Abstract
Setaria viridis was recently described as a new monocotyledonous model species for C4 photosynthesis research and genetic transformation. It has biological attributes (rapid life cycle, small genome, diploid, short stature and simple growth requirements) that make it suitable for use as a model plant. We report an alternative method of S. viridis transformation using floral dip to circumvent the necessity of tissue culture phase for transgenic plant regeneration. S. viridis spikes at boot stage were selected to be immersed in Agrobacterium suspension. T1 seeds could be identified in 1.5–2 months after floral dipping. We demonstrated through molecular analysis and RFP expression that seeds and resulting plants from dipped inflorescences were transformed. Our results suggest the feasibility of S. viridis floral dip transformation as a time-saving and cost-effective compared with traditional methods. To our knowledge, this is the first report using floral dip in S. viridis as an Agrobacterium-mediated transformation method.
Keywords: Model plant, Green millet, Red fluorescent protein
Grass crop plants are the major sources of human food–feed–fiber–fuel in the world. The most agriculturally important grasses are rice, wheat, maize, sorghum and sugarcane. Setaria viridis belongs to the Poaceae family, subfamily Panicoideae that includes the most agronomically important grass crops. S. viridis has a number of characteristics that makes it interesting as a model plant for genetics and functional genomics studies [1]. Plant transformation typically requires refined regeneration methods that involve multiples steps from in vitro culture to greenhouse conditions. Agrobacterium-mediated transformation by vacuum infiltration of Arabidopsis inflorescences was first reported by Bechtold et al. [2] and further modified as floral dip method by Clough and Bent [3]. Floral dip method has been used in genetic engineering strategies as it directly produces genetically modified seeds bypassing the laborious tissue culturing procedures. In addition, tissue culture could generate transformed plants harboring undesirable mutations from somaclonal variation [4]. This method, indubitably, accelerated Arabidopsis thaliana studies as a model plant, quickly advancing our understanding of several other dicotyledonous crops. For that reason, the development of a simple and rapid transformation method for S. viridis as a C4 model for monocotyledonous plants could help to achieve results comparable with those in A. thaliana.
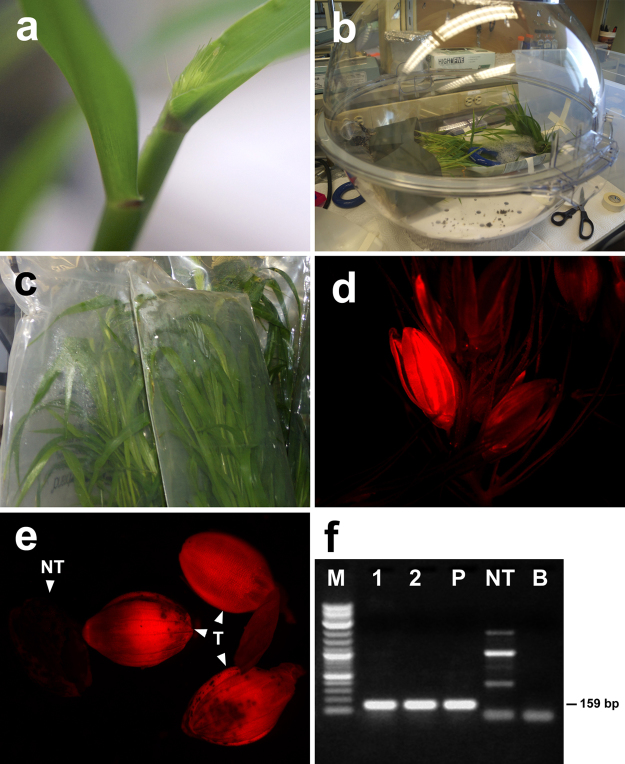
Three seeds of S. viridis (accession A10.1) were cultivated in ten square plastic pots (230 ml) with Supersoil® (Scotts Miracle-Gro Company) and grown in an air-conditioned greenhouse with a 26 °C/22 °C day/night cycle with a 14 h photoperiod. About 26–30 days after planting, S. viridis spikes at the boot stage [5] were used for floral dipping (Fig. 1a).
Fig. 1.
Setaria viridis floral dip transformation. (a) Inflorescence developmental stage (boot stage) selected for transformation. (b) Spikes dipped in Agrobacterium suspension inside a desiccator for vacuum-assisted Agrobacterium infiltration. (c) Spikes covered with plastic bags after infiltration to keep moisten for 24 h. (d) Spike showing RFP-expressing immature seed. (e) RFP-expressing mature seeds (NT – non-transgenic, T – transgenic seeds, indicated by arrow heads). (f) PCR analysis of surviving plants on hygromycin-containing medium (M: molecular weight marker-2-log DNA ladder, lanes 1 and 2: transgenic plants showing expected 159 bp porpRFP-specific band, NT: non-transgenic plant, P: positive control–plasmid pANIC 6 A, B: polymerase chain reaction amplification without DNA template).
In order to transform Setaria plants, Agrobacterium tumefaciens (AGL1 strain) cultures containing the vector pANIC 6A [6] was prepared. The bacteria were cultured in 500 ml of LB medium containing 50 mg/l kanamycin at 30 °C to OD600 = 1.0, and centrifuged at 5000 × g for 15 min, the pellet was washed with 10 ml of MM buffer (10 mM MES, 10 mM MgCl2, pH 5.6) and resuspended in 50 ml of fresh MM buffer. The addition of tobacco (Nicotiana tabacum) extract is known to improve the gene transfer by agrobacteria in monocots [7]. Therefore, tobacco leaf extract was prepared as following: 40 g of fresh leaves were cut into 2 cm2 pieces in 1 l of MM buffer and stirred gently for 1 h. The solution was sieved and the liquid phase was collected. Prior to inoculation, the solution (1 l) was supplemented with 1 ml of 50 mM acetosyringone, 200 μl of Silwet-L77 and 50 g of sucrose. The final solution was mixed with the bacterial suspension used for the floral dip procedure. Spikes of one month-old S. viridis plants at boot stage (Fig. 1a) were infiltrated with bacterial suspension carrying the pANIC 6 A. Briefly, the aerial part of plants containing the spikes were immersed in Agrobacterium suspension inside of a Secador® Techni-dome® 360 vacuum desiccator and subjected to vacuum infiltration (80 kPa) for 10 min (Fig. 1b). The spikes were covered with plastic bags for 24 h (Fig. 1c). The co-cultivation was performed for 3 days under greenhouse conditions without irrigation. After 1.5–2 months seeds were screened for red fluorescence using a Leica MZ16F stereo microscope equipped with an XCite EXFO fluorescence illumination source and a filter combination for fluorescence in the red region of the spectrum (excitation: 558 nm, emission 580 nm). Digital images were collected using a digital camera (Retiga 2000 Fast Cooled 12-bit) (Fig. 1d and e). Mature seeds were collected and germinated in half-strength semi-solid MS medium containing 30 mg/l hygromycin B for selection of transformed seedlings. DNA was extracted from plants surviving on hygromycin-containing medium and processed according to Doyle and Doyle [8]. The molecular analysis of the transformed plants was performed by PCR. Each reaction (20 μl) contained 20 mM Tris–HCl (pH 8.4), 50 mM KCl, 2 mM MgCl2, 200 μM of each dNTP, 1.0 U of Taq polymerase, 50 ng DNA and 5 μM of each oligonucleotide specific to pporRFP gene. The pporRFP primer sequences were: 5′-GGCCATTATACGTGCGACTT-3′ and 5′-CAACGAGAAATTGCACATGC-3′ to generate 159 bp PCR product (Fig. 1f). The samples were submitted to the following amplification cycles in a Bio-Rad iCycler at 95 °C for 30 s, followed by 40 cycles of 95 °C for 30 s, 50 °C for 45 s, 68 °C for 40 s, 68 °C for 5 min. The amplified fragments were subjected to electrophoresis in 1% agarose gel containing 0.5 μg/ml ethidium bromide and visualized in UV transilluminator.
The appropriate stage for dipping appears to be from the mid to late uninucleate microspore stage when the spike has not emerged from the sheath (boot stage) [9]. This stage coincides with the highest frequency uninucleate microspore in the anther [10]. In addition, in rice, the anther/microspores were the preferential targets of the floral-dip transformation [11]. To date, the target tissue for the T-DNA transfer to S. viridis has not been determined. The visual screening in RFP-expressing seeds could not discriminate which part of the seed was transformed (i.e., endosperm, embryo etc.). Therefore, mature seeds were germinated on selective medium and the surviving seedlings were analyzed by PCR to confirm that transgenic plants were generated. From one thousand seeds, 190 survived on hygromycin containing medium. The PCR analysis showed that among them six plants were confirmed to be transformed (transformation efficiency of 0.6% – number of transformants/number of seed set). To our knowledge, this is the first report using floral dip in S. viridis as an Agrobacterium-mediated transformation method. Therefore, we confirmed the feasibility of floral dip for S. viridis. This method could accelerate genetic and genomics studies of important food–feed–fiber–fuel crops such as sugarcane, Miscanthus, elephant grass, Brachiaria, Sorghum and corn. Currently, we are working on the optimization of different steps involved in this method to improve the transformation efficiency.
Acknowledgments
The authors gratefully acknowledge Dr. George Chuck (USDA-ARS Plant Gene Expression Center) for providing the plasmid pANIC 6A, Dr. James Thomson (USDA-ARS Western Regional Research Center) for providing the vacuum desiccator, Dr. Delilah Wood and Dr. Tina Williams (USDA-ARS Western Regional Research Center) for helping with light microscopy. Dr. Carlos Eduardo Lazarini da Fonseca, Embrapa LABEX USA Coordinator. Dr. Polyana K. Martins was supported by CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) fellowship. This work was supported by the Embrapa LABEX USA program.
Footnotes
Available online 4 March 2015
References
- 1.Diao X., Schnable J., Bennetzen J., Li J. Initiation of Setaria as a model plant. Front. Agric. Sci. Eng. 2014;1:16–20. [Google Scholar]
- 2.Bechtold N., Ellis J., Pelletier G. In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C.R. Acad. Sci. 1993;316:1194–1199. [Google Scholar]
- 3.Clough S.J., Bent A. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 4.Bent A.F. Arabidopsis in planta transformation. uses mechanisms, and prospects for transformation of other species. Plant Physiol. 2000;124:1540–1547. doi: 10.1104/pp.124.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blaise M.O., Girardin P., Millet B. Developmental stages and floral ontogenesis of foxtail millet Setaria italica (L) P Beauv. Agronomie. 1992;12:141–156. [Google Scholar]
- 6.Mann D.G.J., LaFayette P.R., Abercrombie L.L., King Z.R., Mazarei M., Halter M.C., Poovaiah C.R., Baxter H., Shen H., Dixon R.A., Parrott W.A., Stewart C.N., Jr Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotech. J. 2012;10:226–236. doi: 10.1111/j.1467-7652.2011.00658.x. [DOI] [PubMed] [Google Scholar]
- 7.Fursova O., Pogorelko G., Zabotina O.A. An efficient method for transient gene expression in monocots applied to modify the Brachypodium distachyon cell wall. Ann. Bot. 2012;110:47–56. doi: 10.1093/aob/mcs103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doyle J.J., Doyle J.L. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- 9.Zale J.M., Agarwal S., Loar S., Steber C.M. Evidence for stable transformation of wheat by floral dip in Agrobacterium tumefaciens. Plant Cell Rep. 2009;28:903–913. doi: 10.1007/s00299-009-0696-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu W., Zheng M.Y., Polle E.A., Konzak C.F. Highly efficient doubled-haploid production in wheat (Triticum aestivum L.) via induced microspore embryogenesis. Crop Sci. 2002;42:686–692. [Google Scholar]
- 11.Rod-in W., Sujipuli K., Ratanasut K. The floral-dip method for rice (Oryza sativa) transformation. J. Agric. Technol. 2014;10:467–474. [Google Scholar]