Abstract
Gene-disruptive mutations contribute to the biology of neurodevelopmental disorders (NDDs), but most pathogenic genes are not known. We sequenced 208 candidate genes from >11,730 patients and >2,867 controls. We report 91 genes with an excess of de novo mutations or private disruptive mutations in 5.7% of patients, including 38 novel NDD genes. Drosophila functional assays of a subset bolster their involvement in NDDs. We identify 25 genes that show a bias for autism versus intellectual disability and highlight a network associated with high-functioning autism (FSIQ>100). Clinical follow-up for NAA15, KMT5B, and ASH1L reveals novel syndromic and non-syndromic forms of disease.
Neurodevelopmental disorders (NDDs) are a heterogeneous collection of psychiatric and clinical diagnoses that encompass autism spectrum disorders (ASD), intellectual disability/developmental delay (ID/DD), attention-deficit/hyperactivity, motor and tic disorders, and language communication disorders1. Although each diagnosis is distinctive based on the Diagnostic and Statistical Manual of Mental Disorders, 5th edition (DSM-5)1, NDDs often co-occur. Twin studies have shown that NDDs have a heritable component2. In addition to phenotypic overlaps, copy number variant (CNV) studies have shown that risk genotypes overlap between NDDs3. While these data strongly suggest that common genetic etiologies underlie a subset of broadly defined NDDs, there has been criticism that gene discovery efforts have failed to distinguish ID/DD genes from those contributing to ASD without ID4.
Large numbers of potentially pathogenic mutations have been identified based on exome sequencing of NDD cohorts, but in most cases only a single occurrence of a DN mutation in a particular gene has been discovered. Significantly larger numbers of cases and controls are required to prove the statistical significance of individual genes. Leveraging samples and data from multiple comorbid conditions, in principle, can increase our sensitivity to identify risk genes. Phenotypic follow-up of cases broadly drawn from NDDs has previously allowed us to explore specific clinical phenotypes in a genotype-first manner5, such as in the case of genes like CHD86, DYRK1A7, and ADNP8.
Using single-molecule molecular inversion probes (smMIPs)9,10, we sequenced the coding and splicing portions of 208 potential NDD risk genes in over 11,730 ASD, ID, and DD cases. smMIPs provide a highly sensitive, specific, and inexpensive approach to target sequence the protein-coding portions of a moderate number of candidate genes in a large number of patients. Samples were collected as part of an international consortium termed the ASID (Autism Spectrum/Intellectual Disability) network that involved 15 centers across seven countries and four continents. The collection has the advantage over many others in that subsequent phenotypic follow-up is possible for a large fraction of the patients.
RESULTS
Mutation discovery
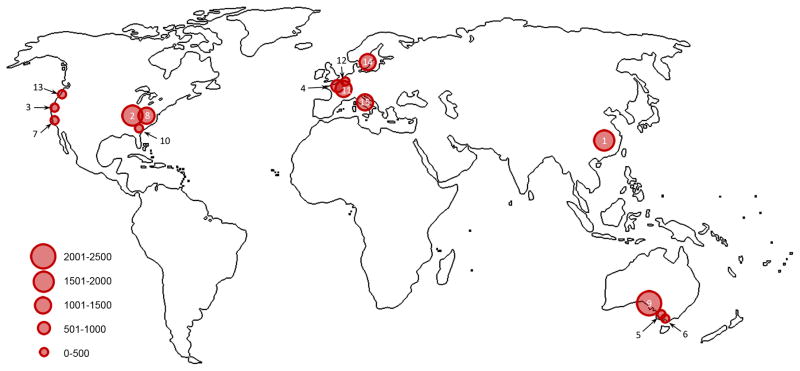
We selected 208 candidate NDD (ASD, ID and DD) disease-risk genes based on published sequencing studies11–17 (Supplementary Tables 1–3) using denovo-db (http://denovo-db.gs.washington.edu)18. Genes were selected and ranked based on the number of published DN recurrences, overlap with a CNV morbidity map19, pathway connectivity20, and absence of DN variants in 1,909 published unaffected sibling control exomes12,13. We designed 12,016 smMIPs distributed across four smMIP pools to cover all annotated RefSeq coding exons as well as five base pairs of flanking intronic sequence (Supplementary Tables 4–7; Methods). We targeted these genes for sequencing in 15 large cohorts of patients (some including unaffected siblings) with a primary ascertainment diagnosis of ASD, ID, or DD where exome sequencing had not previously been performed (Supplementary Tables 8 and 9). The set includes 6,342 patients with a primary diagnosis of ASD and 7,065 patients diagnosed with ID/DD, representing a large international collaboration between research and clinical investigators from the United States, Belgium, The Netherlands, Sweden, Italy, China, and Australia (Fig. 1).
Figure 1. ASID patient network.
13,475 probands with a primary diagnosis of ASD, ID, or DD collected from 15 international groups were screened using smMIPs. Circle size corresponds to the number of samples screened for each cohort. Cohort numbers (1–15) correspond to Supplementary Table 8.
After quality control (QC; Methods; Supplementary Table 10; Supplementary Fig. 1–4), we identified 61,315 QC-passing variants, excluding common dbSNP variants (http://www.ncbi.nlm.nih.gov/SNP/; Methods; Supplementary Table 10). 2,185 were private (i.e., found in only one family in the study; Supplementary Tables 10 and 11) and potentially deleterious (e.g., nonsense, stop-gain, start-loss, frameshift, or disruptive splicing mutations) or missense events with a combined annotation dependent depletion (CADD) score > 30 (MIS30). The number of private, high-impact events identified in probands was significantly greater than in unaffected siblings in the study (false discovery rate (FDR) corrected p = 1.44x10−9; Fisher’s exact test; Supplementary Fig. 5a,b), as expected9,11. This signal was driven primarily by LGD (corrected p = 9.20x10−15) and not MIS30 (corrected p = 0.83) events. We validated 1,125 variants, including all private LGD events as well as 25% of the private MIS30 events by Sanger sequencing (validation rate > 97%; Supplementary Table 11).
Genes with an excess of severe de novo mutation
We assessed inheritance for 286 of the private variants, 35% of which were determined to be sporadic mutations (Supplementary Tables 12 and 13; Supplementary Fig. 5c). The set represents 91 private DN mutations—82 LGD and 9 MIS30—among cases and 9 DN mutations—3 LGD and 6 MIS30—among unaffected siblings and includes 35 recently reported events11,12,21 (Supplementary Table 12; Supplementary Fig. 5d). Allowing for an allele count (AC) ≤3, we identify an additional 32 DN LGD and 15 DN MIS30 events in probands, for a total of 138 DN proband events (114 LGD and 24 MIS30; Supplementary Table 12). Using a probabilistic model derived from human–chimpanzee divergence and an expected rate of 1.5 DN mutations per exome9,22, we calculate the overall probability of detecting 114 or more DN LGD and 24 DN MIS30 variants in our panel of 208 genes, as p = 1.6x10−22 (binomial test) with an odds ratio (OR) of 2.62 (95% CI 2.2–3.09). Combining these results (Supplementary Table 12) with published exome datasets (Supplementary Tables 1 and 2), we identify a total of 393 DN LGD and 98 DN MIS30 events in 208 screened genes, increasing the significance (p = 1.28x10−218; OR = 6.46; [95% CI 5.89–7.06]). Excluding known high-risk NDD genes (Supplementary Table 3), we recalculated the probability of identifying 136 or more DN LGD and 13 DN MIS30 variants among the 84 unknown genes (Supplementary Table 3) where at least one DN LGD mutation has been identified. The frequency of DN mutations is significantly enriched in probands (p = 1.32x10−55, OR = 5.12 [95% CI 4.33–6.01]) suggesting that many of these remaining genes contribute to NDD pathology.
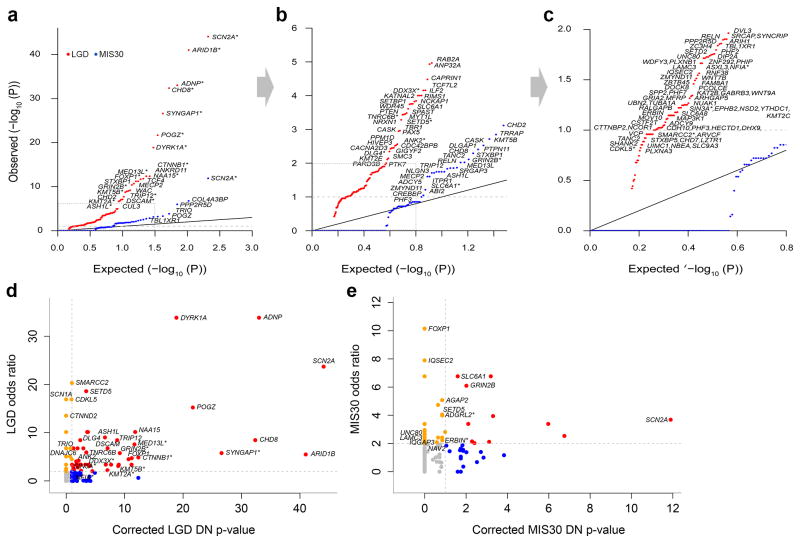
Combining both smMIPs and exome sequence data, we identify 68 genes that reach DN significance for LGD mutations and 23 genes for MIS30 mutations at the level of the individual gene (q < 0.1 by binomial test and more than one LGD or MIS30 event in probands; Table 1; Fig. 2a–c; Supplementary Fig. 6; Supplementary Table 14). Thirteen genes were significant for both DN LGD and MIS30 genes; thus, 78 unique genes show an excess of DN mutations in patients (Table 1). Ten (13%) of these genes are unique to the MIS30 category for probands: TANC2, TRIO, COL4A3BP, TBL1XR1, PPP2R5D, DLGAP1, SRGAP3, PTPN11, ADCY5, and ITPR1 (Table 1; Supplementary Table 15). Thirty-nine of the DN LGD and seven of the DN MIS30 significant genes had been previously linked to NDDs in the literature (Supplementary Table 15; Table 1). Of the 78 DN significant genes, 32 have not been previously described as associated with NDD phenotypes in the literature. The most significant of these genes are TRIP12, KMT5B, and ASH1L (Fig. 3), which were significant for both DN LGD and MIS30 mutations, and NAA15 and DSCAM11 for DN LGD mutations only (corrected p < 1x10−6). The most frequently DN mutated genes in this study were SCN2A, ADNP, CHD8, DYRK1A, and POGZ (Supplementary Table 13). DN mutations in NAA15 were also seen as frequently as DYRK1A and POGZ (Supplementary Table 13). No genes reach DN LGD significance in unaffected sibling controls; although one gene, TRRAP, reaches DN MIS30 significance among the controls. While it is possible that DN mutation of this gene is protective, it is more likely that TRRAP represents a false positive possibly due to an elevated mutation rate compared to expectations based on our statistical model.
Table 1.
Genes that reach de novo (DN) significance.
| smMIP screening | Published exomes | Total | FDR-corrected DN p-valueα | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||
| Gene | DN LGD | DN MIS30 | Probands screened | DN LGD | DN MIS30 | Probands screened | DN LGD | DN MIS30 | LGD | MIS30 | Study |
| SCN2A | 10 | 1 | 13407 | 11 | 6 | 5237 | 21 | 7 | 8.45E-45 | 1.27E-12 | GOLD |
| CHD8 | 5 | 0 | 13407 | 12 | 2 | 6158 | 17 | 2 | 4.26E-33 | 9.48E-03 | GOLD |
| POGZ | 4 | 1 | 13407 | 5 | 1 | 5237 | 9 | 2 | 2.06E-22 | 4.95E-04 | GOLD |
| MED13L | 2 | 0 | 13407 | 6 | 2 | 6158 | 8 | 2 | 1.98E-12 | 1.40E-02 | GOLD |
| STXBP1 | 1 | 0 | 13407 | 5 | 2 | 6158 | 6 | 2 | 2.03E-11 | 7.73E-03 | GOLD |
| GRIN2B | 2 | 1 | 13407 | 4 | 1 | 6158 | 6 | 2 | 6.39E-10 | 9.56E-03 | GOLD |
| KMT5B | 2 | 1 | 12192 | 3 | 1 | 5237 | 5 | 2 | 1.21E-09 | 1.93E-03 | ASD4 |
| TRIP12 | 2 | 0 | 13407 | 4 | 2 | 6158 | 6 | 2 | 1.84E-09 | 1.80E-02 | GOLD |
| CHD2 | 0 | 3 | 13407 | 7 | 1 | 6158 | 7 | 4 | 1.98E-08 | 7.66E-04 | GOLD |
| ASH1L | 2 | 2 | 13407 | 3 | 0 | 5237 | 5 | 2 | 2.26E-07 | 1.80E-02 | GOLD |
| SLC6A1 | 2 | 0 | 13407 | 1 | 2 | 6158 | 3 | 2 | 1.75E-04 | 2.49E-02 | GOLD |
| CASK | 0 | 0 | 12192 | 2 | 2 | 5237 | 2 | 2 | 1.09E-03 | 2.98E-03 | ASD4 |
| RELN | 2 | 1 | 13407 | 1 | 2 | 5237 | 3 | 3 | 1.27E-02 | 1.40E-02 | GOLD |
| ARID1B | 6 | 0 | 13407 | 16 | 0 | 5237 | 22 | 0 | 9.84E-42 | 1.00 | GOLD |
| ADNP | 7 | 0 | 13407 | 9 | 0 | 6158 | 16 | 0 | 9.57E-34 | 1.00 | GOLD |
| SYNGAP1 | 1 | 0 | 13407 | 16 | 1 | 6158 | 17 | 1 | 2.52E-27 | 0.19 | GOLD |
| DYRK1A | 2 | 0 | 13407 | 8 | 0 | 6158 | 10 | 0 | 1.32E-19 | 1.00 | GOLD |
| CTNNB1 | 0 | 0 | 13407 | 7 | 0 | 6158 | 7 | 0 | 4.34E-13 | 1.00 | GOLD |
| ANKRD11 | 0 | 0 | 12192 | 10 | 0 | 5237 | 10 | 0 | 4.34E-13 | 1.00 | ASD5 |
| NAA15 | 4 | 0 | 12192 | 2 | 0 | 5237 | 6 | 0 | 1.52E-12 | 1.00 | ASD4 |
| FOXP1 | 2 | 0 | 13407 | 4 | 0 | 5237 | 6 | 0 | 5.90E-12 | 1.00 | GOLD |
| TCF4 | 4 | 0 | 12192 | 3 | 1 | 5237 | 7 | 1 | 6.26E-12 | 0.22 | ASD4 |
| MECP2 | 2 | 0 | 11731 | 3 | 1 | 5237 | 5 | 1 | 4.12E-11 | 1.94E-02 | ASD6 |
| WAC | 1 | 0 | 13407 | 4 | 0 | 6158 | 5 | 0 | 9.63E-10 | 1.00 | GOLD |
| DSCAM | 2 | 0 | 13407 | 4 | 0 | 6158 | 6 | 0 | 7.89E-08 | 1.00 | GOLD |
| KMT2A | 0 | 0 | 11731 | 6 | 0 | 5237 | 6 | 0 | 8.57E-08 | 1.00 | ASD6 |
| CUL3 | 2 | 0 | 13407 | 2 | 0 | 5237 | 4 | 0 | 1.63E-07 | 1.00 | GOLD |
| TCF7L2 | 0 | 0 | 13407 | 3 | 0 | 6158 | 3 | 0 | 6.97E-05 | 1.00 | GOLD |
| ILF2 | 0 | 0 | 11731 | 2 | 0 | 5237 | 2 | 0 | 7.01E-05 | 1.00 | ASD6 |
| DDX3X | 0 | 0 | 13407 | 3 | 1 | 5237 | 3 | 1 | 9.95E-05 | 0.15 | GOLD |
| RIMS1 | 1 | 0 | 12192 | 2 | 0 | 5237 | 3 | 0 | 9.95E-05 | 1.00 | ASD4 |
| KATNAL2 | 1 | 1 | 13407 | 2 | 0 | 5237 | 3 | 1 | 1.00E-04 | 0.19 | GOLD |
| NCKAP1 | 1 | 0 | 12192 | 2 | 0 | 6158 | 3 | 0 | 1.43E-04 | 1.00 | ASD5 |
| SETBP1 | 0 | 0 | 13407 | 3 | 0 | 5237 | 3 | 0 | 1.50E-04 | 1.00 | GOLD |
| WDR45 | 0 | 0 | 11731 | 2 | 0 | 5237 | 2 | 0 | 1.75E-04 | 1.00 | ASD6 |
| SPAST | 0 | 0 | 12192 | 2 | 0 | 5237 | 2 | 0 | 1.80E-04 | 1.00 | ASD4 |
| PTEN | 0 | 0 | 13407 | 2 | 0 | 6158 | 2 | 0 | 2.46E-04 | 1.00 | GOLD |
| MYT1L | 1 | 0 | 11731 | 2 | 0 | 6158 | 3 | 0 | 3.29E-04 | 1.00 | ASD6 |
| TNRC6B | 1 | 0 | 13407 | 2 | 0 | 5237 | 3 | 0 | 3.49E-04 | 1.00 | GOLD |
| SETD5 | 0 | 1 | 13407 | 3 | 0 | 6158 | 3 | 1 | 3.61E-04 | 0.14 | GOLD |
| NRXN1 | 0 | 0 | 11731 | 3 | 0 | 6158 | 3 | 0 | 3.69E-04 | 1.00 | ASD6 |
| TBR1 | 0 | 0 | 13407 | 2 | 0 | 6158 | 2 | 0 | 5.21E-04 | 1.00 | GOLD |
| PAX5 | 0 | 0 | 13407 | 2 | 0 | 6158 | 2 | 0 | 1.13E-03 | 1.00 | GOLD |
| PPM1D | 0 | 0 | 13407 | 2 | 0 | 6158 | 2 | 0 | 2.73E-03 | 1.00 | GOLD |
| ANK2 | 0 | 1 | 12192 | 4 | 1 | 5237 | 4 | 2 | 2.93E-03 | 0.19 | ASD5 |
| HIVEP3 | 2 | 0 | 13407 | 1 | 0 | 5237 | 3 | 0 | 3.07E-03 | 1.00 | GOLD |
| CDC42BPB | 0 | 0 | 12192 | 2 | 0 | 5237 | 2 | 0 | 3.51E-03 | 1.00 | ASD4 |
| CACNA2D3 | 0 | 0 | 11731 | 2 | 0 | 5237 | 2 | 0 | 3.54E-03 | 1.00 | ASD6 |
| GIGYF2 | 1 | 0 | 13407 | 1 | 0 | 5237 | 2 | 0 | 3.57E-03 | 1.00 | GOLD |
| DLG4 | 1 | 0 | 12192 | 1 | 0 | 5237 | 2 | 0 | 3.77E-03 | 1.00 | ASD5 |
| SMC3 | 1 | 0 | 12192 | 1 | 1 | 5237 | 2 | 1 | 4.51E-03 | 0.19 | ASD4 |
| KMT2E | 0 | 0 | 12192 | 2 | 0 | 5237 | 2 | 0 | 7.16E-03 | 1.00 | ASD4 |
| PARD3B | 1 | 0 | 12192 | 1 | 0 | 5237 | 2 | 0 | 7.79E-03 | 1.00 | ASD5 |
| PTK7 | 1 | 0 | 12192 | 1 | 0 | 5237 | 2 | 0 | 9.99E-03 | 1.00 | ASD4 |
| SRCAP | 1 | 0 | 12192 | 1 | 0 | 5237 | 2 | 0 | 1.25E-02 | 1.00 | ASD4 |
| PHF2 | 0 | 0 | 12192 | 2 | 0 | 5237 | 2 | 0 | 1.60E-02 | 1.00 | ASD5 |
| ZC3H4 | 0 | 0 | 13407 | 2 | 0 | 5237 | 2 | 0 | 1.75E-02 | 1.00 | GOLD |
| SETD2 | 0 | 0 | 13407 | 2 | 0 | 5237 | 2 | 0 | 1.77E-02 | 1.00 | GOLD |
| DIP2A | 0 | 0 | 13407 | 2 | 0 | 5237 | 2 | 0 | 1.79E-02 | 1.00 | GOLD |
| UNC80 | 1 | 0 | 12192 | 1 | 0 | 5237 | 2 | 0 | 1.83E-02 | 1.00 | ASD5 |
| ZNF292 | 1 | 0 | 11731 | 1 | 0 | 5237 | 2 | 0 | 1.88E-02 | 1.00 | ASD6 |
| PHIP | 1 | 0 | 13407 | 1 | 0 | 5237 | 2 | 0 | 1.88E-02 | 1.00 | GOLD |
| WDFY3* | 0 | 0 | 12192 | 2 | 0 | 5237 | 2 | 0 | 1.93E-02 | 1.00 | ASD5 |
| PLXNB1 | 1 | 1 | 12192 | 1 | 0 | 5237 | 2 | 1 | 1.93E-02 | 0.40 | ASD5 |
| ASXL3 | 0 | 0 | 11731 | 2 | 0 | 5237 | 2 | 0 | 2.17E-02 | 1.00 | ASD6 |
| LAMC3 | 0 | 0 | 13407 | 2 | 0 | 5237 | 2 | 0 | 2.73E-02 | 1.00 | GOLD |
| DOCK8 | 1 | 0 | 11731 | 1 | 0 | 5237 | 2 | 0 | 5.40E-02 | 1.00 | ASD6 |
| KMT2C | 0 | 0 | 11731 | 2 | 0 | 5237 | 2 | 0 | 7.15E-02 | 1.00 | ASD6 |
| COL4A3BP | 0 | 0 | 13407 | 0 | 4 | 5237 | 0 | 4 | 1.00 | 1.75E-07 | GOLD |
| PPP2R5D | 0 | 0 | 13407 | 1 | 3 | 6158 | 1 | 3 | 1.40E-02 | 1.04E-06 | GOLD |
| TRIO | 0 | 1 | 13407 | 1 | 3 | 5237 | 1 | 4 | 0.18 | 1.44E-04 | GOLD |
| TBL1XR1 | 0 | 0 | 13407 | 1 | 2 | 6158 | 1 | 2 | 1.45E-02 | 6.37E-04 | GOLD |
| PTPN11 | 0 | 0 | 12192 | 0 | 2 | 5237 | 0 | 2 | 1.00 | 3.93E-03 | ASD4 |
| DLGAP1 | 0 | 1 | 12192 | 0 | 1 | 5237 | 0 | 2 | 1.00 | 5.10E-03 | ASD4 |
| TANC2 | 0 | 1 | 12192 | 1 | 1 | 5237 | 1 | 2 | 0.11 | 1.36E-02 | ASD5 |
| SRGAP3 | 0 | 1 | 12192 | 0 | 1 | 5237 | 0 | 2 | 1.00 | 1.47E-02 | ASD5 |
| ITPR1 | 0 | 1 | 11731 | 0 | 2 | 5237 | 0 | 3 | 1.00 | 1.80E-02 | ASD6 |
| ADCY5 | 0 | 0 | 12192 | 0 | 2 | 5237 | 0 | 2 | 1.00 | 1.94E-02 | ASD5 |
An LGD variant was identified in this gene using previously published smMIPs; therefore, the LGD count differs compared to Supplementary Table 11 to avoid duplicate counting.
FDR corrections were based on the number of samples for which parental DNA could be tested.
Figure 2. Targeted sequencing highlights genes that reach significance for DN mutations and private disruptive variant burden.
(a–c) Quantile-quantile plots comparing the probability (FDR-corrected, inverse log transformed) of recurrent DN mutation for individual genes among proband samples compared to a uniform distribution given the number of genes tested (dashed gray line = significance threshold). Black dashed box (panels (a) and (b)) are zoomed in (panels (b) and (c), respectively). *Genes that reached significance for mutation burden. (d–e) Scatterplots depict the odds ratio (OR) for private variants compared to unaffected controls from ExAC (y-axis) versus the FDR-corrected DN p-value (x-axis; values have been inverse log transformed for plotting) by gene. Gray lines indicate the significance threshold for the DN p-value (horizontal) and an OR of two (vertical). Genes are classified as DN significant and OR > 2 (red dots), OR > 2 only (orange), and those that show a significant DN p-value only (blue). Gene name labels indicate a significant burden (FDR q < 0.1, simulation test) of either private LGD (d) or MIS30 (e) mutations in probands (Table 2; Methods). *Genes in which no control counts were observed where the 95% lower confidence bound was used as the most conservative OR estimate. See Supplementary Table 14 for underlying data.
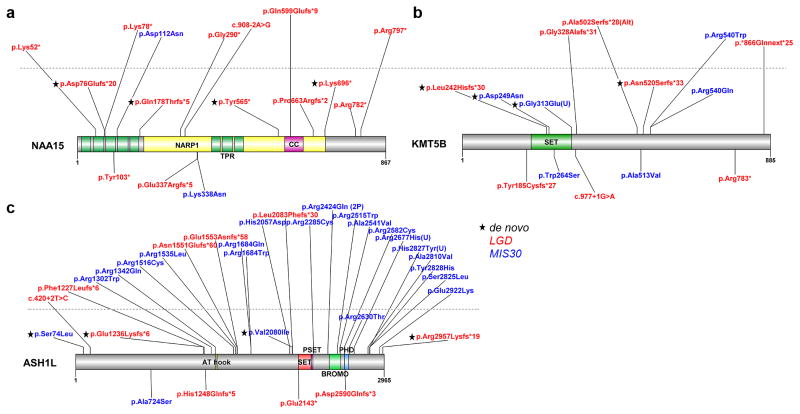
Figure 3. Protein location of private disruptive variants in new NDD candidate risk genes.
(a–c) Protein diagrams of (a) NAA15, (b) KMT5B, and (c) ASH1L with novel private LGD and MIS30 mutations identified in this study and published DN variants indicated in HGVS format. Annotated protein domains are shown (colored blocks) for the largest protein isoforms. Previously published DN variants (below protein structure, Supplementary Table 2) are compared to new variants in this study (above). Variants above the dashed line are of unknown inheritance; variants below the line have been validated for inheritance. Domain abbreviations: NARP1, NMDA receptor-regulated protein 1; CC, coiled coil; TRP, tetratrico peptide repeat region; PHD, plant homeodomain.
Inherited mutations and burden
The majority of validated LGD and MIS30 private variants were inherited (65%) either from mother (33.2%) or father (31.8%) (Supplementary Table 13; Supplementary Fig. 5e). Combined with additional ultra-rare (AC ≤ 3) inherited events (Supplementary Table 16) and published private inherited counts from exome sequencing of the Simons Simplex Collection (SSC)13 (Supplementary Table 17), we observe a nominally significant maternal transmission bias for LGD (but not MIS30) events (p = 0.037, binomial test where sex chromosome events were excluded). Although currently underpowered at the single-gene level to detect specific genes, several showed an elevated number of maternal transmissions (≥3:1 ratio; i.e., AHNAK, DSCAM, NRXN1, NISCH, UIMC1, PLXNB1, PROX2, CHD1, TNRC18, PTK7, and MOV10; Supplementary Fig. 7).
We also estimated the burden of private LGD and MIS30 variants at the single-gene level regardless of inheritance status by comparison with controls from the ExAC database where neuropsychiatric cases had been excluded (n = 45,376; http://exac.broadinstitute.org/; Supplementary Table 18). Separate simulations of LGD and MIS30 events identify 30 and 13 genes with a significant LGD burden and MIS30 burden, respectively (1x106 simulations with Benjamini-Hochberg correction; Table 2; Fig. 2d,e). Four genes—FOXP1, GRIN2B, SCN2A, and SETD5—were significant for both LGD and MIS30 burden and are well-established NDD genes. Interestingly, 18 genes—8 LGD and 10 MIS30—had a significant burden of private disruptive mutations in this study but did not reach DN significance likely due to our inability to test inheritance for all events. Although DN mutations in some of these genes have already been implicated in other NDD studies (e.g., FOXP123, TRIO24, SCN1A25, SIN3A26, and IQSEC227), for others—CTNND2, NAV2, and UNC80—many of the severe mutations in pedigrees are inherited (Supplementary Tables 11 and 16). Given their involvement in neuronal function, axonal projection, dendrite spine formation and oligodendrocyte differentiation28–31, these genes likely begin to define a class of inherited high-impact risk factors.
Table 2.
Genes that carry a significant burden of private disruptive variation in cases.
| LGD | MIS30 | |||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Gene | case count | control count | Corrected burden p-value | DN LGD significant | case count | control count | Corrected burden p-value | DN MIS30 significant |
| SCN2A | 14 | 2 | 8.80E-05 | YES | 12 | 11 | 4.14E-02 | YES |
| GRIN2B | 8 | 0 | 2.26E-04 | YES | 9 | 5 | 3.66E-02 | YES |
| FOXP1 | 7 | 5 | 5.84E-02 | YES | 6 | 2 | 4.34E-02 | NO |
| SETD5 | 11 | 2 | 2.26E-04 | YES | 12 | 10 | 3.66E-02 | NO |
| ARID1B | 13 | 8 | 2.63E-03 | YES | 14 | 34 | 0.56 | NO |
| CHD8 | 10 | 4 | 2.62E-03 | YES | 9 | 22 | 0.63 | YES |
| SYNGAP1 | 8 | 0 | 2.26E-04 | YES | 3 | 10 | 0.90 | NO |
| ADNP | 10 | 1 | 2.20E-04 | YES | 1 | 3 | 0.92 | NO |
| DYRK1A | 10 | 1 | 2.20E-04 | YES | 3 | 8 | 0.83 | NO |
| MED13L | 10 | 0 | 8.80E-05 | YES | 7 | 16 | 0.63 | YES |
| POGZ | 9 | 2 | 1.32E-03 | YES | 7 | 6 | 0.15 | YES |
| NAA15 | 12 | 4 | 3.08E-04 | YES | 1 | 9 | 1.00 | NO |
| CTNNB1 | 7 | 0 | 6.06E-04 | YES | 3 | 9 | 0.83 | NO |
| ASH1L | 8 | 3 | 7.04E-03 | YES | 17 | 35 | 0.35 | YES |
| DDX3X | 5 | 0 | 7.04E-03 | YES | 2 | 0 | 0.30 | NO |
| SMARCC2 | 6 | 1 | 8.26E-03 | NO | 2 | 4 | 0.76 | NO |
| CTNND2 | 4 | 1 | 7.04E-02 | NO | 8 | 31 | 0.98 | NO |
| SIN3A | 5 | 0 | 7.04E-03 | NO | 7 | 14 | 0.56 | NO |
| DSCAM | 6 | 3 | 4.76E-02 | YES | 12 | 39 | 0.83 | NO |
| SCN1A | 5 | 1 | 2.68E-02 | NO | 5 | 11 | 0.66 | NO |
| TNRC6B | 7 | 4 | 3.67E-02 | YES | 7 | 18 | 0.70 | NO |
| KMT2A | 4 | 0 | 2.68E-02 | YES | 3 | 9 | 0.83 | NO |
| TRIP12 | 5 | 2 | 5.84E-02 | YES | 6 | 19 | 0.83 | YES |
| DLG4 | 5 | 2 | 5.84E-02 | YES | 4 | 9 | 0.70 | NO |
| TRIO | 6 | 3 | 4.76E-02 | NO | 9 | 26 | 0.76 | YES |
| NFIA | 3 | 0 | 7.15E-02 | NO | 2 | 2 | 0.58 | NO |
| KMT5B | 5 | 0 | 7.04E-03 | YES | 3 | 0 | 0.13 | YES |
| CDKL5 | 5 | 1 | 2.68E-02 | NO | 0 | 0 | NA | NO |
| DNAJC6 | 6 | 4 | 7.28E-02 | NO | 10 | 15 | 0.30 | NO |
| ANK2 | 9 | 9 | 7.04E-02 | YES | 21 | 58 | 0.62 | NO |
| IQSEC2 | 2 | 0 | 0.20 | NO | 7 | 3 | 4.14E-02 | NO |
| IQGAP3 | 11 | 21 | 0.33 | NO | 21 | 34 | 9.87E-02 | NO |
| SLC6A1 | 1 | 0 | 0.48 | YES | 10 | 5 | 2.30E-02 | YES |
| LAMC3 | 8 | 19 | 0.53 | YES | 15 | 19 | 7.03E-02 | NO |
| UNC80 | 5 | 13 | 0.68 | YES | 30 | 38 | 1.21E-02 | NO |
| ADGRL2 | 1 | 0 | 0.48 | NO | 6 | 0 | 1.21E-02 | NO |
| ERBIN | 1 | 0 | 0.48 | NO | 4 | 0 | 4.34E-02 | NO |
| NAV2 | 1 | 9 | 1.00 | NO | 42 | 74 | 3.63E-02 | NO |
| AGAP2 | 0 | 2 | 1.00 | NO | 9 | 6 | 4.14E-02 | NO |
P-values were calculated by simulating the number of private LGD or MIS30 events found in the study compared to 45,375 ExAC controls and were Benjamini-Hochberg corrected for the number of genes screened in the study where at least one private mutation was found in cases or controls (n = 176). Genes found in this table are labeled in Figure 2d–e. Corrected p-values < 0.1 were considered significant.
Autism versus intellectual disability and developmental delay
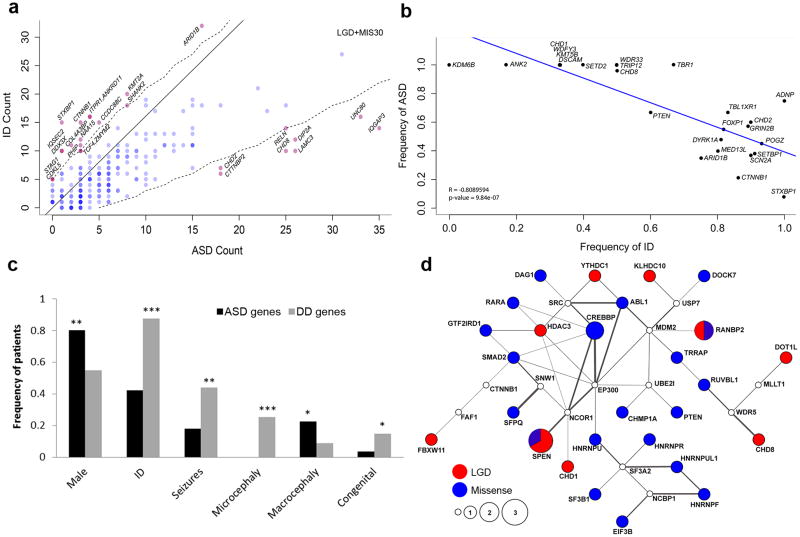
To determine whether individual genes show a bias for clinical phenotype, we performed a separate burden analysis by primary ascertainment diagnosis (i.e., ASD or ID (including DD diagnoses per DSM-5 criteria)) combined with data from previous NDD studies (Supplementary Tables 2, 11, and 17)13. We identify 25 genes that show a bias for primary diagnosis (two one-tailed binomial tests, p < 0.025 for either ASD or ID/DD cases) considering both LGD and MIS30 (Fig. 4a). Eight genes have an ASD bias—CHD2, CTTNBP2, CHD8, LAMC3, DIP2A, RELN, UNC80, and IQGAP3. Of these, only CHD8, CHD2, and DIP2A have been previously implicated as high-risk ASD loci32. Of the 17 ID/DD biased genes, NAA15, ZMYM2, PHIP, and STAG1 have not been previously linked to these phenotypes. We further separated the LGD and MIS30 events and identified additional significant genes for each mutation type, notably a bias for CDH10 LGD and NEMF MIS30 mutations in ASD (Supplementary Fig. 8a) and SCN1A LGD and NRXN1 MIS30 mutations in ID/DD (Supplementary Fig. 8b). Most genes, however, are mutated in both conditions further highlighting the substantial genetic overlap between these comorbid conditions.
Figure 4. ASD versus ID genes.
(a) Probands were categorized based on primary ascertainment either ASD or ID (including DD) and the combined number of LGD and MIS30 events per gene (published and this study) shown. Genes were tested for a bias to one phenotype (ASD or ID) by two one-tailed binomial tests (p < 0.025 for either bias). The solid line indicates equal proportions of mutations corrected for the screened population size. Significantly biased genes (red) are indicated with respect to the threshold (dashed line) and insignificant genes (blue). Darker shades of red or blue indicate multiple genes. (b) Scatterplot shows a negative correlation (Pearson’s correlation) between ASD and ID diagnosis by gene (Table 3). (c) Bar graph compares phenotypic features of patients where genes are associated primarily with ASD diagnosis (>95%, black bars) compared to all other genes (gray bars) in Table 3. Significance was calculated by Fisher’s two-tailed exact test, and p-values were FDR corrected. Exact p values: seizures (p = 1.20x10−4), congenital abnormalities (p = 1.88x10−2), microcephaly (p = 1.79x10−7), macrocephaly (p = 5.25x10−3), males (p = 1.65x10−4). *p < 0.05, **p < 0.001, ***p < 0.0001. (d) SSC probands with ASD and an FSIQ > 100 were selected for pathway enrichment. Node size indicates the mutation score (calculated by MAGI based upon the number of DN mutations), and the color of the node indicates the number of DN LGD (red) and DN missense (no CADD cut-off; blue) mutations have been observed in affected probands, respectively. For SPEN, 2 LGD and 1 missense mutation have been observed and for RANBP2, 1 LGD and 1 missense mutation. White nodes indicate no DN mutations have been observed. Gray lines connect genes with both protein-protein interactions and brain co-expression (Pearson’s correlation coefficient r2 > 0.37, Methods). Thicker lines correspond to more highly co-expressed gene pairs.
Phenotypic assessment of new risk genes
We recontacted individuals with mutations in NAA15, KMT5B, and ASH1L for further follow-up. We identified 12 LGD and one MIS30 private (Supplementary Table 11) variants in NAA15 through our smMIP screening (Fig. 3a) and determined that four LGD mutations were sporadic while two LGD variants, including a C-terminal mutation, were inherited (Supplementary Table 11). NAA15 shows a burden of LGD events in cases (Table 2) as well as an excess of DN LGD variants (Table 1). The gene NAA15 encodes a protein that is a component of the NatA N-acetyltransferase complex, which includes NAA10—a protein associated with Ogden syndrome as well as non-syndromic DD33 and is thought to tether the complex to the ribosome for posttranslational modification of proteins as they exit the ribosome34. In order to identify additional patients for clinical recontact, we relaxed our variant filter to allow for ultra-rare (AC ≤ 3) alleles. In total, we were able to collect clinical information for ten probands with private and three probands with ultra-rare variants in NAA15 (Supplementary Table 19). Patients in our study with NAA15 LGD and MIS30 mutations share phenotypic features, including ID (10/11 patients [91%]), speech delay (5/6 patients [83%]), ASD diagnosis (formal diagnosis in 5/8 patients [63%] with ASD-like traits observed in two additional patients), and nonspecific growth abnormalities (e.g., microcephaly, macrocephaly, and hypertelorism) (Supplementary Table 19). Given the incidence of DD (5.12%) in the general population35,36, we estimate the penetrance of LGD NAA15 mutations to be significant at 35.3% (95% CI 15.7%–63.6%).
Both genes—KMT5B and ASH1L—encode histone-lysine N-methyltransferase proteins thought to be important in chromatin modification, occupancy and gene regulation. Although the role of these genes in NDDs has not been established, a paralog of ASH1L—SETBP1—has been shown to be mutated in NDD cases and associated with ID and loss of expressive language19. We identified two DN LGD and two DN MIS30 mutations in KMT5B in this study (Supplementary Table 12) in addition to three DN LGD and one DN MIS30 published variants12,14 (Supplementary Table 2; Fig. 3b). We were able to collect clinical information from three probands with private variants and four probands with ultra-rare variants in KMT5B (Supplementary Table 20). Patients with disruptive variation in KMT5B shared features such as ID/DD (7/7 patients [100%]), ASD diagnosis (5/6 patients [83%]), language delay (3/4 patients [75%]), motor delay (3/5 patients [60%]), and febrile seizures (3/5 patients [60%]). Attention deficits were also observed in three of these patients (Supplementary Table 20). For ASH1L, we identified two DN LGD and two DN MIS30 mutations in addition to the three DN LGD previously published mutations13,14. We identified many additional LGD and MIS30 variants in ASH1L where parental DNA was not available (Fig. 3c) and found that mutations cluster around the known annotated protein domains. We were able to obtain clinical information for two probands carrying private and three probands carrying ultra-rare ASH1L variants. Individuals with ASH1L disruptive variation in this study had ID (5/5 patients [100%]), ASD (2/3 patients [67%]), and evidence of seizures (2/3 patients [67%]) (Supplementary Table 21).
Phenotypic comparisons and a high-functioning ASD network
We selected patients with DN LGD mutations in 25 of our top-ranked genes in an effort to more broadly compare phenotypic features. Of the recontacted individuals, 70% (88/125) agreed to participate in a more comprehensive phenotypic evaluation (Supplementary Methods). To increase our power to detect differences between patients grouped by gene, 215 case reports from the published literature were combined with the findings collected as part of our recontact study. We assessed the general severity of each NDD using a modified de Vries scale (Supplementary Table 22) and summarized phenotypic features collected during follow-up (Table 3: including rates of ASD, ID, seizures, macro/microcephaly, and congenital abnormalities as well as mean IQ measures and ASD severity).
Table 3.
Key phenotypic traits across participants with gene-disrupting mutations.
| Gene | Total Cases |
Cases evaluated in-depth |
Mean Testing Age in months |
Gender (% male) |
Overall Severity |
ASD | ID | Seizures | Micro- cephaly |
Macro- cephaly |
Congenital Abnormality |
VIQβ | NVIQγ | ASD severityδ |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||||||||||||||
| modified De Vriesα |
N | Rate | N | Rate | N | Rate | N | Rate | N | Rate | N | Rate | N | Mean | N | Mean | N | Mean | N | |||||
|
| ||||||||||||||||||||||||
| KDM6B | 4 | 1 | 136 | 100% | 3.00 | 2 | 100% | 4 | 0% | 4 | 25% | 4 | 0% | 4 | 50% | 4 | 0% | 4 | 97.00 | 4 | 95.75 | 4 | 5.25 | 4 |
| ANK2 | 6 | 1 | 141 | 83% | 5.00 | 2 | 100% | 6 | 17% | 6 | 0% | 6 | 0% | 6 | 17% | 6 | 17% | 6 | 80.50 | 6 | 81.17 | 6 | 6.83 | 6 |
| DSCAM | 3 | 2 | 133 | 100% | 6.50 | 2 | 100% | 3 | 33% | 3 | 0% | 3 | 0% | 3 | 33% | 3 | 0% | 3 | 71.33 | 3 | 55.33 | 3 | 9.00 | 3 |
| KMT5B | 3 | 1 | 115 | 67% | 7.00 | 1 | 100% | 3 | 33% | 3 | 33% | 3 | 0% | 3 | 33% | 3 | 0% | 3 | 47.33 | 3 | 54.67 | 3 | 8.33 | 3 |
| WDFY3 | 3 | 2 | 181 | 100% | 6.00 | 3 | 100% | 3 | 33% | 3 | 33% | 3 | 0% | 3 | 67% | 3 | 0% | 3 | 57.33 | 3 | 77.00 | 3 | 8.00 | 3 |
| CHD1 | 3 | 1 | 123 | 100% | 5.00 | 1 | 100% | 3 | 33% | 3 | 0% | 3 | 0% | 3 | 0% | 3 | 0% | 2 | 83.00 | 3 | 97.67 | 3 | 7.00 | 3 |
| SETD2 | 5 | 2 | 190 | 60% | 4.25 | 3 | 100% | 4 | 40% | 5 | 40% | 5 | 0% | 5 | 40% | 5 | 0% | 5 | 100.67 | 3 | 94.67 | 3 | 7.67 | 3 |
| WDR33 | 2 | 1 | 129 | 100% | 7.00 | 1 | 100% | 2 | 50% | 2 | 50% | 2 | 0% | 2 | 0% | 2 | 0% | 1 | 67.00 | 1 | 66.00 | 2 | 8.50 | 2 |
| TRIP12 | 6 | 1 | 116 | 67% | 5.50 | 2 | 100% | 6 | 50% | 6 | 17% | 6 | 0% | 6 | 0% | 6 | 0% | 5 | 62.67 | 6 | 64.17 | 6 | 6.50 | 6 |
| TBR1 | 6 | 1 | 102 | 50% | 5.50 | 2 | 100% | 6 | 67% | 6 | 17% | 6 | 0% | 6 | 0% | 6 | 0% | 4 | 62.25 | 4 | 60.40 | 5 | 7.20 | 5 |
| CHD8 | 25 | 8 | 131 | 84% | 6.13 | 11 | 96% | 25 | 50% | 24 | 17% | 24 | 0% | 25 | 64% | 25 | 0% | 18 | 60.76 | 17 | 68.40 | 20 | 7.81 | 21 |
| ADNP | 20 | 4 | 81 | 65% | 7.40 | 14 | 75% | 20 | 100% | 20 | 25% | 20 | 10% | 19 | 10% | 19 | 44% | 18 | 33.25 | 4 | 36.00 | 5 | 6.71 | 7 |
| PTEN | 15 | 2 | 62 | 60% | 4.50 | 6 | 67% | 35 | 60% | 15 | 7% | 15 | 0% | 15 | 100% | 15 | 8% | 12 | 71.33 | 6 | 74.57 | 7 | 7.00 | 6 |
| TBL1XR1 | 13 | 1 | 139 | 54% | 5.00 | 2 | 67% | 6 | 83% | 12 | 33% | 6 | 23% | 13 | 15% | 13 | 54% | 11 | 52.33 | 3 | 55.00 | 3 | 6.00 | 3 |
| CHD2 | 12 | 4 | 134 | 50% | 5.75 | 8 | 60% | 10 | 90% | 10 | 83% | 12 | 10% | 10 | 10% | 10 | 25% | 8 | 71.33 | 6 | 63.33 | 6 | 8.17 | 6 |
| GRIN2B | 22 | 3 | 146 | 55% | 4.80 | 8 | 57% | 14 | 89% | 18 | 23% | 21 | 16% | 19 | 5% | 19 | 13% | 15 | 58.00 | 6 | 56.83 | 6 | 8.17 | 6 |
| FOXP1 | 11 | 2 | 160 | 64% | 7.00 | 9 | 55% | 11 | 82% | 11 | 18% | 11 | 0% | 9 | 33% | 9 | 20% | 10 | 53.50 | 2 | 48.00 | 2 | 7.50 | 2 |
| DYRK1A | 21 | 6 | 195 | 57% | 7.76 | 17 | 48% | 21 | 81% | 21 | 57% | 21 | 90% | 21 | 0% | 21 | 33% | 21 | 44.29 | 7 | 51.57 | 7 | 7.43 | 7 |
| POGZ | 44 | 1 | 109 | 57% | 6.12 | 24 | 45% | 44 | 93% | 44 | 11% | 44 | 33% | 43 | 5% | 43 | 13% | 39 | 72.20 | 5 | 68.80 | 5 | 8.60 | 5 |
| MED13L | 15 | 1 | 112 | 53% | 5.44 | 9 | 40% | 10 | 80% | 15 | 20% | 15 | 20% | 15 | 7% | 15 | 36% | 14 | 57.33 | 3 | 73.67 | 3 | 6.67 | 3 |
| SETBP1 | 24 | 1 | 98 | 58% | 6.20 | 5 | 38% | 8 | 91% | 23 | 71% | 24 | 9% | 11 | 0% | 11 | 18% | 11 | 60.33 | 3 | 69.67 | 3 | 6.00 | 3 |
| SCN2A | 55 | 2 | 75 | 53% | 5.14 | 13 | 37% | 35 | 90% | 52 | 75% | 55 | 25% | 44 | 2% | 44 | 21% | 23 | 52.38 | 8 | 57.25 | 8 | 6.50 | 8 |
| ARID1B | 28 | 3 | 137 | 43% | 6.45 | 22 | 35% | 26 | 75% | 28 | 32% | 28 | 7% | 28 | 14% | 28 | 21% | 28 | 67.14 | 7 | 67.00 | 7 | 7.00 | 7 |
| CTNNB1 | 30 | 1 | 121 | 43% | 6.19 | 21 | 21% | 24 | 86% | 29 | 13% | 30 | 75% | 28 | 0% | 28 | 31% | 29 | 51.75 | 4 | 56.00 | 4 | 6.50 | 4 |
| STXBP1 | 49 | 1 | 107 | 51% | 4.37 | 49 | 8% | 49 | 100% | 49 | 86% | 49 | 8% | 49 | 0% | 49 | 0% | 44 | 30.00 | 1 | 30.00 | 1 | 3.00 | 1 |
In order to maximize the number of cases for each assessment, the number of cases considered for calculated variables differs. The N for each variable is listed.
See Supplementary Table 22 for modified de Vries scoring criteria.
Mean verbal IQ (VIQ) has a mean of 100 and standard deviation of 15.
Mean nonverbal IQ (NVIQ) has a mean of 100 and standard deviation of 15.
Mean ASD severity is derived from the ADOS-2 Calibrated Severity Score (CSS) and ranges from 1–10 with scores between 4–10 representing symptoms within ASD, with 10 being the most severe.
Several specific and global patterns emerged from this combined dataset, in particular, an inverse relationship between ASD and ID diagnoses by gene (Pearson’s R = −0.81, p = 9.84x10−7; Fig. 4b). We partitioned patients into two categories: those most strongly associated with ASD (diagnostic rate > 95%) versus those more strongly associated with DD. Patients with mutations in ASD genes show significantly lower rates of seizures (p = 1.20x10−4), congenital abnormalities (p = 1.88x10−2), and microcephaly (p = 1.79x10−7), but higher rates of macrocephaly (p = 5.25x10−3) compared to comorbid ASD and ID genes and strong ID genes (two-tailed Fisher’s exact test; Fig. 4c). In addition, the ASD-dominated genes show a significant difference with respect to gender. ASD genes are more likely (p = 1.65x10−4) to affect males with an overall ratio of 4:1 when compared to other genes (1.2:1 male to female ratio). Although the number of individual patients per gene is still low, it is interesting that several genes show an exclusive male bias (KDM6B, DSCAM, WDFY3, CHD1, and WDR33; Fig. 4c; Table 3). Pathway analysis of these 11 ASD genes indicates a functional enrichment for chromatin remodeling (corrected p = 4.72x10−3; Enrichr tool (http://amp.pharm.mssm.edu/Enrichr/))37 implicating a functional network associated specifically with ASD individuals without ID.
To identify additional genes that may be associated specifically with high-functioning ASD, we revisited the deep phenotyping data collected as part of the SSC and applied the MAGI network-building tool, which compares the spectrum of DN mutations in probands and unaffected siblings to identify co-expression and protein-interaction networks enriched in patients12,13,20. We specifically selected patients with a full-scale IQ (FSIQ) > 100 (n = 668 SSC probands; male bias 9:1) to construct a protein-interaction network based on genes with DN variants in this subset (Supplementary Methods). One statistically significant model emerged (p < 0.01, simulation test; Fig. 4d), including 40 genes and DN mutations in 31 individuals with FSIQ > 100. Although mostly comprised of DN missense mutations, the network shows that DN LGD mutations in FBXW11, CHD1, CHD8, DOT1L, HDAC3, YTHDC1, and KLHDC10 may be important in this patient subset. Both CHD1 and CHD8 individuals were included in our large-scale patient recontact and showed high specificity for ASD diagnosis (Table 3). A pathway analysis for this specific set of genes implicates, once again, chromatin remodeling (p = 0.0003 adjusted p-value) as well as mRNA splicing (p = 0.00026) and Wnt signaling (p = 0.03) as potentially important for ASD without ID.
Functional characterization of candidate genes in Drosophila
In order to provide additional functional evidence especially with respect to nervous system function and behavior, we performed a pilot study investigating 21 genes in Drosophila melanogaster (Supplementary Table 23). For 11 of these genes, DN LGD mutations are significantly enriched in NDD patients. For the other ten genes, there are indications that they may be associated with ASD, such as a higher mutation rate in ASD cohorts or a central position in ASD gene interaction networks20. Others, such as NCKAP1 and WDFY3, are at the cusp of statistical significance. We used the UAS-Gal4 system and inducible RNA interference (RNAi) lines to specifically knockdown these genes in Drosophila neurons. Whenever their locomotor function and overall vigor allowed (Supplementary Table 23), we subjected these knockdown flies to an ASD- and ID-relevant behavioral assay measuring light-off jump habituation, which has been shown to be affected in a number of ASD- or ID-related Drosophila models38–43. In this assay, flies suppress their startle (jump) response to a repeated nonthreatening stimulus (light-off) as a result of experience. Their response thus gradually wanes (Fig. 5a,b). As the most fundamental evolutionarily conserved form of learning, habituation is thought to represent a prerequisite for higher cognitive functions44. Beyond that, a number of studies showed defective habituation of neural activity or behavior in ASD45–48, and it has been proposed that disturbed habituation mechanisms could substantially contribute to defective filtering and other ASD features49,50.
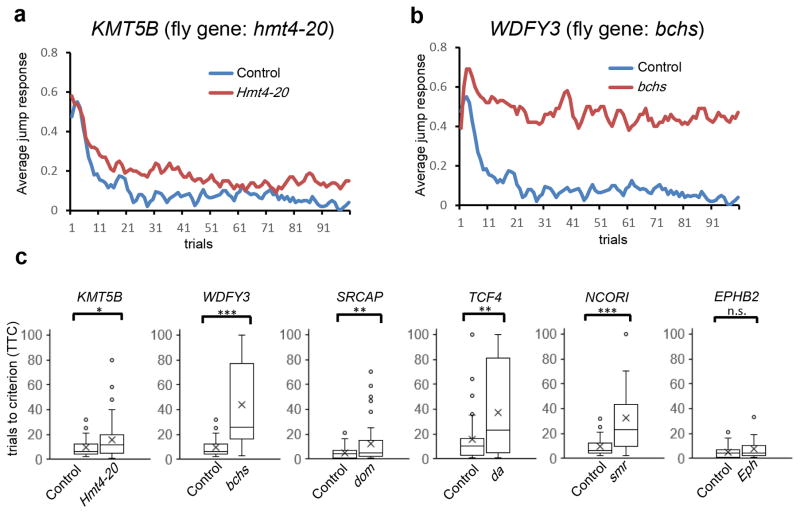
Figure 5. Habituation deficits in Drosophila knockdown models.
(a–b) Representative jump response curves for (a) hmt4-20 (ortholog of KMT5B) and (b) bchs (ortholog of WDFY3) panneuronal knockdown flies. The ratios of flies that responded to light-off stimuli are plotted over 100 trials (64 individual flies were tested for each genotype). Controls are plotted in blue and knockdowns are plotted in red. (c) Distribution of trials to no-jump criterion (TTC, Methods) of knockdowns versus corresponding control flies are plotted (cross, mean; middle line, median; box boundaries, upper and lower quartile; end of whiskers, maximum and minimum; dots, outliers). * p < 0.05, ** p < 0.01, *** p < 0.001 (linear regression model; 64 flies tested for each genotype; exact p values in Supplementary Table 23).
We first examined genes that showed a significant excess of DN LGD mutations—NAA15, KMT5B, ASH1L, and TCF4—and where human phenotypic data strongly support a role in NDDs. For the Nat1 (ortholog of NAA15, VDRC #110689) knockdown, we observe erect wing, impaired locomotor activity, and adult early lethality (within 1–2 days after eclosion). Upon knockdown of Nat1 using a second presumably weaker RNAi line (VDRC #17571), flies exhibit normal morphology and locomotion; however, when challenged in the light-off jump paradigm, their initial response is impaired (19% frequency of initial jumping), precluding proper assessment of habituation (Supplementary Table 23). These results support Nat1’s crucial role in nervous system development. Ash1 (ortholog of ASH1L) neuronal knockdown flies also showed reduced fitness, and, like Nat1 flies, could not be scored in the habituation paradigm. Hmt4-20 (ortholog of KMT5B) and da (ortholog of TCF4) flies, in contrast, were overall healthy but showed specific and significant habituation deficits (Fig. 5a,c), suggesting that both genes play important roles in the molecular machinery that regulates habituation learning.
We observed habituation defects following knockdown of fly homologs for several other significant DN genes, including SYNGAP1, GRIN2B, and SRCAP (Fig. 5c, Supplementary Table 23). In addition, dom (SRCAP) and da (TCF4) flies showed significant morphological abnormalities at the neuromuscular junction (NMJ), a well-studied synaptic model system (Supplementary Fig. 9). NCKAP1, WDFY3, and GIGYF2 are among the tested genes with borderline significance based on our human genetic data. Significant habituation defects were observed for hem (ortholog of NCKAP1), bchs (ortholog of WDFY3), and CG11148 (ortholog of GIGYF2) knockdown flies (Fig. 5c; Supplementary Table 23). Due to the paucity of patients, little is known regarding the clinical phenotypes associated with loss-of-function mutations of these genes; however, these functional studies suggest an important role in neuronal and cognitive function.
DISCUSSION
Targeted sequencing of candidate genes in a large NDD cohort has identified three overlapping categories of high-risk genes. First, we identify 68 genes that now reach DN LGD mutation significance, 39 of which have previously been described. Due to limited availability of parental samples, this estimate is likely conservative. Second, we highlight 24 genes with a significant excess of DN missense mutations in NDD patients; 63% (15/24) overlap genes with DN LGD significance (e.g., SCN2A, STXBP1, CHD2, and CASK) while others are significant based only on an excess of DN MIS30 mutations (e.g., COL4A3BP, TRIO, TBL1XR1, and PPP2R5D; Table 1), similar to the Noonan-syndrome gene (PTPN11)51. Finally, 39 genes reach statistical significance based on a case-control burden testing (Table 2). Of interest are the 13 genes without DN mutation significance (SMARCC2, CTNND2, SIN3A, SCN1A, NFIA, CDKL5, DNAJC6, IQSEC2, IQGAP3, ADGRL2, ERBIN, NAV2, and AGPA2; Table 2) suggesting potential inherited risk factors13,31,52. In total, 44% (91/208) of our candidate genes reach locus-specific significance for disruptive mutations in 5.7% of patients, closely matching empirical expectations12. However, mutation of these genes may not be necessary and sufficient to result in disease. We note, for example, nine families (Supplementary Fig. 10) with disruptive mutations in two or more of the candidate genes.
Three genes without previous phenotype information reach a high level of DN significance (NAA15, KMT5B, and ASH1L). NAA15 was originally identified as an N-methyl-D-aspartate (NMDA) glutamate receptor-regulated gene through screens of NMDAR1 knockout mice53. Knockdown of Naa15 in Drosophila neurons caused severe locomotor defects and lethality. Missense mutations in the NAA15 interacting gene NAA10 are known to cause Ogden syndrome, an X-linked disorder of infancy that can result in severe DD, craniofacial anomalies, hypotonia, cardiac arrhythmias, and in some cases death54. This is consistent with the DD observed in our patients and the fact that DN LGD mutations have been identified in congenital heart disease patients with NDDs55. KMT5B and ASH1L highlight the importance of histone methyltransferases, like DNMT3 and EHMT56, in ID and NDD. Mouse studies have shown that Ash1l protein represses nrxn1α protein in neurons—a known presynaptic adhesion molecule required for synaptic formation57; mutations in NRXN1 have been associated with ASD58. Even less is known about the role the protein encoded by KMT5B plays in the developing brain. However, studies suggest that the H4K20me3 mark established by the KMT5B protein may be involved in cell cycle regulation in baboon neural stem progenitor cells59. Our own analyses in Drosophila support a role for NAA15 and ASH1L in neuronal development and for KMT5B and TCF4 in habituation learning (Supplementary Table 23) consistent with patient phenotypes (Supplementary Tables 18–20).
We designed the study such that approximately half of the patients were ascertained based on a primary diagnosis of ASD while the other half were diagnosed initially as ID/DD in an effort to test the diagnostic specificity of particular genes. While most genes are clearly risk factors for NDD broadly4, secondary analyses of both the genetic burden and subsequent patient follow-up for 25 genes in 303 patients did highlight genes with a statistical bias toward ASD versus ID/DD diagnosis (Table 3; Fig. 4c). We find that patients with mutations in genes enriched for ASD show significantly lower rates of seizures, congenital abnormalities, and microcephaly, but higher rates of macrocephaly compared to comorbid ASD and ID genes and strong ID genes (Fig. 4c). The latter is interesting in light of the observation of increased brain size and/or weight at early ages in subtypes of ASD when compared to ID or typical toddlers60–63. While the number of exome-sequenced patients with a DN mutation are few (4.6% or 31/668 patients), the data highlight a co-expression and protein-interaction network statistically enriched in high-functioning autism patients (FSIQ > 100) when compared to unaffected siblings. This network is biased for DN missense compared to LGD mutations (2:1) indicating that less severe mutations may be playing a role in ASD without ID. The network highlights both mRNA splicing as well as genes important in chromatin remodeling. The latter implicate early developmental programs that regulate cell proliferation, neural patterning and differentiation, axonal guidance consistent with cellular ASD models64–66, and ASD postmortem63,67,68, genomic69,70 and developmental imaging60–62 studies.
ONLINE METHODS
Patient samples
Whole-blood or cell line DNA from patients with ASD, ID, or DD diagnosis were collected from 15 international clinical and research cohorts (Fig. 1). Only DNA samples from The Autism Simplex Collection (TASC) and Autism Genetic Resource Exchange (AGRE) cohorts were derived from cell lines. Clinical workup, including diagnostic evaluation, medical examination, and neuropsychological assessment, was made available for many patients upon request, specifically for patients with mutations in NAA15, KMT5B, and ASH1L (case reports in Supplementary Data). Best estimate clinical DSM-51 diagnoses were made by experienced, licensed clinicians using all available information collected during the evaluation. For a description, the number of individuals represented, and the primary ascertainment criteria for each cohort in this study, refer to Supplementary Table 8. In addition, 2,867 unaffected sibling control individuals were collected for genetic and phenotypic comparison (Supplementary Table 9). All experiments carried out on these individuals were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national), and proper informed consent was obtained for sequencing, recontact for inheritance testing, and phenotypic workup. All sequencing of patient samples was performed at the University of Washington, Seattle, WA, USA.
Detailed descriptions of clinical cohorts
Autism Clinical and Genetic Resources in China (ACGC)
This cohort has been described previously21.
Autism Genetic Resource Exchange (AGRE)
This cohort has been described previously87.
Leuven
The Leuven cohort consists of patients with ASD as diagnosed by the multidisciplinary team in the Expert Centre for Autism Leuven according to DSM-IV-TR (American Psychiatric Association, 2000) criteria. All patients have been examined by a clinical geneticist. Patients with known monogenic conditions have been excluded after a routine genetics workup.
Melbourne & Murdoch
All participants had a DSM-IV or DSM-5 diagnosis of ASD. Diagnoses were community-based performed by a multidisciplinary team (pediatrician, psychologist, and speech pathologist). Diagnoses were confirmed for research purposes through ascertainment of previous ASD and cognitive assessments and a telephone interview with the parents. Information about the pregnancy and birth, developmental milestones, comorbidities, medications, and general health was collected during the interview and from medical records. Current ASD symptomatology was measured via the Social Responsiveness Scale (SRS) or Social Responsiveness Scale – 2nd edition (SRS-2). Pedigrees were obtained for each family detailing the family’s history of medical conditions, mental health disorders, intellectual impairment, ASD diagnoses and ASD traits. DNA was collected from blood or saliva from probands and their parents. Most probands underwent molecular karyotyping for CNVs and single-nucleotide polymorphisms and fragile X DNA testing; older participants had routine karyotyping.
The Autism Simplex Collection (TASC)
This cohort has been described previously88.
Adelaide
Individuals with intellectual disability or developmental delay were recruited who were referred but negative for molecular testing for fragile X and large CNVs by array comparative genomic hybridization (CGH). The vast majority were also singletons and recently clinically diagnosed/ascertained patients for recontact purposes.
Leiden
The cohort consists of patients with developmental delay with or without autistic features. Clinical microarrays to detect CNVs were run on all index patients, and identification of a likely causal CNV was an exclusion criterion. No formal DSM criteria were used in the diagnosis. All patients were seen by experienced clinical geneticists and if indicated specific gene tests were requested. Parents of the patients gave verbal consent for inclusion in this study.
Stockholm
Array CGH was performed for all cases using the Agilent platform with a 180k genome-wide design. The cases were referred for genetic investigation due to a diagnosis of ID/ASD but did not use the DSM-5 guidelines systematically.
Candidate gene selection
Candidate genes with DN mutations were identified from whole-exome and targeted sequencing studies of ASD, ID, and DD based on previously published studies and included 4,874 probands with ASD9,11–14, 151 probands with ID15,16, and 1,133 probands with DD17 (Supplementary Table 1). Genes were ranked based on the following criteria: (1) presence of two or more LGD mutations, (2) presence of multiple missense mutations and at least one LGD mutation, (3) presence of at least one LGD mutation overlapping a region of interest in our published DD CNV morbidity map19, and (4) presence of at least one LGD mutation with network connectivity to either chromatin remodeling/transcription or long-term potentiation as described previously20. Genes with expression in the brain (based on the BrainSpan Atlas of the Developing Human Brain [http://www.brainspan.org/] and GTEx [http://www.gtexportal.org/] databases89) were prioritized. We eliminated genes associated with likely unrelated disorders in OMIM (http://www.omim.org/) and genes that were deemed highly mutable (determined based on data from 6,503 control individuals in the NHLBI Exome Sequencing Project [ESP; http://evs.gs.washington.edu/EVS/]). Finally, we filtered genes based on the number of DN mutations by whole-exome sequencing among unaffected siblings from the SSC11–13. See Supplementary Table 3 for more details on selection criteria.
smMIP sequencing and variant validation
smMIP sequencing was performed as previously described9,10. We targeted the coding portions of all RefSeq annotated transcripts for these 208 genes as well as five base pairs into each exon-adjacent intron in order to capture variation at splice-donor/acceptor sites resulting in the design of 12,016 smMIPs. smMIPs were split into four pools (Gold, ASD4, ASD5, and ASD6; see Supplementary Tables 4–7 for gene breakdown), and each pool was rebalanced so that poorer performing smMIPs were spiked in at a concentration of 10X or 50X. Approximately 192 samples were barcoded and sequenced per lane of Illumina HiSeq 2000 as previously described11, and data analysis was performed using the MIPgen suite of tools (http://shendurelab.github.io/MIPGEN/). Variant calling of smMIP data was performed on each sequencing lane using FreeBayes v0.9.14 with default settings and the hg19 reference. For each of the four pools, all FreeBayes output was combined using GATK. Allele counts per genotype (AC) and the total number of alleles per genotype (AN) counts were recalibrated on the combined variant set using VCFtools. Multi-allelic sites were split into separate entries using vcflib (vcfbreakmulti) and sequencing error repeats and common single-nucleotide polymorphisms (all of dbSNP v129 and variants found in dbSNP v141 at a minor allele frequency ≥0.01 in at least one major population with at least two unrelated individuals having the minor allele) were removed. From the individual genotypes with sequencing depth (DP) of greater than 8X and a quality score (QUAL) of greater than 20, a private filter (found in only one family in the study) was applied to each pooled dataset (i.e., ASD4, ASD5, ASD6, or Gold). These variants were annotated using the Ensembl Variant Effect Predictor tool for GRCh37 (http://grch37.ensembl.org/Homo_sapiens/Tools/VEP/) and with CADD scores90 (http://cadd.gs.washington.edu/; v1.3). All private LGD variants and a portion of MIS30 variants were validated by Sanger sequencing. Specifically, a CADD > 30 was chosen for validation as these events are very rare (<0.1% of all missense events in control genomes90) and more likely to be pathogenic21. Where available, parents were also Sanger sequenced to determine the inheritance status of these variants. In total, we targeted >16,000 unique samples including 13,475 probands and 2,867 unaffected siblings using each of the four smMIP pools. 1,744 of the DNA samples did not have sufficient DNA for all four pools; gene sets were prioritized based on potential disease significance (Gold>ASD4>ASD5>ASD6; Supplementary Table 10) with 13,475 probands (Gold), 12,260 probands (ASD4&5), and 11,731 probands (ASD6). In addition, we sequenced 2,867 unaffected sibling samples with each of the four pools. To determine the performance of each smMIP pool, 10 plates of unaffected siblings (960 samples) were compared in each pool plotting the frequency of these 960 samples that reached at least 8X sequencing coverage for each individual smMIP in the study (Supplementary Table 3). Each of these data points were plotted by gene within each pool (i.e., Gold, ASD4, ASD5, and ASD6 shown in Supplementary Fig. 1–4, respectively). 165 genes passed all QC metrics (75% of smMIPs by gene reached at least 8X coverage in ≥80% of controls), but some exons, due to their size or GC composition, failed to pass these thresholds. For those regions that did not pass QC, we chose to consider variant genotypes identified in samples if they were of high quality (read depth (DP) > 8, phred scaled quality score (QUAL) > 20); however, these variants were not considered for assessments of mutation burden.
Clinical recontact and phenotyping
To systematically compare the effect of particular LGD mutations of targeted genes on phenotype, we recontacted individuals with identified LGD mutations and conducted a comprehensive phenotypic workup assessing function across multiple domains. Per our human subjects approval, we only recontacted individuals that had consented to be approached about future studies during their original assessment. Families were invited to participate in a comprehensive clinical workup that included diagnostic evaluation, medical examination, and neuropsychological assessment (see Supplementary Methods for test battery and procedures). Importantly, all assessments were conducted by examiners naïve to the individual’s genetic event, thereby reducing clinician bias in rendering diagnostic dispositions. In order to make comparisons across groups with similar LGD mutations, each participant was scored according to a modified version of the de Vries scale as a proxy for the overall severity of the phenotype92–94. The modified de Vries scale included presence of facial dysmorphisms, congenital abnormalities, postnatal head growth abnormalities, ID/DD, and the number of DSM-5 diagnoses and medical diagnoses conferred, allowing a score ranging from zero to twelve (Supplementary Table 22). Data collected from these patients were combined with published case reports to increase our power to detect enrichments among patients sharing DN LGD mutations in the same gene or pathway. A total of 323 case reports of individuals with DN LGD mutations of interest and relevant data were included. The relevant phenotype data extracted from cases in the published literature were combined with information collected from individuals that were able to complete the in-person comprehensive evaluation. The modified de Vries scale score for individuals with the same disrupted gene were averaged and then rank-ordered to estimate the impact of the gene mutation on phenotype. Only genes with six or more study participants and published case reports were included in the analysis. Patients that were considered had LGD mutations in one of the following genes: ADNP, ARID1B, CHD2, CHD8, CTNNB1, DYRK1A, FOXP1, GRIN2B, MED13L, POGZ, PTEN, SCN2A, SETBP1, STXBP1, or TBL1XR1.
Patient workups
Comprehensive clinical workup included diagnostic evaluation, medical examination, and neuropsychological assessment. Best estimate clinical DSM-51 diagnoses were made by experienced, licensed clinicians using all available information collected during the research evaluation. The battery included autism-specific diagnostic measures, the Autism Diagnostic Observation Schedule95 and the Autism Diagnostic Interview – Revised96, both administered by research-reliable clinicians. The battery also included assessment of cognitive ability (Differential Ability Scales, DAS97), language ability (Peabody Picture Vocabulary Test – 4th Edition, PPVT; Expressive Vocabulary Test-2nd Edition, EVT), adaptive functioning (Vineland Adaptive Behavior Scales-2nd Edition) and motor ability (Movement ABC; Purdue Pegboard), as well as behavioral and psychiatric disorders (Child and Adolescent Symptom Inventory, 5th Edition (CASI-5); Child Behavior Checklist (CBCL), Aberrant Behavior Checklist (ABC)). Medical diagnoses were assessed using the SSC medical history interview98 and by physical examination conducted by a developmental pediatrician conducting a standardized medical examination.
Participants undergoing comprehensive phenotypic assessment and published case reports in the literature were scored using an adapted de Vries scale as a proxy for the overall severity of the phenotype75,76. Modifications included the removal of stature and prenatal onset growth retardation, the inclusion of medical and psychiatric diagnoses, revision of weighting of intellectual disability into 3 points, and an increase to a total score of 12. Borderline intellectual disability or general delays were rated with 1 point, mild to moderate intellectual disability scored with 2 points, and severe-profound intellectual disability scored with 3 points. Psychiatric and medical diagnoses were tallied and scored 1 if an individual had one diagnosis in these domains and scored 2 if the child had two or more diagnoses in these domains.
DSM-5 diagnoses included: ASD (299.00), attention-deficit hyperactivity disorders (314.01, 314.00), language disorder (315.39), speech sound disorder (315.39), developmental coordination disorder (315.4), anxiety disorders (309.21, 300.29, 300.01, 300.02, 300.09), behavior disorders (313.81, 312.34, 312.81, 312.9), mood disorders (311.0, 296.99, 300.4), elimination disorders (307.6, 307.7). Intellectual disability (319, 315.8) was not tallied in the DSM-5 diagnosis domain. Medical diagnoses were tallied by system: cardiac, gastrointestinal, genital, neurological, pulmonary, renal, and visual and auditory. In order to not double code diagnoses, microcephaly, macrocephaly and congenital abnormalities were not tallied under the medical diagnoses domain.
Relevant phenotypic data were extracted from published case reports of individuals with DN LGD mutations to increase our power to detect enrichments among patients sharing DN LGD mutations in the same gene. A total of 323 case reports of individuals with DN LGD mutations of interest and relevant data were identified and 215 case reports had sufficient data to incorporate in the de Vries scale. LGD mutations included: ADNP8,17,99, ARID1B17,23,100, CHD217,23,101, CHD86,102, CTNNB115,17,103–105, DYRK1A7,17, FOXP117,106,107, GRIN2B15,17,23,101,108–111, MED13L17,23,112,113, POGZ17,38,114,115, PTEN101,116,117, SCN2A15,17,101,118–122, SETBP117,19,23,123–125, STXBP117,101,126, and TBL1XR117,127.
Network analysis
We investigated modules significantly disrupted in high-functioning autism samples (full-scale IQ > 100). We applied MAGI20 on all of the samples from the ASD probands in SSC with FSIQ above 100. This subset of samples covers over 500 total missense DN mutations and 100 LGD mutations. We applied the MAGI tool for module discovery on these variants utilizing protein-interaction networks, gene co-expression networks and severe mutations reported in control population from ESP (http://evs.gs.washington.edu/EVS/; n = 6,500 individuals). The protein-interaction network used was a combination of networks from HPRD128 and String129 databases and the co-expression network was built using BrainSpan Atlas resource. Note that the exact same training networks were used in our previous analysis for autism module discovery20. The parameters were that the pairwise gene co-expression inside modules be, on average, at least 0.415 and the average protein interaction density be 0.085. MAGI found one module of 40 genes significantly enriched in DN mutations (p < 0.01 using 100 random mutation permutation tests).
Drosophila knockdown models
Drosophila orthologs of the genes of interest were determined using Ensembl, Unigene, and flybase databases130,131. Their expression was knocked down using the UAS-Gal4 system132 to induce conditional RNAi. The panneuronal promoter line w1118; 2xGMR-wIR; elav-Gal4, UAS-Dicer-242 and two independent RNAi constructs per gene were used whenever available (www.vdrc.at)133 that fulfilled stringent specificity criteria (s19 value ≥ 0.98)134. Strains containing identical genetic background to the RNAi constructs (#60000, #60100) were crossed to the driver line and used as controls. No effects in our assays were seen when crossing the ‘40DUAS’ line135 (containing UAS repeats but no functional short hairpin RNA, a potential source for dominant phenotypes due to an integration locus of the VDRC KK library135,136) to our panneuronal Gal4 driver. Flies were cultured according to standard procedures. Experimental randomization was not applicable to the Drosophila experiments in this study.
Drosophila light-off jump reflex habituation assay
The Drosophila light-off jump habituation assay was performed as previously described137. Briefly, flies were reared at 25°C and 70% humidity, in a 12:12h light/dark cycle. For all healthy lines, at least 64 3-to-7-day-old male flies were tested per genotype, in at least two independent experiments. Flies were transferred into individual vials of two 16-unit habituation systems (Aktogen ltd., Hungary) and, after 5 min adaptation, exposed to 100 light-off pulses (15 ms each) with 1-second inter-pulse interval. Their jump responses were recorded by two sensitive microphones placed in each vial. A carefully chosen threshold was applied to distinguish the jump responses from the background noise. Data from 64 individual flies per genotype were collected (two independent experiments) and analyzed by a custom Labview Software (National Instruments). Genotypes were blinded during the experiments and automatically analyzed. Flies that jumped at least once in the first five trials were evaluated for habituation (pre-established criterion). Initial jumping responses to light-off pulse decreased with number of pulses and flies were considered habituated when they stopped jumping for five consecutive trials (no-jump criterion). Habituation was scored as the number of trials to the no-jump criterion (TTC). The main effect of genotype, corrected for testing day and 16-unit system on TTC values, was determined using linear model regression analysis with R statistical software (v.3.0.0).
Drosophila NMJ experiments
Flies were reared at 28°C, 60% humidity and a 12:12h light/dark cycle. Type 1b NMJs of muscle 4 were analyzed. Wandering 3rd instar larvae were collected, dissected and fixed in 3.7% PFA for 30 min. Preparations were rinsed with PBS and permeabilized with 0.3% Triton X-100 in PBS for 2h at room temperature. Discs large protein (Dlg1) was visualized using primary antibody anti-Dlg1 (1:25) (Dlg1-4D6, Developmental Studies Hybridoma Bank) conjugated with the Zenon Alexa Fluor 568 Mouse IgG1 labelling kit (Invitrogen), according to the manufacturer’s protocol. The preparations were incubated for 1.5h at room temperature, extensively washed and mounted in ProLong Gold Antifade Mountant (Thermo Fisher Scientific). Genotypes were blinded during the experiments and automatically analyzed. Based on previous experience38–40 fluorescence images were acquired per genotype using an automated Leica DMI6000B high-content microscope. Morphometric analysis was performed in FIJI138 using the Drosophila NMJ Morphometrics macro139. Resulting images were visually inspected for accurate image segmentation. Inaccurately segmented parameters were excluded as previously described139. NMJ boutons number in hmt4-20 knockdown flies was manually assessed, blinded to genotype, by two independent researchers and averaged to obtain the final counts. Statistical analysis was performed using GraphPad PRISM. Area, perimeter, length, longest branch length, and boutons were analyzed by Student’s t-test. Branches, branching points, and islands were analysed by Mann-Whitney U test.
Statistical analyses
To calculate the significance and penetrance of LGD and MIS30 mutations in ASD/ID, we compared our smMIP data to two control datasets: the first included the 2,867 unaffected sibling controls that were sequenced through the same smMIP pipeline (see above), and the second included mutation data from the ExAC database91 where neuropsychiatric cases were removed (ExAC v0.3), representing 45,376 samples. The .vcf file for these 45,376 ExAC samples was annotated using VEP and CADD (as described above). The ExAC dataset was filtered by the same pipeline as the smMIP data (see above) including only “PASS” variants. Private variants were filtered as an AC = 1 for burden statistics. In order to compare the ExAC control counts to the smMIP data, only LGD and MIS30 events were considered. LGD and MIS30 counts by gene for ExAC can be found in Supplementary Table 18. To calculate the significance of private LGD or MIS30 mutation burden in our smMIP dataset compared to unaffected (ExAC) controls, we performed a simulation by shuffling the labels of private case and control observations 1x106 times and calculated the probability of observing at least the number of LGD or MIS30 events seen among our cases. These p-values were corrected (Benjamini-Hochberg) for the number of genes in the study (n = 208). Penetrance and its confidence bounds were calculated using the model described previously36:
where D = disease, G = genotype (the presence of the specific type of event in the gene), and D̄ = absence of disease. The general population incidence of ID/DD in our cohort was assumed to match that described in Rosenfeld et al.35,36 (P(D) = 5.12%), as our cohort composition has a similar representation of youth onset diseases with an important genetic component with broad exclusion of chromosomal disorders. DN significance was calculated as previously described9 using a statistical framework that considers the length of the gene and divergence between chimpanzee and human. In order to calculate DN significance for MIS30 variants, we modified the model to separately enumerate prior probabilities for CADD < 30 and CADD >= 30 missense sites using CADD v1.3.
To compare clinical phenotypes, a Pearson’s correlation coefficient and p value were used to compare overall ASD versus ID diagnosis by genic event. Phenotypic rates were compared between individuals carrying variants in ASD versus DD genes using a two-tailed Fisher’s exact test.
A linear model regression analysis was performed using 64 flies per genotype collected through two independent experiments for the habituation data calculations. For the NMJ experiments, area, perimeter, length, longest branch length, and boutons were analysed by two-tailed Student’s t-test (degrees of freedom for SRCAP (dom) experiments: Area = 60, Length = 65, Boutons = 73, Perimeter = 60; degrees of freedom for TCF4 (da) experiments: Area = 63, Length = 68, Branches = 68, Branching = 68). Branches, branching points, and islands were analysed by Mann-Whitney U test.
Supplementary Material
Acknowledgments
We thank the individuals and their families for participation in this study. We acknowledge the Vienna Drosophila Resource Center and Bloomington Drosophila stock center (NIH P40OD018537). This research was supported, in part, by the following: Simons Foundation Autism Research Initiative (SFARI 303241) and NIH (R01MH101221) to E.E.E., VIDI and TOP grants (917-96-346, 912-12-109) from the Netherlands Organization for Scientific Research and Horizon 2020 Marie Sklodowska-Curie European Training Network (MiND, 643051) to A.S., the NHGRI Interdisciplinary Training in Genome Science Grant (T32HG00035) to H.A.F.S. and T.N.T., Australian NHMRC grants 1091593 and 1041920 and Channel 7 Children’s Research Foundation support to J.G., the National Basic Research Program of China (2012CB517900) and the National Natural Science Foundation of China (81330027, 81525007 and 31400919) to K.X., the China Scholarship Council (201406370028) and the Fundamental Research Funds for the Central Universities (2012zzts110) to T.W., the National Health and Medical Research Council of Australia Project Grants (556759) and (1044175) to I.E.S., P.J.L. and M.B.D., Practitioner Fellowship (1006110) to I.E.S., grants from the Jack Brockhoff Foundation and Perpetual Trustees, the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS, the Swedish brain foundation, the Swedish Research Council, the Stockholm County Council, grants (KL2TR00099 and 1KL2TR001444) from the University of California, San Diego Clinical and Translational Research Institute to T.P., and the Research Fund - Flanders (FWO) to R.F.K. and G.V.D.W. We are grateful to all of the families at the participating Simons Simplex Collection (SSC) sites, as well as the principal investigators (A. Beaudet, R. Bernier, J. Constantino, E. Cook, E. Fombonne, D. Geschwind, R. Goin-Kochel, E. Hanson, D. Grice, A. Klin, D. Ledbetter, C. Lord, C. Martin, D. Martin, R. Maxim, J. Miles, O. Ousley, K. Pelphrey, B. Peterson, J. Piggot, C. Saulnier, M. State, W. Stone, J. Sutcliffe, C. Walsh, Z. Warren, E. Wijsman). We appreciate obtaining access to phenotypic data on SFARI Base. We gratefully acknowledge the resources provided by the Autism Genetic Resource Exchange (AGRE) Consortium and the participating AGRE families. AGRE is a program of Autism Speaks and is supported, in part, by grant 1U24MH081810 from the National Institute of Mental Health to Clara M. Lajonchere (PI). Approved researchers can obtain the SSC population dataset described in this study (http://sfari.org/resources/simons-simplex-collection) by applying at https://base.sfari.org. We thank N. Brown, K. Pereira, T. Vick, T. Desai, C. Green, A. L. Doebley, and L. Grillo for their valuable contributions as well as T. Brown for assistance in editing this manuscript. H.P. is a Senior Clinical Investigator of The Research Foundation-Flanders (FWO). E.E.E. is an investigator of the Howard Hughes Medical Institute.
Abbreviations
- ASD
autism spectrum disorders
- CNV
copy number variant
- ID
intellectual disability
- DD
developmental delay
- NDD
neurodevelopmental disorder
- DN
de novo
- LGD
likely gene-disruptive
- smMIP
single-molecule molecular inversion probe
- CADD
combined annotation dependent depletion
- MIS30
missense mutations with a CADD score > 30
- SSC
Simons Simplex Collection
- FDR
false discovery rate
- NMJ
neuromuscular junction
- TTC
trials to no-jump criterion
Footnotes
DATA AVAILABILITY
The smMIP sequencing data for this study can be downloaded from the NIMH data repository National Database for Autism Research (NDAR) at http://dx.doi.org/10.5072/1324821 and is available to all qualified researchers after data use certification. The URLs for data utilized herein are as follows: NHLBI Exome Sequencing Project (ESP) Exome Variant Server, http://evs.gs.washington.edu/EVS; UCSC Genome Browser, http://genome.ucsc.edu; MIPgen, http://shendurelab.github.io/MIPGEN/; CADD Score, http://cadd.gs.washington.edu; NCBI Gene, http://www.ncbi.nlm.nih.gov/gene; Exome Aggregation Consortium (ExAC), http://exac.broadinstitute.org.
AUTHOR CONTRIBUTIONS
E.E.E., H.A.F.S., B.X., and B.P.C. designed the study; H.A.F.S., B.X., T.W., K.H., L.V., and J.L. performed the experiments; B.P.C. helped with smMIP design and data analysis; F.H. performed the gene network analysis; R.A.B., J.G., and S.T. analyzed the patient data; B.X., M.F., B.H., and A.C.N. performed and analyzed the Drosophila data; other authors participated in the sample collection and DNA extraction and/or preparation. E.E.E., H.A.F.S., B.P.C., B.X., A.S., M.F., and R.A.B. wrote the manuscript with input from all authors.
CONFLICT OF INTERESTS
E.E.E. is on the scientific advisory board (SAB) of DNAnexus, Inc. and was an SAB member of Pacific Biosciences, Inc. (2009–2013) and SynapDx Corp. (2011–2013); E.E.E. is a consultant for Kunming University of Science and Technology (KUST) as part of the 1000 China Talent Program.
References
- 1.Diagnostic and statistical manual of mental disorders. 5. American Psychiatric Association; 2013. [Google Scholar]
- 2.Posthuma D, Polderman TJ. What have we learned from recent twin studies about the etiology of neurodevelopmental disorders? Curr Opin Neurol. 2013;26:111–21. doi: 10.1097/WCO.0b013e32835f19c3. [DOI] [PubMed] [Google Scholar]
- 3.Torres F, Barbosa M, Maciel P. Recurrent copy number variations as risk factors for neurodevelopmental disorders: critical overview and analysis of clinical implications. J Med Genet. 2015 doi: 10.1136/jmedgenet-2015-103366. [DOI] [PubMed] [Google Scholar]
- 4.Matson JL, Shoemaker M. Intellectual disability and its relationship to autism spectrum disorders. Res Dev Disabil. 2009;30:1107–14. doi: 10.1016/j.ridd.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 5.Stessman HA, Bernier R, Eichler EE. A genotype-first approach to defining the subtypes of a complex disease. Cell. 2014;156:872–7. doi: 10.1016/j.cell.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernier R, et al. Disruptive CHD8 Mutations Define a Subtype of Autism Early in Development. Cell. 2014;158:263–76. doi: 10.1016/j.cell.2014.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Bon BW, et al. Disruptive de novo mutations of DYRK1A lead to a syndromic form of autism and ID. Mol Psychiatry. 2016;21:126–32. doi: 10.1038/mp.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Helsmoortel C, et al. A SWI/SNF-related autism syndrome caused by de novo mutations in ADNP. Nat Genet. 2014;46:380–4. doi: 10.1038/ng.2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Roak BJ, et al. Multiplex targeted sequencing identifies recurrently mutated genes in autism spectrum disorders. Science. 2012;338:1619–22. doi: 10.1126/science.1227764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hiatt JB, Pritchard CC, Salipante SJ, O’Roak BJ, Shendure J. Single molecule molecular inversion probes for targeted, high-accuracy detection of low-frequency variation. Genome Res. 2013;23:843–54. doi: 10.1101/gr.147686.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Roak BJ, et al. Recurrent de novo mutations implicate novel genes underlying simplex autism risk. Nat Commun. 2014;5:5595. doi: 10.1038/ncomms6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iossifov I, et al. The contribution of de novo coding mutations to autism spectrum disorder. Nature. 2014;515:216–21. doi: 10.1038/nature13908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krumm N, et al. Excess of rare, inherited truncating mutations in autism. Nat Genet. 2015;47:582–8. doi: 10.1038/ng.3303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Rubeis S, et al. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature. 2014;515:209–15. doi: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Ligt J, et al. Diagnostic exome sequencing in persons with severe intellectual disability. N Engl J Med. 2012;367:1921–9. doi: 10.1056/NEJMoa1206524. [DOI] [PubMed] [Google Scholar]
- 16.Rauch A, et al. Range of genetic mutations associated with severe non-syndromic sporadic intellectual disability: an exome sequencing study. Lancet. 2012;380:1674–82. doi: 10.1016/S0140-6736(12)61480-9. [DOI] [PubMed] [Google Scholar]
- 17.Deciphering Developmental Disorders, S. Large-scale discovery of novel genetic causes of developmental disorders. Nature. 2015;519:223–8. doi: 10.1038/nature14135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Turner TN, et al. denovo-db: a compendium of human de novo variants. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Coe BP, et al. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46:1063–71. doi: 10.1038/ng.3092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hormozdiari F, Penn O, Borenstein E, Eichler EE. The discovery of integrated gene networks for autism and related disorders. Genome Res. 2015;25:142–54. doi: 10.1101/gr.178855.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang T, et al. De novo genic mutations among a Chinese autism spectrum disorder cohort. Nat Commun. 2016;7:13316. doi: 10.1038/ncomms13316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Turner TN, et al. Genome Sequencing of Autism-Affected Families Reveals Disruption of Putative Noncoding Regulatory DNA. Am J Hum Genet. 2016;98:58–74. doi: 10.1016/j.ajhg.2015.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamdan FF, et al. De novo mutations in FOXP1 in cases with intellectual disability, autism, and language impairment. Am J Hum Genet. 2010;87:671–8. doi: 10.1016/j.ajhg.2010.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ba W, et al. TRIO loss of function is associated with mild intellectual disability and affects dendritic branching and synapse function. Hum Mol Genet. 2016;25:892–902. doi: 10.1093/hmg/ddv618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han S, et al. Autistic-like behaviour in Scn1a+/− mice and rescue by enhanced GABA-mediated neurotransmission. Nature. 2012;489:385–90. doi: 10.1038/nature11356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Witteveen JS, et al. Haploinsufficiency of MeCP2-interacting transcriptional co-repressor SIN3A causes mild intellectual disability by affecting the development of cortical integrity. Nat Genet. 2016;48:877–87. doi: 10.1038/ng.3619. [DOI] [PubMed] [Google Scholar]
- 27.Shoubridge C, et al. Mutations in the guanine nucleotide exchange factor gene IQSEC2 cause nonsyndromic intellectual disability. Nat Genet. 2010;42:486–8. doi: 10.1038/ng.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chan CB, et al. PIKE is essential for oligodendroglia development and CNS myelination. Proc Natl Acad Sci U S A. 2014;111:1993–8. doi: 10.1073/pnas.1318185111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McNeill EM, et al. Nav2 hypomorphic mutant mice are ataxic and exhibit abnormalities in cerebellar development. Dev Biol. 2011;353:331–43. doi: 10.1016/j.ydbio.2011.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stray-Pedersen A, et al. Biallelic Mutations in UNC80 Cause Persistent Hypotonia, Encephalopathy, Growth Retardation, and Severe Intellectual Disability. Am J Hum Genet. 2016;98:202–9. doi: 10.1016/j.ajhg.2015.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turner TN, et al. Loss of delta-catenin function in severe autism. Nature. 2015;520:51–6. doi: 10.1038/nature14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sanders SJ, et al. Insights into Autism Spectrum Disorder Genomic Architecture and Biology from 71 Risk Loci. Neuron. 2015;87:1215–33. doi: 10.1016/j.neuron.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rope AF, et al. Using VAAST to identify an X-linked disorder resulting in lethality in male infants due to N-terminal acetyltransferase deficiency. Am J Hum Genet. 2011;89:28–43. doi: 10.1016/j.ajhg.2011.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liszczak G, et al. Molecular basis for N-terminal acetylation by the heterodimeric NatA complex. Nat Struct Mol Biol. 2013;20:1098–105. doi: 10.1038/nsmb.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baird PA, Anderson TW, Newcombe HB, Lowry RB. Genetic disorders in children and young adults: a population study. Am J Hum Genet. 1988;42:677–93. [PMC free article] [PubMed] [Google Scholar]
- 36.Rosenfeld JA, Coe BP, Eichler EE, Cuckle H, Shaffer LG. Estimates of penetrance for recurrent pathogenic copy-number variations. Genet Med. 2013;15:478–81. doi: 10.1038/gim.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen EY, et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128. doi: 10.1186/1471-2105-14-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stessman HA, et al. Disruption of POGZ Is Associated with Intellectual Disability and Autism Spectrum Disorders. Am J Hum Genet. 2016;98:541–52. doi: 10.1016/j.ajhg.2016.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Esmaeeli-Nieh S, et al. BOD1 Is Required for Cognitive Function in Humans and Drosophila. PLoS Genet. 2016;12:e1006022. doi: 10.1371/journal.pgen.1006022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lugtenberg D, et al. De novo loss-of-function mutations in WAC cause a recognizable intellectual disability syndrome and learning deficits in Drosophila. Eur J Hum Genet. 2016;24:1145–53. doi: 10.1038/ejhg.2015.282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kleefstra T, et al. Disruption of an EHMT1-associated chromatin-modification module causes intellectual disability. Am J Hum Genet. 2012;91:73–82. doi: 10.1016/j.ajhg.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Bon BW, et al. CEP89 is required for mitochondrial metabolism and neuronal function in man and fly. Hum Mol Genet. 2013;22:3138–51. doi: 10.1093/hmg/ddt170. [DOI] [PubMed] [Google Scholar]
- 43.Willemsen MH, et al. GATAD2B loss-of-function mutations cause a recognisable syndrome with intellectual disability and are associated with learning deficits and synaptic undergrowth in Drosophila. J Med Genet. 2013;50:507–14. doi: 10.1136/jmedgenet-2012-101490. [DOI] [PubMed] [Google Scholar]
- 44.Schmid S, Wilson DA, Rankin CH. Habituation mechanisms and their importance for cognitive function. Front Integr Neurosci. 2014;8:97. doi: 10.3389/fnint.2014.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kleinhans NM, et al. Reduced neural habituation in the amygdala and social impairments in autism spectrum disorders. Am J Psychiatry. 2009;166:467–75. doi: 10.1176/appi.ajp.2008.07101681. [DOI] [PubMed] [Google Scholar]
- 46.Dinstein I, et al. Unreliable evoked responses in autism. Neuron. 2012;75:981–91. doi: 10.1016/j.neuron.2012.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pellicano E, Rhodes G, Calder AJ. Reduced gaze aftereffects are related to difficulties categorising gaze direction in children with autism. Neuropsychologia. 2013;51:1504–9. doi: 10.1016/j.neuropsychologia.2013.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ethridge LE, et al. Reduced habituation of auditory evoked potentials indicate cortical hyper-excitability in Fragile X Syndrome. Transl Psychiatry. 2016;6:e787. doi: 10.1038/tp.2016.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cascio CJ, Woynaroski T, Baranek GT, Wallace MT. Toward an interdisciplinary approach to understanding sensory function in autism spectrum disorder. Autism Res. 2016;9:920–5. doi: 10.1002/aur.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramaswami M. Network plasticity in adaptive filtering and behavioral habituation. Neuron. 2014;82:1216–29. doi: 10.1016/j.neuron.2014.04.035. [DOI] [PubMed] [Google Scholar]
- 51.Tartaglia M, et al. Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet. 2001;29:465–8. doi: 10.1038/ng772. [DOI] [PubMed] [Google Scholar]
- 52.Iossifov I, et al. Low load for disruptive mutations in autism genes and their biased transmission. Proc Natl Acad Sci U S A. 2015;112:E5600–7. doi: 10.1073/pnas.1516376112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sugiura N, Patel RG, Corriveau RA. N-methyl-D-aspartate receptors regulate a group of transiently expressed genes in the developing brain. J Biol Chem. 2001;276:14257–63. doi: 10.1074/jbc.M100011200. [DOI] [PubMed] [Google Scholar]
- 54.Myklebust LM, et al. Biochemical and cellular analysis of Ogden syndrome reveals downstream Nt-acetylation defects. Hum Mol Genet. 2015;24:1956–76. doi: 10.1093/hmg/ddu611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Homsy J, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262–1266. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Bokhoven H. Genetic and epigenetic networks in intellectual disabilities. Annu Rev Genet. 2011;45:81–104. doi: 10.1146/annurev-genet-110410-132512. [DOI] [PubMed] [Google Scholar]
- 57.Zhu T, et al. Histone methyltransferase Ash1L mediates activity-dependent repression of neurexin-1alpha. Sci Rep. 2016;6:26597. doi: 10.1038/srep26597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Griswold AJ, et al. Targeted massively parallel sequencing of autism spectrum disorder-associated genes in a case control cohort reveals rare loss-of-function risk variants. Mol Autism. 2015;6:43. doi: 10.1186/s13229-015-0034-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rhodes CT, et al. Cross-species Analyses Unravel the Complexity of H3K27me3 and H4K20me3 in the Context of Neural Stem Progenitor Cells. Neuroepigenetics. 2016;6:10–25. doi: 10.1016/j.nepig.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Courchesne E, et al. Unusual brain growth patterns in early life in patients with autistic disorder: an MRI study. Neurology. 2001;57:245–54. doi: 10.1212/wnl.57.2.245. [DOI] [PubMed] [Google Scholar]
- 61.Shen MD, et al. Early brain enlargement and elevated extra-axial fluid in infants who develop autism spectrum disorder. Brain. 2013;136:2825–35. doi: 10.1093/brain/awt166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schumann CM, et al. Longitudinal magnetic resonance imaging study of cortical development through early childhood in autism. J Neurosci. 2010;30:4419–27. doi: 10.1523/JNEUROSCI.5714-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Redcay E, Courchesne E. When is the brain enlarged in autism? A meta-analysis of all brain size reports. Biol Psychiatry. 2005;58:1–9. doi: 10.1016/j.biopsych.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 64.Marchetto MC, et al. Altered proliferation and networks in neural cells derived from idiopathic autistic individuals. Mol Psychiatry. 2016 doi: 10.1038/mp.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sugathan A, et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc Natl Acad Sci U S A. 2014;111:E4468–77. doi: 10.1073/pnas.1405266111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cotney J, et al. The autism-associated chromatin modifier CHD8 regulates other autism risk genes during human neurodevelopment. Nat Commun. 2015;6:6404. doi: 10.1038/ncomms7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Courchesne E, et al. Neuron number and size in prefrontal cortex of children with autism. JAMA. 2011;306:2001–10. doi: 10.1001/jama.2011.1638. [DOI] [PubMed] [Google Scholar]
- 68.Stoner R, et al. Patches of disorganization in the neocortex of children with autism. N Engl J Med. 2014;370:1209–19. doi: 10.1056/NEJMoa1307491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chow ML, et al. Age-dependent brain gene expression and copy number anomalies in autism suggest distinct pathological processes at young versus mature ages. PLoS Genet. 2012;8:e1002592. doi: 10.1371/journal.pgen.1002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Pramparo T, et al. Cell cycle networks link gene expression dysregulation, mutation, and brain maldevelopment in autistic toddlers. Mol Syst Biol. 2015;11:841. doi: 10.15252/msb.20156108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Geschwind DH, et al. The autism genetic resource exchange: a resource for the study of autism and related neuropsychiatric conditions. Am J Hum Genet. 2001;69:463–6. doi: 10.1086/321292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Buxbaum JD, et al. The Autism Simplex Collection: an international, expertly phenotyped autism sample for genetic and phenotypic analyses. Mol Autism. 2014;5:34. doi: 10.1186/2040-2392-5-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ardlie KG, et al. The Genotype-Tissue Expression (GTEx) pilot analysis: Multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kircher M, et al. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–5. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lek M, et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Feenstra I, et al. Balanced into array: genome-wide array analysis in 54 patients with an apparently balanced de novo chromosome rearrangement and a meta-analysis. Eur J Hum Genet. 2011;19:1152–60. doi: 10.1038/ejhg.2011.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vulto-van Silfhout AT, et al. Clinical significance of de novo and inherited copy-number variation. Hum Mutat. 2013;34:1679–87. doi: 10.1002/humu.22442. [DOI] [PubMed] [Google Scholar]
- 94.de Vries BB, et al. Clinical studies on submicroscopic subtelomeric rearrangements: a checklist. J Med Genet. 2001;38:145–50. doi: 10.1136/jmg.38.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lord C, Rutter M, DiLavore PC, Risi S. Autism Diagnostic Observation Schedule. Western Psychological Services; 2001. [Google Scholar]
- 96.Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–85. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- 97.Elliott CD. Differential Ability Scales (2nd ed.): Introductory and technical manual. Harcourt Assessment; San Antonio, TX: 2007. [Google Scholar]
- 98.Fischbach GD, Lord C. The Simons Simplex Collection: a resource for identification of autism genetic risk factors. Neuron. 2010;68:192–5. doi: 10.1016/j.neuron.2010.10.006. [DOI] [PubMed] [Google Scholar]
- 99.Pescosolido MF, et al. Expansion of the clinical phenotype associated with mutations in activity-dependent neuroprotective protein. J Med Genet. 2014;51:587–9. doi: 10.1136/jmedgenet-2014-102444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Hoyer J, et al. Haploinsufficiency of ARID1B, a member of the SWI/SNF-a chromatin-remodeling complex, is a frequent cause of intellectual disability. Am J Hum Genet. 2012;90:565–72. doi: 10.1016/j.ajhg.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Epi KC, et al. De novo mutations in epileptic encephalopathies. Nature. 2013;501:217–21. doi: 10.1038/nature12439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Merner N, et al. A de novo frameshift mutation in chromodomain helicase DNA-binding domain 8 (CHD8): A case report and literature review. Am J Med Genet A. 2016;170A:1225–35. doi: 10.1002/ajmg.a.37566. [DOI] [PubMed] [Google Scholar]
- 103.Kuechler A, et al. De novo mutations in beta-catenin (CTNNB1) appear to be a frequent cause of intellectual disability: expanding the mutational and clinical spectrum. Human Genetics. 2015;134:97–109. doi: 10.1007/s00439-014-1498-1. [DOI] [PubMed] [Google Scholar]
- 104.Tucci V, et al. Dominant beta-catenin mutations cause intellectual disability with recognizable syndromic features. J Clin Invest. 2014;124:1468–82. doi: 10.1172/JCI70372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Winczewska-Wiktor A, et al. A de novo CTNNB1 nonsense mutation associated with syndromic atypical hyperekplexia, microcephaly and intellectual disability: a case report. BMC Neurol. 2016;16:35. doi: 10.1186/s12883-016-0554-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lozano R, Vino A, Lozano C, Fisher SE, Deriziotis P. A de novo FOXP1 variant in a patient with autism, intellectual disability and severe speech and language impairment. Eur J Hum Genet. 2015;23:1702–7. doi: 10.1038/ejhg.2015.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sollis E, et al. Identification and functional characterization of de novo FOXP1 variants provides novel insights into the etiology of neurodevelopmental disorder. Hum Mol Genet. 2016;25:546–57. doi: 10.1093/hmg/ddv495. [DOI] [PubMed] [Google Scholar]
- 108.Adams DR, et al. Three rare diseases in one Sib pair: RAI1, PCK1, GRIN2B mutations associated with Smith-Magenis Syndrome, cytosolic PEPCK deficiency and NMDA receptor glutamate insensitivity. Mol Genet Metab. 2014;113:161–70. doi: 10.1016/j.ymgme.2014.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Endele S, et al. Mutations in GRIN2A and GRIN2B encoding regulatory subunits of NMDA receptors cause variable neurodevelopmental phenotypes. Nat Genet. 2010;42:1021–6. doi: 10.1038/ng.677. [DOI] [PubMed] [Google Scholar]
- 110.Freunscht I, et al. Behavioral phenotype in five individuals with de novo mutations within the GRIN2B gene. Behav Brain Funct. 2013;9:20. doi: 10.1186/1744-9081-9-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Lemke JR, et al. GRIN2B mutations in West syndrome and intellectual disability with focal epilepsy. Ann Neurol. 2014;75:147–54. doi: 10.1002/ana.24073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Cafiero C, et al. Novel de novo heterozygous loss-of-function variants in MED13L and further delineation of the MED13L haploinsufficiency syndrome. Eur J Hum Genet. 2015;23:1499–504. doi: 10.1038/ejhg.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.van Haelst MM, et al. Further confirmation of the MED13L haploinsufficiency syndrome. Eur J Hum Genet. 2015;23:135–8. doi: 10.1038/ejhg.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Fukai R, et al. A case of autism spectrum disorder arising from a de novo missense mutation in POGZ. J Hum Genet. 2015;60:277–9. doi: 10.1038/jhg.2015.13. [DOI] [PubMed] [Google Scholar]
- 115.White J, et al. POGZ truncating alleles cause syndromic intellectual disability. Genome Med. 2016;8:3. doi: 10.1186/s13073-015-0253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Busa T, et al. Clinical presentation of PTEN mutations in childhood in the absence of family history of Cowden syndrome. Eur J Paediatr Neurol. 2015;19:188–92. doi: 10.1016/j.ejpn.2014.11.012. [DOI] [PubMed] [Google Scholar]
- 117.Buxbaum JD, et al. Mutation screening of the PTEN gene in patients with autism spectrum disorders and macrocephaly. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:484–91. doi: 10.1002/ajmg.b.30493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Baasch AL, et al. Exome sequencing identifies a de novo SCN2A mutation in a patient with intractable seizures, severe intellectual disability, optic atrophy, muscular hypotonia, and brain abnormalities. Epilepsia. 2014;55:e25–9. doi: 10.1111/epi.12554. [DOI] [PubMed] [Google Scholar]
- 119.Dhamija R, Wirrell E, Falcao G, Kirmani S, Wong-Kisiel LC. Novel de novo SCN2A mutation in a child with migrating focal seizures of infancy. Pediatr Neurol. 2013;49:486–8. doi: 10.1016/j.pediatrneurol.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 120.Dimassi S, et al. Whole-exome sequencing improves the diagnosis yield in sporadic infantile spasm syndrome. Clin Genet. 2016;89:198–204. doi: 10.1111/cge.12636. [DOI] [PubMed] [Google Scholar]
- 121.Nakamura K, et al. Clinical spectrum of SCN2A mutations expanding to Ohtahara syndrome. Neurology. 2013;81:992–8. doi: 10.1212/WNL.0b013e3182a43e57. [DOI] [PubMed] [Google Scholar]
- 122.Tavassoli T, et al. De novo SCN2A splice site mutation in a boy with Autism spectrum disorder. BMC Med Genet. 2014;15:35. doi: 10.1186/1471-2350-15-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Herenger Y, et al. Long term follow up of two independent patients with Schinzel-Giedion carrying SETBP1 mutations. Eur J Med Genet. 2015;58:479–87. doi: 10.1016/j.ejmg.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 124.Miyake F, et al. West syndrome in a patient with Schinzel-Giedion syndrome. J Child Neurol. 2015;30:932–6. doi: 10.1177/0883073814541468. [DOI] [PubMed] [Google Scholar]
- 125.Takeuchi A, et al. Progressive brain atrophy in Schinzel-Giedion syndrome with a SETBP1 mutation. Eur J Med Genet. 2015;58:369–71. doi: 10.1016/j.ejmg.2015.05.006. [DOI] [PubMed] [Google Scholar]
- 126.Stamberger H, et al. STXBP1 encephalopathy: A neurodevelopmental disorder including epilepsy. Neurology. 2016;86:954–62. doi: 10.1212/WNL.0000000000002457. [DOI] [PubMed] [Google Scholar]
- 127.Heinen CA, et al. A specific mutation in TBL1XR1 causes Pierpont syndrome. J Med Genet. 2016;53:330–7. doi: 10.1136/jmedgenet-2015-103233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Keshava Prasad TS, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Szklarczyk D, et al. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561–8. doi: 10.1093/nar/gkq973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Wheeler DL, et al. Database resources of the National Center for Biotechnology. Nucleic Acids Res. 2003;31:28–33. doi: 10.1093/nar/gkg033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Attrill H, et al. FlyBase: establishing a Gene Group resource for Drosophila melanogaster. Nucleic Acids Res. 2016;44:D786–92. doi: 10.1093/nar/gkv1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 133.Dietzl G, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature. 2007;448:151–6. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 134.Oortveld MA, et al. Human intellectual disability genes form conserved functional modules in Drosophila. PLoS Genet. 2013;9:e1003911. doi: 10.1371/journal.pgen.1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Green EW, Fedele G, Giorgini F, Kyriacou CP. A Drosophila RNAi collection is subject to dominant phenotypic effects. Nat Methods. 2014;11:222–3. doi: 10.1038/nmeth.2856. [DOI] [PubMed] [Google Scholar]
- 136.Vissers JH, Manning SA, Kulkarni A, Harvey KF. A Drosophila RNAi library modulates Hippo pathway-dependent tissue growth. Nat Commun. 2016;7:10368. doi: 10.1038/ncomms10368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kramer JM, et al. Epigenetic regulation of learning and memory by Drosophila EHMT/G9a. PLoS Biol. 2011;9:e1000569. doi: 10.1371/journal.pbio.1000569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Nijhof B, et al. A New Fiji-Based Algorithm That Systematically Quantifies Nine Synaptic Parameters Provides Insights into Drosophila NMJ Morphometry. PLoS Comput Biol. 2016;12:e1004823. doi: 10.1371/journal.pcbi.1004823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.