Abstract
Purpose
We aimed to characterize common genetic variants that influence saturated fatty acid concentrations in East Asians.
Methods
Meta-analysis of genome-wide association studies for circulating SFAs was conducted in two population-based cohorts comprising 3,521 participants of Chinese ancestry.
Results
We identified two novel 14:0-associated loci at LMX1A (LIM homeobox transcription factor 1) and AMPD3 (AMP deaminase 3) (P=5.08×10−9 and P=4.33×10−8, respectively), and a novel 20:0-associated locus at CERS4(ceramide synthase 4) (P=1.73×10−10). We also confirmed the previously reported association of FADS1/2-rs102275 with 18:0 (P=1.115×10−5). In addition, the A alleles of rs11042834 in AMPD3 and rs17159388 in CERS4 also exhibited evidence of associations with high density lipoprotein cholesterol (P=0.0162 and P=0.0161, respectively).
Conclusions
To our knowledge, this is the first GWAS analysis to examine SFA concentrations in East Asian populations. Our findings provide novel evidence that genetic variations of several genes from multiple pathways are associated with SFA concentrations in human body.
Keywords: myristic acid, arachidic acid, genome-wide association study, Chinese, lipids
Introduction
Dietary saturated fatty acids (SFAs) have long been considered independently as major risk factors for coronary heart disease (CHD) [1]. Moreover, plasma or erythrocyte SFAs, as surrogates of their intakes, have also been shown to be associated with risks of CHD [2, 3], type 2 diabetes [4], heart failure [5], ischemic stroke [6]and Alzheimer’s disease [7]. However, individual SFAs seem to have distinctive pathophysiological effects. For example, plasma phospholipid fatty acids 16:0 and 18:0 are associated with increased risk of CHD [3], but no association between 14:0 and cardiovascular diseases is observed [3, 8], and effects of long-chain SFAs, 20:0, 22:0, and 24:0, on type 2 diabetes and CHD remain to be known.
Circulating SFA concentrations are determined by dietary intake, absorption and endogenous synthesis. Therefore, genetic variants that alter uptake, absorption or synthesis of fatty acids may all contribute to the variation in blood SFA concentrations. So far, there is only one genome-wide association study (GWAS) in European ancestry populations identified three novel loci that are associated with circulating concentrations of 16:0 and 18:0 [9], and no studies have investigated the associations between genetic variants and circulating concentration of other SFAs, including 14:0 and long-chain SFAs. Moreover, compared with Europeans, East Asian populations have different dietary pattern and genetic background. A recent genetic adaptation analyses also demonstrated that the effect of fatty acid desaturase (FADS) haplotype on the efficiency of long-chain fatty acids synthesis ability differed between European and African descent, due to a shift in diet polyunsaturated fatty acids intake [10]. Therefore, it is of interest to investigate the transferability of reported genetic associations for de novo lipogenesis fatty acids to other ethnic populations. In the current analysis, we aimed to identify novel genetic variants for individual SFAs, and to examine whether previously identified loci are also associated with circulating concentrations of 16:0 and 18:0 in East Asians from the Nutrition and Health of Aging Population in China (NHAPC) and the Multi-Ethnic Study of Atherosclerosis (MESA), two cohort studies from the Cohorts from Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium.
Subjects and methods
Study cohorts
All data for this study were obtained from two cohort studies in the CHARGE Consortium, the NHAPC and MESA study. The NHAPC study is a population-based cohort study among 3,210 Chinese Hans, who aged 50 to 70 years during recruitments in Beijing and Shanghai. The study design, methods and measurements of this cohort study have been described in detail elsewhere [11]. Briefly, data on demographic variables, health status and physical activity was collected using a standardized questionnaire and anthropometric measurements and overnight fasting blood samples were collected using a standardized protocol. Total cholesterol, LDL-c, HDL-c and triglyceride concentrations were measured enzymatically on an automatic analyzer (Hitachi7080, Japan) with reagents purchased from WakoPure Chemical Industries (Osaka, Japan). The MESA Study is a study of the characteristics of subclinical cardiovascular disease and risk factors that predict progression to clinically overt cardiovascular disease or progression of the subclinical disease [12]. MESA study is a diverse, population-based sample of 6,814 asymptomatic men and women aged 45 to 84 years, with about 12 percent Asians, predominantly of Chinese descent. Written informed consent was obtained from all participants in both studies.
Fatty acid measurements
The total erythrocyte fatty acids were measured in NHAPC samples, while plasma phospholipid fatty acids were measured in the MESA samples. The method for erythrocyte fatty acid measurement in the NHAPC study has been previously described [13]. After being extracted by hexane and iso-propanol, erythrocyte fatty acids were incubated with a mixture of methanol and sulfuric acid for fatty acid methyl esters (FAMEs). FAMEs were then separated and identified by gas chromatography coupled with positive chemical ionization(Agilent 6890 GC-5975B) using methane as reagent gas. Fatty acids were obtained for a subset of 712 Chinese with genotypes available through MESA SHARe. Fatty acids of plasma collected with EDTA and frozen at −70°C, were measured using methods previously described by Cao et al [14]. Lipids are extracted from the plasma using a chloroform/methanol extraction method, and cholesterol esters, triglycerides, phospholipids and free fatty acids are separated by thin layer chromatography. The fatty acid methyl esters are obtained from the phospholipids and are detected by gas chromatography flame ionization. In both studies, relative amount of each fatty acid was calculated as the percentage of total fatty acids. Among the individual SFAs, 14:0, 16:0, 18:0, 20:0, and 22:0 were available in both cohorts and 24:0 was available only in the NHAPC study.
Genotyping and quality control
Details on genotyping and imputation for each cohort are provided in Supplemental Table 1. Briefly, sampleswere genotyped using high-density single nucleotide polymorphism (SNP) marker platforms, Illumina Human660W (Illumina, Inc., San Diego, California) in NHPAC and Affymetrix Genome-Wide Human SNP Array 6.0 in MESA (Santa Clara, California). Samples with call rate< 97% (NHAPC)or ≤ 95% (MESA) were excluded. The samples in the NHAPC study that passed all QC criteria were then used to impute for the ungenotyped or missing SNPs from the HapMap phase II CHB+JPT (release #22- NCBI Build 36) reference panel, and samples in MESA SHARe Asian were imputed from the HapMap Phase I and II CEU+YRI+CHB+JPT (release #22- NCBI Build 36) reference panel using IMPUTE software [15]. We removed all SNPs with MAF <5% (after meta-analysis), HWEP<10−6, or poor imputation quality, defined as the info measure ≤ 0.5 (NHAPC) or an observed divided by expected variance ≤ 0.8 (MESA).
Statistical analysis
Genome-wide association analysis for each individual fatty acid was conducted separately in NHAPC and MESA, and the summary statistics were then combined by inverse-variance weighted meta-analysis using METAL software (www.sph.umich.edu/csg/abecasis.metal). In cohort-specific GWAS, linear regression analysis was applied to examine the association of each genotyped and imputed SNP with individual SFA level under an additive genetic model using ProbABEL(version 0.4.5) [16]. All analyses were adjusted for age, sex, site of recruitment, and first two principal components and used robust standard errors. P values were adjusted for genomic control inflation factor (λGC) [17]. Genomic control inflation factors in NHAPC and MESA were 1.043and 1.017 for 14:0, 1.011and 1.056 for 16:0, 1.013and 1.013 for 18:0, 1.001 and 1.023 for 20:0, and 1.047 and1.026 for 22:0, respectively, while inflation factor was 1.046 for 24:0 in NHAPC, suggesting minimal population stratification for each cohort. We then combined summary statistics of the associations from NHAPC and MESA cohorts using fixed-effect meta-analyses and tested for the heterogeneity between these two studies. P-values for heterogeneity and I2 were estimated. P-values less than 5×10−8 were considered significant. Generalized linear regression was applied to examine associations between SNPs with P≤5×10−8and plasma lipid concentrations in NHAPC study, adjusting for age, sex, recruiting site, and first two principal components under an additive genetic model using R software (version 2.15). All P values were two-sided, and P< 0.05 was considered to be statistically significant.
Results
The characteristics of two populations are shown in Table 1. Mean concentrations of circulating SFAs are 0.38% and 0.23% for 14:0, 21.91% and 25.71% for 16:0, 14.53% and 13.14% for 18:0, 0.32% and 0.25% for 20:0, and 1.29% and 0.62% for 22:0 for NHAPC and MESA samples, respectively. Mean concentration of SFA 24:0 is 4.21% of total fatty acids for NHAPC samples.
Table 1.
CHARGE cohort characteristics
| NHAPC | MESA | |
|---|---|---|
| Country | China | USA |
| Ethnicity | Chinese Hans | Chinese |
| Study design | Population-based | Population-based |
| Sample (n) | 2865 | 656 |
| Females (%) | 56.8% | 51.4% |
| Age (yrs) | 58.6±6.0 | 62.5 ±10.3 |
| Saturated fatty acids (%)a | ||
| 14:0 | 0.38 ±0.35 | 0.23 ±0.07 |
| 16:0 | 21.91 ±2.57 | 25.71 ±1.47 |
| 18:0 | 14.53 ±1.71 | 13.14 ±1.62 |
| 20:0 | 0.32±0.05 | 0.25 ±0.08 |
| 22:0 | 1.29 ±0.28 | 0.61 ±0.08 |
| 24:0 | 4.21 ±1.78 | - |
| Number of significant SNPs (P≤ 5×10−8) | ||
| 14:0 | 11 | 3 |
| 16:0 | 0 | 0 |
| 18:0 | 0 | 0 |
| 20:0 | 1 | 0 |
| 22:0 | 0 | 5 |
| 24:0 | 0 | - |
Data are mean ±SD except where indicated otherwise.
NHAPC Nutritional Health and Aging Population of Chinese, MESA Multi-Ethnic Study of Atherosclerosis
Fatty acids were measured in erythrocyte (NHAPC)and plasma phospholipid (MESA)
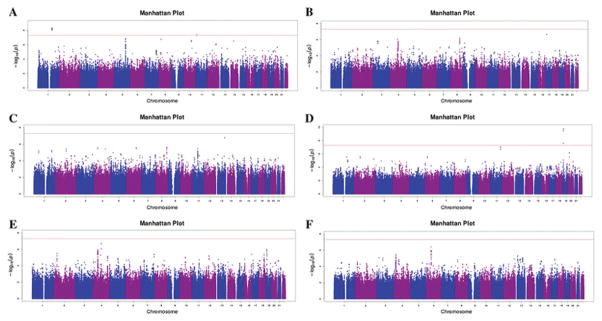
As shown in Fig. 1 and Table 2, multiple SNPs at LMX1A, AMPD3 and CERS4 loci reached genome-wide significance (P values<5×10−8) for associations with SFA (Table 2). Supplemental Tables 2–7 presented the associations of top SNPs (P<5×10−6) with each SFAs in overall individuals and Supplemental Table 8 presented the genome-wide significant SNPs in cohort-specific GWASs.
Fig. 1.
Manhattan plot for meta-analysis of genome-wide associations with saturated fatty acids. The –log10P values from pooled analysis adjusted for age, gender, region and the first two principle components are presented in the figure.
Table 2.
SNPs reaching genome-wide significance in the meta-analysis
| Fatty acid | Nearest Gene | SNP | CHR | Position | Coded allele/non-coded allele | CAF | NHAPC | MESA | Combined | I2 | Pheter | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||
| CAF | beta (SE) | P | CAF | beta (SE) | P | beta (SE) | P | |||||||||
| 14:0 | LMX1A | rs11589386 | 1 | 163270086 | C/T | 0.948 | 0.930 | 0.042 (0.013) | 1.52E-03 | 0.952 | 0.032 (0.006) | 4.78E-07 | 0.034 (0.006) | 5.08E-09 | 0 | 0.52 |
| 14:0 | AMPD3 | rs11042834 | 11 | 10458998 | A/G | 0.914 | 0.912 | 0.027 (0.012) | 2.46E-02 | 0.914 | 0.032 (0.006) | 3.92E-07 | 0.031 (0.006) | 4.33E-08 | 0 | 0.69 |
| 20:0 | CERS4 | rs17159388 | 19 | 8226555 | A/G | 0.191 | 0.191 | −0.011 (0.002) | 1.76E-10 | NA | NA | NA | −0.011 (0.002) | 1.76E-10 | NA | NA |
| Previously reported loci | ||||||||||||||||
| 16:0 | ALG14 | rs2391388 | 1 | 95258413 | C/A | 0.820 | 0.823 | 0.127 (0.086) | 0.140 | 0.816 | 0.137 (0.112) | 0.218 | 0.131 (0.069) | 0.058 | 0 | 0.94 |
| 18:0 | ALG14 | rs6675668 | 1 | 95288225 | T/G | 0.818 | 0.820 | −0.007 (0.058) | 0.905 | 0.809 | −0.113 (0.106) | 0.289 | −0.031 (0.051) | 0.543 | 0 | 0.38 |
| 18:0 | LPGAT1 | rs11119805 | 1 | 209984867 | A/T | 0.145 | 0.137 | −0.071 (0.062) | 0.248 | 0.173 | −0.002 (0.112) | 0.984 | −0.055 (0.054) | 0.310 | 0 | 0.59 |
| 18:0 | FADS1/2 | rs102275 | 11 | 61314379 | C/T | 0.406 | 0.358 | −0.159 (0.048) | 1.01E-03 | 0.596 | −0.316 (0.096) | 9.79E-04 | −0.191 (0.043) | 1.12E-05 | 0.53 | 0.15 |
SNP single nucleotide polymorphism, CHR chromosome, CAF coded allele frequency, NA not available, I2 I square of heterogeneity, P heter heterogeneity of effect sizes between NHAPC and MESA studies
Each copy of the minor T-allele of SNP rs11589386, near LMX1A gene, was associated with 0.034 lower percentage of 14:0 (P=5.08×10−9) (Fig. 2A). AMPD3(at 11p15) is another locus (rs11042834 is the index SNP) that showed genome-wide significance for association with 14:0 (P=4.33×10−8), and each copy of minor allele of this SNP was associated with 0.031 percent decreased in 14:0 (Fig. 2B).
Fig. 2.
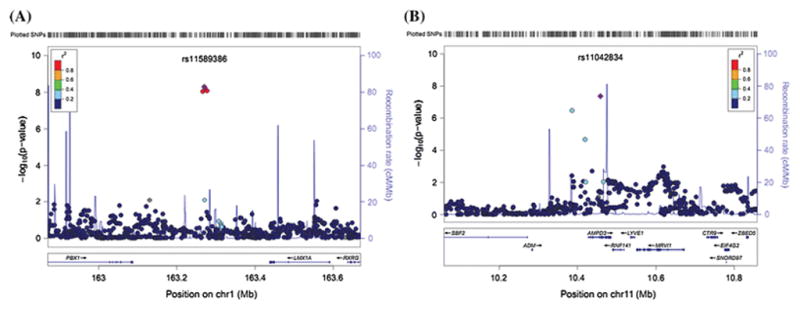
Regional plots of two novel loci for 14:0. Imputed SNPs were estimated by MACH software (see URLs) using LD information from 194 Asians (including 68 CHB, 25 CHS, 84 JPT and 17 MXL) in 1000 Genome 2010.08 release as references. P values were from meta-analysis adjusting for age, gender, region and the first two principle components. The regional plots for the 400kb region centered on index SNPs were generated by using LocusZoom (see URLs). The –log10 P values of SNPs were plotted against their genomic position (NCBI Build 37). The positions of genes were annotated from the UCSC Genome Browser by using GRCh37 assembly. The Index SNPs are purple colored. Linkage disequilibrium (LD) is indicated by color scale in relationship to the Index SNPs, with red for strong LD (r2≥0.8) and blue for lower LD.
Several SNPs at the CERS4 locus were associated with erythrocyte fatty acid 20:0 in the NHAPC study, and rs17159388 is the most significant SNP (P=1.73×10−10, Table 2 and Fig. 3). Moreover, SNP rs651821 in APOA5and rs168622 in SPTLC3 also showed suggestive evidence for association with erythrocyte fatty acid 20:0 (P≤6.48×10−7) (Supplemental Table 5), whereasrs9349666 in ELOVL5 gene that involves in fatty acid metabolism showed suggestive evidence for association with fatty acid 24:0 (P=2.10×10−7)in the NHAPC study (Supplemental Table 7).
Fig. 3.
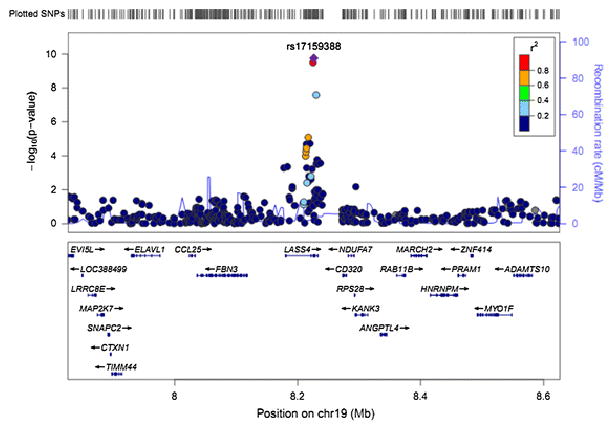
Regional plots of novel locus for 20:0. Imputed SNPs were estimated by MACH software (see URLs) using LD information from 194 Asians (including 68 CHB, 25 CHS, 84 JPT and 17 MXL) in 1000 Genome 2010.08 release as references. P values were from meta-analysis adjusting for age, gender, region and the first two principle components. The regional plots for the 400kb region centered on index SNP was generated by using LocusZoom (see URLs). The –log10 P values of SNPs were plotted against their genomic position (NCBI Build 37). The positions of genes were annotated from the UCSC Genome Browser by using GRCh37 assembly. The Index SNP is purple colored. Linkage disequilibrium (LD) is indicated by color scale in relationship to the Index SNP, with red for strong LD (r2≥0.8) and blue for lower LD.
For loci that have been previously reported to be associated with SFA (16:0 and 18:0), the association between 16:0 and ALG14-rs6675668 (P=0.035), in high linkage disequilibrium with rs2391388 (r2=0.90), was confirmed (number of nominal significant SNPs=10; Supplemental Table 8), and multiple SNPs in FADS1/2 also exhibited significant associations with 18:0 (i.e. P=1.115×10−5 for rs102275; number of nominal significant SNPs=36)(Table 1 and Supplemental Table 9) in this study.
We also examined the three novel genome-wide significant loci for associations with total cholesterol, low density lipoprotein cholesterol(LDL-c), high density lipoprotein cholesterol(HDL-c) and triglycerides in 2,865 participants from NHAPC study (Supplemental Table 10). Minor alleles of AMPD3-rs11042834 and CERS4-rs17159388 were significantly associated with lower HDL-c concentrations (P=0.0162 and 0.0161, respectively), after adjustment of age, gender, region and the first two principle components. Further controlling for 14:0 did not attenuate the effect of minor G-allele of AMPD3-rs11042834 on HDL-c, while adjustment for 20:0 abolished the association between minor A-allele of CERS4-rs17159388and HDL-c.
Discussion
In this study of 3,521 individuals of Chinese descent from two cohorts, three novel variants showed associations with blood SFA concentrations at genome-wide significance. Genetic variants in/or near AMPD3 and LMX1A genes were significantly associated with concentrations of 14:0, while genetic variants in CERS4 gene were significantly associated with concentrations of 20:0. AMPD3-rs11042834 and CERS4-rs17159388 were also nominally associated with plasma HDL-c concentration.
The strongest signal for association with 14:0is rs11589386-C, which is nearLMX1A gene (160kb) encoding a homeodomain and LIM-domain containing protein [18]. LMX1Ais widely expressed in pancreas, skeletal muscle, adipose tissue, kidney, brain, and pituitary [19]. Previous studies have suggested that LIM-homeodomain proteinLmx1 acts as a positive regulator of insulin gene transcription by cooperating with the basic helix-loop-helix (bHLH) protein E47/Pan-1 [20], and existing evidence also shown positive correlation between insulin sensitivity and 14:0 in cross-sectional studies [21, 22]. However, the mechanism underlying the association between LMX1-rs11589386 and 14:0 remains unknown.
Another genetic variant associated with 14:0 is rs11042834 in AMPD3 gene, which encodes erythrocyte adenosine monophosphate deaminase. The same variant is also associated with HDL-c, independent of erythrocyte 14:0 concentration. Consistent with our findings, a prior GWAS observed inverse association of rs2923084-A (not in linkage disequilibrium with rs11042834) nearAMPD3with HDL-c in populations of European origin (beta:−0.41mg/dL; equals to −0.0106 mmol/L) [23]. The protein AMPD3 is an erythrocyte-specific enzyme that catalyzes the hydrolytic deamination of adenosine monophosphate to inosine monophosphate, in the adenylate catabolic pathway [24], and is widely expressed in tissues, including those of brain. Lanaspa et al. reported that the activation of AMP deaminase led to increased production of uric acid and generation of mitochondrial oxidative stress, which further stimulated de novo lipogenesis and activated ATP-citrate lyase and long chain saturated fatty acids syntheses [25]. Further studies are needed to investigate whether genetic variation of AMP gene influence de novo synthesis of 14:0.
We found variants in CERS4 were associated with 20:0 concentrations at genome-wide significance concentration, with the most significant SNP being rs17159388. The saturated fatty acid 20:0 is an important component of sphingolipids. Our observation is consistent with prior GWAS in European participants which showed that several noncoding genetic variants in CERS4, although having non-linkage disequilibrium with rs17159388 (r2<0.05), were associated with circulating plasma concentrations of sphingomyelins species 18:0, 18:1, 20:0, 20:1, ceramides 20:0, and ratios of sphingolipids [26, 27]. More recently, genetic variants in CERS4 associated with 20:0 were also reported in European populations [28]. The most significant SNP rs2100944 was not in LD with SNP rs17159388 identified in the current GWAS (r2=0.012) and the P value of rs2100944 in Chinese populations was 0.040. CERS4is expressed in most tissues including those of adipose and liver and involved in sphingolipid synthesis, especially longer chain ceramides [29]. The 20:0-increasing allele ofCERS4-rs17159388 was also associated with higher HDL-c. But the association was abolished after further adjustment for 20:0, suggesting that the effect of CERS4 on HDL-c is likely to be mediated through an increased concentration of 20:0.
In cohort-specific GWAS, nine loci reached genome-wide significance: six (FECHP, FAM110B, A1CF, JAM3, RASSF8 and MACROD2) for 14:0 and one (CERS4) for 20:0 in the NHAPC cohort, and one (LOC100128956) for 14:0 and one (ST8SIA5) for 22:0 in the MESA cohort (Supplemental Table 8). To avoid false positive results from single GWAS, we conducted meta-analyses of GWAS study with larger sample size and only reported genome-wide significant SNPs in meta-analyses. SNPs at CERS4 remained significant after meta-analysis and was presented in Table 2. Other SNPs showed suggestive significance after meta-analyses. Furthermore, we applied genomic control corrections in each study before meta-analysis to minimize potential confounding by population stratification[17].
The current study highlights the strength of pooling GWAS data from different cohorts for identification of additional novel loci associated with SFA levels. Potential limitations should also be considered. Environmental factors, including dietary intake and lifestyles, may exert confounding effects on the SNP-SFA associations. However, we did not observe any heterogeneity between these two studied cohorts for the significant SNPs identified in the meta-analyses. Future research focusing on the interactions between genetic variants and environmental factors on SFA levels are needed.
In conclusion, we identified three novel loci that were genome-wide significantly associated with saturated fatty acids, mainly with 14:0 and 20:0. These loci also affected concentrations of HDL-c and LDL-c. Our findings expand our knowledge of genes involved in the determination of saturated fatty acids concentrations and provide new insights for future research.
Supplementary Material
Acknowledgments
The authors thank the participants of the MESA study, the Coordinating Center, MESA investigators, and study staff for their valuable contributions. A full list of participating MESA investigators and institutions can be found at http://www.mesa-nhlbi.org. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We are grateful to all participants of the NHAPC, and also thank our colleagues at the laboratory and local CDC staffs of Beijing and Shanghai for their assistance with data collection. This study is supported by the National High Technology Research and Development Program (863 Program 2009AA022704), the National Basic Research Program of China (973 Program 2012CB524900), the National Natural Science Foundation of China (30930081, 81170734 and 81021002), and the Chinese Academy of Sciences (KSCX2-EW-R-10 and SIBS2008006). MESA and the MESA SHARe project are conducted and supported by the National Heart, Lung and Blood Institute (NHLBI) in collaboration with MESA investigators. Support for MESA is provided by contracts N01-HC-95159, N01-HC-95160, N01-HC-95161, N01-HC-95162, N01-HC-95163, N01-HC-95164, N01-HC-95165, N01-HC-95166, N01-HC-95167, N01-HC-95168, N01-HC-95169, UL1-TR-001079, UL1-TR-000040, and DK063491. Funding for MESA SHARe genotyping wa provided by NHLBI Contract N02-HL-64278. MESA SHARe genotyping was performed as Affimetrix (Santa Clara, California, USA) and the Broad Institute of Harvard and MIT (Boston, Massachusetts, USA) using the Affimetrix Genome-Wide Human SNP Array 6.0.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Ethical standards
Both NHAPC and MESA cohorts have been approved by ethics committees and have been performed in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. Written informed consent was obtained from all participants in both studies.
References
- 1.Menotti A, Puddu PE. Coronary heart disease differences across Europe: a contribution from the Seven Countries Study. J Cardiovasc Med. 2013;14:767–72. doi: 10.2459/JCM.0b013e3283628dff. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, Folsom AR, Eckfeldt JH. Plasma fatty acid composition and incidence of coronary heart disease in middle aged adults: the Atherosclerosis Risk in Communities (ARIC) Study. Nutr Metab Cardiovasc Dis: NMCD. 2003;13:256–66. doi: 10.1016/S0939-4753(03)80029-7. [DOI] [PubMed] [Google Scholar]
- 3.Khaw KT, Friesen MD, Riboli E, Luben R, Wareham N. Plasma phospholipid fatty acid concentration and incident coronary heart disease in men and women: the EPIC-Norfolk prospective study. PLoS Med. 2012;9:e1001255. doi: 10.1371/journal.pmed.1001255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kroger J, Zietemann V, Enzenbach C, Weikert C, Jansen EH, Doring F, Joost HG, Boeing H, Schulze MB. Erythrocyte membrane phospholipid fatty acids, desaturase activity, and dietary fatty acids in relation to risk of type 2 diabetes in the European Prospective Investigation into Cancer and Nutrition (EPIC)-Potsdam Study. Am J Clin Nutr. 2011;93:127–42. doi: 10.3945/ajcn.110.005447. [DOI] [PubMed] [Google Scholar]
- 5.Yamagishi K, Nettleton JA, Folsom AR Investigators AS. Plasma fatty acid composition and incident heart failure in middle-aged adults: the Atherosclerosis Risk in Communities (ARIC) Study. Am Heart J. 2008;156:965–74. doi: 10.1016/j.ahj.2008.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yaemsiri S, Sen S, Tinker LF, Robinson WR, Evans RW, Rosamond W, Wasserthiel-Smoller S, He K. Serum fatty acids and incidence of ischemic stroke among postmenopausal women. Stroke. 2013;44:2710–7. doi: 10.1161/STROKEAHA.111.000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang DC, Sun CH, Liu LY, Sun XH, Jin XW, Song WL, Liu XQ, Wan XL. Serum fatty acid profiles using GC-MS and multivariate statistical analysis: potential biomarkers of Alzheimer’s disease. Neurobiol Aging. 2012;33:1057–66. doi: 10.1016/j.neurobiolaging.2010.09.013. [DOI] [PubMed] [Google Scholar]
- 8.de Oliveira Otto MC, Nettleton JA, Lemaitre RN, Steffen LM, Kromhout D, Rich SS, Tsai MY, Jacobs DR, Mozaffarian D. Biomarkers of dairy fatty acids and risk of cardiovascular disease in the multi-ethnic study of atherosclerosis. J Am Heart Assoc. 2013;2:e000092. doi: 10.1161/JAHA.113.000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu JH, Lemaitre RN, Manichaikul A, Guan W, Tanaka T, Foy M, Kabagambe EK, Djousse L, Siscovick D, Fretts AM, et al. Genome-wide association study identifies novel loci associated with concentrations of four plasma phospholipid fatty acids in the de novo lipogenesis pathway: results from the Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) consortium. Circ Cardiovasc Genet. 2013;6:171–83. doi: 10.1161/CIRCGENETICS.112.964619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ameur A, Enroth S, Johansson A, Zaboli G, Igl W, Johansson AC, Rivas MA, Daly MJ, Schmitz G, Hicks AA, et al. Genetic adaptation of fatty-acid metabolism: a human-specific haplotype increasing the biosynthesis of long-chain omega-3 and omega-6 fatty acids. Am JHumGenet. 2012;90:809–20. doi: 10.1016/j.ajhg.2012.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ye X, Yu Z, Li H, Franco OH, Liu Y, Lin X. Distributions of C-reactive protein and its association with metabolic syndrome in middle-aged and older Chinese people. J Am Coll Cardiol. 2007;49:1798–805. doi: 10.1016/j.jacc.2007.01.065. [DOI] [PubMed] [Google Scholar]
- 12.Bild DE, Bluemke DA, Burke GL, Detrano R, Diez Roux AV, Folsom AR, Greenland P, Jacob DR, Jr, Kronmal R, Liu K, et al. Multi-ethnic study of atherosclerosis: objectives and design. Am J Epidemiol. 2002;156:871–81. doi: 10.1093/aje/kwf113. [DOI] [PubMed] [Google Scholar]
- 13.Zong G, Ye X, Sun L, Li H, Yu Z, Hu FB, Sun Q, Lin X. Associations of erythrocyte palmitoleic acid with adipokines, inflammatory markers, and the metabolic syndrome in middle-aged and older Chinese. Am J Clin Nutr. 2012;96:970–6. doi: 10.3945/ajcn.112.040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao J, Schwichtenberg KA, Hanson NQ, Tsai MY. Incorporation and clearance of omega-3 fatty acids in erythrocyte membranes and plasma phospholipids. Clin Chem. 2006;52:2265–72. doi: 10.1373/clinchem.2006.072322. [DOI] [PubMed] [Google Scholar]
- 15.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat Genet. 2007;39:906–13. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 16.Aulchenko YS, Struchalin MV, van Duijn CM. ProbABEL package for genome-wide association analysis of imputed data. BMC Bioinformatics. 2010;11:134. doi: 10.1186/1471-2105-11-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Devlin B, Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 18.German MS, Wang J, Fernald AA, Espinosa R, 3rd, Le Beau MM, Bell GI. Localization of the genes encoding two transcription factors, LMX1 and CDX3, regulating insulin gene expression to human chromosomes 1 and 13. Genomics. 1994;24:403–4. doi: 10.1006/geno.1994.1639. [DOI] [PubMed] [Google Scholar]
- 19.Thameem F, Wolford JK, Wang J, German MS, Bogardus C, Prochazka M. Cloning, expression and genomic structure of human LMX1A, and variant screening in Pima Indians. Gene. 2002;290:217–25. doi: 10.1016/S0378-1119(02)00582-6. [DOI] [PubMed] [Google Scholar]
- 20.Johnson JD, Zhang W, Rudnick A, Rutter WJ, German MS. Transcriptional synergy between LIM-homeodomain proteins and basic helix-loop-helix proteins: the LIM2 domain determines specificity. Mol Cell Biol. 1997;17:3488–96. doi: 10.1128/mcb.17.7.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roberts R, Hodson L, Dennis AL, Neville MJ, Humphreys SM, Harnden KE, Micklem KJ, Frayn KN. Markers of de novo lipogenesis in adipose tissue: associations with small adipocytes and insulin sensitivity in humans. Diabetologia. 2009;52:882–90. doi: 10.1007/s00125-009-1300-4. [DOI] [PubMed] [Google Scholar]
- 22.Ebbesson SO, Tejero ME, Lopez-Alvarenga JC, Harris WS, Ebbesson LO, Devereux RB, MacCluer JW, Wenger C, Laston S, Fabsitz RR, et al. Individual saturated fatty acids are associated with different components of insulin resistance and glucose metabolism: the GOCADAN study. Int J Circumpolar Health. 2010;69:344–51. doi: 10.3402/ijch.v69i4.17669. 1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–13. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sims B, Mahnke-Zizelman DK, Profit AA, Prestwich GD, Sabina RL, Theibert AB. Regulation of AMP deaminase by phosphoinositides. J Biol Chem. 1999;274:25701–7. doi: 10.1074/jbc.274.36.25701. [DOI] [PubMed] [Google Scholar]
- 25.Lanaspa MA, Sanchez-Lozada LG, Choi YJ, Cicerchi C, Kanbay M, Roncal-Jimenez CA, Ishimoto T, Li N, Marek G, Duranay M, et al. Uric acid induces hepatic steatosis by generation of mitochondrial oxidative stress: potential role in fructose-dependent and -independent fatty liver. J Biol Chem. 2012;287:40732–44. doi: 10.1074/jbc.M112.399899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hicks AA, Pramstaller PP, Johansson A, Vitart V, Rudan I, Ugocsai P, Aulchenko Y, Franklin CS, Liebisch G, Erdmann J, et al. Genetic determinants of circulating sphingolipid concentrations in European populations. PLoS Genet. 2009;5:e1000672. doi: 10.1371/journal.pgen.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demirkan A, van Duijn CM, Ugocsai P, Isaacs A, Pramstaller PP, Liebisch G, Wilson JF, Johansson A, Rudan I, Aulchenko YS, et al. Genome-wide association study identifies novel loci associated with circulating phospho- and sphingolipid concentrations. PLoS Genet. 2012;8:e1002490. doi: 10.1371/journal.pgen.1002490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lemaitre RN, King IB, Kabagambe EK, Wu JH, McKnight B, Manichaikul A, Guan W, Sun Q, Chasman DI, Foy M, et al. Genetic loci associated with circulating levels of very long-chain saturated fatty acids. JLipid Res. 2015;56:176–84. doi: 10.1194/jlr.M052456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizutani Y, Kihara A, Igarashi Y. Mammalian Lass6 and its related family members regulate synthesis of specific ceramides. Biochem J. 2005;390:263–71. doi: 10.1042/BJ20050291. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.