Abstract
The production and use of sugarcane in Brazil is very important for bioenergy production and is recognized as one of the most efficient in the world. In our laboratory, Setaria viridis is being tested as a model plant for sugarcane. S. viridis has biological attributes (rapid life cycle, small genome, diploid, short stature and simple growth requirements) that make it suitable for use as a model system. We report a highly efficient protocol for Agrobacterium-mediated genetic transformation of S. viridis. The optimization of several steps in tissue culture allowed the rapid regeneration of plants and increased the rate of transformation up to 29%. This protocol could become a powerful tool for functional genomics in sugarcane.
Keywords: Model system, C4 metabolism, Bioenergy, Sugarcane, Green millet
Sugarcane (Saccharum spp.) is an important biofuel feedstock because of its ability to accumulate high quantities of biomass and sucrose and is one of the most photosynthetically efficient C4 plants [1]. Breeding programs has been the main approach towards sugarcane improvement. However, the time required for a new variety (13 years) and the complexity of the genome of modern sugarcane varieties are major constraints [2]. Since the early 1990s, much progress was achieved in the biotechnological manipulation of sugarcane [3]. Nevertheless, stable transformation and plant regeneration for genomics studies is a time consuming for this crop. Model plants for genetic transformation like Arabidopsis thaliana are used in the proof of concept for many traits in various important crops. Therefore, there is still a need for additional model plants to decode and translate traits that are absent in these species [4]. In order to assist this endeavor, Setaria viridis was recently describe as a new monocotyledonous model species for C4 photosynthesis research and genetic transformation [5]. S. viridis belongs to the Poaceae family, subfamily Panicoideae that is one of the most agronomically important grass, including sugarcane. S. viridis has a number of characteristics that makes it interesting as a model plant [4]. Therefore, there is a need for a simple and highly efficient protocol for S. viridis genetic transformation. Here we report a simple protocol using Agrobacterium-mediated transformation with reporter genes gus (β-glucuronidase) and gfp (green fluorescent protein).
Seeds of S. viridis (accession A10.1) were planted in soil and grown in phytotron chambers under 16 h photoperiod, 26 ± 2 °C, 65% relative humidity and light intensity of 400 μmol m−2 s−1. Mature seeds were used for embryogenic callus induction. Seeds were disinfested after removal the lemmas and paleas with a solution of 10% sodium hypochlorite and 0.1% Tween 20® for 3 min followed by 5 rinses in sterile distilled water. After blotted on sterile filter paper the dehulled mature seeds were placed on callus induction medium (CIM) that consisted of MS salts [6], 1 mg/L d-biotin, 0.5 mg/L pyridoxine HCl, 0.5 mg/L nicotinic acid, 100 mg/L myo-inositol, 0.1 mg/L thiamine-HCl, 0.6 mg/L CuSO4, 30 g/L sucrose, 2 mg/L 2,4-dichlorophenoxyacetic acid, 0.5 mg/L kinetin and 4 g/L Phytagel™. The pH of the medium was adjusted to 5.8. After 3 to 5 weeks of incubation in the dark at 25 ± 2 °C, the callus was divided into small explants and subcultured onto fresh CIM. After 4–5 days these explants are ready to transformation step.
The expression vectors used for transformation of S. viridis are listed in Table 1. These vectors contain the reporter genes gus and gfp both with intron (DNA Cloning Service, Germany). A primary culture of Agrobacterium tumefaciens (EHA 105) was prepared by inoculating a single colony from a freshly streaked YEB agar plate in 5 mL of autoclaved YEB containing 100 mg/L spectinomycin and 50 mg/L rifampicin. The bacterial culture was incubated for 16 h in an orbital shaker at 180 rpm in dark at 28 °C. Secondary culture was initiated by inoculating 50 μL of primary culture into 25 mL YEB supplemented with 200 μM of acetosyringone with the same antibiotics and grown under the same conditions. Bacterial suspension was centrifuged and the pellet was resupended in a liquid CIM medium without CuSO4 to OD600 = 0.6. Approximately 50 calli were incubated for 5 min in Agrobacterium suspensions that contained 200 μM of acetosyringone and 10 μL of a 10% Synperonic® PE/F68 (Sigma–Aldrich) solution per 1 mL of the suspension. The calli were blotted on sterile filter paper and co-cultivated on a fresh CIM medium supplemented with 200 μM of acetosyringone for 3 days in the dark at 22 °C. Following co-cultivation, the calli were subcultured in CIM medium supplemented with 150 mg/L Timetin® for one week in the dark at 25 ± 2 °C and then transferred to CIM selective medium containing 30 mg/L hygromicin B or 3 mg/L glufosinate ammonium (Table 1) and 150 mg/L Timetin® following the same cultivation conditions. One week later, the explants were transferred to a selective regeneration medium (SRM) consisting of MS salts, 1 mg/L d-biotin, 0.5 mg/L pyridoxine HCl, 0.5 mg/L nicotinic acid, 100 mg/L myo-inositol, 0.1 mg/L thiamine HCl, 20 g/L sucrose, 2 mg/L kinetin, 150 mg/L Timetin®, 30 mg/L hygromicin B or 3 mg/L glufosinate ammonium, 2 g/L Phytagel™ with pH 5.8 adjusted before autoclaving. The cultures were incubated in growth chamber at 25 ± 2 °C, 16 h photoperiod using cool white fluorescent light (75 μmol m−2 s−1). In order to promote shoot and root elongation, the surviving seedlings were placed into Magenta™ box containing MS basal medium with the same selective agents and 150 mg/L Timetin®. Regenerated shoots ≥50 mm long were transplanted into pots for acclimatization. For each construct, the transformation efficiency (%) was calculated as the total number of PCR positive plants/total number of inoculated callus ×100. The PCR reaction with gus-specific primers sequences 5′-TTTGTTGATGTGCAGGTGGT-3′ and 5′-CTGCCCAATCCAACATCTCT-3′ and gfp-specific primers sequences 5′-ACCCTCGTCACCACTTTCAC-3′ and 5′-CATGTGGTCCCTCTTCTCGT-3′ were used to generate 424 bp and 669 bp PCR product, respectively (Fig. 1c). Transgenic plants were histochemically assayed for GUS activity (Fig. 1d), as described by Jefferson et al. [7]. GFP-specific fluorescence in transgenic seeds of S. viridis was visualized using a Zeiss Axio Imager Z2 Upright microscope assembled with a FITC/Alexa Fluor 488 filter (Chroma Technology Corp.). The GFP-fluorescence in seedlings was visualized using the Typhoon FLA 9000 laser scanner (GE Healthcare) using LPB filter (Fig. 1e). Segregation analysis was performed in T1 seeds. In order to overcome the dormancy, seeds were treated with concentrated sulfuric acid for 15 min and washed thoroughly in water and then disinfested with a solution of 2% sodium hypochlorite and 0.1% Tween 20® for 5 min followed by 5 rinses in sterile distilled water.
Table 1.
Summary of transgenic plants production by embryogenic calli.
| Vectorsa | Number of explants | Selectable marker | Number of PCR positive plants | Transformation efficiency (%)b |
|---|---|---|---|---|
| p6i | 225 | hpt | 32 | 14.22 |
| p6mD#1 | 218 | hpt | 30 | 13.76 |
| p6mD#2 | 55 | hpt | 16 | 29.09 |
| p6 | 77 | hpt | 6 | 7.79 |
| p7U | 50 | bar | 3 | 6.0 |
| p6mD#3 | 83 | hpt | 11 | 13.25 |
| p6mD#4 | 33 | hpt | 4 | 12.12 |
hpt – hygromycin phosphotransferase gene with an intron, bar – phosphinothricin acetyl transferase gene with an intron.
Vectors were purchased from DNA Cloning Service (www.dna-cloning.com). p6mD vector contains different genes of interest.
For each construct, the transformation efficiency was calculated as the total number of PCR positive plants/total number of inoculated callus ×100.
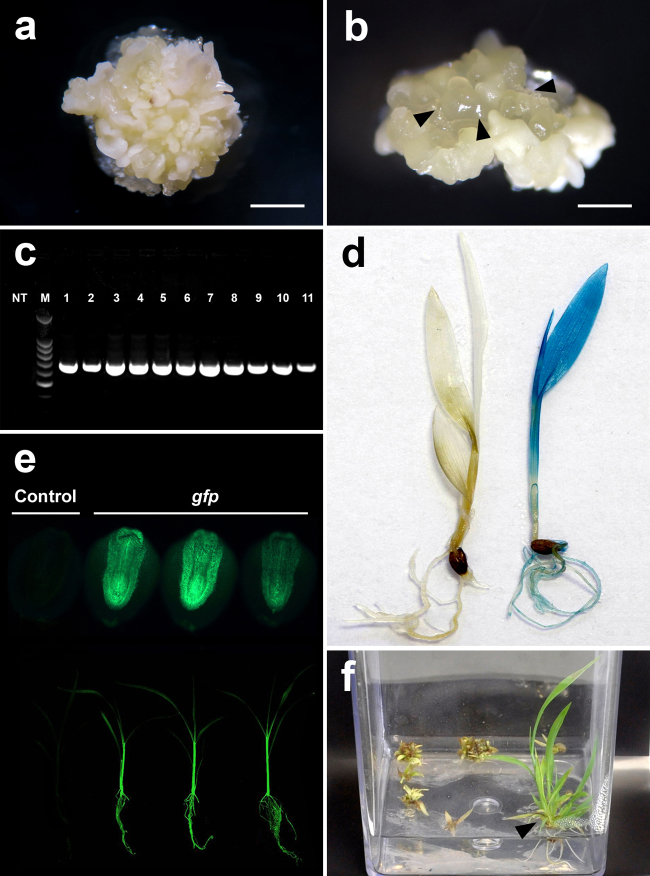
Fig. 1.
Agrobacterium tumefaciens-mediated transformation of Setaria viridis. (a) embryogenic callus after 5 weeks in CIM medium (bar = 2 mm). (b) translucent embryogenic callus most suitable for transformation (arrow heads, bar = 0.5 mm). (c) PCR analysis of the transgenic plants with gfp specific primer (NT: non-transgenic plant, lanes 1–11 transgenic plants, M: molecular weight marker – 100 bp DNA Ladder). (d) Transgenic plant expressing GUS (right) and non-transgenic (left). (e) Transgenic T1 seeds expressing GFP (top) and T1 seedlings expressing GFP (bottom). (f) Regenerated transgenic plantlet in hygromicin-containing selective MS medium (arrow head).
Plant transformation has become a core research tool for crop improvement as well as studies on gene function in plants. Various methodologies of plant transformation have been developed to increase the efficiency of transformation and to achieve stable expression of transgenes in plants [8]. Therefore, this substantially simplified and improved new protocol for S. viridis transformation will significantly contribute to its adoption as a model plant. Callus quality is considered as one of the most important parameters for an efficient tissue culture and in vitro transformation system [9]. In our study, callus formation was induced on CIM from dehulled mature seeds. After 5 weeks of culture, approximately 90% of them became white/yellow embryogenic calli. This callus (Fig. 1a) was divided into small pieces and transferred to fresh CIM. After 4–5 days, a formation of a translucent embryogenic callus was observed (Fig. 1b). Such callus was highly regenerable and very competent for transformation. The regenerated plants were transferred to soil after 3–4 weeks. Seeds were harvested after 6–7 weeks. The whole protocol from seed to seed was around 15 weeks. In addition, the protocol showed high efficiency independently of the vector backbone or selective marker used (Table 1). We also observed that the number of subcultures is critical for transformation and plant regeneration. After two subcultures, a 50% decrease in the transformation efficiency was observed. Moreover, after three or more subcultures no transformed plants could be obtained. Therefore, after transformation, we maintained the calli just for one week in CIM plus Timentin® and then one more week in CIM containing the selective agents. After these two weeks the calli were transferred to regeneration media containing the appropriated selection agent and Timentin®. At this phase, the calli may contain non-transgenic sectors, hence, it is important to keep the selection pressure. After 1–2 weeks in regeneration media the surviving plantlets were transferred to MS medium keeping the selective agents at respective concentration (Fig. 1f). Analysis of gene expression (RT-qPCR) was performed for events of two out of six constructs and showed high levels of transcripts abundance ranging from 45 to 2050-fold in comparison with non-transgenic plants (data not shown). Segregation analysis showed that more than 80% of the transgenic events contained a single-site insertion. Table 2 illustrates a Chi-square test (P < 0.05) of five events from one vector (p6mD#2) showing segregation ratio of 3:1, indicating a single-site insertion of the transgene. The transformation method described here, with up to 29% transformation frequency, with an average of approximately 15%, can provide a valuable tool for genomics and physiological studies. Such transformation efficiency is three times higher than previously reported [10]. Considering that S. viridis and sugarcane belongs to the same family with C4 metabolism (NADP-ME subtype) and similar cell wall composition, the high efficiency of our genetic transformation protocol indicate that S. viridis might be effectively used as a model plant for sugarcane applied research, including those related to abiotic stress tolerance and improved biomass for second generation (2 G) ethanol production.
Table 2.
Segregation ratios in the T1 generation of plants transformed with p6mD#2.
| Event | Resistanta | Sensitiveb | χ2 value for 3:1c | Fits 3:1 ratiod |
|---|---|---|---|---|
| 28 | 27 | 11 | 0.59 | Y |
| 29 | 27 | 3 | 3.60 | Y |
| 35 | 21 | 8 | 0.10 | Y |
| 49 | 29 | 9 | 0.03 | Y |
| 91 | 34 | 19 | 3.32 | Y |
Number of seedlings survived on the medium containing 50 mg/L hygromycin.
Number of seedlings sensitive on the medium containing 50 mg/L hygromycin.
Significantly different at P < 0.05.
Ratio of resistant versus sensitive seedlings on hygromycin.
Acknowledgments
We thank Dr. Frank G. Harmon (USDA-ARS Plant Gene Expression Center) for providing the S. viridis seeds. Dr. Polyana K. Martins was supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) fellowship. The authors also thank Embrapa (Brazilian Agriculture Research Corporation) for research grant (MP2-02.12.01.008.00.00).
Footnotes
This is an open-access article distributed under the terms of the Creative Commons Attribution-NonCommercial-No Derivative Works License, which permits non-commercial use, distribution, and reproduction in any medium, provided the original author and source are credited.
References
- 1.Taparia Y., Fouad W.M., Gallo M., Altpeter F. Rapid production of transgenic sugarcane with the introduction of simple loci following biolistic transfer of a minimal expression cassette and direct embryogenesis. In vitro. Cell. Dev. Biol. Plant. 2011;48:15–22. [Google Scholar]
- 2.Lima M.L.A., Garcia A.A.F., Oliveira K.M., Matsuoka S., Arizono H., Jr., de Souza C.L., de Souza A.P. Analysis of genetic similarity detected by AFLP and coefficient of parentage among genotypes of sugar cane (Saccharum spp.) Theor. Appl. Genet. 2002;104:30–38. doi: 10.1007/s001220200003. [DOI] [PubMed] [Google Scholar]
- 3.Bower R., Birch R.G. Transgenic sugarcane plants via microprojectile bombardment. Plant J. 1992;2:409–416. [Google Scholar]
- 4.Diao X., Schnable J., Bennetzen J., Li J. Initiation of Setaria as a model plant. Front. Agr. Sci. Eng. 2014;1:16–20. [Google Scholar]
- 5.Petti C., Shearer A., Tateno M., Ruwaya M., Nokes S., Brutnell T., Debolt S. Comparative feedstock analysis in Setaria viridis L. as a model for C4 bioenergy grasses and Panicoid crop species. Front Plant Sci. 2013;4:181. doi: 10.3389/fpls.2013.00181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murashige T., Skoog F. A revised for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 1962;15:473–479. [Google Scholar]
- 7.Jefferson R.A. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol. Biol. 1987;5:387–405. [Google Scholar]
- 8.Anami S.N.E., Coussens G., Aesaert S., Van Lijsebettens M. Higher plant transformation: principles and molecular tools. Int. J. Dev. Biol. 2013;57:483–494. doi: 10.1387/ijdb.130232mv. [DOI] [PubMed] [Google Scholar]
- 9.Păcurar D.I., Thordal-Christensen H., Nielsen K.K., Lenk I. A high-throughput Agrobacterium-mediated transformation system for the grass model species Brachypodium distachyon L. Transgenic Res. 2008;5:965–975. doi: 10.1007/s11248-007-9159-y. [DOI] [PubMed] [Google Scholar]
- 10.Van Eck J., Swartwood K. Setaria viridis. In: Wang K., editor. 3rd ed. Vol. 1. Springer Science + Business Media; New York: 2015. pp. 57–67. (Agrobacterium Protocols). [Google Scholar]