Abstract
IMPORTANCE
In neoadjuvant trials, treatment of human epidermal growth factor receptor 2 (HER2)-positive breast cancers with dual HER2 blockade resulted in increased pathologic complete response (pCR) rates compared with each targeted agent alone. Amplification and/or overexpression of HER2 currently remains the only biomarker for therapeutic decisions, but it is insufficient to explain the heterogeneous response to anti-HER2 agents.
OBJECTIVE
To investigate the ability of clinically and biologically relevant genes and gene signatures (GSs) measured by RNA sequencing to predict the efficacy of anti-HER2 agents.
DESIGN, SETTING, AND PARTICIPANTS
The neoadjuvant NeoALTTO trial randomized 455 women with HER2-positive early-stage breast cancer to trastuzumab, lapatinib, or the combination for 6 weeks followed by the addition of weekly paclitaxel for 12 weeks, followed by 3 cycles of fluorouracil, epirubicin, and cyclophosphamide after surgery. The present substudy, which was planned in the NeoALTTO main protocol, evaluated the association of pretreatment gene expression levels defined using RNA sequencing with pCR and event-free survival (EFS).
MAIN OUTCOMES AND MEASURES
Gene expression–based biomarkers using RNA sequencing were examined for their association with response to anti-HER2 therapy and long-term outcome.
RESULTS
Sequencing data were available for 254 (56%) of the NeoALTTO participants (mean [SD] age of substudy participants, 48.8 [11.2] years). The expression of ERBB2/HER2 was the most significant predictor of pCR, followed by HER2-enriched subtype, ESR1, treatment arm, ER immunohistochemical analysis scores, Genomic Grade Index, immune, proliferation, and AKT/mTOR GSs. Adjusting for clinicopathological variables and treatment arms, ERBB2/HER2, HER2-enriched subtype, ESR1, and Genomic Grade Index remained significant. Immune GSs were associated with higher pCR only in the combination arm (odds ratio, 2.1; 95%CI, 1.2–4.0; interaction test P = .01), while the stroma GSs were significantly associated with higher pCR in the single arms and with lower pCR in the combination arm (odds ratio, 0.46; 95%CI, 0.25–0.84; P = .009). None of the evaluated variables was associated with EFS after correction for multiple testing, but this analysis was underpowered.
CONCLUSIONS AND RELEVANCE
High levels of ERBB2/HER2 and low levels of ESR1 were associated with pCR in all treatment arms. In the combination arm, high expression of immune and stroma GSs were significantly associated with higher and lower pCR rates, respectively, and should be further explored as candidate predictive markers.
TRIAL REGISTRATION
clinicaltrials.gov Identifier: NCT00553358
The combination of trastuzumab with either lapatinib or pertuzumab and chemotherapy has been shown to be effective for the treatment of patients with human epidermal growth factor receptor 2 (HER2)-positive breast cancer. Following the improvement in survival observed in patients with metastatic disease,1,2 the drug combination has been investigated in early breast cancer.3–8 Whereas superior efficacy compared with standard trastuzumab therapy has not been shown yet in the adjuvant setting,7 in neoadjuvant trials the dual blockade generally resulted in increased pathologic complete response (pCR) rates compared with each targeted agent alone.3,4
Despite their clinical activity, anti-HER2 treatments do not exert the same effect on all patients with HER2-positive breast cancer. To date, several candidate predictive biomarkers have been explored, among which are activation of the PI3K pathway,9–13 the presence of a truncated form of HER2 receptor (p95HER2),14,15 HER2 serum levels,16 and tumor infiltrating lymphocytes (TILs).17 These studies have given inconsistent results, and none of these biomarkers has demonstrated clinical utility so far. For the time being, amplification and/or overexpression of HER2 remains the only biomarker for therapeutic decision making,18 even though it has been shown inadequate to explain the heterogeneous response to anti-HER2 agents.19
In this study, we took the unique opportunity to profile the transcriptome of pretreatment frozen lesions obtained from patients enrolled in the NeoALTTO trial,3,20 and attempted to explore biomarkers associated with treatment response and long-term outcome. For this purpose, we defined a series of clinically relevant single genes and gene expression signatures previously associated with either response or resistance to trastuzumab treatment,21–26 and we correlated them with pCR and survival information.
Methods
Patient Population
The NeoALTTO trial was a multicenter, randomized phase 3 trial in which 455 patients with HER2-positive early breast cancer were randomly assigned to receive lapatinib 1500 mg/d, trastuzumab (4 mg/kg loading dose followed by 2 mg/kg), or the combination of lapatinib 1000 mg/d and the same dose of trastuzumab for 6 weeks followed by the addition of paclitaxel (80 mg/m2) for 12 weeks before surgery. Lapatinib doses were reduced during the paclitaxel administration. After surgery, all patients received 3 cycles of fluorouracil, epirubicin, and cyclophosphamide and then continued the same anti-HER2–targetedtherapyas administered in the neoadjuvant setting to complete a total duration of 1 year. Patients were recruited between January 5, 2008, and May 27, 2010. Eligible patients had to have a HER2-positive primary breast cancer with a minimum tumor size of 2 cm and adequate cardiac function. The HER2 status was defined either in local accredited laboratories or in a central laboratory while the estrogen receptor (ER) status was defined according to local guidelines (for additional details refer to the eMethods in the Supplement). The primary end point was pCR according to either the National Surgical Adjuvant Breast and Bowel Project (NSABP) criteria (absence of invasive tumor cells in the breast) or the Food and Drug Administration criteria (absence of invasive tumor cells in the breast and in the axillary lymph nodes). Event-free survival (EFS) was the main secondary end point and was defined as the time from randomization to first event. For women who received surgery for breast cancer, events were defined as breast cancer relapse after surgery, second primary malignant neoplasm, or death without recurrence. For women who did not undergo surgery for breast cancer, events were death during clinical follow-up or noncompletion of any neoadjuvant investigational product because of disease progression. Additional details can be found in the original publications.3,20 In this substudy, we used pCR rates as defined by the NSABP criteria.27
The NeoALTTO trial was approved by the ethics committee and relevant health authorities of all the participating sites. Written informed consent was obtained from all patients at study entry, which also covered future biomarker research. This substudy was part of the NeoALTTO main protocol.
Samples Collection and Processing
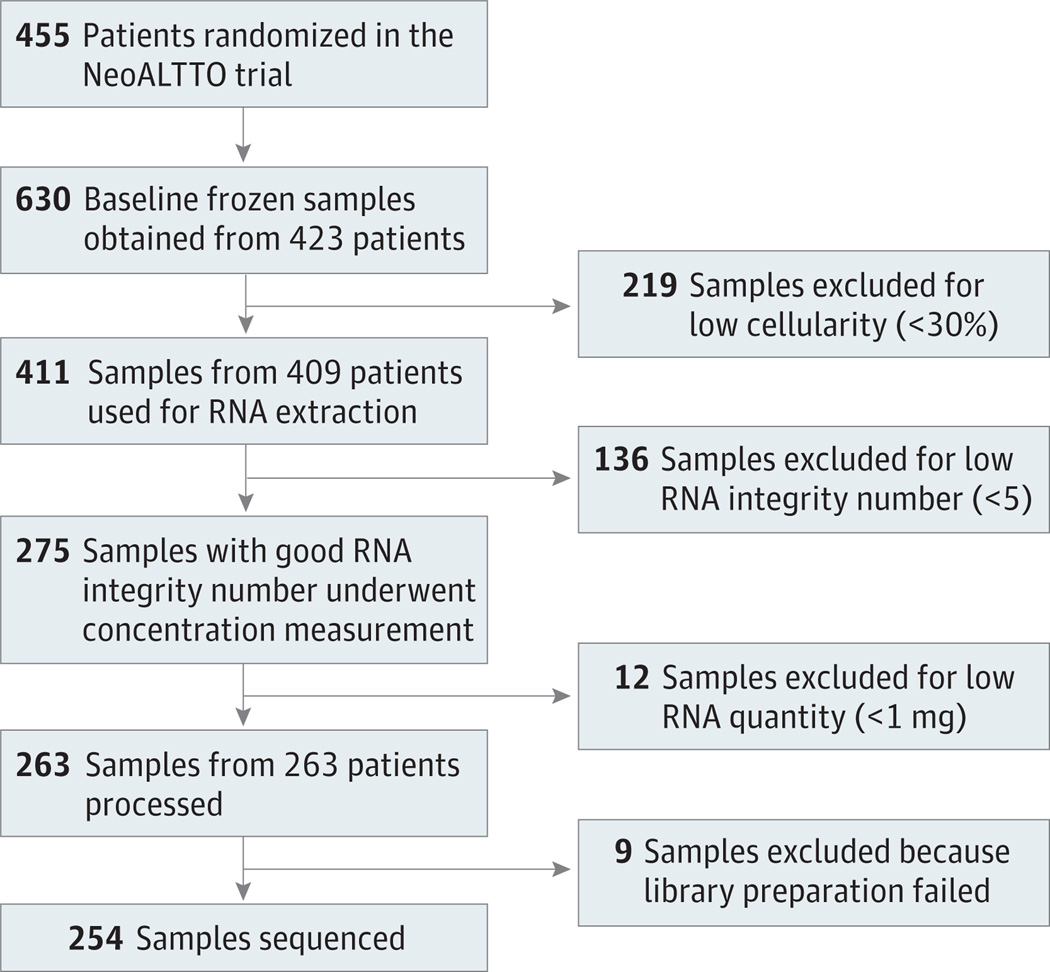
To participate in the trial, patients were asked to provide 2 cores of snap frozen tissue of their primary tumor prior to the initiation of neoadjuvant therapy. At least 1 baseline biopsy was obtained from 423 of the 455 patients enrolled in the trial. As shown in Figure 1, RNA of enough quality and quantity was obtained for 263 patients. From these RNA samples, strand-specific complementary DNA libraries were constructed using the NEB Next Ultra directional RNA library Preparation Kit for Illumina paired-end sequencing on the HiSeq 2500 system (Illumina) following the internal standard operating procedures of GATC Biotech AG. Nine samples failed the library construction step, while the remaining 254 were sequenced. Additional details on the samples processing and sequencing can be found in the eMethods in the Supplement.
Figure 1. NeoALTTO Secondary Analysis Flow Diagram of Patients and Samples.
RNA Sequencing Data Processing
Read pairs were trimmed using Trimmomatic.28 Alignment was performed using STAR.29 The number of reads mapping to each gene was then assessed with the R statistical software30 with the Rsamtools package.31 Fragments per kilobase of transcript per million mapped reads (FPKM) were defined as the number of fragments (1 or both members of a read pair) mapping a gene per kilobase of transcript per million mapped reads, using the most common gene isoform as the transcript. Gene expression levels were corrected for library batch effects using ComBat.32
Besides the expression of the ERBB2/HER2, ESR1, and androgen receptor (AR) genes, we evaluated the following gene expression signatures: 3 immune signatures (Immune1,21 Immune2,22 Immune323); 2 proliferation signatures (Genomic Grade Index [GGI],24 aurka22); an AKT/mammalian target of rapamycin (mTOR) pathway signature25; and 2 stroma signatures (Stroma126; Stroma222). The signature scores were calculated as a weighted sum of the log-expressions of their genes, with gene-specific weights equal to +1 or −1 depending on the direction of their association with the phenotype in the original publication.
The expression levels were made comparable to those of HER2-positive samples from The Cancer Genome Atlas (TCGA)33 by using the cross-studies normalization of the R package genefu34 (R package version 2.3.0). A merged data set was obtained by adding the renormalized NeoALTTO samples to all TCGA samples. PAM50 subtypes were then determined using genefu on the merged data sets.35
Statistical Analysis
The relationship between pCR, EFS, and the expression of genes and gene signatures was assessed using logistic regressions and Cox proportional hazard models adjusted for age (as continuous variable), ER status (positive vs negative), tumor size (≥T3 vs T2), nodal status (N0 vs N1–N3), histological grade (1–2 vs 3), and treatment arm. All statistical analyses were performed using R.30 All interaction and multivariate tests were done using analysis of variance to compare the models with and without the extra term. The reported odds and/or hazard ratios come from the fit of the complete model. Correction for multiple testing was done using the qvalue package.36 P values (after multiple testing correction if needed) less than .05 were considered significant. The segmentation of ESR1 and ERBB2/HER2 expression in 2 groups was done using the C-Means algorithm on each feature separately on the merged TCGA/NeoALTTO data set.
Results
Sequenced Cohort Not Significantly Different From the Whole Study Cohort
A total of 455 patients were enrolled in the NeoALTTO trial3; for 254 of them (55.8%) RNA sequencing data were obtained (Figure 1). Eighty-nine patients (35.0%) were enrolled in the lapatinib arm, 79 (31.1%) in the trastuzumab arm, and 86 (33.9%) in the combination arm. No significant differences in terms of patient characteristics were observed between the substudy and whole NeoALTTO population (eTable 1 in the Supplement).
Clustering of Breast Cancer Samples in Function of Their ESR1 and ERBB2/HER2 Expression Levels
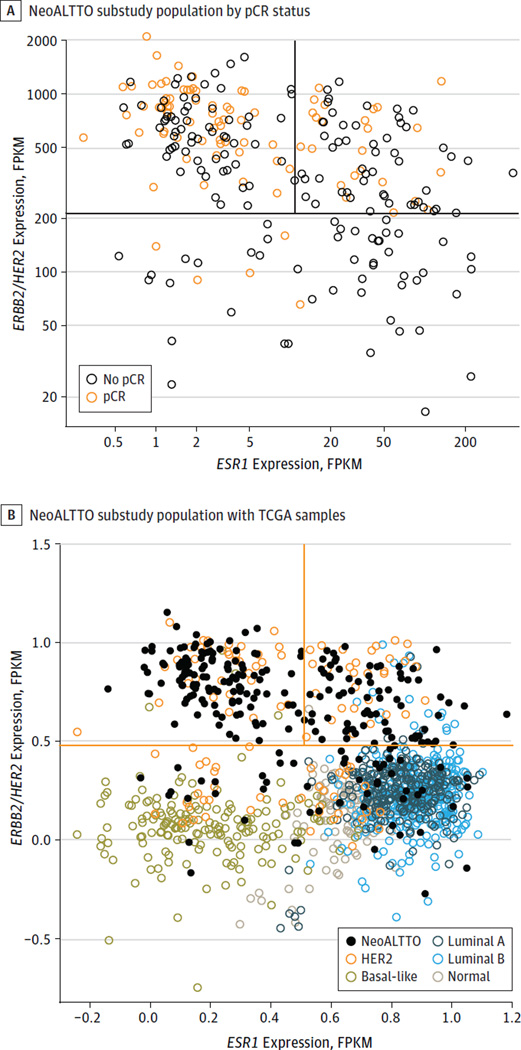
When the samples are plotted in 2 dimensions in function of their mRNA expression levels of ESR1 and ERBB2/HER2 (Figure 2A), they appear to cluster in 3 groups: a high ERBB2/HER2 and low ESR1 group (approximately 45% of the samples), a high ERBB2/HER2 and high ESR1 group (approximately 35% of the samples), and a low ERBB2/HER2 group (approximately 20% of the samples). As shown in Figure 2A, most of the observed pCRs (red dots) cluster in the group of patients with high ERBB2/HER2 and low ESR1 expression levels. In fact, 59 (47%) of these patients had a pCR, while only 24 of 76 (32%) of patients in the high ERBB2/HER2 with high ESR1 group and 5 of 53 (9%) of those in the low ERBB2/HER2 group achieved a pCR (P < .001 for the pCR difference between the 3 groups; P = .04 between high ERBB2/HER2 with low ESR1 and high ERBB2/HER2 with high ESR1 with Fisher exact test). When our samples were plotted together with TCGA samples classified according to their PAM50 profile, roughly 20% of HER2-enriched breast cancers show lower ERBB2/HER2 expression levels, similar to what we found in our cohort (Figure 2B).
Figure 2. Distribution of NeoALTTO Samples and Comparison With Samples From the Cancer Genome Atlas (TCGA).
A, Samples of the NeoALTTO substudy population classified according to their ERBB2/HER2 and ESR1 gene expression levels defined by RNA sequencing and colored according to their pathologic complete response (pCR) status.
B, Samples of the NeoALTTO substudy population shown together with TCGA samples classified according to PAM50. FPKM indicates fragments per kilobase of transcript per million mapped reads.
Comparison Between Standard Testing and RNA Sequencing Data
We also compared the mRNA levels of ERBB2/HER2 and ESR1 with the HER2 and ER status defined by immunohistochemical analysis (IHC) and/or fluorescence in situ hybridization (FISH) before study entrance. As shown in eFigure 1 in the Supplement, the expression levels of ERBB2/HER2 showed a weak correlation with the percentage of stained cells at IHC (ρ = 0.25) and the FISH ratios (ρ = 0.34). Because in the NeoALTTO study the HER2 status could be defined either locally or centrally, we also compared the expression levels of ERBB2/HER2 between the 2 groups. As shown in eFigure 2 in the Supplement, the expression levels of ERBB2/HER2 were significantly higher for samples evaluated centrally compared with locally (P = .02), but both groups contained cases with low ERBB2/HER2 expression levels. When the expression levels of ESR1 were compared between the ER-positive and ER-negative populations as defined by local IHC, even though the expression levels in the 2 groups were different, a proportion of cases were overlapping (eFigure 3 in the Supplement).
pCR Rates According to the PAM50 Classification
We next defined the subtype of our substudy population using the PAM50 classifier. As presented in eTable 2 in the Supplement, 110 (43%) of our population was classified as HER2 enriched. The remaining cases were classified as luminal A (23%), luminal B (16%), basal-like (9%), or normal (8%). Fifty-seven of the 88 (65%) pCRs observed in our substudy were achieved by patients whose tumors were classified as HER2 enriched, while the remainder were distributed among the other 4 subtypes (P < .001).
Clustering of Gene Signatures in a Few Groups
eFigure 4 in the Supplement shows the correlation between the single genes and gene signatures that were analyzed in this study. As expected, a high correlation was observed between the 3 immune-related signatures (ρ = 0.84, 0.89, and 0.92), the 2 proliferation-related signatures (ρ = 0.95), and the 2 stroma-related signatures (ρ = 0.88). Moderate positive correlation was observed between the AKT/mTOR signature and the 2 proliferation-related ones (ρ = 0.49). A moderate, inverse correlation was found between the stroma and the proliferation signatures (ρ = −0.38 to −0.62).
Association of Single Genes, Gene Signatures, Treatment, and Clinicopathologic Parameters With pCR
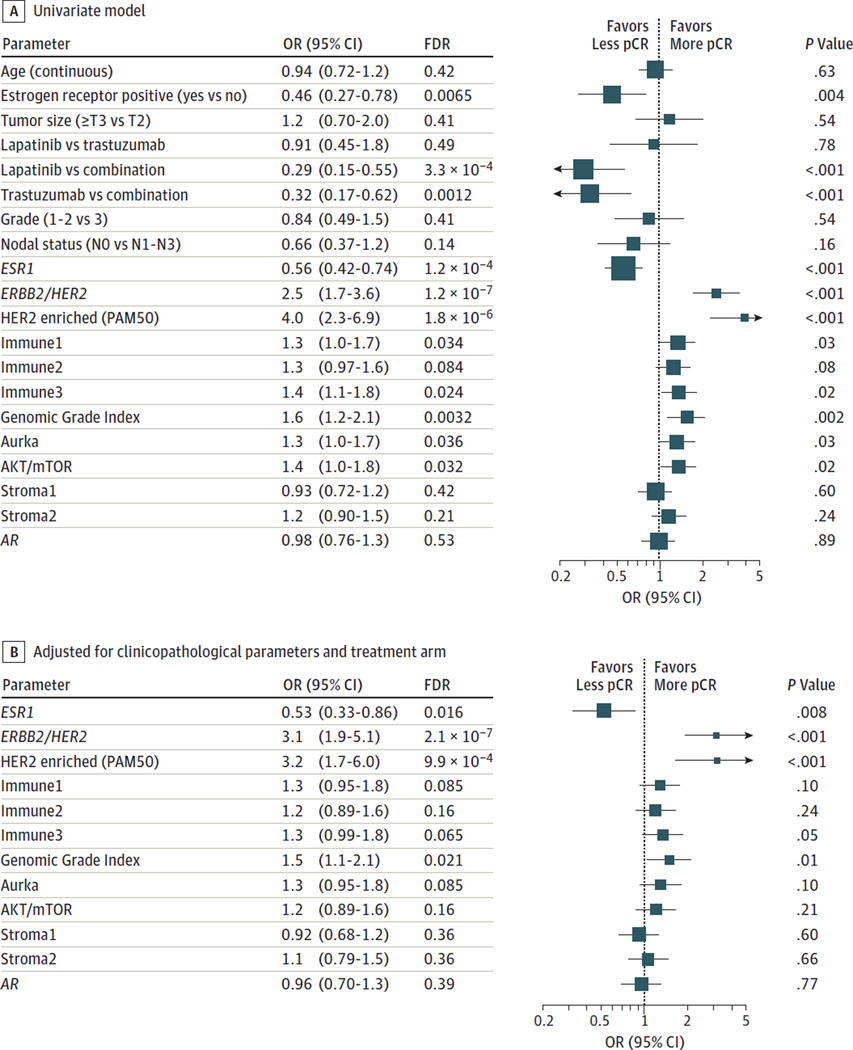
In a univariate model, the association between pCR and single genes, gene signatures, treatment, and clinicopathologic parameters was tested using logistic regression. In the full study cohort (Figure 3A), the main drivers of pCR were ERBB2/HER2 levels, HER2-enriched subtype based on PAM50 classification, ESR1 mRNA levels, ER IHC scores, and the treatment arm. The GGI, proliferation, AKT-mTOR, and 2 immune signatures were also significant at a false discovery rate level of 5%. When each arm was considered separately, immune signatures were associated with pCR only in the combination arm, while the GGI remained significant only in the lapatinib arm (eFigure 5 in the Supplement). One of the stroma signatures was significantly associated with lower pCR in the combination arm.
Figure 3. Association of Single Genes, Gene Expression Signatures, Treatment, and Clinicopathologic Variables With Pathologic Complete Response (pCR).
A, Effect of single genes, gene expression signatures, treatment, and clinicopathologic variables on pCR in a univariate model. B, Effect of single genes and gene expression signatures on pCR adjusting for clinicopathological parameters and treatment arms. A false discovery rate (FDR) < .05 is considered significant. Symbols indicate odds ratio (OR), and error bars, 95% confidence interval (CI). Symbol size is inversely proportional to the size of the CI.
We then assessed the independence of the predictive power of single genes and gene signatures adjusting for clinicopathological parameters and treatment arms (Figure 3B). ESR1, ERBB2/HER2, HER2-enriched subtype, and the GGI were associated with pCR. Interestingly, the level of expression of ESR1 remained significant after accounting for the ER status determined using IHC; the same was found for dichotomized ESR1 expression levels (P = .01). Conversely, ER status measured by IHC was not significant when controlling for ESR1 mRNA expression, both as a continuous (P = .73) and a dichotomized variable (P = .91). The HER2-enriched subtype and the ERBB2/HER2 gene expression levels were independently associated with pCR, probably because the HER2-enriched subtype classification takes into account the levels of expression of the ESR1 gene and proliferation. ERBB2/HER2 expression levels remain predictive even among the high ERBB2/HER2 group as defined in Figure 2A (P = .02).
When adding a treatment interaction term in the model, we noticed no significant differences between the lapatinib and trastuzumab arms (data not shown). We then performed an interaction test considering the combination arm vs the 2 single arms together (eFigure 6 in the Supplement). We observed that the immune signatures were significantly associated with higher pCR rates in the combination arm only (odds ratio, 2.1; 95% CI, 1.2–4.0; interaction test P = .01), while the stroma signatures were significantly associated with higher pCR rates in the single arms and with low pCR rates in the combination arm (odds ratio, 0.46; 95% CI, 0.25–0.84; P = .009). These associations were not significant when considering the combination vs the trastuzumab arm (eFigure 7 in the Supplement). As the effect sizes remain similar, we attribute this loss of significance to the smaller number of patients after the lapatinib arm was removed.
Association of Single Genes, Gene Signatures, Treatment, and Clinicopathologic Parameters With EFS
We then explored the association of single genes, gene signatures, treatment, and clinicopathologic parameters with EFS. In a univariate model, only nodal status and AR expression were associated with EFS but not after correction for multi-testing (eFigure 8 in the Supplement). None of the markers was associated with EFS using a test for interaction with treatment (eFigure 9 in the Supplement). We acknowledge that these analyses were underpowered.
Discussion
This study represents one of the largest analyses from a randomized clinical trial that evaluated the expression of single genes and gene signatures associated with pCR and outcome in patients with HER2-positive breast cancer. Our results confirm that in the neoadjuvant setting, the expression levels of both ERBB2/HER2 and ESR1 genes remain the most important determinants of pCR as compared with standard tests in this patient population. Lower pCR rates were found in the tumors with low ERBB2/HER2 gene expression levels, although we cannot exclude that some of those tumors might be falsely HER2 positive. The predictive value of ERBB2/HER2 expression levels emerged in other neoadjuvant trials using either trastuzumab or its combination with lapatinib or pertuzumab,8,37,38 independently of the technology used including RNA sequencing. Additional evidence, however, is needed before implementation of RNA sequencing in the clinical setting.39
An inverse correlation between the expression levels of the ESR1 gene and pCR was recently observed in the neoadjuvant setting.8 The fact that in our study the level of expression of ESR1 remained significant after accounting for the ER status determined using IHC, and not the opposite, suggests that also in this case ESR1 mRNA levels are more predictive than its protein levels; however, we cannot exclude that this finding is due to the ER testing being performed only in local laboratories in the NeoALTTO trial.
Using a univariate analysis, gene expression signatures representing T-cell–driven immune response and proliferation were significantly associated with pCR. Association between immune signals and response to anti-HER2 treatment has been recently observed in other neoadjuvant8,40 and adjuvant trials.23 The consistent finding in both settings by independent groups places the immune response at the forefront of biomarkers worthy of being applied in clinical practice, as well as for future drug development in the field of HER2-positive breast cancer.17,41,42 In our study, we did not find a significant correlation between immune signatures and EFS, contrary to what was reported for TILs.17 We found a moderate correlation between immune signatures and stromal TILs (ρ = 0.37–0.42), which might explain this discrepancy. A plausible explanation might be that TILs and immune signatures capture different aspects of the host and/or tumor response against cancer. Similar discrepancies were reported by Perez et al23,43 in the adjuvant setting.
Interestingly, in our study, the predictive value of the immune signatures showed an interaction only with the combination arm. The mechanisms explaining the synergy between lapatinib and trastuzumab are not fully elucidated. One of the proposed explanations is that the synergistic inhibitory effect of HER2 signaling determined by the 2 drugs can enhance apoptosis and results in a superior effect of the drug combination compared with each single drug.44,45 Another suggested mechanism is that when used in combination with trastuzumab, lapatinib increases the trastuzumab-mediated antibody-dependent cytotoxicity as it determines accumulations of inactive HER2 receptors at the cell surface.46,47
The 2 stroma signatures that we evaluated showed instead a peculiar behavior as they predicted higher pCR rates in the single arms but lower pCR rates in the combination arm. The signature developed by Farmer and colleagues,26 which captures the activation state of the tumor stroma, predicted poor response to anthracycline-based neoadjuvant chemotherapy in ER-negative patients and in HER2-positive patients treated with chemotherapy only.48 High expression of the PLAU signature was reported by Desmedt et al22 to be associated with poor clinical outcome in patients with HER2-positive, untreated breast cancer. To our knowledge, this is the first time that the predictive value of these signatures has been evaluated in a cohort of patients receiving anti-HER2 treatment, and their opposite role in modulating the response to single vs combined agents needs to be further explored.
In our study, none of the markers that we evaluated (including those associated with pCR) were associated with EFS. The NeoALTTO trial was not originally powered to evaluate the difference between the treatment arms in terms of EFS, and our findings could be weakened by the low statistical power.
Our study has various strengths. It is a preplanned analysis of a randomized phase 3 trial in which the analysis was performed on prospectively collected frozen tissue samples using the latest technology to evaluate gene expression. This strengthens the reliability of the obtained results. On the other hand, only 60% of the collected samples were evaluable, mostly due to low cellularity, which is always a challenge when evaluating tumor biopsies rather than whole surgical specimens. This, however, has not prevented us from finding valuable pCR-associated biomarkers.
Conclusions
The results of this study support the existence of significant molecular heterogeneity among HER2-positive breast cancers and the influence of both estrogen signaling and tumor microenvironment in the response to anti-HER2 therapies. Future studies should take this knowledge into account and aim to determine how these factors could be used to individualize the treatment of patients with HER2-positive disease.
Key Points.
Question
Can we identify biomarkers associated with treatment response in patients with human epidermal growth factor receptor 2 (HER2)-positive early breast cancer treated with neoadjuvant anti-HER2 therapy in the NeoALTTO trial?
Findings
This secondary analysis of the NeoALTTO trial used RNA sequencing data obtained from pretreatment specimens. Across treatment arms, pathological complete response rate was associated with high expression of ERBB2/HER2 and low expression of ESR1. In the combination arm, pathologic complete response rate was associated with high expression of immune gene signatures and low expression of stroma gene signatures.
Meaning
These findings support the relevant role of immune and stroma signals in determining the response to anti-HER2 treatments.
Acknowledgments
Drs Piccart-Gebhart and Sotiriou are co-inventors of the Genomic Grade Index. Dr de Azambuja has received travel grants from GlaxoSmithKline and Roche and honoraria from Roche outside this work. Dr Brase is a Novartis employee. Drs Baselga and Piccart-Gebhart have received personal fees from Roche.
Funding/Support: The NeoALTTO study and this substudy were sponsored by GlaxoSmithKline; lapatinib is an asset of Novartis AG as of March 2, 2015. Dr Venet’s activity is partly supported by a grant of the RégionWallonne.
Role of the Funder/Sponsor: GlaxoSmithKline participated in the collection of biological samples but had no role in the design and conduct of this substudy; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. Novartis reviewed and approved the final version of the manuscript.
Footnotes
Author Contributions: Drs Sotiriou and Venet had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Drs Fumagalli and Venet contributed equally to this work. Drs Loi and Sotiriou are joint senior authors.
Study concept and design: Fumagalli, Venet, de la Peña, Huober, Baselga, Loi, Sotiriou.
Acquisition, analysis, or interpretation of data: Fumagalli, Venet, Ignatiadis, Azim, Maetens, Rothé, Salgado, Bradbury, Pusztai, Harbeck, Gomez, Chang, Coccia-Portugal, Di Cosimo, de Azambuja, Nuciforo, Brase, Piccart-Gebhart, Loi, Sotiriou.
Drafting of the manuscript: Fumagalli, Venet, Azim, Gomez, Loi, Sotiriou.
Critical revision of the manuscript for important intellectual content: Fumagalli, Venet, Ignatiadis, Azim, Maetens, Rothé, Salgado, Bradbury, Pusztai, Harbeck, Chang, Coccia-Portugal, Di Cosimo, de Azambuja, de la Peña, Nuciforo, Brase, Huober, Baselga, Piccart-Gebhart, Loi, Sotiriou.
Statistical analysis: Venet, Bradbury, Loi, Sotiriou.
Obtained funding: Sotiriou.
Administrative, technical, or material support: Venet, Maetens, Rothé, Salgado, Harbeck, Gomez, Chang, Di Cosimo, de Azambuja, de la Peña, Nuciforo, Loi, Sotiriou.
Study supervision: Fumagalli, Ignatiadis, Harbeck, Gomez, de la Peña, Baselga, Piccart-Gebhart, Loi, Sotiriou.
Conflict of Interest Disclosures: No other disclosures are reported.
REFERENCES
- 1.Baselga J, Cortés J, Kim S-B, et al. CLEOPATRA Study Group. Pertuzumab plus trastuzumab plus docetaxel for metastatic breast cancer. N Engl J Med. 2012;366(2):109–119. doi: 10.1056/NEJMoa1113216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blackwell KL, Burstein HJ, Storniolo AM, et al. Overall survival benefit with lapatinib in combination with trastuzumab for patients with human epidermal growth factor receptor 2-positive metastatic breast cancer: final results from the EGF104900 Study. J Clin Oncol. 2012;30(21):2585–2592. doi: 10.1200/JCO.2011.35.6725. [DOI] [PubMed] [Google Scholar]
- 3.Baselga J, Bradbury I, Eidtmann H, et al. NeoALTTO Study Team. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): a randomised, open-label, multicentre, phase 3 trial. Lancet. 2012;379(9816):633–640. doi: 10.1016/S0140-6736(11)61847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gianni L, Pienkowski T, Im YH, et al. Efficacy and safety of neoadjuvant pertuzumab and trastuzumab in women with locally advanced, inflammatory, or early HER2-positive breast cancer (NeoSphere): a randomised multicentre, open-label, phase 2 trial. Lancet Oncol. 2012;13(1):25–32. doi: 10.1016/S1470-2045(11)70336-9. [DOI] [PubMed] [Google Scholar]
- 5.Robidoux A, Tang G, Rastogi P, et al. Lapatinib as a component of neoadjuvant therapy for HER2-positive operable breast cancer (NSABP protocol B-41): an open-label, randomised phase 3 trial. Lancet Oncol. 2013;14(12):1183–1192. doi: 10.1016/S1470-2045(13)70411-X. [DOI] [PubMed] [Google Scholar]
- 6.Schneeweiss A, Chia S, Hickish T, et al. Pertuzumab plus trastuzumab in combination with standard neoadjuvant anthracycline-containing and anthracycline-free chemotherapy regimens in patients with HER2-positive early breast cancer: a randomized phase II cardiac safety study (TRYPHAENA) Ann Oncol. 2013;24(9):2278–2284. doi: 10.1093/annonc/mdt182. [DOI] [PubMed] [Google Scholar]
- 7.Piccart-Gebhart M, Holmes E, Baselga J, et al. Adjuvant lapatinib and trastuzumab for early human epidermal growth factor receptor 2-positive breast cancer: results from the randomized phase III adjuvant lapatinib and/or trastuzumab treatment optimization trial. J Clin Oncol. 2016;34(10):1034–1042. doi: 10.1200/JCO.2015.62.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carey LA, Berry DA, Cirrincione CT, et al. Molecular heterogeneity and response to neoadjuvant human epidermal growth factor receptor 2 targeting in CALGB 40601, a randomized phase III trial of paclitaxel plus trastuzumab with or without lapatinib. J Clin Oncol. 2016;34(6):542–549. doi: 10.1200/JCO.2015.62.1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dave B, Migliaccio I, Gutierrez MC, et al. Loss of phosphatase and tensin homolog or phosphoinositol-3 kinase activation and response to trastuzumab or lapatinib in human epidermal growth factor receptor 2-overexpressing locally advanced breast cancers. J Clin Oncol. 2011;29(2):166–173. doi: 10.1200/JCO.2009.27.7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berns K, Horlings HM, Hennessy BT, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12(4):395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- 11.Nagata Y, Lan K-H, Zhou X, et al. PTEN activation contributes to tumor inhibition by trastuzumab, and loss of PTEN predicts trastuzumab resistance in patients. Cancer Cell. 2004;6(2):117–127. doi: 10.1016/j.ccr.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 12.Loibl S, von Minckwitz G, Schneeweiss A, et al. PIK3CA mutations are associated with lower rates of pathologic complete response to anti-human epidermal growth factor receptor 2 (HER2) therapy in primary HER2-overexpressing breast cancer. J Clin Oncol. 2014;32(29):3212–3220. doi: 10.1200/JCO.2014.55.7876. [DOI] [PubMed] [Google Scholar]
- 13.Majewski IJ, Nuciforo P, Mittempergher L, et al. PIK3CA mutations are associated with decreased benefit to neoadjuvant human epidermal growth factor receptor 2-targeted therapies in breast cancer. J Clin Oncol. 2015;33(12):1334–1339. doi: 10.1200/JCO.2014.55.2158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guarneri V, Frassoldati A, Bottini A, et al. Preoperative chemotherapy plus trastuzumab, lapatinib, or both in human epidermal growth factor receptor 2-positive operable breast cancer: results of the randomized phase II CHER-LOB study. J Clin Oncol. 2012;30(16):1989–1995. doi: 10.1200/JCO.2011.39.0823. [DOI] [PubMed] [Google Scholar]
- 15.Scaltriti M, Nuciforo P, Bradbury I, et al. High HER2 expression correlates with response to the combination of lapatinib and trastuzumab. Clin Cancer Res. 2015;21(3):569–576. doi: 10.1158/1078-0432.CCR-14-1824. [DOI] [PubMed] [Google Scholar]
- 16.Witzel I, Loibl S, von Minckwitz G, et al. Predictive value of HER2 serum levels in patients treated with lapatinib or trastuzumab—a translational project in the neoadjuvant GeparQuinto trial. Br J Cancer. 2012;107(6):956–960. doi: 10.1038/bjc.2012.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Salgado R, Denkert C, Campbell C, et al. Tumor-infiltrating lymphocytes and associations with pathological complete response and event-free survival in HER2-positive early-stage breast cancer treated with lapatinib and trastuzumab: a secondary analysis of the NeoALTTO trial. JAMA Oncol. 2015;1(4):448–454. doi: 10.1001/jamaoncol.2015.0830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guiu S, Mouret Reynier MA, Toure M, Coudert B. Predictive factors of response in HER2-positive breast cancer treated by neoadjuvant therapy. J Oncol. 2013;2013:854121. doi: 10.1155/2013/854121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rimawi MF, Schiff R, Osborne CK. Targeting HER2 for the treatment of breast cancer. Annu Rev Med. 2015;66:111–128. doi: 10.1146/annurev-med-042513-015127. [DOI] [PubMed] [Google Scholar]
- 20.de Azambuja E, Holmes AP, Piccart-Gebhart M, et al. Lapatinib with trastuzumab for HER2-positive early breast cancer (NeoALTTO): survival outcomes of a randomised, open-label, multicentre, phase 3 trial and their association with pathological complete response. Lancet Oncol. 2014;15(10):1137–1146. doi: 10.1016/S1470-2045(14)70320-1. [DOI] [PubMed] [Google Scholar]
- 21.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8(8):R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Desmedt C, Haibe-Kains B, Wirapati P, et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin Cancer Res. 2008;14(16):5158–5165. doi: 10.1158/1078-0432.CCR-07-4756. [DOI] [PubMed] [Google Scholar]
- 23.Perez EA, Thompson EA, Ballman KV, et al. Genomic analysis reveals that immune function genes are strongly linked to clinical outcome in the North Central Cancer Treatment Group n9831 Adjuvant Trastuzumab trial. J Clin Oncol. 2015;33(7):701–708. doi: 10.1200/JCO.2014.57.6298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sotiriou C, Wirapati P, Loi S, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98(4):262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 25.Majumder PK, Febbo PG, Bikoff R, et al. mTOR inhibition reverses Akt-dependent prostate intraepithelial neoplasia through regulation of apoptotic and HIF-1-dependent pathways. Nat Med. 2004;10(6):594–601. doi: 10.1038/nm1052. [DOI] [PubMed] [Google Scholar]
- 26.Farmer P, Bonnefoi H, Anderle P, et al. A stroma-related gene signature predicts resistance to neoadjuvant chemotherapy in breast cancer. Nat Med. 2009;15(1):68–74. doi: 10.1038/nm.1908. [DOI] [PubMed] [Google Scholar]
- 27.Fisher ER, Wang J, Bryant J, Fisher B, Mamounas E, Wolmark N. Pathobiology of preoperative chemotherapy: findings from the National Surgical Adjuvant Breast and Bowel (NSABP) protocol B-18. Cancer. 2002;95(4):681–695. doi: 10.1002/cncr.10741. [DOI] [PubMed] [Google Scholar]
- 28.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dobin A, Davis CA, Schlesinger F, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 31.Morgan M, Pagès H, Obenchain V, Hayden N. Rsamtools: Binary alignment (BAM), FASTA, variant call (BCF), and tabix file import. R package version 1.24.0. [Accessed August 31, 2016]; http://bioconductor.org/packages/release/bioc/html/Rsamtools.html. [Google Scholar]
- 32.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28(6):882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.The Cancer Genome Atlas. FireHose website. [Accessed October 19, 2014]; http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/BRCA/20160128/gdac.broadinstitute.org_BRCA.Merge_rnaseqv2__illuminahiseq_rnaseqv2__unc_edu__Level_3__RSEM_genes__data.Level_3.2016012800.0.0.tar.gz.
- 34.Gendoo DMA, Ratanasirigulchai N, Schröder MS, et al. Genefu: an R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics. 2016;32(7):1097–1099. doi: 10.1093/bioinformatics/btv693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parker JS, Mullins M, Cheang MCU, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Storey JD. A direct approach to false discovery rates. J R Stat Soc Series B Stat Methodol. 2002;64:479–498. [Google Scholar]
- 37.Denkert C, Huober J, Loibl S, et al. HER2 and ESR1 mRNA expression levels and response to neoadjuvant trastuzumab plus chemotherapy in patients with primary breast cancer. Breast Cancer Res. 2013;15(1):R11. doi: 10.1186/bcr3384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneeweiss A, Chia S, Hegg R, et al. Evaluating the predictive value of biomarkers for efficacy outcomes in response to pertuzumab- and trastuzumab-based therapy: an exploratory analysis of the TRYPHAENA study. Breast Cancer Res. 2014;16(4):R73. doi: 10.1186/bcr3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fumagalli D, Blanchet-Cohen A, Brown D, et al. Transfer of clinically relevant gene expression signatures in breast cancer: from Affymetrix microarray to Illumina RNA-Sequencing technology. BMC Genomics. 2014;15:1008. doi: 10.1186/1471-2164-15-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bianchini G, Pusztai L, Pienkowski T, et al. Immune modulation of pathologic complete response after neoadjuvant HER2-directed therapies in the NeoSphere trial. Ann Oncol. 2015;26(12):2429–2436. doi: 10.1093/annonc/mdv395. [DOI] [PubMed] [Google Scholar]
- 41.Savas P, Salgado R, Denkert C, et al. Clinical relevance of host immunity in breast cancer: from TILs to the clinic. Nat Rev Clin Oncol. 2016;13(4):228–241. doi: 10.1038/nrclinonc.2015.215. [DOI] [PubMed] [Google Scholar]
- 42.Bianchini G, Gianni L. The immune system and response to HER2-targeted treatment in breast cancer. Lancet Oncol. 2014;15(2):e58–e68. doi: 10.1016/S1470-2045(13)70477-7. [DOI] [PubMed] [Google Scholar]
- 43.Perez EA, Ballman KV, Tenner KS, et al. Association of stromal tumor-infiltrating lymphocytes with recurrence-free survival in the n9831 adjuvant trial in patients with early-stage HER2-positive breast cancer. JAMA Oncol. 2016;2(1):56–64. doi: 10.1001/jamaoncol.2015.3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xia W, Gerard CM, Liu L, Baudson NM, Ory TL, Spector NL. Combining lapatinib (GW572016), a small molecule inhibitor of ErbB1 and ErbB2 tyrosine kinases, with therapeutic anti-ErbB2 antibodies enhances apoptosis of ErbB2-overexpressing breast cancer cells. Oncogene. 2005;24(41):6213–6221. doi: 10.1038/sj.onc.1208774. [DOI] [PubMed] [Google Scholar]
- 45.Konecny GE, Pegram MD, Venkatesan N, et al. Activity of the dual kinase inhibitor lapatinib (GW572016) against HER-2-overexpressing and trastuzumab-treated breast cancer cells. Cancer Res. 2006;66(3):1630–1639. doi: 10.1158/0008-5472.CAN-05-1182. [DOI] [PubMed] [Google Scholar]
- 46.Scaltriti M, Verma C, Guzman M, et al. Lapatinib, a HER2 tyrosine kinase inhibitor, induces stabilization and accumulation of HER2 and potentiates trastuzumab-dependent cell cytotoxicity. Oncogene. 2009;28(6):803–814. doi: 10.1038/onc.2008.432. [DOI] [PubMed] [Google Scholar]
- 47.Maruyama T, Mimura K, Izawa S, et al. Lapatinib enhances herceptin-mediated antibody-dependent cellular cytotoxicity by up-regulation of cell surface HER2 expression. Anticancer Res. 2011;31(9):2999–3005. [PubMed] [Google Scholar]
- 48.Ignatiadis M, Singhal SK, Desmedt C, et al. Gene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: a pooled analysis. J Clin Oncol. 2012;30(16):1996–2004. doi: 10.1200/JCO.2011.39.5624. [DOI] [PubMed] [Google Scholar]