87–91. doi: 10.1038/nature09880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee WC, et al. Anatomy and function of an excitatory network in the visual cortex. Nature. 2016;532:370–374. doi: 10.1038/nature17192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cossell L, et al. Functional organization of excitatory synaptic strength in primary visual cortex. Nature. 2015;518:399–403. doi: 10.1038/nature14182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ko H, et al. The emergence of functional microcircuits in visual cortex. Nature. 2013;496:96–100. doi: 10.1038/nature12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gabriel JP, Trivedi CA, Maurer CM, Ryu S, Bollmann JH. Layer-specific targeting of direction-selective neurons in the zebrafish optic tectum. Neuron. 2012;76:1147–1160. doi: 10.1016/j.neuron.2012.12.003. [DOI] [PubMed] [Google Scholar]
- 42.Lowe AS, Nikolaou N, Hunter PR, Thompson ID, Meyer MP. A systems-based dissection of retinal inputs to the zebrafish tectum reveals different rules for different functional classes during development. J Neurosci. 2013;33:13946–13956. doi: 10.1523/JNEUROSCI.1866-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nikolaou N, Meyer MP. Imaging circuit formation in zebrafish. Dev Neurobiol. 2012;72:346–357. doi: 10.1002/dneu.20874. [DOI] [PubMed] [Google Scholar]
- 44.Robles E, Filosa A, Baier H. Precise lamination of retinal axons generates multiple parallel input pathways in the tectum. J Neurosci. 2013;33:5027–5039. doi: 10.1523/JNEUROSCI.4990-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogasawara K, McHaffie JG, Stein BE. Two visual corticotectal systems in cat. J Neurophysiol. 1984;52:1226–1245. doi: 10.1152/jn.1984.52.6.1226. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmann KP, Straschill M. Influences of cortico-tectal and intertectal connections on visual responses in the cat’s superior colliculus. Exp Brain Res. 1971;12:120–131. doi: 10.1007/BF00234310. [DOI] [PubMed] [Google Scholar]
- 47.Michael CR. Integration of retinal and cortical information in the superior colliculus of the ground squirrel. Brain Behav Evol. 1970;3:205–209. doi: 10.1159/000125472. [DOI] [PubMed] [Google Scholar]
- 48.Zhao X, Liu M, Cang J. Visual cortex modulates the magnitude but not the selectivity of looming-evoked responses in the superior colliculus of awake mice. Neuron. 2014;84:202–213. doi: 10.1016/j.neuron.2014.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gale SD, Murphy GJ. Distinct representation and distribution of visual information by specific cell types in mouse superficial superior colliculus. J Neurosci. 2014;34:13458–13471. doi: 10.1523/JNEUROSCI.2768-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhao X, Liu M, Cang J. Sublinear binocular integration preserves orientation selectivity in mouse visual cortex. Nat Commun. 2013;4:2088. doi: 10.1038/ncomms3088. ncomms3088 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tada M, Takeuchi A, Hashizume M, Kitamura K, Kano M. A highly sensitive fluorescent indicator dye for calcium imaging of neural activity in vitro and in vivo. Eur J Neurosci. 2014;39:1720–1728. doi: 10.1111/ejn.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brainard DH. The Psychophysics Toolbox. Spat Vis. 1997;10:433–436. [PubMed] [Google Scholar]
- 53.Niell CM, Stryker MP. Highly selective receptive fields in mouse visual cortex. J Neurosci. 2008;28:7520–7536. doi: 10.1523/JNEUROSCI.0623-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ferenczi EA, et al. Optogenetic approaches addressing extracellular modulation of neural excitability. Sci Rep. 2016;6:23947. doi: 10.1038/srep23947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wei W, Elstrott J, Feller MB. Two-photon targeted recording of GFP-expressing neurons for light responses and live-cell imaging in the mouse retina. Nat Protoc. 2010;5:1347–1352. doi: 10.1038/nprot.2010.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study and the custom Matlab code are available from the corresponding author upon reasonable request.