Abstract
AIM
The purpose of this study was to evaluate the diagnostic value of trefoil factor family 3 (TFF3) for the early detection of colorectal cancer (CC).
METHODS
Serum TFF3 and carcino-embryonic antigen (CEA) were detected in 527 individuals, including 115 healthy control (HC), 198 colorectal adenoma (CA), and 214 CC individuals in the training group.
RESULTS
Serum TFF3 showed no significant correlation with age, gender, or tumor location but showed significant correlation with the tumor stage. Serum TFF3 in the CC group was significantly higher than in the HC or CA group. The AUC values of TFF3 for discriminating between HC and CC and between CA and CC were 0.930 (0.903, 0.958) and 0.834 (0.796, 0.873). A multivariate model combining TFF3 and CEA was built. Compared to TFF3 or CEA alone, the multivariate model showed significant improvement (P < 0.001). For discriminating between HC and CC, HC and early stage CC, HC and advanced stage CC, CA and CC, CA and early stage CC, and CA and advanced stage CC in the training group, the sensitivities were 92.99%, 91.46%, 93.18%, 73.83%, 76.83%, and 81.82%, and the specificities were 91.30%, 91.30%, 93.91%, 88.38%, 77.27%, and 88.38%, respectively. After validation, the sensitivities were 89.39%, 85.71%, 90.79%, 72.73%, 71.43%, and 78.95%, and the specificities were 87.85%, 87.85%, 2.52%, 87.85%, 80.77%, and 87.50%, respectively.
CONCLUSION
The multivariate diagnostic model that included TFF3 and CEA showed significant improvement over the conventional biomarker CEA and might provide a potential method for the early detection of CC.
Keywords: Trefoil factor family 3, Colorectal cancer, Colorectal adenoma, Multivariate model, Diagnostic value
Core tip: Serum level of trefoil factor family 3 (TFF3) was used for evaluation the diagnostic value of for the early detection of colorectal cancer (CC). A multivariate model combining TFF3 and carcino-embryonic antigen (CEA) was built. Compared to TFF3 or CEA alone, the multivariate model showed significant improvement. The multivariate diagnostic model that included TFF3 and CEA showed significant improvement over the conventional biomarker CEA and might provide a potential method for the early detection of CC.
INTRODUCTION
Colorectal cancer (CC) is one of the most common cancers worldwide. An estimated 131700 new colorectal cancer patients (69090 male and 62610 female) are estimated to have occurred in the United States in 2015[1], and in China, the incidence of colorectal cancer showed a clearly increased tendency. The prognosis of CC is strongly related to the tumor stage. The 5-year relative survival ratio ranges from greater than 90% in patients with stage I to slightly greater than 10% in patients with stage IV[2]. Although various detection methods are used in clinical practice, such as colonoscopy, fecal occult blood testing, stool DNA testing, and carcino-embryonic antigen (CEA), their diagnostic value is limited by disadvantages, and they cannot meet the needs of clinical detection[3]. A detection method with high sensitivity and specificity, easy availability and low cost is urgently needed for the early detection of CC in clinical practice.
The trefoil factor family proteins (TFFs), secreted by the mammalian gastrointestinal tract, are small and stable molecules. They include three thermo stable and protease-resistant proteins (TFF1, TFF2, and TFF3) and are widely distributed in the gastrointestinal tract[4]. Studies have demonstrated that they play important roles in the mucosal protection and repair of epithelial surfaces and are involved in the development and progression of various types of cancer. TFF levels in plasma were found to be heightened in advanced prostate cancer[5] but reduced in the oral mucosal tissues of oral squamous cell carcinoma patients[6]. The levels of TFF3 in the serum and lung tissues were also increased and indicated that TFF3 might serve as a promising biomarker of lung cancer[7]. TFF3 was also found to be expressed in hepatocellular carcinoma, and its expression correlates with tumor grade[8]. In addition to these kinds of cancers, current studies of TFF3 focus mainly on gastric cancer. The level of TFF3 in serum was found to be a better marker of gastric cancer than pepsinogen, and the combination of the levels of serum pepsinogen and TFF3 could improve screening for gastric cancer[9,10], possibly becoming applicable for the chemoprevention of gastrointestinal cancer associated with chronic persistent inflammation[11].
As described above, although many studies have been performed to evaluate the diagnostic value for different kinds of cancers, there are only a handful of studies evaluating the clinical value of TFF3 for CC, and they focused mainly on metastasis and therapy effect. They found that TFFs may be potential serum biomarkers in patients with metastatic colorectal cancer. Compared to CEA and CA19-9, TFF3 showed higher sensitivity and the same specificity, and it was strongly correlated with the extent of liver disease and seemed to have prognostic value[12]. It was also demonstrated to be a risk factor for early recurrence[13]. In addition, serum TFF3 was found to be an effective biomarker for the detection of tumor stages and distant metastasis and as a predictor of responses to chemotherapy in colorectal cancer[14]. However, to date, there has been no study evaluating the clinical diagnostic value of TFF3 for the early detection of colorectal cancer.
In our study, we aimed to evaluate the diagnostic value of TFF3 for the detection of CC and to build a multivariate diagnostic model that might improve the diagnostic value compared to the indicator alone. It may serve as a potential assistant detection method.
MATERIALS AND METHODS
Patients
Written consent was obtained from all participants enrolled in this study. Our study was approved by the Ethics Committee of Tianjin Medical University Cancer Institute and Hospital. Serum samples of 527 individuals, including 115 healthy control (HC), 198 colorectal adenoma (CA), and 214 CC individuals, were collected for the training group. After the training group, an independent 343 individuals, including 107 HC, 104 CA, and 132 CC individuals, were collected to validate the diagnostic value of the training group. Serum samples were collected before surgery, chemotherapy, radiation therapy or immunotherapy. Age-matched healthy controls were enrolled based on their negative results on the blood biomarker test, computed tomography examination and fecal occult blood testing. The patients enrolled in our study were confirmed by histopathological analysis. The tumor stage was categorized according to the Dukes staging system. Duke stages A and B were categorized as early stage colorectal cancer. Dukes stages C and D were categorized as advanced stage CRC[15]. The clinical characteristics of the patients are shown in Table 1.
Table 1.
Clinical characteristic of the individuals in our study n (%)
| Clinical characteristics |
Colorectal cancer group |
Colorectal adenoma group |
Healthy control group |
|||
| Training (n = 214) | Validation (n = 132) | Training (n = 198) | Validation (n = 104) | Training (n = 115) | Validation (n = 107) | |
| Age, yr | ||||||
| Median | 58 | 60 | 57 | 56 | 55 | 53 |
| Range | 43-72 | 41-76 | 41-70 | 36-73 | 38-68 | 39-62 |
| Sex | ||||||
| Male | 128 (59.81) | 84 (63.64) | 107 (54.04) | 62 (59.62) | 61 (53.04) | 56 (52.34) |
| Female | 86 (40.19) | 48 (36.36) | 91 (45.96) | 42 (40.38) | 54 (46.96) | 51 (47.66) |
| Location | ||||||
| Colon | 102 (47.66) | 64 (48.48) | 104 (52.53) | 56 (53.85) | - | - |
| Rectum | 112 (52.34) | 68 (51.52) | 94 (47.47) | 48 (46.15) | - | - |
| Differentiation grade | ||||||
| Well + moderately | 126 (58.88) | 68 (51.52) | - | - | - | - |
| Poorly | 88 (41.12) | 64 (48.48) | - | - | - | - |
| Stage | ||||||
| A + B | 82 (38.32) | 56 (42.42) | - | - | - | - |
| C + D | 132 (61.68) | 76 (57.58) | - | - | - | - |
CRC: Colorectal cancer; CA: Colorectal adenoma; HC: Healthy control; CEA: Carcino-embryonic antigen.
Serum collection
Ten milliliters of peripheral blood was collected in a tube that contained separating gel and clot activator, and then the tube was centrifuged at 3400 rpm for 7 min. The supernatant was transferred into another new tube. The sample serum was stored in aliquots at -80 °C until detection. No freeze-thawing was allowed prior to cytokine detection.
Detection of TFF3 and CEA
The levels of TFF3 (Item ID: E-EL-H1108c) in serum were detected by ELISA kits, which were provided by Elabscience Biotechnology Co., Ltd. (Wuhan, China). The detection protocol was performed according to the manufacturer’s instructions. Briefly, 100 μL of standard, blank, or sample was added per well. Solutions were added to the bottom of the well and incubated for 90 min at 37 °C. The liquid was removed and 100 μL of biotinylated detection Ab working solution added to each well, followed by incubation for 1 h at 37 °C. The liquid was aspirated and the wells washed three times. Then, any remaining wash buffer was removed, and 100 μL of HRP conjugate working solution was added, followed by incubation for 30 min at 37 °C. The wash process was repeated five times, and then 90 μL of substrate solution was added. Incubation was performed for 15 min at 37 °C followed by the addition of 50 μL of stop solution to each well. The optical density (OD value) of each well at 450 nm was measured by a Bio-Rad iMark Microplate Absorbance Reader (Bio-Rad Laboratories Inc.). The level of TFF3 was calculated according to the standard curve. The coefficient of variation of all kits was less than 10%. The levels of CEA in serum were detected by a Roche Modular Analytics E 170 instrument (Roche Diagnostics, Mannheim, Germany). The detection assays were provided by Roche Diagnostics, United States.
Statistical analysis
All the data were analyzed using MedCalc 12.7.0.0 (MedCalc Software, Mariakerke, Belgium) and SPSS 19.0 (SPSS, Brussels, Belgium). The levels of TFF3 and CEA between groups were compared by one-way analysis of variance with the Bonferroni correction. Binary logistic regression analysis was used to establish the multivariate diagnostic model. Receiver operating characteristic curves were used to evaluate the diagnostic value, and the areas under the curves (AUC) were compared by Z-scores[16]. The Youden index was used to choose the cutoff value that determined the sensitivity and specificity. A two-sided P value of less than 0.05 was considered statistically significant.
RESULTS
Correlation of TFF3 with clinical characteristics and comparison of level in groups
The serum level of TFF3 showed no significant correlation with age, gender, or tumor location but showed a significant correlation with tumor stage. As shown in Figure 1, compared to the healthy control group, the levels of TFF3 in the HC, CA, and CC groups were 14.10 (11.28, 23.19), 23.08 (18.72, 29.09), and 37.66 (29.87, 47.61) pg/mL, respectively. Compared to the HC group, the levels of TFF3 in both the CA group and the CC group showed significant increases (P < 0.001). Compared to the CA group, the level of TFF3 in the CC group showed a significant increase (P < 0.001).
Figure 1.
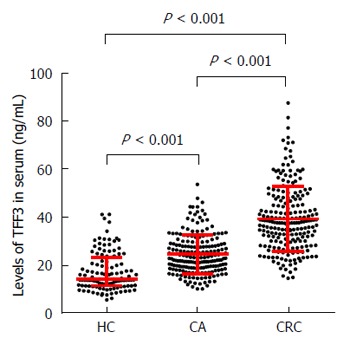
Comparisons of serum trefoil factor family 3 levels in colorectal cancer, colorectal adenoma, and healthy control groups. CRC: Colorectal cancer; CA: Colorectal adenoma; HC: Healthy control groups.
Diagnostic evaluation of TFF3 and CEA for discriminating HC and CC groups
We first analyzed the diagnostic value of TFF3 or CEA alone for discriminating between the HC and CC groups, and then we analyzed the diagnostic value of the combination of TFF3 and CEA. The diagnostic values are given in Supplementary Table 1.
For discriminating between HC and CC, as shown in Figure 2A, the AUC of TFF3 was 0.930 (0.903, 0.958), and at the cutoff value of 21.30 pg/mL, the sensitivity and specificity were 94.86% and 73.91%, respectively. As shown in Figure 2D, the AUC of CEA was 0.850 (0.809, 0.890), and at the cutoff value of 3.09 U/mL, the sensitivity and specificity were 70.56% and 92.17%, respectively. Then, TFF3 and CEA were combined for analysis by binary logistic regression analysis to build the multivariate diagnostic model. The formula of the model was Y=logit(P)=-6.498+0.189XTTF3+0.651XCEA. As shown in Figure 2G, the AUC of the multivariate model was 0.968 (0.951, 0.984), and at the cutoff value of 0.60, the sensitivity and specificity were 92.99% and 91.30%, respectively.
Figure 2.
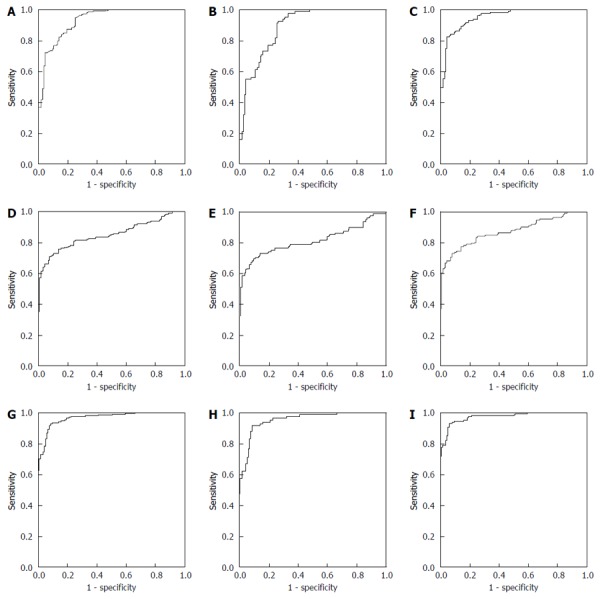
Analysis the Trefoil factors family and carcino-embryonic antigen diagnostic evaluation for discriminating the healthy control and colorectal cancer groups by receiver operating characteristic method in the training group. A: ROC of TFF3 for discriminating HC and CC; B: ROC of TFF3 for discriminating HC and early stage CC; C: ROC of TFF3 for discriminating HC and advanced stage CC; D: ROC of CEA for discriminating HC and CC; E: ROC of CEA for discriminating HC and early stage CC; F: ROC of CEA for discriminating HC and advanced stage CC; G: ROC of multivariate model for discriminating HC and CC; H: ROC of multivariate model for discriminating HC and early stage CC; I: ROC of multivariate model for discriminating HC and advanced stage CC. TFF3: Trefoil factors family; ROC: Receiver operating characteristic; HC: Healthy control; CC: Colorectal cancer.
For discriminating between HC and early stage CC, as shown in Figure 2B, the AUC of TFF3 was 0.892 (0.849, 0.935), and at the cutoff value of 21.30 pg/ml, the sensitivity and specificity were 92.68% and 73.91%, respectively. As shown in Figure 2E, the AUC of CEA was 0.814 (0.745, 0.882), and at the cutoff value of 3.00 U/mL, the sensitivity and specificity were 69.51% and 90.43%, respectively. As shown in Figure 2H, the AUC of the multivariate model built to discriminate between HC and CC was 0.953 (0.926, 0.981), and at the cutoff value of 0.60, the sensitivity and specificity were 91.46% and 91.30%, respectively.
For discriminating between HC and advanced stage CC, as shown in Figure 2C, the AUC of TFF3 was 0.954 (0.931, 0.977), and at the cutoff value of 31.77 pg/mL, the sensitivity and specificity were 81.82% and 95.65%, respectively. As shown in Figure 2F, the AUC of CEA was 0.872 (0.828, 0.917), and at the cutoff value of 3.09 U/mL, the sensitivity and specificity were 72.73% and 92.17%, respectively. As shown in Figure 2I, the AUC of the multivariate model built to discriminate between HC and CC was 0.976 (0.961, 0.992), and at the cutoff value of 0.72, the sensitivity and specificity were 93.18% and 93.91%, respectively.
Diagnostic evaluation of TFF3 for discriminating between CA and CC groups in the training group
After discriminating between the HC and CC groups, we analyzed the diagnostic value of TFF3 and CEA alone or in combination for discriminating between the CA and CC groups. The diagnostic value is shown in Table 2, and the AUCs are shown in Supplementary Figure 1.
Table 2.
Diagnostic evaluation of trefoil factors family, carcino-embryonic antigen alone or combination for discriminating colorectal adenoma and colorectal cancer in the training group
| Indicator | Groups | AUC (95%CI) | Cutoff value | Sensitivity | Specificity |
| TFF3 | CA vs CRC | 0.834 (0.796, 0.873) | 29.89 | 75.23% | 78.28% |
| CA vs early stage CRC | 0.751 (0.691, 0.812) | 29.89 | 58.54% | 78.28% | |
| CA vs advanced CRC | 0.886 (0.849, 0.923) | 34.07 | 75.00% | 89.39% | |
| CEA | CA vs CRC | 0.683 (0.630, 0.737) | 4.96 | 57.01% | 85.86% |
| CA vs early CRC | 0.648 (0.563, 0.734) | 4.53 | 56.10% | 81.31% | |
| CA vs advanced stage CRC | 0.705 (0.641, 0.770) | 4.96 | 60.61% | 85.86% | |
| TFF3+CEA | CA vs CRC | 0.883 (0.851, 0.915) | 0.57 | 73.83% | 88.38% |
| CA vs early stage CRC | 0.823 (0.768, 0.878) | 0.41 | 76.83% | 77.27% | |
| CA vs advanced stage CRC | 0.919 (0.888, 0.951) | 0.57 | 81.82% | 88.38% |
TFF3: Trefoil factors family; CA: Colorectal adenoma; CRC: Colorectal cancer; AUC: Area under curve.
For discriminating between CA and CC, the AUC of TFF3 was 0.834 (0.796, 0.873), and at the cutoff value of 29.89 pg/mL, the sensitivity and specificity were 75.23% and 78.28%, respectively. The AUC of CEA was 0.683 (0.630, 0.737), and at the cutoff value of 4.96 U/mL, the sensitivity and specificity were 57.01% and 85.86%, respectively. Then, TFF3 and CEA were combined by binary logistic regression analysis to build the multivariate diagnostic model. The formula of the model was Y=logit(P)=-5.478+0.139XTTF3+0.265XCEA. The AUC of the multivariate model was 0.883 (0.851, 0.915), and at the cutoff value of 0.57, the sensitivity and specificity were 73.83% and 88.38%, respectively. Compared to TFF3 or CEA alone, the AUC of the multivariate model showed significant improvement (P < 0.001 and P < 0.001).
For discriminating between CA and early stage CC, the AUC of TFF3 was 0.751 (0.691, 0.812), and at the cutoff value of 29.89 pg/mL, the sensitivity and specificity were 58.54% and 78.28%, respectively. The AUC of CEA was 0.648 (0.563, 0.734), and at the cutoff value of 4.53 U/mL, the sensitivity and specificity were 56.10% and 81.31%, respectively. The AUC of the multivariate model built to discriminate between CA and CC was 0.823 (0.768, 0.878), and at the cutoff value of 0.41, the sensitivity and specificity were 76.83% and 77.27%, respectively.
For discriminating between CA and advanced stage CC, the AUC of TFF3 was 0.886 (0.849, 0.923), and at the cutoff value of 34.07 pg/mL, the sensitivity and specificity were 75.00% and 89.39%, respectively. The AUC of CEA was 0.705 (0.641, 0.770), and at the cutoff value of 4.96 U/mL, the sensitivity and specificity were 60.61% and 86.86%, respectively. The AUC of the multivariate model built to discriminate between CA and CC was 0.919 (0.888, 0.951), and at the cutoff value of 0.57, the sensitivity and specificity were 81.82% and 88.38%, respectively.
Validation of the multivariate model for discriminating between HC and CC and between CA and CC in the validation group
After building the multivariate models to discriminate between HC and CC and between CA and CC, independent HC, CA and CC individuals were chosen to validate the diagnostic value of the multivariate models, as shown in Supplementary Table 2.
For discriminating between the HC and CC groups, as shown in Figure 3A, the AUC was 0.941 (0.912, 0.970), and at the cutoff value of 0.60, the sensitivity and specificity were 89.39% and 87.85%, respectively. For discriminating between the HC and early stage CC groups, as shown in Figure 3B, the AUC was 0.910 (0.856, 0.965), and at the cutoff value of 0.60, the sensitivity and specificity were 85.71% and 87.85%, respectively. For discriminating between the HC and advanced stage CC groups, as shown in Figure 3C, the AUC was 0.961 (0.938, 0.991), and at the cutoff value of 0.72, the sensitivity and specificity were 90.79% and 92.52%, respectively. Compared to TFF3 or CEA alone, the AUC of the multivariate model showed significant improvement (P < 0.001 and P < 0.001).
Figure 3.

Analysis the multivariate model diagnostic evaluation by receiver operating characteristic method in the validation group. A: ROC of multivariate model for discriminating HC and CC; B: ROC of multivariate model for discriminating HC and early stage CC; C: ROC of multivariate model for discriminating HC and advanced stage CC; D: ROC of multivariate model for discriminating CA and CC; E: ROC of multivariate model for discriminating CA and early stage CC; F: ROC of multivariate model for discriminating CA and advanced stage CC. TFF3: Trefoil factors family; ROC: Receiver operating characteristic; CA: Colorectal adenoma; CC: Colorectal cancer.
For discriminating between the CA and CC groups, as shown in Figure 3D, the AUC was 0.850 (0.799, 0.902), and at the cutoff value of 0.57, the sensitivity and specificity were 72.73% and 87.50%, respectively. For discriminating between the HC and early stage CC groups, as shown in Figure 3E, the AUC was 0.814 (0.741, 0.887), and at the cutoff value of 0.41, the sensitivity and specificity were 71.43% and 80.77%, respectively. For discriminating between the HC and advanced stage CC groups, as shown in Figure 3F, the AUC was 0.877 (0.824, 0.929), and at the cutoff value of 0.57, the sensitivity and specificity were 78.95% and 87.50%, respectively.
DISCUSSION
TFF3, also called intestinal trefoil factor, consists of 59 amino acid peptides and occurs mainly in the gastrointestinal tract and in the serum. TFF3 expression is elevated during gastrointestinal adenoma progression and has been shown to promote mucosal wound healing. The induction of mucinous metaplasia was observed in mice with high TFF3 expression[17]. The TFFs can be used as biomarkers in various human cancers[18]. For gastric cancer, the serum TFF3 level may be a better biomarker of gastric cancer than the pepsinogen test. When combined with the serum pepsinogen test, TFF3 showed better diagnostic value for the screening of gastric cancer[9,10] and might be a potential non-endoscopic detection method for the screening of gastric cancer[19]. It also acted as an angiogenic factor and functions as a promoter to enhance tumor progression in mammary carcinoma[20]. In addition, the Cytosponge-TFF3 test is a safe and acceptable approach to identify patients with reflux symptoms who warrant endoscopy to diagnose Barrett’s esophagus[21]. TFF3 plays an important role in the development of Barrett’s metaplasia and may have diagnostic value for the early stages of Barrett’s esophagus[22]. Although many studies have been performed to evaluate its diagnostic value for different cancers, few studies have evaluated the diagnostic value of TFF3 for the early detection of CC.
In our study, serum TFF3 showed significant correlation with tumor stage. This result was consistent with previous studies. The relationship between serum TFF3 and lymph node metastases of CC may make it a potentially useful marker for predicting the lymph node metastases[23], and it may also serve as a potential biomarker for the prediction of CC metastasis[24]. TFF3 up-regulation after neoadjuvant chemoradiotherapy for rectal cancer is associated with a higher risk of relapse[25]. Serum TFF3 can potentially be used as a biomarker to assess mucosal healing in ulcerative colitis patients[26]. In our study, compared to the HC and CA groups, serum TFF3 in the CC group showed a significant increase. It may contribute to the development of CC. In previous studies, TFF3 was demonstrated to contribute to the malignant behavior of colon cancer cells[27], and it was up-regulated in mucosal protection and repair. Its levels were increased in correlation with disease activity indices[28]. TFF3 level was also found to correlate with an aggressive phenotype in rat colon cancer cells. These findings provide evidence that TFF3 contributes to the malignant behavior of cancer cells[29]. There are some proposed mechanisms by which TFF3 participates in the development of CC. Signal transducer and activator of transcription (STAT) 3 has been demonstrated to be over expressed in most types of human cancers and classified as an oncogene. TFF3 may exert potent invasive activity through STAT3 signaling in human colorectal cancer cells[30]. In addition, TFF may also promote the proliferation and migration of gastric mucosal epithelial cells by activation of the PI3K/Akt signaling pathway, which has been demonstrated to be strongly related to the development of various cancers[31,32]. IL4-induced Stat6 signaling is active in various cell types, included immune cells and cancer cells. STAT6 activation mediates a transcriptional enhancement of TFF3 by de novo induction, which plays an important role in host protective immunity against the infection synthesized protein in goblet cells[33]. TFF3 has been found to inhibit the TLR4/NF-kappaB signaling pathways, with potential treatment value for the inflammatory bowel disease[34]. Perturbation of the E-cadherin/catenin complex at intercellular junctions appears to be a functional pathway through which TFF2 and TFF3 promote cell migration[35]. In our study, for discriminating between HC and CC, the multivariate model showed significant improvement compared to CEA alone; however, because the prevalence of colorectal cancer was not taken into consideration, the diagnostic value of our study could be biased, and the disparity in the number of patients recruited in our study for the training group may also cause some bias in the diagnostic value. For discriminating between CA and CC, the multivariate model also showed significant improvement compared to CEA, as a method based on non-invasive discrimination. It was better than the conventional non-invasive method. In future research, the multivariate model should be compared with other discrimination methods, such as colonoscopy and fecal occult blood testing.
There are some limitations in our study. First, the number of individuals in the training group was relatively small, causing some bias in the results of our study. A larger sample size and multi-center sampling should be used to validate the diagnostic value of TFF3 and the multivariate diagnostic model. Second, although the diagnostic value of the multivariate model for discriminating between HC and CC was high, the diagnostic value for other kinds of cancers was not evaluated. The multivariate model built in our study currently can only be recognized as an assistant detection method that should be combined with the detection methods used in clinical practice, such as colonoscopy, fecal occult blood testing, and stool DNA testing. Third, in our study, we only evaluated the diagnostic value of TFF3 for the early detection of CC. The levels of the TFF3 after surgery, chemotherapy, radiotherapy, and other kinds of therapy methods were not evaluated. In future research, we will analyze TFF3 for evaluation of the effect of therapy or its correlation with prognosis.
In conclusion, we evaluated the diagnostic value of TFF3 for differentiating between the HC and CC and between the CA and CC groups, and we evaluated a multivariate diagnostic model that included TFF3 and CEA for differentiating between the HC and CC and between the CA and CC groups. Compared to the conventional biomarker CEA, the multivariate diagnostic model showed significant improvement. It could be used as an assistant detection method alongside the conventional screening methods for colorectal cancer, and it could also be used as a potentially effective diagnostic method for discriminating between CA and CC patients in clinical detection.
COMMENTS
Background
Colorectal cancer is one of the most common cancers worldwide. Although various detection methods are used in clinical practice, their diagnostic value is limited by disadvantages, and they cannot meet the needs of clinical detection. A detection method with high sensitivity and specificity, easy availability and low cost is urgently needed for the early detection in clinical practice.
Research frontiers
Although many studies have been performed to evaluate the diagnostic value of trefoil factor family 3 (TFF3) for different kinds of cancers, such as, however, to date, there has been no study evaluating the clinical diagnostic value of TFF3 for the early detection of colorectal cancer.
Innovations and breakthroughs
Serum level of TFF3 was used for evaluation the diagnostic value of for the early detection of colorectal cancer. A multivariate model combining TFF3 and carcino-embryonic antigen (CEA) was built. Compared to TFF3 or CEA alone, the multivariate model showed significant improvement.
Applications
The multivariate diagnostic model that included TFF3 and CEA showed significant improvement over the conventional biomarker CEA and might provide a potential method for the early detection of colorectal cancer.
Terminology
TFFs play important roles in the mucosal protection and repair of epithelial surfaces and are involved in the development and progression of various types of cancer.
Peer-review
This study is an interesting study about the diagnostic value evaluation of trefoil factors family 3 for the early detection of colorectal cancer. The multivariate diagnostic model which included TFF3 and CEA showed significant improvement when compared to the conventional biomarker CEA, and may provide a potential method for the early detection of colorectal cancer. Overall, this study is well designed and the manuscript is well written.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
Institutional review board statement: The study was reviewed and approved by the 302 Hospital of People’s Liberation Army and Institutional Review Board.
Informed consent statement: All study participants or their legal guardians provided written informed consent prior to study enrollment.
Conflict-of-interest statement: We declare that we have no financial or personal relationships with other individuals or organizations that can inappropriately influence our work and that there is no professional or other personal interest of any nature in any product, service and/or company that could be construed as influencing the position presented in or the review of the manuscript.
Data sharing statement: The technical appendix, statistical code, and dataset are available from the corresponding author at guoztj@126.com and hmwang302@126.com. The study participants provided informed consent for data sharing. No additional data are available.
Peer-review started: February 17, 2017
First decision: February 28, 2017
Article in press: March 17, 2017
P- Reviewer: Raisch KP, Sato H S- Editor: Qi Y L- Editor: A E- Editor: Wang CH
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383:1490–1502. doi: 10.1016/S0140-6736(13)61649-9. [DOI] [PubMed] [Google Scholar]
- 3.Gonzalez-Pons M, Cruz-Correa M. Colorectal Cancer Biomarkers: Where Are We Now? Biomed Res Int. 2015;2015:149014. doi: 10.1155/2015/149014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lafontaine PO, Arnal M, Buron N, Solary E, Lizard S, Bron A, Bara J, Gespach C, Creuzot-Garcher C. [Trefoil factor family gene and peptide expression in pterygium] J Fr Ophtalmol. 2003;26:1007–1014. [PubMed] [Google Scholar]
- 5.Vestergaard EM, Borre M, Poulsen SS, Nexø E, Tørring N. Plasma levels of trefoil factors are increased in patients with advanced prostate cancer. Clin Cancer Res. 2006;12:807–812. doi: 10.1158/1078-0432.CCR-05-1545. [DOI] [PubMed] [Google Scholar]
- 6.Chaiyarit P, Utrawichian A, Leelayuwat C, Vatanasapt P, Chanchareonsook N, Samson MH, Giraud AS. Investigation of trefoil factor expression in saliva and oral mucosal tissues of patients with oral squamous cell carcinoma. Clin Oral Investig. 2012;16:1549–1556. doi: 10.1007/s00784-011-0667-z. [DOI] [PubMed] [Google Scholar]
- 7.Qu Y, Yang Y, Ma D, Xiao W. Increased trefoil factor 3 levels in the serum of patients with three major histological subtypes of lung cancer. Oncol Rep. 2012;27:1277–1283. doi: 10.3892/or.2012.1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Khoury T, Chadha K, Javle M, Donohue K, Levea C, Iyer R, Okada H, Nagase H, Tan D. Expression of intestinal trefoil factor (TFF-3) in hepatocellular carcinoma. Int J Gastrointest Cancer. 2005;35:171–177. doi: 10.1385/IJGC:35:3:171. [DOI] [PubMed] [Google Scholar]
- 9.Aikou S, Ohmoto Y, Gunji T, Matsuhashi N, Ohtsu H, Miura H, Kubota K, Yamagata Y, Seto Y, Nakajima A, et al. Tests for serum levels of trefoil factor family proteins can improve gastric cancer screening. Gastroenterology. 2011;141:837–845.e1-7. doi: 10.1053/j.gastro.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang Z, Zhang X, Lu H, Wu L, Wang D, Zhang Q, Ding H. Serum trefoil factor 3 is a promising non-invasive biomarker for gastric cancer screening: a monocentric cohort study in China. BMC Gastroenterol. 2014;14:74. doi: 10.1186/1471-230X-14-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Katoh M. Trefoil factors and human gastric cancer (review) Int J Mol Med. 2003;12:3–9. [PubMed] [Google Scholar]
- 12.Vocka M, Langer D, Petrtyl J, Vockova P, Hanus T, Kalousova M, Zima T, Petruzelka L. Trefoil factor family (TFF) proteins as potential serum biomarkers in patients with metastatic colorectal cancer. Neoplasma. 2015;62:470–477. doi: 10.4149/neo_2015_056. [DOI] [PubMed] [Google Scholar]
- 13.Morito K, Nakamura J, Kitajima Y, Kai K, Tanaka T, Kubo H, Miyake S, Noshiro H. The value of trefoil factor 3 expression in predicting the longterm outcome and early recurrence of colorectal cancer. Int J Oncol. 2015;46:563–568. doi: 10.3892/ijo.2014.2755. [DOI] [PubMed] [Google Scholar]
- 14.Xiao L, Liu YP, Xiao CX, Ren JL, Guleng B. Serum TFF3 may be a pharamcodynamic marker of responses to chemotherapy in gastrointestinal cancers. BMC Clin Pathol. 2014;14:26. doi: 10.1186/1472-6890-14-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong JM, Yen MF, Lai MS, Duffy SW, Smith RA, Chen TH. Progression rates of colorectal cancer by Dukes’ stage in a high-risk group: analysis of selective colorectal cancer screening. Cancer J. 2004;10:160–169. doi: 10.1097/00130404-200405000-00005. [DOI] [PubMed] [Google Scholar]
- 16.Pengjun Z, Xinyu W, Feng G, Xinxin D, Yulan L, Juan L, Xingwang J, Zhennan D, Yaping T. Multiplexed cytokine profiling of serum for detection of colorectal cancer. Future Oncol. 2013;9:1017–1027. doi: 10.2217/fon.13.71. [DOI] [PubMed] [Google Scholar]
- 17.Ge H, Gardner J, Wu X, Rulifson I, Wang J, Xiong Y, Ye J, Belouski E, Cao P, Tang J, et al. Trefoil Factor 3 (TFF3) Is Regulated by Food Intake, Improves Glucose Tolerance and Induces Mucinous Metaplasia. PLoS One. 2015;10:e0126924. doi: 10.1371/journal.pone.0126924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.May FE. The potential of trefoil proteins as biomarkers in human cancer. Biomark Med. 2012;6:301–304. doi: 10.2217/bmm.12.22. [DOI] [PubMed] [Google Scholar]
- 19.Kaise M, Miwa J, Tashiro J, Ohmoto Y, Morimoto S, Kato M, Urashima M, Ikegami M, Tajiri H. The combination of serum trefoil factor 3 and pepsinogen testing is a valid non-endoscopic biomarker for predicting the presence of gastric cancer: a new marker for gastric cancer risk. J Gastroenterol. 2011;46:736–745. doi: 10.1007/s00535-011-0396-8. [DOI] [PubMed] [Google Scholar]
- 20.Lau WH, Pandey V, Kong X, Wang XN, Wu Z, Zhu T, Lobie PE. Trefoil Factor-3 (TFF3) Stimulates De Novo Angiogenesis in Mammary Carcinoma both Directly and Indirectly via IL-8/CXCR2. PLoS One. 2015;10:e0141947. doi: 10.1371/journal.pone.0141947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ross-Innes CS, Debiram-Beecham I, O’Donovan M, Walker E, Varghese S, Lao-Sirieix P, Lovat L, Griffin M, Ragunath K, Haidry R, et al. Evaluation of a minimally invasive cell sampling device coupled with assessment of trefoil factor 3 expression for diagnosing Barrett’s esophagus: a multi-center case-control study. PLoS Med. 2015;12:e1001780. doi: 10.1371/journal.pmed.1001780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dunn LJ, Jankowski JA, Griffin SM. Trefoil Factor Expression in a Human Model of the Early Stages of Barrett’s Esophagus. Dig Dis Sci. 2015;60:1187–1194. doi: 10.1007/s10620-014-3440-8. [DOI] [PubMed] [Google Scholar]
- 23.Huang YG, Li YF, Wang LP, Zhang Y. Aberrant expression of trefoil factor 3 is associated with colorectal carcinoma metastasis. J Cancer Res Ther. 2015;9:376–380. doi: 10.4103/0973-1482.119308. [DOI] [PubMed] [Google Scholar]
- 24.Xue H, Lü B, Zhang J, Wu M, Huang Q, Wu Q, Sheng H, Wu D, Hu J, Lai M. Identification of serum biomarkers for colorectal cancer metastasis using a differential secretome approach. J Proteome Res. 2010;9:545–555. doi: 10.1021/pr9008817. [DOI] [PubMed] [Google Scholar]
- 25.Casado E, Garcia VM, Sánchez JJ, Gómez Del Pulgar MT, Feliu J, Maurel J, Castelo B, Moreno Rubio J, López RA, García-Cabezas MÁ, Burgos E, de Castro J, Belda-Iniesta C, López-Gómez M, Gómez-Raposo C, Zambrana F, Sereno M, Fernández-Martos C, Vázquez P, Lacal JC, González-Barón M, Cejas P. Upregulation of trefoil factor 3 (TFF3) after rectal cancer chemoradiotherapy is an adverse prognostic factor and a potential therapeutic target. Int J Radiat Oncol Biol Phys. 2012;84:1151–1158. doi: 10.1016/j.ijrobp.2012.01.083. [DOI] [PubMed] [Google Scholar]
- 26.Srivastava S, Kedia S, Kumar S, Pratap Mouli V, Dhingra R, Sachdev V, Tiwari V, Kurrey L, Pradhan R, Ahuja V. Serum human trefoil factor 3 is a biomarker for mucosal healing in ulcerative colitis patients with minimal disease activity. J Crohns Colitis. 2015;9:575–579. doi: 10.1093/ecco-jcc/jjv075. [DOI] [PubMed] [Google Scholar]
- 27.Babyatsky M, Lin J, Yio X, Chen A, Zhang JY, Zheng Y, Twyman C, Bao X, Schwartz M, Thung S, et al. Trefoil factor-3 expression in human colon cancer liver metastasis. Clin Exp Metastasis. 2009;26:143–151. doi: 10.1007/s10585-008-9224-9. [DOI] [PubMed] [Google Scholar]
- 28.Grønbaek H, Vestergaard EM, Hey H, Nielsen JN, Nexø E. Serum trefoil factors in patients with inflammatory bowel disease. Digestion. 2006;74:33–39. doi: 10.1159/000096591. [DOI] [PubMed] [Google Scholar]
- 29.Yio X, Zhang JY, Babyatsky M, Chen A, Lin J, Fan QX, Werther JL, Itzkowitz S. Trefoil factor family-3 is associated with aggressive behavior of colon cancer cells. Clin Exp Metastasis. 2005;22:157–165. doi: 10.1007/s10585-005-6615-z. [DOI] [PubMed] [Google Scholar]
- 30.Rivat C, Rodrigues S, Bruyneel E, Piétu G, Robert A, Redeuilh G, Bracke M, Gespach C, Attoub S. Implication of STAT3 signaling in human colonic cancer cells during intestinal trefoil factor 3 (TFF3) -- and vascular endothelial growth factor-mediated cellular invasion and tumor growth. Cancer Res. 2005;65:195–202. [PubMed] [Google Scholar]
- 31.Sun Z, Liu H, Yang Z, Shao D, Zhang W, Ren Y, Sun B, Lin J, Xu M, Nie S. Intestinal trefoil factor activates the PI3K/Akt signaling pathway to protect gastric mucosal epithelium from damage. Int J Oncol. 2014;45:1123–1132. doi: 10.3892/ijo.2014.2527. [DOI] [PubMed] [Google Scholar]
- 32.Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, González-Barón M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30:193–204. doi: 10.1016/j.ctrv.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 33.Blanchard C, Durual S, Estienne M, Bouzakri K, Heim MH, Blin N, Cuber JC. IL-4 and IL-13 up-regulate intestinal trefoil factor expression: requirement for STAT6 and de novo protein synthesis. J Immunol. 2004;172:3775–3783. doi: 10.4049/jimmunol.172.6.3775. [DOI] [PubMed] [Google Scholar]
- 34.Teng X, Xu LF, Zhou P, Sun HW, Sun M. Effects of trefoil peptide 3 on expression of TNF-alpha, TLR4, and NF-kappaB in trinitrobenzene sulphonic acid induced colitis mice. Inflammation. 2009;32:120–129. doi: 10.1007/s10753-009-9110-x. [DOI] [PubMed] [Google Scholar]
- 35.Buda A, Jepson MA, Pignatelli M. Regulatory function of trefoil peptides (TFF) on intestinal cell junctional complexes. Cell Commun Adhes. 2012;19:63–68. doi: 10.3109/15419061.2012.748326. [DOI] [PubMed] [Google Scholar]


