Abstract
To study the significance of signal transducer and activator of transcription (Stat) 3 in lung epithelial development of fetal mice, we examined fetal mouse lungs, focusing on the expression of Clara cell secretory protein (CCSP), Forkhead box protein J1 (Foxj1), calcitonin gene-related peptide (CGRP), phosphorylated Stat3 (Tyr705), and hairy/enhancer of split (Hes) 1, and observed cultured fetal lungs upon treatment with IL-6, a Stat3 activator, or cucurbitacin I, a Stat3 inhibitor. Moreover, the interaction of Stat3 signaling and Hes1 was studied using Hes1 gene-deficient mice. Phosphorylated Stat3 was detected in fetal lungs and, immunohistochemically, phosphorylated Stat3 was found to be co-localized in developing Clara cells, but not in ciliated cells. In the organ culture studies, upon treatment with IL-6, quantitative RT-PCR revealed that CCSP mRNA increased with increasing Stat3 phosphorylation, while cucurbitacin I decreased Hes1, CCSP, Foxj1 and CGRP mRNAs with decreasing Stat3 phosphorylation. In the lungs of Hes1 gene-deficient mice, Stat3 phosphorylation was not markedly different from wild-type mice, the expression of CCSP and CGRP was enhanced, and the treatment of IL-6 or cucurbitacin I induced similar effects on mouse lung epithelial differentiation regardless of Hes1 expression status. Stat3 signaling acts in fetal mouse lung development, and seems to regulate Clara cell differentiation positively. Hes1 could regulate Clara cell differentiation in a manner independent from Stat3 signaling.
Keywords: Stat3, Hes1, Clara cell, ciliated cell, neuroendocrine cell
I. Introduction
Signal transducer and activator of transcription (Stat) 3 is one of the main intracellular signaling molecules that mediate proinflammatory interleukin 6 (IL-6) [1, 5, 13, 26]; it plays critical roles in several biological functions, including proliferation, migration, survival and differentiation [13]. The significance of Stat3 signaling in development and differentiation has been reported in various organs and cells, including kidney [23], mammary gland [4], skin keratinocytes [15] and embryonic stem cells [27]. However, direct studies of Stat3 in lung development and lung epithelial cell differentiation remain limited. A high concentration of IL-6, an activator of Stat3, has been reported to promote lung branching morphogenesis [19], and leukemia inhibitory factor inhibits lung branching morphogenesis accompanied by the downregulation of Stat3 phosphorylation [20]. In addition, Stat3 signaling is reported to be important in the process of repair of injured bronchiolar epithelium as defect of Stat3 gene disturbs bronchiolar epithelial cell proliferation [11]. Important molecular pathways for lung development are recaptured during injury and regeneration [12], and, the pathways such as Stat3 for lung regeneration, vice versa, should be important during development.
Lung branching morphogenesis and epithelial cell differentiation during the fetal developmental period proceed with epithelial-mesenchymal interaction regulated by many secreted factors and their downstream effectors [3, 17, 18]. One of the critical systems determining lung epithelial cell differentiation is a network of differentiation-promoting and -suppressing basic helix-loop-helix transcription (bHLH) factors [6, 7]. In mice lacking the gene for mammalian homologue of achaete scute complex (Mash) 1, one of the differentiation-promoting bHLHs, pulmonary neuroendocrine (PNE) cells are not detected, while in mice lacking the gene for hairy/enhancer of split (Hes) 1, one of the recessive bHLHs, many PNE cells appear in the lung at the expense of Clara cells [7]. The latter observation suggests that Hes1 can suppress the neuroendocrine differentiation of lung epithelial cells and could play roles in Clara cell differentiation. Hes1 expression is regulated positively by Notch signaling [8]. Using various Notch receptor gene-deficient mice, Morimoto et al. [16] found that Notch2 makes a dominant contribution to Clara cell fate determination and Notch1 and Notch3 make modest contributions to Clara cell selection. On the other hand, Foxj1-positive ciliated cells are increased in Notch gene-deficient conditions, and so Notch signaling could contribute to Clara cell/ciliated cell selection [16]. According to Xing et al. [29], transforming growth factor beta signaling via ALK5 positively regulates Hes1 expression by suppressing PTEN, and could promote Clara cell differentiation. And, Notch1 is required to regeneration of Clara cells after naphthalene injury with stimulation of Hes5 and Pax6 [28].
Kamakura et al. [9] presented that Hes1 and Hes5 are associated with Janus-activated kinase (Jak) 2 and Stat3 and promote Stat3 activation in neuroepithelial cells. In addition, direct protein-protein interactions that coordinate cross-talk between the Notch1-Hes1 and Jak-Stat pathways are suggested [9]. Moreover, in neural tissues, Hes1 is reported to be coupled with Stat3 activation during neural differentiation [14, 15, 30]. However, relationship between Notch1-Hes1 and Stat3 signaling pathways has not been studied in lung development.
In the present study, we applied immunohistochemical staining to determine the distribution of phosphorylated Stat3-positive cells in the developing lung epithelium of fetal mice, and the involvement of Stat3 signaling in Clara cell differentiation, by observing cultured fetal mouse lungs treated with Stat3 activator (IL-6) or inhibitor (cucurbitacin I). Stat3 is phosphorylated at various phosphorylated sites, and we studied phosphorylation at Tyr705 as phosphrylation at Tyr705 regulates STAT3 dimerization, nuclear translocation and DNA binding [22, 24]. Furthermore, we cultured the fetal lungs of Hes1 gene-deficient mice treated with these Stat3 modulators in order to reveal the relationship between Hes1 and Stat3 signaling in mouse lung development.
II. Materials and Methods
Animals
ICR mice were purchased from Japan SLC (Shizuoka, Japan). They were kept with free access to water and food, and were maintained on a 12-hr light/dark cycle under pathogen-free conditions. This study was approved by the Animal Care Committee of Kumamoto University (#25-076). To obtain fetal lungs, mice were mated during the night, and the day of discovery of a vaginal plug was counted as fetal day (e)0. In this study, lungs of ICR mice from fetal days 12.5–18.5 (e12.5, e14.5, e16.5, e18.5) were used, and for organ culture, lungs from e12.5 mice were used and cultured for 72 hr.
Western blotting for Stat3
For the Western blotting analysis, lung samples were obtained from five fetuses at e14.5 and one fetus each at e16.5 and e18.5, and three sets of lung tissues for Western blotting analyses were made from 9 pregnant mice. The fetal lungs of ICR mice at e14.5, e16.5 and e18.5 were homogenized on ice with ice-cold homogenizing buffer [5 mM Tris, 0.25 M sucrose, 2 mM EDTA, 2 mM ethylene glycol bis(2-aminoethyl ether)-tetraacetic acid (EGTA)] containing the protease inhibitors phenylmethane sulfonyl fluoride (PMSF, 0.5 mM), dithiothreitol (DTT, 0.5 mM) and leupeptin (5 μg/ml), clarified by centrifugation and resuspended in lysate buffer (50 mM Tris, 150 mM NaCl, 25 mM NaF, 25 mM glycerophosphate, 2 mM EDTA, 2 mM EGTA, 0.3% NP-40, 0.5 mM PMSF, 0.5 mM DTT, 5 μg/ml leupeptin). The lysate was centrifuged and the concentration of the supernatant protein extract was determined using the Bio-Rad protein assay kit (Bio-Rad Laboratories, CA, USA). Aliquots of 30 μg of protein were subjected to SDS-PAGE (in 8–12% gels), and the protein was then transferred to nitrocellulose membranes. The blots were blocked with 5% non-fat milk in phosphate-buffered saline solution. Rabbit antibodies against Stat3 (Cell Signaling, Beverly, MA) and phosphorylated Stat3 (Tyr705; Cell Signaling) were used for the Western blotting analysis. The filters were incubated with each primary antibody and then with horseradish peroxidase-conjugated secondary antibody, and the stained proteins were visualized with an enhanced chemiluminescence detection kit (Amersham Pharmacia Biotech, Uppsala, Sweden). β-actin was also detected in each sample. Comparative quantitation of phosphorylated Stat3 protein in the cultured fetal lung tissues and Hes1-gene deficient mouse lungs was performed by densitometric analysis of blotting bands using an image analysis software (CS Analyzer, ATTO Corporation, Tokyo), and the ratio values of phosphorylated Stat3 were determined relative to values obtained for Stat3. Each of the ratio values was tested by oneway ANOVA using GraphPad Prism statistical software, and the significance was determined using Newman-Keuls method at the 0.05 level.
Immunohistochemistry
Fetal lungs at e14.5, e16.5 and e18.5 (6 on each day) were isolated, fixed in phosphate-buffered 4% paraformaldehyde solution and embedded in paraffin. Paraffin-embedded sections were rehydrated through xylene and graded ethanol solutions. After treatment with 0.3% hydrogen peroxide, the sections were heated at 95°C for 40 min in antigen retrieval solution (pH 8; Nichirei, Tokyo, Japan). After treatment with 4% Blockace in PBS (Dai-Nippon-Pharmaceutical, Suita, Japan) for 20 min, the sections were treated with goat anti-Clara cell secretory protein (CCSP; Santa Cruz Biotech, Santa Cruz, CA), mouse anti-Foxj1 (Santa Cruz), goat anti-calcitonin gene-related peptide (CGRP; Abcam, Cambridge, MA), rabbit anti-Hes1 (Santa Cruz) or rabbit anti-phosphorylated Stat3 (Tyr705) (Cell Signaling) antibody overnight at 4°C. After washing, the sections were treated with anti-goat, -rabbit or -mouse IgG conjugated with HRP-polymer (ImmPress Reagent; Vector Laboratories, Burlingame, CA) for 30 min at room temperature. The sections were further treated with diaminobenzidine-hydrogen peroxide solution, and counterstained with hematoxylin.
Double immunostaining for phosphorylated Stat3 and either CCSP or Foxj1, and for Hes1 and either CCSP or Foxj1
Paraffin-embedded sections were rehydrated through xylene and graded ethanol solutions. After treatment with 0.3% hydrogen peroxide, the sections were heated at 95°C for 40 min in antigen retrieval solution (Nichirei) for phosphorylated Stat3 immunostaining or Hes1 immunostaining. After treatment with 4% Blockace in PBS (Dai-Nippon-Pharmaceutical) for 20 min at room temperature, the sections were treated with either rabbit anti-phosphorylated Stat3 (Tyr705, Cell Signaling) or rabbit anti-Hes1 (Santa Cruz) antibody overnight at 4°C. After washing, the sections were treated with the secondary antibodies conjugated with HRP polymer for the corresponding animal IgGs (ImmPress Reagent, Vector) for 30 min at room temperature. For phosphorylated Stat3 or Hes1 immunostaining, diaminobenzidine-hydrogen peroxide solution was applied.
After washing in PBS three times, the sections were again heated at 95°C for 20 min in the antigen retrieval solution for CCSP immunostaining (DAKO REAL Target Retrieval Solution, pH6; DAKO, Carpinteria, CA) or in the solution (pH8, Nichirei) for Foxj1. After treatment with 4% Blockace in PBS (Dainippon Pharmaceutical) for 20 min at room temperature, the sections were treated with either goat anti-CCSP (Santa Cruz) or mouse anti-Foxj1 (Santa Cruz) antibody for 1 hr at room temperature.
After washing, the sections were treated with secondary antibodies conjugated with HRP polymer or AP polymer for the corresponding animal IgGs (ImmPress Reagent, Vector) for 30 min at room temperature. For CCSP immunostaining, Vulcan FastTM Red Chromogen Kit (Biocare Medical, Concord, CA) was applied, and for Foxj1 immunostaining, Vina GreenTM Chromogen Kit (Biocare Medical) was applied, and counterstained with hematoxylin.
Quantitative RT-PCR analysis
mRNA from the fetal lungs of ICR mice at e12.5, e14.5, e16.5 and e18.5 was isolated using the ISOGEN reagent (Nippon Gene, Tokyo, Japan) according to the manufacturer’s instructions. Sample lungs were analyzed from one fetus on fetal days e12.5, e14.5, e16.5 and e18.5. The experiments were repeated using three sets of fetal lungs. cDNA was synthesized using the PrimeScript® RT reagent Kit (Takara-Bio, Shiga, Japan). Quantitative RT-PCR was performed on a LightCycler Nano Real-Time PCR System (Roche Diagnostics, Mannheim, Germany), using a Fast Start Essential DNA Green Master (Roche) for CCSP, Foxj1, CGRP, Hes1, Mash1, Notch1, Notch2, Notch3 and β-actin. Data analysis was performed using the LightCycler Nano software, version 1.0 (Roche). All reactions were normalized to β-actin. Results were plotted as relative expression compared with the result of e12.5, which was scaled to 1. The PCR primer pairs used in this study are listed in Table 1. The mean ± SD for each experimental group is expressed in the bar graphs. Differences between the means were tested by oneway ANOVA followed by Newman-Keuls method for multiple comparisons using GraphPad Prism statistical software. P value, 0.05 was considered significant.
Table 1. .
Target gene primer sequence (5'–3')
| CCSP | TACCATGAAGATCGCCATCACAAGGCTTCAGGGATGCCACATAAC |
| Foxj1 | CACGTGAAGCCACCCTACTCCTAGAGGCACTTTGATGAAGCACTTG |
| CGRP | GTGCAGGACTATATGCAGATGAAAGCACAGGTGGCAGTGTTGCAG |
| Hes1 | AAAGACGGCCTCTGAGCACGGTGCTTCACAGTCATTTCCA |
| Mash1 | AAGAGCTGCTGGACTTTACCAACTGATTTGACGTCGTTGGCGAGA |
| Notch1 | TGCCTTGAGTGTGCTGGAATGATTCTGCCACAGGCGTATACTTGA |
| Notch2 | CCCATTCAAGTTCACTGAATCAACAAGCTTTAGGGCAGACAGTCACCA |
| Notch3 | TCCTCACTTCACTGCATTCCAGATGGAGTTGAGGCTTTGAGCAGA |
| β-actin | CATCCGTAAAGACCTCTATGCCAACATGGAGCCACCGATCCACA |
Organ culture
The e12.5 embryos were dissected from pregnant mice. The lungs with the trachea were isolated from each embryo and cultured on a 6-well tissue culture plate, with an 8-μm pore size cell culture insert (Falcon/Becton Dickinson, NJ). Cultures were maintained in serum-free, chemically defined Fitton-Jackson modification BGJb medium (Life Technologies, Grand Island, NY) containing 50 units/ml penicillin and 50 μg/ml streptomycin. For experimental treatments, the medium was supplemented with one of the following: recombinant mouse IL-6 (R&D, Minneapolis, MN, USA) at 10 ng/ml or 50 ng/ml, or curcubitacin I (Calbiochem, San Diego, CA) at 1 μM or 5 μM. Control explants with IL-6 or cucurbitacin I treatment were incubated in medium containing dimethyl sulfoxide (DMSO, Sigma) at 2.5 μl/ml. A total of 16–18 pairs of lungs were used for each experimental group, and each insert supported up to 6 pairs of lung explants. Each medium was changed every 24 hr. We performed 3 independent studies.
To clarify the effects of IL-6 and cucurbitacin I, phase contrast micrographs were taken at the beginning (day0) and at the end (day3) of culture. The number of the terminal buds in the left lung was counted on each day, and the branching ratio (the number of buds at the end of the culture/the number of buds at the beginning of the culture) was calculated. The mean ± SD for each experimental group is expressed in the graphs.
Differences between the means were tested by oneway ANOVA using GraphPad Prism statistical software, and the significance was determined using Newman-Keuls method at the 0.05 level.
A total of 10–12 pairs of lungs were harvested for Western blotting analysis for phosphorylated Stat3 (Tyr705), three pairs of lungs were obtained for detecting CCSP, FoxJ1, CGRP and Hes1 mRNAs by quantitative RT-PCR, and three to four pairs were used for immunohistochemistry for CCSP, FoxJ1, Hes1 and phosphorylated Stat3 (Tyr705) after 4% paraformaldehyde fixation and paraffin embedding.
To determine the relationship between Stat3 phosphorylation and Hes1 gene expression, Hes1 gene knockout mice were used for the following studies. First, the phosphorylation of Stat3 expressed in E18.5 fetal mouse lungs from wild-type and Hes1 knockout mice was analyzed by Western blotting. Moreover, E12.5 fetal lungs from wild-type and Hes1 knockout mice were used for organ culture studies. The genotyping of Hes1 in the mice was as described in our previous study [7]. The lungs were applied to organ culture for 3 days with IL-6 treatment at a concentration of 50 ng/ml or cucurbitacin I treatment at a concentration of 5 μg/ml. After cultivation, three pairs of lungs were obtained for detecting CCSP, FoxJ1, CGRP and Hes1 mRNAs by quantitative RT-PCR.
III. Results
Modulation of expression patterns in fetal mouse lungs
In the present study, immunohistochemistry for CCSP was used for detecting Clara cells, FoxJ1 for ciliated cells and CGRP for neuroendocrine cells [10, 21].
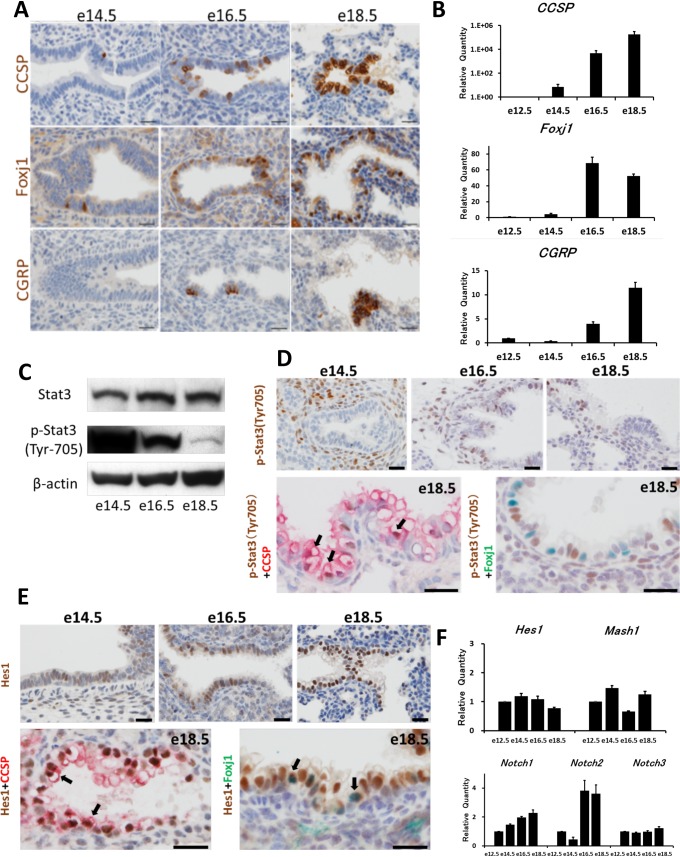
Immunohistochemically, CCSP-positive Clara cells appeared on e14.5 and increased with development, and FoxJ1-positive ciliated cells showed similar staining pattern to CCSP-positive cells (Fig. 1A). CGRP-positive neuroendocrine cells were fewer than CCSP- or FoxJ1-positive cells, and often formed clusters (Fig. 1A). Quantitative RT-PCR analysis supported the immunohistochemical study, and revealed that the expression of differentiation markers, such as CCSP, Foxj1 and CGRP, generally increased with the growth of the lungs (Fig. 1B).
Fig. 1.
Expression of lung epithelial markers, Stat3, and Notch-related molecules in the developing lungs of fetal mice. A: Immunohistochemically, Clara cell secretory protein (CCSP)-positive cells and Forkhead box protein J1 (Foxj1)-positive cells appear on e14.5 and then increase. Calcitonin gene-related peptide (CGRP)-positive cells are a few. Counterstained with hematoxylin. Bar = 20 mm. B: Real-time PCR analysis reveals that CCSP, Foxj1 and CGRP mRNAs increase with development. C: Western blotting reveals that phosphorylated Stat 3 (Tyr 705; p-Stat3) decreases over the developmental period. D (upper column): Immunohistochemically, p-Stat3-positive epithelial cells are abundant in e16.5 and e18.5. Bar = 20 mm. D (lower column): Double immunostaining for p-Stat3 (brown) with CCSP (red) or Foxj1 (green) reveals that p-Stat3 is evident in Clara cells (arrows), but not in ciliated cells. Counterstained with hematoxylin. Bar = 20 μm. E: Double immunostaining for Hairy and enhancer of split 1 (Hes1; brown) with CCSP (red) or Foxj1 (green) reveals that Hes1 is positive in both Clara cells (arrows) and ciliated cells (arrows). Counterstained with hematoxylin. Bar = 20 μm. F: Real-time PCR analysis reveals that Hes1 and mammalian achaete scute complex homolog 1 (Mash1) mRNAs do not vary markedly in the fetal lungs. Notch1 mRNA increases gradually over the development period, and Notch2 mRNA increases remarkably in the late fetal period.
Regrading to Stat3 expression in the fetal lungs, Western blotting analyses revealed that the expression of Stat3 did not vary markedly, but phosphorylated Stat3 (Tyr705) was decreased with development (Fig. 1C). Immunohistochemically, cells positive for phosphorylated Stat3 (Tyr705) were detected among developing epithelial cells and mesenchymal cells in e14.5 lungs, and were scattered among developing airway cells thereafter (Fig. 1D). Double immunostaining for phosphorylated Stat3 (Tyr705) with CCSP or Foxj1 was also attempted. Developing Clara cells showed positive staining for phosphorylated Stat3 (Tyr705), but ciliated cells showed negative staining for it (Fig. 1D).
Meanwhile, the immunohistochemical study revealed that Hes1 was positive in bronchiolar epithelial cells, and more distinct on e18.5 (Fig. 1E). Double immunostaining for CCSP and Hes1 or Foxj1 and Hes1 revealed that both cell types were positive for Hes1 (Fig. 1E). Hes1 mRNA did not change remarkably until e18.5, and Notch1 and 2 mRNAs increased with growth (Fig. 1F).
Organ culture
To reveal the significance of Stat3 in lung development, fetal lung tissue explants were treated with Stat3 activator (IL-6) or inhibitor (cucurbitacin I). The culturing of explant lung tissues showed progressive branching morphogenesis. IL-6 treatment enhanced branching morphogenesis, but cucurbitacin I suppressed branching of the lung explants (Fig. 2A).
Fig. 2.
Effects of IL-6 and cucurbitacin I on the growth pattern, p-Stat3, lung epithelial markers and Hes1 in cultured embryonic lungs (e12.5) for 3 days. A: Serial photographs of the same explants at 0 day and 3 days of cultivation. The cultured lung explants show that lung tissues grow with treatment of IL-6 and that cucurbitacin I suppresses growth. Bar = 500 μm. B: Western blotting for p-Stat3 reveals that IL6 increases p-Stat3 and cucurbitacin I decreases p-Stat3 in cultured fetal lung tissues. Comparative quantitation of p-Stat3 in the cultured lung explants treated with IL-6 or cucurbitacin I compared to the control tissues is shown as a bar graph. Immunostaining for pStat-3 supports the Western blotting studies. Counterstained with hematoxylin. Bar = 20 μm. C: Immunostaining of the cultured lung explants reveals that CCSP-positive Clara cells and FoxJ1-positive ciliated cells increase with the treatment of IL-6, but decrease with the treatment of cucurbitacin I. Cucurbitacin I depresses Hes1 expression in the explants. Counterstained with hematoxylin. Bar = 20 μm. D: Real-time PCR analysis reveals that CCSP mRNA is increased but FoxJ1, CGRP and Hes1 mRNAs are not altered with the treatment of IL-6, while CCSP, Foxj1 and Hes1 mRNAs are decreased with the treatment of cucurbitacin I.
Western blotting study of the cultured lungs revealed that the phosphorylated form of Stat3 (Tyr705) was increased with the treatment of IL-6 and decreased with cucurbitacin I (Fig. 2B). Immunohistochemical studies revealed that the nuclear expression of phosphorylated Stat3 (Tyr705) increased with the treatment of IL-6, but Cucurbitacin I decreased the immunostaining for phosphorylated Stat3 (Tyr705) (Fig. 2B).
Immunohistochemically, CCSP- and Foxj1-positive cells appeared more with the treatment of IL-6, but Hes1 immunostaining was not altered (Fig. 2C). Cucurbitacin I decreased the immunostaining for CCSP, Foxj1, and Hes1 (Fig. 2C). Quantitative RT-PCR study revealed that IL-6 enhanced mildly the expression of CCSP mRNA, but CGRP and Hes1 mRNAs were not altered (Fig. 2D). On the other hand, cucurbitacin I decreased CCSP, Foxj1 and Hes1 mRNAs in a dose-dependent manner (Fig. 2D).
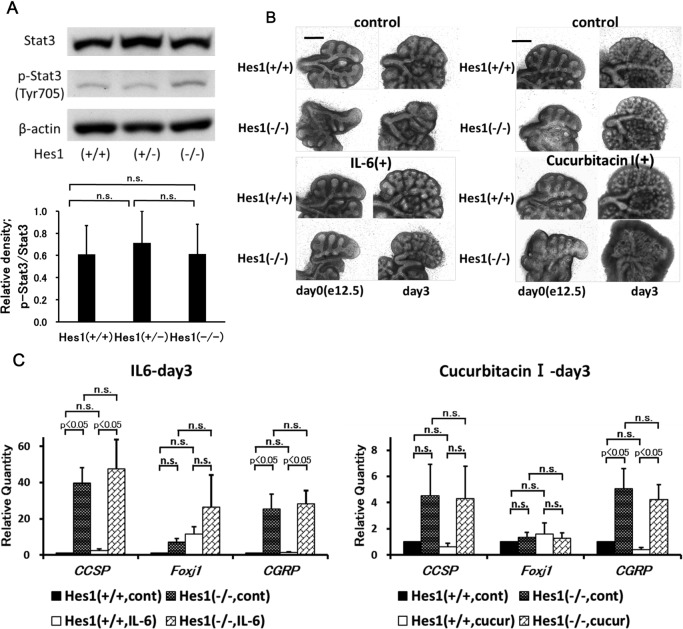
Hes1 gene knockout mouse study
As Stat3 signaling, coupled with Notch-Hes1-signaling pathway, is reported to work in neuroepithelial differentiation [8], relationship between Stat3 and Hes1 was studied with use of Hes1 gene knockout mice and organ culture of Hes1 gene knockout mouse lung was attempted. The size of the lungs from Hes1 gene knockout mice was smaller than that from wild-type mice at e12.5 [7]. The expression of phosphorylated Stat3 (Tyr705) in e18.5 mouse lung did not differ markedly, regardless of the Hes1 gene expression conditions (Fig. 3A). Similar to that in wild-type mouse lungs, in the lung explants from Hes1 gene knockout mice, IL-6 enhanced growth and cucurbitacin I inhibited growth (Fig. 3B). Quantitative RT-PCR study revealed that Hes1 gene deficiency increased CCSP and CGRP markedly, but not Foxj1 (Fig. 3C). IL-6 induced mildly increased expression of CCSP in wild-type mouse lungs, but its effects on CCSP expression in Hes1 gene knockout mouse lungs were not remarkable. In Hes1 gene knockout mouse lungs, IL-6 did not enhance Foxj1 expression significantly. IL-6 did not alter CGRP expression in Hes1 gene knockout mouse lungs. The effects of cucurbitacin I on Hes1 gene knockout mouse lungs seemed to be similar to those on wild-type mouse lungs (Fig. 3C).
Fig. 3.
Expression of p-Stat3 (Tyr705) in Hes1 gene-deficient (−/−) mice (e18.5), and effects of IL-6 or cucurbitacin I on the cultured embryonic lungs of Hes1 gene-deficient (−/−) mice (e12.5) for 3 days. A: Western blotting for Stat3 and p-Stat3 in embryonic lungs from wild-type (+/+), heterozygous (+/−) and homozygous (−/−) Hes1 gene-deficient mice (e18.5) was done. Comparative quantitation of p-Stat3 compared to Stat3 in the mouse lungs with various Hes1 gene conditions shows no remarkable difference in phosphorylation of Stat3 among them. B: Organ culture studies reveal that Hes1 gene expression condition does not modify the effects on the cultured lung explants by IL-6 and cucurbitacin I. Bar = 500 μm. C: Real-time PCR analysis of mRNAs. The lungs from Hes1 (−/−) mice show increased expression of CCSP and CGRP mRNAs. The effects of IL-6 or cucurbitacin I on the cultured lung tissues seem similar, regardless of the Hes1 gene expression conditions.
IV. Discussion
In the present study, we surveyed the modulation of phosphorylated Stat3 (Tyr705) in developing fetal mouse lungs; phosphorylated Stat3 (Tyr705) decreased before birth, which was confirmed by immunohistochemistry. The pattern of decreased phosphorylation of signaling molecules and enhanced lung epithelial cell differentiation during lung development has also been found in AKT and ERK [25]. Immunohistochemical studies showed that phosphorylated Stat3 (Tyr705) was positive in immature lung epithelial and mesenchymal cells, but after differentiation to diverse lung epithelial cells, positive staining was localized mainly to developing Clara cells. This suggests that Clara cell differentiation might be regulated by Stat3 signaling, and the progenitor function of Clara cells [10] could be associated with Stat3 signaling. A report showing the importance of Stat3 signaling in bronchiolar epithelial repair and cell migration [11] suggests similar functional significance of Stat3 in the development and reparative processes of the lung. On the other hand, immunohistochemical study revealed that ciliated cells were negative for phosphorylated Stat3 (Tyr705). This suggests that the differentiation of ciliated cells from Clara cells could be accompanied by the dephosphorylation of Stat3, and the molecular dynamics of post-transcriptional modification of Stat3 might act to determine the cell fate decision in the lung epithelial systems.
In order to clarify the role of Stat3 signaling, IL-6 and cucurbitacin I were applied in experiments involving organ culture of fetal lung explants. IL-6 is one of the representative Stat3 activators [5], and Western blotting and immunohistochemical analyses revealed increased phosphorylated Stat3 (Tyr705) in cultured lung tissues. According to Nogueira-Silva et al. [19], IL-6 is expressed in the developing rat lungs in the fetal period, and the addition of exogenous IL-6 enhances the branching of fetal rat lung explants, which is true in the present mouse study. Upon the treatment of IL-6, CCSP expression was enhanced, and, in vivo, Stat3 phosphorylation appeared to be identical in CCSP-positive cells. These observations susgest that positive relationship between Stat3 phosphorylation and Clara cell differentiation should occur in developing fetal lungs. Cucurbitacin I is a highly selective inhibitor of Jak/Stat3 [2]. The present organ culture study shows that cucurbitacin I suppresses the phosphorylation of Stat3, delays lung growth and also inhibits bronchiolar epithelial differentiation; it also suggests that Stat3 signaling should be important in lung growth and epithelial cell differentiation.
In our previous study, the incidence of Clara cells was lower in the lung of Hes1 gene-deficient mice than in the lung of wild-type mice, and we imagined that Hes1 could play roles in Clara cell differentiation [7]. Our present study demonstrated the enhanced expression of CCSP and CGRP in Hes1 gene-deficient mice. Taking the findings from these studies together, Hes1 could modulate Clara cell differentiation as a suppressive factor, like in the case of neuroendocrine cell differentiation. The number of Clara cells in Hes1 gene-deficient mice appeared to be decreased [7], but this would not always imply positive regulation of Hes1 for Clara cell differentiation as the influx to neuroendocrine cell differentiation could be stronger than the influx to Clara cell differentiation in Hes1 gene-deficient mice. According to Kamakura et al. [9], the Notch-Hes1 pathway is associated with JAK2/Stat3 and promotes the activation of Stat3. In Hes1 gene-deficient mouse lungs, the phosphorylation of Stat3 (Tyr705) was not suppressed. Exogenous IL-6 did not affect the expression of Hes1 in the cultured lungs from wild-type mice, and the conditions of the expression of Hes1 did not modify the effects on cell differentiation due to IL-6. Cucurbitacin I suppresses Hes1, but its effects on cell differentiation do not seem to differ between the lungs from wild-type and Hes1 gene-deficient mice. These observations suggest that Clara cell differentiation could be promoted by IL-6, and the enhanced expression of CCSP seen in Hes1 gene-deficient mice could be independent of Stat3 activation.
The present study has obtained various novel findings concerning the involvement of Stat3 signaling and Hes1 in mouse bronchiolar epithelial differentiation. During the fetal development of mouse lungs, phosphorylated Stat3 (Tyr705) was seen in Clara cells. In organ culture systems, exogenous IL-6 up-regulated Stat3 phosphorylation accompanied by increased CCSP expression, and cucurbitacin I down-regulated Stat3 phosphorylation accompanied by the decreased expression of Hes1, CCSP and Foxj1. In the lungs of Hes1 gene-deficient mice, phosphorylated Stat3 (Tyr705) did not differ from that in the wild-type mouse lungs, and CCSP expression was greater than in wild-type mouse lungs. Thus, the present study suggests that Stat3 activation and Hes1 could play important roles in determining the fate of mouse bronchiolar epithelial cells, but Stat3 and Hes1 do not seem to co-operate with each other. Further analyses are necessary to conclude on this observation by knockdown or mutation experiments of these molecules.
V. Acknowledgments
The authors thank Ms. Takako Maeda for her excellent technical assistance, and thank Dr. Koki Hasegawa and Dr. Kanako Niimori for their suggestions. The authors also thank Prof. Ryoichiro Kageyama of Kyoto University for his help with the Hes1 gene-deficient mice. This study was supported in part by a grant from the Smoking Research Foundation and by a Grant-in-Aid for Scientific Research (C; No. 23220010) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
VI. References
- 1.Akira S., Nishio Y., Inoue M., Wang X. J., Wei S., Matsusaka T., Yoshida K., Sudo T., Naruto M. and Kishimoto T. (1994) Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell 77; 63–71. [DOI] [PubMed] [Google Scholar]
- 2.Blaskovich M. A., Sun J., Cantor A., Turkson J., Jove R. and Sebti S. M. (2003) Discovery of JSI-124 (cucurbitacin I), a selective janus kinase/signal transducer and activator of transcription 3 signaling pathway inhibitor with potent antitumor activity against human and murine cancer cells in mice. Cancer Res. 63; 1270–1279. [PubMed] [Google Scholar]
- 3.Gaillard, D. and Puchelle, E. (1999) Differentiation and maturation of airway epithelial cells: Role of extracellular matrix and growth factors. In “Lung Development”, ed. by C. Gaultier, J. Bourbon and M. Post. Springer, New York, pp. 46–76. [Google Scholar]
- 4.Gurusamy D., Ruiz-Torres S. J., Johnson A. L., Smith D. A. and Waltz S. E. (2014) Hepatocyte growth factor-like protein is a positive regulator of early mammary gland ductal morphogenesis. Mech. Dev. 133; 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hirano T. (1998) Interleukin 6 and its receptor: Ten years later. Int. Rev. Immunol. 16; 249–284. [DOI] [PubMed] [Google Scholar]
- 6.Ito T. (1999) Differentiation and proliferation of pulmonary neuroendocrine cells. Prog. Histochem. Cytochem. 34; 247–322. [DOI] [PubMed] [Google Scholar]
- 7.Ito T., Udaka N., Yazawa T., Okudela K., Hayashi H., Sudo T., Guillemot F., Kageyama R. and Kitamura H. (2000) Basic helix-loop-helix transcription factors regulate the neuroendocrine differentiation of fetal mouse pulmonary epithelium. Development 127; 3913–3921. [DOI] [PubMed] [Google Scholar]
- 8.Jarriault S., Brou C., Logeat F, Schroeter E. H., Kopan R. and Israel A. (1995) Signalling downstream of activated mammalian Notch. Nature 377; 355–358. [DOI] [PubMed] [Google Scholar]
- 9.Kamakura S., Oishi K., Yoshimatsu T., Nakafuku M., Masuyama N. and Gotoh Y. (2004) Hes binding to STAT3 mediates crosstalk between Notch and JAK–STAT signalling. Nat. Cell Biol. 6; 547–554. [DOI] [PubMed] [Google Scholar]
- 10.Kameyama H., Kudoh S., Udaka N., Kagayama M., Hassan W., Hasegawa K., Niimori-Kita K. and Ito T. (2014) Bromodeoxyuridine (BrdU)-label-retaining cells in mouse terminal bronchioles. Histol. Histopathol. 29; 659–668. [DOI] [PubMed] [Google Scholar]
- 11.Kida H., Mucenski M. L., Thitoff A. R., Le Cras T. D., Park K. S., Ikegami M., Müller W. and Whitsett J. A. (2008) GP130-STAT3 regulates epithelial cell migration and is required for repair of the bronchiolar epithelium. Am. J. Pathol. 172; 1542–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kotton D. N. and Morrisey E. E. (2014) Lung regeneration: mechanisms, applications and emerging stem cell populations. Nat. Med. 20; 822–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy D. E. and Darnell J. E. (2002) Stats: Transcriptional control and biological impact. Nat. Rev. Mol. Cell Biol. 3; 651–662. [DOI] [PubMed] [Google Scholar]
- 14.Li Y., Zhuang P., Shen B., Zhang Y. and Shen J. (2012) Baicalin promotes neuronal differentiation of neural stem/progenitor cells through modulating p-stat3 and bHLH family protein expression. Brain Res. 1429; 36–42. [DOI] [PubMed] [Google Scholar]
- 15.Mii S., Murakumo Y., Asai N., Jijiwa M., Hagiwara S., Kato T., Asai M., Enomoto A., Ushida K., Sobue S., Ichihara M. and Takahashi M. (2012) Epidermal hyperplasia and appendage abnormalities in mice lacking CD109. Am. J. Pathol. 181; 1180–1189. [DOI] [PubMed] [Google Scholar]
- 16.Morimoto M., Nishinakamura R., Saga Y. and Kopan R. (2012) Different assemblies of Notch receptors coordinate the distribution of the major bronchial Clara, ciliated and neuroendocrine cells. Development 139; 4365–4373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morrisey E. E. and Hogan B. L. (2010) Preparing for the first breath: Genetic and cellular mechanisms in lung development. Dev. Cell 18; 8–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nogawa H. and Ito T. (1995) Branching morphogenesis of embryonic mouse lung epithelium in mesenchyme-free culture. Development 121; 1015–1022. [DOI] [PubMed] [Google Scholar]
- 19.Nogueira-Silva C., Santos M., Baptista M. J., Moura R. S. and Correia-Pinto J. (2006) IL-6 is constitutively expressed during lung morphogenesis and enhances fetal lung explant branching. Pediatr. Res. 60; 530–536. [DOI] [PubMed] [Google Scholar]
- 20.Nogueira-Silva C., Piairo P., Carvalho-Dias E., Peixoto F. O., Moura R. S. and Correia-Pinto J. (2012) Leukemia inhibitory factor in rat fetal lung development: Expression and functional studies. PLoS One 7; e30517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rawlins E. L. and Hogan B. L. (2006) Epithelial stem cells of the lung: privileged few or opportunities for many? Development 133; 2455–2465. [DOI] [PubMed] [Google Scholar]
- 22.Reich N. C. and Liu L. (2006) Tracking STAT nuclear traffic. Nat. Rev. Immunol. 6; 602–612. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt-Ott K. M., Yang J., Chen X., Wang H., Paragas N., Mori K., Li J.-Y., Lu B., Costantini F., Schiffer M., Bottinger E. and Barasch J. (2005) Novel regulators of kidney development from the tips of the ureteric bud. J. Am. Soc. Nephrol. 16; 1993–2002. [DOI] [PubMed] [Google Scholar]
- 24.Sehgal P. B. (2008) Paradigm shifts in cell biology of STAT signaling. Semin. Cell Dev. Biol. 19; 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang J., Ito T., Udaka N., Okudela K., Yazawa T. and Kitamura H. (2005) PI3K–AKT pathway mediates growth and survival signals during development of fetal mouse lung. Tissue Cell 37; 25–35. [DOI] [PubMed] [Google Scholar]
- 26.Wegenka U. M., Lütticken C., Buschmann J., Yuan J., Lottspeich F., Müller-Esterl W., Schindler C., Roeb E., Heinrich P. C. and Horn F. (1994) The interleukin-6-activated acute-phase response factor is antigenically and functionally related to members of the signal transducer and activator of transcription (STAT) family. Mol. Cell. Biol. 14; 3186–3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie X., Chan K. S., Cao F., Huang M., Li Z., Lee A., Weissman I. L. and Wu J. C. (2009) Imaging of STAT3 signaling pathway during mouse embryonic stem cell differentiation. Stem Cells Dev. 18; 205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xing Y., Li A., Borok Z., Li C. and Minoo P. (2012) NOTCH1 is required for regeneration of Clara cells during repair of airway injury. Stem Cells 30; 946–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xing Y., Li C., Li A., Sridurongrit S., Tiozzo C., Bellusci S., Borok Z., Kaartinen V. and Minoo P. (2010) Signaling via Alk5 controls the ontogeny of lung Clara cells. Development 137; 825–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X., Smith A. J., Waterhouse A., Blin G., Malaguti M., Lin C. Y., Osorno R., Chambers I. and Lowell S. (2013) Hes1 desynchronizes differentiation of pluripotent cells by modulating STAT3 activity. Stem Cells 31; 1511–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]