Abstract
Gain-of function mutations in some genes underlie neurodegenerative conditions whereas loss-of-function mutations have distinct phenotypes. Such appears to be the case with the protein ataxin 1 (ATXN1), which forms a transcriptional repressor complex with capicua (CIC). Gain-of-function of the complex leads to neurodegeneration, but ATXIN1-CIC is also essential for survival. We set out to understand the functions of ATXN1-CIC in the developing forebrain and found that losing the complex results in hyperactivity, impaired learning and memory, and abnormal maturation and maintenance of upper layer cortical neurons. We also found that CIC modulates social interactions in the hypothalamus and medial amygdala. Informed by these neurobehavioral features in mouse mutants, we identified five patients with de novo heterozygous truncating mutations in CIC that share similar clinical features, including intellectual disability, attention deficit/hyperactivity disorder (ADHD), and autism spectrum disorder. Our study demonstrates that loss of ATXN1-CIC complexes causes a spectrum of neurobehavioral phenotypes.
Introduction
Ataxin 1 (ATXN1) has been slow to yield its functions to investigation. Expansion of its polyglutamine tract was discovered in 1993 to produce the adult-onset disease spinocerebellar ataxia type 1, but deletion of the gene in Atxn1−/− mice produces only mild defects in learning and memory without neurodegeneration1–5. We subsequently discovered that mutant ataxin 1 alters transcription6, and we searched for protein interactors that contribute to its activities. We found that the transcriptional repressor capicua (CIC) forms a co-repressor complex (ATXN1-CIC) in vivo with ATXN1 and with its paralog ataxin 1-like (ATXN1L), and that CIC anchors the complex to DNA to repress the target genes of CIC7–9.
While these studies provided insights into ATXN1-CIC function, the consequences of losing ATXN1-CIC in the developing brain remains unknown. In mice, the ATXN1-CIC complex is essential for survival, as mice either lacking both Atxn1 and Atxn1l or homozygous knockout allele of Cic that reduces CIC levels by more than 80% (Cic-L) die shortly after birth9,10. Atxn1−/−; Atxn1l−/− mice exhibit hydrocephalus, abdominal wall closure defect, and lung defects9. Cic-L−/− mice phenocopy Atxn1−/−; Atxn1l−/− mice, confirming that these three proteins indeed function as a complex in vivo9. Interestingly, large deletions spanning ATXN1 on chromosome 6p22 have been reported in patients with autism spectrum disorder (ASD), developmental delay/intellectual disability (DD/ID), seizures, and attention deficit/hyperactivity disorder (ADHD)11–13, but it is unclear whether the heterozygous loss of ATXN1 or other genes in the deleted region lead to the clinical presentation. In the case of CIC, two different missense mutations have been reported in two isolated cases of intellectual disability14,15, but here again it is unclear whether these variants are causative, because there have been no functional studies to confirm the pathogenicity of the reported missense variants in CIC.
We set out to investigate the neurodevelopmental consequences of losing the ATXN1-CIC complex. To bypass the early lethality caused by defects in peripheral organs9, we utilized Cre-loxP technology to generate conditional knockouts of either Atxn1-Atxn1l or Cic. We first focused our study on the developing forebrain, a region known to contribute prominently to the clinical symptoms of patients with neurodevelopmental disorders16,17, using Emx1-Cre18. We discovered that forebrain deletion of ATXN1-CIC caused learning/memory deficits and hyperactivity that responded to low-dose amphetamine. Emx1-Cre conditional mutant neurons exhibited histological and cellular defects. Loss of ATXN1-CIC complex outside of Emx1-Cre domains using Otp-Cre, which was expressed in the hypothalamus and medial amygdala, led to social interaction defects. Informed by the behavioral phenotypes, we searched for patients that have truncating mutations in CIC. We found five patients with de novo heterozygous truncating mutations in CIC that are predicted to undergo nonsense-mediated decay. These patients present with ASD, DD/ID, seizures, and ADHD, features overlapping with those uncovered by the mouse studies. Altogether these results demonstrate the necessity of ATXN1-CIC complex for normal brain development and reveal the region-specific contributions of dysfunctional ATXN1-CIC complex to intellectual disability, hyperactivity, and social behavioral deficits.
Results
Deleting the ATXN1-CIC complex in the developing forebrain results in multiple behavioral abnormalities
The stable ATXN1-CIC complex is present in many brain regions, including the cerebellum7,8, cortex and hypothalamus (Supplementary Fig. 1). Because constitutive knockouts were not viable, we first utilized Cic+/− mice for our initial effort to study the roles of ATXN1-CIC complex in the brain. A battery of behavioral tests showed that Cic+/− mice behaved similarly to their wildtype littermate controls in all the tests except for the open field assay, where they exhibited mild hyperactivity (Supplementary Fig. 2). To further study the functions of the ATXN1-CIC complex in the developing brain, we generated conditional knockouts of either Atxn1-Atxn1l or Cic in the developing forebrain through breeding with Emx1-Cre mice, in which the Cre recombinase is expressed in progenitor cells in the dorsal telencephalon starting at embryonic day 10.5 (E10.5), and will delete the conditional alleles in forebrain excitatory neurons and glia18. We confirmed the knockout efficiency in the mutant mice by western blot and immunofluorescent staining, which demonstrated marked reduction of either ATXN1L or CIC protein levels (Supplementary Fig. 3a–c). Upregulation of the known CIC target genes Etv1, Etv4 and Etv5 in the mutant cortex further demonstrated efficient removal of the complex (Supplementary Fig. 3d).
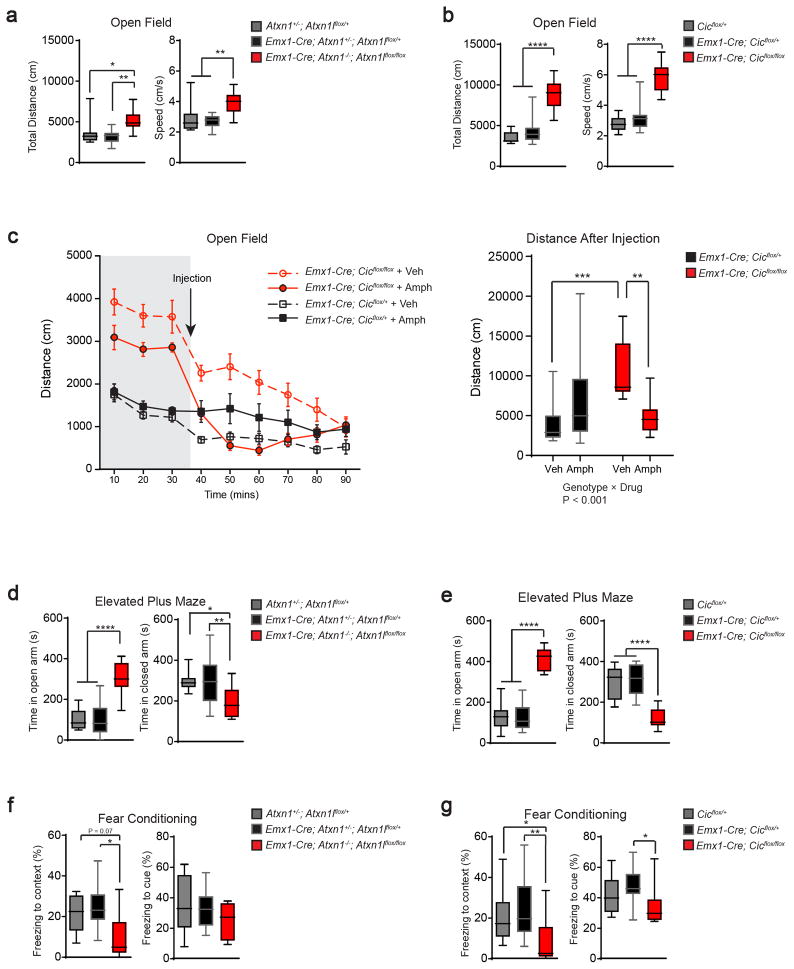
We performed a series of behavioral tests on the Atxn1-Atxn1l and Cic conditional knockout mice to assess general activity, anxiety, learning/memory, and social behavior. Both mouse lines covered greater distance at greater speed in the open-field test than their littermate controls (Fig. 1a, b). Given the ADHD symptoms in patients with deletions spanning ATXN111, we tested the effects of low-dose amphetamine on the mutant mice. As has been shown in some ADHD models, low-dose amphetamine exerted a paradoxical calming effect on the mutant animals19. Emx1-Cre; Cicflox/flox groups treated with vehicle or amphetamine exhibited different baseline activities before the drug injection (time points 10–30 mins). We reasoned that this was probably due to chance and expected normal variability as the experimenters were blinded to the genotypes and treatments. Importantly, we found a significant interaction between genotype and drug (Fig. 1c, right panel), demonstrating the paradoxical calming effect of low-dose amphetamine on mutant mice. In the elevated plus-maze test, conditional knockout mice spent more time in the open-arm and less time in the closed-arm, which could be due to reduced anxiety (Fig. 1d, e). We used the fear conditioning assay to examine learning and memory in the conditional mutants, and discovered that they exhibited reduced freezing, especially in the test of spatial context, indicating that they have impaired learning and memory (Fig. 1f, g). Finally, we assessed social interaction behavior using the three-chamber test and found that the mutant mice behaved in a similar way to the control mice. (Supplemental Fig. 4).
Figure 1. Deleting Atxn1-Atxn1l or Cic in the developing forebrain results in behavioral abnormalities.
(a, b) In the open field test, both Atxn1-Atxn1l and Cic conditional knockout mice showed increased exploratory activities as demonstrated by increased total distance travelled and increased movement speed (n = 9–16, 12–13 weeks old). (c) After recording 30 minutes of baseline activity, we administered low-dose amphetamine (2 mg/kg body weight) and monitored locomotion for 60 minutes. Low-dose amphetamine calmed the Cic conditional knockout mice (n = 10–14, 12–13 weeks old). (d, e) In the elevated plus maze test, both mutant mice spent more time in the open arm and less time in the closed arm (n = 11–18, 12–13 weeks old). (f, g) Both mutant mouse lines had reduced freezing in the fear conditioning test (n = 8–13, 16–20 weeks old). Data are represented as mean ± s.e.m. in the left panel of 1c. All Other data are presented in box-and-whisker plots, with center lines represent median, box limits represent interquartile range, and whiskers represent minimum to maximum data range. *: P < 0.05; **: P < 0.01; ***: P < 0.001; ****: P < 0.0001
It is worth noting that conditional knockouts of either Atxn1-Atxn1l or Cic displayed similar behavioral deficits, which is consistent with their role as constituents of a protein complex9. Interestingly, the protein levels of CIC were significantly reduced in the Atxn1-Atxn1l conditional knockout tissue, but the levels of ATXN1 and ATXN1L were not reduced in the Cic conditional knockouts (Supplementary Fig. 3a, b), suggesting that the observed mutant phenotypes are mostly driven by the loss of CIC functions and disruption of the complex.
Deleting the ATXN1-CIC complex causes a postnatal reduction in the cortical thickness and the number of CUX1+ cells
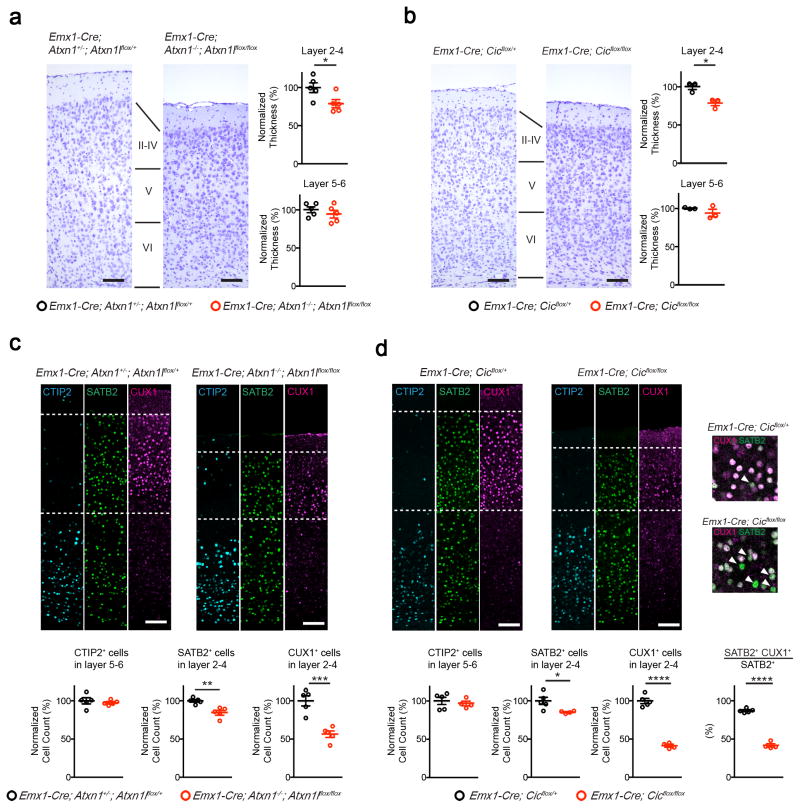
To determine whether the behavioral deficits in the mutant mice arise from abnormal anatomical development, we examined brain sections from 5-week old mice. We found that the structures of amygdala and hippocampus were similar between control and Emx1-Cre conditional knockout animals (Supplementary Fig. 5 and 6). The conditional knockout mice, however, had reduced thickness of cortical layer 2–4, while the thickness of layer 5–6 remained normal (Fig. 2a, b). At 20 weeks of age, the structures of amygdala remained comparable between control and mutant mice (Supplementary Fig. 7a and b). We observed a similar reduction in the thickness of cortical layer 2–4 in the mutants, whereas the thickness of layer 5–6 was not affected (Supplementary Fig. 7c). The hippocampal dentate gyrus from 20-week-old mutant mice showed decreased thickness, while the thickness of CA1 and CA3 regions were normal (Supplementary Fig. 7d). The dentate gyrus is critical for receiving and processing neuronal input from other brain regions, such as the entorhinal cortex, before sending signals to CA320. The pathology in the dentate gyrus, which manifested when the mutant mice were older, might contribute to the learning and memory deficits in the contextual fear conditioning test (Fig. 1f, g). We further examined the synaptic plasticity in the hippocampus and found that, while basal synaptic transmission and long-term synaptic plasticity were preserved in the Emx1-Cre Cic knockout mice, there was a decrease in the probability of presynaptic neurotransmitter release (Supplementary Fig. 8).
Figure 2. Deleting Atxn1-Atxn1l or Cic in the developing forebrain results in reduced thickness of upper cortical layers.
(a, b) By Nissl staining, both mutant mouse lines had reduced thickness of layer 2–4, but not layer 5–6 (n = 3–5, scale bar = 100 μm). (c, d) Immunofluorescent staining of the cortex from 5-week old control and mutant animals. The number of CTIP2+ cells in layer 5–6 did not change, but the numbers of SATB2+ and CUX1+ cells were reduced in the mutant cortex (n = 4–5, scale bar = 100 μm). (d, inset) Mutant mice showed fewer layer 2–4 SATB2+ cells coexpressing CUX1. (Arrowhead: SATB2 single positive cells.) Data are represented as mean ± s.e.m. *: P < 0.05; **: P < 0.01; ***: P < 0.001; ****: P < 0.0001
We next examined the cortex using markers specific to layer 2–4 (CUX1), layer 5–6 (CTIP2, also known as BCL11B), and layer 2–6 (SATB2). Consistent with the Nissl staining results, the number of CTIP2+ cells remained unchanged (Fig. 2c, d). Interestingly, while the number of SATB2+ neurons in layer 2–4 was only slightly reduced, the number of layer 2–4 CUX1+ neurons was dramatically reduced in the mutants (Fig. 2c, d). In control layer 2–4 neurons, most of the SATB2+ cells co-expressed CUX1, in contrast to the mutants where only half of layer 2–4 SATB2+ cells were CUX1-positive (Fig. 2d). These findings suggest that the reduction in CUX1+ cells was primarily driven by the loss of CUX1 expression in layer 2–4 neurons. In support of this, Cux1 expression was reduced in the mutant cortex (Supplementary Fig. 9a). The expression levels of Cux2, which is a paralog of Cux1 and a marker for layer 2–4 pyramidal neurons, was however unaffected in the mutant cortex, suggesting there is no global defect in the fate of layer 2–4 neurons (Supplementary Fig. 9b). Because the majority of the callosal projection neurons (CPNs) reside in the upper cortical layers, and the projections of CPN form the corpus callosum21, we examined the formation of the corpus callosum with Luxol Fast Blue staining, which stains myelin. The formation of the corpus callosum in the mutant brains was not disrupted, nor was the thickness of the corpus callosum significantly altered (Supplementary Fig. 10). Since Atxn1-Atxn1l conditional mutants phenocopied Cic conditional mutants, we next focused on Cic conditional mutants to further dissect the cortical defects resulting from the loss of function of the complex.
One possible explanation for the observed reduction in the number of layer 2–4 neurons is defective progenitor proliferation. To address this, we first assessed whether CIC is expressed in the progenitor cells in the embryonic cortex and found that to be the case (Supplementary Fig. 11a). Since layer 2–4 neurons are generated primarily around E14.5-E16.5 in mice22, we assessed the proliferation of the cortical progenitors at E14.5 and E16.5 with a two-hour pulse-labeling of thymidine analog EdU (Ethynyl-2′-deoxyuridine). By combining EdU labeling with specific markers (TBR2 for intermediate progenitors and PAX6 for radial glia), we found that the numbers of progenitors and their proliferation rates (estimated from cells that were double-positive for EdU) remained unchanged in the mutant cortex (Supplementary Fig. 11b).
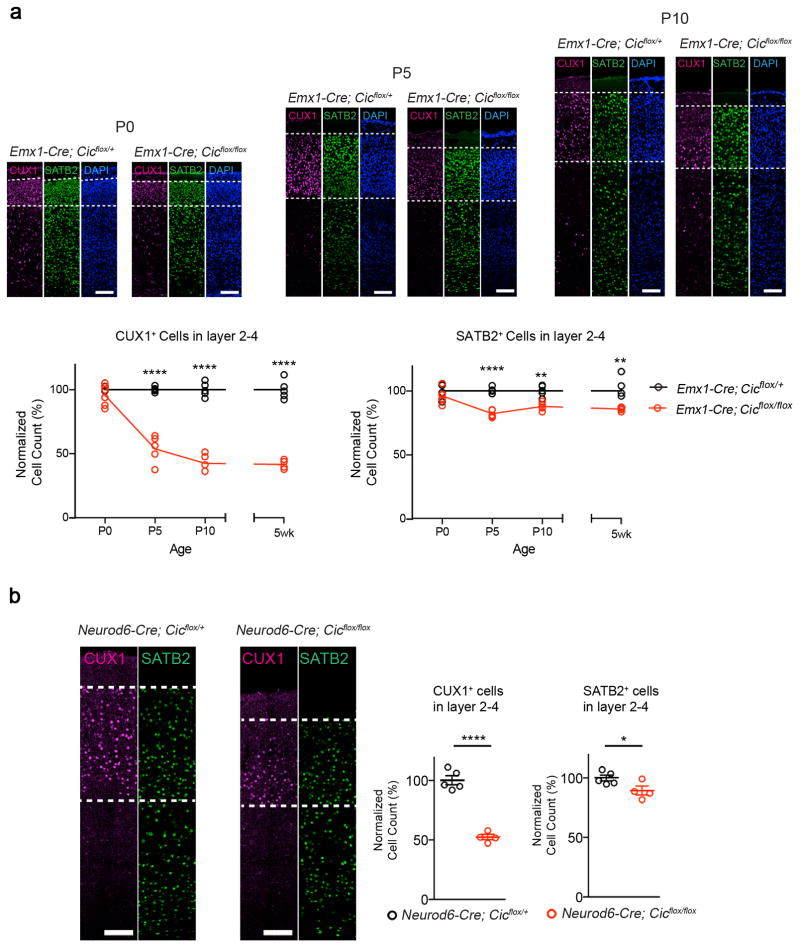
We next hypothesized that the reduction in the number of CUX1+ cells is due to postnatal defects in neuronal maturation or maintenance. Indeed, the numbers of CUX1+ and SATB2+ neurons in layer 2–4 in the mutant cortex were similar to those of the controls at P0, but the numbers declined postnatally, and by P10 the numbers were comparable to those of 5-week old animals (Fig. 3a). Furthermore, we generated Cic conditional knockout mice using Neurod6-Cre (NEX-Cre), which has a spatial-temporal expression pattern similar to Emx1-Cre but is selective for post-mitotic excitatory neurons23. When we examined the cortex in post-mitotic conditional mutant mice, we still observed a reduction in the number of cells expressing CUX1 and SATB2 in layer 2–4 (Fig. 3b). These experiments suggest that the reduction in CUX1- and SATB2-expressing cells was caused by defects in post-mitotic neurons in the mutant cortex.
Figure 3. The histological defects in Emx1-Cre Cic mutant animals occur postnatally in post-mitotic neurons.
(a) The numbers of CUX1+ cells and SATB2+ cell in layer 2–4 diminished postnatally. Immunofluorescent staining with CUX1 and SATB2 revealed that the number of CUX1+ cells and SATB2+ cells at P0 were comparable among control and mutant animals. The number gradually decreased in the mutant mice, and by P10 the numbers were comparable to those from 5-week old animals (n = 4–6, scale bar = 100 μm). (The 5-week old data are from Figure 2.) (b) Immunofluorescent staining with CUX1 and SATB2 revealed that the numbers of layer 2–4 CUX1+ and SATB2+ cells were reduced in Neurod6-Cre conditional mutants, which removes Cic in post-mitotic neurons (n = 4–5, scale bar = 100 μm). Data are represented as mean ± s.e.m. *: P < 0.05; ****: P < 0.0001
Increased apoptosis of upper layer neurons offers a potential explanation for the postnatal decline of SATB2+ cells in layer 2–4 of mutant mice. To address this, we monitored cell death in P5 and P10 mutant cortex using activated caspase-3 (CASP3) immunostaining. We found more CASP3+ cells in the upper layers of Emx1-Cre mutant cortex at P5, while the numbers of CASP3+ cells were similar between mutant and control cortex at P10 (Supplemental Fig. 12). These results suggest a transient upregulation of apoptosis in the upper layers of P5 mutant cortex, consistent with the observed initial decline of SATB2+ cells in layer 2–4 of mutant mice at P5.
CIC is critical for neuronal dendritic branching
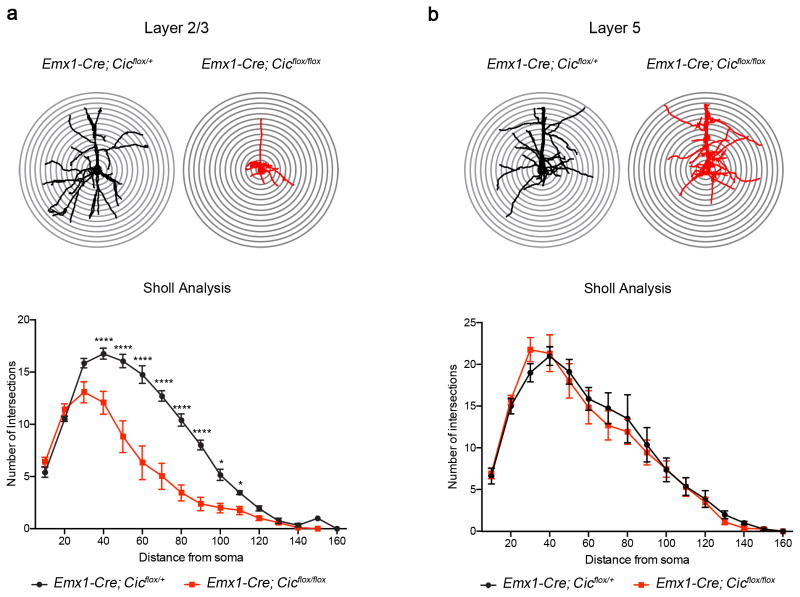
Because CUX1 has roles in promoting dendritic branching24,25 and CUX1 levels were reduced in the mutant cortex, we set out to examine the dendritic morphology of layer 2/3 neurons. To this end, we sparsely labeled P0 neurons using low-titer AAV virus with YFP 26. We harvested brains when the animals were five weeks old and examined dendritic branching patterns of pyramidal neurons. Using Sholl analysis, we discovered that the mutant layer 2/3 pyramidal neurons displayed significantly reduced dendritic branching complexity compared to controls (Fig. 4a). The dendritic branching complexity of layer 5 pyramidal neurons did not differ from that of controls (Fig. 4b).
Figure 4. Morphological defects in layer 2/3 pyramidal neurons in the Emx1-Cre Cic conditional knockout mice.
(a) Sholl analysis showed that mutant layer 2/3 pyramidal neurons had reduced dendritic complexity as compared with control animals. (b) The dendritic complexity in layer 5 neurons was not affected in mutant mice (n = 4–6 animals, 2–4 cells per animal). Data are represented as mean ± s.e.m. *: P < 0.05; ****: P < 0.0001
Deleting Cic in the hypothalamus and medial amygdala causes abnormal social interaction in mice
We next explored the neurobehavioral consequences of deleting the ATXN1-CIC complex outside of the Emx1-expressing domain. We utilized a Cre line that targets hypothalamic and medial amygdala neurons using the promoter from orthopedia homeobox (Otp)27,28. Using the ROSA26fsTRAP reporter line to label Cre-positive cells29. We found that Cre was expressed in the medial amygdala (MeA) and multiple hypothalamic regions, including the paraventricular nucleus (PVN), supraoptic nucleus (SON), lateral hypothalamic area (LHA), as well as dorsomedial and ventromedial hypothalamic nuclei (DMH and VMH) (Supplementary Fig. 13). The expression pattern of Otp-Cre was consistent with the endogenous expression pattern of Otp, covering neuroendocrine neurons (e.g., PVN and SON) as well as regions that underlie social and emotional responses (VMH and MeA)27,30–32. We then generated Otp-Cre Cic conditional knockout mice and validated the knockout efficiency of Cic by confirming that the protein levels of CIC were greatly reduced in Cre-positive cells (Supplementary Fig. 14a).
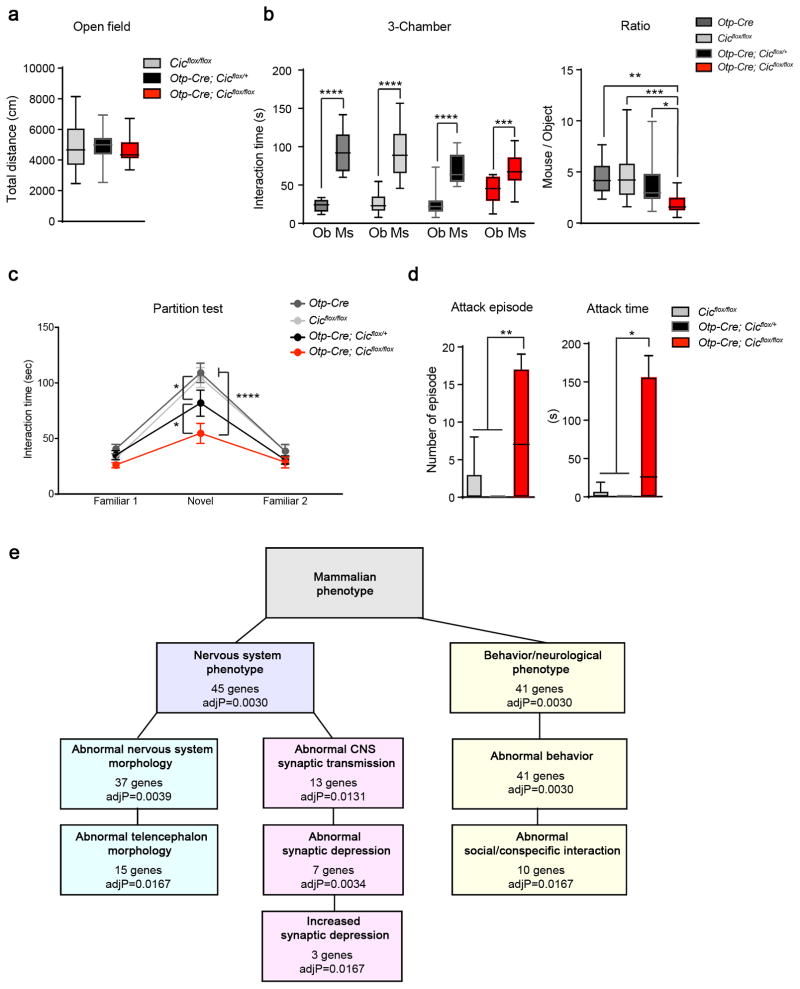
To investigate whether CIC functions in Otp-lineage neurons to modulate neurobehavior, we performed a battery of behavioral tests. Interestingly, the Otp-Cre conditional mutant mice showed distinct behavioral defects in comparison to Emx1-Cre conditional mutants. The Otp-Cre conditional knockouts did not have abnormal behavior in the open-field, elevated plus-maze, or fear-conditioning assays (Fig. 5a and Supplementary Fig. 15), but they did show prominent social interaction deficits. In the three-chamber test, the conditional knockout mice interacted less with other mice (Fig. 5b). In the partition test, the conditional knockout mice spent less time interacting with novel mice, while the Otp-Cre; Cicflox/+ mice showed an intermediate phenotype (Fig. 5c). In the resident-intruder test, the conditional knockout mice behaved more aggressively towards intruders, with more and longer attack episodes than controls (Fig. 5d). These results suggest that CIC has important functions in the medial amygdala and hypothalamus to modulate social interaction.
Figure 5. Deleting Cic from the hypothalamus and medial amygdala results in abnormal social behavior.
(a) The Otp-Cre conditional knockout mice showed normal exploratory activities in the open-field test (n = 16–28, 8–12 weeks old). (b) In the three-chamber test, the Otp-Cre conditional mutant animals still preferred mouse (Ms) over object (Ob), but they spent less time with mouse as compare to controls (n = 15–19, 8–12 weeks old). (c) In the partition test, the Otp-Cre; Cicflox/flox mice spent less time interacting with the novel partner mice. The Otp-Cre; Cicflox/+ mice had an intermediate phenotype (n = 14–19, 8–12 weeks old). (d) The Otp-Cre conditional knockout males were more aggressive in the resident-intruder test than controls, showing more and longer attacks toward a male intruder (n = 9–11, 8–12 weeks old). (e) Gene set enrichment analysis of differentially expressed genes between Otp-Cre; Cicflox/flox; ROSAfsTRAP and Otp-Cre; ROSAfsTRAP neurons. Data are represented as mean ± s.e.m. in c. All other data are presented in box-and-whisker plots, with center lines represent median, box limits represent interquartile range, and whiskers represent minimum to maximum data range. *: P < 0.05; **: P < 0.01; ***: P < 0.001; ****: P < 0.0001
In contrast to the pronounced cortical layering defects in the Emx1-Cre knockout animals, we did not observe any gross histological abnormalities or altered cellularity of the Otp-lineage neurons in the Otp-Cre conditional mutants (Supplementary Fig. 14b and 16). Expression levels and patterns of several hypothalamic nuclei markers were also unaltered in mutant mice (Supplementary Fig. 17). Since CIC is a transcriptional regulator, we next examined gene expression profiles in Otp-lineage neurons to investigate the molecular changes leading to the social interaction deficits in the Otp-Cre mutant mice. To isolate RNAs specifically from these neurons, we applied the translating ribosome affinity purification (TRAP) approach using the Cre-dependent ROSAfsTRAP mouse line29, where the ribosomes are tagged with GFP in Cre-expressing cells. TRAP and RNA isolation was performed when animals were 8–10 weeks old33. RNAseq and gene expression analysis showed that a total of 207 genes were differentially expressed between Otp-Cre; Cicflox/flox; ROSAfsTRAP and Otp-Cre;ROSAfsTRAP neurons, with 123 upregulated genes and 84 downregulated genes (FDR<0.05; Supplementary Fig. 18 and Supplementary Table 1). Known CIC targets Etv4 and Etv5 were among the most upregulated genes. Gene set analysis revealed that differentially expressed genes is significantly enriched for abnormal nervous system morphology, abnormal synaptic transmission, and abnormal social/conspecific interaction (Fig. 5e). Notably, ten genes associated with abnormal synaptic transmission and eight genes associated with abnormal social/conspecific behavior showed more than 1.5-fold changes (Supplementary Table 2). We further validated a subset of genes from each group using quantitative real-time PCR (Supplementary Fig. 19). Dysregulation in the expression of these genes might contribute to the social behavioral deficits observed in the Otp-Cre mutant mice.
Heterozygous CIC truncating mutations cause a neurodevelopmental syndrome in human patients
Prior to our study, it was unclear whether loss-of-function mutations in ATXN1 and CIC cause neurodevelopmental disorders, as no isolated truncating alleles had been identified for either gene. Our studies showed that the ATXN1-CIC complex plays critical roles in brain development that affect behavior, and that ATXN1 and ATXN1L execute their functions mainly through forming a complex with CIC. Guided by this knowledge and our prediction that homozygous loss-of-function in CIC would lead to premature lethality, we sought to identify patients with heterozygous loss-of-function mutations in CIC. It is also worth noting that CIC is predicted to be loss-of-function intolerant in the ExAC database (pLI = 1.00, Version 0.3.1)34. Although a 19:42796882 G/GC loss-of-function variant was reported in multiple individuals in the ExAC database, we found that the high frequency of this variant probably arises from sequencing/reporting errors or very low-grade mosaicism in the individuals. Thus the occurrence of this variant does not contradict the loss-of-function intolerance prediction from ExAC and our genetic findings below.
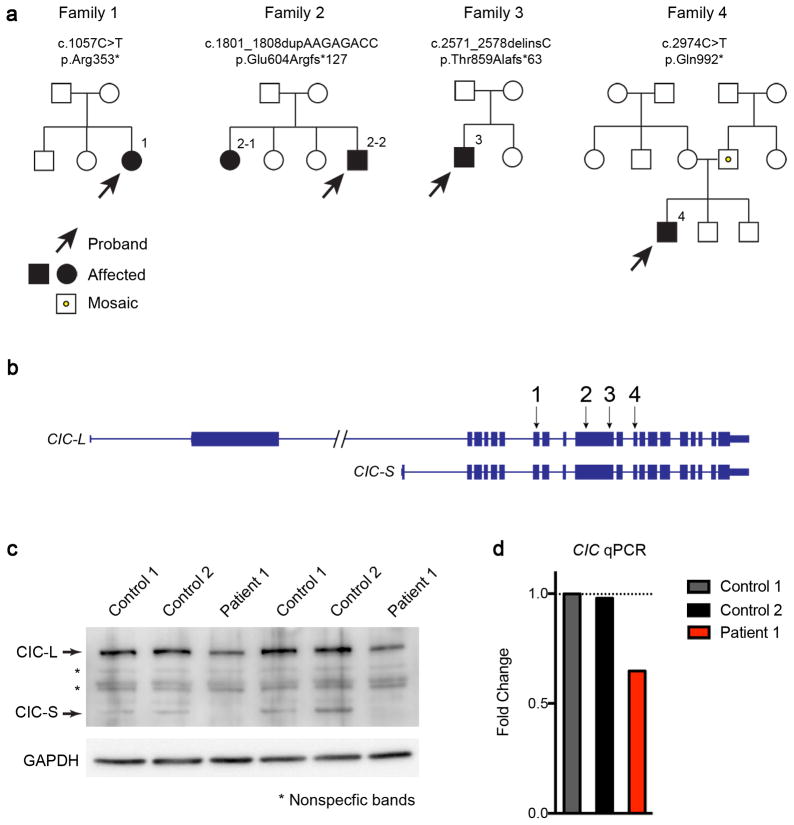
Through collaborative efforts, we identified four individuals from three families with heterozygous de novo truncating mutations in CIC, and one individual with an inherited nonsense variant from a father who is low-grade mosaic (see below) for the variant (Fig. 6a, b). Individuals 1 and 2-2 were part of exome trios. We additionally performed whole exome sequencing on individual 2-1. Individuals 3 and 4 were sequenced by whole exome sequencing with de novo variants confirmed through targeted Sanger sequencing of the parents. The CIC variants in all affected individuals were confirmed by Sanger sequencing. Besides the CIC variants, no variants were identified in known disease-causing genes that accounted for the clinical phenotypes. The five individuals had presented clinically with a spectrum of phenotypes, including developmental delay/intellectual disability (DD/ID), attention-deficit/hyperactivity disorder (ADHD), autism spectrum disorder (ASD), and seizures (Table 1, Supplementary Note).
Figure 6. Identification of heterozygous CIC truncating mutations in four families.
(a) Pedigrees of four families with CIC truncating mutations. Both patients in the second family have the same mutation, but neither parent harbors the mutation in their somatic DNA; one of the parents is presumed to be germline mosaic. Sanger sequencing of the CIC variant regions in the two unaffected siblings revealed they do not carry the same mutations in CIC. The father in the fourth family is a low-grade somatic mosaic as demonstrated by Sanger sequencing. (b) Genomic locus of human CIC showing the location of the mutations found in the four families. All four mutations are predicted to create premature stop codons. (c) Western blot analysis of fibroblasts from patient 1 and two controls showing that CIC protein levels were reduced in the patient. Images were cropped for better presentation. (d) qPCR of CIC from patient 1 and control fibroblasts showed that the RNA levels of CIC were reduced in fibroblasts from the patient. The expression levels were normalized to control 1, and fold changes were plotted. Five to six technical repeats were performed for each sample.
Table 1.
Characteristics of patients with CIC mutations.
| 1 | 2-1 | 2-2 | 3 | 4 | |
|---|---|---|---|---|---|
| Age at latest evaluation (years) | 16 | 27 | 9 | 4 | 15 |
| Gender | Female | Female | Male | Male | Male |
| Nucleotide change (NM_015125.4) | c.1057C>T | c.1801_1808dupAAGAGACC | c.2571_2578delinsC | c.2974C>T | |
| Amino acid change | p.Arg353* | p.Glu604Argfs*127 | p.Thr859Alafs*63 | p.Gln992* | |
| Inheritance | De novo | De novo (presumed germline mosaicism) | De novo | Paternally inherited (father mosaic) | |
| ASD/Autistic features | + | − | − | + | + |
| Attention-deficit/hyperactivity | − | − | + | − | + |
| DD/ID | + | + | + | + | + |
| Seizures | + | + | + | − | − |
| Dysmorphic features | − | − | − | − | − |
| Head circumference at latest exam (cm) | 57.5 | N/A | 54 | 50.2 | 54 |
| Head circumference at latest exam (percentile) | 99th | N/A | 90th | 32nd | 27th |
| Latest MRI results | Multiple punctate foci of T2 hyperintensity within the subcortical white matter. | Normal brain MRI. | Normal brain MRI. | Single punctate focus of T2 hyperintensity within the right frontal lobe white matter. | Several periventricular T2 hyperintensities in the white matter, more concentrated in the parieto-occipital region. |
| Neurological symptoms at last exam | Normal muscle tone; normal deep tendon reflexes; no dyskinesia; no ataxia. | Normal muscle tone; normal deep tendon reflexes; normal gait; no ataxia. | Truncal hypotonia; normal deep tendon reflexes; normal gait; no ataxia. | Mild diffuse hypotonia; normal reflexes; no dyskinesia; no ataxia. | Normal muscle tone; hyperreflexia; no pyramidal syndrome; no ataxia. |
| Other | ALL, diastasis recti, mild telangiectasia. | Pulmonary stenosis. | Heart murmur, mild cutis marmorata telangiectasia. | Marfanoid habitus, stereotypic movements. | |
ASD: autism spectrum disorder, DD: developmental delay, ID: intellectual disability, ALL: acute lymphoblastic leukemia, N/A: data not available.
Family 2 was notable for having two apparently affected siblings, and subsequent interrogation of the mutant allele in all six family members by Sanger sequencing found that the genotype segregated faithfully with the phenotype, suggesting germline mosaicism for the mutant allele in one of the parents (Supplementary Fig. 20). The father in family 4 is mosaic for the mutant allele (~15% mosaicism). By investigating the mutant allele in all family members using Sanger sequencing, we found that only the proband is heterozygous for the mutant allele, while the two unaffected brothers have only the wildtype allele (Supplementary Fig. 21). We examined the fibroblasts from patient 1 and found that both the protein and RNA levels of CIC were reduced to almost 50% (Fig. 6c, d, Supplementary Fig. 22), supporting the idea that the mutation in patient 1 leads to nonsense-mediated decay and causes loss-of-function.
Discussion
In this study, we examined the roles of ATXN1-CIC complex in the developing nervous system and discovered a cell-type- and region-specific role for this complex in neurobehavioral functions. First, within the cortex, ATXN1-CIC complex is more important for upper layer neurons. Although CIC is expressed in all cortical layers and the protein was effectively removed from all layers in the Emx1-Cre conditional knockouts, the reduction of cortical thickness and cellular morphological defects were evident in the upper cortical layers while lower layers appeared intact. One explanation is that the ATXN1-CIC complex might have distinct downstream target genes in different cortical layers; it’s also possible that other transcription factors compensate for the loss of CIC in the lower cortical layers. There has been rapid advancement in single-cell transcriptomic technologies, and studying the transcriptomic changes in different cell types at specific time points may provide insight into the possible mechanisms for the selective phenotype we observed.
The upper layer defects observed in Emx1-Cre conditional mutants are most likely driven by defects in postnatal maturation or maintenance of these neurons, as reduction in SATB2+ neurons was evident only after postnatal day 5 (P5). This is further supported by our analyses of the Neurod6-Cre conditional knockout mice, suggesting that the reduction was due to defects in the post-mitotic neurons but not in their fate specification. Indeed, at P5, we observed a transient increase in the number of layer 2–4 neurons undergoing apoptosis in the Emx1-Cre mutant cortex, which could contribute to the postnatal reduction of SATB2+ cells in layer 2–4. In addition, SATB2+ cells in layer 2–4 from mutant cortex also expressed less Cux1 RNA and the protein. The underlying mechanism leading to CUX1 reduction remains elusive, but evidence suggest that CUX1 dosage is critical for dendritic morphology of cortical neurons. Layer 2–3 neurons from Cux1−/− mice showed fewer dendritic branches and spines24, whereas overexpression of Cux1 in cultured cortical neurons increased spine density25. Therefore, in our studies, reduced CUX1 levels in Emx1-Cre mutant cortex could contribute to the observed dendritic arborization defects of upper layer neurons. Very recently, homozygous mutations in a human accelerated region (HAR) that could increase the expression of human CUX1 were identified in patients with ASD and ID25. Our findings resonate with previous studies and highlight the importance of CUX1 dosage in modulating cognitive and social behavior outcome.
In some ADHD patients, brain imaging studies have identified cortical abnormalities16, and there is a reduction in cortical thickness and a marked delay in the maturation across different brain regions35,36. Our Emx1-Cre conditional mutant animals provide evidence for the potential consequences of disrupting upper cortical layers in causing hyperactivity and learning deficit phenotypes. This provides an exciting opportunity to understand the contribution of specific cortical circuit defects to the clinical phenotypes of patients with ADHD. Upper layer neurons project axons that contribute to the corpus callosum, but the formation and thickness of the corpus callosum was unaffected in Emx1-Cre mutant mice. This suggests that the formation of long range projections by upper layer neurons is anatomically normal, but it remains possible that they are functionally compromised.
Although the Emx1-Cre conditional mutants recapitulated the hyperactivity and learning/memory deficits seen in patients with heterozygous loss of CIC, the Otp-Cre knockout mice displayed phenotypes resembling ASD. The ventromedial hypothalamic (VMH) nuclei and medial amygdala (MeA), both targeted by the Otp-Cre, are activated in both aggressive and affiliative behavior31,32. Mis-expression of genes that influence synaptic transmission and social interaction might interfere with the synaptic plasticity and disrupt social brain circuits at the cellular level. For example, Shank3, which encodes a dosage-sensitive synaptic scaffold protein whose dysfunction is involved in human neuropsychiatric disorders, is significantly reduced in Otp-Cre mutant neurons. In Otp-Cre mutant neurons, the combinatorial effects of Shank3 and other dysregulated genes that have reached their dysfunction thresholds might compromise healthy neuronal activity under social stimuli. Although the Emx1-Cre and Otp-Cre conditional knockouts together reproduced many clinical features of patients with CIC loss-of-function mutations, we did not observe overt seizures in any of our mutant mice by visual examinations (Cic+/−, Emx1-Cre mutants, or Otp-Cre mutants). We also did not find any abnormalities in the electroencephalograms (EEGs) during two 8-hour recordings from Cic+/− or Emx1-Cre mutants (data not shown). We have, however, observed seizures in Cic-L−/− mice on a highly mixed background (Huda Y Zoghbi, personal observation). It is possible that seizures in the patients arise from dysfunction of forebrain inhibitory neurons, a cell type not covered by Emx1-Cre. Future studies with additional Cre drivers can help pinpoint the responsible cell types for seizures and elucidate possible underlying mechanisms.
In conclusion, studying the roles of ATXN1-CIC complex in the mouse brain has led us to identify new mutations underlying neuropsychiatric disease in humans. Our findings highlight distinct brain regions underlying the clinical symptoms in the patients and provide tractable models to further dissect the circuitries contributing to ADHD, learning deficits, and social behavior abnormalities. Our strategy underscores the value of model organisms in uncovering new syndromes and their pathophysiology. It is the findings in the Emx1-Cre and Otp-Cre driven Cic deletion mice that led us to search for patients with CIC loss-of-function mutations and informed us of the breadth of CIC haploinsufficiency manifestations. Two-way research between mouse models and human patients continues to reveal the neuronal and molecular bases of complex neurobehavioral disorders.
Online Methods
Human subjects
Patients were identified either by direct contact from family (family 1) or connections through GeneMatcher (family 2–4)37. Individuals 1 and 2-2 were part of exome trios (proband plus two biological parents). Individual 2-1 was subsequently enrolled in whole-exome sequencing study and her results were compared with the parents and individual 2-2. Probands 3 and 4 were sequenced by whole exome sequencing with de novo variants confirmed through targeted Sanger sequencing of the parents. The CIC variants in all affected individuals were confirmed by Sanger sequencing. Buccal swab DNAs from patient 2-2’s three siblings were sent from GeneDx to Dr. Zoghbi’s lab for the testing of CIC variants using Sanger sequencing. Fibroblasts from patient 1 were generated by Dr. Finnell’s laboratory. Family 1 and 2 were enrolled under a research protocol approved by the Institutional Review Board of Baylor College of Medicine. Family 3 was enrolled under a research protocol approved by the Institutional Review Board of Mayo Clinic. All individuals or their guardians consented to participation in publication.
Statistical tests
Statistical tests were performed with GraphPad Prism. The details of the statistical tests and results are reported in Supplementary Table 3. For behavioral tests, values in all groups were assumed to be normally distributed. Welch’s correction was used for t-tests when variances between groups were significantly different by F-test. Geisser–Greenhouse correction was used for one-way ANOVA
DNA Sanger sequencing of the proband’s siblings in family 2
Buccal swab DNA samples in NaOH were retrieved from GeneDx and PCRs were performed using these sample. After PCR, the products amplified from the two unaffected siblings’ DNAs were purified and then sent for Sanger sequencing. The PCR products amplified from the affected sibling’s DNA were added with A-tails using a Taq polymerase and ligated into pGEM-T easy vectors (Promega). The ligation mixture was transformed into E. coli DH5α and colonies were isolated followed by plasmid purification (Nucleospin, Macherey Nagel).
Mouse models
All procedures in mice were approved by the Institutional Animal Care and Use Committee of Baylor College of Medicine. Atxn1 null3, Atxn1lflox9, Emx1-Cre18, Neurod6-Cre23, and ROSAfsTRAP29 have been described previously. To generate the Otp-Cre allele, bacterial artificial chromosomes (BAC) containing Otp (RP24-337G4) was modified using recombineering to replace the Otp coding sequence with Cre38. The modified BAC DNA was isolated and digested with BsiWI to release the transgene for pronuclear injection. Pups derived from the microinjected embryos were screened for the presence of the transgene. Transgenic founders were then mated to WT FVB mice and the offspring were screened for germline transmission. Transgenic mice with germline transmission were then backcrossed to the C57BL/6 background for more than seven generations.
Generation of Cicflox and Cicnull alleles were carried out as follow. ES cells with Cictm1a(KOMP)Wtsi allele were obtained from EUCOMM and were injected into C57BL/6 blastocysts. The Cicflox mice were then generated by breeding to Actin-FLP mice to remove the genetrap cassette, and the Cicnull allele was generated by crossing to CMV-Cre mice (Supplementary Fig. 23). All mice were maintained on a C57BL/6 background. Day of plug was denoted E0.5 for timed-pregnant experiment. Primers for genotyping are reported in Supplementary Table 4. Both males and females were used, and we did not observe any gender difference in the phenotypes.
Antibodies
The following primary antibodies were used for western blot: rabbit polyclonal anti-ATXN12, rabbit polyclonal anti-ATXN1L8, mouse monoclonal anti-VCL (Sigma, V9131), mouse monoclonal anti-GAPDH (Advanced ImmunoChemical Inc., 2-RGM2), and guinea pig polyclonal anti-CIC7. The primary antibodies used in immunofluorescence studies were: Rabbit anti-CUX1 (Santa Cruz, sc-13024), Rat anti-CTIP2 (Abcam, ab18465), Mouse anti-SATB2 (Abcam, ab51502), Rabbit anti-CIC (Generated in house using antigen described in7), Rabbit anti-PAX6 (Covance, PRB-278P), Rabbit anti-TBR2 (Abcam, ab23345), Chicken anti-GFP (Abcam, ab13970), Rabbit anti-CASP3 (Cell Signaling Technology #9661).
Behavioral tests
The open-field test, elevated plus maze test39, low-dose amphetamine treatment40, and fear conditioning41 were performed as previously described. The three-chamber test was performed as previously described39, with the modification that ambient light was maintained at 700–800 lux and white noise was used. The partition test was performed as previously described39, with the modification that test mice were individually housed in standard housing cages separated into two compartments by a clear perforated Plexiglas barrier for one day. The resident-intruder test was performed as previously described39 with the modification that male mice were singly housed for four weeks before testing to establish dominance. Tests were video-recorded and the episodes and time of attacks were analyzed.
Novel object recognition test was performed with the following protocol. During habituation, the test mouse was placed in the habituation arena and allowed to explore for 30 min. Following habituation, the test mouse was then introduced into the test arena where two identical objects (Legos) were placed with equal distance to the walls of the arena. The test mouse was allowed to explore the arena for 5 min (trial 1). After the first trial, the test mouse was returned to the habituation arena and the test arena and objects were cleaned with ethanol. On trials 1 to 3, identical objects were placed in the arena for the test mouse to explore and familiarize. On trial 4, a novel object, which was a Lego with a different color and shape than the familiar objects, was introduced into the test arena to replace one of the familiar objects. The test mouse was then allowed to explore the arena for 5 min. On trial 5, the familiar object replaced the novel object and was re-introduced back into the test arena. The amount of time that the test mouse spent on investigating (sniffing, climbing, and touching) the objects was recorded using the AnyMaze video tracking system. The test mouse was allowed to rest for at least 10 min between each trial.
Sample size in behavioral studies was estimated based on our previous experience with these behavioral tests. All behavioral tests were performed with the investigators blinded to the genotypes or treatment. In the low-dose amphetamine treatments, mice were assigned to either vehicle or amphetamine treatments using a list randomizer.
Hippocampal slice preparation and electrophysiology
Mice were anaesthetized by isoflurane inhalation and decapitated. The brain was isolated and immersed in chilled (2–5°C) cutting solution containing (in mM) 220 sucrose, 2.5 KCl, 0.5 CaCl2, 7 MgCl2, 1.25 NaH2PO4, 25 NaHCO3, 7 D-glucose. Transverse hippocampal slices (400 μm) were prepared with a vibratome (Leica Microsystems Inc., Buffalo Grove, IL). The slices were incubated at 37°C for 30 minutes in artificial cerebrospinal fluid (ACSF) containing (in mM) 125 NaCl, 2.5 KCl, 2 CaCl2, 1 MgCl2, 1.25 NaH2PO4, 25 NaHCO3, and 14.83 D-glucose, and then transferred to room temperature for another 30 minutes before recording.
Extracellular stimuli were administered along the Schaffer collaterals using a bipolar tungsten stimulation electrode (WPI), field excitatory postsynaptic potentials (fEPSPs) were recorded in stratum radiatum. The recording pipettes were filled with 2 M NaCl (1–2 MΩ) and their distance to the stimulating electrode were kept constant (~300 μm). Input/output (I/O) curves were generated using incremental stimulus intensities and were used to assess baseline synaptic transmission. All subsequent stimuli were set to an intensity that evoked a 30–40% of the maximum fEPSP slope. Paired pulse facilitation was assessed via systematic stimuli with different interval (25, 50, 100, 200, and 400 mSec). The paired pulse ratio was obtained by dividing the rising slope of the second fEPSP with the rising slope of the first fEPSP. For LTP experiments, stable baseline fEPSPs were recorded for 20 min, and then LTP was induced by 2 trains of high-frequency stimulation (HFS, 100 Hz for 1 s) with 20 s intertrain interval. The magnitude of potentiation was determined by measuring the rising slope of the fEPSP.
The recordings were made using Multiclamp 700B amplifiers (Molecular Devices, Union City, CA). Data acquisition and analysis were performed using digitizer DigiData 1440A and analysis software pClamp 10 (Molecular Devices). Signals were filtered at 2 kHz and sampled at 10 kHz. All recordings were performed at 30 ± 1°C by using an automatic temperature controller (Warner Instrument, Hamden, CT).
Co-immunoprecipitation (Co-IP)
The cortex or hypothalamus were dissected from an 8-week old wildtype mouse and homogenized in 500 μL of extraction buffer (75 mM NaCl, 5 mM MgCl2, 50 mM Tris pH 8.0, 0.5% Triton X-100, supplemented with fresh protease inhibitors and phosphatase inhibitors). The homogenates were then centrifuged at 13,200 rpm for 15 min at 4 °C. The supernatants were diluted five times using the extraction buffer without any detergent. For each IP, 20 μL of prepared Dynabeads Protein A (Novex, Life Technology) was added to 500 μL of the diluted extracts, together with 1 μL of rabbit anti-CIC antibody or normal rabbit IgG. IP reactions were performed overnight at 4 °C with gentle head-to-tail rotations. The beads were then washed three times with the extraction buffer (with detergent) and proteins bound to the beads were eluted with SDS sample loading buffer. Co-IP samples were analyzed using western blot.
Column fractionation and analysis
Gel-filtration (sizing) chromatography was performed as previously described42 using cortex or hypothalamus from five 8-week old animals. For each column chromatography, 4 mg of total proteins were loaded to the column for analysis.
Histology
Samples were dissected and fixed in 4% paraformaldehyde in PBS or 10% neutral buffered formalin for 24–48 hours. The tissues were then incubated in 70% ethanol for 24 hours, 95% ethanol overnight, 100% ethanol for 4 hours, chloroform overnight, and paraffin for four hours twice. The tissues were then embedded in paraffin and sectioned at 8-μm thickness. Nissl (cresyl violet) staining and luxol fast blue staining were performed using standard protocols.
Immunofluorescence Staining and TUNEL assay
For Emx1-Cre and Neurod6-Cre mutant studies, the animals were anesthetized and transcardialy perfused with 4% paraformaldehyde in PBS. Tissues were collected and fixed in 4% paraformaldehyde in PBS at 4°C overnight. The tissues were cryoprotected in 15% and 30% sucrose in PBS, and then imbedded and frozen in Tissue-Tek® OCT compound. Tissues were sectioned at 25-μm thickness and mounted on Superfrost Plus Microscope Slides and air-dried, followed by rehydration in PBS. For Otp-Cre studies, brain tissues were directly frozen in Tissue-Tek® OCT compound and sectioned at 25-μm thickness and mounted on Superfrost Plus Microscope Slides. Prior to staining, the slides were fixed in 10% neutral buffered formalin for 15 mins, and washed three times with PBS. All slides were blocked in blocking buffer at room temperature for 1 hour, following by incubating with primary antibodies in blocking buffer at 4°C overnight. The slides were washed with 0.1% Triton X-100 in PBS three times, and incubated with secondary antibodies in blocking buffer at room temperature for two hours. The slides were then washed with 0.1% Triton X-100 in PBS two times, counterstained with DAPI, washed with 0.1% Triton X-100 in PBS once and PBS once. Slides were mounted with coverslips with ProLong Gold or ProLong Diamond (Molecular Probes). When needed, antigen retrieval was performed before blocking using Antigen Unmasking Solution (Vector Laboratory, Citric Acid Based) at 95°C for 20–60 minutes and cool for at least 20 minutes at room temperature. The slides were then washed in PBS three times and then blocked and stained as described above.
TUNEL assays were performed using the Click-iT® Plus TUNEL assay kit (with Alexa Fluor 488 Dye, Molecular Probes) with minor modifications: 25-μm thick, air-dried cryo-sections mounted on the slides were used for the assay; in addition to the proteinase K treatment recommended by the manufacturer, an additional 700 μg of proteinase K (at 1 mg/ml) was added to each slide and the slides were incubated for 70 min at room temperature; after the TUNEL assay, the sections were counter-stained with DAPI.
EdU (5-ethynyl-2′-deoxyuridine) labeling
50 mg/kg of EdU (Santa Cruz, sc-284628) was injected intraperitoneally into timed-pregnant female mice. Two hours later, injected mice were euthanized, and embryonic brains were dissected and fixed overnight. The tissues were then cryo-protected and sectioned as described in the Immunofluorescent Staining section. EdU was labeled with Click-iT® EdU Alexa Fluor® Imaging Kit (Thermo) before immunofluorescent staining.
Image acquisition and analysis
Confocal images were acquired with Leica TCS SP8 or ZEISS LSM 880. Epifluorescence images were acquired with Leica DM4000 microscope. Light microscopy images were acquired with Leica DM4000 or Zeiss Axio Scan.Z1 slide scanner. For cell counting, images from parietal cortices were cropped at 200-μm bins and were quantified with Fiji using the Cell Counter plugin43. The thickness of pyramidal layers of CA1 and CA3, as well as the thickness of granule layer of DG were measured using Fiji. All analyses were performed with the experimenter blinded to the genotype.
Dendritic branching analysis
AAV-YFP (AAV8 with CBA-YFP-2A-YFP-2A-YFP) was generated and injected into lateral ventricles as previously described26. To achieve sparse labeling, 4.8×107 viral particles were injected into each ventricle. At 5 weeks of age, the animals were anesthetized and transcardialy perfused with 4% PFA in PBS. Brains were collected and fix in 4% PFA in PBS at 4°C overnight. 100 μm sections were obtained with Leica vibratome. The sections were mounted on Superfrost Plus Microscope Slides and coverslipped with VECTASHIELD (Vector Laboratories). Pyramidal neurons were identified based on their morphology and location. Z-stack images were taken with ZEISS LSM 880. Neuronal structure reconstruction and Scholl analysis was performed with Neurolucida 360 software (MBF Bioscience).
In situ hybridization
RNA in situ hybridization was performed on adult mouse brain sections. The DNA templates for the in situ hybridization probes were amplified from mouse brain cDNA and cloned into pGEMT-Easy vectors. To generate DIG-labeled mRNA probes in vitro transcription was performed using a kit from Roche according to the manufacturer’s instruction. The primers and further information about the probes are listed in Supplementary Table 4. Adult mouse brains were collected and immediately embedded and frozen in Tissue-Tek® OCT compound. Coronal sections were collected from the somatosensory cortex at 25-μm thickness. In situ hybridization was performed as previously described44 and the stained slides were imaged using a Zeiss Axio Scan.Z1 slide scanner.
Quantitative reverse transcription PCR
RNA was extracted with TRIzol and reverse transcribed using M-MLV Reverse Transcriptase (Thermo). Real-time PCR was performed with PerfeCTa SYBR Green FastMix ROX (Quanta Biosciences) or iTaq™ Universal SYBR® Green (BIO-RAD). Primers are listed in Supplementary Table 4. Rps16 was used as the reference gene, and fold changes were calculated using the 2−ΔΔCt method45,46. In short, the mean of the threshold cycle (Ct) of three technical replicates was used as the Ct of a given animal.
ANOVA or t test was performed on the ΔCt values46. The confidence intervals of ΔΔCt values were calculated, and the confidence intervals of 2−ΔΔCt values were plotted in the figures and reported in Supplementary Table 346.
Translating ribosome affinity purification (TRAP) and RNA profiling
TRAP profiling was performed as previously described29,33. Briefly, hypothalamus and adjacent amygdala regions were dissected from 8–10 weeks-old male mice. Tissues were homogenized in 3 mL of homogenization buffer and 60 μg each of 19C8 and 19F7 was used for every immunoprecipitation experiment. The subsequent sample quality check and processing of the purified RNA was performed by the Genomic and RNA Profiling Core at Baylor College of Medicine. Using NuGEN Ovation RNA-Seq System v2, purified double-stranded cDNA was generated. For validation, real-time PCR was performed with iTaq™ Universal SYBR® Green (BIO-RAD). Primers are listed in Supplementary Table 4. Rps16, Hprt, and Gapdh were used as reference genes, and fold changes were calculated using the 2−ΔΔCt method45,46.
RNAseq data analysis
For each sample, about 100 million pairs of 100 bp pair-end reads were generated. Raw reads were trimmed before mapping to reference genome. Trimmed reads were then aligned to the Mus musculus genome (UCSC mm10) using TopHat v2.0.947 with default parameters. The mappability for all 8 samples was about 50%. HTSeq48 was used to obtain read counts. Using the obtained read counts, differential gene analysis was carried out using the DESeq package in the R environment. A gene was identified as significantly changed if falls under the FDR of 5%.
Phenotype association analyses were performed using the WebGestalt (WEB-based GEne SeT AnaLysis) Toolkit49,50.
Supplementary Material
Acknowledgments
We thank the families for their participation in the study and Dr. Hui Zheng for providing Neurod6-Cre (NEX-Cre) mice. The project was support by grants NIH/NICHD R01 HD081216 and HD083809 to R.H.F.; NIH/NHGRI 1UM1HG008898-01 to M.N.B.; NIH/NINDS R37 NS22920 to H.T.O.; NIH/NINDS R01 NS089664 to R.V.S., NIH/NINDS R01 NS027699-26 and R37 NS027699-28 to H.Y.Z. Q.T. was supported by NIH/NINDS F32 NS083091 and M.W.C.R. received support from Canadian Institutes of Health Research Fellowship 201210MFE-290072–173743. H.Y.Z. is an investigator of the Howard Hughes Medical Institute. We thank the RNA In Situ Hybridization Core (supported by a Shared Instrumentation grant 1S10OD016167), and the Microscopy Core, and the Neuropathology Core at Baylor College of Medicine. All three cores are supported by the NIH IDDRC grant U54HD083092 from the Eunice Kennedy Shriver National Institute of Child Health & Human Development. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Eunice Kennedy Shriver National Institute of Child Health & Human Development or the National Institutes of Health. This project was partly supported by the Genomic and RNA Profiling Core at Baylor College of Medicine and the expert assistance of the core director, Dr. Lisa D. White, Ph.D. We would like to thank the Mayo Clinic Center for Individualized Medicine Investigative and Functional Genomics Program for funding and support.
Footnotes
Data Availability
The gene expression data that support the findings of this study are available in the GEO (Gene Expression Omnibus) repository with the accession code GSE83243.
Author Contributions
H.L., Q.T., and H.Y.Z. conceived and designed the experiments. H.T.O. and R.V.S. provided critical input to the project. H.L., Q.T., M.WC.R., W.W., J.K., R.R., S.Y., J.M.P., T.L., and M.C.L. performed the experiments. X.L., Y.L., J.D.F., J.H., and M.C. assisted in generating the mouse models. Y.W. and Z.L. performed RNAseq analysis. M.E.Z.J. cultured fibroblasts for patient 1. Q.T. performed human fibroblast RNA and protein analysis. H.L., Q.T., R.H.F., Y.L., P.A., K.A-Y., L.V.M., D.L., N.J-M., A-L.M-B., J.T., M.A.C., D.B., B.L., E.W.K., N.A., M.N.B., C.P.S., and H.Y.Z. recruited patients, acquired clinical data, or analyzed whole-exome or Sanger sequencing results. H.L., Q.T., and H.Y.Z. drafted the original manuscript, and all authors assisted in editing the manuscript.
Competing Financial Interests
None declared.
References
- 1.Orr HT, et al. Expansion of an unstable trinucleotide CAG repeat in spinocerebellar ataxia type 1. Nat Genet. 1993;4:221–6. doi: 10.1038/ng0793-221. [DOI] [PubMed] [Google Scholar]
- 2.Burright EN, et al. SCA1 transgenic mice: a model for neurodegeneration caused by an expanded CAG trinucleotide repeat. Cell. 1995;82:937–48. doi: 10.1016/0092-8674(95)90273-2. [DOI] [PubMed] [Google Scholar]
- 3.Matilla A, et al. Mice lacking ataxin-1 display learning deficits and decreased hippocampal paired-pulse facilitation. J Neurosci. 1998;18:5508–16. doi: 10.1523/JNEUROSCI.18-14-05508.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Watase K, et al. A long CAG repeat in the mouse Sca1 locus replicates SCA1 features and reveals the impact of protein solubility on selective neurodegeneration. Neuron. 2002;34:905–19. doi: 10.1016/s0896-6273(02)00733-x. [DOI] [PubMed] [Google Scholar]
- 5.Asher M, Johnson A, Zecevic B, Pease D, Cvetanovic M. Ataxin-1 regulates proliferation of hippocampal neural precursors. Neuroscience. 2016 doi: 10.1016/j.neuroscience.2016.02.011. [DOI] [PubMed] [Google Scholar]
- 6.Lin X, Antalffy B, Kang D, Orr HT, Zoghbi HY. Polyglutamine expansion down-regulates specific neuronal genes before pathologic changes in SCA1. Nat Neurosci. 2000;3:157–63. doi: 10.1038/72101. [DOI] [PubMed] [Google Scholar]
- 7.Lam YC, et al. ATAXIN-1 interacts with the repressor Capicua in its native complex to cause SCA1 neuropathology. Cell. 2006;127:1335–47. doi: 10.1016/j.cell.2006.11.038. [DOI] [PubMed] [Google Scholar]
- 8.Bowman AB, et al. Duplication of Atxn1l suppresses SCA1 neuropathology by decreasing incorporation of polyglutamine-expanded ataxin-1 into native complexes. Nat Genet. 2007;39:373–9. doi: 10.1038/ng1977. [DOI] [PubMed] [Google Scholar]
- 9.Lee Y, et al. ATXN1 protein family and CIC regulate extracellular matrix remodeling and lung alveolarization. Dev Cell. 2011;21:746–57. doi: 10.1016/j.devcel.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fryer JD, et al. Exercise and genetic rescue of SCA1 via the transcriptional repressor Capicua. Science. 2011;334:690–3. doi: 10.1126/science.1212673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Celestino-Soper PB, et al. Deletions in chromosome 6p22.3-p24.3, including ATXN1, are associated with developmental delay and autism spectrum disorders. Mol Cytogenet. 2012;5:17. doi: 10.1186/1755-8166-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baroy T, et al. Haploinsufficiency of two histone modifier genes on 6p22.3, ATXN1 and JARID2, is associated with intellectual disability. Orphanet J Rare Dis. 2013;8:3. doi: 10.1186/1750-1172-8-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Di Benedetto D, et al. 6p22.3 deletion: report of a patient with autism, severe intellectual disability and electroencephalographic anomalies. Mol Cytogenet. 2013;6:4. doi: 10.1186/1755-8166-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vissers LE, et al. A de novo paradigm for mental retardation. Nat Genet. 2010;42:1109–12. doi: 10.1038/ng.712. [DOI] [PubMed] [Google Scholar]
- 15.Athanasakis E, et al. Next generation sequencing in nonsyndromic intellectual disability: from a negative molecular karyotype to a possible causative mutation detection. Am J Med Genet A. 2014;164A:170–6. doi: 10.1002/ajmg.a.36274. [DOI] [PubMed] [Google Scholar]
- 16.Giedd JN, Rapoport JL. Structural MRI of pediatric brain development: what have we learned and where are we going? Neuron. 2010;67:728–34. doi: 10.1016/j.neuron.2010.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ecker C, Bookheimer SY, Murphy DG. Neuroimaging in autism spectrum disorder: brain structure and function across the lifespan. Lancet Neurol. 2015;14:1121–34. doi: 10.1016/S1474-4422(15)00050-2. [DOI] [PubMed] [Google Scholar]
- 18.Gorski JA, et al. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J Neurosci. 2002;22:6309–14. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sotnikoca TD, Gainedinov RR. Attention-decifit hyperactivity disorder. In: Pietropaolo S, Sluyter F, Crusio WE, editors. Behavioral Genetics of the Mouse: Volume 2, Genetic Mouse Models of Neurobehavioral Disorders. Vol. 2. Cambridge University Press; 2014. pp. 164–72. [Google Scholar]
- 20.Jonas P, Lisman J. Structure, function, and plasticity of hippocampal dentate gyrus microcircuits. Frontiers in Neural Circuits. 2014;8 doi: 10.3389/fncir.2014.00107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fame RM, MacDonald JL, Macklis JD. Development, specification, and diversity of callosal projection neurons. Trends Neurosci. 2011;34:41–50. doi: 10.1016/j.tins.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hevner RF, et al. Beyond laminar fate: toward a molecular classification of cortical projection/pyramidal neurons. Dev Neurosci. 2003;25:139–51. doi: 10.1159/000072263. [DOI] [PubMed] [Google Scholar]
- 23.Goebbels S, et al. Genetic targeting of principal neurons in neocortex and hippocampus of NEX-Cre mice. Genesis. 2006;44:611–21. doi: 10.1002/dvg.20256. [DOI] [PubMed] [Google Scholar]
- 24.Cubelos B, et al. Cux1 and Cux2 regulate dendritic branching, spine morphology, and synapses of the upper layer neurons of the cortex. Neuron. 2010;66:523–35. doi: 10.1016/j.neuron.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Doan RN, et al. Mutations in Human Accelerated Regions Disrupt Cognition and Social Behavior. Cell. 2016;167:341–354. e12. doi: 10.1016/j.cell.2016.08.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JY, et al. Viral transduction of the neonatal brain delivers controllable genetic mosaicism for visualising and manipulating neuronal circuits in vivo. Eur J Neurosci. 2013;37:1203–20. doi: 10.1111/ejn.12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acampora D, et al. Progressive impairment of developing neuroendocrine cell lineages in the hypothalamus of mice lacking the Orthopedia gene. Genes Dev. 1999;13:2787–800. doi: 10.1101/gad.13.21.2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Diaz C, Morales-Delgado N, Puelles L. Ontogenesis of peptidergic neurons within the genoarchitectonic map of the mouse hypothalamus. Front Neuroanat. 2014;8:162. doi: 10.3389/fnana.2014.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou P, et al. Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proc Natl Acad Sci U S A. 2013;110:15395–400. doi: 10.1073/pnas.1304124110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez-Bolado G, Grinevich V, Puelles L. Editorial: Development of the hypothalamus. Front Neuroanat. 2015;9:83. doi: 10.3389/fnana.2015.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim Y, et al. Mapping social behavior-induced brain activation at cellular resolution in the mouse. Cell Rep. 2015;10:292–305. doi: 10.1016/j.celrep.2014.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kas MJ, Modi ME, Saxe MD, Smith DG. Advancing the discovery of medications for autism spectrum disorder using new technologies to reveal social brain circuitry in rodents. Psychopharmacology (Berl) 2014;231:1147–65. doi: 10.1007/s00213-014-3464-y. [DOI] [PubMed] [Google Scholar]
- 33.Mellen M, Ayata P, Dewell S, Kriaucionis S, Heintz N. MeCP2 binds to 5hmC enriched within active genes and accessible chromatin in the nervous system. Cell. 2012;151:1417–30. doi: 10.1016/j.cell.2012.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–291. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shaw P, et al. Attention-deficit/hyperactivity disorder is characterized by a delay in cortical maturation. Proc Natl Acad Sci U S A. 2007;104:19649–54. doi: 10.1073/pnas.0707741104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shaw P, et al. Longitudinal mapping of cortical thickness and clinical outcome in children and adolescents with attention-deficit/hyperactivity disorder. Arch Gen Psychiatry. 2006;63:540–9. doi: 10.1001/archpsyc.63.5.540. [DOI] [PubMed] [Google Scholar]
- 37.Sobreira N, Schiettecatte F, Valle D, Hamosh A. GeneMatcher: a matching tool for connecting investigators with an interest in the same gene. Hum Mutat. 2015;36:928–30. doi: 10.1002/humu.22844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warming S, Costantino N, Court DL, Jenkins NA, Copeland NG. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 2005;33:e36. doi: 10.1093/nar/gni035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chao HT, et al. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468:263–9. doi: 10.1038/nature09582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han K, et al. SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature. 2013;503:72–7. doi: 10.1038/nature12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Crespo-Barreto J, Fryer JD, Shaw CA, Orr HT, Zoghbi HY. Partial loss of ataxin-1 function contributes to transcriptional dysregulation in spinocerebellar ataxia type 1 pathogenesis. PLoS Genet. 2010;6:e1001021. doi: 10.1371/journal.pgen.1001021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Park J, et al. RAS-MAPK-MSK1 pathway modulates ataxin 1 protein levels and toxicity in SCA1. Nature. 2013;498:325–331. doi: 10.1038/nature12204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yaylaoglu MB, et al. Comprehensive expression atlas of fibroblast growth factors and their receptors generated by a novel robotic in situ hybridization platform. Dev Dyn. 2005;234:371–86. doi: 10.1002/dvdy.20441. [DOI] [PubMed] [Google Scholar]
- 45.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–8. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 46.Yuan JS, Reed A, Chen F, Stewart CN., Jr Statistical analysis of real-time PCR data. BMC Bioinformatics. 2006;7:85. doi: 10.1186/1471-2105-7-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–9. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res. 2013;41:W77–83. doi: 10.1093/nar/gkt439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–8. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.