Highlights
-
•
Precursor concentration affected cell viability and resveratrol yields.
-
•
Cell viability and culture conditions influenced resveratrol yields.
-
•
Results show the relevance of process monitoring for bioprocess optimization.
Keywords: Resveratrol, Cell physiology, Plasmid stability, Flow Cytometry, Real-time qPCR
Abstract
Resveratrol is a plant secondary metabolite commonly found in peanuts and grapevines with significant health benefits. Recombinant organisms can produce large amounts of resveratrol and, in this work, Escherichia coli BW27784 was used to produce resveratrol in bioreactors while monitoring cell physiology and plasmid stability through flow cytometry and real-time qPCR, respectively. Initially, the influence of culture conditions and precursor addition was evaluated in screening assays and the data gathered was used to perform the bioreactor assays, allowing the production of 160 μg/mL of resveratrol. Cellular physiology and plasmid instability affected the final resveratrol production, with lower viability and plasmid copy numbers associated with lower yields. In sum, this study describes new tools to monitor the bioprocess, evaluating the effect of culture conditions, and its correlation with cell physiology and plasmid segregational stability, in order to define a viable and scalable bioprocess to fulfill the need for larger quantities of resveratrol.
1. Introduction
Resveratrol (3,5,4′-trans-hydroxystilbene) is a phytoalexin and a polyphenolic compound that belongs to the stilbene family [1]. This natural occurring and multi-biofunctional chemical [2] exists in both cis- and trans- isomeric forms due to its two phenol rings linked by a styrene double bond [3]. Resveratrol is produced by plants in response to biotic and abiotic stress and has been used as a folk remedy to treat various ailments. Several biological properties have been associated with the use of resveratrol, namely cardio and neuroprotective effects [4], [5], anticancer, and antimicrobial [6], [7] as well as the ability to prolong lifespan [8]. Based on its presumed properties, the interest in resveratrol by the pharmaceutical, nutraceutical, and cosmetic industries is increasing [9]. Resveratrol used by these industries is generally chemically synthesized through several routes [10]. As chemical synthesis is a time-consuming process [10] that may be affected by the low reactivity of reagents, more sustainable alternatives to chemical synthesis are in demand for resveratrol production. In order to overcome these hurdles, new biological-based processes using plant cell systems and recombinant microorganisms are being evaluated to produce resveratro [19]. Despite the high resveratrol amounts produced by Saccharomyces cerevisiae [11], Escherichia coli is the recombinant microorganism of choice due to its ability to quickly produce this compound [9], sometimes in large amounts, as has been described in previous studies [12]. Process productivity can be severely affected by cell physiology and plasmid stability [14], due to decreased cell growth, as a result of lower cell viability, or due to lower enzyme quantities, as a result of decreased plasmid copy number or gene expression [15]. So, in order to optimize resveratrol production and to guarantee the maximal output of the process, the assessment of cultivation conditions and other process variables effect in cell physiology and plasmid segregational stability is of vital importance [13]. The present work describes resveratrol production in bioreactor using E. coli BW27784 transformed with pAC-4CL1 and pUC-STS plasmids while monitoring cell physiology and plasmid segregational stability through flow cytometry and real-time qPCR, respectively, in order to evaluate whole process performance.
2. Materials and methods
2.1. Plasmid, bacterial strain, and growth conditions
The bacterial host E. coli BW27784 (E. coli Genetic Stock Center, New Haven, CT, USA) was transformed with pAC-4CL1 plasmid (Addgene plasmid 35,947, Cambridge, MA, USA) encoding for 4-coumaroyl CoA ligase from Arabidopsis thaliana and pUC-STS plasmid (Addgene plasmid 35,949, Cambridge, MA, USA) encoding for stilbene synthase from Arachis hypogaea [16]. Plasmid pAC-4CL1 has a p15A origin with the genes coded by the plasmid being constitutively expressed. pUC-STS has a pBR322 origin of replication and the genes carried by this plasmid were also constitutively expressed from the lac promoter [16]. E. coli was genetically manipulated using transformation by the heat shock protocol. Briefly, the competent cells were generated by addition of magnesium chloride (100 mM) and calcium chloride (100 mM in the first step and 85 mM in the second step of the protocol) to E. coli cells obtained from a LB medium cultivated at 37 °C. Then, the suspension was incubated on ice for 25 min and the pellet was collected. The transformation was performed by addition of 1 μL of each plasmid, followed by incubation on ice for 30 min, heating at 42 °C for 30 s and subsequent transfer to ice. 200 μL of SOC medium were added to the previous suspension and incubated at 37 °C. For selection of transformants, this suspension was spread in LB plates containing 50 μg/mL chloramphenicol and 100 μg/mL ampicillin. The expression system was cultivated in M9 medium (per 1 L of water: 6.779 g of Na2HPO4, 3 g of KH2PO4, 0.5 g of NaCl, 1 g of NH4Cl, 1.25 g of yeast extract, 5 g of glycerol, 2 mL of MgSO4·7H2O 1 M, and 0.1 mL of CaCl2·2H2O 1 M) [16]. All cultures were started with an OD600 of 0.05, grown in 250 mL shake flasks containing 62.5 mL of medium, with 50 μg/mL chloramphenicol, and 100 μg/mL ampicillin, at 250 rpm and 30 °C.
2.2. Screening assays
In order to establish working ranges for further experiments, four factors were tested in screening assays: precursor (p-coumaric acid) concentration (0–20 mM), OD600 at time of precursor addition (0.1–1), temperature (25–42 °C), and pH (5–9). p-Coumaric acid was dissolved in DMSO to a final concentration of 1 M and sterilized by using a 0.22 μm pore size filter. Growth was suspended after 48 h of fermentation.
2.3. Bioreactor assays
E. coli was cultivated in four 0.5 L working volume parallel bioreactor (Infors HT, Bottmingen, Switzerland) containing 250 mL of M9 medium. The bioreactors were operated with strictly controlled parameters including pH, temperature, airflow, agitation (250 rpm) and dissolved oxygen (30%). The pH was maintained through the automatic addition of 1 M NaOH and 1 M H2SO4. All the parameters were monitored continuously using the IRIS software (Infors HT, Bottmingen, Switzerland) and all cultures were performed under subdued light in order to avoid trans-resveratrol isomerization to cis-resveratrol. Fermentations were carried out for 30 h and samples were taken aseptically at 22 and 30 h of fermentation to control growth and to evaluate resveratrol production, cell physiology and plasmid stability. The dry cell weight was calculated based on the previous established relation between OD600 and dry cell weight where one unit of OD600 was found to correspond to a dry cell weight of 0.25 g/L [17].
2.4. Analytical chromatography
Prior to injection, resveratrol was extracted from cell-free culture supernatant using a liquid–liquid extraction with ethyl acetate. Briefly, 1 mL of culture broth was centrifuged at 13,000 rpm for 5 min. The resulting supernatant was mixed with 50 μL of hydrochloric acid and carbamazepine (internal standard (IS), 100 μg/mL final concentration) and extracted with 1 mL of ethyl acetate. The extraction mixture was dried at 30 °C under a nitrogen gas stream, dissolved in 100 μL of mobile phase [18] and filtered through a 0.22 μm pore size filter. Each sample was analyzed in triplicate and all samples were stored at −20 °C prior to HPLC analysis to prevent resveratrol degradation. Ten μL of extract were applied to a Zorbax 300SB-C18 reverse-phase analytical column (4.6 mm ID × 150 mm, Agilent Technologies, Santa Clara, CA, USA) using an Agilent 1200 UPLC system equipped with a diode array detector. The process was performed as described in Paulo et al. [18], with a flow rate of 1 mL/min. Standard curves were constructed by plotting the area ratio between resveratrol and IS versus resveratrol concentration. All resveratrol analyses were performed in triplicate at each fermentation time.
2.5. Flow cytometry
Samples were analyzed on a CyAn ADP (Beckman Coulter, Brea, CA, USA) flow cytometer equipped with a 20 mW semiconductor laser at 488 nm. Fluorescence (FL1 and FL3 bandpass filters) and light scatter (FSC and SSC) signals were acquired logarithmically. Acquisition was performed with Summit 4.3 (Beckman Coulter, Brea, CA, USA) software. To reduce electronic and small particle noise, threshold levels were set on SSC. For the evaluation of cell viability, a bis-(1,3-dibutylbarbituric acid) trimethine oxonol (BOX, 2.5 μg/mL final concentration) and propidium iodide (PI, 10 μg/mL final concentration) dual staining was performed as previously described [13]. The fluorescence signals were collected by FL1 (BOX) and FL3 (PI) bandpass filters and 5000 events/cells were acquired for each sample.
2.6. Real-time qPCR
Fermentation samples for real-time qPCR were prepared as previously described [13]. Specific primers (Stab Vida, Lisboa, Portugal) for chloramphenicol resistance gene (forward: 5′-ACCGTAACACGCCACATCTT-3′; reverse: 5′-TTCTTGCCCGCCTGATGAAT-3′) and ampicillin resistance gene (forward: 5′-TCCTTGAGAGTTTTCGCCCC-3′; reverse: 5′-TTCATTCAGCTCCGGTTCCC-3′) were used to amplify fragments in each of the two plasmids used. Real-time qPCR efficiency was determined for this primer set using standard solutions of known plasmid copy number. Real-time qPCR (IQ5 Biorad, Hercules, CA, USA) reactions were performed using 3 μL of sample for a 20 μL reaction containing 10 μL of Maxima™ SYBR Green qPCR Master Mix (Fermentas, Burlington, ON, Canada) and, 400 nM of pAC-4CL1 or 200 nM of pUC-STS primer set. Regarding pUC-STS, reactions were incubated at 95 °C for 3 min, followed by 30 cycles of 10 s at 95 °C and 30 s at 58 °C. For pAC-4CL1, reactions were incubated at 95 °C for 3 min, followed by 30 cycles of 10 s at 95 °C and 30 s at 60 °C. The amplified PCR fragments were checked by melting curves: reactions were heated from 55 to 95 °C with 10 s holds at each temperature (0.05 °C/s). Bacterial cell concentration was kept constant at 3 × 104 cells/reaction and for each fermentation sample, triplicate measurements were performed. PCN standards for calibration curve were made according to a previously described method [13]. Acquisition and analysis were performed in BioRad IQ 5 Software, Hercules, CA, USA.
3. Results and discussion
3.1. Screening assays
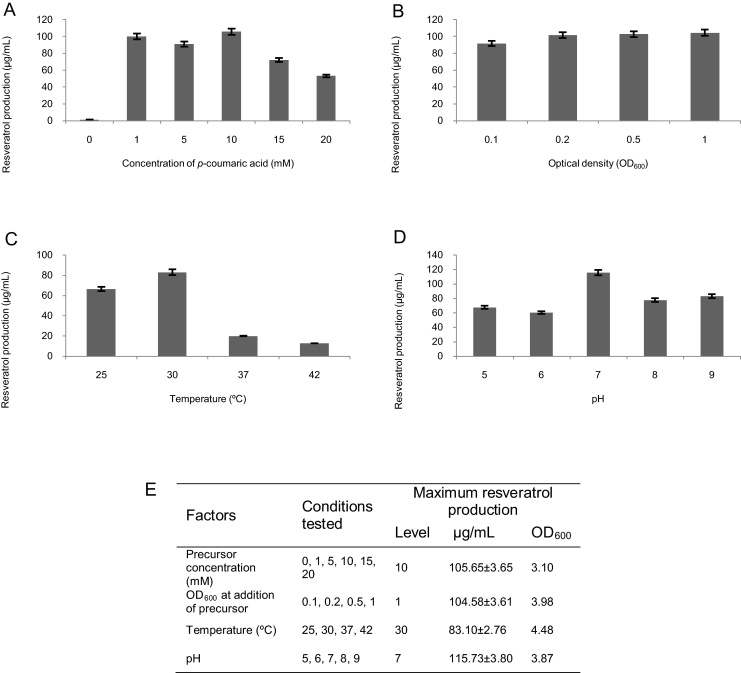
In order to determine the most relevant parameters and their ranges for resveratrol production in bioreactors, a set of screening assays was performed and the results are summarized in Fig. 1. In these assays, four parameters were evaluated: concentration of p-coumaric acid added, optical density (OD600) of the culture when this addition was performed, incubation temperature, and pH.
Fig. 1.
Influence of p-coumaric acid concentration (A), optical density (OD600) at addition of precursor (B), temperature (C), pH (D) on resveratrol production in the screening assays and maximum production obtained (E), with the level and optical density corresponding to the maximum obtained in each factor, in a 48 h fermentation. The values are presented as mean ± SD of three measurements.
The strategy used in these screening assays was based on a selection of baseline set of levels for each factor (1 mM of precursor added at OD600 of 0.1 in M9 medium at 30 °C, pH 7, and 250 rpm). Then, successively, each factor was varied over its range, while keeping the other factors constant. These screening assays allowed the attainment of a maximum yield of approximately 100 μg/mL of resveratrol.
Six concentrations of p-coumaric acid were tested ranging from 0 to 20 mM. These concentrations were selected based on previous experiments [16]. Due to the limited aqueous solubility of p-coumaric acid, its maximum concentration was chosen in order to allow a proper dissolution in the aqueous culture medium [16]. It was observed that, if p-coumaric acid was above a concentration of 10 mM, resveratrol production and cell growth started to decrease, which could be associated with the possible inhibitory effect on cell functions produced by higher p-coumaric acid concentrations [19]. The addition of 1 mM to 10 mM of p-coumaric acid yielded the highest results; however, low concentrations may be preferable in this situation due to the detrimental effects of p-coumaric acid in both production and growth. Regarding the OD600 of the culture at the time of precursor addition, the highest resveratrol concentrations were obtained between an OD600 of 0.5 and 1, which means that the addition of precursor in the early stages of growth may affect E. coli growth at lag phase. Lou et al. [20] observed that Gram negative bacteria treated with p-coumaric acid presented slight leakages of cellular cytoplasmic contents only 90 min after treatment, which may consequently affect resveratrol production.
Finally, with respect to the culture conditions evaluated, the best temperature for trans-resveratrol production seemed to be 30 °C, as higher temperatures (37 and 42 °C), although allowing higher cell growth, yielded lower resveratrol concentrations. This decrease in trans-resveratrol production at higher temperatures might be associated with the possible degradation of this compound if subjected to higher temperatures [21], as shown in a previous study [22] that demonstrated trans-resveratrol degradation for temperatures over 35 °C. Regarding the initial pH, a value of 7.0 allowed the achievement of the highest resveratrol yield. Taking into account that resveratrol is stable in a wide pH range [23], up to a pH of 9.0, above which the deprotonating of resveratrol occurs [24], the highest yield obtained at a pH of 7.0 may be related with the fact that this is the optimal pH for E. coli growth.
3.2. Bioreactor assays
Table 1 lists the conditions used in the assays of resveratrol production scale-up performed in bioreactor. The values for the conditions tested were established considering the results obtained in the screening assays. The range of values was established in order to assess the influence of each parameter in final resveratrol production and cell physiology. The influence of the conditions tested on resveratrol yield and productivity, cell growth and viability and plasmid segregational stability can be seen on Table 2.
Table 1.
Description of bioreactor conditions: temperature, pH, precursor (p-coumaric acid) concentration, and optical density at time of precursor addition (OD600) for each of the fermentation assays.
| Assay | Temperature (°C) | pH | Precursor concentration (mM) | OD600 |
|---|---|---|---|---|
| 1 | 25 | 7 | 8 | 0.575 |
| 2 | 28 | 6.5 | 4 | 0.35 |
| 3 | 28 | 6.5 | 4 | 0.8 |
| 4 | 28 | 6.5 | 12 | 0.35 |
| 5 | 28 | 6.5 | 12 | 0.8 |
| 6 | 28 | 7.5 | 4 | 0.35 |
| 7 | 28 | 7.5 | 4 | 0.8 |
| 8 | 28 | 7.5 | 12 | 0.35 |
| 9 | 28 | 7.5 | 12 | 0.8 |
| 10 | 31 | 6 | 8 | 0.575 |
| 11 | 31 | 7 | 0 | 0.575 |
| 12 | 31 | 7 | 8 | 0.125 |
| 13 | 31 | 7 | 8 | 0.575 |
| 14 | 31 | 7 | 8 | 1.025 |
| 15 | 31 | 7 | 16 | 0.575 |
| 16 | 31 | 8 | 8 | 0.575 |
| 17 | 34 | 6.5 | 4 | 0.35 |
| 18 | 34 | 6.5 | 4 | 0.8 |
| 19 | 34 | 6.5 | 12 | 0.35 |
| 20 | 34 | 6.5 | 12 | 0.8 |
| 21 | 34 | 7.5 | 4 | 0.35 |
| 22 | 34 | 7.5 | 4 | 0.8 |
| 23 | 34 | 7.5 | 12 | 0.35 |
| 24 | 34 | 7.5 | 12 | 0.8 |
| 25 | 37 | 7 | 8 | 0.575 |
Table 2.
Values of optical densities, resveratrol production yields, cellular viability (PI and BOX positive cells), and plasmid copy number obtained in each of the bioreactor assays described in Table 1 after 22 and 30 h of fermentation.
| Assay | OD600 |
Resveratrol volumetric yield (μg/mL) |
Resveratrol productivity (mg/gh−1) |
Cellular viability |
Plasmid copy number (PCN) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PI+(%) |
BOX+(%) |
pAC-4CL1 |
pUC-STS |
|||||||||||
| 22 h | 30 h | 22 h | 30 h | 22 h | 30 h | 22 h | 30 h | 22 h | 30h | 22 h | 30 h | 22 h | 30 h | |
| 1 | 1.84 | 2.53 | 57.94 ± 2.44 | 69.17 ± 1.34 | 5.73 ± 0.16 | 3.65 ± 0.31 | 1.48 | 1.43 | 23.31 | 29.7 | 26 ± 10.60 | 28 ± 0.96 | 25 ± 4.09 | 30 ± 10.15 |
| 2 | 2.77 | 4.43 | 84.1 ± 9.49 | 153.73 ± 1.08 | 5.52 ± 1.27 | 4.63 ± 1.90 | 5.56 | 7 | 21.32 | 15.02 | 115 ± 47.14 | 130 ± 14.93 | 20 ± 2.18 | 26 ± 5.44 |
| 3 | 2.72 | 4.19 | 86.3 ± 6.41 | 159.96 ± 2.86 | 5.77 ± 0.48 | 5.09 ± 0.96 | 4.3 | 7.06 | 21.66 | 18.08 | 215 ± 84.94 | 242 ± 36.44 | 72 ± 16.10 | 89 ± 15.53 |
| 4 | 2.24 | 2.73 | 23.51 ± 0.31 | 46.07 ± 0.65 | 1.91 ± 0.52 | 2.25 ± 0.27 | 0.28 | 1.48 | 30.23 | 27.68 | 47 ± 15.48 | 77 ± 9.61 | 95 ± 46.53 | 86 ± 11.48 |
| 5 | 2.35 | 2.9 | 39.38 ± 3.02 | 64.72 ± 2.24 | 3.05 ± 0.02 | 2.98 ± 0.03 | 1.18 | 3.78 | 18.84 | 20.38 | 29 ± 7.25 | 77 ± 36.28 | 22 ± 11.15 | 35 ± 9.34 |
| 6 | 2.76 | 4.44 | 39.48 ± 1.47 | 65.54 ± 0.63 | 2.60 ± 0.44 | 1.97 ± 0.88 | 1.88 | 3.1 | 18.58 | 20.5 | 28 ± 23.60 | 40 ± 15.39 | 4 ± 0.71 | 4 ± 1.25 |
| 7 | 2.76 | 4.42 | 41.71 ± 2.89 | 97.56 ± 5.61 | 2.75 ± 0.06 | 2.94 ± 0.08 | 2.08 | 3.7 | 16.08 | 14.08 | 30 ± 12.76 | 47 ± 21.25 | 23 ± 3.87 | 14 ± 6.76 |
| 8 | 2.13 | 3.35 | 40 ± 1.08 | 79.32 ± 3.62 | 3.41 ± 0.37 | 3.16 ± 0.55 | 3.12 | 3.7 | 9.9 | 13.94 | 51 ± 14.66 | 62 ± 5.71 | 15 ± 6.66 | 14 ± 2.27 |
| 9 | 2.56 | 2.86 | 28.57 ± 0.28 | 50.68 ± 1.88 | 2.03 ± 0.32 | 2.36 ± 0.09 | 1.17 | 1.26 | 30.95 | 26.7 | 33 ± 7.63 | 36 ± 17.88 | 45 ± 6.66 | 62 ± 2.27 |
| 10 | 2.04 | 3.16 | 28.66 ± 5.05 | 32.53 ± 3.64 | 2.55 ± 0.33 | 1.37 ± 0.16 | 5.62 | 5.1 | 19.86 | 16.14 | 218 ± 54.82 | 394 ± 74.85 | 91 ± 9.32 | 64 ± 11.60 |
| 11 | 4.5 | 4.9 | 1.53 ± 0.27 | 3.11 ± 2.32 | 0.06 ± 0.01 | 0.08 ± 0.01 | 3.12 | 2.4 | 21.3 | 27.34 | 117 ± 73.19 | 53 ± 16.91 | 31 ± 22.22 | 62 ± 30.71 |
| 12 | 2.68 | 3.02 | 56.45 ± 3.06 | 69.87 ± 5.27 | 3.83 ± 0.97 | 3.08 ± 0.50 | 1.37 | 2.13 | 23.42 | 27.08 | 51 ± 24.16 | 82 ± 11.03 | 128 ± 41.38 | 206 ± 57.15 |
| 13 | 2.78 | 3.9 | 74.34 ± 1.92 | 100.59 ± 7.83 | 4.86 ± 0.75 | 3.44 ± 1.35 | 2.3 | 2.42 | 24.42 | 18.8 | 34 ± 6.40 | 44 ± 7.86 | 78 ± 21.13 | 160 ± 73.50 |
| 14 | 2.54 | 3.41 | 23.71 ± 4.15 | 78.01 ± 5.08 | 1.70 ± 0.43 | 3.05 ± 0.47 | 1.24 | 2.83 | 27.63 | 33.04 | 64 ± 4.93 | 81 ± 4.69 | 177±35.29 | 239 ± 34.75 |
| 15 | 2.24 | 2.31 | 52.24 ± 9.92 | 109.28 ± 2.44 | 4.24 ± 0.15 | 6.31 ± 0.69 | 1.87 | 2.94 | 24.9 | 31.11 | 48 ± 10.12 | 67 ± 13.21 | 103 ± 53.40 | 135 ± 81.07 |
| 16 | 1.42 | 2.82 | 22.21 ± 1.72 | 26.32 ± 0.86 | 2.84 ± 0.28 | 1.24 ± 0.85 | 3.76 | 6.13 | 19.82 | 30.87 | 81 ± 6.62 | 113 ± 9.87 | 44 ± 14.56 | 69 ± 7.81 |
| 17 | 4.65 | 4.64 | 32.37 ± 1.92 | 35.06 ± 2.80 | 1.27 ± 0.03 | 1.01 ± 0.16 | 3.9 | 5.9 | 22.8 | 43.2 | 64 ± 2.01 | 94 ± 18.35 | 69 ± 78.15 | 59 ± 24.94 |
| 18 | 4.04 | 3.97 | 58.52 ± 0.53 | 61.99 ± 0.99 | 2.63 ± 0.03 | 2.08 ± 0.42 | 4.58 | 8.28 | 23.98 | 32.9 | 55 ± 30.94 | 96 ± 8.80 | 28 ± 11.43 | 33 ± 7.82 |
| 19 | 1.82 | 3.1 | 9.32 ± 0.75 | 13.41 ± 0.97 | 0.93 ± 0.21 | 0.58 ± 0.46 | 3.14 | 4.32 | 10.02 | 16.78 | 84 ± 27.46 | 44 ± 5.94 | 40 ± 4.00 | 21 ± 4.95 |
| 20 | 3.09 | 3.59 | 17.51 ± 1.22 | 15.90 ± 0.17 | 1.03 ± 0.12 | 0.59 ± 0.19 | 1.94 | 4.07 | 21.19 | 39.07 | 48 ± 16.73 | 36 ± 2.86 | 10 ± 8.92 | 5 ± 0.93 |
| 21 | 3.04 | 3.87 | 13.81 ± 1.72 | 23.22 ± 1.96 | 0.83 ± 0.33 | 0.80 ± 0.85 | 2.93 | 5.88 | 8.82 | 18.8 | 17 ± 3.60 | 14 ± 1.10 | 48 ± 12.05 | 39 ± 20.97 |
| 22 | 3.85 | 3.66 | 21.17 ± 1.69 | 20.18 ± 1.15 | 1.00 ± 0.11 | 0.74 ± 0.08 | 2.78 | 4.48 | 18.34 | 13.32 | 53 ± 16.21 | 31 ± 8.37 | 33 ± 6.93 | 14 ± 5.75 |
| 23 | 3.15 | 4.76 | 11.69 ± 0.52 | 14.90 ± 2.23 | 0.67 ± 0.21 | 0.42 ± 0.39 | 5.1 | 3.5 | 38.58 | 50.16 | 75 ± 34.75 | 107 ± 42.68 | 71 ± 13.66 | 88 ± 19.65 |
| 24 | 3.74 | 3.84 | 16.13 ± 1.27 | 15.77 ± 2.82 | 0.78 ± 0.07 | 0.55 ± 0.24 | 4.3 | 5.72 | 27.4 | 21.76 | 42 ± 5.80 | 55 ± 5.07 | 32 ± 4.50 | 25 ± 4.56 |
| 25 | 3.84 | 4.07 | 13.59 ± 2.63 | 31.72 ± 3.47 | 0.64 ± 0.25 | 1.04 ± 0.03 | 2.42 | 4.6 | 11.51 | 21.78 | 3 ± 0.43 | 2 ± 0.10 | 14 ± 2.86 | 40 ± 31.24 |
As expected, if the concentration of precursor added was 0 mM (assay 11), the production is approximately null. It was also observed that low concentrations of resveratrol were generally associated with higher concentrations of precursor, as a concentration of 12 mM of p-coumaric acid allowed the attainment of a resveratrol productivity of 2.98 mg/gh−1 (assay 5) while a concentration of 4 mM allowed an almost two-fold increase of resveratrol productivity to 5.09 mg/gh−1 (assay 3), with the same correlation being obtained in terms of resveratrol volumetric yields. It can also be observed that p-coumaric acid seemed to have a detrimental effect on cellular growth, as higher concentrations of p-coumaric acid added resulted in lower OD600 values (assays 4, 5, 8, 9, and 15) when compared to assays without or with lower concentrations of p-coumaric acid (assays 2, 3, 6, 7, and 11). The influence of temperature can be seen by the resveratrol yield analysis when observing the assays results for 25, 31, and 37 °C with the other variables constant (assay 1, 13 and 25, respectively). It was observed that for the lowest (25 °C) and highest (37 °C) tested temperatures, resveratrol production was low, with the best results, both in terms of volumetric yield and productivity being achieved for assays at 28 and 31 °C (assays 2–16), thus corroborating the results obtained for this parameter in the screening assays. However, at 25 °C (assay 1, Table 2), E. coli did not produce high amounts of resveratrol as 25 °C is not within the E. coli optimal growth range, which can result in slower transport processes and growth [25], and consequently lower resveratrol production. Although 37 °C is the temperature closer to the optimum E. coli growth temperature [25] this temperature may lead to trans-resveratrol degradation [22], since it is an easily degradable compound [21], which resulted in lower production levels. Regarding the pH, a pH around 6.5–7.0 seemed to be an optimal value to produce resveratrol, since the production tripled from 32.53 μg/mL, at a pH of 6.0 (assay 10), to 100.59 μg/mL, at a pH of 7.0 (assay 13) and then decreased again to 26.32 μg/mL (assay 16), at a pH of 8.0. The same trend was also observed for resveratrol specific values that almost tripled from 1.37 (pH 6.0) to 3.44 (pH 7.0) and then decreased again to 1.24 (pH 8.0). This pH influence on resveratrol production could be related with the optimal pH for E. coli growth as seen in the screening assays. In these assays, the OD600 at the time of induction had a slight impact on final production. This can be seen in assays 12–14, where the highest and lowest OD600 were tested (1.025 and 0.125, respectively) and also an intermediate value (0.575, assay 13). Although resveratrol production in assays 12 and 14 did not differ much from each other, after 30 h of growth, in assay 13, higher values of resveratrol production were achieved, highlighting the fact that the precursor should be added at the beginning of the exponential phase of growth to prevent early leakages, ruptures, and general damage to the membrane [20] and consequent decrease in resveratrol production. It can be seen that the best resveratrol productivity (6.31 mg/gh−1, assay 15) was obtained at 31 °C, pH 7.0, with a precursor concentration of 16 mM added at an OD600 of 0.575, which highlights the relevance of extending the range of conditions. On the other hand, the highest resveratrol production (159.96 μg/mL, assay 3) was achieved at 28 °C, pH 6.5, with a precursor concentration of 4 mM added at an OD600 of 0.8. These discrepancies in resveratrol production yields can be partially explained by the very distinct OD600 values obtained for assays 3 and 15 (4.19, and 2.31, respectively). However, the assay with the most similar conditions to those achieved in the screening assays (assay 13) still exhibited a value (100.59 μg/mL) close to the one obtained in the screening assays and in another study [16], indicating that this is a very reproducible process, which is of vital importance when designing an industrial fermentation process.
Since process productivity can be affected by plasmid segregational stability and physiological states of cells [14] due to decrease plasmid and/or protein levels and cellular growth, these two parameters were monitored for each of these bioreactor assays.
3.3. Cellular viability
In order to assess cell physiology, a PI/BOX dual-staining was performed. BOX was used to evaluate membrane potential, since it accumulates intracellularly when the cytoplasmic membrane is depolarized, and PI was used to verify the membrane integrity, as it only enters the cell if the membrane is injured. Overall, the percentage of healthy cells decreased throughout the fermentation, as the percentage of depolarized (BOX-positive) cells globally showed a marked increase from 22 to 30 h of fermentation (Table 2). Although the vast majority of the cells was in a healthy state, this percentage is smaller when compared to the values obtained in other bioprocess monitoring studies [13]. The higher values of depolarized cells may be due to the fact that M9 medium is a minimal medium [26], which limits nutrient availability and causes an increase in cell depolarization due to nutrient starvation [13]. With respect to the influence of cellular viability on growth, lower percentages of healthy cells seem to correspond to lower optical density values, indicative of slower growth. In general, lower resveratrol production yields were obtained when the cells are more depolarized, as can be seen in assays 20 and 23 (Table 2), as 39.07% and 50.16% of depolarized cells yielded 15.90 μg/mL and 14.90 μg/mL of resveratrol, respectively that also corresponded to low resveratrol specific productivities, 0.59 and 0.42 mg/gh−1, respectively. Nevertheless, there were some exceptions to this fact, meaning that resveratrol production was also dependent on the growth conditions. This assumption can be observed in assay 15, where despite the high values of depolarization (31.1%), 109.28 μg/mL (6.31 mg/gh−1) of resveratrol were obtained, which can be explained by the possible trans-resveratrol degradation in culture medium due to the growth conditions [27].
Temperature, as one of the most important factors in cell growth, also influenced cellular viability, as half of the assays with more than 30% of depolarized cells were performed either at 34 °C (assays 17, 18, 20, 23). Apparently, precursor concentration seemed to affect cellular viability, as can be seen in assay 15, where the addition of 16 mM of p-coumaric acid caused an increase in the percentage of depolarized cells. This decreased cellular viability could be due to the higher concentration of p-coumaric acid added to the culture, which may cause a destabilization of the cell membrane [28] by altering the dynamics of phospholipid chains [28]. However, other factors may also be involved in the increase of the percentage of cells with depolarized membranes, since some assays the raise in precursor concentration was not associated with this behaviour (Table 2). The results obtained showed that culture conditions could affect cellular viability which, in turn, affected resveratrol production, as lower percentage of healthy cells yielded lower resveratrol production at the end of fermentations.
3.4. Plasmid segregational stability
In a production bioprocess, the aim is to fully exploit the host cell’s capacity for recombinant protein synthesis. According to Grabherr et al. [15], protein production is based on appropriate gene expression, high copy number plasmids, and optimized growth conditions during the process.
Based on this, measuring plasmid segregational stability through PCN variation throughout the fermentation may also provide new insights and allow a more comprehensive understanding of resveratrol production, helping to define the best conditions to obtain the highest yield. In the majority of these assays, PCN increases both in pAC-4CL1 and pUC-STS from 22 to 30 h (Table 2), which could partially explain the higher resveratrol production yields also obtained in the samples taken after 30 h of fermentation. Absolute PCN values for pUC-STS (high copy number plasmid) are also lower in comparison with pAC-4CL1 (low copy number plasmid) values, indicating that the production of stilbene synthase could be the limiting step of this resveratrol production process, since high copy number plasmids perform a deficient regulation of gene expression, sometimes resulting in a residual production of protein [29]. The PCN values reported in this work are lower than the ones described by other studies using E. coli recombinant systems [13]. This fact could be related with the metabolic burden imposed to the E. coli cell by the maintenance and replication of two plasmids which resulted in lower cell growth and PCN values, indicating a possible increase in plasmid segregational instability, which may lead to plasmid loss [14]. Although in some assays, it is possible to observe a positive correlation between total PCN values and resveratrol specific productivity (assays 2, 3, 13, and 25), there are others where the opposite relation is observed (assays 10 and 15). Therefore, it was not possible to establish a relation between PCN and resveratrol productivity which can be due to the fact that this is a dual plasmid system and that resveratrol, being produced as an extracellular product, can be deteriorated by the culture conditions used as already discussed above.
4. Conclusions
This study describes resveratrol production by E. coli BW27784 containing pAC-4CL1 and pUC-STS plasmids and the assessment of physiological states and plasmid segregational stability during bioreactor cultivation. Resveratrol yield was greatly influenced by culture conditions as a result of the possible interactions established between the culture conditions on opposite to a linear variation for each condition tested and resveratrol yields. Cellular viability also showed to impact resveratrol production since growth conditions influenced physiological states. p-Coumaric acid played a critical role in resveratrol production, since it influenced the cellular viability due to interactions with the cell membrane, which affected the percentages of healthy cells and consequent resveratrol volumetric yields.
Monitoring resveratrol production is also important due to its ability to influence cellular viability caused by its inherent antimicrobial properties. The presence of two plasmids within the same cell influenced the final yield, because the metabolic burden generated might result in decreased cellular viability. Plasmid segregational stability evaluation revealed that no apparent relationship was obtained between plasmid copy number and resveratrol yields. In sum, this study indicates that these monitoring tools might be considered for a comprehensive application to resveratrol bioprocesses, in order to optimize and choose the most suitable design to create a valuable alternative to chemical synthesis.
Acknowledgments
This work was partially funded by FEDER funds through Programa Operacional Factores de Competitividade–COMPETE and by National Funds through FCT - Fundação para a Ciência e Tecnologia within the scope of Project “PTDC/AGR-ALI/121876/2010”. Susana Ferreira and Filomena Silva acknowledge doctoral (SFRH/BD/66857/2009) and post-doctoral (SFRH/BPD/79250/2011) fellowships from Fundação para a Ciência e Tecnologia within the scope of QREN–POPH–Advanced Formation programs co-funded by Fundo Social Europeu and MEC. The authors would like to thank Professor Claudia Schmidt-Dannert, for her donation of plasmids pAC-4CL1 and pUC-STS through Addgene.
Footnotes
Available online 30 October 2014
References
- 1.Burns J., Yokota T., Ashihara H., Lean M.E.J., Crozier A. J. Agric. Food Chem. 2002;50:3337–3340. doi: 10.1021/jf0112973. [DOI] [PubMed] [Google Scholar]
- 2.Lee D.G., Jung H.J., Seu Y.B. J. Microbiol. Biotechnol. 2007;17:1324–1329. [PubMed] [Google Scholar]
- 3.Sales J.M., Resurrección A.V.A. Crit. Rev. Food Sci. 2014;54:734–770. doi: 10.1080/10408398.2011.606928. [DOI] [PubMed] [Google Scholar]
- 4.Quideau S., Deffieux D., Douat-Casassus C., Pouységu L. Angew. Chem. Int. Ed. 2011;50:586–621. doi: 10.1002/anie.201000044. [DOI] [PubMed] [Google Scholar]
- 5.Sinha K., Chaudhary G., Gupta Y.K. Life Sci. 2002;71:655–665. doi: 10.1016/s0024-3205(02)01691-0. [DOI] [PubMed] [Google Scholar]
- 6.Jeandet P., Delaunois B., Aziz A., Donnez D., Vasserot Y., Cordelier S., Courot E. J. Biomed. Biotechnol. 2012;2012:1–14. doi: 10.1155/2012/579089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paulo L., Ferreira S., Gallardo E., Queiroz J.A., Domingues F., World J. Microbiol. Biotechnol. 2010;26:1533–1538. [Google Scholar]
- 8.Marienhagen J., Bott M. J. Biotechnol. 2013;163:166–178. doi: 10.1016/j.jbiotec.2012.06.001. [DOI] [PubMed] [Google Scholar]
- 9.Donnez D., Jeandet P., Clémont C., Courot E. Trends Biotechnol. 2009;27:706–713. doi: 10.1016/j.tibtech.2009.09.005. [DOI] [PubMed] [Google Scholar]
- 10.Botella L., Nájera C. Tetrahedron. 2004;50:5563–5570. [Google Scholar]
- 11.Wang Y., Yu O. J. Biotechnol. 2012;157:258–260. doi: 10.1016/j.jbiotec.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Lim C.G., Fowler Z.L., Hueller T., Schaffer S., Koffas M.A.G. Appl. Environ. Microbiol. 2011;77:3451–3460. doi: 10.1128/AEM.02186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva F., Lourenço O., Maia C., Queiroz J.A., Domingues F.C. Process Biochem. 2011;46:174–181. [Google Scholar]
- 14.Silva F., Queiroz J.A., Domingues F. Biotechnol. Adv. 2012;30:691–708. doi: 10.1016/j.biotechadv.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 15.Grabherr R., Nilsson E., Striedner G., Bayer K. Biotechnol. Bioeng. 2002;77:142–147. doi: 10.1002/bit.10104. [DOI] [PubMed] [Google Scholar]
- 16.Watts K., Lee P.C., Schmidt-Dannert C. BMC Biotechnol. 2006;6:22–34. doi: 10.1186/1472-6750-6-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Silva F., Passarinha L., Sousa F., Queiroz J.A., Domingues F.C. J. Microbiol. Biotechnol. 2009;19:1408–1414. doi: 10.4014/jmb.0805.329. [DOI] [PubMed] [Google Scholar]
- 18.Paulo L., Domingues F., Queiroz J.A., Gallardo E. J. Agric. Food Chem. 2011;59:2157–2168. doi: 10.1021/jf105004y. [DOI] [PubMed] [Google Scholar]
- 19.Alves M.J., Ferreira I.C.F.R., Froufe H.J.C., Abreu R.M.V., Martins A., Pintado M. J. Appl. Microbiol. 2013;115:346–357. doi: 10.1111/jam.12196. [DOI] [PubMed] [Google Scholar]
- 20.Lou Z., Wang H., Rao S., Sun J., Ma C., Li J. Food Control. 2012;25:550–554. [Google Scholar]
- 21.Liazid A., Palma M., Brigui J., Barroso C.G. J. Chromatogr. A. 2007;1140:29–34. doi: 10.1016/j.chroma.2006.11.040. [DOI] [PubMed] [Google Scholar]
- 22.Chung I.M., Kim J.J., Lim J.D., Yu C.Y., Kim S.H., Hahn S.J. Environ. Exp. Bot. 2006;54:4–53. [Google Scholar]
- 23.Trela B., Waterhouse A.L. J. Agric. Food Chem. 1996;44:1253–1257. [Google Scholar]
- 24.López-Nicolás J.M., García-Carmona F. Food Chem. 2008;109:868–875. doi: 10.1016/j.foodchem.2008.01.022. [DOI] [PubMed] [Google Scholar]
- 25.Madigan M.T., Martinko J.M., Stahl D.A., Clark D.P. In: Brock Biology of Microorganisms. Espinoza D., editor. Pearson Education Inc.; San Francisco: 2012. pp. 134–135. [Google Scholar]
- 26.Yun M., Park C.-G., Kim J.-Y., Rock C.O., Jackowski S., Park H.-W. Biol. Chem. 2000;275:28093–28099. doi: 10.1074/jbc.M003190200. [DOI] [PubMed] [Google Scholar]
- 27.Yang N.-C., Lee C.-H., Song T.-Y. Biosci. Biotechnol. Biochem. 2010;74:63–68. doi: 10.1271/bbb.90549. [DOI] [PubMed] [Google Scholar]
- 28.Borges A., Ferreira C., Saavedra M.J., Simões M. Microbiol. Drug Resist. 2013;19:256–265. doi: 10.1089/mdr.2012.0244. [DOI] [PubMed] [Google Scholar]
- 29.Gruber D.F., Pieribone V.A., Porton B., Kao H.-T. Protein Express. Purif. 2008;60:53–57. doi: 10.1016/j.pep.2008.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]