Abstract
Tomato is a model species for fleshy fruit development. The shapes and sizes of tomato (Solanum lycopersicum L.) are mainly controlled by several loci, including locule number (lc). Two single nucleotide polymorphisms were found downstream of WUSCHEL (SlWUS) in a putative tomato CArG cis-regulatory element. The lc mutation may affect the binding of AGAMOUS(AG), cause the up-regulation of SlWUS and result in increased locule numbers. In this study, tissue expression levels showed that SlWUS is expressed in young floral buds and shoot apexes. Silencing SlWUS on an lc mutant genetic background with an RNA interference (RNAi) strategy resulted in smaller flowers and fruit than those of the wild-type plants, with decreased locule number. Further study revealed that the SlWUS RNAi lines exhibited altered expression levels of the TAG1 and SlCLV3 genes that participate in the regulation of tomato flower and fruit locule development. In conclusion, this study provides the first genetic evidence that SlWUS may be the candidate gene of the lc locus and reveals the function of SlWUS in flower development.
Keywords: SlWUS, flower organogenesis, fruit size, locule number, tomato
Introduction
Tomato (Solanum lycopersicum L.) is an ideal model plant for fruit development research (Klee and Giovannoni, 2011; Consortium, 2012). Fruit size is the primary characteristic of commercial tomato varieties and an important goal for tomato domestication. Domesticated tomato fruit is enlarged 1000 times compared to its wild progenitors, which is an extreme case. In this dramatic transition, both carpel cell division and carpel number determine the final size of tomato fruit (Tanksley, 2004). A relatively small number of genes are involved in these two processes. In the first process, negative regulation of FRUIT WEIGHT 2.2 (FW2.2) (Frary et al., 2000), which located in the plasma membrane, is required for the control of carpel cell number (Liu and Tanksley, 2003). Hence, low expression levels of the large-fruited allele of FW2.2 stimulate cell division, which leads to increases in the final size of tomato fruit. Nonetheless, increased locule number has the greatest effect on tomato fruit size, which is derived from the carpels in tomato flowers. Almost all wild tomatoes and several small-fruited tomato cultivars produce fruits with only two to four locules. However, most cultivars consumed today produce fruits with six or more locules (Tanksley, 2004). Therefore, increased locule number contributes as much as 50% variance to fruit enlargement and is believed to represent a late step in the substantial increase in tomato fruit size during domestication (Lippman and Tanksley, 2001; Tanksley, 2004). An increase in locule number is highly associated with an increase in the number of floral organs, especially the carpel, and this trait is controlled by multiple quantitative trait loci (QTL) (Barrero and Tanksley, 2004), and a few of these loci have been identified (Grandillo et al., 1999; Fernández-Lozano et al., 2015; Illa-Berenguer et al., 2015). To date, the locule number has been shown to be controlled by two loci-fascinated (fas) and locule number (lc) (Lippman and Tanksley, 2001; Knaap and Tanksley, 2003; Barrero et al., 2006). Significant epistatic interactions have been detected between lc and fas (Lippman and Tanksley, 2001; Barrero and Tanksley, 2004). The positive epistatic interaction between lc and fas suggests that they are part of a single regulatory network that controls the size of floral meristems (van der Knaap et al., 2014; Swinnen et al., 2016).
The mutation in fas results in increased carpel numbers with more significant effects on carpel number than lc (Lippman and Tanksley, 2001). The fas mutant phenotype was previously reported to be caused by loss of expression of a YABBY-like transcription factor (Cong et al., 2008). However, recent studies that were carried out to further dissect the contribution of the fas locus; these studies identified a 294-kb inversion on chromosome 11 with breakpoints in intron 1 of a YABBY-like gene and 1 kb upstream of the tomato CLAVATA3 (SlCLV3), which demonstrated that a regulatory change in SlCLV3 underlies the fas mutant phenotype (Huang and van der Knaap, 2011).
The lc locus regulates the number of locules, and lc mutation leads to a tomato fruit with more than the two to four locules (Barrero et al., 2006; Muños et al., 2011). The lc locus is located within a non-coding region, but the lc locus may correspond to two single nucleotide polymorphisms (SNPs), which are located 1,080 bp downstream of the tomato ortholog of WUS (Muños et al., 2011) that encodes a transcription factor that is essential to maintain stem cell identity in the shoot apical meristems (SAM) (Mayer et al., 1998; Clark, 2001). In Arabidopsis, increased expression of WUS results in increased floral organ number, which resembles the phenotype of the lc mutant (Mayer et al., 1998; Clark, 2001). Although the function of these two SNPs has not yet been identified, they may be involved in the regulation of the expression of SlWUS or other genes that play a major role in floral development (Muños et al., 2011).
The homeobox gene WUSCHEL (WUS) plays an important role in maintaining the balance between the proliferation and differentiation of stem cells in the meristems of Arabidopsis thaliana. WUS positively regulates the expression of AGAMOUS (AG), which is part of the MADS box transcription factor (Lenhard et al., 2001; Lohmann et al., 2001). AG plays a key role in regulating stamen and gynoecium determinacy (Yanofsky et al., 1990). Thus, WUS-induced expression of AG links meristem activities to organ identity processes. In reverse, AG down-regulates the expression of WUS, which provides the mechanism for promoting the cells at the center of the flower to differentiate into carpels (Lohmann et al., 2001; Liu et al., 2011). In Arabidopsis, WUS down-regulation is mediated by two downstream CArG cis-regulatory elements that bind to AG and lead to the epigenetic silencing of WUS (Tilly et al., 1998; Liu et al., 2011). Interestingly, the two SNPs located downstream of tomato WUSCHEL (SlWUS) are located in a putative tomato CArG cis-regulatory element (van der Knaap et al., 2014). It has been reported that TAG1 silencing lines show defects in stamen and carpel development in tomatoes (Pan et al., 2010). Although floral determinacy in Arabidopsis relies on a negative autoregulatory mechanism involving AG and WUS, the interaction between TAG1 and the SlWUS in tomato is unknown. Additional key components of the WUS signaling pathway are the CLAVATA (CLV) proteins (Schoof et al., 2000; Clark, 2001; Lenhard and Laux, 2003). Furthermore, the WUS and CLV3 feedback loop is closely connected to the control of meristem size in Arabidopsis (Schoof et al., 2000). The fas mutant phenotype is caused by a loss of expression of a clv3-like ortholog (SlCLV3) and changes in the regulation of SlWUS that underlie the lc locus (Muños et al., 2011).
The lc locus is reported to be a key determining factor of the final carpel number in tomato fruit. SlWUS has been proposed as a candidate gene for lc. To address the potential role of SlWUS in tomato locules, ribonucleic acid interference (RNAi) transgenic plants were produced, which lead to the down-regulation of the SlWUS gene in tomato. The flower and fruit phenotype in the SlWUS RNAi lines revealed the involvement of SlWUS in the control of tomato floral organs and fruit locule number. Further genetic analysis indicated that the expression levels of the TAG1 and SlCLV3 genes were altered in the SlWUS RNAi lines. These results suggested that SlWUS plays an important role in tomato flower and fruit development.
Materials and Methods
Plant Materials, DNA Extraction, and Marker Development for the LC Gene
Three tomato (S. lycopersicum L.) lines, ‘MLK1,’ ‘FL1,’ and ‘Zhongshu6’ were planted in May 2014 at Shenyang Agriculture University, Shenyang, China. The ‘MLK1’ fruit were large with 10 or more locules, the ‘FL1’ fruit were small with 2–4 locules and the ‘Zhongshu6’ fruit were large with 6–8 locules. The cultivated allele of lc is present in ‘MLK1’ and ‘Zhongshu 6,’ and the wild-type allele of lc is present in ‘FL1.’ With the exception of the differences in locule number, the sepals, petals, and other agronomic traits are similar among these three variants. Genomic DNA was isolated from the leaves of the above mentioned plants using a DNA Extraction Kit (Tiangen Biotech, Beijing, China) in accordance with the manufacturer’s instructions. The PCR-based markers developed by Rodríguez et al. (2011) were used to identify the recessive high-locule-number allele at the LC locus used in this work. The primers lc-F1 (5′-GTCTCTTGGATGATGACTATTGCACTTT-3′) and lc-R1 (5′-AAAGTAGTACGAATTGTCCAATCAGTCAG-3′) were used to amplify the low-locule-number allele, while lc-F2 (5′-CTTTTCCTAAAAGATTTGGCATGAGGT-3′) and lc-R2 (5′-TCAGCGCCTCATTTTCTATAGTATTTGT-3′) were used to amplify the recessive high-locule-number allele. When the cultivated allele is present, lc-F1 and lc-R1 will amplify a band of 533 bp; when the wild-type allele is present, lc-F2 and lc-R2 will amplify a band of 395 bp. The two primers were used in the same PCR master mix following the method described by Rodríguez et al. (2011).
Construction of RNAi and Overexpression Vectors and Tomato Transformation
The cDNA of SlWUS was amplified from ‘Zhongshu6’ by real-time polymerase chain reaction (RT–PCR) using specific primers from tomato (GenBank accession number AJ538329) (Muños et al., 2011). The RNAi-specific primers used for the SlWUS gene are as follows: SlWUS-for (5′-CAACGAGCGATCAGATAAGAATA-3′) and SlWUS-rev (5′-ATGGACACTGAACACCTGGATTA-3′). The amplified fragment was transferred into the pCR8/GW/TOPO entry vector by means of a TOPO reaction (Invitrogen). The nucleotide sequence was verified, and the fragment was then introduced to the silencing vector pB7GWIWG2 with an LR recombinant reaction. The constructs were then introduced into Agrobacterium tumefaciens LBA4404 by electroporation.
The seeds of S. lycopersicum ‘Zhongshu6’ were surface sterilized, sown on 1/2 MS culture medium and germinated with a 16 h day/8 h night regime at 25°C for approximately 6–8 days. Cotyledon explants were cut and preincubated for 2 day on KCMS in the dark at 25°C. After preculturing, the cotyledon explants were cultured with an A. tumefaciens LBA4404 suspension liquid for 4 min with lightly shaking. After infection, the explants were returned to the premedium for 2 days in the dark at 25°C. Then, the cotyledon explants were transferred to selection medium including MS salts, 3% sucrose, 7 g/L agar, 2 mg/L 6-BA, 0.2 mg/L IAA, 400 mg/L cephalosporin and 0.5 mg/L glufosinate-ammonium to induce shoot generation. The cotyledon explants that regenerated new shoots were then transferred to rooting culture medium, which included MS salt, 3% sucrose, 5 g/L agar, 400 mg/L cephalosporin and 0.05 mg/L NAA. The non-transgenic lines and transgenic plants were planted in a greenhouse.
RNA Extraction and Gene Expression Analysis by qRT–PCR
Samples were collected from 10 to 15 tomato plants and immediately frozen in liquid nitrogen (Cong et al., 2008) (Supplementary Figure 1). Total RNA was extracted using an RNA extraction kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. Total RNA was treated by DNase I to remove any genomic DNA contamination. The first-strand cDNA was reverse transcribed from 2 mg of total RNA using the Omniscript kit (Qiagen) according to the manufacturer’s instructions. The real-time PCR analysis was performed as described by Jain et al. (2006). The cDNA samples were used as the template and mixed with 200 nmol of each primer and the SYBR Green PCR Master Mix (Tiangen, Beijing, China) for real-time PCR analysis using an ABI 7500 Real-Time PCR System and Software 7500 ver. 2.0.3 (Applied Biosystems, USA) according to the manufacturer’s instructions. The temperature procedure was: 95°C for 15 min, 40 cycles of 95°C for 30 s, 57°C for 30 s, and 70°C for 1 min. The fluorescence signal was measured during the extension at 70°C of each cycle. The Ubiquitin3 gene from tomato was used as the control for normalization. The sequences of all primers that were used in this study are listed in Supplementary Table 1.
All values are expressed as the mean ± standard deviation of three independent experiments, and the data were analyzed using Origin 8.0. Analysis of variance was performed using one-way ANOVA with SPSS 13.0 software followed by Duncan’s multiple range tests for each experiment at P < 0.05.
Phenotypic Analysis of Tomato Flowers and Fruit
For flowers at the anthesis stage, 20 measurements (2 samples × 10 plants) of the widest diameter were calculated via a caliper gauge, and the sepal and petal areas were calculated. The number of sepals, petals, stamens, and carpels of the non-transgenic lines and transgenic plants was evaluated in tripped flowers. In addition, five mature fruits of the non-transgenic lines and transgenic plants were used to calculate the mean fruit weight (g), fruit width (mm), fruit length (mm), and the number of locules. All values are expressed as the mean ± standard deviation. The data were further subjected to analysis of variance, and least significant difference test was used to compare the average values. A probability of P < 0.05 was considered statistically significant.
Results
SlWUS Shows High Transcript Accumulation during Tomato Flower Development
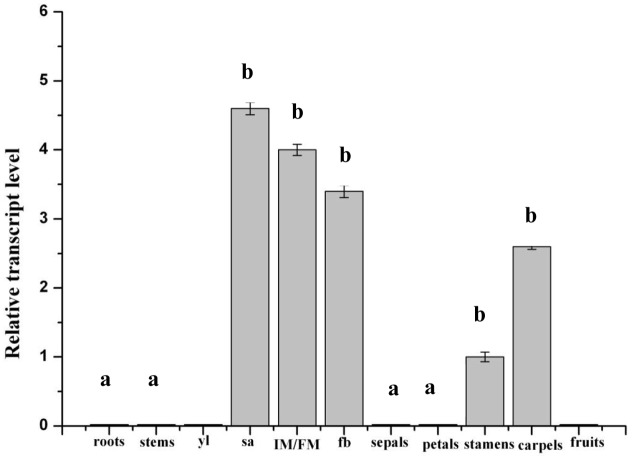
Previous studies have reported that SlWUS is expressed in rapidly growing organs (Laux et al., 1996; van der Knaap et al., 2014). To more explicitly characterize this pattern of expression, RT-PCR analyses were carried out using SlWUS gene-specific primers. SlWUS expression was detected in the young flower buds, shoot apexes, and inflorescence meristem/floral meristem (IM/FM); expression was undetectable in the roots, stems, leaves, and fruits. In the flowers, SlWUS transcripts were detected in the stamens and carpels; expression was undetectable in the sepals and petals (Figure 1). An increase in locule number is highly associated with an increase in the number of floral organs, especially the carpel. Our results showed that SlWUS was expressed during flower development, which indicates an important function for SlWUS in the development of tomato flowers.
FIGURE 1.
Profiling of SlWUS transcript accumulation in wild-type tomato plant tissues monitored by qRT-PCR. Samples were collected from the Solanum pimpinellifolium ‘Zhongshu 6.’ The level for stamens was used as a reference. yl, young leaves; sa, shoot apexes; IM/FM, infloresence meristem/floral meristem; yfb, young floral buds. The data are the mean values corresponding to three independent experiments. The error bars represent the standard errors. Values followed by the same letter (a or b) are not significantly different (P > 0.05, Duncan’s multiple range test).
Higher Expression of SlWUS Is Found in Plants on an lc Mutation Background
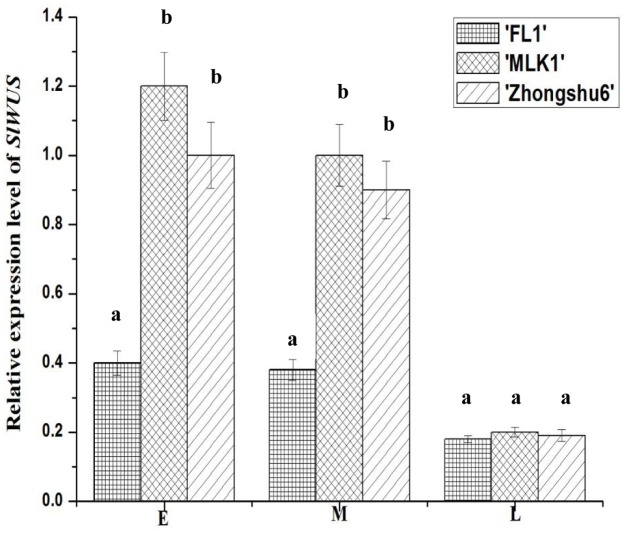
To elucidate the effect of the lc locus on the expression of SlWUS, we examined the expression of SlWUS in the flower buds of three tomato lines, including ‘MLK1,’ ‘FL1,’ and ‘Zhongshu6’. The ‘MLK1’ fruit were large with 10 or more locules, the ‘FL1’ fruit were small with 2–4 locules and the ‘Zhongshu6’ fruits were large with 6–8 locules. The cultivated allele of lc is present in ‘MLK1’ and ‘Zhongshu 6,’ and the wild-type allele of lc is present in ‘FL1.’ The results showed that SlWUS abundance decreased gradually during flower bud development. Meanwhile, at each developmental stage of the flower buds, the expression levels of SlWUS were higher in ‘MLK1’ and ‘Zhongshu6’ than those in ‘FL1’ (Figure 2), which indicated that the expression of SlWUS was the difference between the wild-type and mutant alleles of lc.
FIGURE 2.
Expression analysis of SlWUS in ‘FL1,’ ‘MLK1,’ and ‘Zhongshu6’ plants assessed by real-time PCR. E, early development before the initiation of carpel primordial; M, mid-stage development shortly after carpel initiation; L, late-stage development; ‘MLK1’ and ‘Zhongshu6,’ the mutant alleles of lc; ‘FL1,’ the wild-type alleles of lc. The data are presented as the mean (±SE) values corresponding to three independent experiments. Values followed by the same letter (a or b) are not significantly different (P > 0.05).
The Silencing of SlWUS Affects Tomato Flower Development and Decreases Tomato Fruit Locules
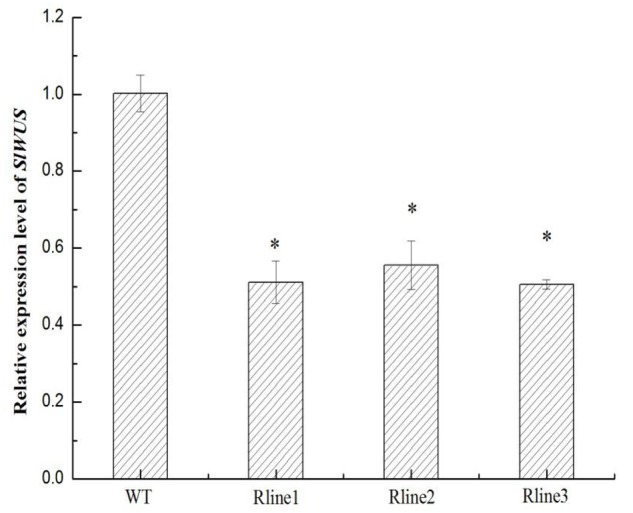
To further understand the function of SlWUS in tomato, we used a gene-specific region from the 3′ end of the cDNA (see “Materials and Methods”) to construct an RNA interference (RNAi) vector. The construct was introduced into Zhongshu6 by Agrobacterium-mediated transformation. There independent homozygous transgenic plants were produced for the construct. The gene expression analysis by real-time PCR showed that the expression levels of SlWUS were significantly reduced in the RNAi lines compared with those in the non-transgenic lines (Figure 3). All three of the SlWUS RNAi lines retained 50% of the control mRNA level. Therefore, one SlWUS RNAi line, Rline1, was selected for further study. The effects of SlWUS silencing on flower and fruit development were then investigated in the SlWUS RNAi lines. To characterize the flower development of the SlWUS RNAi lines, floral organ numbers at the anthesis stage were scored. The wild-type flowers were included in four whorls of floral organs and consisted of 5–6 sepals, 5–6 petals, approximately 6–7 stamens, and 6–8 carpels. In contrast, the SlWUS RNAi line flowers consisted of 4–5 sepals, 4–5 petals, 5–6 stamens, and 3–4 carpels. The number of carpels was significantly lower in the SlWUS RNAi lines than in WT (Figures 4A,D), which indicates that SlWUS gene function is essential to control carpel number during flower development. In addition, the width of the flowers were measured at the anthesis stage and revealed significant decreases in the SlWUS RNAi lines (Figure 4F). In addition, the SlWUS RNAi line flowers showed narrower sepals and petals, with remarkably decreased lamina area (Figures 4B,C,E). Therefore, the SlWUS RNAi lines differed significantly from WT with respect to the flower anthesis stage.
FIGURE 3.
Characterization of three independent SlWUS RNAi lines. Transcript levels of SlWUS in young flower buds of three independent SlWUS RNAi lines (Rline1, Rline2, and Rline3) relative to those of wild type (WT). The data are presented as the mean (±SE) values corresponding to three independent experiments. ∗Indicates significant differences between transgenic lines and WT plants at P < 0.05, according to Duncan’s multiple range test.
FIGURE 4.
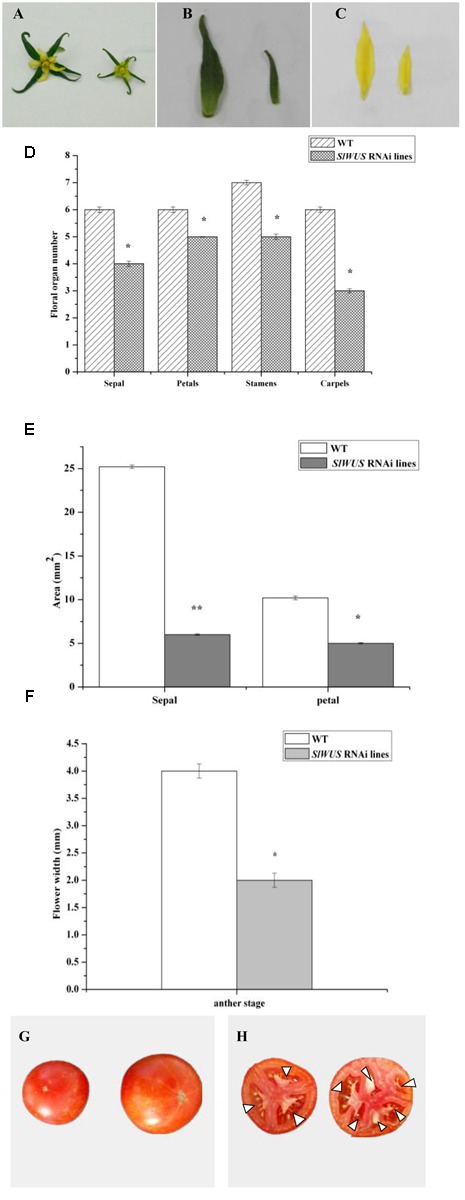
Alteration of flower and fruit morphometrical characteristics in SlWUS RNAi plants. (A) WT (left) and SlWUS RNAi plant flowers at anthesis. (B) WT (left) and SlWUS RNAi plants sepals at anthesis. (C) WT (left) and SlWUS RNAi plants petals at anthesis. (D) Mean comparison of the organ number in the four floral whorls at the anthesis stage. (E) Quantification of sepal and petal area in the control and SlWUS RNAi plant flowers at anthesis. The bars represent the standard error (n ≥ 100). (F) Mean comparison of flower width at the anthesis stage. (G) WT (right) and SlWUS RNAi plant mature fruit. (H) WT (right) and SlWUS RNAi plant fruit locule number. The data are presented as the mean (±SE) values corresponding to three independent experiments. ∗Indicates significant differences between transgenic lines and WT plants at P < 0.05, according to Duncan’s multiple range test. ∗∗Indicates significant differences between transgenic lines and WT plants at P < 0.01.
An increase in locule number is highly associated with an increase in the number of carpels in tomato. We observed that the wild-type plants yielded fruit with 6–8 locules, while the SlWUS RNAi lines yielded fruit with 3–4 locules (Figure 4H). Together with this decrease in locule number, there was a significant decrease in the weight and size of the fruit of the SlWUS RNAi lines (Figure 4G and Table 1). Altogether, the results indicated that the final size of the fruit of the SlWUS RNAi lines is determined by the decrease in the number of carpels that occurs during floral development.
Table 1.
Comparison of mature fruits among the wild-type (WT) and SlWUS RNAi plants.
| Weight (g) | Number of locules | Fruit size |
||
|---|---|---|---|---|
| Width (mm) | Length (mm) | |||
| WT | 174.9 ± 8.7a | 6.7 ± 0.7a | 86.7 ± 3.5a | 72.8 ± 1.4a |
| SlWUS RNAi lines | 85.9 ± 3.2b | 3.3 ± 0.3b | 63.0 ± 1.7b | 58 ± 1.2b |
Values followed by the same letter (a or b) are not significantly different (P > 0.05, Duncan’s multiple range test).
Expression Levels of a Carpel Development Gene Are Altered in the SlWUS RNAi Lines
To further illustrate the mechanisms that are involved in controlling the flower and fruit locule number phenotypes observed in the SlWUS RNAi lines, the expression levels of the genes involved in the control of floral organ number and fruit size that were modified in the transgenic lines were analyzed. Thus, the transcript levels of YABBY, TAG1, SlCLV3, and FW2.2, genes were evaluated in the WT and SlWUS RNAi lines during flower development. The results showed that the expression levels of the FW2.2 gene, a negative regulatory factor associated with carpel cell number (Frary et al., 2000), were not significantly different among the WT and SlWUS RNAi lines during flower development (Figure 5A). In addition, similar levels of YABBY gene expression were observed in the WT and SlWUS RNAi lines during flower development (Figure 5B). The expression levels of TAG1 were significantly (P < 0.05) down-regulated in the SlWUS RNAi lines (Figure 5C). Similarly, the expression levels of SlCLV3 were significantly (P < 0.05) down-regulated in the SlWUS RNAi lines during flower development (Figure 5D).
FIGURE 5.
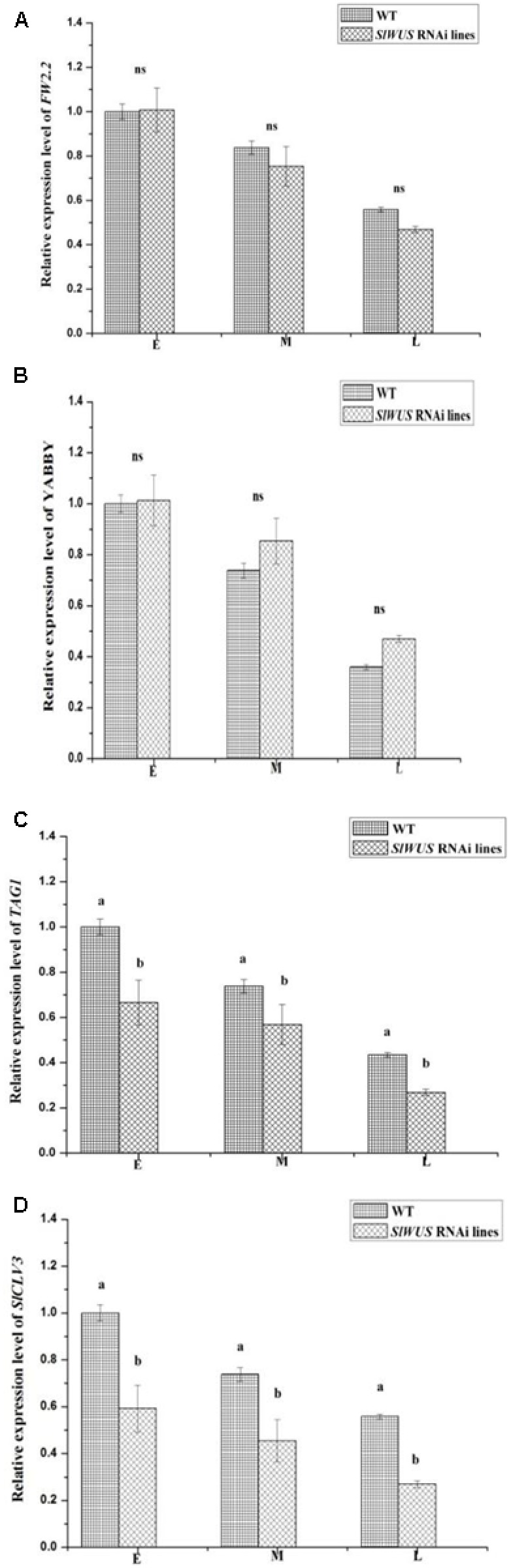
Expression analysis of (A) FW2.2, (B) YABBY, (C) TAG1, and (D) SlCLV3 during flower development. E, early development before the initiation of carpel primordial; M, mid-stage development shortly after carpel initiation; and L, late-stage development. The data are presented as the mean (±SE) values corresponding to three independent experiments. ns, no statistically significant differences. Values followed by the same letter (a or b) are not significantly different (P > 0.05).
Discussion
In tomato fruit, locule (cavities that develop from carpels) number ranges from 2 to 10 or more. The locules develop directly from the carpels in the tomato flower. It is known that lc is one of the key determinants of the final locule number in fruit, and lc mutation leads to a tomato fruit with more than two and four locules (Barrero et al., 2006; Muños et al., 2011). The lc locus is a non-coding region, which is located 1,080 bp downstream of the presumptive tomato homolog of WUS (Muños et al., 2011) that encodes a transcription factor that is essential to maintain stem cell identity in SAM (Mayer et al., 1998; Clark, 2001). Increased expression of WUS in Arabidopsis results in increased floral organ number, which is similar to the phenotype of the lc mutant (Mayer et al., 1998; Clark, 2001). Therefore, based on its predicted function, SlWUS is the most likely candidate gene for the lc locus (Barrero et al., 2006; van der Knaap et al., 2014). WUS encodes a homeodomain transcription factor that plays an important role in maintaining the balance between the proliferation and differentiation of stem cells in the meristems of A. thaliana (Laux et al., 1996; Mayer et al., 1998; Clark, 2001). The analysis of SlWUS revealed a similar expression pattern to that of WUS in Arabidopsis, which was detected in the young floral buds, SAM and the IM/FM (Figure 1). Furthermore, the SlWUS transcripts were detected in the stamens and carpels (Figure 1), which suggest a putative role for SlWUS in development of stamens and carpels in tomato. In addition, the phenotypic analysis of the stamens and carpels from the SlWUS lines revealed that SlWUS plays a role in the number of stamens and carpels (Figures 4D,H).
The tomato lc allele is associated with two SNPs downstream of SlWUS. These two SNPs were responsible for the increase in locule number, and they might participate in the regulation of SlWUS expression (Muños et al., 2011). In this study, the expression of SlWUS was the underlying difference between the wild-type and mutant alleles of lc, and higher expression of SlWUS was detected in the lc mutation background plants (Figure 2). These results suggest that an lc mutation may permit elevated expression of SlWUS. The maintenance of larger stem cells leads to increased locule numbers. Down-regulating SlWUS expression in the tomato transgenic plants resulted in a decreased fruit size (Figure 4H and Table 1). Fruit size is the primary characteristic of commercial tomato varieties and an important goal for tomato domestication. It is known that locule number largely affects the final fruit size in tomato. This increased locule number contributes as much as 50% variance to fruit enlargement (Lippman and Tanksley, 2001; Tanksley, 2004). The character analysis of tomato fruits from the SlWUS lines revealed that the decrease in fruit size was related to a decreased locule number (Figures 4D,H).
During tomato fruit domestication, both carpel cell division and carpel number determine the final size of tomato fruit (Tanksley, 2004). A relatively small number of genes were involved in the two processes. The FW2.2 gene is responsible for the first process (Frary et al., 2000; Cong et al., 2002). Tomato fruit size is quantitatively controlled, and several QTLs were identified in this process (Paterson et al., 1991; Grandillo et al., 1999). Among these loci, the FW2.2 gene largely governs fruit size (Frary et al., 2000; Ariizumi et al., 2013). The SlWUS RNAi lines did not show altered expression of the FW2.2 gene during floral development (Figure 5A), which is involved in the negative control of the carpel cell division that is associated with carpel cell number (Frary et al., 2000). The result indicated that the decreased tomato fruit size in the SlWUS RNAi lines was not a result of the cell division process regulated by FW2.2. Whereas, the fas gene is the main determinant of the floral organ number of the second process, and mutations in this gene are considered major contributors that result in the increase in fruit size in modern cultivated species by increasing the locule number from two to more than six (Lippman and Tanksley, 2001; Cong et al., 2008). The expression pattern expression of the fas gene is not altered in the SlWUS RNAi lines during floral development (Figure 5B), and we hypothesize that SlWUS function is required downstream of or parallel to fas function. Moreover, WUS positively regulates the expression of AG (Lenhard et al., 2001; Lohmann et al., 2001), and AG is critical in determining stamen and gynoecium identity (Yanofsky et al., 1990). It has been reported that TAG1 silencing lines show defects in stamen and carpel development (Pan et al., 2010). In our SlWUS RNAi lines, the number of stamens and carpels decreased (Figure 4D). In addition, the expression of TAG1 was significantly down-regulated (Figure 5C). Thus, these results suggest that a negative autoregulatory mechanism that involves TAG1 and SlWUS in tomato is similar to that in Arabidopsis, and the number of carpels in tomato may be regulated through a pathway involving the SlWUS and TAG1 genes. Other key components of the WUS signaling pathway are provided by the CLAVATA (CLV) proteins (Schoof et al., 2000; Clark, 2001; Lenhard and Laux, 2003). The WUS and CLV3 feedback loop is especially closely connected to the control of meristem size in Arabidopsis (Schoof et al., 2000). It is known that mutations in the CLV signaling pathway genes cause meristems to enlarge, which can lead to extra organs in the flowers and fruits. In addition, a regulatory change in SlCLV3 underlies the fas mutant phenotype (Xu et al., 2015). In the SlWUS RNAi lines, the CLV-WUS feedback loop was also mildly affected. The expression level of SlCLV3 was significantly down-regulated (Figure 5D). These results indicated that tomato domestication relied on changes in the interaction of the CLV-WUS feedback loop. YABBY expression is not altered in the RNAi-SlWUS lines, which is further evidence that fas is not encoded by YABBY (Xu et al., 2015).
Conclusion
The results presented here demonstrate that the down-regulation of SlWUS on an lc mutant background affects fruit size via decreasing the number of locules. This finding suggests that SlWUS plays a role in tomato fruit size, and the identification of other transcription factors that interact with SlWUS is still required. The SlWUS RNAi lines produced in our study provide a suitable tool to better illustrate the molecular mechanisms by which SlWUS may control locule number. Furthermore, further functional studies are required to investigate how these two SNPs affect the molecular function of SlWUS in tomato locules using CRIPSR/Cas9 gene editing technology (Brooks et al., 2014; Doudna and Charpentier, 2014; Belhaj et al., 2015).
Author Contributions
HL and MQ designed and carried out the experiments, analyzed the results, and wrote the manuscript. MS, YiL, YuL, TX, and YaL provided scientific advice, and revised the manuscript. TL conceived the research area, provided scientific advice, and supervised the project.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00457/full#supplementary-material
References
- Ariizumi T., Shinozaki Y., Ezura H. (2013). Genes that influence yield in tomato. Breed. Sci. 63 3–13. 10.1270/jsbbs.63.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrero L. S., Cong B., Wu F., Tanksley S. D. (2006). Developmental characterization of the fasciated locus and mapping of Arabidopsis candidate genes involved in the control of floral meristem size and carpel number in tomato. Genome 49 991–1006. 10.1139/g06-059 [DOI] [PubMed] [Google Scholar]
- Barrero L. S., Tanksley S. D. (2004). Evaluating the genetic basis of multiple-locule fruit in a broad cross section of tomato cultivars. Theor. Appl. Genet. 109 669–679. 10.1007/s00122-004-1676-y [DOI] [PubMed] [Google Scholar]
- Belhaj K., Chaparro-Garcia A., Kamoun S., Patron N. J., Nekrasov V. (2015). Editing plant genomes with CRISPR/Cas9. Curr. Opin. Biotechnol. 32 76–84. 10.1016/j.copbio.2014.11.007 [DOI] [PubMed] [Google Scholar]
- Brooks C., Nekrasov V., Lippman Z. B., Van Eck J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166 1292–1297. 10.1104/pp.114.247577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. E. (2001). Cell signalling at the shoot meristem. Nat. Rev. Mol. Cell Biol. 2 276–284. 10.1038/35067079 [DOI] [PubMed] [Google Scholar]
- Cong B., Barrero L. S., Tanksley S. D. (2008). Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 40 800–804. 10.1038/ng.144 [DOI] [PubMed] [Google Scholar]
- Cong B., Liu J., Tanksley S. D. (2002). Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Natl. Acad. Sci. U.S.A. 99 13606–13611. 10.1073/pnas.172520999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium T. G. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485 635–641. 10.1038/nature11119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudna J. A., Charpentier E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096 10.1126/science.1258096 [DOI] [PubMed] [Google Scholar]
- Fernández-Lozano A., Yuste-Lisbona F. J., Pérez-Martín F., Pineda B., Moreno V., Lozano R., et al. (2015). Mutation at the tomato EXCESSIVE NUMBER OF FLORAL ORGANS (ENO) locus impairs floral meristem development, thus promoting an increased number of floral organs and fruit size. Plant Sci. 232 41–48. 10.1016/j.plantsci.2014.12.007 [DOI] [PubMed] [Google Scholar]
- Frary A., Nesbitt T. C., Frary A., Grandillo S., Van Der Knaap E., Cong B., et al. (2000). fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289 85–88. 10.1126/science.289.5476.85 [DOI] [PubMed] [Google Scholar]
- Grandillo S., Ku H., Tanksley S. (1999). Identifying the loci responsible for natural variation in fruit size and shape in tomato. Theor. Appl. Genet. 99 978–987. 10.1007/s001220051405 [DOI] [Google Scholar]
- Huang Z., van der Knaap E. (2011). Tomato fruit weight 11.3 maps close to fasciated on the bottom of chromosome 11. Theor. Appl. Genet. 123 465–474. 10.1007/s00122-011-1599-3 [DOI] [PubMed] [Google Scholar]
- Illa-Berenguer E., Van Houten J., Huang Z., Van Der Knaap E. (2015). Rapid and reliable identification of tomato fruit weight and locule number loci by QTL-seq. Theor. Appl. Genet. 128 1329–1342. 10.1007/s00122-015-2509-x [DOI] [PubMed] [Google Scholar]
- Jain M., Kaur N., Tyagi A. K., Khurana J. P. (2006). The auxin-responsive GH3 gene family in rice (Oryza sativa). Funct. Integr. Genomics 6 36–46. 10.1007/s10142-005-0142-5 [DOI] [PubMed] [Google Scholar]
- Klee H. J., Giovannoni J. J. (2011). Genetics and control of tomato fruit ripening and quality attributes. Annu. Rev. Genet. 45 41–59. 10.1146/annurev-genet-110410-132507 [DOI] [PubMed] [Google Scholar]
- Knaap E. V. D., Tanksley S. D. (2003). The making of a bell pepper-shaped tomato fruit: identification of loci controlling fruit morphology in Yellow Stuffer tomato. Theor. Appl. Genet. 107 139–147. 10.1007/s00122-003-1224-1 [DOI] [PubMed] [Google Scholar]
- Laux T., Mayer K., Berger J., Jurgens G. (1996). The WUSCHEL gene is required for shoot and floral meristem integrity in Arabidopsis. Development 122 87–96. [DOI] [PubMed] [Google Scholar]
- Lenhard M., Bohnert A., Jurgens G., Laux T. (2001). Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between WUSCHEL and AGAMOUS. Cell 105 805–814. 10.1016/S0092-8674(01)00390-7 [DOI] [PubMed] [Google Scholar]
- Lenhard M., Laux T. (2003). Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130 3163–3173. 10.1242/dev.00525 [DOI] [PubMed] [Google Scholar]
- Lippman Z., Tanksley S. D. (2001). Dissecting the genetic pathway to extreme fruit size in tomato using a cross between the small-fruited wild species Lycopersicon pimpinellifolium and L. esculentum var. Giant Heirloom. Genetics 158 413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J., Tanksley S. D. (2003). Generation and analysis of an artificial gene dosage series in tomato to study the mechanisms by which the cloned quantitative trait locus fw2.2 controls fruit size. Plant Physiol. 132 292–299. 10.1104/pp.102.018143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Kim Y., Müller R., Yumul R. E., Liu C., Pan Y., et al. (2011). AGAMOUS terminates floral stem cell maintenance in Arabidopsis by directly repressing WUSCHEL through recruitment of polycomb group proteins. Plant Cell 23 3654–3670. 10.1105/tpc.111.091538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohmann J. U., Hong R. L., Hobe M., Busch M. A., Parcy F., Simon R., et al. (2001). A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 105 793–803. 10.1016/S0092-8674(01)00384-1 [DOI] [PubMed] [Google Scholar]
- Mayer K. F., Schoof H., Haecker A., Lenhard M., Jürgens G., Laux T. (1998). Role of WUSCHEL in regulating stem cell fate in the Arabidopsis shoot meristem. Cell 95 805–815. 10.1016/S0092-8674(00)81703-1 [DOI] [PubMed] [Google Scholar]
- Muños S., Ranc N., Botton E., Bérard A., Rolland S., Duffé P., et al. (2011). Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 156 2244–2254. 10.1104/pp.111.173997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan I. L., Mcquinn R., Giovannoni J. J., Irish V. F. (2010). Functional diversification of AGAMOUS lineage genes in regulating tomato flower and fruit development. J. Exp. Bot. 61 1795–1806. 10.1093/jxb/erq046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Damon S., Hewitt J. D., Zamir D., Rabinowitch H. D., Lincoln S. E., et al. (1991). Mendelian factors underlying quantitative traits in tomato: comparison across species, generations, and environments. Genetics 127 181–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez G. R., Muños S., Anderson C., Sim S.-C., Michel A., Causse M., et al. (2011). Distribution of SUN, OVATE, LC, and FAS in the tomato germplasm and the relationship to fruit shape diversity. Plant Physiol. 156 275–285. 10.1104/pp.110.167577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K. F. X., Jürgens G., Laux T. (2000). The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL Genes. Cell 100 635–644. 10.1016/S0092-8674(00)80700-X [DOI] [PubMed] [Google Scholar]
- Swinnen G., Goossens A., Pauwels L. (2016). Lessons from domestication: targeting Cis-Regulatory elements for crop improvement. Trends Plant Sci. 21 506–515. 10.1016/j.tplants.2016.01.014 [DOI] [PubMed] [Google Scholar]
- Tanksley S. D. (2004). The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell 16 181–189. 10.1105/tpc.018119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilly J. J., Allen D. W., Jack T. (1998). The CArG boxes in the promoter of the Arabidopsis floral organ identity gene APETALA3 mediate diverse regulatory effects. Development 125 1647–1657. [DOI] [PubMed] [Google Scholar]
- van der Knaap E., Chakrabarti M., Chu Y. H., Clevenger J. P., Illa-Berenguer E., Huang Z., et al. (2014). What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 5:227 10.3389/fpls.2014.00227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C., Liberatore K. L., Macalister C. A., Huang Z., Chu Y.-H., Jiang K., et al. (2015). A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 47 784–792. 10.1038/ng.3309 [DOI] [PubMed] [Google Scholar]
- Yanofsky M. F., Ma H., Bowman J. L., Drews G. N., Feldmann K. A., Meyerowitz E. M. (1990). The protein encoded by the Arabidopsis homeotic gene agamous resembles transcription factors. Nature 346 35–39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.